Abstract
Small Ubiquitin-Like Modifiers (SUMO) are posttranslational modifiers that regulate target protein activity in diverse ways. The most common group of SUMO substrates is transcription factors, whose transcriptional activity can be altered positively or negatively as a result of SUMOylation. DLX3 is a homeodomain transcription factor involved in placental development, in the differentiation of structures involving epithelial-mesenchymal interactions, such as hair, teeth and nails, and in bone mineralization. We identified two potential SUMOylation sites in the N-terminal domain of DLX3 at positions K83 and K112. Among the six members of the Distal-less family, DLX3 is the only member containing these sites, which are highly conserved among vertebrates. Co-expression experiments demonstrated that DLX3 can be SUMOylated by SUMO1. Site-directed mutagenesis of lysines 83 and 112 to arginines (K83R and K112R) demonstrated that only K112 is involved in SUMOylation. Immunocytochemical analysis showed that SUMOylation does not affect DLX3 subcellular localization. Moreover, using electrophoresis mobility shift assay, we found that DLX3 is still able to bind DNA when SUMOylated. Using luciferase reporter assays, we showed that DLX3K112R exhibits a significantly lower transcriptional activity compared to DLX3WT, suggesting that SUMOylation has a positive effect on DLX3 activity. We identified a new level of regulation in the activity of DLX3 that may play a crucial role in the regulation of hair, teeth and bone development.
Keywords: DLX3, SUMOylation, SUMO1, Nuclear localization, DNA binding, Transcriptional activity
INTRODUCTION
Post-translational modifications occur to a majority of proteins to regulate their activity as a result of a particular stimulus. The small ubiquitin-like modifier, SUMO, is a posttranslational modifier with distinct effects on a wide variety of targets. It can increase protein stability [Desterro et al., 1998], influence interactions between distinct proteins [Seeler and Dejean, 2001], change subcellular localization [Morita et al., 2005; Wilson and Rangasamy, 2001], and affect nuclear trafficking [Pichler and Melchior, 2002]. In mammalian cells, four SUMO isoforms (SUMO1-4) have been identified. The human SUMO1 gene encodes a 101-amino acid polypeptide related to ubiquitin. SUMO1 is known to share ~50% sequence identity with SUMO2/3, whereas Sumo2 and Sumo3 share 87% sequence identity. While SUMO4 expression appears to be tissue-specific, SUMO1-3 are widely expressed and show distinct substrate and de-sumoylating protease specificity. Albeit such differences, all SUMOs undergo a series of enzymatic reactions at the C terminus to become covalently bonded to their targets.
Thus far, the most common group of SUMO substrates is transcription factors, whose transcriptional activity is altered as a result of SUMOylation. Previous studies have revealed both positive and negative regulation of transcription factor activity. Proteins whose transcriptional response is modulated as a result of SUMOylation include p53 [Gostissa et al., 1999], Dorsal [Bhaskar et al., 2002], HSF2 [Goodson et al., 2001; Tateishi et al., 2009], c-Jun [Muller et al., 2000], Androgen receptor [Poukka et al., 2000], Sox2 [Tsuruzoe et al., 2006], and several members of the DExD/H box RNA helicases family, i.e. Dhx5 and Dhx20 [Fuller-Pace et al., 2007; Jacobs et al., 2007].
Here we investigate the role of SUMOylation on DLX3, which belongs to the superfamily of homeodomain transcription factors known to be widely involved in the patterning of the developing embryo. In mouse and human, there are six DLX genes organized into three pairs of inverted, convergently transcribed genes (DLX1-2, DLX3-4, and DLX5-6) [Morasso and Radoja, 2005]. DLX3 is linked to DLX4 on chromosome 11 in mouse and on chromosome 17 in humans. Furthermore, DLX3 is expressed in the placenta early during embryonic development [Morasso et al., 1999], while it is later found in skin as well as in structures involving epithelial-mesenchymal interactions, such as the middle and inner ear, teeth and hair follicles [Robinson and Mahon, 1994]. Using a conditional knockout approach, we showed that Dlx3 plays a crucial role in hair development [Hwang et al., 2008]. There is also clinical evidence that DLX3 plays a significant role in the patterning of hair, teeth, and bone. In fact, a 4-G deletion occurring three base pairs downstream of the DLX3 homeodomain leads to a frameshifted C-terminal domain whose sequence differs completely from that of wildtype DLX3. This truncated protein is manifested in an ectodermal dysplasia called Tricho-Dento-Osseous syndrome, an automosomal dominant disorder characterized by defects in hair (kinky), teeth (enamal hypoplasia and taurodontism), and bone (increased thickness and density of craniofacial bone) [Price et al., 1998].
Here, we identify and characterize for the first time a SUMOylation site in DLX3. We show that SUMOylation does not affect DLX3 subcellular localization and DNA binding activity but potentially promotes DLX3 transcriptional activity.
