This work demonstrates that the deubiquitinating enzyme AMSH3 interacts with two ESCRT-III subunits, VPS2.1 and VPS24.1, and that they colocalize in class E compartments. Characterization of VPS2.1 shows that it is essential for embryogenesis and that its localization is dependent on both the ATPase activity of the AAA-ATPase SKD1 and the deubiquitinating activity of AMSH3.
Abstract
Ubiquitination and deubiquitination regulate various cellular processes. We have recently shown that the deubiquitinating enzyme Associated Molecule with the SH3 domain of STAM3 (AMSH3) is involved in vacuole biogenesis and intracellular trafficking in Arabidopsis thaliana. However, little is known about the identity of its interaction partners and deubiquitination substrates. Here, we provide evidence that AMSH3 interacts with ESCRT-III subunits VPS2.1 and VPS24.1. The interaction of ESCRT-III subunits with AMSH3 is mediated by the MIM1 domain and depends on the MIT domain of AMSH3. We further show that AMSH3, VPS2.1, and VPS24.1 localize to class E compartments when ESCRT-III disassembly is inhibited by coexpression of inactive Suppressor of K+ transport Defect 1 (SKD1), an AAA-ATPase involved in the disassembly of ESCRT-III. We also provide evidence that AMSH3 and SKD1 compete for binding to VPS2.1. Furthermore, we show that the loss of AMSH3 enzymatic activity leads to the formation of cellular compartments that contain AMSH3, VPS2.1, and VPS24.1. Taken together, our study presents evidence that AMSH3 interacts with classical core ESCRT-III components and thereby provides a molecular framework for the function of AMSH3 in plants.
INTRODUCTION
Ubiquitination and deubiquitination are functionally reciprocal processes that regulate a broad range of cellular events. Deubiquitinating enzymes (DUBs) and ubiquitinating enzymes play active roles in the regulation of substrate stability, activity, and cellular behavior (reviewed in Clague and Urbé, 2006; Reyes-Turcu et al., 2009). DUBs are also essential for ubiquitin precursor processing and the recycling of ubiquitin molecules, and they thereby contribute to the maintenance of the free ubiquitin pool. Recent studies have shown that DUB function is also required for various aspects of plant development (Yan et al., 2000; Doelling et al., 2001, 2007; Sridhar et al., 2007; Liu et al., 2008; Luo et al., 2008; Schmitz et al., 2009).
MPR1, PAD1 N-terminal+ (MPN+) domain metalloproteases comprise one class of eukaryotic DUBs in addition to four families of Cys proteases, the ubiquitin-specific proteases, ubiquitin C-terminal hydrolases, ovarian tumor proteins, and Machado Joseph disease domain proteins (Komander et al., 2009). Associated Molecule with the SH3 domain of STAM (AMSH) proteins are MPN+ domain DUBs that are widely conserved (Maytal-Kivity et al., 2002). Arabidopsis thaliana has three AMSH homologs, named AMSH1, AMSH2, and AMSH3, that were identified by the homology of their catalytic MPN+ domain to the MPN+ domain of the human AMSH (Maytal-Kivity et al., 2002; Isono et al., 2010). We recently showed that AMSH3 is an active DUB that hydrolyzes both K48- and K63-linked ubiquitin chains (Isono et al., 2010). amsh3 null mutants are seedling lethal and show vacuole biogenesis and intracellular trafficking defects.
Endosomal sorting complex required for transport (ESCRT) -0, I, II, and III are involved in intracellular trafficking and are important for the proper sorting of ubiquitinated membrane proteins to the intraluminal vesicles of the multivesicular bodies (MVBs) (reviewed in Williams and Urbé, 2007; Raiborg and Stenmark, 2009; Hurley and Hanson, 2010). This process is crucial for the proper delivery of the cargo for degradation in the vacuole/lysosome. The endocytosis of transmembrane proteins is triggered by ubiquitination. ESCRT-0, I, and II subunits recruit the ubiquitinated cargos on endosomal membranes via their ubiquitin binding motifs. ESCRT-III then sorts the cargos into the intraluminal vesicles of the MVB by exerting membrane bending, scission, and fusion.
ESCRT-III is best studied in yeast and consists of a core complex comprised of four subunits: vacuolar protein sorting 2 (Vps2p)/charged MVB proteins or chromatin modifying protein 2 (CHMP2), Vps20p/CHMP6, Vps24p/CHMP3, and Sucrose Non-Fermenting 7 (Snf7p)/VPS32/CHMP4. There are further ESCRT-III–associated proteins like Vps46p/CHMP1/Doa4-independent degradation 2 (Did2p) and Vps60p/CHMP5 that were shown to function together with the ESCRT-III core complex (Babst et al., 2002). Vps20p is a membrane-anchored subunit that recruits Snf7p to the endosomal membrane, which then oligomerizes on the membrane. The binding of Vps24p to the Snf7p filament stops the oligomerization. Vps2p binds to Vps24p and together with Vps46p and Vps60p then recruits the AAA-ATPase Vps4p/suppressor of K+ transport growth defect 1 (SKD1) to ESCRT-III (Teis et al., 2008; Saksena et al., 2009). Vps4p/SKD1 forms an oligomeric complex that disassembles ESCRT-III upon binding. The disassembly of ESCRT-III is essential for the completion of the sorting of cargos to the intraluminal vesicles of MVBs (Babst et al., 1998; Ghazi-Tabatabai et al., 2008; Lata et al., 2008). The regulated assembly and disassembly of ESCRT-III is a prerequisite for proper MVB formation, and in a yeast vps4 mutant, ESCRT-III components aggregate in so-called class E compartments while cargo sorting is blocked (Babst et al., 1997).
ESCRT-III subunits are highly conserved in eukaryotes, and homologs of all ESCRT components, except those of ESCRT-0, can be detected in the Arabidopsis genome (Winter and Hauser, 2006; Schellmann and Pimpl, 2009). Although as yet poorly understood, accumulating evidence suggests that Arabidopsis ESCRT proteins also constitute an important part of the MVB sorting pathway and contribute to proper plant development. A recent study of Arabidopsis CHMP1A/B or VPS46 has shown that they are required for the delivery of PIN1, PIN2, and AUX1 to the vacuole (Spitzer et al., 2009). It was also shown that an ESCRT-I subunit ELCH/VPS23 is involved in cytokinesis (Spitzer et al., 2006), and the characterization of SKD1 indicated that it is involved in MVB formation and vacuole maintenance (Haas et al., 2007). In contrast with yeast and cultured mammalian cells, SKD1 is an essential gene in Arabidopsis (Haas et al., 2007). Despite their presumed central role in endocytosis, the analysis of Arabidopsis ESCRT proteins at the biochemical and cell biological level has just started.
Human AMSH was originally identified as an interactor of the signal transducing adaptor molecule (STAM) that is one of the ESCRT-0 proteins (Tanaka et al., 1999) and is implicated in intracellular trafficking. Further studies have revealed the interaction of human AMSH with multiple ESCRT-0 and ESCRT-III components. A Xenopus laevis AMSH protein was shown to interact with an ESCRT-III binding protein, apoptosis-linked gene 2 interacting protein X/Bro1p (McCullough et al., 2004, 2006; Agromayor and Martin-Serrano, 2006; Tsang et al., 2006; Kyuuma et al., 2007; Ma et al., 2007). It was also shown that the binding of STAM enhances the DUB activity of human AMSH in vitro (McCullough et al., 2006). The interaction of human AMSH with the ESCRT proteins suggests two possibilities for AMSH function at different steps in endocytosis. First, human AMSH may deubiquitinate endocytosed cargo through the interaction with ESCRT-0 and thereby promote recycling of the cargo to the plasma membrane. Second, human AMSH may serve to recycle ubiquitin molecules from cargos prior sorting into the MVB. However, despite intensive studies, whether and how AMSH regulates ESCRT assembly and disassembly and whether AMSH3 DUB activity is required for proper ESCRT-III formation have not been resolved.
Although the catalytic MPN+ domain is conserved in Arabidopsis AMSH3, the N terminus of the protein has only low homology to human AMSH (Isono et al., 2010). While the human AMSH possesses three characterized protein interaction domains, the microtubule interacting and trafficking (MIT), clathrin binding, and STAM binding domains (Tanaka et al., 1999; McCullough et al., 2006; Nakamura et al., 2006; Tsang et al., 2006), Arabidopsis AMSH3 has only the MIT domain. Together with the fact that apparent ESCRT-0 homologs are absent in Arabidopsis, this suggests that Arabidopsis AMSH3 may have different interacting partners than its mammalian homolog.
To date, no direct interactors of Arabidopsis AMSH3 have been identified. In this study, we therefore aimed at identifying interacting proteins of AMSH3 to understand the role and mode of AMSH3 function. Here, we report that AMSH3 interacts with ESCRT-III subunits VPS2.1 and VPS24.1 in vitro and that they colocalize and interact in class E compartments when the disassembly of ESCRT-III is inhibited by the overexpression of an inactive SKD1. In addition, two MIT domain–containing proteins, AMSH3 and SKD1, compete for binding to VPS2.1. We also show that the loss of AMSH3 DUB activity causes the formation of class E–like compartments that contain VPS2.1 and VPS24.1 and thereby provide the molecular framework for AMSH3 and ESCRT-III interaction in plants.
