This work finds that mechanosensitive channels play an important role in the division of Arabidopsis chloroplasts and contribute to division site placement in E. coli cells, linking membrane tension to the assembly of FtsZ rings in chloroplasts and bacteria.
Abstract
Chloroplasts must divide repeatedly to maintain their population during plant growth and development. A number of proteins required for chloroplast division have been identified, and the functional relationships between them are beginning to be elucidated. In both chloroplasts and bacteria, the future site of division is specified by placement of the Filamentous temperature sensitive Z (FtsZ) ring, and the Min system serves to restrict FtsZ ring formation to mid-chloroplast or mid-cell. How the Min system is regulated in response to environmental and developmental factors is largely unstudied. Here, we investigated the role in chloroplast division played by two Arabidopsis thaliana homologs of the bacterial mechanosensitive (MS) channel MscS: MscS-Like 2 (MSL2) and MSL3. Immunofluorescence microscopy and live imaging approaches demonstrated that msl2 msl3 double mutants have enlarged chloroplasts containing multiple FtsZ rings. Genetic analyses indicate that MSL2, MSL3, and components of the Min system function in the same pathway to regulate chloroplast size and FtsZ ring formation. In addition, an Escherichia coli strain lacking MS channels also showed aberrant FtsZ ring assembly. These results establish MS channels as components of the chloroplast division machinery and suggest that their role is evolutionarily conserved.
INTRODUCTION
Chloroplasts are specialized organelles responsible for many essential metabolic reactions, most notably photosynthesis. Like all plastids, chloroplasts are not created de novo but are differentiated from a preexisting population of proplastids present in meristematic cells (reviewed in Lopez-Juez and Pyke, 2005). As plant cells expand, established chloroplasts must undergo a series of divisions to maintain the appropriate population density. Modern day chloroplasts arose from an endosymbiotic relationship between a heterotrophic protozoan and a cyanobacterium and, reminiscent of their bacterial origins, divide by binary fission (Cavalier-Smith, 2000; Miyagishima et al., 2003). Given this evolutionary lineage, bacterial cell division is often used as a framework for identifying the components and understanding the mechanisms of chloroplast division. This approach has led to the identification of many homologs of essential bacterial division proteins within the plant genome, and a strong evolutionary conservation of division mechanisms has now been documented (Yang et al., 2008; reviewed in Nakanishi et al., 2009b).
Cell division has been well characterized in the gram-negative bacterium Escherichia coli. Division at mid-cell is specified by the polymer-forming GTPase Filamentous temperature sensitive Z (FtsZ), which forms a structure known as the Z-ring on the cytoplasmic surface of the inner membrane (Bi and Lutkenhaus, 1991; de Boer et al., 1992a; Mukherjee et al., 1993; Mukherjee and Lutkenhaus, 1994). The Z-ring is thought to act as a scaffold for the assembly of the division apparatus and contribute some of the force required for fission (Osawa et al., 2009; reviewed in de Boer, 2010). Two nonredundant families of FtsZ genes are found in seed plants (Osteryoung and Vierling, 1995; Osteryoung et al., 1998; Stokes and Osteryoung, 2003; Schmitz et al., 2009), and both FtsZ1 and FtsZ2 localize to a ring-like structure at the division site (McAndrew et al., 2001; Vitha et al., 2001; Fujiwara et al., 2008). Plant and bacterial lines under- or overexpressing FtsZ exhibit enlarged chloroplasts and filamentous cells, respectively, though plants with altered levels of FtsZ are otherwise phenotypically normal (Dai and Lutkenhaus, 1992; Stokes et al., 2000; El-Kafafi et al., 2005, 2008; Liu et al., 2007; Yoder et al., 2007; Schmitz et al., 2009; Karamoko et al., 2011). Arabidopsis thaliana FtsZ proteins have GTPase activity and are capable of forming filaments in vitro (El-Kafafi et al., 2005; Olson et al., 2010; Smith et al., 2010).
Proper placement of the Z-ring ensures segregation of cytoplasmic contents and is tightly regulated in E. coli by the Min system (Bi et al., 1991; reviewed in de Boer, 2010). Minicell C (MinC) and MinD form a complex that negatively regulates FtsZ polymerization, while MinE acts as a topological specificity factor by inhibiting MinCD function (de Boer et al., 1989, 1991, 1992b; Huang et al., 1996). All three proteins oscillate from pole to pole such that MinCD activity is restricted to the ends of the cell and FtsZ polymerization is allowed to progress only at mid-cell (Hu and Lutkenhaus, 1999; Raskin and de Boer, 1999a, 1999b; Rowland et al., 2000; Fu et al., 2001; Hale et al., 2001). Arabidopsis homologs of MinD and MinE have been identified and appear to have some conserved functions in regulating Z-ring placement. Arabidopsis MinD antisense lines and a loss-of-function allele of MinD, accumulation and replication of chloroplasts11 (arc11), exhibit enlarged chloroplasts with multiple Z-rings, suggesting that At-MinD behaves as a Z-ring assembly inhibitor (Colletti et al., 2000; Kanamaru et al., 2000; Vitha et al., 2003; Fujiwara et al., 2004). Conversely, At-MinE antisense suppression lines and two mutant alleles of At-MinE, minE1 and arc12, exhibit grossly enlarged chloroplasts containing numerous short FtsZ filaments (Glynn et al., 2007; Fujiwara et al., 2008). In summary, multiple lines of evidence indicate that, similar to their bacterial homologs, At-MinD and At-MinE antagonistically regulate Z-ring assembly (Itoh et al., 2001; Maple et al., 2002; Reddy et al., 2002; Fujiwara et al., 2004; Glynn et al., 2007; Maple and Møller, 2007b; Fujiwara et al., 2008).
Several aspects of FtsZ ring assembly in chloroplasts are specific to plants (Nakanishi et al., 2009b). Arabidopsis ARC3, a stromal protein that contains an incomplete FtsZ-like region, has been reported to interact directly with MinD, MinE, and FtsZ and copurify with FtsZ1, FtsZ2, and ARC6 (Shimada et al., 2004; Maple et al., 2007; McAndrew et al., 2008). arc3 mutants resemble arc11 mutants in that they exhibit enlarged chloroplasts and multiple FtsZ rings (Pyke and Leech, 1994; Shimada et al., 2004; Glynn et al., 2007; Maple et al., 2007). No MinC homolog has been identified in plants, though FtsZ assembly is sensitive to overexpression of E. coli MinC, suggesting that the system is capable of responding to MinC activity (Tavva et al., 2006). Instead, it has been proposed that ARC3 has taken on the role of MinC in land plants and directly inhibits FtsZ polymerization by actively competing with FtsZ monomers (Maple and Møller, 2007b; Maple et al., 2007; Yang et al., 2008; Nakanishi et al., 2009b). Other plant-specific genes required for normal FtsZ assembly in Arabidopsis chloroplasts have been identified, including Multiple Chloroplast Division Site1 (MCD1), ARC6, and Paralog of ARC6 1 (PARC6/CDP) (Vitha et al., 2003; Glynn et al., 2009; Nakanishi et al., 2009a; Zhang et al., 2009).
Two additional candidates for regulators of chloroplast division and Z-ring placement are MscS-Like 2 (MSL2) and MSL3. MSL2 and MSL3 are redundantly required for normal chloroplast size and colocalize with MinE at the poles of plastids (Haswell and Meyerowitz, 2006). Along with the other eight MSL proteins encoded in the Arabidopsis genome, MSL2 and MSL3 share homology with the bacterial mechanosensitive (MS) ion channel MscS, a largely nonselective channel that is thought to serve as an osmotic safety valve under conditions of hypoosmotic stress. MscS and another MS channel are referred to as MscL open in response to increased membrane tension, permitting the release of osmolytes and preventing cellular lysis (Levina et al., 1999; Sukharev, 2002; Pivetti et al., 2003; Haswell, 2007). Here, we add to our understanding of how Z-ring placement may be regulated by characterizing the role played by MscS-Like channels in both chloroplast division and bacterial cell fission.
