UKL1 and UKL2 encode redundant plastid isoforms of uridine kinase. Loss of function of either reduces leaf starch and cellulose biosynthesis and enhances lignin biosynthesis. Mutation in UKL1 or UKL2 also affects seed composition, resulting in higher storage protein and lower oil content. We show that plastid uridine salvage activity is important for photoassimilate allocation and partitioning.
Abstract
Nucleotides are synthesized from de novo and salvage pathways. To characterize the uridine salvage pathway, two genes, UKL1 and UKL2, that tentatively encode uridine kinase (UK) and uracil phosphoribosyltransferase (UPRT) bifunctional enzymes were studied in Arabidopsis thaliana. T-DNA insertions in UKL1 and UKL2 reduced transcript expression and increased plant tolerance to toxic analogs 5-fluorouridine and 5-fluorouracil. Enzyme activity assays using purified recombinant proteins indicated that UKL1 and UKL2 have UK but not UPRT activity. Subcellular localization using a C-terminal enhanced yellow fluorescent protein fusion indicated that UKL1 and UKL2 localize to plastids. The ukl2 mutant shows reduced transient leaf starch during the day. External application of orotate rescued this phenotype in ukl2, indicating pyrimidine pools are limiting for starch synthesis in ukl2. Intermediates for lignin synthesis were upregulated, and there was increased lignin and reduced cellulose content in the ukl2 mutant. Levels of ATP, ADP, ADP-glucose, UTP, UDP, and UDP-glucose were altered in a light-dependent manner. Seed composition of the ukl1 and ukl2 mutants included lower oil and higher protein compared with the wild type. Unlike single gene mutants, the ukl1 ukl2 double mutant has severe developmental defects and reduced biomass accumulation, indicating these enzymes catalyze redundant reactions. These findings point to crucial roles played by uridine salvage for photoassimilate allocation and partitioning.
INTRODUCTION
In higher plants, the net product of photosynthesis in a leaf chloroplast (triose phosphate) is primarily used for transient starch biosynthesis in the chloroplast or for Suc biosynthesis in the cytoplasm (Gifford and Evans, 1981). Leaf Suc is exported to sink organs to sustain growth, and leaf starch is catabolized during dark periods for plant growth. Therefore, synthesis of Suc and starch can be considered competing processes (Preiss, 1982). There are two different models to explain the partitioning of Suc and starch. The program model suggests that partitioning of photoassimilate is programmed such that a certain portion of the photoassimilate is always directed for starch biosynthesis in the chloroplast (Schulze et al., 1991; Huber and Hanson, 1992). The overflow model posits that triose phosphate is preferably used for Suc biosynthesis when its production is low, and triose phosphate will be diverted for starch biosynthesis only after Suc biosynthesis is saturated (Eichelmann and Laisk, 1994).
There is longstanding interest in understanding the mechanisms that regulate photoassimilate allocation and partitioning because of its fundamental role in plant life and because of its potential applications for biomass improvement at both the quality and quantity levels. Previous studies have demonstrated that some key metabolites (e.g., orthophosphate, triose phosphate [cytosol-to-chloroplast concentration ratio], and fructose 2,6-bisphosphate) collectively regulate the partitioning process (Jelitto et al., 1992; Huber, 1986; Fujita et al., 2003). At the biochemical level, starch and Suc biosynthesis in leaves uses different precursors. Formation of ADP-glucose commits Glc for starch biosynthesis in the chloroplast; by contrast, formation of UDP-glucose directs Glc for Suc biosynthesis in the cytoplasm or for cellulose biosynthesis at the interface between cell wall and cell membrane. Since biosynthesis of ADP-glucose and UDP-glucose requires ATP or UTP, respectively, for activation, it is possible that nucleotide availability also could affect photoassimilate allocation and partitioning.
It has been demonstrated that adenylate supply to the plastid is fundamentally important for starch biosynthesis in storage organs, such as potato (Solanum tuberosum) tubers. Overexpression of the amyloplast ATP:ADP translocator results in increased starch accumulation, and antisense inhibition of the same protein results in reduced starch yield (Tjaden et al., 1998). Antisense inhibition of plastid adenylate kinase in potato results in a substantial increase in adenylate pools; the transgenic plants also show higher starch and tuber yield than the wild type (Regierer et al., 2002). Subsequent studies revealed that pyrimidine nucleotides play a crucial role in regulating carbohydrate metabolism in sink organs. Specifically, downregulation of uridine 5′-monophosphate synthase by antisense technology reduces de novo pyrimidine synthesis and stimulates the uridine salvage pathway in potato tubers (Geigenberger et al., 2005). This shift in the pathway of UMP synthesis is accompanied by increased levels of tuber uridine nucleotides, increased flux of Suc to starch and cell wall synthesis, and increased amounts of starch and cell wall components in the tubers (Geigenberger et al., 2005). Thus, uridine nucleotides derived from de novo biosynthesis or salvage pathways affect carbon flux quite differently in sink organs.
Recent evidence suggests that uracil salvage is necessary for vegetative growth, such as early seedling development. A T-DNA knockout of plastidial uracil phosphoribosyltransferase (UPP gene), which is responsible for almost all uracil phosphoribosyltransferase (UPRT) activity in Arabidopsis thaliana, causes a light-dependent dramatic pale-green to albino phenotype, dwarfism, and an inability to produce viable progeny; plastid biogenesis and starch accumulation are also affected (Mainguet et al., 2009). Potato tubers have been used as a system to study nucleotide metabolism in storage organs, ultimately revealing crosstalk between nucleotide and carbon metabolism (Geigenberger et al., 1998, 2005; Loef et al., 1999; Giermann, et al., 2002; Katahira and Ashihara, 2002). Although also a sink tissue, seeds have very different developmental programs and reserve composition compared with potato tubers. Thus, it is unclear to what extent the knowledge gained from potato tubers can be transferred to the developing seed. Moreover, although several studies have been performed on purine and pyrimidine metabolism in plants (Stasolla et al., 2003; Zrenner et al., 2006), it is unclear if uridine salvage contributes to photoassimilate allocation and partitioning in leaves.
To gain a better insight how uridine salvation affects photoassimilate partitioning and seed composition, two isozymes, UKL1 and UKL2, which encode plastid uridine kinases (UKs), were characterized in detail. Knockout of plastid uridine scavenging results in seed composition changes (i.e., lower lipid and higher storage protein) without significantly affecting total seed weight per plant. Many leaf metabolites also are affected. Our data demonstrate that plastid uridine salvage is an important aspect of photoassimilate partitioning.
RESULTS
The ukl1 and ukl2 Mutants Display Increased Tolerance to Toxic Nucleoside Analogs 5-Fluorouracil and 5-Fluorouridine
In Arabidopsis, five homologous genes contain both a UPRT domain and an N-terminal UK domain and are herein referred to as UK-like1 (UKL1), UKL2, UKL3, UKL4, and UKL5. Unlike UKL3, UKL4, and UKL5, UKL1 and UKL2 have transit peptides at the N terminus. Individual knockout lines for UKL1 and UKL2 were identified by RT-PCR (see Supplemental Figure 1 online). Three individual T-DNA lines (SALK_108468, SALK_026938, and SALK_147532) were identified for UKL1 (see Supplemental Figure 1A online, top panel). Transcript expression is completely abolished in the SALK_108468 line, and transcript levels are unaffected in the other two lines (see Supplemental Figure 1B online, left). Two individual T-DNA insertion lines (SALK_058257 and CS821451) contain inserts within the UKL2 gene (see Supplemental Figure1A online, bottom panel). The T-DNA insertion in the CS821451 line does not affect UKL2 expression, whereas in the SALK_058257 line, UKL2 expression is undetectable (see Supplemental Figure 1B online, right). Therefore, the SALK_108468 and SALK_058257 lines were renamed ukl1 and ukl2, respectively. These results are in agreement with previous reports (Islam et al., 2007; Mainguet et al., 2009).
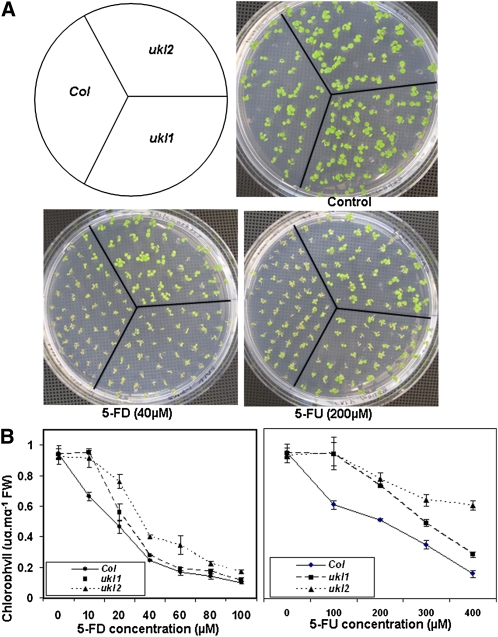
Pyrimidine salvage pathways offer multiple potential targets to incorporate cytotoxic analogs, such as 5-fluorouracil (5-FU) and 5-fluorouridine (5-FD), into cellular nucleotide pools. 5-FU and 5-FD are converted to 5-fluoro-uridine monophosphate by UPRT and UK, respectively. 5-Fluoro-uridine monophosphate is further converted to 5-fluoro-uridine triphosphate (5-FUTP) by a two-step route. In humans, cytotoxicity of 5-FU and 5-FD is varied: (1) 5-FUTP can be incorporated into RNA instead of UTP; (2) 5-FU is activated by the formation of 5-FdUMP, an inhibitor of thymidylate synthase that is involved in the synthesis of DNA; (3) 5-FU and 5-FD can be incorporated into DNA; and (4) 5-FUTP can be conjugated to sugars and incorporated into membranes, thus altering membrane functions. However, the cytotoxic mechanisms of 5-FU and 5-FD in plants are less studied, and it remains unclear if one or more of these mechanisms is the mode of action. To determine if sensitivity to 5-FU and 5-FD is affected in the ukl1 and ukl2 mutants, seeds were germinated on half-strength Murashige and Skoog (MS) medium supplemented with 5-FU or 5-FD at specified concentrations. After 5 d of growth, chlorophyll content was measured (Figure 1). The ukl1 mutant showed resistance to 10 μM 5-FD; when the concentration was increased to 20 μM, ukl1 no longer displayed any significant difference in sensitivity compared with the wild type. In contrast, the ukl2 mutant showed higher tolerance to 5-FD up to 80 μM. Compared with 5-FD, the ukl1 and ukl2 mutants showed a much higher range of tolerance to 5-FU. Even in the presence of 400 μM 5-FU, the ukl1 and ukl2 mutants showed differences in chlorophyll accumulation from the wild type. Overall, the ukl2 mutant showed a higher degree of tolerance to 5-FU and 5-FD than the ukl1 mutant. Since the 5-FU and 5-FD toxic effects are caused by the remaining total UK and UPRT activity, respectively, in the ukl1 or ukl2 mutant, tolerance to 5-FU or 5-FD indicates less UK or UPRT activity in the gene knockout lines. The observation that ukl2 is more tolerant to 5-FU and 5-FD than the ukl1 mutant suggests that UKL2 has higher activity than UKL1 in vivo. To achieve a similar suppression of seedling greening using 5-FU, higher concentrations were needed (compared with 5-FD) for both the ukl1 and ukl2 mutants, which suggests that 5-FD is the preferred substrate for the UKL1 and UKL2 proteins.