MATERIALS AND METHODS
Plasmids
The bidirectional vector pBi4 was used to simultaneously express the reporter protein EGFP with V5DLX3 (pBi-V5DLX3/GFP), under control of a unique Tetracycline Responsive Element (TRE). Site-Directed Mutagenesis was then utilized to mutate the two lysines (83 and 112) that are potentially involved in DLX3 SUMOylation into arginines. These mutants were obtained by introducing A to G point mutations in DLX3 cDNA (A248G and A335G, respectively). At position 248, A was mutated into G using the following primers: Sense-GCTTACTCGCCCAGGTCGGAATATACC; Antisense-GGTATATTCCGACCTGGGCGAGTAAGC (mutated base in bold). The resulting construct was named pBi-V5DLX3K83R. At position 112, A was mutated into G using the following primers: Sense-CCAGTGTCGGTGAGAGAGGAGCCGGAA; Antisense-TTCCGGCTCCTCTCTCACCGACACTGG. The resulting construct was named pBi-V5DLX3K112R. The double mutant was also generated and the construct was named pBi-V5DLX32K.
SUMO1 and SUMO1-ΔGG, a mutated form of SUMO1 lacking the C-terminal double glycine which forms an isopeptide bond with the target protein, were tagged at the N-terminus with a His tag and a c-Myc tag, and cloned into pTRE2 for tetracycline inducible expression [Li et al., 2006].
Cell culture and transfections
Saos2 human osteosarcoma cells expressing the tetracycline inducible transactivator rTA (Saos2-TetOFF, Clontech) were grown in DMEM (10% fetal bovine serum, 1% penicillin/streptomycin, and 1 ug/ml G418). For transfections, the cells were grown to at least 70% confluence. 2 × 106 cells were used per transfection with each construct (Amaxa Nucleofactor).
Ni column pull-down
48 hours after transfection with DNA, Saos2-TetOFF cells were lysed in 1ml Buffer A (100mM NaH2PO4, 10mM Tris-Cl, 6M GuHCl, pH=8.0) supplemented with 10mM N-ethyl maleimide and 7% β-mercaptoethanol. The lysates were then sonicated, spun down for 15 min., and incubated with 40μl of Ni-NTA agarose beads (Qiagen) for 1 hour. The beads were then washed with Buffer A once, Buffer B (100mM NaH2PO4, 10mM Tris-Cl, 8M urea, pH=8.0) twice, and Buffer C (100mM NaH2PO4, 10mM Tris-Cl, 8M urea, pH=6.3) twice. 50mM TRIS was used to wash beads, which were then resuspended in 2X NuPAGE LDS-sample buffer and 250mM imidazole.
Cell lysis and Western blot analysis
48 hours after transfection, GFP was visualized and the cultured cells were rinsed with phosphate-buffered saline, scraped with lysis buffer (50mM Tris-HCl pH 7.5, 1mM EDTA, 200mM NaCl, 0.1% Nonidet P-40,) supplemented with a protease inhibitor cocktail (Complete Mini, EDTA-free, Roche), 15mM N-ethyl maleimide, 20mM iodoacetamide, and sonicated. 4X NuPAGE® LDS Sample Buffer (Invitrogen) was added to these lysates after normalization of protein concentrations. These protein extracts were run on 4-12% Bis-Tris gels and MOPS SDS running buffer. Proteins were transferred onto PVDF membranes and blocked in 5% nonfat powdered milk in TBS/Tween at room temperature for 1 hour. The blots were probed with primary antibody diluted in 5% nonfat powdered milk in TBS/Tween and then with secondary antibody diluted in TBS/Tween, both at room temperature for one hour. Primary antibodies used were anti-V5 (1:2000, Serotec), anti-cMyc (1:1000, Santa Cruz). Secondary antibody used were goat anti-mouse horseradish peroxidase (1:3000, Bio-Rad). After application of each antibody, the blots were rinsed three times with TBS/Tween under similar conditions. The blots were developed by ECL (Pierce).
Immunocytochemistry
Transfected cells were seeded on glass coverslips coated with 0.1% gelatin. 48 hours after transfection, cells were washed three times in PBS and fixed with 4% paraformaldehyde in PBS for 15 minutes at room temperature. A 5 minute incubation in 0.2% Triton in PBS was used to permeabilize the cells before blocking unspecific sites using 3% BSA in PBS for 1 hour. Primary antibodies diluted in blocking solution were applied for 1 hour. Primary antibodies used: anti-V5 (1:100, Serotec), anti-cMyc (1:100, Santa Cruz). Secondary antibodies diluted in blocking solution were applied for 30 minutes. Secondary antibodies used: Alexa Fluor® 543 goat anti-mouse IgG and Alexa Fluor® 488-conjugated anti-rabbit IgG (1:400, Invitrogen). Nuclei were stained using DAPI and coverslips were mounted on glass slides using Mowiol (Calbiochem). Images were acquired using a Zeiss 510 META confocal microscope.
Electrophoresis Mobility Shift Assays
Nuclear extracts were prepared using a Nuclear Extract Kit (Active Motif). Recombinant SUMO-DLX3 fusion protein was produced using the SUMOpro kit (LifeSensors). A probe containing the DLX3 consensus binding site (GGGGGATAATTGCTGG), was radiolabeled using the High Prime DNA Labeling Kit (Roche) and [γ-32P]dCTP. Nuclear extracts or recombinant proteins were pre-incubated in 1X gel shift binding buffer (Promega) for 15 minutes at 4°C, with an excess of unlabeled probe for competition assays or with appropriate antibody (anti-DLX3 or anti-cMyc) for supershift assays. After this pre-incubation, each sample was supplemented with 5×104 DPM of radiolabeled probe and incubated for 30 minutes at 4°C. The binding reactions were resolved on 6% DNA retardation gels (Invitrogen). The gels were dried and DNA-protein complexes were visualized by autoradiography.