RESULTS
MSL2 and MSL3 Are Required for Normal Z-Ring Placement
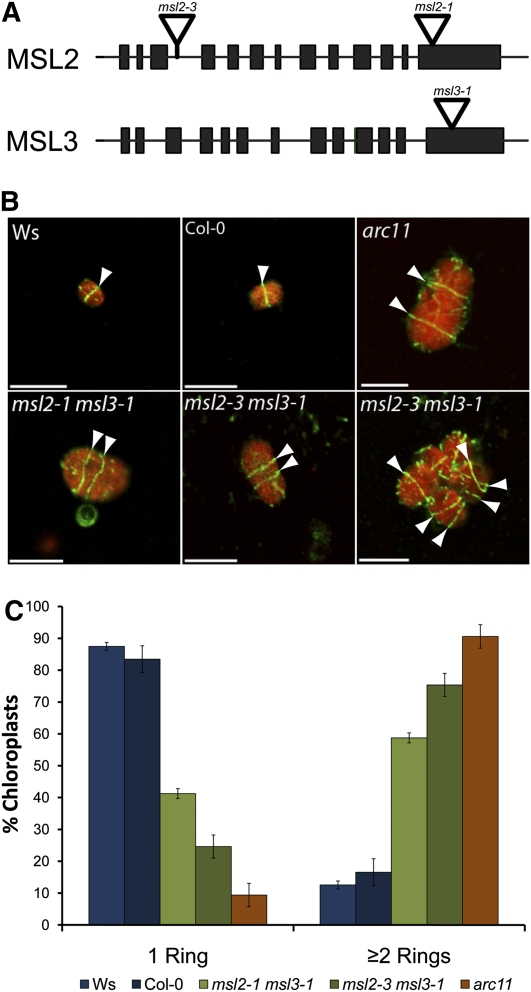
We previously observed that msl2-1 msl3-1 double mutant plants exhibit heterogeneously sized chloroplasts, as well as other plastidic and whole-plant phenotypes (Haswell and Meyerowitz, 2006). Single msl2-1 and msl3-1 mutants appear wild type, suggesting that MSL2 and MSL3 function redundantly. In this study, we extend our characterization of the msl2-1 msl3-1 double mutant and add another allele of MSL2 (msl2-3) to our analysis. The T-DNA insertion in the msl2-3 allele introduces a stop codon at amino acid 98 and is therefore likely to be a null allele, whereas the insertions in msl2-1 and msl3-1 alleles are located in the last exon of each gene and are likely to be partial loss-of-function alleles (Figure 1A; see Supplemental Figure 1 online). Though msl2-3 single mutants do not exhibit appreciably enlarged chloroplasts, msl2-3 msl3-1 mutants have enlarged chloroplasts similar to those observed in the msl2-1 msl3-1 double mutant (see Figure 3).
Figure 1.
msl2 msl3 Double Mutant Chloroplasts Contain Multiple Z-Rings.
(A) Representation of MSL2 and MSL3 coding regions. Exons are indicated by black bars and introns by lines. The T-DNA insertion sites in msl2-3, msl2-1, and msl3-1 alleles are indicated.
(B) Immunofluorescence micrographs of FtsZ1 ring placement in chloroplasts isolated from rosette leaves. Arrowheads indicate Z-rings, shown in green. Chlorophyll autofluorescence is pseudocolored red. Bars = 10 μm.
(C) Quantitative analysis of isolated chloroplasts containing single or multiple Z-rings. The average of three independent experiments ± the se of the means is presented. n ≥ 100 chloroplasts, per replicate.
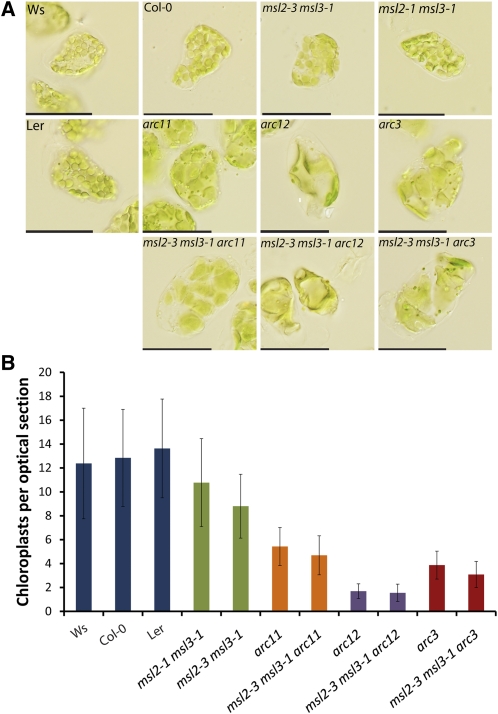
Figure 3.
Chloroplast Number in Single, Double, and Triple msl and arc Mutants.
(A) Chloroplasts from mature leaf mesophyll cells. Leaves were fixed with glutaraldehyde prior to imaging. Bars = 50 μm. Ler, Landsberg erecta.
(B) Quantitative analysis of chloroplast number per optical section in the indicated genotypes. The mean value ± sd is presented from data collected in two independent experiments. n ≥ 125 cells per genotype.
The enlarged chloroplast phenotype of msl2-1 msl3-1 and msl2-3 msl3-1 double mutant plants suggested that chloroplast division might be altered in these mutants. To test this hypothesis, we characterized Z-ring placement in wild-type and mutant chloroplasts by immunofluorescence microscopy (IFM). The primary antibody used in these experiments was an anti-FtsZ antibody raised against FtsZ from Bacillus subtlilis (a gift of P. Levin, Washington University). This antibody recognized FtsZ1, but not FtsZ2-1 or FtsZ2-2, in an immunoblot of Arabidopsis proteins (see Supplemental Figure 2 online). Protoplasts isolated from the rosette leaves of 3-week-old plants were fixed and chloroplasts released onto poly-lysine–treated slides by tapping on the cover slip. After incubation with anti-FtsZ primary and fluorescein isothiocyanate (FITC)-conjugated secondary antibodies, confocal laser scanning microscopy was used to capture images of representative chloroplasts from each genotype (Figure 1B). Most wild-type chloroplasts contained only one Z-ring, though occasionally more were observed (Figure 1C). By contrast, msl2 msl3 mutant chloroplasts frequently contained multiple Z-rings, some with a disorganized, forked appearance, similar to those observed in the arc11, arc3, mcd1, and parc6 mutants but distinct from those observed in other plastid division mutants, such as arc5, plastid division1 plastid division2 (pdv1 pdv2), or arc2 (Vitha et al., 2003; Miyagishima et al., 2006; Glynn et al., 2007, 2009; Fujiwara et al., 2008; Nakanishi et al., 2009a; Suzuki et al., 2009).
Z-ring number, like chloroplast size, is highly heterogeneous in msl2 msl3 mutants (Figure 1B, bottom right panel). We therefore quantified the number of complete Z-rings per chloroplast in wild-type, msl2 msl3, and arc11 plants. After IFM, the number of intact Z-rings was counted; complete Z-rings were identified by adjusting the focal plane to visualize both the top and bottom of each ring. Figure 1C presents data compiled from three independent experiments. In wild-type plants, 83 to 87% of chloroplasts had only one Z-ring, whereas arc11 mutants were at the opposite extreme, containing 91% of chloroplasts with two or more Z-rings. The msl2-1 msl3-1 and msl2-3 msl3-1 mutants exhibited a phenotype similar to the arc11 mutant, with 59 and 75% of the chloroplasts containing two or more Z-rings, respectively. Thus, not only are MSL2 and MSL3 required for normal Z-ring assembly, but the defect in Z-ring placement in msl2 msl3 mutants is nearly as severe as that observed in the absence of functional MinD, a primary component of the plastid division machinery.