RESULTS
AMSH3 Interacts with Subunits of ESCRT-III
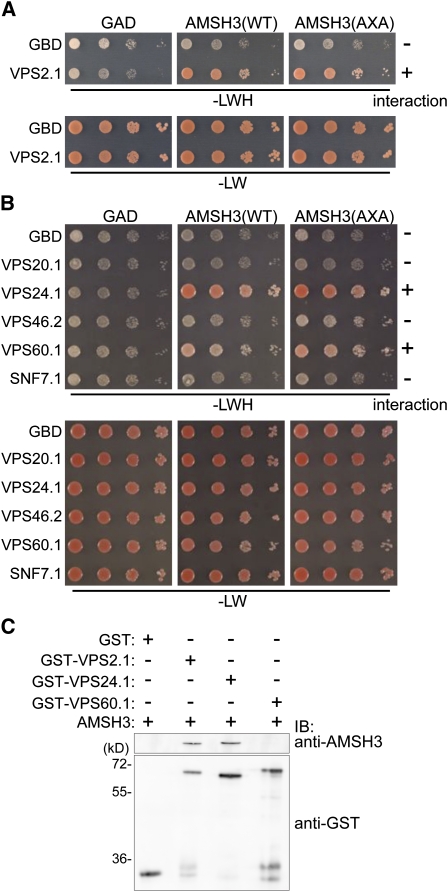
We recently showed that the Arabidopsis DUB AMSH3 is involved in vacuole biogenesis and intracellular trafficking (Isono et al., 2010). To further understand the molecular function of AMSH3, we wanted to identify interacting proteins of AMSH3, and conducted a yeast two-hybrid (Y2H) screen with a GAL4 DNA binding domain (GBD) fused to an enzymatically inactive AMSH3(AXA) variant (Isono et al., 2010) and an Arabidopsis cDNA library (Kim et al., 1997). We chose the GBD-AMSH3(AXA) fusion as bait, as the wild-type GBD-AMSH3 showed autoactivation in our Y2H assay. Since AMSH3 is implicated in vacuole biogenesis and intracellular trafficking, we decided to focus on interactor candidates with known function in intracellular trafficking and found gene fragments coding for VPS2.1, which is a homolog of yeast VPS2 (Winter and Hauser, 2006). When the full-length open reading frame (ORF) of Arabidopsis VPS2.1 was cloned and tested for interaction with AMSH3 in a Y2H assay, the interaction was confirmed (Figure 1A; see Supplemental Figure 1A online).
Figure 1.
AMSH3 Interacts with ESCRT-III Homologs VPS2.1, VPS24.1, and VPS60.1.
(A) and (B) Y2H analysis of GAD-AMSH3(WT) and GAD-AMSH3(AXA) with GBD-VPS2.1 (A) and GBD-fused ESCRT-III components VPS20.1, VPS24.1, VPS46.2, VPS60.1, or SNF7.1 (B). Transformants were grown in liquid culture and diluted to A600 = 1, and 10-fold serial dilutions were spotted on SC-LW or SC-LWH plates to test their auxotrophic growth. The presence or absence of the interaction is indicated on the right of each panel. The expression of all fusion proteins was verified by immunoblotting (see Supplemental Figures 1A and 1B online).
(C) In vitro binding assay using purified GST, GST-VPS2.1, GST-VPS24.1, and GST-VPS60.1 with AMSH3. After the GST pull-down, bead-bound materials were analyzed by immunoblotting with anti-AMSH3 and anti-GST antibodies.
[See online article for color version of this figure.]
In mammals and yeast, Vps2p/CHMP2 is one of the core components of ESCRT-III. Since VPS2 proteins in other organisms are part of the multiprotein complex ESCRT-III, we next tested whether other predicted Arabidopsis ESCRT-III subunits also interact with AMSH3. ESCRT-III consists of a so-called core complex and ESCRT-III–associated or –related proteins (Babst et al., 2002). We therefore conducted a directed Y2H analysis of AMSH3 with Arabidopsis ESCRT-III core subunit homologs (VPS20.1, VPS24.1, and SNF7.1) and ESCRT-III–related protein homologs (VPS46.2/CHMP1B and VPS60.1) (Winter and Hauser, 2006). Among the tested components, two additional ESCRT-III proteins, VPS24.1 and VPS60.1, showed interaction with AMSH3, regardless of its DUB activity, since the enzymatically inactive AMSH3(AXA) also showed interaction (Figure 1B; see Supplemental Figure 1B online).
To test whether the interactions between AMSH3 and ESCRT-III components are direct, we next performed an in vitro glutathione S-transferase (GST) pull-down assay. For this assay, AMSH3, GST fusion proteins of VPS2.1, VPS24.1, VPS60.1, and the GST moiety alone were expressed and purified from Escherichia coli. AMSH3 was pulled down when incubated together with GST-VPS2.1 or GST-VPS24.1, but not with GST or GST-VPS60.1 (Figure 1C), indicating that VPS2.1 and VPS24.1, but not VPS60.1, directly interact with AMSH3. This result suggests that AMSH3 and the ESCRT-III interaction is mediated by at least two different subunits.
VPS2.1 but Not VPS2.2 or VPS2.3 Interact with AMSH3
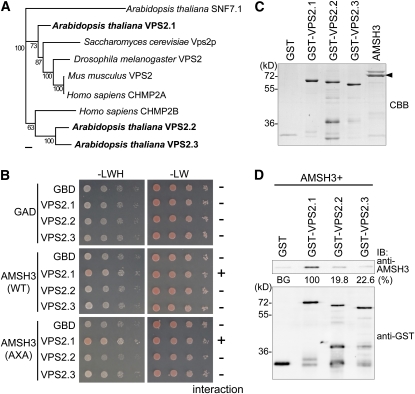
Most of the ESCRT-III subunits have multiple sequence homologs in Arabidopsis. There are three VPS2 homologs and two VPS24 homologs (Winter and Hauser, 2006) (see Supplemental Figures 2 and 3 online). Among the three Arabidopsis VPS2 proteins, VPS2.1, VPS2.2, and VPS2.3, VPS2.1 is more closely related to the yeast, Drosophila melanogaster, mouse, and human VPS2 proteins than to the VPS2.2 and VPS2.3 proteins (Figure 2A; see Supplemental Data Set 1 and Supplemental Figure 2 online).
Figure 2.
VPS2.1, but Not VPS2.2 and VPS2.3, Interacts with AMSH3.
(A) Phylogenetic analysis of Arabidopsis VPS2.1, VPS2.2, and VPS2.3, budding yeast Vps2p, fruit fly VPS2, mouse CHMP2, and human CHMP2A and CHMP2B using the Geneious Tree Builder. The accession numbers are provided in Methods. AtSNF7.1 was used as an outgroup. Bootstrap values are indicated beside each branch. Bar = 0.1 amino acid substitutions per site.
(B) Y2H analysis of GAD-AMSH3(WT) or GAD-AMSH3(AXA) and GBD, GBD-VPS2.1, GBD-VPS2.2, or GBD-VPS2.3. Transformants were tested for growth on SC-LWH medium as in Figures 1A and 1B. The presence or absence of the interaction is indicated on the right of each panel. Note that only VPS2.1 interacts with AMSH3. The expression of all fusion proteins was verified by immunoblotting (see Supplemental Figure 1C online).
(C) Purified GST, GST-VPS2.1, GST-VPS2.2, GST-VPS2.3, and AMSH3 were resolved by SDS-PAGE and stained by Coomassie blue to show equal loading. The arrowhead indicates AMSH3.
(D) In vitro binding assay of AMSH3 with GST-fused Arabidopsis VPS2 proteins. After GST pull down, bead-bound proteins were analyzed by immunoblotting using anti-AMSH3 and anti-GST antibodies. The efficiency of the GST pull-down assay was assessed as a percentage of the bound AMSH3 after subtraction of signal intensity of the GST alone sample (BG, background) using Multi Gauge software. The average value of three independent experiments is shown. The percentage of the VPS2.1 sample was set to 100%.
[See online article for color version of this figure.]
The observation that the three Arabidopsis VPS2 proteins belong to different clusters raised the question as to whether the three proteins have distinct functions. To investigate this possibility, we first wanted to know whether VPS2.2 and VPS2.3 can also interact with AMSH3 and performed a Y2H assay. While VPS2.1 interacted with AMSH3, neither VPS2.2 nor VPS2.3 did (Figure 2B; see Supplemental Figure 1C online). To further examine this interaction, we next performed an in vitro binding assay with purified AMSH3 and GST-fused VPS2.1, VPS2.2, and VPS2.3 using GST as a negative control (Figure 2C). GST-VPS2 proteins migrate higher than the calculated molecular mass (~50 kD) (Figure 2C) like their yeast homolog (Babst et al., 2002), probably owing to the protein properties. While GST-VPS2.1 directly interacted with AMSH3, only 19.8 and 22.6% of the amount of AMSH3 bound to GST-VPS2.1 was bound to GST-VPS2.2 and GST-VPS2.3, respectively (Figure 2D), indicating that VPS2.1, VPS2.2, and VPS2.3 have indeed differential binding affinities for AMSH3.
VPS2.1 Interacts with AMSH3 through Its MIM1 Domain
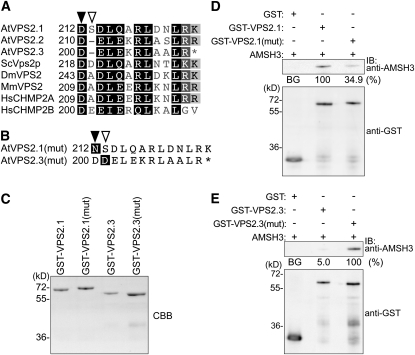
To further examine the molecular cause for the differential interaction of the Arabidopsis VPS2 proteins with AMSH3, we analyzed the amino acid sequence of VPS2.1, VPS2.2, and VPS2.3 more closely. VPS2 is conserved from yeast to humans, and all VPS2/CHMP2 proteins have a Microtubule Interacting and Transport (MIT)-Interacting Motif (MIM) at their C terminus (Figure 3A; see Supplemental Figure 2 online). The MIM domain is defined by its interaction with the MIT domain (Obita et al., 2007; Stuchell-Brereton et al., 2007), and two variations of the MIM domain have been identified in different ESCRT-III components from yeast: MIM1 in VPS2 and VPS24, and MIM2 in VPS20 and SNF7 (Obita et al., 2007; Shim et al., 2007; Stuchell-Brereton et al., 2007; Kieffer et al., 2008).
Figure 3.
Differences in the MIM1 Sequence Account for the Differential Interaction of VPS2 Proteins and AMSH3.
(A) MIM1 sequence alignment of At VPS2.1, VPS2.2, and VPS2.3 and their budding yeast (Sc), fruit fly (Dm), mouse (Mm), and human (Hs) homologs. The filled arrowhead indicates the position of the conserved Asp, and the open arrowhead indicates the gap present in Arabidopsis VPS2.2 and VPS2.3 MIM1.
(B) Mutations introduced to create Arabidopsis VPS2.1(mut) and VPS2.3(mut). For AtVPS2.1(mut), the Asp (D212) (filled arrow, Figure 3A) was mutated to an Asn, and for AtVPS2.3 (mut), the gap between D200 and D201 (open arrow, Figure 3A) was filled with an Asp.
(C) Purified GST-VPS2.1, GST-VPS2.1(mut), GST-VPS2.3, and GST-VPS2.3(mut) were subjected to SDS-PAGE and stained with Coomassie blue to show equal loading.
