Reverse genetics approaches were conducted to elucidate the function of the plant-specific calcium binding protein CAS (calcium sensor) that is localized in chloroplast thylakoid membranes. Our findings demonstrate that CAS and Ca2+ are critically involved in the regulation of the high-light response and particularly in the regulation of LHCSR3 expression in green alga Chlamydomonas reinhardtii.
Abstract
The plant-specific calcium binding protein CAS (calcium sensor) has been localized in chloroplast thylakoid membranes of vascular plants and green algae. To elucidate the function of CAS in Chlamydomonas reinhardtii, we generated and analyzed eight independent CAS knockdown C. reinhardtii lines (cas-kd). Upon transfer to high-light (HL) growth conditions, cas-kd lines were unable to properly induce the expression of LHCSR3 protein that is crucial for nonphotochemical quenching. Prolonged exposure to HL revealed a severe light sensitivity of cas-kd lines and caused diminished activity and recovery of photosystem II (PSII). Remarkably, the induction of LHCSR3, the growth of cas-kd lines under HL, and the performance of PSII were fully rescued by increasing the calcium concentration in the growth media. Moreover, perturbing cellular Ca2+ homeostasis by application of the calmodulin antagonist W7 or the G-protein activator mastoparan impaired the induction of LHCSR3 expression in a concentration-dependent manner. Our findings demonstrate that CAS and Ca2+ are critically involved in the regulation of the HL response and particularly in the control of LHCSR3 expression.
INTRODUCTION
Photosynthetic organisms must acclimate to their light environment to optimize photosynthesis and minimize photooxidative damage. In low light (LL), excitation energy is collected to fuel photochemical work, while in high light (HL), excess photons have to be quenched to avoid photodamage. The most rapid response to excess light is provided by a mechanism called nonphotochemical quenching (NPQ). NPQ can be divided in three components: qE, qT, and qI. qT and qI are linked to state transitions and photoinhibitory processes, respectively. The most important constituent of NPQ is qE, which regulates the thermal dissipation of excess absorbed light energy and operates at a time scale of seconds to minutes, allowing for effective photoprotection.
In vascular plants, the photosystem II (PSII) protein PSBS is essential for qE (Li et al., 2000), whereas qE induction in the green alga Chlamydomonas reinhardtii is mediated by LHCSR3, an ancient light-harvesting protein that is missing in vascular plants (Peers et al., 2009). Interestingly, the moss Physcomitrella patens, which possesses genes encoding for PSBS and LHCSR proteins, uses both types of regulatory proteins to operate qE (Alboresi et al., 2010). This suggests that land plants evolved a novel PSBS-dependent qE mechanism before losing the ancestral LHCSR-dependent qE found in algae. While qE is constitutive in vascular plants, it is induced upon acclimation to HL in green algae (Peers et al., 2009). Despite mechanistic description of qE-dependent processes, the signal transduction processes that drive light acclimation in algae, in particular those that control the expression of LHCSR proteins, are poorly understood (Li et al., 2009). In addition to protective mechanisms that are directly related to incident light and the capacity of photosynthesis, fast phototactic movements contribute to photoprotection by avoiding excess exposure to light. Phototaxis involves both light-induced H+ and Ca2+ signaling events that are associated with the eyespot region of Chlamydomonas and mediate the regulation of flagellar bending and appropriate swimming direction (Nagel et al., 2002; Berthold et al., 2008). In C. reinhardtii, many aspects of flagellar function are Ca2+ dependent (Smith, 2002; Patel-King et al., 2004; DiPetrillo and Smith, 2010). In general, defined changes in cytoplasmic Ca2+ concentration are a common biological response to various abiotic and biotic stresses, and such calcium signatures trigger downstream signal transduction and changes in gene expression (Dodd et al., 2010). Stimulus-specific Ca2+ signatures are decoded by Ca2+ binding proteins that function as Ca2+ sensors (Kudla et al., 2010). Upon Ca2+ binding, these proteins change their conformation and interact with their respective target proteins and regulate their biological activity.
The CAS (Ca2+-sensing receptor) protein from Arabidopsis thaliana has initially been described as a cell surface receptor residing in the plasma membrane that mediates extracellular Ca2+ sensing in guard cells (Han et al., 2003). However, functional proteomics approaches revealed the presence of CAS in thylakoid membranes from Arabidopsis (Peltier et al., 2004) and C. reinhardtii (Allmer et al., 2006; Terashima et al., 2010), and recent studies established that CAS is exclusively localized in the thylakoid membrane (Nomura et al., 2008; Vainonen et al., 2008; Weinl et al., 2008). Here, CAS exerts crucial functions for proper stomatal regulation in response to elevations of external Ca2+ through the modulation of cytoplasmic Ca2+ dynamics (Nomura et al., 2008; Weinl et al., 2008). Moreover, dark-induced increases of stromal Ca2+ levels precede the generation of cytosolic Ca2+ transients in tobacco (Nicotiana tabacum) leaf cells (Sai and Johnson, 2002), further suggesting that the chloroplast represents an element of the cellular Ca2+ network and contributes to the cytosolic Ca2+ signaling (Bussemer et al., 2009; McAinsh and Pittman, 2009; Murchie et al., 2009; Kudla et al., 2010).
Besides its role in signal transduction, Ca2+ is an essential cofactor in PSII-driven oxygenic photosynthesis (Boussac et al., 1989; Krieger et al., 1993). Crystal structures of PSII from Thermosynechococcus elongatus obtained at 3.5- and 3.0-Å resolution indeed showed calcium and manganese as integral components of the water-splitting complex (Ferreira et al., 2004). The PSII subunits PSBP and PSBO are capable of Ca2+ binding, but molecular details about Ca2+ delivery into PSII are unknown (Suorsa and Aro, 2007).
Here, we employed reverse genetics approaches to investigate the function of CAS in the unicellular green alga C. reinhardtii. We report that Ca2+ and CAS are essential for induction of HL-dependent expression of LHCSR3 and correspondingly efficient qE quenching to dissipate excess energy. Moreover, the data indicate that the presence of CAS is required for efficient PSII activity and recovery after light stress. These findings identify Ca2+ and CAS as crucial components of acclimation and adaptation to HL. Taken together, our work reveals that HL acclimation and control of LHCSR3 expression require the proper function of CAS and its interconnection with the cellular Ca2+ network.
RESULTS
CAS Protein Expression Is Enhanced in HL
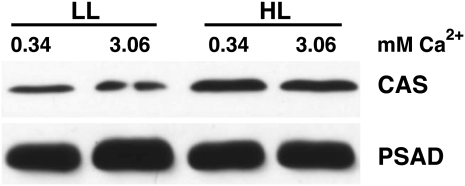
Recent proteomic data revealed that CAS protein expression is induced threefold under iron deficiency compared with iron-replete conditions (Naumann et al., 2007). To investigate whether CAS protein expression is influenced by light and/or calcium, wild-type C. reinhardtii cells were grown in photoautotrophic medium (high-salt medium) containing either 0.34 or 3.06 mM Ca2+ under LL (60 μE m−2 s−1) and HL (200 μE m−2 s−1). Isolated thylakoid membranes were fractionated by SDS-PAGE, and CAS and PSAD protein amounts were analyzed by immunoblotting, using anti-CAS and anti-PSAD antibodies. The data presented in Figure 1 reveal that the CAS protein is twofold induced in HL compared with LL, while PSAD amounts remained unchanged. Moreover, the results of these experiments indicated that CAS protein expression is not altered by a 10-fold increase in Ca2+ concentration in the growth medium.
Figure 1.
CAS Protein Expression Is Enhanced in HL.
Quantification of CAS amounts in thylakoids isolated from LL- and HL-treated wild-type (CC-124) cells upon SDS-PAGE fractionation and immunoblot analysis (5 μg of chlorophyll per lane; PSAD served as the loading control). Cells were grown in HSM and LL and were shifted to HL for 24 h in HSM medium containing the indicated amount of Ca2+ in the form of its chloride salt. Starting cell density of the cultures was 1 × 106 cells/mL. Densitometric analyses of the CAS and PSAD signals showed that CAS is twofold upregulated in HL.
Reduction of CAS Protein Accumulation Renders C. reinhardtii HL Sensitive and Impairs HL-Induced LHCSR3 Expression
To assess the function of the CAS protein in C. reinhardtii, artificial microRNA (amiRNA) and RNA interference (RNAi) approaches were employed. We isolated several amiRNA and RNAi transformants in which the accumulation of the CAS protein in comparison to the wild type was reduced to different degrees. The reduction in CAS protein amounts ranged from twofold up to more than 20-fold (Figures 2A,3A, and 8A; see Supplemental Figures 1A and 5A online).
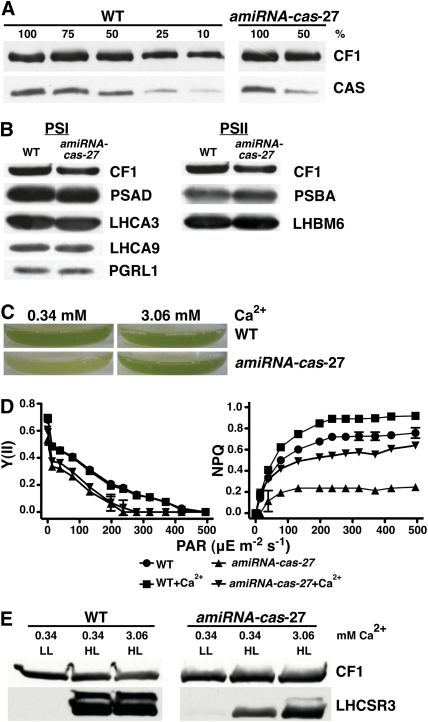
Figure 2.
Knockdown of CAS Leads to Light Sensitivity and to Decreased NPQ Induction and LHCSR3 Expression.
(A) Quantification of CAS amounts in thylakoids isolated from the wild type (WT; cw15-arg7) and amiRNA-cas-27. Cells were grown in TAP under LL upon SDS-PAGE fractionation and immunoblot analysis (100% equals 4 μg of chlorophyll per lane; CF1 served as the loading control).
(B) Quantification of PSI and PSII subunits in the wild type (cw15-arg7) and amiRNA-cas-27 grown in TAP under LL. Four micrograms of chlorophyll of whole-cell extracts was fractionated on a 13% SDS-PAGE, and several protein abundance were analyzed by immunoblots. PSI-related proteins: PSAD, LHCA3, LHCA9, and PGRL1; PSII-related proteins: PSBA and LHCBM6; loading control: CF1.
(C) Growth phenotype of the wild type (cw15-arg7) and cas-kd strain amiRNA-cas-27 after 18 h under HL. Cells were grown in 80% HSM containing the indicated amount of Ca2+ in the form of its chloride salt. Starting cell density of the cultures was 1 × 106 cells/mL. The picture shows the bottom of Erlenmeyer flasks containing the cell cultures.