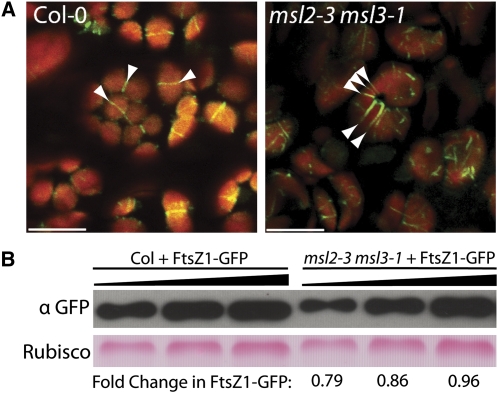
To characterize Z-ring formation in live cells, we obtained a transgenic line expressing FtsZ1-GFP (green fluorescent protein) under the control of the FtsZ1-1 upstream region (M. Fujiwara, RIKEN). This and other FtsZ-GFP fusion proteins have been used in several studies of chloroplast division in Arabidopsis petioles and pollen (Vitha et al., 2001; Fujiwara et al., 2008, 2009, 2010). In wild-type lines with low levels of FtsZ1-GFP expression, a single ring of GFP fluorescence was observed in cauline leaf chloroplasts (arrows, left panel, Figure 2A). However, when the FtsZ1-GFP transgene was crossed into the msl2-3 msl3-1 background, grossly enlarged chloroplasts containing multiple Z-rings were observed (arrows, right panel, Figure 2A). Many of these enlarged chloroplasts also contained partial or branched rings.
Figure 2.
Multiple Rings of FtsZ1-GFP Signal Are Observed in msl2 msl3 Chloroplasts.
(A) FtsZ1-GFP localization in cauline leaves. Arrowheads indicate Z-rings present in Col-0 and msl2-3 msl3-1 plants, respectively. GFP (green) and chlorophyll fluorescence (red) are pseudocolored. Bars = 10 μm.
(B) Quantitative immunoblot of FtsZ1-GFP protein levels in wild-type and mutant backgrounds. Three 1.5-fold dilutions of each protein extract were loaded. Top panel, GFP signal; bottom panel, Ponceau S staining of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco). Estimates of FtsZ1-GFP protein levels in msl2-3 msl3-1 mutants relative to those in Col-0 are shown below each lane.
To avoid the well-documented phenotypic effects of At-FtsZ overexpression (Stokes et al., 2000; Vitha et al., 2001; Fujiwara et al., 2008), live imaging analysis was performed only on lines expressing FtsZ1-GFP at low levels. To establish further that the increased Z-ring number observed in the msl2 msl3 mutant chloroplasts cannot be attributed to overexpression of FtsZ1-GFP, quantitative immunoblotting of protein extracts from the plants imaged in Figure 2A was performed. Similar levels of FtsZ1-GFP were present in both mutant and wild-type backgrounds (Figure 2B).
MSL2 and MSL3 Function in the Same Pathway as MinD, MinE, and ARC3
We next wished to determine whether MSL2 and MSL3 regulate Z-ring formation through the previously characterized Min pathway. To address this question, we generated and characterized triple msl2 msl3 arc11, msl2 msl3 arc3, and msl2 msl3 arc12 mutant plants. The arc11 and arc3 alleles can be considered null as MinD and ARC3 proteins are undetectable in arc11 and arc3 mutants, respectively (Shimada et al., 2004; Nakanishi et al., 2009a). The arc12 allele is also likely to be null, as it introduces a stop codon upstream of the conserved regions required for dimerization and interaction with MinD (Itoh et al., 2001; Glynn et al., 2007; Maple and Møller, 2007a). We hypothesized that if MSL2 and MSL3 function in the same pathway as components of the Min system to regulate Z-ring assembly, triple mutants should exhibit the phenotype of the most severe parental mutant. However, if MSL2 and MSL3 act in a separate pathway, the triple mutants should display an additive or even synergistic chloroplast phenotype.
Fixed mesophyll cells from arc11, arc12, and arc3 mutants contain a small number of greatly enlarged chloroplasts, whereas msl2 msl3 mutants contain relatively more, heterogeneously sized chloroplasts (Figure 3A). At this level of analysis, all three triple msl2 msl3 arc mutants resembled their arc mutant parent with respect to chloroplast size and number. To compare these mutants in a quantitative manner, we counted the number of chloroplasts in optical sections of mesophyll cells from each genotype. Confocal images were taken of mesophyll tissue at the tips of fully expanded rosette leaves that had been exposed to strong light to induce chloroplast movement to the sides of the cell. Representative images are presented in Supplemental Figure 3 online. The number of chloroplasts per section at the widest circumference of each cell was counted, and the results from two independent experiments compiled (Figure 3B). Wild-type accessions had an average of 12 to 14 chloroplasts per optical section, while the msl2 msl3 double mutant had an average of 9. We found that msl2 msl3 arc11, msl2 msl3 arc12, and msl2 msl3 arc3 triple mutants closely resembled the single arc11, arc12, and arc3 mutants, with five, two, and four chloroplasts per optical section, respectively (Figure 3B). The small but statistically significant decrease (Student’s t test, P > 0.01; less than one chloroplast per optical section) in chloroplast number seen in the msl2 msl3 arc11 and msl2 msl3 arc3 triple mutants is likely due to the variability in cell size seen in the msl2 msl3 mutant background. Cell size is linearly correlated with chloroplast number (Pyke and Leech, 1992).
We next analyzed Z-ring formation in the chloroplasts of single, double, and triple msl and arc mutant plants. FtsZ IFM revealed multiple Z-rings in chloroplasts isolated from msl2 msl3 arc11 plants, similar to those observed in the msl2 msl3 and arc11 genotypes (Figure 4). Consistent with the number of chloroplasts per cell, no statistically significant difference in the percentage of chloroplasts containing two or more rings was observed in msl2 msl3 arc11 triple mutants compared with single arc11 mutants (Student’s t test, P > 0.2). Similarly, msl2 msl3 arc3 triple mutants exhibited no significant increase in Z-ring number over the arc3 single mutant (Student’s t test, P > 0.2; see Supplemental Figure 4 online).
Figure 4.
msl2 msl3 arc11 Triple Mutants Are Phenotypically Indistinguishable from Single arc11 Mutants.
(A) Immunofluorescence micrographs of Z-ring placement in chloroplasts isolated from rosette leaves of the indicated genotypes. FITC and chlorophyll fluorescence are pseudocolored green and red, respectively. Bars = 10 μm.
(B) Quantitative analysis of Z-rings in isolated chloroplasts subjected to IFM. The average values from two independent experiments ± se of the means are presented. n ≥ 100 chloroplasts per replicate.
(C) The distribution of Z-ring number in chloroplasts of the indicated genotypes. Z-rings of isolated chloroplasts were visually counted and percentage of chloroplasts containing one to nine Z-rings was calculated. n ≥ 50 chloroplasts per genotype.
In the analysis presented in Figure 4B, chloroplasts were put into two categories: one Z-ring or more than one Z-ring. As the latter category contained chloroplasts with Z-ring numbers ranging from 2 to 9, we analyzed the distribution of Z-ring number in the chloroplasts of single, double, and triple mutants to determine if a difference between arc11 and msl2 msl3 arc11 mutants could be observed (Figure 4C). Most chloroplasts from wild-type plants had one Z-ring; <18% contained two or three Z-rings and none had more than four. By contrast, 56 and 51% of the chloroplasts from msl2 msl3 double mutants contained two or three Z-rings, and numbers as high as 7 were observed. In the arc11 mutant, the distribution of Z-ring number was shifted further toward increasing numbers of rings per chloroplast, with 62% of the chloroplasts showing ring numbers of 3, 4, and 5. The distribution of ring number in both msl2 msl3 arc11 triple mutant lines was strikingly similar to that observed in the arc11 single mutants, suggesting that the introduction of msl2 and msl3 mutant alleles did not enhance the defect in Z-ring formation already present in the arc11 mutant.
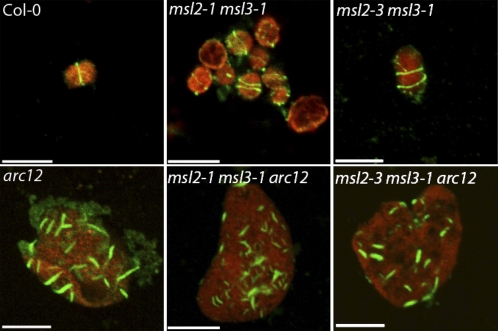
MinE Is Required for Multiple Z-Rings to Form in the msl2 msl3 Background
We next analyzed Z-ring assembly in msl2 msl3 arc12 triple mutant plants. MinE, a component of the Arabidopsis Min system, is thought to oppose the action of MinD and ARC3, acting as a positive regulator of FtsZ assembly. As previously shown (Glynn et al., 2007), Z-rings were absent in chloroplasts isolated from arc12 plants. Instead, small fragments of FtsZ polymers were detected throughout the enlarged chloroplasts (bottom left panel, Figure 5). The same phenotype was observed in chloroplasts isolated from both msl2 msl3 arc12 triple mutants (bottom middle and right panels, Figure 5). This result suggests that the formation of multiple Z-rings in msl2 msl3 double mutants requires MinE and lends further support to a model where MSL2 and MSL3 act in the same pathway as the Min system to regulate chloroplast division.