Figure 1.
ukl1 and ukl2 Show Reduced Sensitivity to 5-FU and 5-FD Inhibition of Seedling Growth.
(A) ukl1 and ukl2 are more tolerant to 5-FU and 5-FD inhibition of seedling growth. The wild type and mutants were germinated on half-strength MS medium supplemented with 5-FU or 5-FD at specified concentrations for 5 d. Col, Columbia ecotype.
(B) Quantification of the chlorophyll content of the wild type and mutants grown on MS medium containing different concentrations of 5-FU or 5-FD for 5 d. Data are expressed as mean ± se for three measurements. FW, fresh weight.
[See online article for color version of this figure.]
Recombinant UKL1 and UKL2 Proteins Display Only UK Activity
The above-mentioned 5-FU and 5-FD feeding experiments provide only indirect evidence that the UKL1 and UKL2 proteins posses both UK and UPRT activity. Since there are differences in the uptake and subsequent metabolism of uridine and uracil, direct confirmation of UK and UPRT activity was necessary. Therefore, the UKL1 and UKL2 proteins were expressed in Escherichia coli (see Supplemental Figures 2A and 2B online), and the purified proteins assayed for UK, UPRT, uridine-ribohydrolase (URH), and UMP kinase activity in vitro. The experiments demonstrated that recombinant UKL1 and UKL2 proteins have UK activity, with a Vmax of 14.8 and 243.9 μmol·h−1·mg−1 and a Km for uridine of 0.34 and 1.18 mM for UKL1 and UKL2, respectively (see Supplemental Figures 2C and 2D online). UPRT, URH, and UMP kinase activities were undetectable with both enzymes.
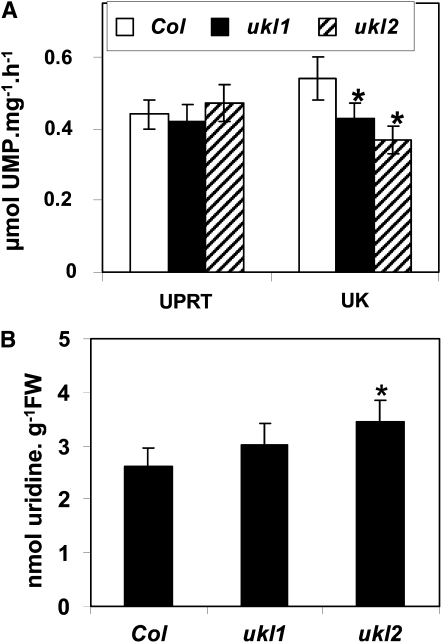
To confirm reduced activity in the ukl1 and ukl2 mutants, UK and UPRT activity was quantified from 4-week-old aerial leaves. ukl1 and ukl2 mutants showed 20.3 and 31.5% lower UK activity, respectively, compared with wild-type plants (Figure 2A). There was no significant difference in UPRT activity, which supports the recombinant protein results that the UKL1 and UKL2 proteins have UK but not UPRT activity. The uridine and uracil content in 1-week-old seedlings also was measured. The uridine content in the ukl2 mutant was 32% higher than the wild type, and the ukl1 mutant showed a small but insignificant increase in uridine content (Figure 2B). Uracil was undetectable in the wild-type plants and in the ukl1 and ukl2 mutants. The lack of UPRT activity for UKL1 and UKL2 raises the question: Why do the ukl1 and ukl2 mutants show increased tolerance toward 5-FU compared with the wild type? One possible answer is that nucleoside phosphorylase reversibly converts 5-FU and Rib into 5-FD, although this activity has never been reported in plants. Another possible answer is that uridine, uracil, or other nucleosides or nucleobases accumulate in the ukl1 and ukl2 mutants, thus diluting the specific activity of the toxic intermediates.
Figure 2.
UK Activity Is Lower and Uridine Content Higher in ukl1 and ukl2 Mutants Compared with the Wild Type.
(A) UK and UPRT activity assays from 4-week-old aerial leaves of wild-type and ukl1 and ukl2 mutant plants (n = 4). Col, Columbia ecotype.
(B) Uridine and uracil content were quantified from wild-type and ukl1 and ukl2 mutant plants (n = 3). Plants were germinated on MS plates, and tissues were harvested after 1 week of growth for uridine and uracil isolation. Data are expressed as mean ± se. Asterisk represents significant difference (P < 0.05). FW, fresh weight.
Starch Accumulation in the ukl1 and ukl2 Mutants Is Reduced
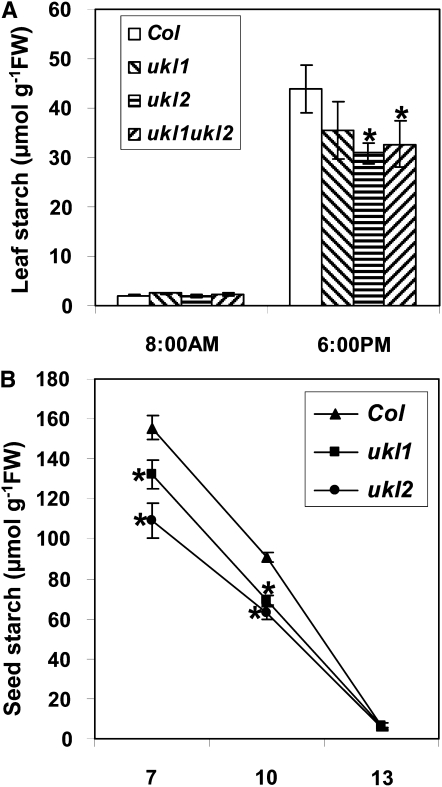
To observe if leaf starch biosynthesis is affected by the ukl1 or ukl2 mutation, whole plants were harvested before a light cycle and then again late in the light cycle. At the beginning of a light cycle, we observed little leaf starch and no differences among the wild type, ukl1, and ukl2 plants, which indicates that leaf starch degradation during the dark period is not affected (Figure 3A). However, late in the light cycle, the ukl2 mutant had accumulated 30% less leaf starch (Figure 3A) than the wild type on a fresh weight basis. The ukl1 mutant also accumulated less leaf starch compared with wild-type plants, but the reduction was statistically insignificant (Figure 3A).
Figure 3.
Leaf and Seed Starch Content in ukl1 and ukl2 Mutants Is Reduced.
(A) Quantification of the leaf starch content of the wild type and mutants. Five individual plants were used. Data are expressed as means ± se. P values are 0.59, 0.004, and 0.036 for the ukl1 ukl2 single mutants and the ukl1 ukl2 double mutant, respectively. Col, Columbia ecotype; FW, fresh weight.
(B) Quantification of the seed starch content during different developmental stages (n = 4). Siliques were harvested at 7, 10, and 13 DAF; seeds were dissected from siliques for starch quantization. Student’s t tests were performed on the ukl1 and ukl2 mutants: at 7 DAF, the P values were 0.032 and 0.012, respectively; and at 10 DAF, the P values were 0.045 and 0.039, respectively. Asterisk represents significant difference (P < 0.05).
To observe if the UKL1 or UKL2 gene mutations affect starch levels in seeds, developing seeds were harvested at 7, 10, and 13 d after fertilization (DAF) and their starch content quantified. At 7 DAF, seeds from ukl1 and ukl2 mutant plants accumulated 15 and 30% less starch, respectively, than the wild-type plants (Figure 3B). Starch content sharply decreased thereafter, and starch content reached its lowest level at 13 DAF, at which point no differences were observed between wild-type and mutant plants (Figure 3B). These results indicate that seed starch accumulation is reduced in the ukl1 and ukl2 mutants but that starch degradation is largely unaffected.
External Application of Orotic Acid Enhances Leaf Starch Accumulation in the ukl2 Mutant
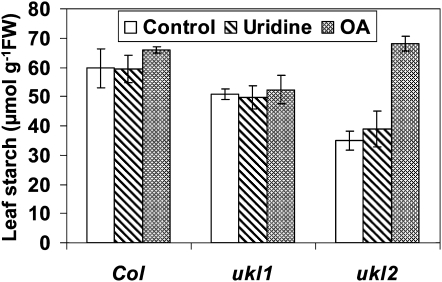
Orotic acid (OA) and uridine are substrates for de novo and salvage pathways of uridine nucleotide synthesis, respectively. Externally supplied OA leads to a specific increase in uridine nucleotide levels and a stimulation of Suc degradation and starch synthesis in discs from growing potato tubers (Loef et al., 1999). To investigate if exogenously applied uridine or OA affects leaf starch accumulation in planta, wild-type and ukl1 and ukl2 mutant plants were sprayed at 9 am daily with 1 mM uridine or 1 mM OA for a period of 1 month. Leaves from the plants were subsequently harvested in late afternoon and starch quantified. Application of uridine did not significantly affect leaf starch content in the wild-type plants or in the ukl1 and ukl2 mutant plants. Application of OA, on the other hand, did increase leaf starch content by 94% in the ukl2 mutant plants compared with a water-treated ukl2 control but had no affect on the leaf starch content in wild-type and ukl1 mutant plants (Figure 4).
Figure 4.
OA Application Affects Leaf Starch Content in ukl2 mutant plants.
Plants were grown under long-day conditions for 30 d and then sprayed with 1 mM uridine or OA daily after germination. Leaves were harvested at 6:00 pm for starch quantification. Data are expressed as mean ± se (n = 5). Col, Columbia ecotype; FW, fresh weight.
Metabolic Profiling of the ukl2 Mutant Reveals Changes in Multiple Pathways
To better understand why starch content is reduced in the ukl2 mutant and its relationship with nucleotide metabolism, metabolites were comparatively profiled. Metabolic profiling of the wild type and the ukl2 mutant identified 283 compounds, 42 of which are unknown (Table 1; see Supplemental Data Set 1 online). Ten amino acids were detected. Compared with the wild type, the ukl2 mutant had higher levels of l-Asp (twofold higher), l-Glu (40% higher), l-Phe (43% higher), and Gly (4% higher). N-acetyl Glu content was about half in the ukl2 mutant (Table 1).