(D) Irradiance dependence of quantum yield of PSII (YII) and of NPQ in the wild type (cw15arg7) and amiRNA-cas-27. Cells were grown in TAP under LL and then exposed for 2 h under HL in 80% HSM. Values plotted are the means of three samples ± sd.
(E) Quantification of LHCSR3 amounts in the wild type (cw15-arg7) and amiRNA-cas-27. Cells were grown in TAP under LL and then shifted for 2 h to HL (200 μE m−2 s−1) in 80% HSM containing 0.34 or 3.06 mM CaCl2 concentration. Five micrograms of chlorophyll of whole-cell extracts was fractionated on a 13% SDS-PAGE, and LHCSR3 abundance was analyzed by immunoblots. CF1 signal served as a loading control.
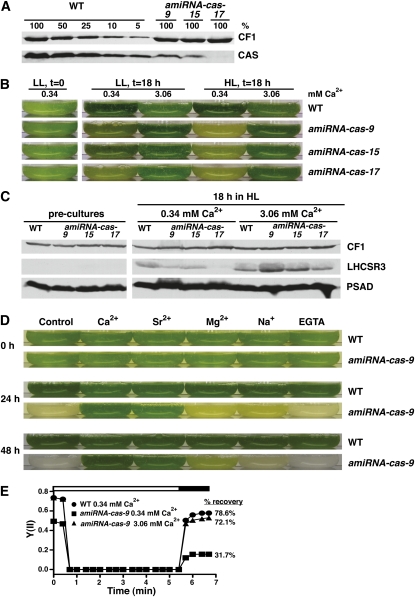
Figure 3.
Three Additional CAS Knockdown Lines Confirm the amiRNA-cas-27 HL Growth Phenotype and the Impact on LHCSR3 Expression.
(A) Quantification of CAS amounts in thylakoids isolated from the wild type (WT; CC-124) and amiRNA-cas-9, -15, and -17. Cells were grown in TAP under LL and analyzed by SDS-PAGE fractionation and immunoblotting (100% equals 10 μg of chlorophyll per lane; CF1 signal served as loading control).
(B) Growth phenotype of wild-type (CC-124) and cas knockdown strains (amiRNA-cas-9, -15, and -17) after an 18-h shift from LL TAP to LL or HL 80% HSM containing 0.34 or 3.06 mM CaCl2. Starting cell density of the cultures was 1 × 106 cells/mL. The picture shows the bottom of Erlenmeyer flasks containing the cell cultures.
(C) Quantification of LHCSR3 in wild-type (CC-124) and cas knockdown strains. Five micrograms of chlorophyll of whole-cell extracts derived from the experiment of Figure 3B were fractionated on a 13% SDS-PAGE and PSAD, and LHCSR3 and CF1 abundance was analyzed by immunoblots. CF1 signal served as a loading control.
(D) Growth phenotype of the wild type (CC-124) and amiRNA-cas-9 24 and 48 h after being shifted from LL TAP to HL 80% HSM with the addition either of the indicated ions in their chloride salt form (final concentration 3.06 mM) or of EGTA (2 mM). Control contained 0.34 mM Ca2+, 0.41 mM Mg2+, and 0.27 mM Na+. Starting cell density of the cultures was 1 × 106 cells/mL.
(E) Recovery of PSII activity in wild-type and amiRNA-cas-9 cells. amiRNA-cas-9 cells were grown for 16 h under LL in HSM medium containing the indicated amounts of Ca2+. Cells were then dark adapted for 20 min, and the recovery of their PSII activity was recorded during a short dark period (black bar), which followed a 5-min light period of 800 μE m−2 s−1 (white bar). Values plotted are the means of three measurements ± sd of single biological samples of a representative experiment.
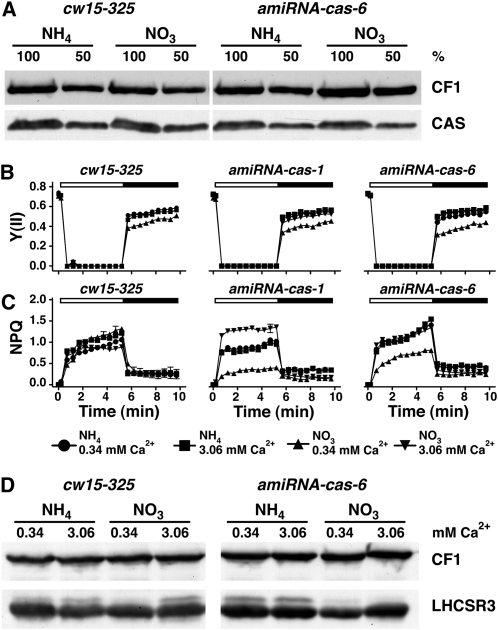
Figure 8.
amiRNA-cas-1 and -6 Show Unaltered Fv/Fm Compared with the Wild Type but Diminished NPQ Induction and PSII Recovery, Which Can Be Rescued by Calcium.
(A) Quantification of CAS amounts in thylakoids isolated from the wild type (cw15-325) and amiRNA-cas-6 grown under LL in TAP-NH4 (amiRNA expression repressed) or TAP-NO3 (amiRNA expression activated) upon SDS-PAGE fractionation and immunoblot analysis (100% equals 70 μg of protein per lane; CF1 signal served as loading control). Densitometric analyses showed that CAS levels in amiRNA-cas-6 are reduced by 40% compared with the wild-type levels (in NO3).
(B) and (C) Determination of PSII quantum yield and recovery and NPQ. The wild type, amiRNA-cas-1, and amiRNA-cas-6 were initially shifted from LL TAP-NH4 to LL TAP-NO3 for 40 h to activate the amiRNA expression and subsequently shifted to HL in either HSM-NH4 or HSM-NO3 containing 0.34 or 3.06 mM Ca2+ for 24 h. After 20 min of dark adaptation, the quantum yields of PSII (B) and NPQ (C) were recorded during 5.2 min of illumination at 800 μE m−2 s−1 (white bar) followed by 4.3 min of darkness (black bar), during which recovery of PSII and relaxation of NPQ could be followed. Values plotted are the means of three measurements ± sd.
(D) Quantification of LHCSR3 amounts in the wild type (cw15-325) and amiRNA-cas-6. Cells were initially shifted from LL TAP-NH4 to LL TAP-NO3 for 40 h to activate the amiRNA expression and subsequently shifted to HL in either HSM-NH4 or HSM-NO3 containing 0.34 or 3.06 mM Ca2+ for 24 h. Before exposure to HL, the cultures were set to 2.5 μg chlorophyll/mL. Whole-cell extracts (2.5 μg of chlorophyll) were fractionated on a 13% SDS-PAGE, and LHCSR3 abundance was analyzed by immunoblots. CF1 signal served as a loading control.
A twofold reduction in CAS levels, as observed in the amiRNA-cas-27 and RNAi-cas-9 strains grown photoheterotrophically (Tris-Acetate-Phosphate [TAP] medium, 17.4 mM acetate) in LL, did not impact the abundance of PSII nor of the CF1 ATPase subcomplex as assessed by immunoblot analyses (Figure 2B; see Supplemental Figure 1B online). Immunoblotting further revealed that photosystem I (PSI) polypeptides remained unaltered in amiRNA-cas-27 (Figure 2B), while in RNAi-cas-9, LHCA3 is slightly downregulated while LHCA4 and LHCA9 were induced (see Supplemental Figure 1B online). For RNAi-cas-9, quantitative RT-PCR confirmed that cas mRNA abundance was likewise downregulated as observed for protein amounts.
However, downregulation of CAS in these lines led to severe light sensitivity when cells were shifted from photoheterotrophic growth in LL (TAP medium) to HL and to a medium where the need for photosynthetic activity was enforced by lowering the acetate concentration to 20% (HSM/TAP 80/20 [v/v]) (Figure 2C; see Supplemental Figure 1C online). The light-sensitive phenotype could be rescued by a 10-fold increase in the Ca2+ concentration from 0.34 to 3.06 mM in the growth medium. To evaluate the photosynthetic performance of the amiRNA-cas-27 and RNAi-cas-9 strains in comparison to the wild type, cells were shifted for 2 h to HL and from photoheterotrophic growth conditions to primarily photoautotrophic growth conditions (HSM/TAP 80/20 [v/v]). The decrease in CAS amount diminished the PSII-dependent quantum efficiency Y(II) by ~20%. This decrease was independent of the Ca2+ concentration in the medium. By contrast, NPQ was found to be threefold lower in ami-RNA-cas-27 compared with the wild type (Figure 2D; see Supplemental Table 1 online) but could be rescued by a 10-fold increase of Ca2+ in the growth medium.
In an independent experiment, the RNAi-cas-9 strain was challenged for 16 h in HL in the same medium as described above. In line with the results found for the ami-RNA strain, the NPQ levels in the RNAi-cas-9 strain were twofold lower than in the wild type (see Supplemental Figure 1D online). Here, the quantum efficiency of PSII was slightly diminished in the RNAi-cas-9 strain in regard to the wild type. However, NPQ and quantum efficiency of PSII could be rescued by a 10-fold increase in the Ca2+ concentration (see Supplemental Figure 1D online). Immunoblot analyses revealed that the impact in NPQ coincided with significantly diminished levels of the light-dependent induction of LHSCR3 protein in the mutant strains after the shifting of cells into HL for 2 or 24 h (Figure 2E; see Supplemental Figure 1E online). Most remarkably, increasing the Ca2+ concentration in the growth medium from 0.34 to 3.06 mM restored the NPQ capabilities and reconstituted proper induction of LHCSR3 (Figures 2D and 2E; see Supplemental Figures 1D and 1E online). Since NPQ is dependent on the presence of LHCSR3 (Peers et al., 2009), these data imply that the restored induction of LHCSR3 upon additional supply of Ca2+ is responsible for the rescued NPQ phenotype. Taken together, these results indicate that already a reduction of CAS protein accumulation by 50% impairs NPQ and the corresponding LHSCR3 expression induction, identifying CAS as critical factor in these processes.
Severe Downregulation of CAS Enhances HL Phenotypes and Reveals Impaired PSII Recovery
To further investigate the HL-induced expression of LHCSR3 and its dependence on the presence of CAS, three additional amiRNA-cas strains that possessed 10% or less of the wild-type CAS protein levels (Figure 3A) were investigated. Strong downregulation of CAS caused a growth phenotype in the amiRNA-cas strains already under LL conditions (Figure 3B), which was dramatically increased after an 18-h shift into HL. This phenotype again could be rescued by an increased Ca2+ concentration in the growth medium from 0.34 to 3.06 mM Ca2+ (Figure 3B). It should be pointed out that in contrast with the amiRNA-cas strains, the addition of 3.06 mM Ca2+ to the wild type had an adverse effect on its growth (Figure 3B).