Figure 5.
msl2 msl3 arc12 Triple Mutants Fail to Form Complete Z-Rings.
IFM analysis of Z-ring placement in chloroplasts isolated from rosette leaves. FITC and chlorophyll fluorescence are pseudocolored green and red, respectively. Representative images from two independent experiments are presented. Bars = 10 μm.
MS Channels Are Required for Normal Z-Ring Placement in E. coli
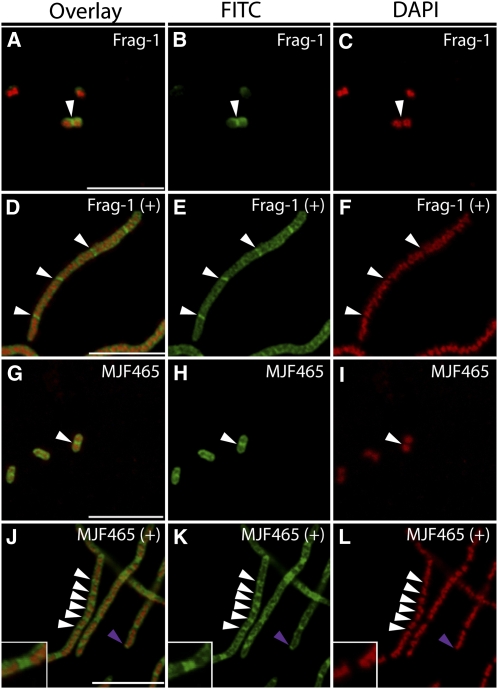
We next considered that MS channels might also play a role in the regulation of Z-ring placement in bacteria. To test this hypothesis, IFM using an antibody raised against E. coli FtsZ was used to visualize Z-rings in the MJF465 strain, which lacks three MS channels, MscS, MscL, and MscK, and is unable to survive extreme hypoosmotic downshock (Levina et al., 1999). No apparent difference between the growth of the wild type (Frag-1) and MJF465 was observed under standard conditions, and IFM revealed Z-rings placed at mid-cell in both backgrounds (Figures 6A to 6C and 6G to 6I). However, when treated with the septation inhibitor cephalexin, MJF465 cells frequently exhibited increased numbers of Z-rings compared with the wild type (arrows, Figures 6D to 6F and 6J to 6L). Additionally, we consistently observed polar Z-rings and double Z-rings (purple arrow and inset, Figures 6J to 6L) in MJF465 cells treated with cephalexin. Approximately half of the MJF465 cells treated with cephalexin showed these types of alterations in Z-ring assembly; in these cells, the average distance between Z-rings was 1.7 ± 0.9 μm (n = 78) compared with 5.83 ± 2.0 μm (n = 55) in Frag-1. This phenotype resembles that of the ΔminB mutant (PB114), in which the Min system operon, including minC, minD, and minE, is deleted (de Boer et al., 1989; Yu and Margolin, 1999). Inhibition of mini-cell formation through treatment with cephalexin revealed that PB114 mutant cells exhibit multiple and polar Z-rings, as previously shown (Adler et al., 1967; Davie et al., 1984; Yu and Margolin, 1999; see Supplemental Figure 5 online). In PB114 cells treated with cephalexin, the distance between Z-rings was 2.1 ± 0.5 μm (n = 67). Thus, MS channels contribute to normal Z-ring placement in E. coli, and Z-ring placement in MJF465 cells resembles that of ΔminB cells.
Figure 6.
Bacterial MS Channels Are Required for Normal FtsZ Ring in the Presence of Cephalexin.
Immunofluorescence micrographs of FtsZ (white arrowheads, pseudocolored green) and 4′,6-diamidino-2-phenylindole (DAPI) staining of nucleoids (pseudocolored red) of wild-type (Frag-1; [A] to [F]) and mscS- mscL- mscK- (MJF465; [G] to [L]) E. coli cells. Addition of 10 mM cephalexin for 1 h prior to fixing ([D] to [F] and [J] to [L]) is indicated by a plus sign. Purple arrowheads indicate polar Z-rings. Double Z-rings were frequently observed in the MJF465 cells (inset, [J] to [L]). Bars = 10 μm. Ler, Landsberg erecta.
DISCUSSION
MSL2 and MSL3 were first identified based on their homology to the well-characterized bacteria MS channel MscS (Martinac and Kloda, 2003; Pivetti et al., 2003; Haswell and Meyerowitz, 2006). Based on the observation that MSL3 can rescue the osmotic stress sensitivity of the E. coli MS channel mutant MJF465, MSL2 and MSL3 are likely candidates for MS channels in Arabidopsis (Levina et al., 1999; Haswell and Meyerowitz, 2006). However, their localization to the envelope of chloroplasts—intracellular organelles without walls—was puzzling. Furthermore, the underlying source of the multiple phenotypes associated with msl2-1 msl3-1 mutant plants remained unknown. In this report, a genetic approach was used to investigate the role MSL2 and MSL3 play in the determination of chloroplast size and the conserved nature of their function.
Defective Chloroplast Division Explains the Enlarged Chloroplasts of msl2 msl3 Double Mutants
The production of enlarged chloroplasts typically results from defects in the initiation or execution of chloroplast division. Many established components of the plastid division apparatus were first identified as mutants with enlarged plastids, including nine arc mutants, cdp1, mcd1, pdv1, and br04 (Pyke and Leech, 1992, 1994; Pyke et al., 1994; Aldridge et al., 2005; Miyagishima et al., 2006; Nakanishi et al., 2009a; Suzuki et al., 2009; Zhang et al., 2009). In this analysis of msl2 msl3 mutants, FtsZ ring formation was used as a cell-based marker for normal chloroplast division. The production of a Z-ring is an early step in division, so mutations that affect FtsZ ring assembly or placement could be distinguished from defects in division downstream of FtsZ ring formation or defects in the perception of size. Both IFM and confocal imaging of an FtsZ1-GFP transgene showed that the chloroplasts in msl2 msl3 mutant plants exhibited multiple Z-rings (Figures 1 and 2). Two independent methods of analysis thus show that the enlarged chloroplasts observed in msl2 msl3 double mutants can be explained by defects in chloroplast division and, more specifically, by overactive Z-ring assembly.
MSL2 and MSL3 Are Components of the Plastid Division Machinery
The multiple Z-ring phenotype of msl2 msl3 mutants closely resembles that of previously identified lesions in factors that negatively regulate FtsZ assembly: MinD, ARC3, MCD1, and PARC6 (Figures 1B and 2A; Glynn et al., 2007, 2009; Fujiwara et al., 2008; Nakanishi et al., 2009a). Analyses of genetic lesions, overexpression studies, and protein–protein assays have demonstrated that MinD, ARC3, MCD1, and PARC6 are dedicated components of the chloroplast Min system (reviewed in Maple and Møller, 2007b, 2010; Yang et al., 2008; Miyagishima and Kabeya, 2010). Given the specific nature of the multiple Z-ring phenotype, MSL2 and MSL3 can also be considered components of the chloroplast division machinery.