(D) In vitro binding assays of GST, GST-VPS2.1, and GST-VPS2.1(mut) with AMSH3. The binding assay was performed as in Figure 2D. Note that GST-VPS2.1(mut) interacts more weakly with AMSH3 than does the wild-type GST-VPS2.1. The efficiency of the GST pull-down assay was assessed as in Figure 2D. The average values of three independent experiments are shown.
(E) In vitro binding assays of GST, GST-VPS2.3, and GST-VPS2.3 (mut) with AMSH3 were performed as in Figure 2D. Note that GST-VPS2.3(mut) interacts much stronger with AMSH3 than GST-VPS2.3. The efficiency of the GST pull-down assay was assessed as a percentage of the bound AMSH3 after subtraction of signal intensity of the GST alone sample (BG, background) using Multi Gauge software. The average percentage value of three independent experiments is shown. The percentage of VPS2.3(mut) was set to 100%.
Since AMSH3 has a MIT domain at its N terminus, we speculated that VPS2.1 and AMSH3 interact through their MIM1 and MIT domains, respectively. Amino acid sequence comparisons of VPS2.1, VPS2.2, and VPS2.3 showed that the conserved residues in MIM1 [(D/E)XXLXXRLXXL(K/R)] are present in all three Arabidopsis VPS2 proteins. We noted, however, that the spacing between two acidic amino acids (Asp-212 and Asp-214 in VPS2.1) is altered in the MIM1 of VPS2.2 and VPS2.3 (Figure 3A). To test whether the amino acid sequence of the MIM1 is important for the interaction of VPS2 proteins and AMSH3, we constructed mutant variants of VPS2.1 and VPS2.3. In VPS2.1(mut), Asp-212 was mutated to an Asn, and in VPS2.3(mut), an additional Asp was inserted after the conserved D200 (Figure 3B). The wild-type and mutant VPS2.1 and VPS2.3 proteins were then expressed as GST fusion proteins in E. coli, and purified proteins were tested in an in vitro binding assay (Figure 3C). While the wild-type VPS2.1 interacted with AMSH3, the mutation in VSP2.1 [VPS2.1(mut)] led to a much weaker interaction with AMSH3 (34.9% of the GST-VPS2.1), suggesting that the Asp-212 residue is required for the interaction of these two proteins (Figure 3D). In turn, to investigate whether an additional amino acid after Asp-200 in VPS2.3 is necessary for the interaction, wild-type and mutant VPS2.3 proteins were tested for their binding to AMSH3. Interestingly, while the wild-type VPS2.3 interacted only weakly with AMSH3, VPS2.3(mut) could strongly interact with AMSH3 (Figure 3E), suggesting that the spacing between the two conserved acidic amino acids in MIM1 is indeed important for the interaction of VPS2 proteins with AMSH3.
The AMSH3 MIT Domain Is Necessary for the Interaction with ESCRT-III Subunits
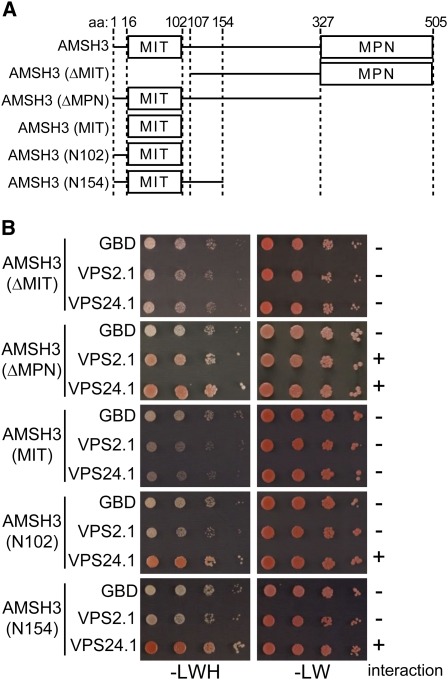
We next wanted to know whether the interactions of VPS2.1 and VPS24.1 with AMSH3 are dependent on the MIT domain of AMSH3. To examine this, we created truncated versions of AMSH3 (ΔMIT, ΔMPN, MIT, N102, and N154) and tested their interaction with VPS2.1 and VPS24.1 by Y2H (Figure 4A). Supporting the notion that AMSH3 and the ESCRT-III subunits interact via the MIT domain of AMSH3, both VPS2.1 and VPS24.1 lost their interaction with AMSH3 when the N-terminal MIT domain was deleted (Figure 4B; see Supplemental Figure 1D online). In turn, we found that the isolated MIT domain (MIT) was not sufficient for the interaction with VPS2.1 or VPS24.1. However, when the N-terminal 16 amino acids were added to the MIT domain of AMSH3 (N102), VPS24.1 interacted with this variant (Figure 4B; see Supplemental Figure 1D online), showing that this extended MIT domain of AMSH3 is sufficient for the interaction with VPS24.1. By contrast, VPS2.1 required a much longer N-terminal region (ΔMPN) for the interaction with AMSH3 (Figure 4B; see Supplemental Figure 1D online), suggesting that the two ESCRT-III subunit homologs may have different interaction surfaces on AMSH3.
Figure 4.
The MIT Domain of AMSH3 Is Necessary for the Interaction of AMSH3 and Putative ESCRT-III Subunits.
(A) Schematic presentation of the AMSH3 truncation constructs used in the Y2H analysis.
(B) Y2H analysis with truncation constructs of AMSH3 with VPS2.1 or VPS24.1. Yeast cells were transformed with the indicated truncation constructs of GAD-AMSH3 and GBD-, GBD-VPS2.1–, or GBD-VSP24.1–containing plasmids. Transformants were tested for their auxotrophic growth on SC-LW and SC-LWH media as in Figures 1A and 1B. The presence or absence of the interaction is indicated on the right of each panel. The expression of the Y2H constructs was verified by immunoblotting (see Supplemental Figure 1D online).
[See online article for color version of this figure.]
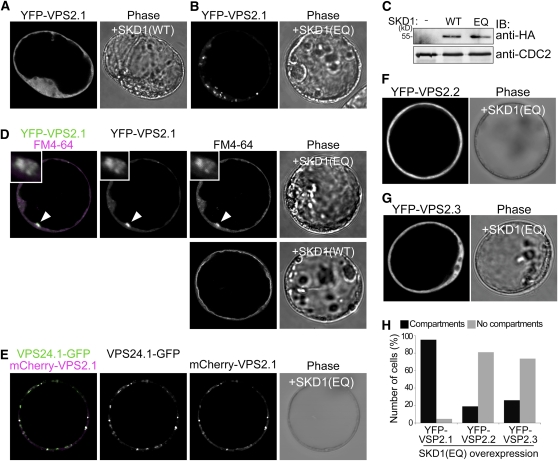
VPS2.1 but Not VPS2.2 and VPS2.3 Localize to SKD1(EQ)-Induced Class E Compartments
Another MIT domain containing protein in the ESCRT pathway is SKD1/VPS4 (Obita et al., 2007) (see Supplemental Figure 4 online). SKD1 is an AAA-ATPase that interacts with ESCRT-III subunits via its MIT domain and whose ATPase activity is required for ESCRT-III disassembly (Babst et al., 1998). It was previously shown that the expression of an enzymatically inactive SKD1 leads to the formation of so-called class E compartments in yeast and Arabidopsis (Babst et al., 1997, 1998; Haas et al., 2007). Class E compartments are aberrant endosomal structures, and in yeast, it was shown that ESCRT-III subunits accumulate on these structures in vps4 mutant cells (Babst et al., 1998).
We thus reasoned that if VPS2.1 was a bona fide Arabidopsis ESCRT-III subunit and if the VPS2.1-containing ESCRT-III complex disassembly was dependent on SKD1, then VPS2.1 should accumulate in class E compartments upon ATPase-defective SKD1(EQ) coexpression. Since the overexpression of SKD1(EQ) had been shown to be toxic to plants (Haas et al., 2007), we had to examine its effect in Arabidopsis cell culture–derived protoplasts. To test our hypothesis, we generated a wild-type SKD1 [SKD1(WT)], and a SKD1(EQ) mutant variant fused to an HA-tag. Upon overexpression of SKD1(EQ) but not SKD1(WT), yellow fluorescent protein (YFP)-VPS2.1 relocated from the cytosol to large punctate structures (Figures 5A to 5C). Since the endocytosis tracer dye FM4-64 also accumulated in these structures (Figure 5D) and since VPS24.1-GFP (green fluorescent protein) and mCherry-VPS2.1 colocalized in these structures specifically upon SKD1(EQ) overexpression (Figure 5E; see Supplemental Figure 5A online), we concluded that these structures are class E compartments that were induced by SKD1(EQ) expression. The localization and relocation observed upon SKD1(EQ) coexpression are not due to the overexpression of VPS2.1 since VPS2.1 expressed under its own promoter showed similar cytosolic localization and reactivity toward SKD1(EQ) overexpression (see Supplemental Figures 6A and 6B online). Furthermore, since VPS2.1 and VPS24.1 colocalized in class E compartments, we concluded that these two proteins are bona fide ESCRT-III subunits that are integrated into the membrane-bound ESCRT-III like their animal and yeast homologs.
Figure 5.
VPS2.1, but Not VPS2.2 and VPS2.3, Localize to SKD1(EQ)-Induced Class E Compartments.
(A) and (B) UBQ10pro:YFP-VPS2.1 was coexpressed with 35Spro:HA-SKD1(WT) (A) and 35Spro:HA-SKD1(EQ) (B) in Arabidopsis protoplasts and analyzed using a confocal laser scanning microscope. Note that instead of the cytosolic localization observed in (A), YFP is localized to intracellular structures in (B).
(C) Expression of HA-SKD1(WT) and HA-SKD1(EQ) was verified by immunoblotting with an anti-HA antibody on total extracts of transformed protoplasts. An anti-CDC2 antibody was used as a loading control.
(D) UBQ10pro:YFP-VPS2.1 was coexpressed with 35Spro:HA-SKD1(EQ), and protoplasts were stained with FM4-64 2 h prior to observation by confocal microscopy (top panel). Arrowheads indicate the class E compartment where YFP-VPS2.1 and FM4-64 signals colocalize. Insets are magnifications of a class E compartment. Note that FM4-64 reaches the tonoplast membrane after 3 h of incubation in a cell overexpressing SKD1(WT) (bottom panel).
(E) Coexpression of 35Spro:VPS24.1-GFP, UBQ10pro:mCherry-VPS2.1, and 35Spro:HA-SKD1(EQ) in Arabidopsis protoplasts. Upon overexpression of SKD1(EQ), VPS24.1-GFP and mCherry-VPS2.1 colocalize in class E compartments.