Growing the amiRNA-cas strains at 0.34 mM Ca2+ in HL for 18 h did not induce a strong increase of LHCSR3 in contrast with the wild type (Figure 3C). This phenotype was particularly evident in strain amiRNA-cas-17, which had the most severe reduction of CAS protein levels and was completely unable to induce LHCSR3 after HL treatment (Figure 3C). It should be noted that C. reinhardtii cells that were precultivated in TAP and LL did not express LHCSR3, as revealed by immunoblotting using specific anti-LHCSR3 antibodies with anti-PsaD and anti-CF1 antibodies for loading controls. Intriguingly, the diminished light-dependent induction of LHCSR3 in all three amiRNA-cas strains could be rescued by increasing the Ca2+ concentration from 0.34 to 3.06 mM in the growth medium (Figure 3C), confirming our data from amiRNA-cas-27 strain.
Remarkably, in the 3.06 mM Ca2+ growth medium after 18 h in HL, the amount of LHCSR3 in the amiRNA-cas-9 and amiRNA-cas-15 strains was higher than in the wild type, also pointing to lack of regulation in LHCSR3 expression in the cas-kd strains.
To address the possibility that the differences in LHCSR3 expression between wild-type and cas-kd strains in 0.34 and 3.06 mM Ca2+ were due to altered growth, the cell densities of the wild type and amiRNA-cas-9 were monitored at 0, 2, and 4 h after the shift to HL. Supplemental Figure 2 online clearly showed that diminished LHCSR3 expression in the cas-kd line cannot be attributed to a slower growth compared with the wild type and that addition of calcium to the growth medium selectively impacts on LHCSR3 expression and not cellular growth.
Addition of Sr2+ also protected amiRNA-cas-9 from HL sensitivity, Mg2+ had only a minimal potential for rescue, whereas Na+ had no effect at all. The Ca2+ chelator EGTA did not render amiRNAcas-9 nor wild-type strains more sensitive to HL (Figure 3D). The Fv/Fm of the amiRNA-cas-9 was found to be 35% less compared with the wild type, when cells were shifted for 16 h from TAP to HSM medium in LL (Figure 3E; see Supplemental Table 2 online). The PSII quantum efficiency of the wild type and amiRNA-cas-9 was recorded during a 5-min exposure to HL (800 μE m−2 s−1) followed by a short (2 min) dark recovery phase. At the end of this dark phase, the PSII quantum efficiency of the wild type recovered to almost 80%, whereas amiRNA-cas-9 recovered only to 30% of its maximal PSII efficiency. It is noteworthy that the presence of a 10-fold higher Ca2+ concentration in the culture medium (16 h in HSM LL) restored both Fv/Fm and recovery of PSII quantum efficiency in the dark of amiRNA-cas-9 to wild-type levels. Taken together, these data support the notion that addition of Ca2+ to the growth media rescued PSII activity and recovery in cas-kd lines.
Measurements of calcium content in the wild type (CC-124), amiRNA-cas-9, and RNAi-cas-9 using atomic absorption spectroscopy revealed that the cellular calcium content was not diminished in cas-kd in regard to wild-type lines (Figure 4). This finding was confirmed by comparing the cellular calcium content in the wild type (cw15-arg7), amiRNA-cas-11, and amiRNA-cas-27 by ion chromatography (see Supplemental Figure 3 online).
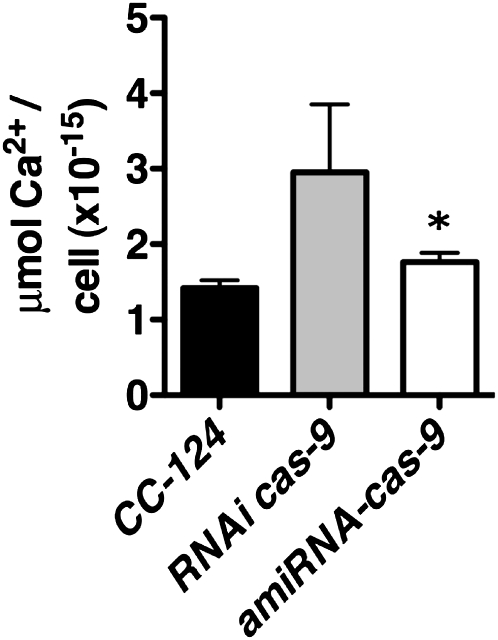
Figure 4.
Downregulation of CAS Does Not Lead to Decrease in Cellular Calcium Content.
Calcium content of the cas-kd strains RNAi-cas-9 (gray bar) and amiRNA-cas-9 (white bar) compared with their wild-type background CC-124 (black bar) as assessed by atomic absorption spectroscopy. Data are presented on a per cell basis. The values are the means ± sd of three biological samples. The asterisk indicates statistically significant difference from wild-type levels (paired t test, 95% confidence level).
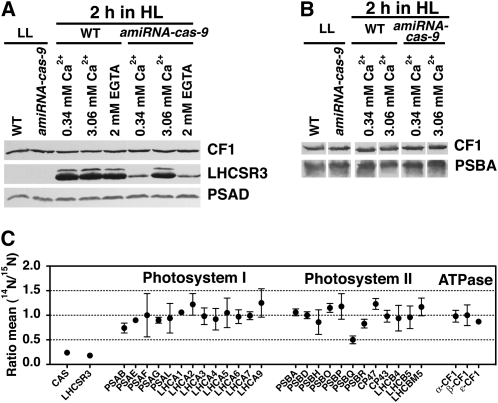
Nevertheless, it is apparent that an increase in Ca2+ concentration in the growth medium restored the light-dependent induction of LHCSR3 in the cas-kd strains. In this context, we addressed whether this restoration occurs in a cas-kd strain with severely downregulated CAS levels after a short shift into HL. To this end, the amiRNA-cas-9 strain was shifted for 2 h into HL using three different growth conditions (0.34 mM Ca2+, 0.34 mM Ca2+ plus 2 mM EGTA, and 3.06 mM Ca2+), and the induction of LHCSR3 was investigated by immunoblotting using anti-LHCSR3 antibodies together with anti-PSAD and anti-CF1 antibodies for loading control (Figure 5A). The experiment clearly showed that the wild type induced LHCSR3 at 0.34 and 3.06 mM Ca2+ as well as in the presence of EGTA in the growth medium. By contrast, the amiRNA-cas-9 strain was capable of inducing LHCSR3 to wild-type levels only in the presence of 3.06 mM Ca2+ in the growth medium, whereas at normal Ca2+ concentration or in the presence of EGTA, induction of LHCSR3 was abolished (Figure 5A). Subsequently, we investigated whether in the amiRNA-cas-9 strain PSII stability was affected after a 2-h HL treatment and whether this provoked the reduction of LHCSR3 accumulation. Immunoblot analyses of whole-cell extracts from wild-type and amiRNA-cas-9 strains demonstrated (Figure 5B) that in the cas-kd strain, the levels of PSBA remained stable under conditions where LHCSR3 failed to accumulate (0.34 mM Ca2+).
Figure 5.
LHCSR3 Induction Is Strongly Reduced in amiRNA-cas-9, While Photosystem Levels Remain Unaltered.
(A) Immunoblot quantitation of LHCSR3. Five micrograms of chlorophyll of whole-cell extracts of wild-type (WT; CC-124) and amiRNA-cas-9 cells were fractionated on a 13% SDS-PAGE gel. LHCSR3, PSAD, and CF1 abundances were analyzed by immunoblots. CF1 signal served as a loading control. The cells were initially grown in LL TAP and were then shifted for 2 h to HL 80% HSM, containing the indicated amount of CaCl2 or EGTA.
(B) Immunoblot analysis of PSBA abundance in whole-cell extracts of the wild type (CC-124) and amiRNA-cas-9 exposed to HL for 2 h in 80% HSM. Five micrograms of chlorophyll were loaded per lane of the SDS-PAGE gel. CF1 signal was used as loading control.
(C) Amounts of CAS, LHCSR3, subunits of PSI and PSII, and ATPase from the wild type (CC-124) and amiRNA-cas-9 exposed to HL for 2 h in 80% HSM were analyzed by a quantitative comparative proteomics approach. In this graph, the ratios of 14N-labeled (amiRNA-cas-9) and 15N-labeled (wild-type CC-124) are presented (mean values and their respective sd).
Quantitative Proteomics Confirms Downregulation of CAS and LHCSR3 in the amiRNA-cas-9 Strain
To further corroborate our protein expression studies and to extend the analysis to other proteins, we performed a quantitative proteomic experiment taking advantage of metabolic 14N/15N labeling. Wild-type cells were labeled with 15N, and amiRNA-cas-9 cells were labeled with 14N. The differentially labeled cells were shifted for 2 h to HL, harvested, mixed based on equal chlorophyll content, and fractionated by SDS-PAGE. After fractionation, the SDS-PAGE bands were excised, digested with trypsin, and analyzed by liquid chromatography–tandem mass spectrometry. The ratio mean 14N/15N (amiRNA-cas-9/wild type) for selected proteins is shown in Figure 5C. As expected, CAS and LHCSR3 were found at low ratios of 0.24 ± 0.02 and 0.3 ± 0.00, respectively, whereas the ratios of PSI and PSII subunits and of ATPase components remained rather unchanged with values between 0.8 and 1.0 (except PsbQ, which was found at a ratio of ~0.5). The downregulation of PSBQ is most likely not of significant functional importance, since in the presence of PSBP and PSBO, PSBQ depletion has no impact on the activity of PSII in Arabidopsis grown under normal light conditions (Yi et al., 2006). It should be noted that LHCA2 and LHCA9 appeared to be slightly upregulated in the mutant versus the wild type, which was also reflected in the immunoblot data of RNAi-cas-9 cells (see Supplemental Figure 1B online). Overall, with respect to CAS, LHCSR3, PSII, and the PSI complex proteins, the quantitative proteomics data confirmed the immunoblot analyses presented in Figures 2, 3, 5, and Supplemental Figure 1 online.