Luciferase reporter assay
To determine the transcriptional activity, a synthetic oligonucleotide containing three tandem copies of the DLX3 responsive element (DRE; GCGATAATTGCGGCGATAATTGCGGCGATAATTGCG) followed by the HSV thymidine kinase proximal promoter region was cloned into the pGL3-promoter vector (pGL3-3XDRE) driving a Firefly luciferase reporter cassette [Duverger et al., 2008]. Saos2-TetOff cells were transiently transfected with pBi-V5DLX3WT or pBi-V5DLX3K112R constructs, together with pTRE2-SUMO1, pGL3-3xDRE and the pRL-TK vectors (Renilla luciferase used for normalization). 24 hours after transfection, relative luciferase activity was measured using the Dual-Luciferase® Reporter Assay System (Promega). Statistical analysis was performed using Prism 5.02.
RESULTS
DLX3 is the only member of the DLX family containing putative SUMOylation sites, one of which is conserved among several vertebrates
The DLX3 amino acid sequence was analyzed and two potential SUMOylation sites ψKXE, where ψ represents a hydrophobic amino acid, X represents any amino acid, were discovered at lysine 83 (pkse) and lysine 112 (vkee) (Fig. 1A). Because proline is less hydrophobic than valine, we hypothesized that lysine 83 is less susceptible to SUMOylation than lysine 112. The DLX3 amino acid sequence in several vertebrates was aligned, and K112 was found to be conserved among human, mouse, chicken, zebrafish, and xenopus (Fig. 1B). An amino acid sequence comparison was made between all human DLX proteins. DLX3 is the only member of the DLX family containing these two potential SUMOylation sites (Fig. 1C). This specificity and the conservation among vertebrates led us to further investigate the effect of SUMOylation on DLX3.
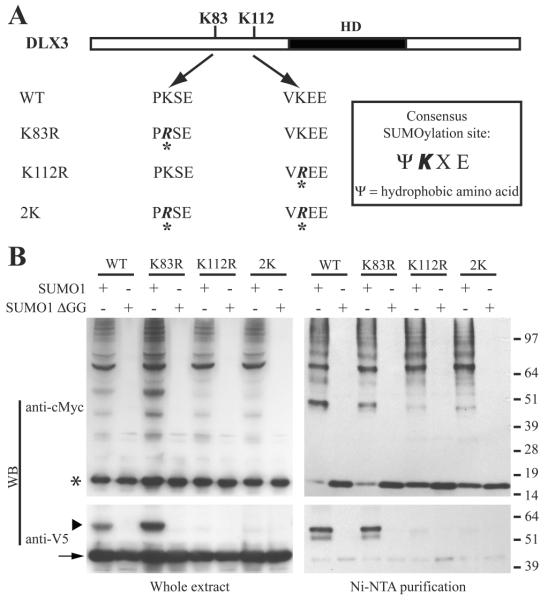
Figure 1. Potential SUMOylation sites in the DLX3 protein sequence.
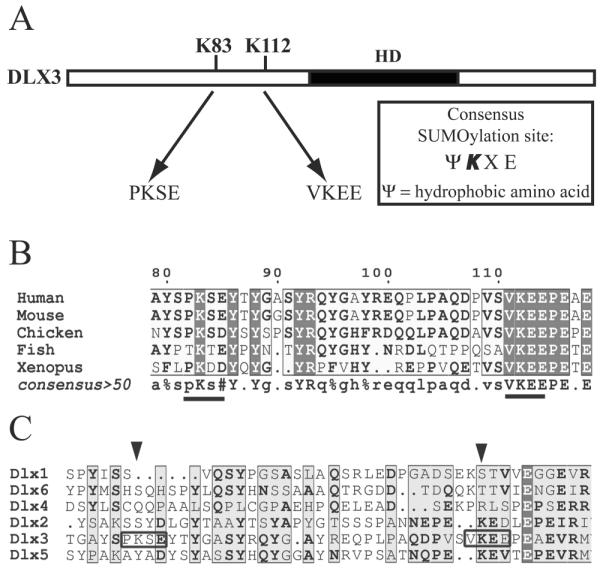
A) Diagram showing the position of the two potential SUMOylation sites in DLX3. B) Alignment of the DLX3 sequence from several Vertebrates showing a conservation of the potential SUMOylation site at position 112. C) Alignment of the sequence of all six human DLX proteins showing that potential SUMOylation sites are present only in DLX3.