Table 1.
Partial List of the Metabolites Identified from Wild-Type and ukl2 Mutant Plants
| Metabolite | ukl2/Col | P |
| Amino acids | ||
| Gly | 1.04 | 0.042 |
| l-Asp (3TMS) | 3.16 | 0.032 |
| l-Glu, N-acetyl-(2TMS) | 0.56 | 0.001 |
| l-Glu (3TMS) | 1.40 | 0.046 |
| l-Phe (2TMS) | 1.43 | 0.049 |
| Acids and hydroxy acids | ||
| Fumaric acid (2TMS) | 1.35 | 0.008 |
| Maleic acid (2TMS) | 2.33 | 0.003 |
| (R*,S*)-3,4-dihydroxybutanoic acid (3TMS) | 1.86 | 0.004 |
| Threonic acid-1,4-lactone (2TMS), trans- | 2.12 | 0.032 |
| Malic acid (3TMS) | 2.39 | 0.031 |
| Picolinic acid (1TMS) | 3.77 | 0.039 |
| Erythronic acid (4TMS) | 1.95 | 0.039 |
| Propanoic acid, 2-oxo-3- (3TMS) | 1.35 | 0.027 |
| Pentonic acid (5TMS) | 1.40 | 0.035 |
| Anthranilic acid | 0.95 | 0.325 |
| Benzoic acid | 2.19 | 0.035 |
| Dehydroascorbic acid dimmer | 4.27 | 0.005 |
| Furo[2,3-H]coumarine | 2.98 | 0.043 |
| Benzoic acid, 2,6-bis(3TMS) | 2.01 | 0.018 |
| Hexadecanoic acid (1TMS) | 1.68 | 0.003 |
| 9,12-Octadecadienoic acid | 1.44 | 0.037 |
| trans-sinapinic acid (2TMS) | 1.49 | 0.011 |
| Butanoic acid | 0.64 | 0.010 |
| Sugars and sugar alcohols | ||
| Fructose methoxyamine {BP} (5TMS) | 1.52 | 0.004 |
| Psicose methoxyamine {BP} (5TMS) | 2.16 | 0.001 |
| Glucose methoxyamine (5TMS) | 1.83 | 0.012 |
| Glycerol (3TMS) | 1.29 | 0.039 |
| Suc (8TMS) | 1.09 | 0.166 |
| l-Mannose | 1.56 | 0.008 |
| Melezitose (11TMS) | 2.28 | 0.033 |
| Galactinol (9TMS) | 2.35 | 0.035 |
| Trehalose (8TMS) | 1.01 | 0.478 |
Plants were grown in growth chambers for 30 d under a long-day photoperiod. Aerial leaves were harvested after 10 h of illumination. Metabolites were extracted and analyzed by gas chromatography–mass spectrometry and identified by library searching. The relative abundance in ukl2 and the wild type was represented by their mean ion intensity ratio (ukl2/Col). Five biological replicates were performed. Student’s t tests were performed to calculate P values. Col, Columbia ecotype.
Of the 53 organic acids detected, intermediates of the tricarboxylic acid (TCA) cycle were the most substantially different in the ukl2 mutant. Malate and fumarate levels in the ukl2 mutant were significantly higher, 140 and 35%, respectively, compared with the wild type. Citrate was 23% higher in the ukl2 mutant than in the wild type, although this elevated level was not statistically significant. Levels of succinate were the same in the wild type as in the ukl2 mutant. Interestingly, maleic acid was elevated 1.3-fold in the ukl2 mutant. Maleic acid can be converted into fumarate by maleate cis-trans-isomerase and converted into malate by hydration catalyzed by malate hydrolase. Two TCA cycle intermediates, oxaloacetate and α-ketoglutarate, can be used for Asp and Glu biosynthesis, respectively, and pool levels of these two amino acids also were elevated in the ukl2 mutant (Table 1). The similar levels of succinate in the ukl2 mutant and the wild type may suggest that only a partial TCA cycle is operating during the light period. Benzoic acid, a product of phenol serial oxidation through benzaldehyde, was 1.2-fold higher in the ukl2 mutant compared with the wild type (Table 1); accordingly, phenol also was elevated by 2.8-fold in the ukl2 mutant (see Supplemental Data Set 1 online, row 224).
Approximately 30 sugars and sugar alcohols were detected. There were no differences in levels of two major transportable forms of sugars, Suc and trehalose, between the wild type and the ukl2 mutant. However, Glc, Fru, and Man were 83, 52, and 56% higher, respectively, in the ukl2 mutant compared with the wild type. The ukl2 mutant, compared with the wild type, also had elevated levels of psicose, a C-3 epimer of d-fructose (1.2-fold higher); melezitose, a nonreducing trisaccharide sugar, (1.3-fold higher); and galactinol (1.4-fold higher) (Table 1). The enhanced concentration of monosaccharides in the ukl2 mutant supports the observation of reduced leaf and seed starch due to reduced biosynthesis (Table 1; see Supplemental Data Set 1 online).
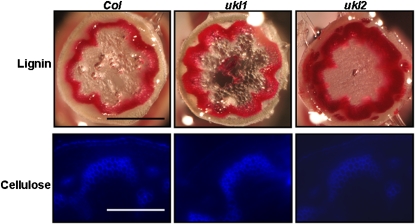
Lignin and Cellulose Biosynthesis Were Perturbed in the ukl2 Mutant
Trans-sinapinic acid and coumarine are used for lignin and lignan biosynthesis in planta (Whetten and Sederoff, 1995). Trans-sinapinic acid and coumarine, two derivatives of l-Phe, were 49 and 200% higher, respectively, in the ukl2 mutant compared with the wild type, which is in accordance with the enhanced pool size of l-Phe in the ukl2 mutant (Table 1). Phe concentration has been demonstrated to profoundly affect flux through the phenylpropanoid pathway (Anterola et al., 2002); Phe feeding to Pinus taeda suspension cells increased p-coumaryl and coniferyl alcohol synthesis and Phe ammonia-lyase, 4-coumarate-CoA ligase, caffeoyl-CoA O-methyltransferase, and cinnamoyl-CoA reductase transcript levels (Anterola et al., 2002). 3,4-Dihydroxybutanoic acid, erythronic acid, and threnoic acid, which are the products of lignin oxidation (Tsutsumi et al., 1990), were 86, 95, and 112% higher, respectively, in the ukl2 mutant compared with the wild type. Dehydroascorbic acid, a reducing form of ascorbic acid, was elevated more than threefold in the ukl2 mutant. Dehydroascorbic acid can be converted into ascorbic acid and hydrogen peroxide by peroxidase (Bolwell et al., 1995). The rising concentration of dehydroascorbic suggests that hydrogen peroxide production is also elevated in the ukl2 mutant. Hydrogen peroxide is required for monolignol oxidation and dehydrogenative polymerization (Ros-Barceló et al., 2002). The collective data suggest that lignin and lignan biosynthesis or polymerization is enhanced in ukl2 mutant. To test this, lignin and cellulose staining were performed. The staining results indicated that lignin content was higher and cellulose content was lower in the ukl2 mutant compared with the wild type. Lignin and cellulose content in the ukl1 mutant was not different from the wild type (Figure 5).
Figure 5.
Lignin and Cellulose Accumulation in the ukl2 Mutant Is Altered.
Cellulose content is reduced and lignin content enhanced in the ukl2 mutant plants. For visualization of secondary walls, tissues were stained with phloroglucinol-HCl for lignin and with fluorescent brightener 28 for cellulose. Scale bars for lignin and cellulose staining are 1 and 2 mm, respectively. Col, Columbia ecotype.
The de Novo Pyrimidine Biosynthesis Pathway Is Transcriptionally Induced in the ukl2 Mutant
Glu and Asp, two precursors for de novo pyrimidine biosynthesis, were 40 and 216% higher, respectively, in the ukl2 mutant compared with the wild type (Table 1). Enhanced levels of these metabolites in the ukl2 mutant raised the possibility that de novo pyrimidine biosynthesis was affected in ukl2 mutant. To test this possibility, RT-PCR was conducted to observe the transcript levels of the genes involved in this pathway. The results indicate that three genes were upregulated during the day period in both mutants compared with the wild type (see Supplemental Figure 3 online). These are a carbamoylphosphate synthase, which catalyzes the first step for de novo pyrimidine biosynthesis, an Asp transcarbamoylase, which catalyzes the first committed step of de novo pyrimidine biosynthesis, and a 1,5-phosphoribosyl-1-pyrophosphate (PRPP) synthase, which converts ribose-5-phosphate into PRPP for de novo pyrimidine biosynthesis.
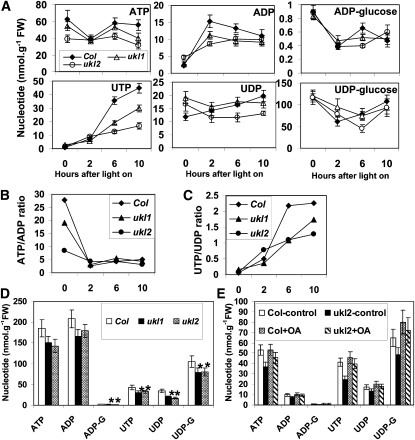
Nucleotide Levels in the ukl1 and ukl2 Mutants Are Affected during the Light Period
Since RT-PCR indicated that de novo pyrimidine biosynthesis pathway is affected in the ukl1 and ukl2 mutants during the light period (see Supplemental Figure 3 online), nucleotide levels were quantified by HPLC to further study this observation. In dark-adapted leaves, ATP content in the ukl2 mutant plant was 27 to 43% lower than in the wild type and ukl1 mutant plants (Figure 6A). This difference dissipated within 2 h after illumination, when ATP was mostly consumed, but reappeared 6 to 10 h after illumination (Figure 6A, top left panel). For ADP, both the ukl1 and ukl2 mutant plants contained less ADP than the wild type from 2 to 6 h after illumination, and this difference was mitigated by 10 h of light cycle as well as dark adaptation (Figure 6A, top middle panel). ADP-glucose content in the ukl1 and ukl2 mutant plants was 21 and 40% lower, respectively, at 6 h after light cycle compared with the wild type; there was no difference at other time points (Figure 6A, top right panel). UTP showed a much different accumulation trend compared with ATP. UTP levels were low in dark-adapted leaves and increased only slightly for the first 2 h after illumination, then increased rapidly between 2 and 6 h of illumination. During the first 2 h of illumination, there was no difference in UTP content among wild-type, ukl1, and ukl2 mutant plants. After 6 to 10 h of light cycle, the ukl1 and ukl2 mutant plants had significantly lower UTP content (34 to 65%) than wild-type plants (Figure 6A, bottom left panel). In dark-adapted leaves, UDP content was higher in the ukl1 and ukl2 mutant plants compared with wild-type plants; 2 h after illumination, no significant difference in UDP content was observed between wild-type and the ukl mutant plants. At 6 and 10 h after illumination, the ukl2 mutant plant had lower UDP levels than both the ukl1 mutant and wild-type plants (Figure 6A, bottom middle panel). UDP-glucose content was 53 and 28% higher in the ukl1 and ukl2 mutant than wild-type plants at 2 h after illumination and 42% lower in the ukl2 mutant than the wild type at 6 h after illumination. No significant differences were observed between wild-type and ukl1 mutant plants at any time for UDP-glucose (Figure 6A, bottom right panel). AMP and UMP content could not be accurately quantified due to their coelution with unknown compounds. The ratios of ATP to ADP and UTP to UDP also were calculated (Figures 6B and 6C). Before illumination, the ukl1 and ukl2 mutant plants showed a lower ratio of ATP to ADP compared with the wild type; this difference disappeared during the light period (Figure 6B). By contrast, the ukl1 and ukl2 mutant plants displayed a lower ratio of UTP to UDP 2 h after illumination (Figure 6C).