(F) and (G) UBQ10pro:YFP-VPS2.2 (F) and 35Spro:YFP-VPS2.3 (G) were coexpressed in Arabidopsis protoplasts with 35Spro:HA-SKD1(EQ). Note that in contrast with YFP-VPS2.1, the cytosolic localization of YFP-VPS2.2 and YFP-VPS2.3 does not change upon SKD1(EQ) overexpression.
(H) Number of cells (percentage) with YFP-VPS2.1 (n = 47), YFP-VPS2.2 (n = 40), and YFP-VPS2.3 (n = 59) positive class E compartments after SKD1(EQ) overexpression. Black bars, cells with class E compartments; gray bars: cells without class E compartments.
Among the three Arabidopsis VPS2 homologs, only VPS2.1 showed in vitro and Y2H interaction with AMSH3, probably owing to differences in the MIM1 sequence. Since SKD1 has a MIT domain, we speculated that these differences in MIM1 could also lead to differential interactions of the VPS2 proteins with SKD1 and, thus, to differential reactivity toward SKD1(EQ) overexpression. Indeed, in contrast with YFP-VPS2.1, where 95% (n = 47) of all cells accumulate the fusion protein on class E compartments upon coexpression of SKD1(EQ), only 18% (n = 40) and 26% (n = 59) of cells accumulated YFP-VPS2.2 and YFP-VPS2.3 in class E compartments after SKD1(EQ) overexpression (Figures 5F to 5H). Upon SKD1(WT) overexpression or without SKD1 coexpression, VPS2 fusion proteins showed cytosolic localization (Figure 5A; see Supplemental Figures 5B and 5C online). Since only VPS2.1 shows interaction with AMSH3, and only VPS2.1 reacts toward the overexpression of dominant-negative SKD1, VPS2.1 may be the only true ESCRT-III subunit among the Arabidopsis VPS2 proteins. It can thus be speculated that VPS2.2 and VPS2.3 may function in a different context outside of the classical ESCRT-III that is not dependent on SKD1 activity.
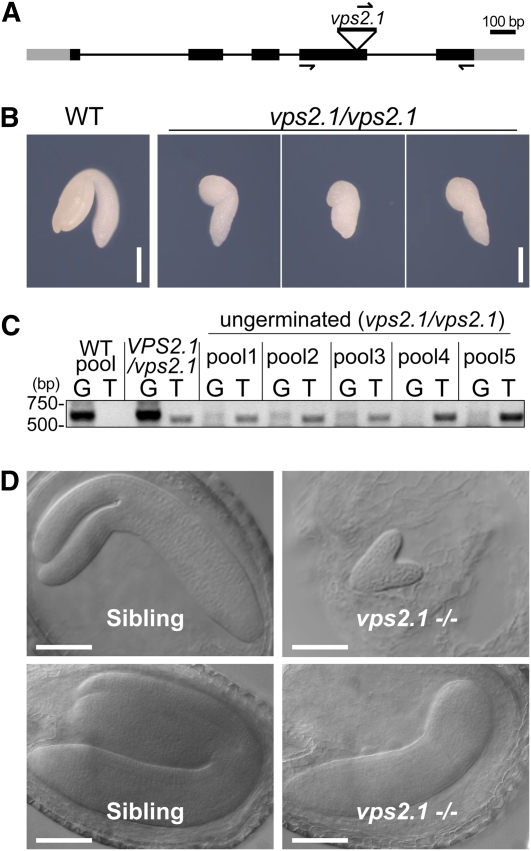
vps2.1 Homozygous Mutants Are Embryo Lethal
To date, mutant analysis of the ESCRT-III core complex subunits has not been reported in Arabidopsis. We therefore obtained and examined a T-DNA insertion mutant of VPS2.1, in which the T-DNA is inserted in the fourth exon (Figure 6A). Since we could not detect any vps2.1 homozygous seedlings among the progeny of a self-pollinated VPS2.1/vps2.1 heterozygous plant, we hypothesized that the homozygous vps2.1 is embryo lethal. Indeed, among the seeds of a VPS2.1/vps2.1 heterozygous plant (total n = 416), we found that 13.7% did not germinate and 7.2% were aborted within the silique (Table 1). Since we found fewer ungerminated seeds (3.3%, n = 183) in a self-pollinated wild-type plant (Table 1), this result suggests that vps2.1 homozygous mutants arrest growth during embryogenesis. To confirm this, we genotyped embryos dissected from ungerminated seeds together with wild-type embryos and a VPS2.1/vps2.1 heterozygous seedlings. Since the DNA yield from individual ungerminated embryos was very low, we analyzed them in pools of four embryos. This experiment revealed that the ungerminated seeds indeed contain vps2.1/vps2.1 homozygous mutant embryos (Figures 6B and 6C). Phenotypic analysis of the dissected mutant embryos showed that these are smaller than those of the wild type and that they arrest development at the walking-stick stage of embryogenesis (Figure 6B). Furthermore, 15% of the embryos (n = 225) of a VPS2.1/vps2.1 self-pollinated plant showed developmental delay in the heart or torpedo stage, suggesting that the loss of VPS2.1 function already has consequences at an early stage of embryogenesis (Figure 6D; see Supplemental Table 1 online). The VPS2.1pro:YFP-VPS2.1 (genomic) construct could partially rescue the developmental arrest of vps2.1 mutant embryos, whereas a Ribosomal Protein S5 (RPS5a):VPS2.1 (cDNA) construct could also rescue the germination defects of vps2.1 homozygous mutants (see Supplemental Figures 7A to 7D and Supplemental Tables 1 and 2 online). These results indicate that the observed vps2.1 phenotypes are indeed due to the misregulation of VPS2.1.
Figure 6.
vps2.1 Homozygous Mutants Are Embryo Lethal.
(A) T-DNA insertion and primer binding site in VPS2.1. Lines indicate introns and boxes indicate exons (black boxes, coding region; gray boxes, untranslated regions). The position of the T-DNA in the vps2.1 mutant is shown.
(B) Photographs of wild-type (WT) and vps2.1 homozygous embryos dissected from dry seeds. Bars = 0.2 mm.
(C) PCR analysis of wild-type and vps2.1 homozygous mutant embryos. Twenty embryos were dissected from the ungerminated seeds of a VPS2.1/vps2.1 self-pollinated plant, and total DNA was extracted from pools of four embryos. DNA extracted from a pool of four wild-type embryos, and DNA from a VPS2.1/vps2.1 heterozygous plant was used as control. PCR was performed with either gene-specific forward and reverse primers (G lanes) or a T-DNA left border primer in combination with the gene-specific reverse primer (T lanes). Note that all ungerminated seeds contain embryos homozygous for the T-DNA insertion. The primers used for genotyping and their sequences are presented in Methods and Supplemental Table 3 online.
(D) vps2.1 homozygous mutant embryos (right panels) are delayed in embryogenesis in comparison to their wild-type-looking siblings (left panels). Bars = 100 μm.
[See online article for color version of this figure.]
Table 1.
Progeny Analysis of Wild-Type and VPS2.1/vps2.1 Heterozygous Plants
| Genotype of the Parent Plant | Germinated | Ungerminated | Aborted | n |
| VPS2.1/VPS2.1 | 96.7% | 3.3% | 0% | 183 |
| VPS2.1/vps2.1 | 79.1% | 13.7% | 7.2% | 416 |
To find out whether the underrepresentation of the homozygous mutants is due to a defect in female or male gametes, we performed reciprocal crosses between a VPS2.1/vps2.1 and a wild-type plant (Table 2). The progeny of the two types of crosses was genotyped. When VPS2.1/vps2.1 was used as the female partner, the ratio of heterozygous to wild-type progeny was with 42.1:57.9%, which is not significantly different from the expected 50:50% ratio. However, when VPS2.1/vps2.1 was used as the male parent, the ratio of heterozygous to wild-type progeny was 29.5:70.5%, which deviates significantly from the expected 50:50% ratio, indicating that vps2.1 pollen are less fertile than the wild type. Together, these results suggest that VPS2.1 is already required during gametogenesis and is necessary for proper embryogenesis. Moreover, the observation that a vps2.1 single mutant develops a severe phenotype indicates that VPS2.1 function is not redundant with that of VPS2.2 and VPS2.3, both of whose homozygous T-DNA insertion mutants show only mild growth defects but not embryo lethality (see Supplemental Figures 8A to 8C online).
Table 2.
Analysis of Genetic Transmission of VPS2.1/vps2.1
| Genotype (Female × Male) | VPS2.1/VPS2.1 | VPS2.1/vps2.1 | n | χ2 P |
| VPS2.1/vps2.1 × Col-0 | 57.9% | 42.1% | 38 | 0.11410 |
| Col-0 × VPS2.1/vps2.1 | 70.4% | 29.5% | 54 | 0.00005 |
AMSH3 and ESCRT-III Components Colocalize in Class E Compartments
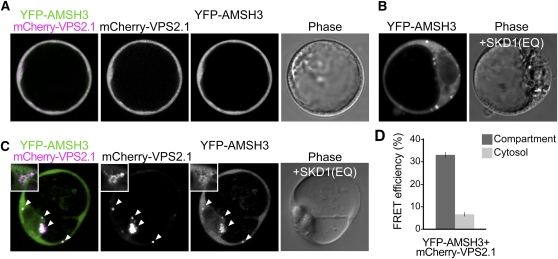
We next wanted to examine whether AMSH3 functions together with the ESCRT-III subunits in vivo. For this aim, we first tried to coimmunoprecipitate AMSH3 and VPS2.1. However, VPS2.1 turned out to be a very unstable protein, and we could not detect YFP-VPS2.1 with an anti-GFP or with a generated specific anti-VPS2.1 antibody (see Supplemental Figures 9A and 9B online) in the total extract precleared for immunoprecipitation (see Supplemental Figure 9C online). Therefore, we decided to examine the colocalization of AMSH3 and VPS2.1. Both AMSH3 and VPS2.1 fusion proteins showed an apparent cytoplasmic localization in Arabidopsis cell culture–derived protoplasts (Figure 7A) as well as in the root epidermal cells of transgenic Arabidopsis seedlings (see Supplemental Figure 5E online). The localization of both AMSH3 and VPS2.1 was not affected by treatments of the commonly used endocytosis inhibitors Brefeldin A or Wortmannin A (see Supplemental Figure 5E online), suggesting that both proteins are not stably associated with endosomes or organelles that are sensitive to these treatments.
Figure 7.