DLX3 is SUMOylated by SUMO1
To determine the functionality of the two potential SUMOylation sites in DLX3, we co-expressed V5DLX3 (DLX3 tagged with a V5 epitope) with wildtype SUMO1 in Saos2-TetOFF cells. As a control, we also co-expressed V5DLX3 with a mutated form of SUMO1 lacking the C-terminal double glycine which forms an isopeptide bond with the target protein (SUMO1-ΔGG). SUMO1 and SUMO1-ΔGG are tagged with both a His tag and a c-Myc tag. Western blot was performed to analyze the size pattern of c-Myc tagged and V5-tagged proteins in these cells (Fig. 2A). Results monitored with anti-cMyc showed a low molecular weight band, around 15 kDa, corresponding to the monomer of SUMO1 or SUMO1-ΔGG, as well as a smear corresponding to all endogenous proteins SUMOylated by SUMO1, but not SUMO1-ΔGG (Fig. 2A, left panel). The anti-V5 blot revealed two major bands for V5DLX3 when co-expressed with SUMO1: the lower band corresponding to the normal size of V5DLX3, and the upper corresponding to a V5DLX3 derivative with additional 15-20 kDa (Fig. 2A, right panel). This upper band was not detected with SUMO1-ΔGG. These observations suggest that V5DLX3 is SUMOylated by SUMO1. To confirm that the upper complex detected is indeed SUMOylated V5DLX3, we performed the same western blot analysis after purifying the protein extracts with a Ni-column (Ni-NTA, binding His-tagged SUMO1) to pull-down all SUMOylated proteins. Using this approach, we detected SUMOylated V5DLX3 among the proteins purified on the Ni-column (Fig. 2B). These results demonstrate that V5DLX3 is SUMOylated by SUMO1. Moreover, the fact that we detected only one band for SUMOylated V5DLX3 suggests that only one lysine is involved in DLX3 SUMOylation.
Figure 2. Detection of DLX3 SUMOylation by SUMO1.
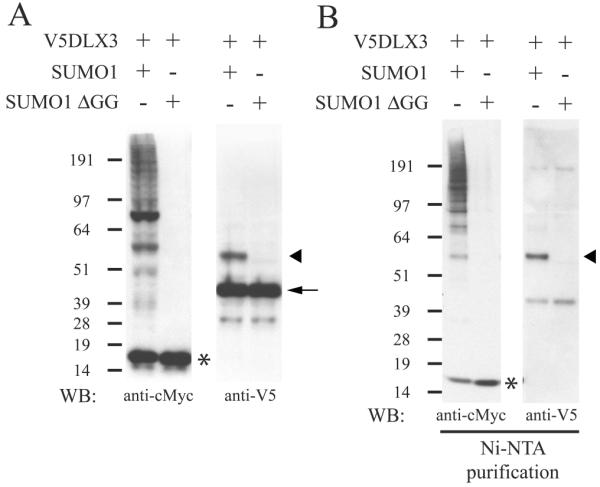
A) Western blot performed on cells expressing V5DLX3 and SUMO1, or V5DLX3 and SUMO1-ΔGG. SUMO1 and SUMO1- ΔGG were tagged with both a cMyc tag and a His tag. Anti-cMyc was used to detect all SUMO1-conjugated proteins and SUMO1-ΔGG. Anti-V5 was used to detect V5DLX3 and SUMOylated V5DLX3. B) Same Western blot as in A performed after purification of the protein extracts on Ni-NTA beads (pulls down all SUMO1-conjugated proteins and SUMO1-ΔGG that are His-tagged). Arrow: V5DLX3; arrowhead: SUMOylated V5DLX3; asterisk: monomeric SUMO1.
DLX3 is SUMOylated by SUMO1 on K112, but not K83
To test which of the two potential SUMOylation sites in DLX3 is actually involved in SUMOylation, we mutated the lysine residues at positions 83 and 112 into arginines (Fig. 3A). Thus, we generated two single mutants (DLX3K83R and DLX3K112R) as well as a double mutant in which the two lysine residues were mutated (DLX32K). In Saos2-TetOff cells, we co-expressed SUMO1 or SUMO1-ΔGG with DLX3WT, DLX3K83R, DLX3K112R or DLX32K, respectively. Western blot analysis was performed using anti-cMyc and anti-V5 antibodies, both on whole extracts and on Ni-column purified protein fraction (Fig. 3B). This assay revealed that mutation in position 83 does not preclude SUMOylation of V5DLX3, since a band corresponding to SUMOylated DLX3 appears for both DLX3WT and DLX3K83R. This band, however, was absent for DLX3K112R. As predicted, the double mutant DLX32K is not SUMOylated. These observations demonstrate that lysine K112 is the only SUMOylation site in DLX3.
Figure 3. Identification of the lysine involved in DLX3 SUMOylation.
A) Diagram displaying the different mutants that were produced to determine which is the lysine involved in DLX3 SUMOylation: mutation of lysine 83 into an arginine (K83R), mutation of lysine 112 into an arginine (K112R), and mutation of both lysines into arginines (2K). B) Same approach as in Fig. 2 was used to analyze the SUMOylation capacity of DLX3 and the three mutants described in A. Arrow: V5DLX3; arrowhead: SUMOylated V5DLX3; asterisk: monomeric SUMO1.