Calcium and Calmodulin Signaling Are Crucial for Appropriate LHCSR3 Induction
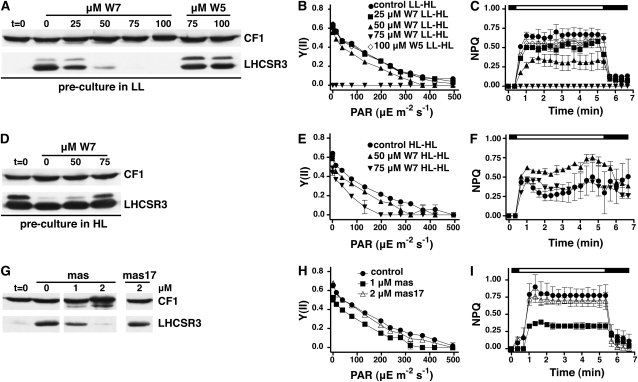
To obtain mechanistic insights into the calcium signaling mechanisms involved in the light-induced expression of LHCSR3, we conducted experiments with a calmodulin antagonist N-(6-aminohexyl)-5-chloro-1-naphthelene-sulfonamid-hydrochloride (W7) (Hidaka et al., 1981). We also used the biological inactive analog W5 [N-(6-aminohexyl)-1-naphthelene-sulfonamid-hydrochloride], which represents a negative control for W7-dependent calmodulin inhibition experiments. Wild-type cells grown in LL and TAP were shifted for 2 h to HL in 80% HSM (HSM/TAP 80/20 [v/v]) medium in the presence or absence of W7 or W5. Figure 6A depicts the impact of W7 and W5 on light-dependent induction of LHCSR3 in the wild type, as assessed by immunoblot analyses. At a concentration of 25 μM W7, a clear decrease in induction of LHCSR3 was observed, whereas at 50 μM W7, LHCSR3 levels were barely detectable. At 75 μM W7, no PSII activity nor any NPQ was measureable (Figures 6B and 6C; see Supplemental Table 3 online). In sharp contrast, the W5 control did not show such an impact, even at concentrations as high as 100 μM. Moreover, at 50 μM W7, the PSII quantum efficiency Y(II) was only slightly diminished in the wild type (Figure 6B). Together, these results indicate that the impaired induction of LHCSR3 expression that was also reflected in the absence NPQ induction (Figure 6C), specifically results from the effects of W7, suggesting that calmodulin and/or calmodulin-affected Ca2+ transients (Kaplan et al., 2006) play a role in the signal transduction involved in the light-dependent regulation of LHCSR3 expression.
Figure 6.
W7 and Mastoparan Drug Studies Point to the Involvement of Calcium and Calmodulin in the Light-Dependent Regulation of LHCSR3 Expression.
(A) Wild-type cells (CC-124) were shifted for 2 h from LL TAP to HL 80% HSM containing the indicated concentrations of the calmodulin antagonist W7 and its inactive analog W5 (which served as negative control). Chlorophyll (2.5 μg) of whole-cell extracts was fractionated on a 13% SDS-PAGE, and LHCSR3 abundance was analyzed by immunoblots. CF1 signal served as a loading control.
(B) and (C) Irradiance dependence of quantum yield of PSII (B) and NPQ induction (C) in the wild-type (CC-124) cells of the experiment described in (A). The white bar indicates irradiation with white light at 890 μE m−2 s−1, and the black bar indicates darkness. Values plotted are the means of three measurements ± sd.
(D) As described in (A) with the difference that wild-type cells were grown in HL and TAP for 12 h before their 2-h HL treatment in 80% HSM.
(E) and (F) As described in (B) and (C) but referring to the experiment of (D).
(G) Wild-type cells (CC-124) grown in LL and TAP were exposed to HL for 2 h in 80% HSM containing the indicated concentrations of mastoparan and its inactive analog mas-17 (which served as negative control). Whole-cell extracts (2.5 μg of chlorophyll) were fractionated on a 13% SDS-PAGE, and LHCSR3 abundance was analyzed by immunoblots. CF1 signal served as a loading control.
(H) and (I) Irradiance dependence of quantum yield of PSII (H) and NPQ induction (I) of the wild-type (CC-124) cells of the experiment described in (G). The white bar indicates irradiation with light at 890 μE m−2 s−1, and the black bar indicates darkness. Values plotted are the means of three samples ± sd.
The experiment was repeated with a preculture that had been exposed to HL for 12 h, which led to full induction of LHCSR3 (Figure 6D). In contrast with the effects observed after preculture in LL (Figures 6B and 6C), the addition of 75 μM W7 to the culture for 2 h under HL did not eradicate the photosynthetic performance [measured as Y(II)] nor abolish their capability to induce NPQ at wild-type levels (Figures 6E and 6F; see Supplemental Table 4 online). This differential effect of W7 to HL-treated (LHCSR3 induced) and LL-treated (LHCSR3 not induced) wild-type cells indicates that the priming of cells by growth in HL diminished the impact of W7, pointing to the fact that calmodulin-dependent functions might be particularly important for the transition from LL to HL adaptation. As a result, gene expression due to a LL-to-HL shift could be altered as exemplified by the impact of W7 on LHCSR3 expression. In a related experiment, we treated wild-type C. reinhardtii cells with mastoparan, a wasp venom tetradecapeptide that is a potent activator of G proteins (Higashijima et al., 1990). In Chlamydomonas, mastoparan causes accumulation of inositol 1,4,5-trisphosphate (IP3) (Quarmby et al., 1992), causing an artificial rise in intracellular-free Ca+2 (Berridge and Irvine, 1989). Mastoparan inhibited the accumulation of LHCSR3 (Figure 6G) and the induction of NPQ by more than 50% at the concentration of 1 μM with a PSII quantum efficiency Y(II) being largely unaffected (Figures 6H and 6I; see Supplemental Table 5 online). By contrast, mas-17 (an inactivated analog of mastoparan) at the concentration of 2 μΜ did neither affect LHCSR3 expression nor the photosynthetic performance (Figures 6G and 6H).
Photosynthetic Electron Transfer Is Crucially Involved in the Regulation of the Light-Dependent Expression of LHCSR3
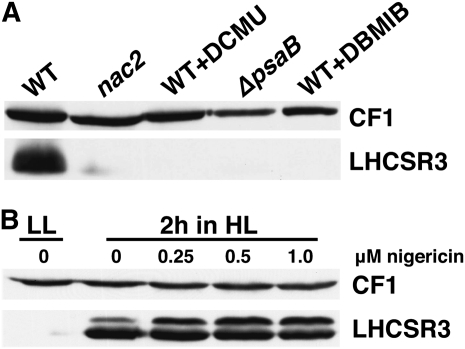
In the course of experiments with the PSII-deficient mutant nac2, we noticed that LHCSR3 was barely expressed after 18 h exposure of the mutant cells to HL (Figure 7A). The same was observed with a PSI-deficient mutant strain (ΔpsaB). Moreover, we found that DCMU (at 20 μM) and DBMIB (20 μM) inhibited LHCSR3 expression in wild-type cells exposed to HL for 2 h. DCMU blocks photosynthetic electron transfer at the Qb site of PSII, whereas DBMIB inhibits electron transfer at the Qo site of the cytochrome b6f complex (Trebst, 2007). In consequence, both inhibitors will hinder light-driven NADPH production, decreasing the redox poise of the chloroplast stroma. Similarly, deletion of PSI or PSII components impairs light-driven NADPH production and reduces stroma redox poise. Interestingly, application of the proton gradient uncoupler nigericin that repressed NPQ by ~80%, but not the electron transfer processes (see Supplemental Figure 4 and Supplemental Table 8 online), did not affect LHCSR3 induction after a 2-h HL exposure (Figure 7B), supporting the notion that despite the presence of LHCSR3, a proton gradient is required for efficient qE (Niyogi et al., 1997). These data indicate that the ΔpH can be significantly diminished without affecting the induction of LHCSR3 expression. Consequently, induction of LHCSR3 expression does also depend on active photosynthetic electron transfer.
Figure 7.
Photosynthetic Electron Transfer Is Involved in the Regulation of LHCSR3 Expression.
(A) Expression of LHCSR3 requires active photosynthetic electron transfer. Wild-type cells (WT; cw15-arg7) in the presence and absence of DCMU (20 μM) or DBMIB (20 μM) as well as nac2 (PSII-deficient mutant; Kuchka et al., 1989) and Δpsab (PSI-deficient mutant; Redding et al., 1998) mutant cells were exposed in HL for 2 h in 80% HSM. Subsequently, whole-cell extracts (2.5 μg of chlorophyll) were fractionated on a 13% SDS-PAGE gel, and LHCSR3 abundance was analyzed by immunoblots. CF1 signal served as a loading control.
(B) Impact of nigericin (0.25 to 1.0 μM) in LHCSR3 expression in wild-type cells (CC-124) during a 2-h HL shift experiment in 80% HSM. Protein analyses were performed as described in (A).
The Low LHCSR3 Expression in cas-kd Lines Cannot Be Attributed to a Diminished Electron Transfer but Rather to a Ca2+/CAS-Dependent Signaling Defect
LHCSR3 expression depends on the efficiency of photosynthetic electron transfer (Figure 7A). CAS depletion also has a slight impact on the quantum efficiency of PSII as revealed by the diminished Fv/Fm values of the cas-kd strains (Figures 2D and 3E; see Supplemental Figure 1D online). To address whether the impact on LHCSR3 expression is due to impaired signaling or efficiency of photosynthetic electron transfer, we took advantage of an inducible amiRNA expression system (Schmollinger et al., 2010). This inducible amiRNA expression system is under control of the NIT1 promoter and thus is repressed in the presence of ammonium and activated in the presence of nitrate. Four inducible amiRNA-cas lines were generated using the cw15-325 wild-type background (nit1+ nit2+ arg7). When shifted for 40 h from LL TAP-NH4 (amiRNA repressed) to LL TAP-NO3 (amiRNA activated), both amiRNA-cas-1 and amiRNA-cas-6 displayed an about 10-fold reduction of CAS quantities compared with the wild type grown in TAP-NO3 (Figure 8A; see Supplemental Figure 5A online).
The lowered amount of CAS in the two mutant strains did not cause significant differences in their Fv/Fm values after 24 h in HL HSM-NO3 in comparison to the PSII quantum efficiency measured for the transformants in HSM-NH4 and for the wild type in HSM-NH4 and HSM-NO3 (Figure 8B; see Supplemental Table 6 online). However, the NPQ levels of amiRNA-cas-1 and amiRNA-cas-6 in HSM-NO3 were significantly diminished and could be rescued by the addition of calcium to a final concentration of 3.06 mM (Figure 8C). In line with the diminished NPQ induction, accumulation of LHCSR3 in NO3 amiRNA-cas-6 and amiRNA-cas-1 cells were lower compared with NH4 and wild type (NH4 and NO3) and could be rescued by calcium (Figure 8D; see Supplemental Figure 5B online). These results clearly indicate that despite the additional layer of regulation of LHCSR3 expression by photosynthetic electron transfer, the impact in induction of LHCSR3 in response to HL in cas-kd lines is due to altered/defective Ca2+/CAS-dependent signaling caused by the downregulation of CAS.