AMSH3 and VPS2.1 Colocalize and Interact in Class E Compartments.
(A) AMSH3pro:YFP-AMSH3 was cotransformed with UBQ10pro:mCherry-VPS2.1 in Arabidopsis protoplasts. Both fusion proteins show cytoplasmic localization in Arabidopsis cell culture–derived protoplasts.
(B) YFP-AMSH3 localizes to class E compartments upon SKD1(EQ) coexpression.
(C) Coexpression of AMSH3pro:YFP-AMSH3 and UBQ10pro:mCherry-VPS2.1 with 35Spro:HA-SKD1(EQ) in Arabidopsis cell culture–derived protoplasts. Note that a fraction of YFP-AMSH3 colocalizes with mCherry-VPS2.1 in class E compartments. Magnification of a class E compartment is shown in the inset. Arrowheads indicate class E compartments.
(D) FRET analysis by the sensitized emission method between YFP-AMSH3 and mCherry-VPS2.1. Arabidopsis cell culture–derived protoplasts were cotransformed with 35Spro:HA-SKD1(EQ), AMSH3pro:YFP-AMSH3, and UBQ10pro:mCherry-VPS2.1, and the mean FRET efficiency in either the compartments or the cytosol was measured in a total of 16 cells. Dark-gray bar, compartment; light-gray bar, cytosol. Error bars indicate se.
If AMSH3 interacts with ESCRT-III in vivo, its localization should also be affected by SKD1(EQ) overexpression. Indeed, when cotransformed with SKD1(EQ), 31% of YFP-AMSH3 expressing cells (n = 45) had YFP signals in class E compartments (Figure 7B), while cotransformation with wild-type SKD1 did not yield such signals (see Supplemental Figure 5D online). Moreover, YFP-AMSH3 colocalized in these compartments with mCherry-VPS2.1 upon SKD1(EQ) expression (Figure 7C), suggesting that AMSH3 and ESCRT-III closely localize in vivo. The SKD1(EQ)-dependent colocalization of mCherry-VPS2.1 and YFP-AMSH3 was also observed when both fusion proteins were expressed under their native promoter (see Supplemental Figure 6C online). Notably, even after SKD1(EQ) overexpression, part of YFP-AMSH3 remained cytosolic, suggesting that either only a small fraction of AMSH3 interacts with ESCRT-III or that the interaction between AMSH3 and ESCRT-III is transient.
To address the question of whether AMSH3 and VPS2.1 interact in class E compartments, we further analyzed Förster resonance energy transfer (FRET) efficiency in YFP-AMSH3, mCherry-VPS2.1, and HA-SKD(EQ) cotransformed cells. An increase in energy transfer efficiency was expected if AMSH3 and VPS2.1 interact in vivo. Indeed, YFP-AMSH3 and mCherry-VPS2.1 showed an average FRET efficiency of 33.5% in class E compartments, while the FRET efficiency in the cytosol was only 6.7% (Figure 7D), indicating that these proteins interact in vivo specifically on these compartments.
ESCRT-III Does Not Directly Regulate AMSH3 DUB Activity
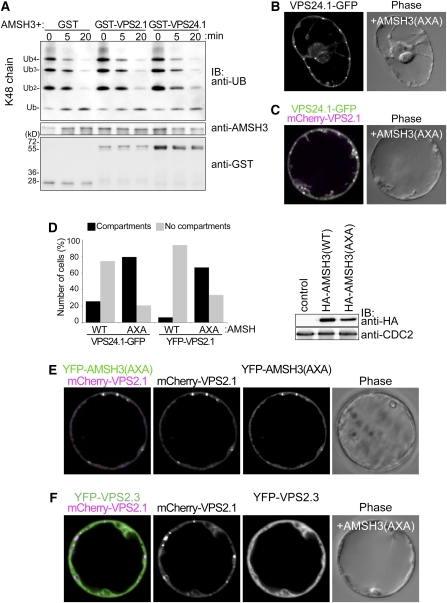
The binding of an ESCRT-0 component STAM to human AMSH was shown to promote the AMSH DUB activity in vitro (Kim et al., 2006; McCullough et al., 2006). To see whether ESCRT-III subunits can affect the enzyme activity of AMSH3, we tested the DUB activity of AMSH3 in an in vitro DUB assay in the presence of GST-VPS2.1 and GST-VPS24.1. In our assay, AMSH3 typically hydrolyzed K48 ubiquitin chains to monoubiquitins after 20 min. This tendency was not affected when AMSH3 was preincubated with either GST-VPS2.1 or GST-VPS24.1 for 20 min prior to the DUB assay (Figure 8A; see Supplemental Figure 10 online), indicating that neither VPS2.1 nor VPS24.1 directly regulate AMSH3 DUB activity in vitro.
Figure 8.
AMSH3 Enzymatic Activity Affects VPS2.1 and VPS24.1 Intracellular Localization.
(A) AMSH3 DUB assay with purified GST, GST-VPS2.1, or GST-VPS24.1 using K48-linked ubiquitin chains. AMSH3 was preincubated with GST fusion proteins prior the addition of ubiquitin chains to the reaction mixture. Reactions were stopped after 0, 5, and 20 min and subjected to immunoblots using the indicated antibodies.
(B) and (C) Coexpression of 35Spro:HA-AMSH3(AXA) with 35Spro:VPS24.1-GFP (B) or UBQ10pro:YFP-VPS2.1 and 35Spro:VPS24.1-GFP (C). Upon expression of AMSH3(AXA), both VPS2.1 and VSP24.1 fusion proteins partially mislocalize on compartments.
(D) Quantification of the effect of 35Spro:HA-AMSH3(WT) and 35Spro:HA-AMSH3(AXA) coexpression on ESCRT-III localization (left panel). Solid bars, cells with compartments; gray bars, cells without compartments. n = 35 and 34 for 35Spro:VPS24.1-GFP expressed with 35Spro:HA-AMSH3(WT) and 35Spro:HA-AMSH3(AXA), respectively, and n = 16 and 30 for UBQ10pro:YFP-VPS2.1 expressed with 35Spro:HA-AMSH3(WT) and 35Spro:HA-AMSH3(AXA), respectively. Expression of HA-AMSH3(WT) and HA-AMSH3(AXA) was verified by immunoblotting with an anti-HA antibody (right panel). An anti-CDC2 antibody was used to show equal loading.
(E) mCherry-VPS2.1 and YFP-AMSH3(AXA) colocalized on the AMSH3(AXA)-induced compartments.
(F) mCherry-VPS2.1, but not YFP-VPS2.3, localizes on AMSH3(AXA)-induced cellular compartments.
AMSH3 DUB Activity Is Required for Proper ESCRT-III Localization
Finally, we wanted to know whether AMSH3 dysfunction in turn can affect ESCRT-III localization. To investigate this possibility, we coexpressed the enzymatically inactive AMSH3(AXA) with YFP-VPS2.1 and VPS24.1-GFP. Interestingly, the cytosolic localization of both YFP-VPS2.1 and VPS24.1-GFP was affected by the coexpression of HA-AMSH3(AXA) (Figure 8B). G/YFP signals accumulated in the intracellular compartments of 79.4% (n = 34) and 66.7% (n = 30) of the VPS24.1-GFP– and YFP-VPS2.1–expressing cells, respectively. By contrast, only 25.7% (n = 35) and 6.25% (n = 16) of cells showed G/YFP signals, respectively, in compartments with coexpression of wild-type AMSH3 (Figure 8D). mCherry-VPS2.1 and VPS24.1-GFP colocalized on these AMSH(AXA)-induced compartments (Figure 8C). Furthermore, in contrast with YFP-AMSH3(WT), YFP-AMSH3(AXA) itself was relocated to these compartments and colocalized with mCherry-VPS2.1 (cf. Figures 8E and 7A). If these compartments are class E–like compartments, VPS2.3, which is resistant to SKD1(EQ) overexpression, should also be resistant to AMSH3(AXA) overexpression. Indeed, when YFP-VPS2.3, mCherry-VPS2.1, and HA-AMSH3(AXA) were coexpressed, only mCherry-VPS2.1 localized on the AMSH3(AXA)-induced compartments, whereas YFP-VPS2.3 remained cytosolic (Figure 8F). These results suggest that the DUB activity of AMSH3 is required for proper localization of AMSH3 as well as of ESCRT-III.
SKD1 and AMSH3 Compete for ESCRT-III Binding
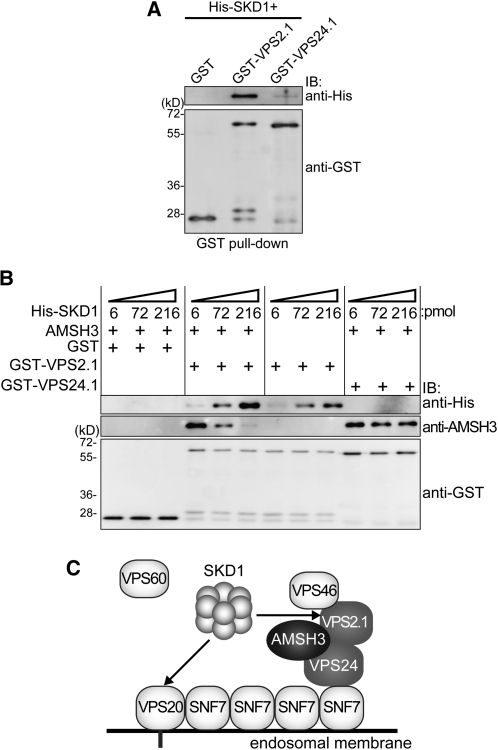
AMSH3 and SKD1 both possess a MIT domain that can serve as an interaction surface for ESCRT-III components. The fact that AMSH3(AXA) overexpression causes a strong mislocalization of ESCRT-III subunits to class E–like compartments suggests that the binding of AMSH3 to ESCRT-III, especially when it is inactive, may partially block the access of SKD1 to ESCRT-III. If this was true, SKD1 and AMSH3 should compete for the binding to ESCRT-III. To test this hypothesis, we first wanted to establish whether SKD1 could interact with VPS2.1 and VPS24.1, two ESCRT-III subunits that interact with AMSH3, since SKD1 and VPS2.1 interaction was not reported in previous interaction studies using Arabidopsis ESCRT proteins (Shahriari et al., 2010). We therefore conducted an in vitro binding assay of SKD1 with VPS2.1 and VPS24.1 and found that VPS2.1 interacts with SKD1 (Figure 9A).
Figure 9.