SUMOylation does not affect DLX3 subcellular localization
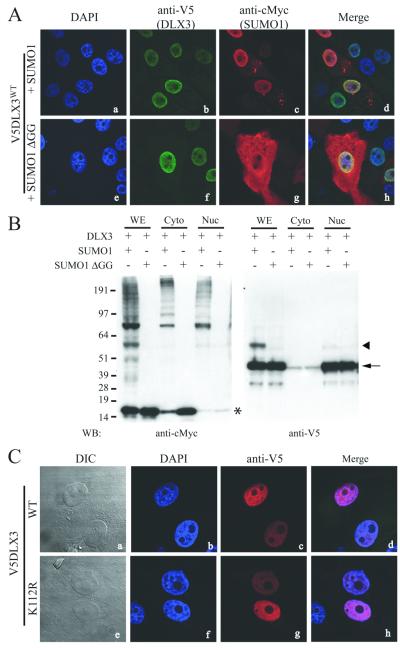
In order to assess the effect of SUMOylation on DLX3 subcellular localization, we performed immunohistochemical analysis on Saos2-TetOFF cells co-transfected with V5DLX3 and SUMO1 or SUMO1-ΔGG (Fig. 4A). Immunostaining using anti-cMyc antibody showed that SUMO1 was present in both the cytoplasm and the nucleus: the nuclear expression was rather diffuse, however, it tended to be stronger at the nuclear periphery, while in the cytoplasm it formed dense aggregates (Fig 4A, c). SUMO1-ΔGG exhibited a completely different distribution pattern: it was distributed throughout the cytoplasm in a filament-like pattern and was also present in the nucleus (Fig 4A, g). The distribution pattern of V5DLX3 in the nucleus appeared the same in the presence of wild-type and mutant SUMO1 (Fig. 4A, b and f). This observation suggests that SUMO1 does not dramatically affect DLX3 subcellular localization. However, we observed a partial co-localization between DLX3 and SUMO1 at the periphery of the nucleus (Fig. 4A, d). We used cell fractionation and western blot analysis to corroborate our immunohistochemical observations (Fig 4B). The anti-cMyc blot revealed that the cytoplasm contained both monomeric SUMO1 and SUMO1 conjugated with target proteins, while in the nucleus SUMO1 was essentially present in its conjugated form. SUMO1-ΔGG, that can only be monomeric, exhibited a much stronger expression in the cytoplasm than in the nucleus. The anti-V5 blot showed that both V5DLX3 and SUMOylated V5DLX3 were accumulated almost exclusively in the nuclear fraction. These data confirm that SUMO1 does not dramatically affect DLX3 subcellular localization. We then investigated whether DLX3 subcellular localization was affected by its inability to be SUMOylated. To address this question, immunohistochemical analysis was carried out with Saos2-TetOff cells transfected with V5DLX3WT or V5DLX3K112R. As shown in Figure 4C, both wild type and K112R mutant were exclusively located in the nucleus. Thus, preventing DLX3 from being SUMOylated does not affect its subcellular localization. Taken together, these data suggest that SUMOylation does not play a significant role in determining the distribution pattern of DLX3 in cells.
Figure 4. Effect of SUMOylation on DLX3 subcellular localization.
A) Immunohistochemical analysis performed on Saos2-TetOff cells expressing V5DLX3 and SUMO1, or V5DLX3 and SUMO1-ΔGG. Anti-V5 was used to detect V5DLX3 and SUMOylated V5DLX3 (b and f). Anti-cMyc was used to detect SUMO1, all SUMO1-conjugated proteins and SUMO1-ΔGG (c and g). DAPI was used to stain nuclei (a and e). Merge images are shown in d and h. B) Western blot on fractionated protein extracts (cytoplasm/nucleus) from Saos2-TetOff cells expressing V5DLX3 and SUMO1, or with V5DLX3 and SUMO1-ΔGG. Anti-cMyc was used to detect all SUMO1-conjugated proteins and SUMO1-ΔGG. Anti-V5 was used to detect V5DLX3 and SUMOylated V5DLX3. WE: whole extract; Cyto: cytoplasmic fraction; Nuc: nuclear fraction; Arrow: V5DLX3; arrowhead: SUMOylated V5DLX3; asterisk: monomeric SUMO1. C) Immunohistochemical analysis performed on Saos2-TetOff cells expressing V5DLX3WT or V5DLX3K112R. Anti-V5 was used to detect V5DLX3WT and V5DLX3K112R distribution (c and g). DIC: differential interference contrast (a and e). DAPI was used to stain nuclei (b and f). Merge images are shown in d and h.
SUMOylated DLX3 is able to bind DNA
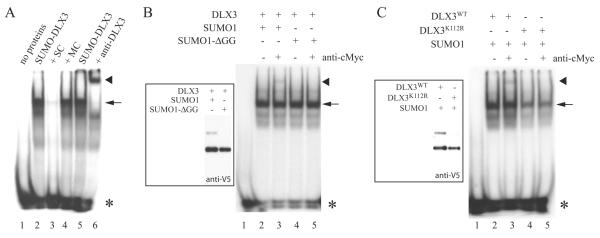
To determine the role of SUMOylation on DLX3 function, we next explored the effect of SUMOylation on DLX3 binding activity. We first asked if SUMOylated DLX3 was able to bind DNA. To address this question, we first generated a recombinant SUMO-DLX3 fusion protein and tested its ability to bind to the DLX3 consensus binding site in an Electrophoresis Mobility Shift Assay (EMSA), using a probe containing a DLX3 binding site. A protein-DNA complex was formed, that could be competed using an excess of non-radioactive probe (self competitor), but not using an excess of a mutated non-radioactive probe (mutant competitor) (Fig. 5A, lanes 2-4). To confirm that this protein-DNA complex contained SUMO-DLX3, we performed a supershift assay using anti-DLX3 antibody and were able to detect a protein-DNA-antibody complex (Fig. 5A, compare lanes 5 and 6). Even though the SUMO-DLX3 fusion protein does not perfectly mimic the tertiary structure of SUMOylated DLX3, this strategy is commonly used in the field [Ouyang et al., 2009] and gives a preliminary suggestion that having SUMO bound to DLX3 does not seem to affect its ability to bind DNA. To test this hypothesis in a more physiologically meaningful context, we performed EMSA using nuclear extracts from cells expressing V5DLX3 with SUMO1 or SUMO1-ΔGG (Fig. 5B, inset). In both cases (wild-type and mutant SUMO1), we could detect a protein-DNA complex formed between DLX3 and the probe (Fig. 5B, arrow). In order to detect if SUMOylated DLX3 is involved in a complex with the probe, we performed a supershift assays using anti-cMyc antibody (recognizing SUMO1 and SUMO1-ΔGG). As shown in Figure 5B, a partial supershift could be detected in the presence of anti-cMyc when DLX3 was expressed with SUMO1, but not with SUMO1-ΔGG (Fig. 5B, arrowhead, compare lanes 3 and 5). We performed a similar experiment in which we prepared nuclear extracts from cells expressing SUMO1 with DLX3WT or DLX3K112R (Fig. 5C, inset), and performed an EMSA as described above. As expected, using anti-cMyc antibody, we could detect a partial supershift for DLX3WT but not for DLX3K112R (Fig. 5C, compare lanes 3 and 5). These observations demonstrate that when SUMO1 is bound to DLX3, DLX3 is still able to bind to its consensus binding site. We also showed that DLX3K112R is able to bind DNA (Fig 5C, arrowhead, lanes 4 and 5), demonstrating that preventing DLX3 from being SUMOylated does not affect its ability to bind DNA.