The Diminished Expression of LHCSR3 Accounts for the HL Phenotype of cas-kd Cells
According to the data obtained for the npq4 mutant (LHCSR3 knockout), the diminished levels of expression of LHCSR3 are not sufficient to explain the HL phenotype of the cas-kd lines since the npq4 totally devoid of LHCSR3 had no pronounced sensitivity to HL after a 24-h treatment (see Supplemental Figure 6A online). To reinvestigate the light sensitivity caused by the depletion of LHCSR3, we generated six LHCSR3 knockdown lines using the inducible amiRNA expression system described above (Schmollinger et al., 2010) and analyzed two of them, amiRNA-lhcsr3-2 and amiRNA-lhcsr3-3 in detail. To quantify the amounts of LHCSR3 in these two amiRNA lines, we shifted cells from LL TAP-NH4 to LL TAP-NO3 for 40 h and applied a 2-h HL treatment in HSM-NH4 and HSM-NO3 containing 0.34 and 3.06 mM calcium. As shown in Figure 9A and Supplemental Figure 5C online, LHCSR3 is significantly reduced in amiRNA-lhcsr3-2 and amiRNA-lhcsr3-3 (in HSM-NO3) compared with LHCSR3 quantities in HSM-NO3 wild type, in both 0.34 and 3.06 mM Ca2+. Interestingly this LHCSR3 downregulation also has an impact to Fv/Fm values and recovery of PSII after HL treatment, correlating with the degree of LHCSR3 downregulation in the two mutant lines (Figure 9B; see Supplemental Table 7 online).
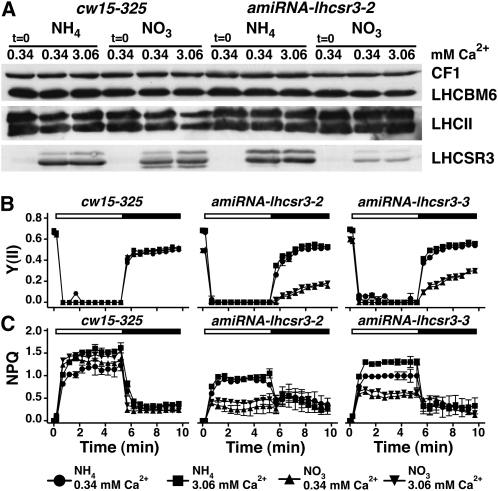
Figure 9.
lhcsr3-kd Lines Show an HL Phenotype Similar to the cas-kd Lines and This Cannot Be Rescued by Calcium.
(A) Quantification of LHCSR3, LHCBM6, and LHCII amounts in whole-cell extracts of the wild type (cw15-325) and amiRNA-lhcsr3-2 upon SDS-PAGE fractionation and immunoblot analysis (100% equals 2.5 μg of chlorophyll per lane; CF1 signal served as loading control). The wild type and amiRNA-lhcsr3-2 were initially shifted from LL TAP-NH4 to LL TAP-NO3 for 40 h to activate the amiRNA expression and subsequently shifted for 2 h to HL in either HSM-NH4 or HSM-NO3 containing 0.34 or 3.06 mM Ca2+. Before exposure to HL, the cultures were set to 2.5 μg chlorophyll/mL.
(B) and (C) Determination of PSII quantum yield and recovery and NPQ. The wild type, amiRNA-lhcsr3-2, and amiRNA-lhcsr3-3 were initially shifted from LL TAP-NH4 to LL TAP-NO3 for 40 h to activate the amiRNA expression and subsequently shifted for 2 h to HL in either HSM-NH4 or HSM-NO3 containing 0.34 or 3.06 mM CaCl2. After 20 min of dark adaptation, the quantum yield of PSII (B) and NPQ (C) were recorded during 5.2 min of illumination at 800 μE m−2 s−1 (white bar) followed by 4.3 min of darkness (black bar), during which recovery of PSII and relaxation of NPQ could be followed. Values plotted are the means of three measurements ± sd.
This phenotype cannot be attributed to altered LHCII quantity in the lhcsr3-kd lines, as evidenced by the very similar levels of LHCBM6 and of the major LHCII proteins in lhcsr3-kd and wild-type strains (Figure 9A; see Supplemental Figure 5C online). As expected, NPQ induction was lower in both amiRNA-lhcsr3-2 and amiRNA-lhcsr3-3 in HSM-NO3 (Figure 9C). It is crucial to note that the HL phenotype of the lhcsr3-kd lines (low Fv/Fm and PSII recovery), also apparent from the growth phenotype of amiRNA-lhcsr3-3 after 24 h in HL HSM-NO3 (see Supplemental Figure 6C online), could not be rescued by addition of calcium in the culture medium. Importantly, the induction of NPQ could not be rescued by a 10-fold higher calcium concentration in the growth medium. This is in stark contrast with the cas-kd phenotype and strengthens the conclusion that a direct link between Ca2+, CAS, and LHCSR3 exists. This supports the notion that the diminishment of LHCSR3 in cas-kd causes the HL phenotype.
DISCUSSION
In this work, we addressed the function of the chloroplast localized calcium sensor CAS in the green alga C. reinhardtii. To study the functional consequences of the downregulation of CAS expression, we took advantage of both amiRNA and RNAi approaches. The different cas-kd lines generated and characterized in this study exhibited a pronounced light sensitivity, a severe impact in LHCSR3 expression, and a prominent defect in PSII recovery compared with wild-type C. reinhardtii cells. The fact that these phenotypes were rescued by a 10-fold increase in Ca2+ concentration in the growth medium (Figures 2, 3, and 8; see Supplemental Figure 1 online) further suggests an involvement of CAS in chloroplast-dependent Ca2+ homeostasis and signaling as it has also been suggested by studies in Arabidopsis (Nomura et al., 2008; Weinl et al., 2008).
The Role of CAS and Ca2+ in Cellular Signaling in Chlamydomonas
How could CAS and Ca2+ function in retrograde signaling? Recent data demonstrated that rapid spatiotemporal patterning of cytosolic Ca2+ transients was functionally interrelated to flagellar excision in C. reinhardtii (Wheeler et al., 2008). Thus, such fast cytosolic Ca2+ transients may rely on CAS function and a CAS-dependent release of Ca2+ from the chloroplast contributing to cellular signaling. Ca2+ signaling across the inner envelope of the chloroplast requires import and export activities for Ca2+. In Pisum sativum, it was shown that Ca2+ transport across the inner plastid envelope membrane occurs and is driven by ATP- and pH-dependent pathways during different light conditions (Roh et al., 1998). Moreover, a Cu+-ATPase, AtHMA1, that is essential for plant survival under HL conditions, is present at the chloroplast envelope membrane (Seigneurin-Berny et al., 2006). Interestingly, AtHMA1 also contributes to high-affinity Ca2+ transport activity and is sensitive to the sarcoplasmic/endoplasmic reticulum Ca2+ATPase inhibitor thapsigargin (Moreno et al., 2008). C. reinhardtii possesses an ortholog of AtHMA1 (C. reinhardtii protein ID 195998), although the protein localization is currently unclear. Besides fluxes across the inner envelope, there is also evidence for the presence of an active Ca2+/H+ antiport system in the thylakoid membrane (Ettinger et al., 1999), suggesting that the activity of the Ca2+/H+ antiporter facilitates the light-dependent uptake of Ca2+ by chloroplasts and reduces stromal Ca2+ levels. Overall, these data substantiate the notion that chloroplast membranes display Ca2+ transporting activity and thereby support a role of chloroplasts in cellular Ca2+ signaling that is brought about by Ca2+ fluxes across the inner envelope and/or the thylakoid membrane. However, it remains to be solved how exactly CAS-dependent Ca2+ binding and release or signaling is mechanistically interconnected to chloroplast Ca2+ import/export activities.
Experiments with a calmodulin antagonist W7 (Figure 6) indicated that calmodulin and/or calmodulin-affected Ca2+ transients (Kaplan et al., 2006) may play a role in the signal transduction involved in the light-dependent regulation of LHCSR3 expression. In regard to signaling, calmodulin proteins are known to interact with calmodulin-dependent kinases (CCaMK) or calmodulin binding transcription activators (CAMTAs), for example. The later CAMTAs appear to be involved in various stress responses (Bouché et al., 2002) and are implicated, for example, in cold regulation and freezing tolerance in Arabidopsis (Doherty et al., 2009). The kinases have important roles in symbiotic root responses where CCaMK converts Ca2+ oscillation signals into a protein phosphorylation readout, leading to changes in nuclear gene expression and modulation of the symbiotic response (Gleason et al., 2006; Tirichine et al., 2006). Momentarily it is unknown whether CAMTAs or CCaMKs are involved in the HL-mediated regulation of LHCSR3 expression. Besides the function of calmodulin in signal transduction, it has been shown that the import of nuclear-encoded chloroplast proteins is regulated via Ca2+ and in particularly via TIC32, a calmodulin binding protein of the TIC translocon (Chigri et al., 2005, 2006). In this regard it might be possible that W7 simply blocks the HL-induced import of LHCSR3, so that LHCSR3 accumulation in the chloroplast fails.
Several important facets of cellular Ca2+ signaling appear to be fundamentally different in C. reinhardtii than in higher plants. Recent biochemical and genomic studies revealed that certain classes of Ca2+ release channels, such as four-domain voltage-dependent Ca2+ channels, transient receptor potential channels, and IP3 receptors, which play central roles in animal physiology, are absent from vascular plants but are present in the C. reinhardtii (Wheeler and Brownlee, 2008). This strongly suggests that Ca2+ signaling via cytosolic Ca2+ transients in C. reinhardtii possibly involves other components and underlies different mechanistic principles that Ca2+ signaling in vascular plants. The observed sensitivity of light-dependent induction of LHCSR3 to mastoparan in Chlamydomonas, which affects IP3 accumulation and IP3 receptor function, leading to a rise in intracellular-free Ca+2 in animal cells (Berridge and Irvine, 1989), further supports the function of a non-plant-like Ca2+ signaling system in this algae. Therefore, it is not surprising that loss of CAS function results in distinct phenotypes in Arabidopsis and C. reinhardtii.
CAS and Ca2+ Function in Acclimation of Photosynthesis to HL
The light sensitivity and the diminished efficiency in PSII recovery represent phenotypes that were not observed in CAS-deficient Arabidopsis (Vainonen et al., 2008). It is very likely that the impact in PSII recovery is due to the lack in the induction of LHCSR3 in cas-kd lines since a strong decrease in PSII recovery and NPQ induction is also observed in amiRNA-lhcsr3 lines in HSM-NO3 using the inducible amiRNA expression system (Figures 8 and 9). In contrast with amiRNA-lhcsr3 lines, however, PSII recovery and NPQ induction in amiRNA-cas strains, harboring the amiRNA-cas construct under the control of the NIT1 promoter, are rescued by the addition of a 10-fold higher Ca2+ concentration to the nitrate-containing medium.