AMSH3 Competes with SKD1 for VPS2.1 Binding.
(A) SKD1 in vitro binding assay. GST, GST-VPS2.1, and GST-VPS24.1 loaded on GST Sepharose 4B beads were incubated with His-SKD1. After extensive washing, the bead-bound materials were analyzed by immunoblotting using anti-His and anti-GST antibodies.
(B) SKD1 competition assay. Binding assay of GST-2.1 or GST-24.1 with 6 pmol of AMSH3 was performed as in Figure 1C in the presence of 6, 72, and 216 pmol of His-SKD1. Note that AMSH3 binding becomes weaker in the presence of an excess amount of SKD1.
(C) Predicted model for the AMSH3–ESCRT-III interaction. AMSH3 associates with ESCRT-III through its binding to VPS2.1 and VPS24.1. AMSH3 competes with SKD1 for the binding to VPS2.1 MIM1, but probably not to VPS20. The timely disassociation of AMSH3 may be necessary for SKD1 to be able to bind efficiently and disassemble ESCRT-III.
We next performed a competition assay with purified His-SKD1, AMSH3, GST-VPS2.1, and GST-VPS24.1. While His-SKD1 did not affect the binding of AMSH3 to VPS24.1, we found that an increasing amount of SKD1 in the binding assay lead to a reduction of AMSH3 binding to GST-VPS2.1 (Figure 9B), suggesting that AMSH3 and SKD1 compete for the binding to VPS2.1. In an assay without AMSH3, SKD1 bound to VPS2.1 in a dosage-dependent manner (Figure 9B). Taken together, our results indicate that SKD1 competes with AMSH3 for binding to VPS2.1 and that AMSH3 activity is probably required for the efficient disassociation of AMSH3 from ESCRT-III and, hence, the recruitment of SKD1 to the complex (Figure 9C).
DISCUSSION
In this study, we identified two ESCRT-III core subunit homologs, VPS2.1 and VPS24.1, as interactors of the DUB AMSH3 in Arabidopsis. Human AMSH was shown to strongly interact with VPS24/CHMP3, VPS46/CHMP1, and SNF7/CHMP4 (Agromayor and Martin-Serrano, 2006; Tsang et al., 2006), indicating a partially different interaction pattern between the human and plant AMSH proteins and the ESCRT-III subunits. This may be due to the difference in the region surrounding the MIT domain of the human and Arabidopsis AMSH proteins. Although bioinformatic analyses have shown that the ESCRT components are conserved in Arabidopsis (Winter and Hauser, 2006), little is known about the ESCRT-III function in plants. Through coexpression studies using SKD1(EQ), we could show that VPS2.1 and VPS24.1 behave like bona fide ESCRT-III subunits and that they interact directly with AMSH3 and colocalize with AMSH3 in class E compartments.
Interestingly, we found that the three VPS2 homologs from Arabidopsis have different characteristics. First, in contrast with VPS2.1, which is the closest Arabidopsis homolog of yeast Vps2p, VPS2.2 and VPS2.3 did not show MIM1-dependent interaction with AMSH3. Second, VPS2.1, but not VSP2.2 and VPS2.3, accumulate in class E compartments induced by the overexpression of the dominant-negative SKD1 and VPS2.3 also did not accumulate in the AMSH3(AXA)-induced compartments. Third, the vps2.1 single homozygous mutant is embryo lethal, implicating the nonredundancy of VPS2.1. VPS2.1, VPS2.2, and VPS2.3 may therefore be integrated into different ESCRT-III complexes that are differently regulated by SKD1. Alternatively, VPS2.2 and VPS2.3 may function independently of the classical ESCRT-III and are therefore not regulated by SKD1. A recent article by Shahriari et al. (2011) describes the investigation of the interaction network of ESCRT proteins by Y2H and bimolecular fluorescence complementation. They propose that VPS2.3 is the VPS2 protein of Arabidopsis that is integrated into ESCRT-III. However, at the same time we noticed that in their system only VPS2.1 shows interaction with VPS24.1. Since yeast vps24Δ mutant analysis has shown that the binding of Vps2p, but of no other ESCRT-III core subunit, is dependent on Vps24p, these subunits probably function closely with each other (Teis et al., 2008). Moreover, since we found VPS2.1 and VPS24.1 to colocalize on SKD1(EQ)-induced class E compartments, we suggest that VPS2.1 is part of ESCRT-III in Arabidopsis. This differential function of the three different Arabidopsis VPS2 homologs will be the theme of future investigations.
In contrast with the binding of STAM, which was reported to promote human AMSH DUB activity (Kim et al., 2006; McCullough et al., 2006), the binding of ESCRT-III subunits to AMSH3 did not regulate its DUB activity directly. We therefore speculate that ESCRT-III may serve as a platform for AMSH3 to deubiquitinate ubiquitinated MVB cargos at the ESCRT-III positive endosomes. Alternatively, AMSH3 may deubiquitinate proteins of the endocytosis machinery itself that are localized in the proximity to ESCRT-III.
Interestingly, the previously reported SKD1(EQ) overexpression phenotype (Shahriari et al., 2010) and the amsh3 mutant (Isono et al., 2010) phenotype have several features in common. First, both have defects in soluble cargo transport to the vacuole, and second, both have defects in vacuole biogenesis or maintenance. These resemblances indicate that the loss of AMSH3 or AMSH3 activity may have the same impact on cellular events as the dominant-negative SKD1. In line with this notion, our results showed that VPS2.1 and VPS24.1 mislocalize upon overexpression of the enzymatically inactive AMSH3(AXA).
In contrast with the wild-type AMSH fusion protein, YFP-AMSH3(AXA) localized on intracellular compartments. The inhibition of AMSH3 activity may therefore prevent the dissociation of AMSH3 from ESCRT-III or the ESCRT-III–containing endosomes, where its substrates reside. The inhibition of AMSH3 DUB activity is predicted to prevent its disassociation from the ubiquitinated substrates (Isono et al., 2010). This, in turn, may mask the SKD1 interaction sites on ESCRT-III and causes incomplete or inefficient disassembly of the complex. Alternatively, the disassociation of AMSH3 from ESCRT-III after the completion of deubiquitination may be required for the efficient binding of the ESCRT-III–associated proteins, such as VPS46/CHMP1/Did2p, which was shown to be involved in SKD1 recruitment (Nickerson et al., 2006). Future studies should seek to identify the major substrates of AMSH3 and determine how AMSH3 deubiquitination contributes to the ESCRT-mediated MVB pathway.
METHODS
Biological Material
All experiments were performed with Arabidopsis thaliana (Columbia-0 [Col-0] background). The T-DNA insertion line of VPS2.1 was obtained from the GABI-Kat collection (GABI_670D06) (Rosso et al., 2003). The T-DNA insertion mutant of VPS2.2 (SALK_059274) and VPS2.3 (SALK_127119) was obtained from the Nottingham Arabidopsis Stock Centre. Genotyping-PCR, RT-PCR, and analysis of the mutants are described in Supplemental Methods online.
Plant transformations were performed using the floral dip method (Clough and Bent, 1998). Seedlings were grown in continuous light on standard Murashige and Skoog growth medium (Duchefa Biochemie) supplemented with 1% Suc, and adult plants were grown in soil.
Cloning Procedures
All of the primers used for cloning are listed in Supplemental Table 3 online.
For the Y2H constructs, the ORFs of VPS2.1, VPS2.3, and VPS24.1 were PCR amplified, and the resulting fragments were cloned into the NdeI and SmaI (VPS2.1), NdeI and EcoRI (VPS2.3), and the NdeI and BamHI (VPS24.1) sites of pGBKT7 vector (Clontech). The amplified ORFs of VPS2.2, VPS20.1, VPS46.1, VPS46.2, VPS60.1, and SNF7.1 were cloned into the EcoRI and BamHI sites of pGBKT7 vector. To yield AD-AMSH3, GAD-AMSH3(AXA) and GBD-AMSH3(AXA), the wild-type and mutant AMSH3 were amplified with primers AK0 and EI14 using G67772 clone (ABRC) and GST:AMSH3(AXA) (Isono et al., 2010) as template, respectively, and cloned into the BamHI-SalI sites of pGADT7 (Clontech) and pGBKT7 vector, respectively. The truncated constructs of AMSH3, GAD-AMSH3-ΔMPN, -MIT, -N102, and -N154 were constructed with fragments amplified with AK0 and At AMSH3 rv, AK70 and AK51, AK0 and AK51, and AK0 and AK97, respectively, and ligated into the BamHI-SalI sites of the pGADT7 vector. The AMSH3-ΔMIT was amplified with AK88 and EI14 and cloned into the XhoI site of pGADT7.
GST-AMSH3 was described previously (Isono et al., 2010). For GST-VPS2.1, GST-2.3, GST-24.1, and GST-60.1, the corresponding ORFs were PCR amplified using primers AK1 and AK2, AK3 and AK4, AK66 and AK67, and AK68 and AK69, respectively, and the resulting fragments were cloned between the XhoI and SalI sites (VPS2.1), EcoRI and SalI sites (VPS2.3), and the BamHI and SalI sites (VPS24.1 and VPS60.1) of pGEX-6P-1 (GE Healthcare). GST-VPS2.2 was obtained by subcloning the VPS2.2 gene from the Y2H construct into the pGEX-6P-1 vector between the EcoRI and SalI sites. For GST-VPS2.1(mut) and GST-VPS2.3(mut) constructs, the mutations were introduced with primers AK44 and AK45 and AK47 and AK48, respectively, using DpnI-based site-directed mutagenesis (Stratagene).