Figure 5. Effect of SUMOylation on DLX3 DNA binding activity.
A) Electrophoresis Mobility Shift Assay (EMSA) using recombinant SUMO-DLX3 fusion protein and a radiolabelled probe containing a consensus DLX3 binding site (lanes 2 and 5). An excess of non-radioactive probe was used to compete with the radioactive DLX3 consensus probe (SC: self competitor; lane 3). An excess of a mutated non-radioactive probe was used as a control (MC: mutant competitor; lane 4). Anti-DLX3 antibody was used to supershift protein-DNA complexes involving SUMO-DLX3 (lane 6). B) EMSA using nuclear extracts from cells expressing V5DLX3 and SUMO1 (lanes 2 and 3), or V5DLX3 and SUMO1-ΔGG (lanes 4 and 5). Anti-cMyc antibody was used to supershift protein-DNA complexes involving SUMOylated DLX3 (lanes 3 and 5). The inset on the left shows a western blot performed on the nuclear extracts using anti-V5 antibody. C) EMSA using nuclear extracts from cells expressing V5DLX3WT and SUMO1 (lanes 2 and 3), or V5DLX3K112R and SUMO1 (lanes 4 and 5). Anti-cMyc antibody was used to supershift protein-DNA complexes involving SUMOylated DLX3 (lanes 3 and 5). The inset on the left shows a western blot performed on the nuclear extracts using anti-V5 antibody. Asterisk: free probe; arrow: protein-DNA complex; arrowhead: antibody-protein-DNA complex.
SUMOylation has a positive effect on DLX3 transcriptional activity
In order to test the effect of SUMOylation on DLX3 transcriptional activity, we performed luciferase reporter assays. The first question we asked was what is the effect of overexpressing SUMO1 on DLX3 transcriptional activity. To address this question, we transfected Saos2-TetOFF cells with pCMV-V5DLX3 (constitutive expression of V5DLX3), pTRE2-SUMO1 (inducible expression of SUMO1), pGL3-3xDRE (3 copies of the DLX3 responsive element upstream of the Firefly luciferase reporter gene), and pRL-TK (constitutive expression of Renilla luciferase gene used for normalization). After transfection, the cells were grown for 24 hours with or without doxycycline, and assayed for Firefly and Renilla luciferase activity. Using this strategy, we found that DLX3 transcriptional activity was slightly higher in the presence of SUMO1 (−Dox) than in its absence (+Dox), but this difference was not statistically significant (data not shown). Considering that this moderate effect of SUMO1 overexpression on DLX3 transcriptional activity could be due to the effect of SUMO1 on other endogenous targets that may interact with DLX3, we decided to focus on the comparison between DLX3WT and DLX3K112R. In this next assay, Saos2-TetOFF cells were transfected with pBi-GFP, pBi-V5DLX3WT/GFP or pBi-V5DLX3K112R/GFP, together with pTRE2-SUMO1, pGL3-3xDRE, and pRL-TK. After transfection, the cells were grown for 24 hours with or without doxycycline, and relative luciferase activity (Firefly/Renilla) was measured (Fig. 6). As expected, in the absence of doxycycline, we detected a significant increase in relative luciferase activity in the presence of DLX3WT compared to the GFP control. The transcriptional activity measured for DLX3K112R was significantly lower than that of DLX3WT, demonstrating that preventing DLX3 SUMOylation in a context where SUMO1 is active significantly reduces its transcriptional activity. In the presence of doxycycline that shuts down the expression of the transgenes in Saos2-TetOff cells, the relative luciferase activity was reduced to a basal level in all conditions, confirming that the transcriptional activities measured are related to the transgenes expression. Taken together, these data suggest that SUMOylation potentially promotes DLX3 transcriptional activity.
Figure 6. Effect of SUMOylation on DLX3 transcriptional activity.