Figure 6.
Uridinylate and Adenylate Levels in Wild-Type, ukl1, and ukl2 Leaves.
(A) Adenylate and uridinylate dynamic changes during light period in Arabidopsis leaves. Plants were grown in growth chambers for 30 d. Aerial leaves were harvested at 0, 2, 6, and 10 h after illumination for nucleotide isolation. Data are expressed as mean ± se of determinations on six individual plants per line. Col, Columbia ecotype; FW, fresh weight.
(B) and (C) The ATP/ADP (B) and UTP/UDP (C) ratios have been calculated to characterize levels of the major adenine and uridine nucleotides.
(D) Silique nucleotide levels in the wild type and ukl1 and ukl2 mutants. Siliques were harvested 5 DAF after 8 h illumination. Asterisk represents significant difference (P < 0.05, n = 3).
(E) OA application affected nucleotide levels in the ukl2 mutant. Plants were grown in growth chambers and sprayed daily with 1 mM orotate for 3 weeks, beginning 1 week after germination. Data are expressed as mean ± se of determinations on six individual plants per line.
Siliques from plants at 5 DAF were harvested 8 h after illumination for nucleotide quantification. The results indicate that the ukl1 and ukl2 mutant plants have significantly lower UTP, UDP, UDP-glucose, and ADP-glucose content compared with the wild type (Figure 6D). This agrees with the low seed starch phenotype for the ukl1 and ukl2 mutant plants.
External application of orotate to the ukl2 mutant plants largely rescued the low leaf starch phenotype (Figure 4). To understand if the changes in leaf starch content accompany a nucleotide level change, the aerial leaves from 4-week-old plants were harvested for nucleotide measurement after 3 weeks of OA application. OA application restored nucleotide levels in ukl2 mutant plants (Figure 6E).
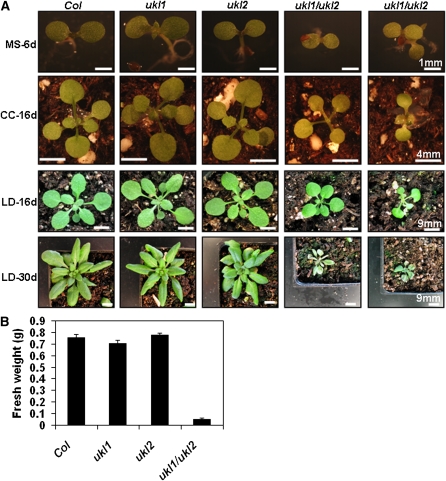
UKL1 and UKL2 Show Redundant Effects on Biomass Accumulation
The lack of any visible growth defects in the ukl1 or ukl2 mutant plants could have two explanations: (1) uridine salvage has a minor role in planta or (2) the UKL1 and UKL2 genes are redundant. To test the second possibility, ukl1 ukl2 double mutant plants were produced and analyzed. When germinated on MS media for 6 d, the ukl1 and ukl2 single mutant plants developed green cotyledons, similar to the wild type. In contrast, the double mutant plants grew much more slowly than both the wild-type and the single mutant plants. Additionally, the cotyledon was pale green (Figure 7, top panel). About 30% of the double mutant plants died at this seedling stage. After being transferred to soil, the ukl1 ukl2 double mutant plants initiated true leaf growth, although at a slower rate compared with the wild type and regardless of whether they were grown under continuous light or long-day conditions (Figure 7, two middle panels). The ukl1 ukl2 double mutant plants showed variable severity in growth retardation and senesced at different developmental stages; in most plants, the apical meristem and the young leaf began to senesce first, followed by the older leaf. This observation agrees with the gene expression data showing that UKL1 and UKL2 are highly expressed in the shoot apex inflorescence (http://jsp.weigelworld.org/expviz.jsp). No ukl1 ukl2 double mutant plants reached reproductive maturity (Figure 7, bottom panel). These observations imply that uridine salvage plays an essential role for meristem maintenance, young leaf growth, and phase transition from vegetative growth to reproduction. Quantitation of biomass from plants grown under long-day conditions showed no difference between wild-type plants and the ukl1 and ukl2 mutant plants; however, the ukl1 ukl2 double mutant plants accumulated only ~7% of the biomass of wild-type plants (Figure 7B). Thus, we conclude that the lack of visible growth defects in the ukl1 and ukl2 mutant plants is due to gene redundancy.
Figure 7.
UKL1 and UKL2 Are Genetically Redundant and Required for Vegetative Growth in Arabidopsis.
(A) The wild type, ukl1 and ukl2 single mutants, and ukl1 ukl2 double mutants (two independent lines) were germinated on MS medium. Seedlings were transferred into soil and grown under continuous white light (CC) or long-day (LD) conditions. Plants were photographed at either 6, 16, or 30 d as noted. Col, Columbia ecotype.
(B) Fresh weight of aerial leaves grown under LD condition for 30 d. Data are expressed as mean ± se (n = 24).
[See online article for color version of this figure.]
Lipid and Protein Composition of ukl1 and ukl2 Mutant Seeds Are Altered
UKL1 and UKL2 are actively expressed during seed development, although with different trends. The UKL1 transcript is highly expressed between developmental stages 3 and 7, while the UKL2 transcript shows only a gradual increase at these developmental stages (see Supplemental Figure 4 online). This expression pattern suggests that uridine salvage plays an important role during seed filling. To explore this possibility, physical and composition traits of the seed were measured in the ukl1 and ukl2 mutant plants (Table 2). No significant differences were observed with respect to total silique and total seed mass per plant, but individual seed mass from the ukl1 and ukl2 mutant plants was 7 and 9% greater, respectively, than wild-type seed. The lipid content of the seeds from the ukl1 and ukl2 mutant plants was 13.5 and 11.5% lower, respectively, compared with the wild type. Seed protein content was 5 and 7% higher in the ukl1 and ukl2 seeds, respectively. No significant differences were recorded in seed starch, Suc, Glc, and Fru content between wild-type and mutant seeds, except that seeds from the ukl1 mutant plant had 18.7 and 74.8% higher starch and Suc content, respectively, than the wild type (Table 2). The seed compositions of the ukl1 and ukl2 mutant plants showed small but statistically significant changes in lipid and protein compared with seeds from wild-type plants, likely due to the functional redundancy of UKL1 and UKL2 in developing seed.
Table 2.
Seed Trait and Composition Analysis of the Wild Type and ukl1 and ukl2 Mutants
| Seed Traits | Col | ukl1 | P | ukl2 | P |
| Total siliques/plant | 193.3 ± 23.0 | 191.2 ± 10.0 | 0.470 | 184.5 ± 11.9 | 0.390 |
| Seed weight (mg)/plant | 112.2 ± 10.5 | 104.4 ± 5.1 | 0.300 | 118.0 ± 8.0 | 0.360 |
| Seed weight (mg)/1000 seeds | 20.4 ± 0.8 | 21.8 ± 1.0 | 0.040 | 22.3 ± 0.6 | 0.047 |
| Lipid (%) | 38.0 ± 1.1 | 32.8 ± 0.5 | 0.003 | 33.6 ± 1.1 | 0.020 |
| Protein (%) | 38.8 ± 0.3 | 40.9 ± 0.7 | 0.010 | 41.5 ± 0.7 | 0.003 |
| Starch (%) | 0.25 ± 0.01 | 0.29 ± 0.01 | 0.010 | 0.27 ± 0.03 | 0.220 |
| Suc (%) | 1.79 ± 0.22 | 2.68 ± 0.28 | 0.040 | 2.18 ± 0.12 | 0.140 |
| Glc (%) | 0.32 ± 0.06 | 0.32 ± 0.07 | 0.410 | 0.32 ± 0.06 | 0.520 |
| Fru (%) | 0.74 ± 0.05 | 0.78 ± 0.08 | 0.070 | 0.75 ± 0.04 | 0.080 |
Plants were grown under identical conditions until seeds reached maturity. Seeds were harvested from individual plants and evenly dried in the presence of anhydrous CaSO4 before analysis. Data are expressed as means ± se (n = 5). Col, Columbia ecotype.