UBQ10pro:YFP-VPS2.1, UBQ10pro:YFP-VPS2.2, and UBQ10pro:mCherry-VPS2.1 were generated with Cre/Lox recombination (NEB) using pUNI-VPS2.1 and pUNI-VPS2.2 for the recombination with pNIGEL7 and pNIGEL17 (Geldner et al., 2009) for YFP and mCherry fusion, respectively. To yield pUNI-VPS2.1 and pUNI-VPS2.2, the ORFs were PCR amplified using primers AK21 and AK22 and AK77 and AK78, respectively, and cloned between the SfiI and NotI (VPS2.1) and EcoRI and SalI (VPS2.3) sites of the pUNI51 vector (ABRC). 35Spro:VPS24.1-GFP was generated in 35S-GW-GFP (J. Parker, Cologne, Germany) by Gateway cloning (Invitrogen) with fragments amplified with AK90 and AK92 and Gateway adaptor primers. For the 35Spro:YFP-VPS2.3 construct, the VPS2.3 coding sequence was transferred by recombination from the G13604 (ABRC) clone to the pExtag-YFP-GW (J. Parker). To generate a VPS2.1pro:YFP-VPS2.1 construct, a VPS2.1 genomic fragments including 195 bp of the teminator region was amplified with AK32 and AK33 as well as Gateway adaptor primers and cloned by Gateway technology into pExtag-YFP-GW (J. Parker). YFP-VPS2.1 was then cut out and subcloned into the XhoI-NotI site of pGreen0229. A 1940-bp promoter region of VPS2.1 was amplified with AK38 and AK41 and first cloned into pCR 2.1-TOPO TA (Invitrogen) to be subcloned into the KpnI-XhoI site of pGreen0229 to obtain VPS2.1pro:YFP-VPS2.1. To generate RPS5apro:VPS2.1, the ORF of VPS2.1 was amplified with primers AK1 and AK2 and cloned into the XhoI site of pGreenII BAR RPS5a-tNOS (D. Weijers, Wageningen, The Netherlands).
For the SKD1 constructs, the ORF of SKD1 was amplified using primers EI189 and EI190 as well as Gateway adaptor primers and first cloned into the Gateway entry vector pDONR207 (Invitrogen). The EQ mutation was introduced using primers EI193 and EI194, and the wild type and EQ mutant ORFs were cloned into pJaWohl 2B HA (J. Parker) and pDEST17 (Invitrogen) to yield 35Spro:HA-SKD1 and His-SKD1, respectively. 35Spro:HA-AMSH3(AXA) (pJaWohl 2B HA) and 35Spro:YFP-AMSH3(AXA) (pExTag YFP) were created using a previously described Gateway entry clone (Isono et al., 2010). UBQ10pro:mCherry-AMSH3 was created by Cre recombinase-based cloning into the pNIGEL17 vector (Geldner et al., 2009). For AMSH3pro:YFP-AMSH3, the AMSH3 promoter and coding region were amplified using primers EI180 and EI181 and EI182 and EI141, respectively, and YFP was inserted between the promoter and the coding region.
Y2H Assays
A Y2H screen was performed as described previously (Schwechheimer and Deng, 2002). An Arabidopsis full-length cDNA library (Kim et al., 1997) was transformed into the yeast strain Y190 containing the pGBKT7-AMSH3(AXA) plasmid. Approximately 2 × 106 transformants were screened on synthetic complete (SC) medium lacking Leu (L), Trp (W), and His (H) and supplemented with 10 mM 3-amino-1,2,4-triazole. The prey plasmids were extracted, transformed into the XL1Blue Escherichia coli strain, and retransformed together with pGBKT7 or pGBKT7-AMSH3(AXA) into Y190 yeast cells for testing the growth and β-galactosidase activity. The positive candidates were sequenced with the GAL4-AD and ACT-rv primers.
pGBKT7 plasmids containing the ESCRT-III subunits VPS2.1, VPS2.2, VPS2.3, VPS20.1, VPS24.1, VPS46.1, VPS46.2, VPS60.1, and SNF7.1 were cotransformed with pGADT7-AMSH3 or pGADT7-AMSH3(AXA) plasmids into the yeast strain Y190. In addition, the truncated versions of AMSH3, GAD-AMSH3-ΔMIT, -ΔMPN, -MIT, -N102, and -N154 were cotransformed with pGBKT7, -VPS2.1, and -VPS24.1. Transformants were selected after 3 d on SC medium lacking Leu and Trp (-LW) at 30°C. To examine Y2H interactions, the transformants were grown in liquid culture to the late log phase, diluted to A600 = 1, and 10-fold serial dilutions were grown on solid SC medium lacking Leu and Trp (SC-LW) or Leu, Trp, and His (-LWH) with 5 mM 3-amino-1,2,4-triazole for 2 d at 30°C.
Protein Extraction, Immunoblotting, and Antibodies
Yeast total proteins were extracted as described previously (Kushnirov, 2000). SDS-PAGE, Coomassie Brilliant Blue staining, and immunoblotting were performed according to the standard protocol. Antibodies used for immunoblotting were anti-AMSH3 (Isono et al., 2010), anti-GST (GE Healthcare), anti-GAL4BD (Santa-Cruz), anti-CDC2(PSTAIRE) (Santa-Cruz), horseradish peroxidase–conjugated anti-HA (Sigma-Aldrich), and anti-His (Sigma-Aldrich).
Protein Purification, in Vitro Binding Assays, and DUB Assays
GST and GST-VPS2.1, GST-VPS2.2, GST-VPS2.3, and His-SKD1 were expressed in the E. coli Rosetta (DE3) strain (Merck Chemicals) and purified with Glutathione-Sepharose 4B beads (GE Healthcare). Proteins were eluted with 0.2 M reduced glutathione. His-SKD1 was purified using Ni Sepharose High Performance beads (GE Healthcare) and eluted with 250 mM imidazole. Eluted proteins were dialyzed against buffer A (50 mM Tris-HCl, 100 mM NaCl, and 10% glycerol, pH 7.5). The purification of AMSH3 has been previously described (Isono et al., 2010). Protein concentration was determined after SDS-PAGE and Coomassie Brilliant Blue staining using the Benchmark Protein Ladder (Invitrogen) as a standard and a LAS-4000 mini (FUJI Film) and the Multi Gauge (FUJI Film) software.
For the in vitro binding assay, Glutathione-Sepharose 4B beads (GE Healthcare) were saturated with GST or GST fusion proteins. Glutathione-Sepharose 4B beads with 1.5 μg of bound GST, GST-VPS2.1, GST-VPS2.2, or GST-VPS2.3 were then incubated with 6 pmol AMSH3 in 100 μL cold buffer A containing 0.2% Triton X-100 for 30 min at 4°C. The beads were then washed three times with cold buffer A containing 0.3% Triton X-100 and 1 mM dithiothreitol and three times more with cold buffer A. Bead-bound proteins were analyzed by immunoblotting. For the competition of SKD1 and AMSH3, the in vitro binding assay was performed with the addition of 6, 72, or 216 pmol of His-SKD1 to each reaction. DUB assays were performed as described previously (Isono et al., 2010), with 250 μg K48-linked UB chains (Biomol) and 1 pmol of AMSH3 preincubated with 2 pmol of GST, GST-VPS2.1, or GST-VPS24.1.
Protoplast Transformation
Protoplasts were isolated from Arabidopsis suspension-cultured cells (Col-0) 3 to 4 d after subculturing by incubation of 2 g of cell culture with 1% Cellulase R-10 (Yakult Pharmaceutical) and 0.25% Macerozyme R-10 (Yakult Pharmaceutical). Typically, 20 μg of plasmid DNA was transformed by polyethylene glycol–mediated transformation to the protoplasts and analyzed after 16 to 20 h of incubation.
Microscopy and FRET Measurements
GFP-, YFP-, and mCherry-fused proteins were analyzed with an FV-1000/IX81 confocal laser scanning microscope (Olympus) using a UPlanSApo ×60/1.20 (Olympus) objective. GFP, YFP, and mCherry were excited using the 488-, 515-, and 559-nm laser line, respectively. Images were obtained using the Fluoview software (Olympus) and processed using Photoshop CS3 (Adobe). FRET efficiency measurement between YFP-AMSH3 and mCherry-VPS2.1 was performed by the sensitized emission method and analyzed using the Fluoview software (Olympus). Acceptor spectral bleed-through and donor spectral bleed-through were determined by capturing images of cells expressing only YFP-AMSH3 or mCherry-VPS2.1. Average FRET efficiency was calculated for 86 regions (compartments) and 57 regions (cytosol) in a total of 16 cells.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: AMSH3 (AT4G16144), CDC2 (AT3G48750), SKD1 (AT2G27600), SNF7.1 (AT4G29160), VPS2.1 (AT2G06530), VPS2.2 (AT5G44560), VPS2.3 (AT1G03950), VPS20.1 (AT5G63880) VPS24.1 (AT5G22950), VPS24.2 (AT3G45000), VPS46.2 (AT1G73030), VPS60.1 (AT3G10640), ScVPS2p (P36108-1), DmVPS2 (Q8SXB1-1), MmVPS2 (Q9DB34-1), CHMP2A (O43633-1), CHMP2B (Q9UQN3-1), HsVPS4A (Q9UN37), HsVPS4B (O75351), MmVPS4A (Q8VEJ9), MmVPS4B (P46467), DmVPS4 (Q9Y162), ScVPS4p (P52917), and AtSKD1(At2g27600).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Verification of Y2H Construct Expression.
Supplemental Figure 2. Alignment of VPS2.1, VPS2.2, and VPS2.3 with Their Counterparts from Human, Mouse, Fruit Fly, and Yeast.
Supplemental Figure 3. Alignment of VPS24.1 and VPS24.2 with Their Counterparts from Human, Mouse, Fruit Fly, and Yeast.
Supplemental Figure 4. Alignment of MIT Domain of SKD1 with Their Counterparts from Human, Mouse, Fruit Fly, and Yeast.
Supplemental Figure 5. Localization of VPS Proteins and AMSH3.
Supplemental Figure 6. Localization of VPS2.1 Expressed under the Native Promoter.
Supplemental Figure 7. Complementation of the vps2.1 Mutant.
Supplemental Figure 8. Phenotype and Complementation of vps2.2 and vps2.3 Mutants.
Supplemental Figure 9. VPS2.1-Specific Antibody.
Supplemental Figure 10. DUB Assay with AMSH3 or GST.
Supplemental Table 1. Embryo Analysis of VPS2.1/vps2.1.
Supplemental Table 2. Complementation of vps2.1.
Supplemental Table 3. Primers Used in This Study.
Supplemental Methods. Genotyping-PCR, RT-PCR, Root Length Measurement, and Anti-VPS2.1 Antibody Production.
Supplemental Data Set 1. Alignment Used to Generate the Phylogeny Presented in Figure 2A.