Given the fact that PSII recovery and NPQ induction observed in the wild type and the nitrate-inducible amiRNA-lhcsr3 lines are comparable in HSM-NH4, it appears that the described phenotypes under nitrate are due to the impact in LHCSR3 expression. This leads to the important question: How could the difference and severity in HL phenotype between the nqp4 knockout (see Supplemental Figure 6A online) and knockdown LHCSR3 lines (see Supplemental Figure 6C online) be explained? It is possible that in the knockout strain, because of the complete absence of LHCSR3, alternative routes for photoprotection have been switched on compared with the knockdown lines where the residual LHCSR3 levels are not sufficient to efficiently photoprotect the cells. It is also possible that different genetic backgrounds are responsible for the difference in phenotype. Targeted gene knockouts in mice are known to reveal strain-dependent phenotypes as evidenced, for example, for targeted disruption of the EGF receptor, which was lethal in some laboratory strains but viable in others (Threadgill et al., 1995). This aspect has certainly to be addressed in further studies.
On the other hand, it has to be noted that the npq4 strain, lacking the genes encoding for LHCSR3, is also more light sensitive compared with wild type when exposed for several days to HL (Peers et al., 2009). Notably, the impact in PSII protection and recovery is also observed in P. patens (Alboresi et al., 2010), pointing to an important role of LHCSR3 in photoprotection. The PSII-deficient mutant nac2 (Kuchka et al., 1989) did not show an obvious phenotype after 24 h of growth in HL (see Supplemental Figure 6B online), indicating that a lack of photoprotection is more deleterious than the absence of PSII in the presence of a carbon source.
As already discussed above, we provide unambiguous evidence that adding a 10-fold higher Ca2+ concentration to the growth medium (Figures 2, 3, 5, and 8; see Supplemental Figures 1 and 5B online) rescues qE and the light-dependent induction of LHCSR3 in the cas-kd lines, suggesting an involvement of CAS and Ca2+ in the regulation of the nuclear encoded LHCSR3 gene expression. It is of note that besides increased Ca2+ concentration, only elevation of Sr2+ content in the growth media resulted in a rescue of the CAS-dependent growth phenotypes. It is known that Sr2+ can functionally replace Ca2+ in the water-splitting complex of PSII (Ghanotakis et al., 1984), thereby underpinning the Ca2+ specificity of the CAS-dependent response in C. reinhardtii. Notably, Sr2+ can bind to the EF hand of apo-calmodulin (Shirran and Barran, 2009) and could thereby functionally rescue the calmodulin-dependent signal transduction pathway leading to expression of LHCSR3. Nevertheless, we cannot exclude that CAS might also have an additional functional role for efficient recovery of PSII activity via the repair cycle or by balancing degradation and regeneration of PSII via Ca2+.
However, our data supports the conclusion that the delay in LHCSR3 expression in the CAS depletion lines can explain the impact in PSII repair and the pronounced light sensitivity of these strains, since these phenotypes were observed in the ami-lhcsr3 lines (Figure 9). Therefore, we propose, as already mentioned above, that LHCSR3 is also important for photoprotection of PSII. It is clear that novel mechanistic data are required to elucidate the precise role of LHCSR3 in PSII photoprotection. Our data also indicate that active photosynthetic electron transfer is required for light-driven induction of LHCSR3 (Figure 7A). We hypothesize that reducing equivalents produced by PSI function as a retrograde signal (Li et al., 2009) linking the redox poise of the stroma to the control of nuclear gene expression.
These findings raise the question whether an impact in PSII quantum efficiency in the CAS depletion lines is responsible for the delay in light-dependent induction of LHCSR3 expression. Differences in NPQ induction and LHCSR3 expression became apparent in the amiRNA-cas-27 strain grown for 2 h in HL at 0.34 or 3.06 mM Ca2+ (Figure 2D). Yet, the PSII quantum efficiency for amiRNA-cas-27 at the two different Ca2+ concentrations remained almost the same (Figure 2D). Moreover, the PSII quantum efficiency in the nitrate-inducible amiRNA-cas lines under a nitrate growth regime was unchanged after a 2-h HL shift in wild-type and amiRNA-cas lines, although NPQ induction was significantly impaired (Figures 8B and 8C; see Supplemental Table 6 online). Thus, differences in the induction of NPQ and of LHCSR3 expression are clearly linked to CAS and Ca2+.
The results presented in this study indicate that CAS function in C. reinhardtii is crucial for light-induced expression of LHCSR3 and, therefore, important for HL acclimation. Our data also indicate that active downregulation of LHCSR3 under HL is deleterious for C. reinhardtii and can explain the HL phenotypes observed in amiRNA-cas lines. The algal-specific responses in respect to CAS and LHCSR3 may be interconnected to the Ca2+ dependent and light-dependent phototactic movements of this organism, which require the chloroplast-localized eyespot. We propose that coordinated modulation of chloroplast-dependent Ca2+ signaling provides an important signaling convergence point, allowing for coordinating gene expression, maintaining PSII activity, recovery, and/or turnover, as well as driving HL acclimation.
METHODS
Strains and Culture Conditions
Wild-type Chlamydomonas reinhardtii strain CC-124, the Arg auxotrophic wall-deficient strains cw15-arg7 and cw15-325 (mt+ arg7 nit1+ nit2+; kindly provided by Michael Schroda Max Planck Institute of Molecular Plant Physiology, Potsdam-Golm, Germany), the ΔpsaB (Redding et al., 1998) and nac2 (Kuchka et al., 1989) mutant strains, as well as the CAS knockdown strains were maintained photoheterotrophically in Tris-Acetate-Phosphate (TAP) medium (Harris, 1989), at 25°C, with 120 rpm shaking and 60 μE m-2s−1 continuous white light if not indicated otherwise. For cultivation on plates, media was supplemented with 1.5% agar. For growth of cw15-arg7 and cw15-325, the medium was supplemented with 100 μg/mL Arg. The ΔpsaB and nac2 mutant strains were kind gifts from K. Redding (Arizona State University, Tempe, AZ) and J. Nickelsen (Ludwig-Maximilians University, Munchen, Germany), respectively, and cw15-arg7 was a kind gift of T. Happe (Ruhr Universtity, Bochum, Germany). HL growth experiments were performed as follows: Precultures were cultivated in TAP at LL (60 μE m−2 s−1) to the early exponential phase, and the cell density was determined using a Fuchs-Rosenthal chamber. Cells were washed one time with HSM/TAP (80/20) medium, resuspended in HSM/TAP (80/20) at a starting cell density of 1 × 106 cells/mL, and supplemented with the indicated salts. Cultures were then exposed to HL (200 μE m−2 s−1) for the indicated time. For metabolic labeling, NH4Cl in solid and liquid media was substituted with 15N-NH4Cl (Cambridge Isotope Laboratories). For the activation of the inducible amiRNA expression system, cells were shifted from TAP to TAP-NO3 medium for 40 h before the onset of further experimental procedures. In TAP-NO3 medium, NH4 was replaced by 4 mM KNO3.
It is of note that the described phenotypes in the amiRNA and RNAi mutant strains were lost after some time (between 2 and 12 months) and that the loss of phenotype correlated with the restoration of CAS protein expression, strongly pointing to the fact that the observed phenotypes are closely linked to CAS expression levels.
Plasmid Construction and Generation of RNAi Lines
Total RNA of Chlamydomonas cells was isolated with TRI reagent (Sigma-Aldrich), and cDNA synthesis was performed with the reverse transcription system (Promega) using the manufacturer’s protocols. A 1478-bp fragment, which includes the coding region of CAS, was amplified from cDNA using the following primers with added restriction sites: FORS-RHO-B (5′-CGGGATCCAGAGCGGTGTGAAGATCCTG-3′, underlined BamHI site) and REV-RHO-X/E (5′-GCTCTAGACGGAATTCACATACACCTTGCC-3′, underlined XbaI and EcoRI sites, respectively). A longer 1729-bp fragment that contained an additional 251 bp of adjacent sequence to function as a spacer in the inverted repeat construct was amplified using the primer FORL-RHO-B (5′-CGGGATCCGAAAATGCAGCTTGCTAACG-3′, underlined BamHI) and REV-RHO-X/E. Both fragments were cloned in antisense orientation into pBluescript SK− (Stratagene) using the added restriction sites. The resulting inverted repeat cassette was excised with EcoRI and cloned into the unique EcoRI site of the RNAi vector pNE537 (Rohr et al., 2004). Biolistic transformation of the RNAi construct into a wild-type strain (CC-124) and the subsequent screening were performed as described (Naumann et al., 2005). Transformed cells were selected and maintained on media containing 7 μM 5-fluoro-indole and 10 μg/mL paromomycin. Liquid cultures of the RNAi strains were supplemented with 10 μg/mL paromomycin.
Generation of amiRNA plasmids was done as described by Molnar et al. (2009). Target gene-specific oligonucleotide sequences were designed using the Web MicroRNA Designer platform (WMD2; http://wmd2.weigelworld.org/cgi-bin/mirnatools.pl; Ossowski et al., 2008). The resulting oligonucleotides amiFor_CAS_3UTR, 5′-ctagtCGCGTAGTGTATATCTCTTTAtctcgctgatcggcaccatgggggtggtggtgatcagcgctaTAAACAGATATACACTACGCGg-3′, and amiRev_CAS_3UTR, 5′-ctagcCGCGTAGTGTATATCTGTTTAtagcgctgatcaccaccacccccatggtgccgatcagcgagaTAAAGAGATATACACTACGCGa-3′ (uppercase letters indicate amiRNA*/amiRNA sequences), that target the 3′ untranslated region of CAS were annealed and ligated into pChlamiRNA3 (SpeI digested). The generated plasmid was transformed into the cell wall–deficient cw15-arg7 strain using glass beads as described (Kindle et al., 1989).
Based on the inducible amiRNA system (Schmollinger et al., 2010), knockdown strains of Chlamydomonas CAS and LHCSR3 protein were generated, respectively. Oligonucleotide sequences were as follows: amiFor_TEF2, 5′-ctagtGAGCCAGTACATGTACCTTAAtctcgctgatcggcaccatgggggtggtggtgatcagcgctaTTAACGTACATGTACTGGCTCg-3′, and amiRev_TEF2 5′-ctagcGAGCCAGTACATGTACGTTAAtagcgctgatcaccaccacccccatggtgccgatcagcgagaTTAAGGTACATGTACTGGCTCa-3′; amiFor_SR3CDS, 5′-ctagtACGGCTCCCTTTGAGGCTGTAtctcgctgatcggcaccatgggggtggtggtgatcagcgctaTACACCCTCAAAGGGAGCCGTg-3′, and amiRev_SR3CDS, 5′-ctagcACGGCTCCCTTTGAGGGTGTAtagcgctgatcaccaccacccccatggtgccgatcagcgagaTACAGCCTCAAAGGGAGCCGTa-3′, targeting the 3′ untranslated region of CAS and the CDS of LHCSR3, respectively.
amiRNA strains were created as described above using the vector pMS539 (Schmollinger et al., 2010) and cw15-325 as recipient strain for glass bead transformation.