We reasoned that the double mutants of ukl1 and ukl2 would show much more significant seed composition changes; however, since the double mutant senesces before bolting, we were unable to test this hypothesis. Undoubtedly, seed composition is affected by reduced photoassimilate supply from the leaf, resulting in lower starch during development. Reduced starch during early seed development would likely contribute to the reduced oil phenotype. Additionally, the ukl1 and ukl2 mutant plants contained lower nucleotide pools, which may contribute to the reduced oil content since de novo fatty acid synthesis requires ATP for carboxylation of acetyl-CoA, the first step in this pathway.
ukl1 and ukl2 Mutants Are Rescued by Expressing Wild-Type UKL1 or UKL2, Respectively
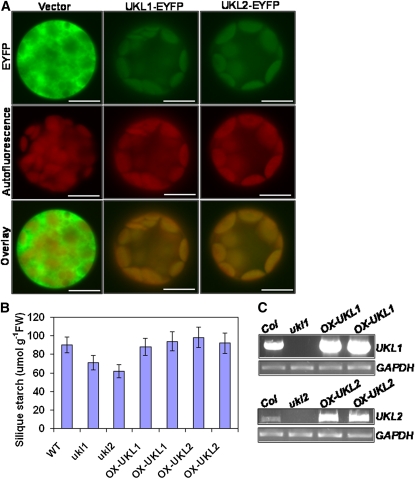
Because only one T-DNA knockout line was identified for UKL1 and UKL2 through the currently available T-DNA line collections (www.Arabidopsis.org), it was necessary to correlate genotype with phenotype by complementation. Transgenic lines were constructed by expressing the coding sequence of UKL1 and UKL2 cDNA with an enhanced yellow fluorescent protein (EYFP) fusion at their C termini in the homozygous mutant background. Because there are no visible growth defects for the ukl1 and ukl2 mutant plants, we used the low seed starch phenotype of ukl1 and ukl2 to confirm their complementation. Siliques from plants at 7 DAF of two individual T1 transgenic lines as well as the wild type and ukl1 and ukl2 homozygous mutants were harvested, and then their starch content quantified. The results demonstrate that expression of a wild-type copy of either UKL1 or UKL2 can rescue the low seed starch phenotype (Figure 8B), indicating that the UKL1 or UKL2 mutation is responsible for this phenotype. To further confirm the expression of UKL1 and UKL2 in the transgenic lines, RT-PCR was conducted. The results confirmed the overexpression of UKL1 and UKL2 in the two individual transgenic lines under their respective mutant background (Figure 8C).
Figure 8.
Subcellular Localization and Complementation of UKL1 and UKL2.
(A) Fluorescence imaging of protoplasts isolated from stably transformed Arabidopsis leaf cells expressing EYFP, UKL1-EYFP, or UKL2-EYFP chimeric proteins. Top panels show EYFP signal in green; middle panels show chlorophyll autofluorescence in red; bottom panels show overlay of the EYFP and autofluorescence signals. Bars = 10 μm.
(B) Silique starch content of the wild type (WT), ukl1 and ukl2 mutants, and individual transgenic lines overexpressing UKL1 or UKL2 in their respective homozygous mutant background. Siliques at 7 DAF were harvested after 9 h illumination for starch quantification. Data are expressed as mean ± se (n = 4). FW, fresh weight.
(C) Overexpression of UKL1 or UKL2 in the transgenic lines confirmed with RT-PCR. T2 seeds from above complemented transgenic lines were germinated on MS plates for 5 d and used for RNA isolation. Seedlings were harvested for total RNA isolation. GAPDH was used as loading control. Col, Columbia ecotype.
UKL1 and UKL2 Localize to the Plastid
The subcellular localizations of UKL1 and UKL2 remain controversial as subcellular prediction algorithms give different results. Loctree, MultiLoc, and TargetP predict that the UKL1 and UKL2 proteins localize to plastids. Subloc and WoLFPSORT predict that the UKL1 and UKL2 proteins localize to the nucleus. A previous green fluorescent protein (GFP) fusion study of UKL1 conducted by Islam et al. (2007) indicated that UKL1 was cytoplasmic. However, the authors did not indicate which binary vector was used, failed to show GFP localization for the empty vector control, and did not clarify if the construct could functionally complement the phenotype in a stable manner. Therefore, it was necessary to reevaluate this result. To observe the protein subcellular localization of UKL1 and UKL2 proteins, the T1 transgenic lines were first screened for the EYFP signal by epifluorescence using a stereomicroscope equipped with an EYFP filter. T2 seeds from EYFP positive lines were germinated on MS plates, and leaves were harvested to prepare protoplasts for fluorescence observation. The results demonstrated that UKL1 and UKL2 localize to plastids based upon colocalization with chlorophyll autofluorescence (Figure 8A).
DISCUSSION
UKL1 and UKL2 Encode Plastid Isoforms of UK
Three lines of evidence support the conclusion that UKL1 and UKL2 encode UKs. First, the ukl1 and ukl2 mutant plants were less sensitive to 5-FD for seedling growth and chlorophyll biosynthesis. Mainguet et al. (2009) stated that they could not reproduce the 5-FU and 5-FD feeding experiments by Islam et al. (2007), possibly due to the fact that the former group included 1% Suc in the growth medium for seedling germination. Since pyrimidine nucleotides are involved in Suc biosynthesis and degradation, the addition of Suc to the growth medium might counteract the 5-FU and 5-FD toxicity toward germinating seedlings. The 5-FU and 5-FD feeding experiments employed in this report use the same MS medium as Islam et al. (2007). However, Islam et al. (2007) first germinated the seeds on regular MS medium for 4 d and then transferred the seedlings into a liquid MS medium containing 5-FU or 5-FD. In contrast, we directly germinated seeds on MS medium supplemented with 5-FU or 5-FD, similar to Mainguet et al. (2009). Despite this minor difference, we obtained similar results as Islam et al. (2007). Considering that active production of pyrimidine nucleotides, following seed imbibition, occurs via the salvage pathway (Ashihara, 1977; Stasolla et al., 2001), our treatment method would allow maximal incorporation of 5-FU and 5-FD into nucleotide pools, thus explaining the more sensitive dosage response (Figure 1). Islam et al. (2007) employed much higher concentrations of 5-FD (0 to 400 μm) for their feeding experiment.
The second line of evidence that UKL1 and UKL2 encode UKs is from the in vitro activity analysis of recombinant protein (see Supplemental Figures 2C and 2D online). The recombinant UKL1 or UKL2 proteins displayed only UK activity; UPRT, URH, and UMP kinase activities were undetectable. Lastly, 4-week-old ukl1 and ukl2 mutant plants have lower UK activity but similar UPRT activity compared with wild-type plants, indicating that UKL1 and UKL2 function as UKs in vivo (Figure 3A). Since the UKL1 and UKL2 proteins do not possess UPRT activity, the function of the UPRT domain is unclear. One possibility is that the UPRT moiety functions as a regulatory domain to sense UMP, uracil, and/or PRPP (Mainguet et al., 2009). If accurate, the increased tolerance to 5-FU in the ukl1 and ukl2 mutant plants might be related to reduced sensing of 5-FU.
In addition to UKL1 and UKL2, the Arabidopsis genome contains three other putative bifunctional UK/UPRT proteins (UKL3, UKL4, and UKL5) as well as other genes putatively encoding UK-like phosphoribulokinase/UK family proteins. The biochemical and physiological functions of most of these proteins remain unknown. Understanding their roles in uridine salvage will offer a more complete picture about nucleotide metabolism in vivo.
Both Uridine and Uracil Salvage Activities in the Plastid Are Required for Early Seedling Growth
Availability of nucleotides during the early phase of imbibition is critical for successful embryo germination (Stasolla et al., 2003). The first phase of seed germination is characterized by high enzyme activity of pyrimidine salvage reactions (Stasolla et al., 2002), which are replaced by de novo synthesis later in development (Ashihara, 1977). Genetic studies in Arabidopsis demonstrated that uracil salvage in the plastid is important for early seedling development. For example, a mutation in plastid UPP, which contributes most of the UPRT activity in vivo, results in a pale-green seedling (Mainguet et al., 2009). Here, we found that ukl1 and ukl2 mutant plants grew normally after germination, but that the ukl1 ukl2 double mutant has pale-green cotyledons at the early seedling stage (Figure 7), which indicates uridine salvage also plays a crucial role for early seedling growth.
There are two different views as to why plants need both UK and UPRT activities for uridine and uracil salvage, respectively, during seed germination. First, uracil and uridine salvage might operate in a temporally distinct manner. For example, in somatic embryos, uridine incorporation into nucleic acids and nucleotides is high in quiescent embryos and then slowly declines during the first 6 d of germination (Stasolla et al., 2001). Second, no URH exists in plastids. URH catalyzes uridine degradation to uracil (Jung et al., 2009). If URH exists in the plastid, uridine can be converted to uracil and salvaged by UPP, the ukl1 ukl2 double mutant plants should not have any dramatic developmental defects. The growth defects of the ukl1 ukl2 double mutant plants, however, support the notion that no URH activity exists in plastids. A β-glucuronidase fusion study revealed that URH1 is expressed mainly in the vascular cells of roots and in root tips, guard cells, and pollen; no signal is detectable in cotyledons (Jung et al., 2009). A URH1-GFP fusion protein localized to the cytosol (Jung et al., 2009). Since disruption of the uridine degradation activity by increasing or decreasing URH1 activity results in germination delay (Jung et al., 2009), these data together suggest that uridine salvage versus degradation must be well balanced and finely regulated in the early phase of plant development.
The roles played by pyrimidine salvage in the cytoplasm remain less studied. However, indirect evidence obtained from the rsw10 mutant analysis suggests that cytoplastic uracil salvage also is important for early seedling growth. RSW10 encodes a cytosolic ribose 5-phosphate isomerase that converts d-ribulose-5-phosphate to d-ribose-5-phosphate, the precursor for PRPP biosynthesis. The rsw10 mutant plants showed ballooning of root trichoblast 2 to 5 d after germination when grown at higher temperature. The visible phenotype was rescued by feeding with uridine, suggesting the phenotype likely linked with a defect in the production of UDPG (Howles et al., 2006). Considering that plastid UPP contributes most of the UPRT activity in planta (Mainguet et al., 2009), the cytoplastic uracil salvage may only be a minor contribution to the overall pool of UMP. However, the rsw10 mutant phenotype suggests that cytosolic uracil salvage plays an important role for certain cell type (e.g., root trichoblast) either under specific developmental stage (e.g., 2- to 5-d germination) or under specific growth conditions (e.g., higher temperature).
Repressing Uridine Salvage Activity Results in the Activation of the Uridine de Novo Biosynthesis Pathway
In plants, nucleotides can be synthesized de novo from PRPP and simple molecules (e.g., CO2, amino acids, and tetrahydrofolate) or can be derived from preformed nucleosides and nucleobases via salvage reactions (Moffatt and Ashihara, 2002). In sink organs, like the potato tuber, inhibition of de novo pyrimidine synthesis leads to a compensatory stimulation of the pyrimidine salvage pathway (Geigenberger et al., 2005), suggesting that pyrimidine de novo synthesis and salvage pathways are regulated by common intermediates. In this study, downregulation of uridine salvage activity in the ukl1 and ukl2 mutant plants resulted in transcriptional induction of the committed step and two other activities of the pyrimidine de novo synthesis pathway (see Supplemental Figure 3 online). Interestingly, repression of pyrimidine scavenging results in the accumulation of Glu and Asp (Table 1), precursors for pyrimidine de novo biosynthesis. Besides the interaction between uridine salvage and de novo pyrimidine biosynthesis, there is also interaction between uracil salvage and uridine salvage. For example, Howles et al. (2006) demonstrated that the uridine feeding can rescue rsw10 mutant phenotype, suggesting that reduced cytosolic uracil salvage in this mutant also can be complemented by enhanced uridine salvage in the cytosolic or plastid.