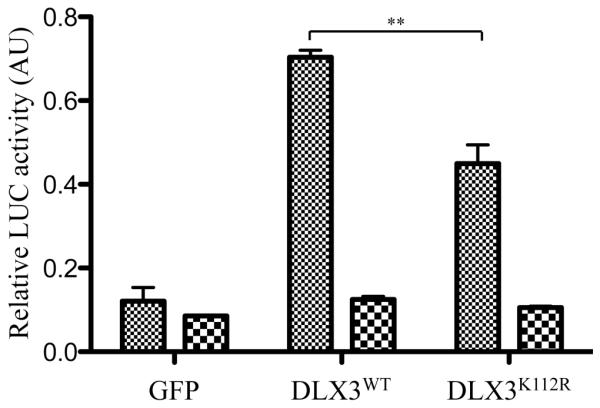
Luciferase reporter assay comparing the transcriptional activity of DLX3WT and DLX3K112R. Saos2-TetOff cells were transfected with pBi-GFP, pBi-V5DLX3WT/GFP or pBi-V5DLX3K112R/GFP, together with pTRE2-SUMO1, the reporter construct pGL3-3xDRE (Firefly luciferase under control of three copies of a DLX3 responsive element) and the normalization vector pRL-TK (Renilla luciferase under control of a TK promoter). Transfected cells were grown for 24 hours in the absence (transgenes ON) or presence (transgenes OFF) of doxycycline and relative luciferase activity (Firefly/Renilla) was measured. In the presence of SUMO1, DLX3K112R transcriptional activity is significantly lower than DLX3WT transcriptional activity (t-Test, p=0.008).
DISCUSSION
In the last decade, the number of proteins identified as being post-transcriptionally modified by SUMOylation has shown an exponential rise. Although these modifiers are related to the ubiquitin family, they are not associated to protein degradation and exhibit a large variety of effects on their target proteins. The knockout of SUMO1 is lethal, and SUMO1 haploinsufficiency has been associated with cleft lip and palate in human [Alkuraya et al., 2006]. Even though these observations do not determine specific targets of SUMOylation and how their function is altered in vivo, they demonstrate that SUMOylation plays an essential role in embryonic development. Altered SUMOylation of p63α has been shown to contribute to the Split-Hand/Split-Foot malformation phenotype [Huang et al., 2004], which further supports the importance of SUMOylation of specific proteins in the regulation of developmental processes. The reports published so far on SUMOylated proteins show that it is difficult to predict the effect of SUMOylation on the activity of a protein. A large majority of the proteins that have been identified as targets of SUMO are transcription factors. In this study, we identified for the first time a SUMOylation site in the homeodomain transcription factor DLX3.
We identified lysine K112 as the unique SUMOylation site in DLX3. This lysine branches SUMO in the N-terminal domain of DLX3, less that 20 amino acids upstream of its nuclear localization signal (NLS) located right before the homeodomain [Bryan and Morasso, 2000]. In spite of the proximity between lysine K112 and the NLS, SUMOylation does not affect DLX3 nuclear localization. Recent reports showed that it is quite frequent to find a SUMOlation site near a nuclear export signal (NES). For example, KLF-5 SUMOylation favors its retention within the nucleus by inhibiting its NES and thus preventing its translocation to the cytoplasm [Du et al., 2008]. SUMO is also involved in the subnuclear localization of protein complexes, particularly in controlling the assembly of PML-nuclear bodies [Heun, 2007]. We did not observe any obvious change in the distribution of DLX3 within the nucleus, neither by inducing nor inhibiting SUMOylation. However, we did observe a partial co-localization between DLX3 and SUMO1 at the periphery of the nucleus.
DNA binding can be affected by SUMOylation. Although the first report of the SUMOylation of the Heat Shock Factor HSF2 suggested a positive effect of SUMO1 on HSF2 DNA binding activity [Goodson et al., 2001], a more recent study showed that HSF2 is unable to bind DNA when bound by SUMO [Tateishi et al., 2009]. The same effect was observed for Sox2 [Tsuruzoe et al., 2006]. Here we show that DLX3 is still able to bind DNA when it is bound to SUMO, and that mutating lysine K112 does not prevent DNA binding, suggesting that SUMOylation does not dramatically affect the ability of DLX3 to bind DNA.
SUMOylation is a very dynamic, reversible and unstable process, which makes it difficult to analyze in a physiological context. Although there has been numerous reports on the SUMOylation of transcription factors in the last decade, it is still unclear how the SUMOylation of a very small proportion of a transcription factor can dramatically affect its transcriptional activity. Although SUMOylation has been associated to both activation and repression of transcriptional activities, a large majority of the reports published so far have shown a repressor effect of SUMO on transcriptional activity [Yang et al., 2003]. Among the transcription factors whose activity is promoted by SUMOylation are two factors involved in muscle differentiation: myocardin and nkx2.5 [Wang et al., 2007; Wang et al., 2008]. Here, we show that, in a context where SUMO1 activity is high, the transcriptional activity of a mutant of DLX3 that cannot be SUMOylated is significantly lower relative to that of wild-type DLX3. This suggests that SUMOylation has a positive effect on DLX3 transcriptional activity. The mechanism responsible for this effect may involve interactions with transactivation partners that remain to be identified. Among the potential candidates are other members of the DLX family, as well as members of the MSX family, since members of these two families have been shown to form homodimers and heterodimers [Zhang et al., 1997]. The interaction and interplay between DLX3, DLX5 and MSX2 has been shown to play an essential role in the regulation of Runx2 and osteocalcin expression during osteoblast differentiation [Hassan et al., 2004; Hassan et al., 2006]. Future studies should investigate the possible involvement of SUMOylation in this process. DLX3 is also involved in ectodermal appendages development such as hair and teeth, where interacting partners potentially have essential regulatory roles as well. The identification of such factors will be an essential pre-requisite to the analysis of the impact of DLX3 SUMOylation in these tissues.