MSL2 and MSL3 Regulate Z-Ring Formation through the Min System
To determine if MSL2 and MSL3 regulate FtsZ assembly through the same pathway as known components of the plastid division apparatus, we compared single, double, and triple mutants. If MSL2 and MSL3 act in the same pathway as MinD, MinE, and ARC3, the phenotype of triple msl2 msl3 arc mutants would be the same as single arc mutants. Alternatively, if MSL2 and MSL3 affect Z-ring assembly in a manner independent of Z-ring assembly, both pathways would be impaired in msl2 msl3 arc triple mutants, and an increase in phenotypic severity should be seen. As shown in Figures 3 to 5 and Supplemental Figure 4 online, msl2 msl3 arc11, msl2 msl3 arc3, and msl2 msl3 arc12 triple mutants contained the same number of chloroplasts and displayed a Z-ring formation phenotype nearly identical to those observed in single arc11, arc3, or arc12 mutants. Importantly, the phenotypes of arc11 and arc3 mutants are not so severe that an additive or even synthetic phenotype would not be discernable in msl2 msl3 arc11 or msl2 msl3 arc3 triple mutants (Figures 3 and 4; see Supplemental Figure 4 online). The lack of enhancement of these phenotypes in the triple mutants suggests that MSL2, MSL3, ARC3, and MinD regulate Z-ring formation through the same pathway. Additional evidence that MSL2 and MSL3 function in the same pathway as the Min system is the observation that the multiple Z-ring phenotype of msl2 msl3 double mutants required the function of MinE. Whereas the msl2 msl3 mutants exhibited chloroplasts with multiple Z-rings, no complete Z-rings were present in the triple msl2 msl3 arc12 mutant, a phenotype identical to that observed in the single arc12 mutant (Glynn et al., 2007; Figure 5).
MS Channels Affect Z-Ring Assembly in Bacteria
MSL2 and MSL3, as well as several established components of the chloroplast Min system, are evolutionarily conserved (Haswell and Meyerowitz, 2006; reviewed in Pivetti et al., 2003; Nakanishi et al., 2009b). It is therefore intriguing that MS channels also play a role in the placement of Z-rings in E. coli. As shown in Figure 6, in the absence of MscS, MscK, and MscL—the major MS channels of the E. coli plasma membrane—cells treated with cephalexin exhibited increased Z-ring number, double rings, and rings located at the poles of cells. These results are consistent with the hypothesis that MS channels in E. coli affect Z-ring placement through the Min system, as they do in plants. As Z-rings are placed solely at the sites between nucleoids (cf. white arrows in Figures 6K and 6L), the defects in Z-ring placement we observed in MJF465 cannot be attributed to a deficit of the nucleoid occlusion pathway (reviewed in Bramkamp and van Baarle, 2009).
MS Channels Provide a Functional Link between Chloroplast Division and Membrane Tension
These results raise the question of the molecular mechanism by which MSL2 and MSL3 affect Z-ring formation. They do not appear to act at the level of gene expression, as mRNA levels of FTSZ1, FTSZ2-1, FTSZ2-2, MinD, ARC3, and MCD1 were the same in msl2 msl3 and wild-type plants (see Supplemental Figure 6 online). Furthermore, quantitative immunoblotting showed that increased levels of FtsZ-GFP are not observed in msl2 msl3 mutants; alterations in FtsZ protein levels thus cannot explain the increased Z-ring number seen in this line (Figure 2B).
As MSL2 and MSL3 each contain an N-terminal chloroplast transit peptide, they are predicted to localize to the inner envelope of the chloroplast (Haswell and Meyerowitz, 2006; Li and Chiu, 2010). Topology predictions place their large C-terminal domain inside the stroma, (ARAMEMNON database; Schwacke et al., 2003), an arrangement analogous to that of MscS within the E. coli cell. This predicted topology raises the possibility that MSL2 and MSL3 might interact directly with stromal components of the plastid division machinery. However, we were unable to detect direct interactions between the C-terminal domain of MSL3 and FtsZ1, FtsZ2-1, MinE, MinD, ARC3, or PARC6 (A. Vijayaraghavan and E.S. Haswell, unpublished data; see Methods for details) and thus favor the hypothesis that MSL2 and MSL3 indirectly regulate the Min system. Like MscS, MSL2 and MSL3 may serve to release membrane tension, perhaps derived from chloroplast expansion, and constriction during division or other stresses. If so, the increased membrane tension present in msl2 msl3 mutant chloroplasts could affect the function of membrane-associated members of the Min system. Both ARC3 and MinD are predicted to interact with the inner chloroplast envelope, ARC3 through its membrane occupation and recognition nexus (MORN) motif and MinD through C-terminal amphipathic helices (Kanamaru et al., 2000; Hu et al., 2002; Szeto et al., 2002; Shimada et al., 2004). Membrane tension could also affect the function of PARC6, a chloroplast inner envelope protein that inhibits FtsZ assembly and interacts directly with ARC3 (Glynn et al., 2009). An alternative model is that MSL2 and MSL3 are required to maintain ion homeostasis in the chloroplast stroma; in their absence, changes in ion concentration could affect the function of chloroplast division proteins. For example, MinD is a calcium-dependent ATPase (Aldridge and Møller, 2005) and its function could be altered by abnormal stromal Ca2+ levels.
Taken together, the data presented here indicate that MS channels play an important role in the division of Arabidopsis chloroplasts and contribute to division site placement in E. coli cells, linking membrane tension to the assembly of FtsZ rings in chloroplasts and bacteria. Future work will uncover the nature of the mechanical signal that is relayed by MSL2 and MSL3 to the Min system to properly regulate FtsZ ring placement.
METHODS
Plant Growth and Mutant Analysis
All plants were grown on soil at 21°C under a 16-h light regime. arc11 (CS281) and arc3 (CS264) mutant lines were obtained from the ABRC (Ohio State University) and are in the Landsberg erecta background. The msl2-3 mutant line (GK-195D11) was identified in the GABI-Kat collection of T-DNA insertion lines (Rosso et al., 2003) and is in the Columbia-0 (Col-0) background. The ftsZ-KO and arc12 mutant lines were a gift of K. Osteryoung and are in the Columbia-0 background (Glynn et al., 2007; Schmitz et al., 2009). msl2-1 and msl3-1 mutants are in the Wassilewskija background (Haswell and Meyerowitz, 2006). Derived cleaved-amplified polymorphic sequence genotyping of the arc mutants was performed as described by Neff et al. (1998) using the following oligo pairs and restriction enzymes: arc11DCAPS.F/arc11DCAPS.R (SphI), arc12DCAPS.F/arc12DCAPS.R (NcoI), and arc3DCAPS.F2/arc3DCAPS.R6 (EcoRV). PCR genotyping of msl mutant alleles was performed using the following oligo pairs: JL-202/2.3′ (msl2-1), JL-202/3.3′ (msl3-1), and LB-GABI/msl2-R4 (msl2-3). Refer to Supplemental Table 1 online for oligo sequences.
Isolation and Immunofluorescence Staining of Chloroplasts
Chloroplast isolation and immunoflorescence staining was performed as described by Strawn et al. (2007) with the following modifications. Leaf fragments were digested in protoplast buffer (400 mM sorbitol, 20 mM MES/KOH, pH 5.2, and 0.5 mM CaCl2) containing cellulase (2.5 mg mL−1) and pectinase (0.8 mg mL−1). Samples were blocked for 20 min in PBS, 5% BSA. The anti-FtsZ antibody (Levin and Losick, 1996) was used at a dilution of 1:5000. Slides were blocked for 20 min in PBS, 5% BSA, followed by a 1-h incubation with FITC-conjugated anti-rabbit secondary antibody (1:160; Sigma-Aldrich). Slides were mounted using the SlowFade Antifade kit (Invitrogen).
Light Microscopy of Chloroplasts
Tissue collection, fixation, and sample preparation were performed as described previously (Haswell and Meyerowitz, 2006). Images were captured with an Olympus DP71 microscope digital camera and processed with DP-BSW software.
Confocal Microscopy
All confocal laser scanning microscopy was performed using a FLUOVIEW FV1000 (Olympus), and images were captured with FVIO-ASW software (Olympus). For imaging of FtsZ-GFP–expressing lines (Fujiwara et al., 2008), FITC and GFP signals were excited at 488 nm and emissions collected with a 505- to 606-nm band-pass filter. Chlorophyll autofluorescence was excited at 635 nm and emissions collected with a 655- to 755-nm band-pass filter. For bacterial images, excitation wavelengths of 488 and 405 nm were used, and emissions were collected using 505- to 605-nm (FITC) and 430- to 470-nm (DAPI) band-pass filters.