Repressed Uridine Salvage Activity Affects Carbon Partitioning into Cellulose and Lignin
Cellulose is the principal component of plant cell walls, accounting for 15 to 30% of the dry mass of all primary cell walls and an even larger percentage of secondary cell walls (Whetten and Sederoff, 1995). After cellulose, lignin, a polymer of aromatic subunits usually derived from Phe, is the second most abundant terrestrial biopolymer, accounting for ~30% of the organic carbon in the biosphere (Boerjan et al., 2003). Cellulose and lignin represent the largest sink for photoassimilates during the vegetative phase of growth. A previous study of rsw10 mutant plants provided indirect evidence that limitation of UDPG supply can reduce cellulose biosynthesis in developing roots (Howles et al., 2006). Our experimental evidence indicates that cellulose and lignin biosynthesis is coordinated partly at the substrate level by controlling the supply of precursors, mainly UDP-glucose and Phe. Uridine salvage supplies UTP for UDP-glucose biosynthesis, thus affecting cellulose biosynthesis (Figure 5). Restricted carbon flow into cellulose biosynthesis in the ukl2 mutant results in increased Glc and Fru levels (Table 1). This likely forces more triose phosphate into the shikimate pathway for Phe biosynthesis. More Phe is then channeled into the phenylpropanoid pathway to produce monolignols, such as trans-sinapinic acid and coumarine for lignin polymer biosynthesis (Table 1), thus altering carbon partitioning into cellulose and lignin (Figure 5). The data also indicate that plants are very flexible in their responses to available precursors for carbon partitioning. The lack of any visible growth defects in the ukl2 mutant plants and the severe phenotype of the ukl1 ukl2 double mutant plants suggest that plant can tolerate the cell wall compositional changes to some degree.
The Uridine Salvage Pathway Is Connected to Both C and N Metabolism
Nucleotides, which contain both carbon and nitrogen synthesized from carbon skeletons and amino acids, are ideally poised to serve as a sensor to coordinate carbon and nitrogen metabolism in planta. When plants are exposed to nitrate, they redirect carbon from starch synthesis to the production of amino acids and organic acids such as malate by controlling the synthesis and activity of key enzymes (Scheible et al., 1997). For example, high nitrate upregulates phophenolpyruvate carboxylase, an enzyme that catalyzes the synthesis of oxaloacetate, a citric acid cycle intermediate that is readily converted to α-ketoglutarate (Sugiharto et al., 1990). By contrast, ADP-glucose pyrophosphorylase, which is required for starch biosynthesis, is downregulated in response to nitrate treatment (Scheible et al., 1997). Interestingly, the ukl2 mutant plant shares several features with plants treated with high concentrations of nitrate, such as reduced leaf starch content (Figure 3A) and increased contents of Glu, Asp, and malate (Table 1), which suggests that uridine metabolism may involve crosstalk with nitrogen status in plants. The ukl2 mutant plants also contain elevated levels of many carbohydrates, including Glc, Fru, and Man. Glc and Fru have been demonstrated to stimulate leaf nitrate and nitrite reductase (Vincentz et al., 1993), thus enhancing nitrogen assimilation in the leaf. Recent characterization of a cytosolic fumarase mutant (fum2) suggests that fumarate accumulation in the light is linked to nitrogen assimilation (Pracharoenwattana et al., 2010). In the fum2 knockout mutant plants, fumarate levels in the leaf are reduced and starch content is elevated. The mutant also has lower levels of many amino acids in leaves during the day. In general, the phenotype of fum2 is opposite of ukl2. For example, in comparison to the fum2 mutant, the ukl2 mutant plants have lower leaf starch and higher fumarate content, and many amino acids accumulate during the light period. Opposite phenotypes between the ukl2 and fum2 mutants suggest that uridine salvage activity can affect photosynthate allocation into fumarate and starch during the light period, thus altering nitrogen assimilation. Asp and Glu are two major transportable forms of organic nitrogen and could be transported from leaf to seed through the phloem, which could explain why the seeds from ukl1 and ukl2 mutant plants have elevated protein content (Table 2). However, it is well known that oil and protein content are inversely correlated in seed. Moreover, it was demonstrated recently that this relationship is independent of maternal carbon or nitrogen supply, as an embryo-specific mutation in de novo fatty acid synthesis produces reduced oil and elevated protein levels in mature seeds (Chen et al., 2009). Therefore, an alternative explanation for the increased protein observed in the ukl2 mutant could be a shift in carbon partitioning, from oil to protein, within the developing embryo.
Uridine Salvage Plays an Important Role in Plant Metabolism
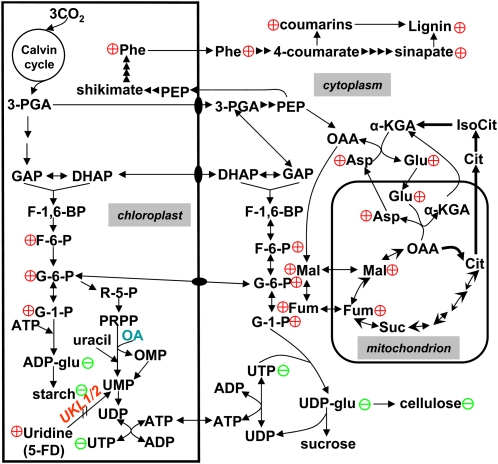
Based on previous studies of pyrimidine metabolism and on this current study of the ukl1 and ukl2 mutant plants, we propose a model to summarize the crosstalk between uridine salvation and primary metabolism in light-adapted leaves (Figure 9). In photosynthetically active leaves, carbon dioxide is fixed to produce 3-phosphoglycerate and eventually β-d-glucose-6-phosphate (G-6-P). A portion of this enters the oxidative pentose phosphate pathway to produce pentose phosphate for nucleotide biosynthesis; some G-6-P is converted to β-d-glucose-1-phosphate and then activated by ATP to form ADP-glucose, which is committed to starch biosynthesis in the plastid. Plastid intermediates (i.e., 3-phosphoglycerate, d-glyceraldehyde-3-phosphate, dihydroxyacetone phosphate, and G-6-P) also can be exported to the cytoplasm, where β-d-glucose-1-phosphate is activated by UTP to form UDP-glucose, the substrate for both Suc and cellulose biosynthesis. In the ukl2 mutant, partial loss of uridine salvage activity results in reduced levels of UTP and UDP after 6 h light as well as reduced UDP-glucose (Figure 6A). It is likely the nucleotide changes are the original event for the varied metabolic changes in the ukl2 mutants. The limitation of UDP-glucose does not significantly affect Suc biosynthesis, but rather cellulose synthesis (Figure 5). Reduced carbon flow into cellulose may explain elevated Glc and Fru pools (Table 1) and eventual buildup of triose phosphate. Excess availability of triose phosphate may have stimulated Phe production and enhanced flux into the shikimate and phenylpropanoid pathways for lignin and lignan biosynthesis, thus altering cell wall composition (Figure 5). Theoretically the buildup of Glc and Fru in the ukl2 mutant would also enhance starch biosynthesis in the plastid; however, the limitation of ATP could restrict ADP-glucose synthesis in the mutant (Figure 6A), thus reducing leaf starch synthesis (Figure 3A). It is unclear why UDP-glucose limitation affects cellulose but not Suc biosynthesis. Considering that UDP-glucose and Suc are both generated in the cytoplasm but UDP-glucose has to be transported to the plasma membrane surface for cellulose microfibrils biosynthesis, this spatial arrangement may give Suc biosynthesis an advantage for access to limited UDP-glucose pools. This may have reproductive relevance since floral organs rely on leaf Suc supply for growth. The ukl2 mutant plants have reduced leaf starch but similar levels of Suc levels compared with the wild type. These observations further support the overflow model (Eichelmann and Laisk, 1994) and unravel a novel role of pyrimidine scavaging to regulate photoassimilate allocation and partitioning in Arabidopsis leaves.
Figure 9.
Model for Interaction of Uridine Salvage in Light-Adapted Leaves.
Plastid pyrimidine salvage activity influences photoassimilate partitioning into starch, cellulose, and lignin. The knockout of UKL2 reduces UDP-glucose synthesis and restricts carbon flow into cellulose biosynthesis. The concomitant buildup of simple carbohydrates appears to channel carbon into lignin biosynthesis as well as other metabolic intermediates. 3-PGA, 3-phosphoglycerate; GAP, d-glyceraldehyde-3-phosphate; DHAP, dihydroxyacetone phosphate; F1,6-P, fructose-1,6-bisphosphate; F-6-P, d-fructose-6-phosphate; G-6-P, β-d-glucose-6-phosphate; G-1-P, β-d-glucose-1-phosphate; ADP-glu, ADP-d-glucose; UDP-glu, UDP-d-glucose; PEP, phosphoenolpyruvate; OAA, oxaloacetate; Cit, citrate; IsoCit, isocitrate; α -KGA, α-ketoglutarate; Suc-CoA, succinyl-CoA; Suc, succinate; Fum, fumarate; Mal, malate; OMP, orotidine 5′-monophosphate. Plus (+), higher in ukl2 than wild type; minus (−), lower in ukl2 mutant than the wild type.
[See online article for color version of this figure.]
METHODS
Plant Materials
T-DNA insertion lines were ordered from the ABRC. Seeds were sown in a 1:1 mixture of water-saturated vermiculite and peat moss–enriched soil and grown in a growth chambers either under long-day conditions (14-h-light/10-h-dark cycle [light period = 8:00 am to 10:00 pm], 23°C day/20°C night, 50% humidity, and light intensity of 200 μmol m−2 s−1) or under continuous white light (20 μmol m−2 s−1, 25°C, 45% humidity).
Assessment of Growth Sensitivity of Arabidopsis Seedlings to 5-FU and 5-FD
Seeds from wild-type and homozygous mutant plants were germinated on half-strength MS medium supplemented with different concentrations of 5-FU and 5-FD (Sigma-Aldrich) for 5 d. Treated plants were photographed. Total chlorophyll content from leaf tissues collected from 5-FU– and 5-FD–treated seedlings was measured by the method described by Lichtenthaler (1987).
Uridine and OA Application
Uridine and OA (Sigma-Aldrich) were dissolved in water at 1 mM. The plants were sprayed daily after germination; the control plants were sprayed with water only at same schedule.