Protein Analysis
Whole cells were analyzed by SDS-PAGE according to Laemmli (1970) and stained with Coomassie Brilliant Blue or blotted on nitrocellulose membrane. After incubation with specific antibodies, the signal was detected by enhanced chemical luminescence. Antibodies against PSAD (Naumann et al., 2005), LHCSR3 (Naumann et al., 2007), LHCA3 (Moseley et al., 2002), LHCBM6 (Hippler et al., 2001), and PGRL1 (Naumann et al., 2007) were used in a 1:1000 dilution. The antibody against CF1 (Moseley et al., 2002) was used in 1:10,000 dilution. Antibodies against PSBA (obtained from Agrisera) and LHCII (P25K; Hippler et al., 2001) were used in a 1:2500 dilution. Antibodies against CAS, LHCA4, and LHCA9 were generated by Eurogentec using the following peptides for immunization of rabbits: CAS (ARADELDSTVESVVG), LHCA4 (AVPENVKEREWIDAWC), and LHCA9 (ARPWLPGLNPPAHLKC). Densitometric analyses of the blots were performed using the software UN-SCAN-IT (Silk Scientific).
Liquid Chromatography–Tandem Mass Spectrometry Analysis of Proteins
Sample preparation and subsequent mass spectrometry analyses were conducted as described by Terashima et al. (2010). Comparative quantitative analyses took advantage of stable isotopic labeling of C. reinhardtii using 15N. The appropriate mass shifts depending on the number of 15N atoms in the respective peptides and the quantitative evaluation of 14N/15N ratios of unlabeled and labeled sister peptides was employed by the quantitation algorithm qTRACE (Terashima et al., 2010).
Chlorophyll Fluorescence
Fluorescence was measured using a Maxi-Imaging PAM chlorophyll fluorometer (Heinz Walz). Samples were dark adapted for 20 min before each measurement. The effective photochemical quantum yield of PSII was measured as PSII yield [Y(II) = (Fm' − F)/Fm'], whereas total NPQ was calculated as (Fm − F’m)/Fm’. The variable fluorescence Fv was calculated as Fv = Fm − Fo, and Fv/Fm was used to evaluate the maximum fluorescence in the dark-adapted state; Fm’ is the maximum fluorescence in any light-adapted state; Fo is the minimal fluorescence in the dark-adapted state.
Measurement of Calcium Content
Cells were grown in TAP under LL, harvested in the exponential phase by centrifugation at 3000g, and washed twice with double-distilled water. The pellet was resuspended in a small volume of double-distilled water, and the cells were broken by passing them through a self-made bionebulizer at a pressure of ~35 p.s.i.
Ion Chromatography
The cell lysates were boiled for 20 min, centrifuged for 20 min at 12,000g, and filtered through a 0.22-μm filter (Millipore). Extracts were measured with an ICS-1000 (Dionex) using a CS12 cation exchange column and AS40 autosampler (Dionex). For each sample, 500 μL extract was applied to rinse the injection loop (25 μL) and ensure complete equilibration. For all measurements, 20 mM methanesulfonic acid was used as eluent and the Chromeleon 6.80 software (Dionex) for data analysis. Calibration line was obtained by dilution of Dionex Six Cation-I standard.
Atomic Absorption Spectroscopy
To determine the total calcium amount, a Shimadzu AA-6200 atomic absorption spectrophotometer with a double beam optical system, running in flame atomic absorption spectrometry mode, was used for validating the samples. Background correction was performed using a deuterium lamp (190- to 430-nm wavelength range).
Digestion/Determination
The samples were directly digested in a 10-mL volumetric flask. A volume of 2 mL of the homogenized samples, 4 mL nitric acid (65%), and 1 mL sulfuric acid (95%) were transferred into the flask and heated in a water bath at 70°C for 2 h under frequent shaking. The samples had to be diluted by a factor of 10, and an aqueous solution of lanthanum (III) chloride (2% [w/v]) was added as modifier. To determine the total analyte concentration, an external calibration was used, in which the acid concentrations were simulated. The limit of detection was determined as 0.01 μg/mL for calcium according to the method of Boumans (Boumans, 1991; Boumans et al., 1991).
W7 and Mastoparan Drug Studies
The calmodulin antagonist W7, its inactive analog W5, the G-protein activator mastoparan, and its inactive analog mas-17 were all purchased from Sigma-Aldrich and Enzo Life Sciences. Stock solutions of the drugs were prepared in water (in the appropriate concentrations). Cells were exposed to the drugs for 10 min in LL before the onset of the HL experiments.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: TEF2 (CAS), EDO96984.1; LHCSR3.1, EDP01013.1; and LHCSR3.2, EDP01087.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analysis of the HL Response of the cas-kd Line RNAi-cas-9.
Supplemental Figure 2. The Differences in LHCSR3 Expression between Wild-Type and cas-kd Strains in 0.34 and 3.06 mM Ca2+ Are Not Due to Altered Growth.
Supplemental Figure 3. Downregulation of CAS Does Not Lead to a Decrease in Cellular Calcium Content in amiRNA-cas-11 and amiRNA-cas-27.
Supplemental Figure 4. Nigericin at 1 μM Selectively Impacts NPQ but Not Photosynthetic Electron Transfer in Wild-Type Cells (CC-124).
Supplemental Figure 5. Characterization of amiRNA-cas-1 and amiRNA-lhcsr3-3 Strains.
Supplemental Figure 6. Growth Phenotypes of npq4, nac2, and amiRNA-lhcsr3-3 after 24 h in HL.
Supplemental Table 1. Fm, Fo, and Fv/Fm Values of Figure 2D.
Supplemental Table 2. Fm, Fo, and Fv/Fm Values of Figure 3E.
Supplemental Table 3. Fm, Fo, and Fv/Fm Values of Figures 6B and 6C.
Supplemental Table 4. Fm, Fo, and Fv/Fm Values of Figures 6E and 6F.
Supplemental Table 5. Fm, Fo, and Fv/Fm Values of Figures 6H and 6I.
Supplemental Table 6. Fm, Fo, and Fv/Fm Values of Figures 8B and 8C.
Supplemental Table 7. Fm, Fo, and Fv/Fm Values of Figures 9B and 9C.
Supplemental Table 8. Fm, Fo, and Fv/Fm Values of Supplemental Figure 4 Online.
Supplementary Material
Acknowledgments
M.H. and J.K. acknowledge support from the Deutsche Forschungsgemeinshaft. M.H. also acknowledges support from the FP7-funded Sunbiopath Project (GA245070). We thank G. Finazzi and C. Fufezan for fruitful discussions and for valuable comments on the manuscript and Mia Terashima for critically reading the manuscript. We thank Jan Offenborn for help with the ion chromatography measurements.
AUTHOR CONTRIBUTIONS
D.P., A.B., and M. Hippler designed the research. D.P., A.B., I.J., K.T., S.V.B., M. Holtkamp, and S.W. performed research. U.K. and J.K. contributed analytical tools. D.P., A.B., S.W., and M. Hippler analyzed data. D.P., A.B., J.K., and M. Hippler wrote the article.
References
- Alboresi A., Gerotto C., Giacometti G.M., Bassi R., Morosinotto T. (2010). Physcomitrella patens mutants affected on heat dissipation clarify the evolution of photoprotection mechanisms upon land colonization. Proc. Natl. Acad. Sci. USA 107: 11128–11133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmer J., Naumann B., Markert C., Zhang M., Hippler M. (2006). Mass spectrometric genomic data mining: Novel insights into bioenergetic pathways in Chlamydomonas reinhardtii. Proteomics 6: 6207–6220 [DOI] [PubMed] [Google Scholar]
- Berridge M.J., Irvine R.F. (1989). Inositol phosphates and cell signalling. Nature 341: 197–205 [DOI] [PubMed] [Google Scholar]
- Berthold P., Tsunoda S.P., Ernst O.P., Mages W., Gradmann D., Hegemann P. (2008). Channelrhodopsin-1 initiates phototaxis and photophobic responses in chlamydomonas by immediate light-induced depolarization. Plant Cell 20: 1665–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N., Scharlat A., Snedden W., Bouchez D., Fromm H. (2002). A novel family of calmodulin-binding transcription activators in multicellular organisms. J. Biol. Chem. 277: 21851–21861 [DOI] [PubMed] [Google Scholar]
- Boumans P.W. (1991). Measuring detection limits in inductively coupled plasma emission spectrometry using the “SBR-RSDB approach”-1. A tutorial discussion of the theory. Spectrochimica Acta 46B: 431–445 [Google Scholar]
- Boumans P.W., Ivaldi J., Slavin W. (1991). Measuring detection limits in inductively coupled plasma emission spectrometry-II. Experimental data and their interpretation. Spectrochimica Acta 46B: 641–665 [Google Scholar]
- Boussac A., Zimmermann J.L., Rutherford A.W. (1989). EPR signals from modified charge accumulation states of the oxygen evolving enzyme in Ca2+-deficient photosystem II. Biochemistry 28: 8984–8989 [DOI] [PubMed] [Google Scholar]
- Bussemer J., Vothknecht U.C., Chigri F. (2009). Calcium regulation in endosymbiotic organelles of plants. Plant Signal. Behav. 4: 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chigri F., Hörmann F., Stamp A., Stammers D.K., Bölter B., Soll J., Vothknecht U.C. (2006). Calcium regulation of chloroplast protein translocation is mediated by calmodulin binding to Tic32. Proc. Natl. Acad. Sci. USA 103: 16051–16056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chigri F., Soll J., Vothknecht U.C. (2005). Calcium regulation of chloroplast protein import. Plant J. 42: 821–831 [DOI] [PubMed] [Google Scholar]
- DiPetrillo C.G., Smith E.F. (2010). Pcdp1 is a central apparatus protein that binds Ca(2+)-calmodulin and regulates ciliary motility. J. Cell Biol. 189: 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd A.N., Kudla J., Sanders D. (2010). The language of calcium signalling. Annu. Rev. Plant Biol. 61: 593–620 [DOI] [PubMed] [Google Scholar]
- Doherty C.J., Van Buskirk H.A., Myers S.J., Thomashow M.F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger W.F., Clear A.M., Fanning K.J., Peck M.L. (1999). Identification of a Ca2+/H+ antiport in the plant chloroplast thylakoid membrane. Plant Physiol. 119: 1379–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira K.N., Iverson T.M., Maghlaoui K., Barber J., Iwata S. (2004). Architecture of the photosynthetic oxygen-evolving center. Science 303: 1831–1838 [DOI] [PubMed] [Google Scholar]