Chloroplast Counting
The second true leaf was removed from 4-week-old soil-grown plants, placed flat on Murashige and Skoog solid medium, and exposed to direct light for 4 h, with an average intensity of 100 μmol m−2 s−1, to induce chloroplast movement. Tissue removed from the tip of leaves was mounted in water, and confocal scans were taken of mesophyll cells at their largest circumference. After imaging, the number of chloroplasts per optical section were counted using ImageJ version 1.44 software (http://imagej.nih.gov/ij/).
Bacterial Growth Conditions and Immunofluorescence
Frag-1 and MJF465 strains (Levina et al., 1999) or PB103 and PB114 (de Boer et al., 1989) were grown in Luria- Bertani medium to an OD600 of 0.4 and then diluted to an OD600 of 0.1 and treated with or without 10 μg mL−1 cephalexin. After 1 to 2 h of growth, cells were fixed in 16% paraformaldehyde/4% glutaraldehyde for 30 min at room temperature followed by 1 h on ice. Cells were washed and resuspended in GTE (50 mM Glc, 25 mM Tris, and 10 mM EDTA) to an OD600 of 0.2. Immunoflorescence was performed as described (Levin, 2002). Slides were incubated overnight at 4°C with rabbit anti-FtsZ antibody (1:4000; Mercer and Weiss, 2002), followed by a conjugated anti-rabbit FITC antibody (1:200; Sigma-Aldrich). Slides were treated with 1 μg mL−1 4′,6-diamidino-2-phenylindole and mounted with the SlowFade Antifade kit. Distance between FtsZ rings was measured in pixels using ImageJ.
Quantitative Immunoblotting
One hundred milligrams of plant tissue was ground in liquid N2, and 200 μL of extraction buffer (150 mM NaCl, 100 mM Tris, pH 7.5, 0.1% Triton X-100, 10% glycerol, and protease inhibitors) was added. After spinning at 4°C for 4 min, equal volumes of extract and 2× SDS sample buffer were mixed and boiled for 5 min. Protein sample dilutions were separated on a 10% SDS-polyacrylamide gel and electroblotted to polyvinylidene fluoride membrane. Membranes were blocked for 1 h with 5% nonfat milk in TBST and probed with mouse anti-GFP (Clontech; 1:5000) or anti-FtsZ antibody (1:5000) (Levin and Losick, 1996) followed by anti-mouse IgG-HRP (Sigma-Aldrich; 1:5000). Chemiluminescent detection was performed with the Thermo-Scientific SuperSignal West Femto detection kit (Pierce). FtsZ1-GFP protein levels were quantified as described by Schmitz et al. (2009) with the following modifications. The calculated density value of msl2-3 msl3-1 bands was divided by the density value of Col-0 bands to determine the FtsZ1-GFP protein levels present in msl2-3 msl3-1 plants relative to FtsZ1-GFP protein levels in Col-0 plants. Bands imaged from lanes containing comparable total protein levels of Col-0 and msl2-3 msl3-1, as determined by Ponceau S staining of ribulose-1,5-bisphosphate carboxylase/oxygenase, indicate similar total protein concentrations at each dilution.
Quantitative RT-PCR
Primer mixes designed to amplify FtsZ1 (FtsZ1.QPCR.F and FtsZ1.QPCR.R), FtsZ2-1 (FtsZ2-1.QPCR.F and FtsZ2-1.QPCR.R), FtsZ2-2 (FtsZ2-2.QPCR.F and FtsZ2-2.QPCR.R), MinD (MinD.QPCR.F and MinD.QPCR.R), ARC3 (ARC3.QPCR.F and ARC3.QPCR.R) MCD1 (MCD1.QPCR.F and MCD1.QPCR.R), and ACTIN2/7/8 (ACTF-QPCR/equal volumes of Actin2.R-QPCR, Actin7.R-QPCR and Actin8.R-QPCR) were added to a cocktail containing 1× SYBR Green PCR Master Mix (Applied Biosciences) and 0.5 μL rosette leaf cDNA to make a final 10 μL reaction. After amplification in the StepOnePlus real-time PCR system, the data was analyzed using StepOne software (Applied Biosciences).
Yeast Two-Hybrid Analysis
We tested for protein–protein interactions between MSL3 and components of the plastid division machinery using the ProQuest two-hybrid system (Invitrogen). cDNA sequence encoding the soluble C terminus of MSL3 (amino acids 286 to 678) was cloned into the pDEST 32 bait vector, while full-length cDNAs for MinD, MinE, FtsZ1, FtsZ2-1, and cDNAs encoding the stromal portions of ARC3 (amino acids 41 to 741) and PARC6 (amino acids 77 to 352) were cloned into the pDEST 22 prey vector. The assays were performed according to the manufacturer’s instructions.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MSL2, At5g10490; MSL3, At1g58200; MinD, At5g24020; MinE, At1g69390; ARC3, At1g75010; FtsZ1, At5g55280; FtsZ2-1, At2g36250; FtsZ2-2, At3g52750; and MCD1, At1g20830.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Characterization of the msl2-3 Allele.
Supplemental Figure 2. An Antibody Raised against Bacillus subtilis FtsZ Recognizes Arabidopsis thaliana FtsZ1.
Supplemental Figure 3. Representative Images of Confocal Scans Used to Determine Chloroplast Number in Single, Double, and Triple msl and arc Mutants.
Supplemental Figure 4. msl2 msl3 arc3 Triple Mutants Are Phenotypically Indistinguishable from Single arc3 Mutants.
Supplemental Figure 5. Z-Ring Placement in the ΔminB Strain of E. coli.
Supplemental Figure 6. Transcript Levels of Genes Involved in Chloroplast Division Are Not Altered in msl2 msl3 Mutants.
Supplemental Table 1. Oligos Used in Genotyping and Quantitative RT-PCR.
Supplementary Material
Acknowledgments
We thank the GABI-KAT project for the msl2-3 mutant line, the ABRC for the arc11 and arc3 mutant lines, Katherine Osteryoung for the arc12 mutant and ftsZ KO lines, and Makoto Fujiwara for the FtsZ1-GFP transgenic line. The anti-EcFtsZ antibody was a kind gift from David Weiss. PB114 and PB103 were provided by Piet de Boer. We thank Petra Levin and her laboratory for expert advice and help and for the anti-BsFtsZ antibody. We also thank Anupama Vijayaraghavan, Mike Dyer, and the rest of the Jeanette Goldfarb Plant Growth Facility staff for their assistance. This research was funded by National Science Foundation Award 0816627.
AUTHOR CONTRIBUTIONS
M.E.W, G.S.J, and E.S.H performed research and contributed new tools. M.E.W. and E.S.H designed research, analyzed data, and wrote the manuscript.