Starch, Protein, Lipid, and Metabolite Extraction and Quantification
Whole leaves from individual plants were harvested after 30 d of growth in growth chambers, measured for fresh weight, immersed in liquid nitrogen, and then placed at 80°C for storage. Samples were prepared within 1 week of storage. To prepare the samples for starch analyses, frozen leaves were homogenized in a mortar and pestle in the presence of liquid nitrogen; 2 mL of 80% ethanol was added into the mortar, and the leaves were ground again. Homogenates were transferred to glass screw-cap tubes and incubated at 100°C for 15 min and 80°C for an additional 40 min, with periodic mixing. Samples were then centrifuged at 2000g for 5 min, and the supernatants were transferred to a fresh tube. Pellets were back-extracted twice with 2 mL of 80% (v/v) ethanol, and supernatants were pooled into one tube. Starch content was measured in the washed pellets. Supernatants were lyophilized for metabolite analysis. The starch fractions were analyzed using the EnzyChrom assay kit (BioAssay Systems). For metabolite analysis, samples were analyzed as described by Fiehn et al. (2000) with minor modifications. Samples were derivatized by methoximation by adding methoxyamine hydrochloride (Fluka) in pyridine (20 mg/mL) at a ratio of 1:2 (μL of methoxyamine hydrochloride:mg fresh weight), followed by heating at 30°C for 90 min. Samples were then silylated by adding 80 μL of N-methyl-N-(trimethylsilyl)trifluoroacetamide (Acros) + 1% trimethylchlorosilane (Aldrich) to 60 μL of the methoximated samples, then incubated at 37°C for 30 min. Derivatized metabolite mixtures were analyzed using a Hewlett Packard 6890 gas chromatograph, 5973 mass selective detector, and 7683 series injector (Agilent Technologies). Samples (2 μL) were injected with a split ratio of 25:1 and resolved on a 30 m × 0.25 mm × 0.25 μm SPB 50 column (Supelco). Injection temperature was 230°C, the interface was set to 250°C, and the ion source was adjusted to 200°C. Helium flow was 1 mL min−1. After a 5-min solvent delay at 80°C, the oven temperature was increased at a rate of 5°C min−1 to 310°C and then held at 310°C for 10 min. Mass spectra were recorded from a mass-to-charge ratio of 50 to 600 at 2.69 scans/s. Following chromatographic separation and data acquisition, raw data files were exported as AIA format and were deconvoluted and searched against the NIST Mass Spectral Database and Golm metabolomics library (http://csbdb.mpimp-golm.mpg.de/csbdb/gmd/msri/gmd_msri.html) by AMDIS software (Stein, 1999). The search generated a list of ion retention time values. The ion intensity values were extracted using MET-IDEA software (Broeckling et al., 2006) and are provided in Supplemental Data Set 1 online.
Protein samples were prepared by following the procedure described by Siloto et al. (2006) and analyzed by a Pierce 660-nm protein assay reagent (Thermo Fischer Scientific). Fatty acids were derivatized using acidified methanol and analyzed by gas chromatography–mass spectrometry (Li et al., 2006).
Nucleotide Extraction and Quantification
The aerial leaves from individual plants were harvested before bolting and their fresh weight determined. Nucleotides were extracted by following the method described by Jelitto et al. (1992) and analyzed by HPLC as described previously (Geigenberger et al., 1994) using an EC 250/4.6 nucleosil 100-10 SB column (Macherey-Nagel) configured to a Waters 2695 separations module and 996 photodiode array detector (Waters). Detection wavelength was set at 254 nm with an automatic spectral scan from 200 to 300 nm. Data acquisition and analyses were performed by Empower 2 software (Waters). Chemical standards were analyzed under the same conditions to develop a calibration curve for quantification. Potassium phosphate was purchased from Calbiochem. For uridine and uracil isolation, seeds were germinated on MS plates for 7 d under continuous white light. The uridine and uracil isolation and quantification was following the method described by Morris (1987).
Double Mutant Construction
To construct the ukl1 ukl2 double mutant, ukl1 was crossed with ukl2. F1 seeds were germinated and self-pollinated. Resultant F2 seeds were grown on soil. Individual plants were genotyped by PCR to identify double mutants.
Histology
The base parts of inflorescence stems from 5-week-old plants were fixed and embedded according to Chen and Thelen (2010). The 1.5-μm-thick sections of stems were stained with 0.01% Fluorescent Brightener 28 (Sigma-Aldrich) and observed with a UV fluorescence microscope to detect cellulose (Hughes and McCully, 1975). Lignin was visualized by staining fresh sections of 60-d-old plants with phloroglucinol-HCl (Acros) (McCarthy et al., 2009).
Enzyme Activity Assay
The activities of UK, UPRT, and UMP kinase were assayed by following the procedure described by Katahira and Ashihara (2002). URH activity was assayed as described by Kurtz et al. (2002). Total leaf protein was prepared as described by Katahira and Ashihara (2002) for enzyme activity assay.
Complementation and Subcellular Localization Study
UKL1 and UKL2 cDNAs were PCR amplified. The BamHI and NotI restriction enzyme sites were added to forward and reverse primers, respectively. All primer sequences are shown in Supplemental Table 1 online. The PCR products were digested by BamHI and NotI and then inserted into the entry vector PE6C such that an EYFP fluorescence tag was in frame with UKL1 or UKL2 at the C terminus (Dubin et al., 2008). The cassette was then moved into the binary vector pSITE-0B using Gateway LR clonase (Chakrabarty et al., 2007). The constructs were transformed into Arabidopsis thaliana using the Agrobacterium tumefaciens–mediated floral dip method. The T1 seeds were germinated on MS plates for 3 d and then screened for EYFP expression under Leica MZFLIII stereomicroscope with an EYFP filter. The expressed T1 transgenic lines were transferred into soil until grown to maturity. Siliques from 7 DAF were harvested for starch content quantification to determine complementation. T2 seeds from transgenic lines were germinated on MS plates; leaves were harvest to prepare protoplast using the method described by Jain et al. (2008). Fluorescence imaging was taken by Olympus BX 61 microscope.
PCR and RT-PCR
DNA for PCR genotyping was prepared by phenol extraction followed by isopropanol precipitation. RNA extraction, first-strand biosynthesis, and PCR reaction were conducted as described (Chen and Thelen, 2010). Band signal intensity was quantified by Image Quant TL 1D version 7.0 (GE Healthcare) and normalized to UBQ internal standard. Three biological replicates were performed for quantification.
Recombinant Protein Expression
UKL1 and UKL2 cDNA were amplified by Pfu polymerase, and PCR fragments were subcloned into the pET200 vector (Invitrogen). All primer sequences are shown in Supplemental Table 1 online. Sequencing results indicated that 5 bp (AGTAG) at the sequence encoding the UKL1 C terminus were deleted during topoisomerase-mediated recombination. To induce protein expression, the constructs were transformed into Escherichia coli BL21 DE3 cells, and protein expression was induced by addition of 1 mM isopropyl-β-D-thiogalactopyranoside. Recombinant proteins were purified by Ni-NTA agarose (Qiagen).
Statistical Analysis
Where differences are described in the text as significant, this means that P < 0.05 in a t test was performed using the two-tailed equal variance algorithm within Microsoft Excel 2003.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers UKL1 (At5g40870), UKL2 (At3g27190), UKL3 (At1g55810), UKL4 (At4g26510), UKL5 (At3g27440), carbamoylphosphate synthase (At1g29900), Asp transcarbamoylase (At1g75330), and PRPP synthase (At1g10700). T-DNA lines corresponding to UKL1 were as follows: SALK_108468 (ukl1), SALK_026938, and SALK_147532; T-DNA lines corresponding to UKL2 were SALK_058257 (ukl2) and CS821451.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ukl1 and ukl2 Mutant Identification.
Supplemental Figure 2. Recombinant UKL1 and UKL2 Protein Kinetic Assay.
Supplemental Figure 3. Transcriptional Expression of Genes in the de Novo Pyrimidine Biosynthesis Pathway Is Altered in the ukl1 and ukl2 Mutants.
Supplemental Figure 4. UKL1 and UKL2 Are Differentially Expressed during Seed Development.
Supplemental Table 1. Primers Used in This Study.
Supplemental Data Set 1. Metabolic Profiling of the Wild Type and the ukl2 Mutant.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Stock Center for providing T-DNA insertion lines. We also thank the three reviewers for their valuable insights and suggestions, Melody Kroll for proofreading the manuscript, and Zhiyong Xiong for fluorescence imaging. Financial support was provided by a Life Science Postdoctoral Fellowship from the University of Missouri and National Science Foundation Plant Genome Research Program Grant DBI-0332418. This investigation was supported by the National Science Foundation–Plant Genome Research Program Young Investigator Award DBI-0332418 and a University of Missouri Life Sciences Fellowship to M.C.
AUTHOR CONTRIBUTIONS
M.C. contributed to experimental design, experiment execution, data analysis, and article writing. J.J.T. contributed to experimental design and article writing.