- Ghanotakis D.F., Babcock G.T., Yocum C.F. (1984). Calcium reconstitutes high rates of oxygen evolution in polypeptide depleted photosystem II preparations. FEBS Lett. 167: 127–130 [Google Scholar]
- Gleason C., Chaudhuri S., Yang T., Muñoz A., Poovaiah B.W., Oldroyd G.E. (2006). Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441: 1149–1152 [DOI] [PubMed] [Google Scholar]
- Han S., Tang R., Anderson L.K., Woerner T.E., Pei Z.M. (2003). A cell surface receptor mediates extracellular Ca(2+) sensing in guard cells. Nature 425: 196–200 [DOI] [PubMed] [Google Scholar]
- Harris E.H. (1989). The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. (San Diego, CA: Academic Press; ). [DOI] [PubMed] [Google Scholar]
- Hidaka H., Sasaki Y., Tanaka T., Endo T., Ohno S., Fujii Y., Nagata T. (1981). N-(6-aminohexyl)-5-chloro-1-naphthalenesulfonamide, a calmodulin antagonist, inhibits cell proliferation. Proc. Natl. Acad. Sci. USA 78: 4354–4357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima T., Burnier J., Ross E.M. (1990). Regulation of Gi and Go by mastoparan, related amphiphilic peptides, and hydrophobic amines. Mechanism and structural determinants of activity. J. Biol. Chem. 265: 14176–14186 [PubMed] [Google Scholar]
- Hippler M., Klein J., Fink A., Allinger T., Hoerth P. (2001). Towards functional proteomics of membrane protein complexes: Analysis of thylakoid membranes from Chlamydomonas reinhardtii. Plant J. 28: 595–606 [DOI] [PubMed] [Google Scholar]
- Kaplan B., Davydov O., Knight H., Galon Y., Knight M.R., Fluhr R., Fromm H. (2006). Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis-elements in Arabidopsis. Plant Cell 18: 2733–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle K.L., Schnell R.A., Fernández E., Lefebvre P.A. (1989). Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J. Cell Biol. 109: 2589–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger A., Weis E., Demeter S. (1993). Low-Ph-induced Ca2+ ion release in the water-splitting system is accompanied by a shift in the midpoint redox potential of the primary quinone acceptor-Q(a). Biochim. Biophys. Acta 1144: 411–418 [Google Scholar]
- Kuchka M.R., Goldschmidt-Clermont M., van Dillewijn J., Rochaix J.D. (1989). Mutation at the Chlamydomonas nuclear NAC2 locus specifically affects stability of the chloroplast psbD transcript encoding polypeptide D2 of PS II. Cell 58: 869–876 [DOI] [PubMed] [Google Scholar]
- Kudla J., Batistic O., Hashimoto K. (2010). Calcium signals: The lead currency of plant information processing. Plant Cell 22: 541–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Li X.P., Björkman O., Shih C., Grossman A.R., Rosenquist M., Jansson S., Niyogi K.K. (2000). A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403: 391–395 [DOI] [PubMed] [Google Scholar]
- Li Z., Wakao S., Fischer B.B., Niyogi K.K. (2009). Sensing and responding to excess light. Annu. Rev. Plant Biol. 60: 239–260 [DOI] [PubMed] [Google Scholar]
- McAinsh M.R., Pittman J.K. (2009). Shaping the calcium signature. New Phytol. 181: 275–294 [DOI] [PubMed] [Google Scholar]
- Molnar A., Bassett A., Thuenemann E., Schwach F., Karkare S., Ossowski S., Weigel D., Baulcombe D. (2009). Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 58: 165–174 [DOI] [PubMed] [Google Scholar]
- Moreno I., Norambuena L., Maturana D., Toro M., Vergara C., Orellana A., Zurita-Silva A., Ordenes V.R. (2008). AtHMA1 is a thapsigargin-sensitive Ca2+/heavy metal pump. J. Biol. Chem. 283: 9633–9641 [DOI] [PubMed] [Google Scholar]
- Moseley J.L., Allinger T., Herzog S., Hoerth P., Wehinger E., Merchant S., Hippler M. (2002). Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO J. 21: 6709–6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie E.H., Pinto M., Horton P. (2009). Agriculture and the new challenges for photosynthesis research. New Phytol. 181: 532–552 [DOI] [PubMed] [Google Scholar]
- Nagel G., Ollig D., Fuhrmann M., Kateriya S., Musti A.M., Bamberg E., Hegemann P. (2002). Channelrhodopsin-1: A light-gated proton channel in green algae. Science 296: 2395–2398 [DOI] [PubMed] [Google Scholar]
- Naumann B., Busch A., Allmer J., Ostendorf E., Zeller M., Kirchhoff H., Hippler M. (2007). Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics 7: 3964–3979 [DOI] [PubMed] [Google Scholar]
- Naumann B., Stauber E.J., Busch A., Sommer F., Hippler M. (2005). N-terminal processing of Lhca3 is a key step in remodeling of the photosystem I-light-harvesting complex under iron deficiency in Chlamydomonas reinhardtii. J. Biol. Chem. 280: 20431–20441 [DOI] [PubMed] [Google Scholar]
- Niyogi K.K., Bjorkman O., Grossman A.R. (1997). Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell 9: 1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura H., Komori T., Kobori M., Nakahira Y., Shiina T. (2008). Evidence for chloroplast control of external Ca2+-induced cytosolic Ca2+ transients and stomatal closure. Plant J. 53: 988–998 [DOI] [PubMed] [Google Scholar]
- Ossowski S., Schwab R., Weigel D. (2008). Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Patel-King R.S., Gorbatyuk O., Takebe S., King S.M. (2004). Flagellar radial spokes contain a Ca2+-stimulated nucleoside diphosphate kinase. Mol. Biol. Cell 15: 3891–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peers G., Truong T.B., Ostendorf E., Busch A., Elrad D., Grossman A.R., Hippler M., Niyogi K.K. (2009). An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462: 518–521 [DOI] [PubMed] [Google Scholar]
- Peltier J.B., Ytterberg A.J., Sun Q., van Wijk K.J. (2004). New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J. Biol. Chem. 279: 49367–49383 [DOI] [PubMed] [Google Scholar]
- Quarmby L.M., Yueh Y.G., Cheshire J.L., Keller L.R., Snell W.J., Crain R.C. (1992). Inositol phospholipid metabolism may trigger flagellar excision in Chlamydomonas reinhardtii. J. Cell Biol. 116: 737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K., MacMillan F., Leibl W., Brettel K., Hanley J., Rutherford A.W., Breton J., Rochaix J.D. (1998). A systematic survey of conserved histidines in the core subunits of Photosystem I by site-directed mutagenesis reveals the likely axial ligands of P700. EMBO J. 17: 50–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh M.H., Shingles R., Cleveland M.J., McCarty R.E. (1998). Direct measurement of calcium transport across chloroplast inner-envelope vesicles. Plant Physiol. 118: 1447–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr J., Sarkar N., Balenger S., Jeong B.R., Cerutti H. (2004). Tandem inverted repeat system for selection of effective transgenic RNAi strains in Chlamydomonas. Plant J. 40: 611–621 [DOI] [PubMed] [Google Scholar]
- Sai J., Johnson C.H. (2002). Dark-stimulated calcium ion fluxes in the chloroplast stroma and cytosol. Plant Cell 14: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmollinger S., Strenkert D., Schroda M. (2010). An inducible artificial microRNA system for Chlamydomonas reinhardtii confirms a key role for heat shock factor 1 in regulating thermotolerance. Curr. Genet. 56: 383–389 [DOI] [PubMed] [Google Scholar]
- Seigneurin-Berny D., Gravot A., Auroy P., Mazard C., Kraut A., Finazzi G., Grunwald D., Rappaport F., Vavasseur A., Joyard J., Richaud P., Rolland N. (2006). HMA1, a new Cu-ATPase of the chloroplast envelope, is essential for growth under adverse light conditions. J. Biol. Chem. 281: 2882–2892 [DOI] [PubMed] [Google Scholar]
- Shirran S.L., Barran P.E. (2009). The use of ESI-MS to probe the binding of divalent cations to calmodulin. J. Am. Soc. Mass Spectrom. 20: 1159–1171 [DOI] [PubMed] [Google Scholar]
- Smith E.F. (2002). Regulation of flagellar dynein by calcium and a role for an axonemal calmodulin and calmodulin-dependent kinase. Mol. Biol. Cell 13: 3303–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suorsa M., Aro E.M. (2007). Expression, assembly and auxiliary functions of photosystem II oxygen-evolving proteins in higher plants. Photosynth. Res. 93: 89–100 [DOI] [PubMed] [Google Scholar]
- Terashima M., Specht M., Naumann B., Hippler M. (2010). Characterizing the anaerobic response of Chlamydomonas reinhardtii by quantitative proteomics. Mol. Cell. Proteomics 9: 1514–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill D.W., et al. (1995). Targeted disruption of mouse EGF receptor: Effect of genetic background on mutant phenotype. Science 269: 230–234 [DOI] [PubMed] [Google Scholar]
- Tirichine L., et al. (2006). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441: 1153–1156 [DOI] [PubMed] [Google Scholar]
- Trebst A. (2007). Inhibitors in the functional dissection of the photosynthetic electron transport system. Photosynth. Res. 92: 217–224 [DOI] [PubMed] [Google Scholar]
- Vainonen J.P., Sakuragi Y., Stael S., Tikkanen M., Allahverdiyeva Y., Paakkarinen V., Aro E., Suorsa M., Scheller H.V., Vener A.V., Aro E.M. (2008). Light regulation of CaS, a novel phosphoprotein in the thylakoid membrane of Arabidopsis thaliana. FEBS J. 275: 1767–1777 [DOI] [PubMed] [Google Scholar]
- Weinl S., Held K., Schlücking K., Steinhorst L., Kuhlgert S., Hippler M., Kudla J. (2008). A plastid protein crucial for Ca2+-regulated stomatal responses. New Phytol. 179: 675–686 [DOI] [PubMed] [Google Scholar]
- Wheeler G.L., Brownlee C. (2008). Ca2+ signalling in plants and green algae—Changing channels. Trends Plant Sci. 13: 506–514 [DOI] [PubMed] [Google Scholar]
- Wheeler G.L., Joint I., Brownlee C. (2008). Rapid spatiotemporal patterning of cytosolic Ca2+ underlies flagellar excision in Chlamydomonas reinhardtii. Plant J. 53: 401–413 [DOI] [PubMed] [Google Scholar]
- Yi X., Hargett S.R., Frankel L.K., Bricker T.M. (2006). The PsbQ protein is required in Arabidopsis for photosystem II assembly/stability and photoautotrophy under low light conditions. J. Biol. Chem. 281: 26260–26267 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.