Supplementary Material
Acknowledgments
We thank Caterina Brancato (University of Tübingen) for the Arabidopsis cell culture and technical advice, Niko Geldner (Lausanne University) for the pNIGEL vectors, and Dolf Weijers (Wageningen University) for the RPS5a promoter containing vector. We also thank Ramon Angel Torres-Ruiz (Technische Universität München), Caroline Höfle (Technische Universität München), and Takashi Ueda and Tomohiro Uemura (University of Tokyo) for technical advice. We thank Marie-Kristin Nagel and Cornelia Kolb (Technische Universität München) for critical reading of the manuscript. We also thank the GABI mutant seed collection and the Nottingham Arabidopsis Stock Centre for providing seeds and ABRC for providing cDNA clones. This work was supported by grants from the Deutsche Forschungsgemeinschaft [SCHW 751/7-1 (SPP1365/1) to C.S. and IS 221/2-2 (SPP1365/2) to E.I.] and by grants from the Austrian Science Fund (P16410-B12 and P17888-B14) to M.-T.H. E.I. was a recipient of a fellowship from the Japan Society for the Promotion of Science, and A.K. was a recipient of a scholarship from the Greek State Scholarship Foundation in frame of the ERASMUS program.
AUTHOR CONTRIBUTIONS
A.K., M.-T.H., C.S., and E.I designed and analyzed the experiments. A.K., F.A., N.S., S.N., and E.I. performed the experiments. A.K. and E.I. wrote the article.
References
- Agromayor M., Martin-Serrano J. (2006). Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo. J. Biol. Chem. 281: 23083–23091 [DOI] [PubMed] [Google Scholar]
- Babst M., Katzmann D.J., Estepa-Sabal E.J., Meerloo T., Emr S.D. (2002). Escrt-III: An endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3: 271–282 [DOI] [PubMed] [Google Scholar]
- Babst M., Sato T.K., Banta L.M., Emr S.D. (1997). Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16: 1820–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Wendland B., Estepa E.J., Emr S.D. (1998). The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17: 2982–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague M.J., Urbé S. (2006). Endocytosis: The DUB version. Trends Cell Biol. 16: 551–559 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Doelling J.H., Phillips A.R., Soyler-Ogretim G., Wise J., Chandler J., Callis J., Otegui M.S., Vierstra R.D. (2007). The ubiquitin-specific protease subfamily UBP3/UBP4 is essential for pollen development and transmission in Arabidopsis. Plant Physiol. 145: 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J.H., Yan N., Kurepa J., Walker J., Vierstra R.D. (2001). The ubiquitin-specific protease UBP14 is essential for early embryo development in Arabidopsis thaliana. Plant J. 27: 393–405 [DOI] [PubMed] [Google Scholar]
- Geldner N., Dénervaud-Tendon V., Hyman D.L., Mayer U., Stierhof Y.-D., Chory J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazi-Tabatabai S., Saksena S., Short J.M., Pobbati A.V., Veprintsev D.B., Crowther R.A., Emr S.D., Egelman E.H., Williams R.L. (2008). Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure 16: 1345–1356 [DOI] [PubMed] [Google Scholar]
- Haas T.J., Sliwinski M.K., Martínez D.E., Preuss M., Ebine K., Ueda T., Nielsen E., Odorizzi G., Otegui M.S. (2007). The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. Plant Cell 19: 1295–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J.H., Hanson P.I. (2010). Membrane budding and scission by the ESCRT machinery: It’s all in the neck. Nat. Rev. Mol. Cell Biol. 11: 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono E., Katsiarimpa A., Müller I.K., Anzenberger F., Stierhof Y.D., Geldner N., Chory J., Schwechheimer C. (2010). The deubiquitinating enzyme AMSH3 is required for intracellular trafficking and vacuole biogenesis in Arabidopsis thaliana. Plant Cell 22: 1826–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer C., Skalicky J.J., Morita E., De Domenico I., Ward D.M., Kaplan J., Sundquist W.I. (2008). Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev. Cell 15: 62–73 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Kim J., Harter K., Theologis A. (1997). Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 94: 11786–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.S., Kim J.A., Song H.K., Jeon H. (2006). STAM-AMSH interaction facilitates the deubiquitination activity in the C-terminal AMSH. Biochem. Biophys. Res. Commun. 351: 612–618 [DOI] [PubMed] [Google Scholar]
- Komander D., Clague M.J., Urbé S. (2009). Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10: 550–563 [DOI] [PubMed] [Google Scholar]
- Kushnirov V.V. (2000). Rapid and reliable protein extraction from yeast. Yeast 16: 857–860 [DOI] [PubMed] [Google Scholar]
- Kyuuma M., Kikuchi K., Kojima K., Sugawara Y., Sato M., Mano N., Goto J., Takeshita T., Yamamoto A., Sugamura K., Tanaka N. (2007). AMSH, an ESCRT-III associated enzyme, deubiquitinates cargo on MVB/late endosomes. Cell Struct. Funct. 31: 159–172 [DOI] [PubMed] [Google Scholar]
- Lata S., Schoehn G., Jain A., Pires R., Piehler J., Gottlinger H.G., Weissenhorn W. (2008). Helical structures of ESCRT-III are disassembled by VPS4. Science 321: 1354–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wang F., Zhang H., He H., Ma L., Deng X.W. (2008). Functional characterization of the Arabidopsis ubiquitin-specific protease gene family reveals specific role and redundancy of individual members in development. Plant J. 55: 844–856 [DOI] [PubMed] [Google Scholar]
- Luo M., Luo M.Z., Buzas D., Finnegan J., Helliwell C., Dennis E.S., Peacock W.J., Chaudhury A. (2008). UBIQUITIN-SPECIFIC PROTEASE 26 is required for seed development and the repression of PHERES1 in Arabidopsis. Genetics 180: 229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.M., Boucrot E., Villén J., Affar B., Gygi S.P., Göttlinger H.G., Kirchhausen T. (2007). Targeting of AMSH to endosomes is required for epidermal growth factor receptor degradation. J. Biol. Chem. 282: 9805–9812 [DOI] [PubMed] [Google Scholar]
- Maytal-Kivity V., Reis N., Hofmann K., Glickman M.H. (2002). MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 3: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J., Clague M.J., Urbé S. (2004). AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 166: 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J., Row P.E., Lorenzo O., Doherty M., Beynon R., Clague M.J., Urbé S. (2006). Activation of the endosome-associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body-sorting machinery. Curr. Biol. 16: 160–165 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Tanaka N., Kitamura N., Komada M. (2006). Clathrin anchors deubiquitinating enzymes, AMSH and AMSH-like protein, on early endosomes. Genes Cells 11: 593–606 [DOI] [PubMed] [Google Scholar]
- Nickerson D.P., West M., Odorizzi G. (2006). Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J. Cell Biol. 175: 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obita T., Saksena S., Ghazi-Tabatabai S., Gill D.J., Perisic O., Emr S.D., Williams R.L. (2007). Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 449: 735–739 [DOI] [PubMed] [Google Scholar]
- Raiborg C., Stenmark H. (2009). The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458: 445–452 [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu F.E., Ventii K.H., Wilkinson K.D. (2009). Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78: 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Saksena S., Wahlman J., Teis D., Johnson A.E., Emr S.D. (2009). Functional reconstitution of ESCRT-III assembly and disassembly. Cell 136: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellmann S., Pimpl P. (2009). Coats of endosomal protein sorting: Retromer and ESCRT. Curr. Opin. Plant Biol. 12: 670–676 [DOI] [PubMed] [Google Scholar]
- Schmitz R.J., Tamada Y., Doyle M.R., Zhang X., Amasino R.M. (2009). Histone H2B deubiquitination is required for transcriptional activation of FLOWERING LOCUS C and for proper control of flowering in Arabidopsis. Plant Physiol. 149: 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C., Deng X.W. (2002). Studying protein-protein interactions with the yeast two-hybid system. In Molecular Plant Biology: A Practical Approach, Gilmartin P.M., Bowler C., (Oxford, UK: Oxford University Press; ), pp. 173–198 [Google Scholar]
- Shahriari M., Keshavaiah C., Scheuring D., Sabovljevic A., Pimpl P., Häusler R.E., Hülskamp M., Schellmann S. (2010). The AAA-type ATPase AtSKD1 contributes to vacuolar maintenance of Arabidopsis thaliana. Plant J. 64: 71–85 [DOI] [PubMed] [Google Scholar]
- Shahriari M., Richter K., Keshavaiah C., Sabovljevic A., Huelskamp M., Schellmann S. (2011). The Arabidopsis ESCRT protein-protein interaction network. Plant Mol. Biol. 76: 85–96 [DOI] [PubMed] [Google Scholar]
- Shim S., Kimpler L.A., Hanson P.I. (2007). Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic 8: 1068–1079 [DOI] [PubMed] [Google Scholar]
- Spitzer C., Reyes F.C., Buono R., Sliwinski M.K., Haas T.J., Otegui M.S. (2009). The ESCRT-related CHMP1A and B proteins mediate multivesicular body sorting of auxin carriers in Arabidopsis and are required for plant development. Plant Cell 21: 749–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C., Schellmann S., Sabovljevic A., Shahriari M., Keshavaiah C., Bechtold N., Herzog M., Müller S., Hanisch F.G., Hülskamp M. (2006). The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development 133: 4679–4689 [DOI] [PubMed] [Google Scholar]
- Sridhar V.V., Kapoor A., Zhang K., Zhu J., Zhou T., Hasegawa P.M., Bressan R.A., Zhu J.K. (2007). Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 447: 735–738 [DOI] [PubMed] [Google Scholar]
- Stuchell-Brereton M.D., Skalicky J.J., Kieffer C., Karren M.A., Ghaffarian S., Sundquist W.I. (2007). ESCRT-III recognition by VPS4 ATPases. Nature 449: 740–744 [DOI] [PubMed] [Google Scholar]
- Tanaka N., Kaneko K., Asao H., Kasai H., Endo Y., Fujita T., Takeshita T., Sugamura K. (1999). Possible involvement of a novel STAM-associated molecule “AMSH” in intracellular signal transduction mediated by cytokines. J. Biol. Chem. 274: 19129–19135 [DOI] [PubMed] [Google Scholar]
- Teis D., Saksena S., Emr S.D. (2008). Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell 15: 578–589 [DOI] [PubMed] [Google Scholar]
- Tsang H.T., Connell J.W., Brown S.E., Thompson A., Reid E., Sanderson C.M. (2006). A systematic analysis of human CHMP protein interactions: Additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics 88: 333–346 [DOI] [PubMed] [Google Scholar]
- Williams R.L., Urbé S. (2007). The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 8: 355–368 [DOI] [PubMed] [Google Scholar]
- Winter V., Hauser M.T. (2006). Exploring the ESCRTing machinery in eukaryotes. Trends Plant Sci. 11: 115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N., Doelling J.H., Falbel T.G., Durski A.M., Vierstra R.D. (2000). The ubiquitin-specific protease family from Arabidopsis. AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol. 124: 1828–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.