References
- Anterola A.M., Jeon J.H., Davin L.B., Lewis N.G. (2002). Transcriptional control of monolignol biosynthesis in Pinus taeda: Factors affecting monolignol ratios and carbon allocation in phenylpropanoid metabolism. J. Biol. Chem. 277: 18272–18280 [DOI] [PubMed] [Google Scholar]
- Ashihara H. (1977). Changes in the activities of the de novo and salvage pathways of pyrimidine nucleotide biosynthesis during germination of black gram (Phaseolus mungo) seeds. Z. Pflanzenphysiol. 81: 199–211 [Google Scholar]
- Boerjan W., Ralph J., Baucher M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bolwell G.P., Butt V.S., Davies D.R., Zimmerlin A. (1995). The origin of the oxidative burst in plants. Free Radic. Res. 23: 517–532 [DOI] [PubMed] [Google Scholar]
- Broeckling C.D., Reddy I.R., Duran A.L., Zhao X.C., Sumner L.W. (2006). MET-IDEA: Data extraction tool for mass spectrometry-based metabolomics. Anal. Chem. 78: 4334–4341 [DOI] [PubMed] [Google Scholar]
- Chakrabarty R., Banerjee R., Chung S.M., Farman M., Citovsky V., Hogenhout S.A., Tzfira T., Goodin M. (2007). PSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: Probing Nicotiana benthamiana-virus interactions. Mol. Plant Microbe Interact. 20: 740–750 [DOI] [PubMed] [Google Scholar]
- Chen M.J., Mooney B.P., Hajduch M., Joshi T., Zhou M., Xu D., Thelen J.J. (2009). System analysis of an Arabidopsis mutant altered in de novo fatty acid synthesis reveals diverse changes in seed composition and metabolism. Plant Physiol. 150: 27–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.J., Thelen J.J. (2010). The plastid isoform of triose phosphate isomerase is required for the postgerminative transition from heterotrophic to autotrophic growth in Arabidopsis. Plant Cell 22: 77–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin M.J., Bowler C., Benvenuto G. (2008). A modified Gateway cloning strategy for overexpression tagged proteins in plants. Plant Methods 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichelmann H., Laisk A.H. (1994). CO2 uptake and electron transport rates in wild-type and a starchless mutant of Nicotiana sylvestris. The role and regulation of starch synthesis at saturating CO2 concentrations. Plant Physiol. 106: 679–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O., Kopka J., Trethewey R.N., Willmitzer L. (2000). Identification of uncommon plant metabolites based on calculation of elemental compositions using gas chromatography and quadrupole mass spectrometry. Anal. Chem. 72: 3573–3580 [DOI] [PubMed] [Google Scholar]
- Fujita K., Okada M., Lei K., Ito J., Ohkura K., Adu-Gyamfi J.J., Mohapatra P.K. (2003). Effect of P-deficiency on photoassimilate partitioning and rhythmic changes in fruit and stem diameter of tomato (Lycopersicon esculentum) during fruit growth. J. Exp. Bot. 54: 2519–2528 [DOI] [PubMed] [Google Scholar]
- Geigenberger P., Hajirezaei M., Geiger M., Deiting U., Sonnewald U., Stitt M. (1998). Overexpression of pyrophosphatase leads to increased Suc degradation and starch synthesis, increased activities of enzymes for Suc-starch interconversions, and increased levels of nucleotides in growing potato tubers. Planta 205: 428–437 [DOI] [PubMed] [Google Scholar]
- Geigenberger P., Merlo L., Reimholz R., Stitt M. (1994). When growing potato tubers are detached from their mother plant there is a rapid inhibition of starch synthesis, involing inhibition of ADP-glucose pyrophosphorylase. Planta 193: 486–493 [Google Scholar]
- Geigenberger P., Regierer B., Nunes-Nesi A., Leisse A., Urbanczyk-Wochniak E., Springer F., van Dongen J.T., Kossmann J., Fernie A.R. (2005). Inhibition of de novo pyrimidine synthesis in growing potato tubers leads to a compensatory stimulation of the pyrimidine salvage pathway and a subsequent increase in biosynthetic performance. Plant Cell 17: 2077–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giermann N., Schröder M., Ritter T., Zrenner R. (2002). Molecular analysis of de novo pyrimidine synthesis in solanaceous species. Plant Mol. Biol. 50: 393–403 [DOI] [PubMed] [Google Scholar]
- Gifford R.M., Evans L.T. (1981). Photosynthesis, carbon partitioning, and yield. Annu. Rev. Plant Physiol. 32: 485–509 [Google Scholar]
- Howles P.A., Birch R.J., Collings D.A., Gebbie L.K., Hurley U.A., Hocart C.H., Arioli T., Williamson R.E. (2006). A mutation in an Arabidopsis ribose 5-phosphate isomerase reduces cellulose synthesis and is rescued by exogenous uridine. Plant J. 48: 606–618 [DOI] [PubMed] [Google Scholar]
- Huber S.C. (1986). Fructose 2,6-bisphosphate as a regulatory metabolite in plant. Annu. Rev. Plant Physiol. 37: 233–246 [Google Scholar]
- Huber S.C., Hanson K.R. (1992). Carbon partitioning and growth of a starchless mutant of Nicotiana sylvestris. Plant Physiol. 99: 1449–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J., McCully M.E. (1975). The use of an optical brightener in the study of plant structure. Stain Technol. 50: 319–329 [DOI] [PubMed] [Google Scholar]
- Islam M.R., Kim H., Kang S.W., Kim J.S., Jeong Y.M., Hwang H.J., Lee S.Y., Woo J.C., Kim S.G. (2007). Functional characterization of a gene encoding a dual domain for uridine kinase and uracil phosphoribosyltransferase in Arabidopsis thaliana. Plant Mol. Biol. 63: 465–477 [DOI] [PubMed] [Google Scholar]
- Jain R., Katavic V., Agrawal G.K., Guzov V.M., Thelen J.J. (2008). Purification and proteomic characterization of plastids from Brassica napus developing embryos. Proteomics 8: 3397–3405 [DOI] [PubMed] [Google Scholar]
- Jelitto T., Sonnewald U., Willmitzer L., Hajirezeai M., Stitt M. (1992). Inorganic pyrophosphate content and metabolites in potato and tobacco plants expressing E. coli pyrophosphatase in their cytosol. Planta 188: 238–244 [DOI] [PubMed] [Google Scholar]
- Jung B., Flörchinger M., Kunz H.H., Traub M., Wartenberg R., Jeblick W., Neuhaus H.E., Möhlmann T. (2009). Uridine-ribohydrolase is a key regulator in the uridine degradation pathway of Arabidopsis. Plant Cell 21: 876–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira R., Ashihara H. (2002). Profiles of pyrimidine biosynthesis, salvage and degradation in disks of potato (Solanum tuberosum L.) tubers. Planta 215: 821–828 [DOI] [PubMed] [Google Scholar]
- Kurtz J.E., Exinger F., Erbs P., Jund R. (2002). The URH1 uridine ribohydrolase of Saccharomyces cerevisiae. Curr. Genet. 41: 132–141 [DOI] [PubMed] [Google Scholar]
- Li Y.H., Beisson F., Pollard M., Ohlrogge J. (2006). Oil content of Arabidopsis seeds: The influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H.K. (1987). Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 148: 350–382 [Google Scholar]
- Loef I., I, Stitt M., Geigenberger P. (1999). Orotate leads to a specific increase in uridine nucleotide levels and a stimulation of Suc degradation and starch synthesis in discs from growing potato tubers. Planta 209: 314–323 [DOI] [PubMed] [Google Scholar]
- Mainguet S.E., Gakière B., Majira A., Pelletier S., Bringel F., Guérard F., Caboche M., Berthomé R., Renou J.P. (2009). Uracil salvage is necessary for early Arabidopsis development. Plant J. 60: 280–291 [DOI] [PubMed] [Google Scholar]
- McCarthy R.L., Zhong R., Ye Z.H. (2009). MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 50: 1950–1964 [DOI] [PubMed] [Google Scholar]
- Moffatt B., Ashihara H. (2002). Purine and pyrimidine nucleotide synthesis and metabolism. In The Arabidopsis Book; 1: e0018, doi/10.1199/tab.0018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris C.E. (1987). Determination of uracil, uridine and formic acid in egg products by high-performance liquid chromatography. J. Chromatogr. A 394: 408–413 [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I., Zhou W., Keech O., Francisco P.B., Udomchalothorn T., Tschoep H., Stitt M., Gibon Y., Smith S.M. (2010). Arabidopsis has a cytosolic fumarase required for the massive allocation of photosynthate into fumaric acid and for rapid plant growth on high nitrogen. Plant J. 62: 785–795 [DOI] [PubMed] [Google Scholar]
- Preiss J. (1982). Regulation of the biosynthesis and degradation of starch. Annu. Rev. Plant Physiol. 33: 431–454 [Google Scholar]
- Regierer B., Fernie A.R., Springer F., Perez-Melis A., Leisse A., Koehl K., Willmitzer L., Geigenberger P., Kossmann J. (2002). Starch content and yield increase as a result of altering adenylate pools in transgenic plants. Nat. Biotechnol. 20: 1256–1260 [DOI] [PubMed] [Google Scholar]
- Ros-Barceló A., Pomar F., López-Serrano M., Martínez P., Balleata M.C., Pedreňo M.A. (2002). Developmental regulation of the H2O2-producing system and of a basic perocidase isoenzyme in the Zinnia elegans lignifying xylem. Plant Physiol. Biochem. 40: 325–332 [Google Scholar]
- Scheible W.R., Gonzalez-Fontes A., Lauerer M., Muller-Rober B., Caboche M., Stitt M. (1997). Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9: 783–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze W., Stitt M., Schulze E.D., Neuhaus H.E., Fichtner K. (1991). A quantification of the significance of assimilatory starch for growth of Arabidopsis thaliana L. Heynh. Plant Physiol. 95: 890–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siloto R.M.P., Findlay K., Lopez-Villalobos A., Yeung E.C., Nykiforuk C.L., Moloney M.M. (2006). The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasolla C., Katahira R., Thorpe T.A., Ashihara H. (2003). Purine and pyrimidine nucleotide metabolism in higher plants. J. Plant Physiol. 160: 1271–1295 [DOI] [PubMed] [Google Scholar]
- Stasolla C., Loukanina N., Ashihara H., Yeung E.C., Thorpe T.A. (2001). Changes in pyrimidine nucleotide biosynthesis during germination of white spruce (Picea glauca) somatic embryos. In Vitro Cell. Dev. Biol. Plant 37: 285–292 [Google Scholar]
- Stasolla C., Loukanina N., Ashihara H., Yeung E.C., Thorpe T.A. (2002). Pyrimidine mucleotide biosynthesis and nucleic acid metabolism in embryos and megagametophytes of white spruce (Picea glauca) during germination. Physiol. Plant. 115: 155–165 [DOI] [PubMed] [Google Scholar]
- Stein S.E. (1999). An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometery data. J. Am. Soc. Mass Spectrom. 10: 770–781 [Google Scholar]
- Sugiharto B., Miyata K., Nakamoto H., Sasakawa H., Sugiyama T. (1990). Regulation of expression of carbon-assimilating enzymes by nitrogen in maize leaf. Plant Physiol. 92: 963–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden J., Möhlmann T., Kampfenkel K., Henrichs G., Neuhaus H.E. (1998). Altered plastidic ATP/ADP-translocator activity influences potato (Solanum tuberosum L.) tuber morphology, yield and composition of tuber starch. Plant J. 16: 531–540 [Google Scholar]
- Tsutsumi Y., Islam A., Anderson C.D., Sarkkanen K.V. (1990). Acidic permanganate oxidations of lignin and model compounds: Comparison with ozonolysis. Holzforschung 44: 59–66 [Google Scholar]
- Vincentz M., Moureaux T., Leydecker M.T., Vaucheret H., Caboche M. (1993). Regulation of nitrate and nitrite reductase expression in Nicotiana plumbaginifolia leaves by nitrogen and carbon metabolites. Plant J. 3: 315–324 [DOI] [PubMed] [Google Scholar]
- Whetten R., Sederoff R. (1995). Lignin biosynthesis. Plant Cell 7: 1001–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrenner R., Stitt M., Sonnewald U., Boldt R. (2006). Pyrimidine and purine biosynthesis and degradation in plants. Annu. Rev. Plant Biol. 57: 805–836 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.