References
- Adler H.I., Fisher W.D., Cohen A., Hardigree A.A. (1967). Miniature Escherichia coli cells deficient in DNA. Proc. Natl. Acad. Sci. USA 57: 321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge C., Maple J., Møller S.G. (2005). The molecular biology of plastid division in higher plants. J. Exp. Bot. 56: 1061–1077 [DOI] [PubMed] [Google Scholar]
- Aldridge C., Møller S.G. (2005). The plastid division protein AtMinD1 is a Ca2+-ATPase stimulated by AtMinE1. J. Biol. Chem. 280: 31673–31678 [DOI] [PubMed] [Google Scholar]
- Bi E., Dai K., Subbarao S., Beall B., Lutkenhaus J. (1991). FtsZ and cell division. Res. Microbiol. 142: 249–252 [DOI] [PubMed] [Google Scholar]
- Bi E.F., Lutkenhaus J. (1991). FtsZ ring structure associated with division in Escherichia coli. Nature 354: 161–164 [DOI] [PubMed] [Google Scholar]
- Bramkamp M., van Baarle S. (2009). Division site selection in rod-shaped bacteria. Curr. Opin. Microbiol. 12: 683–688 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. (2000). Membrane heredity and early chloroplast evolution. Trends Plant Sci. 5: 174–182 [DOI] [PubMed] [Google Scholar]
- Colletti K.S., Tattersall E.A., Pyke K.A., Froelich J.E., Stokes K.D., Osteryoung K.W. (2000). A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr. Biol. 10: 507–516 [DOI] [PubMed] [Google Scholar]
- Dai K., Lutkenhaus J. (1992). The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J. Bacteriol. 174: 6145–6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie E., Sydnor K., Rothfield L.I. (1984). Genetic basis of minicell formation in Escherichia coli K-12. J. Bacteriol. 158: 1202–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P., Crossley R., Rothfield L. (1992a). The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359: 254–256 [DOI] [PubMed] [Google Scholar]
- de Boer P.A. (2010). Advances in understanding E. coli cell fission. Curr. Opin. Microbiol. 13: 730–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P.A., Crossley R.E., Hand A.R., Rothfield L.I. (1991). The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 10: 4371–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P.A., Crossley R.E., Rothfield L.I. (1989). A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56: 641–649 [DOI] [PubMed] [Google Scholar]
- de Boer P.A., Crossley R.E., Rothfield L.I. (1992b). Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J. Bacteriol. 174: 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kafafi S., Karamoko M., Pignot-Paintrand I., Grunwald D., Mandaron P., Lerbs-Mache S., Falconet D. (2008). Developmentally regulated association of plastid division protein FtsZ1 with thylakoid membranes in Arabidopsis thaliana. Biochem. J. 409: 87–94 [DOI] [PubMed] [Google Scholar]
- El-Kafafi S., Mukherjee S., El-Shami M., Putaux J.L., Block M.A., Pignot-Paintrand I., Lerbs-Mache S., Falconet D. (2005). The plastid division proteins, FtsZ1 and FtsZ2, differ in their biochemical properties and sub-plastidial localization. Biochem. J. 387: 669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Shih Y.L., Zhang Y., Rothfield L.I. (2001). The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc. Natl. Acad. Sci. USA 98: 980–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M.T., Hashimoto H., Kazama Y., Abe T., Yoshida S., Sato N., Itoh R.D. (2008). The assembly of the FtsZ ring at the mid-chloroplast division site depends on a balance between the activities of AtMinE1 and ARC11/AtMinD1. Plant Cell Physiol. 49: 345–361 [DOI] [PubMed] [Google Scholar]
- Fujiwara M.T., Hashimoto H., Kazama Y., Hirano T., Yoshioka Y., Aoki S., Sato N., Itoh R.D., Abe T. (2010). Dynamic morphologies of pollen plastids visualised by vegetative-specific FtsZ1-GFP in Arabidopsis thaliana. Protoplasma 242: 19–33 [DOI] [PubMed] [Google Scholar]
- Fujiwara M.T., Nakamura A., Itoh R., Shimada Y., Yoshida S., Møller S.G. (2004). Chloroplast division site placement requires dimerization of the ARC11/AtMinD1 protein in Arabidopsis. J. Cell Sci. 117: 2399–2410 [DOI] [PubMed] [Google Scholar]
- Fujiwara M.T., Sekine K., Yamamoto Y.Y., Abe T., Sato N., Itoh R.D. (2009). Live imaging of chloroplast FtsZ1 filaments, rings, spirals, and motile dot structures in the AtMinE1 mutant and overexpressor of Arabidopsis thaliana. Plant Cell Physiol. 50: 1116–1126 [DOI] [PubMed] [Google Scholar]
- Glynn J.M., Miyagishima S.Y., Yoder D.W., Osteryoung K.W., Vitha S. (2007). Chloroplast division. Traffic 8: 451–461 [DOI] [PubMed] [Google Scholar]
- Glynn J.M., Yang Y., Vitha S., Schmitz A.J., Hemmes M., Miyagishima S.Y., Osteryoung K.W. (2009). PARC6, a novel chloroplast division factor, influences FtsZ assembly and is required for recruitment of PDV1 during chloroplast division in Arabidopsis. Plant J. 59: 700–711 [DOI] [PubMed] [Google Scholar]
- Hale C.A., Meinhardt H., de Boer P.A. (2001). Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J. 20: 1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haswell E.S. (2007). MscS-like proteins in plants. Curr. Top. Membr. 58: 329–359 [Google Scholar]
- Haswell E.S., Meyerowitz E.M. (2006). MscS-like proteins control plastid size and shape in Arabidopsis thaliana. Curr. Biol. 16: 1–11 [DOI] [PubMed] [Google Scholar]
- Hu Z., Gogol E.P., Lutkenhaus J. (2002). Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc. Natl. Acad. Sci. USA 99: 6761–6766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Lutkenhaus J. (1999). Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 34: 82–90 [DOI] [PubMed] [Google Scholar]
- Huang J., Cao C., Lutkenhaus J. (1996). Interaction between FtsZ and inhibitors of cell division. J. Bacteriol. 178: 5080–5085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh R., Fujiwara M., Nagata N., Yoshida S. (2001). A chloroplast protein homologous to the eubacterial topological specificity factor minE plays a role in chloroplast division. Plant Physiol. 127: 1644–1655 [PMC free article] [PubMed] [Google Scholar]
- Kanamaru K., Fujiwara M., Kim M., Nagashima A., Nakazato E., Tanaka K., Takahashi H. (2000). Chloroplast targeting, distribution and transcriptional fluctuation of AtMinD1, a Eubacteria-type factor critical for chloroplast division. Plant Cell Physiol. 41: 1119–1128 [DOI] [PubMed] [Google Scholar]
- Karamoko M., El-Kafafi S., Mandaron P., Lerbs-Mache S., Falconet D. (2011). Multiple FtsZ2 isoforms involved in chloroplast division and biogenesis are developmentally associated with thylakoid membranes in Arabidopsis. FEBS Lett. 585: 1203–1208 [DOI] [PubMed] [Google Scholar]
- Levin P.A. (2002). Light microscopy techniques for bacterial cell biology. In Molecular Cellular Microbiology, Vol. 31, Sansonetti P., Zychlinsky A., (London, New York: Academic Press; ), pp. 115–132 [Google Scholar]
- Levin P.A., Losick R. (1996). Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 10: 478–488 [DOI] [PubMed] [Google Scholar]
- Levina N., Tötemeyer S., Stokes N.R., Louis P., Jones M.A., Booth I.R. (1999). Protection of Escherichia coli cells against extreme turgor by activation of MscS and MscL mechanosensitive channels: Identification of genes required for MscS activity. EMBO J. 18: 1730–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.M., Chiu C.C. (2010). Protein transport into chloroplasts. Annu. Rev. Plant Biol. 61: 157–180 [DOI] [PubMed] [Google Scholar]
- Liu W.Z., Kong D.D., Wang D., Ju C.L., Hu Y., Liu X.L., Sun J.S., He Y.K. (2007). The involvement of NtFtsZ2-1 gene in the regulation of chloroplast division and expansion in tobacco. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 33: 267–276 [PubMed] [Google Scholar]
- Lopez-Juez E., Pyke K.A. (2005). Plastids unleashed: Their development and their integration in plant development. Int. J. Dev. Biol. 49: 557–577 [DOI] [PubMed] [Google Scholar]
- Maple J., Chua N.H., Møller S.G. (2002). The topological specificity factor AtMinE1 is essential for correct plastid division site placement in Arabidopsis. Plant J. 31: 269–277 [DOI] [PubMed] [Google Scholar]
- Maple J., Møller S.G. (2007a). Interdependency of formation and localisation of the Min complex controls symmetric plastid division. J. Cell Sci. 120: 3446–3456 [DOI] [PubMed] [Google Scholar]
- Maple J., Møller S.G. (2007b). Plastid division: Evolution, mechanism and complexity. Ann. Bot. (Lond.) 99: 565–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maple J., Møller S.G. (2010). The complexity and evolution of the plastid-division machinery. Biochem. Soc. Trans. 38: 783–788 [DOI] [PubMed] [Google Scholar]
- Maple J., Vojta L., Soll J., Møller S.G. (2007). ARC3 is a stromal Z-ring accessory protein essential for plastid division. EMBO Rep. 8: 293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinac B., Kloda A. (2003). Evolutionary origins of mechanosensitive ion channels. Prog. Biophys. Mol. Biol. 82: 11–24 [DOI] [PubMed] [Google Scholar]
- McAndrew R.S., Froehlich J.E., Vitha S., Stokes K.D., Osteryoung K.W. (2001). Colocalization of plastid division proteins in the chloroplast stromal compartment establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants. Plant Physiol. 127: 1656–1666 [PMC free article] [PubMed] [Google Scholar]
- McAndrew R.S., Olson B.J., Kadirjan-Kalbach D.K., Chi-Ham C.L., Vitha S., Froehlich J.E., Osteryoung K.W. (2008). In vivo quantitative relationship between plastid division proteins FtsZ1 and FtsZ2 and identification of ARC6 and ARC3 in a native FtsZ complex. Biochem. J. 412: 367–378 [DOI] [PubMed] [Google Scholar]
- Mercer K.L., Weiss D.S. (2002). The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J. Bacteriol. 184: 904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima S.Y., Froehlich J.E., Osteryoung K.W. (2006). PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 18: 2517–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima S.Y., Kabeya Y. (2010). Chloroplast division: Squeezing the photosynthetic captive. Curr. Opin. Microbiol. 13: 738–746 [DOI] [PubMed] [Google Scholar]
- Miyagishima S.Y., Nishida K., Kuroiwa T. (2003). An evolutionary puzzle: Chloroplast and mitochondrial division rings. Trends Plant Sci. 8: 432–438 [DOI] [PubMed] [Google Scholar]
- Mukherjee A., Dai K., Lutkenhaus J. (1993). Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc. Natl. Acad. Sci. USA 90: 1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Lutkenhaus J. (1994). Guanine nucleotide-dependent assembly of FtsZ into filaments. J. Bacteriol. 176: 2754–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H., Suzuki K., Kabeya Y., Miyagishima S.Y. (2009a). Plant-specific protein MCD1 determines the site of chloroplast division in concert with bacteria-derived MinD. Curr. Biol. 19: 151–156 [DOI] [PubMed] [Google Scholar]
- Nakanishi H., Suzuki K., Kabeya Y., Okazaki K., Miyagishima S.Y. (2009b). Conservation and differences of the Min system in the chloroplast and bacterial division site placement. Commun. Integr. Biol. 2: 400–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M.M., Turk E., Kalishman M. (2002). Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 18: 613–615 [DOI] [PubMed] [Google Scholar]
- Olson B.J., Wang Q., Osteryoung K.W. (2010). GTP-dependent heteropolymer formation and bundling of chloroplast FtsZ1 and FtsZ2. J. Biol. Chem. 285: 20634–20643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M., Anderson D.E., Erickson H.P. (2009). Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J. 28: 3476–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K.W., Stokes K.D., Rutherford S.M., Percival A.L., Lee W.Y. (1998). Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K.W., Vierling E. (1995). Conserved cell and organelle division. Nature 376: 473–474 [DOI] [PubMed] [Google Scholar]
- Pivetti C.D., Yen M.R., Miller S., Busch W., Tseng Y.H., Booth I.R., Saier M.H., Jr (2003). Two families of mechanosensitive channel proteins. Microbiol. Mol. Biol. Rev. 67: 66–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K.A., Leech R.M. (1992). Chloroplast division and expansion is radically altered by nuclear mutations in Arabidopsis thaliana. Plant Physiol. 99: 1005–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K.A., Leech R.M. (1994). A genetic analysis of chloroplast division and expansion in Arabidopsis thaliana. Plant Physiol. 104: 201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K.A., Rutherford S.M., Robertson E.J., Leech R.M. (1994). arc6, a fertile Arabidopsis mutant with only two mesophyll cell chloroplasts. Plant Physiol. 106: 1169–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin D.M., de Boer P.A. (1999a). MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol. 181: 6419–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin D.M., de Boer P.A. (1999b). Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA 96: 4971–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy M.S., Dinkins R., Collins G.B. (2002). Overexpression of the Arabidopsis thaliana MinE1 bacterial division inhibitor homologue gene alters chloroplast size and morphology in transgenic Arabidopsis and tobacco plants. Planta 215: 167–176 [DOI] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Rowland S.L., Fu X., Sayed M.A., Zhang Y., Cook W.R., Rothfield L.I. (2000). Membrane redistribution of the Escherichia coli MinD protein induced by MinE. J. Bacteriol. 182: 613–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A.J., Glynn J.M., Olson B.J., Stokes K.D., Osteryoung K.W. (2009). Arabidopsis FtsZ2-1 and FtsZ2-2 are functionally redundant, but FtsZ-based plastid division is not essential for chloroplast partitioning or plant growth and development. Mol. Plant 2: 1211–1222 [DOI] [PubMed] [Google Scholar]
- Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., Frommer W.B., Flügge U.I., Kunze R. (2003). ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H., Koizumi M., Kuroki K., Mochizuki M., Fujimoto H., Ohta H., Masuda T., Takamiya K. (2004). ARC3, a chloroplast division factor, is a chimera of prokaryotic FtsZ and part of eukaryotic phosphatidylinositol-4-phosphate 5-kinase. Plant Cell Physiol. 45: 960–967 [DOI] [PubMed] [Google Scholar]
- Smith A.G., Johnson C.B., Vitha S., Holzenburg A. (2010). Plant FtsZ1 and FtsZ2 expressed in a eukaryotic host: GTPase activity and self-assembly. FEBS Lett. 584: 166–172 [DOI] [PubMed] [Google Scholar]
- Stokes K.D., McAndrew R.S., Figueroa R., Vitha S., Osteryoung K.W. (2000). Chloroplast division and morphology are differentially affected by overexpression of FtsZ1 and FtsZ2 genes in Arabidopsis. Plant Physiol. 124: 1668–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes K.D., Osteryoung K.W. (2003). Early divergence of the FtsZ1 and FtsZ2 plastid division gene families in photosynthetic eukaryotes. Gene 320: 97–108 [DOI] [PubMed] [Google Scholar]
- Strawn M.A., Marr S.K., Inoue K., Inada N., Zubieta C., Wildermuth M.C. (2007). Arabidopsis isochorismate synthase functional in pathogen-induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. J. Biol. Chem. 282: 5919–5933 [DOI] [PubMed] [Google Scholar]
- Sukharev S. (2002). Purification of the small mechanosensitive channel of Escherichia coli (MscS): The subunit structure, conduction, and gating characteristics in liposomes. Biophys. J. 83: 290–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Nakanishi H., Bower J., Yoder D.W., Osteryoung K.W., Miyagishima S.Y. (2009). Plastid chaperonin proteins Cpn60 alpha and Cpn60 beta are required for plastid division in Arabidopsis thaliana. BMC Plant Biol. 9: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto T.H., Rowland S.L., Rothfield L.I., King G.F. (2002). Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc. Natl. Acad. Sci. USA 99: 15693–15698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavva V.S., Collins G.B., Dinkins R.D. (2006). Targeted overexpression of the Escherichia coli MinC protein in higher plants results in abnormal chloroplasts. Plant Cell Rep. 25: 341–348 [DOI] [PubMed] [Google Scholar]
- Vitha S., Froehlich J.E., Koksharova O., Pyke K.A., van Erp H., Osteryoung K.W. (2003). ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15: 1918–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S., McAndrew R.S., Osteryoung K.W. (2001). FtsZ ring formation at the chloroplast division site in plants. J. Cell Biol. 153: 111–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Glynn J.M., Olson B.J., Schmitz A.J., Osteryoung K.W. (2008). Plastid division: Across time and space. Curr. Opin. Plant Biol. 11: 577–584 [DOI] [PubMed] [Google Scholar]
- Yoder D.W., Kadirjan-Kalbach D., Olson B.J., Miyagishima S.Y., Deblasio S.L., Hangarter R.P., Osteryoung K.W. (2007). Effects of mutations in Arabidopsis FtsZ1 on plastid division, FtsZ ring formation and positioning, and FtsZ filament morphology in vivo. Plant Cell Physiol. 48: 775–791 [DOI] [PubMed] [Google Scholar]
- Yu X.C., Margolin W. (1999). FtsZ ring clusters in min and partition mutants: Role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol. Microbiol. 32: 315–326 [DOI] [PubMed] [Google Scholar]
- Zhang M., Hu Y., Jia J., Li D., Zhang R., Gao H., He Y. (2009). CDP1, a novel component of chloroplast division site positioning system in Arabidopsis. Cell Res. 19: 877–886 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.