Interestingly, the SUMOylation site in DLX3 is conserved among vertebrates and none of the five other members of the Distal-less family contains a SUMOylation site. This high specificity suggests that SUMOylation may play a major role in the regulation of DLX3 activity during embryonic development. DLX3 also distinguishes itself from other members of the Distal-less family by its distribution and function during embryonic development. First, it is the only Distal-less member that has not been detected in the mammalian central nervous system. Second, mice that are null for Dlx3 die at E9.5 from placental defects [Morasso et al., 1999], while Dlx1, Dlx2 and Dlx5 were shown to be essential for mouse craniofacial development but not for placental development [Depew et al., 2005]. Furthermore, DLX3 is one of the few transcription factors in which mutations have been linked to a human ectodermal dysplasia, namely Tricho-Dento-Osseous syndrome [Morasso and Radoja, 2005; Price et al., 1998]. These differences suggest that DLX3, while keeping common features with its family members, acquired specific characteristics during evolution, including its potential to be regulated by SUMOylation.
ACKNOWLEDGEMENTS
We would like to thank the Light Imaging Section of the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS). This work was supported by the Intramural Research Program of NIAMS, of the National Institutes of Health.
REFERENCES
- Alkuraya FS, Saadi I, Lund JJ, Turbe-Doan A, Morton CC, Maas RL. SUMO1 haploinsufficiency leads to cleft lip and palate. Science. 2006;313:1751. doi: 10.1126/science.1128406. [DOI] [PubMed] [Google Scholar]
- Bhaskar V, Smith M, Courey AJ. Conjugation of Smt3 to dorsal may potentiate the Drosophila immune response. Mol Cell Biol. 2002;22:492–504. doi: 10.1128/MCB.22.2.492-504.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan JT, Morasso MI. The Dlx3 protein harbors basic residues required for nuclear localization, transcriptional activity and binding to Msx1. Journal of Cell Science. 2000;113:4013–23. doi: 10.1242/jcs.113.22.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew MJ, Simpson CA, Morasso M, Rubenstein JL. Reassessing the Dlx code: the genetic regulation of branchial arch skeletal pattern and development. Journal of Anatomy. 2005;207:501–61. doi: 10.1111/j.1469-7580.2005.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–9. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- Du JX, Bialkowska AB, McConnell BB, Yang VW. SUMOylation regulates nuclear localization of Kruppel-like factor 5. J Biol Chem. 2008;283:31991–2002. doi: 10.1074/jbc.M803612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duverger O, Lee D, Hassan MQ, Chen SX, Jaisser F, Lian JB, Morasso MI. Molecular consequences of a frameshifted DLX3 mutant leading to Tricho-Dento-Osseous syndrome. Journal of Biological Chemistry. 2008;283:20198–208. doi: 10.1074/jbc.M709562200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller-Pace FV, Jacobs AM, Nicol SM. Modulation of transcriptional activity of the DEAD-box family of RNA helicases, p68 (Ddx5) and DP103 (Ddx20), by SUMO modification. Biochem Soc Trans. 2007;35:1427–9. doi: 10.1042/BST0351427. [DOI] [PubMed] [Google Scholar]
- Goodson ML, Hong Y, Rogers R, Matunis MJ, Park-Sarge OK, Sarge KD. Sumo-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J Biol Chem. 2001;276:18513–8. doi: 10.1074/jbc.M008066200. [DOI] [PubMed] [Google Scholar]
- Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwarz SE, Scheffner M, Del Sal G. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 1999;18:6462–71. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan MQ, Javed A, Morasso MI, Karlin J, Montecino M, van Wijnen AJ, Stein GS, Stein JL, Lian JB. Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Molecular & Cellular Biology. 2004;24:9248–61. doi: 10.1128/MCB.24.20.9248-9261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan MQ, Tare RS, Lee SH, Mandeville M, Morasso MI, Javed A, van Wijnen AJ, Stein JL, Stein GS, Lian JB. BMP2 commitment to the osteogenic lineage involves activation of Runx2 by DLX3 and a homeodomain transcriptional network. Journal of Biological Chemistry. 2006;281:40515–26. doi: 10.1074/jbc.M604508200. [DOI] [PubMed] [Google Scholar]
- Heun P. SUMOrganization of the nucleus. Curr Opin Cell Biol. 2007;19:350–5. doi: 10.1016/j.ceb.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Huang YP, Wu G, Guo Z, Osada M, Fomenkov T, Park HL, Trink B, Sidransky D, Fomenkov A, Ratovitski EA. Altered sumoylation of p63alpha contributes to the split-hand/foot malformation phenotype. Cell Cycle. 2004;3:1587–96. doi: 10.4161/cc.3.12.1290. [DOI] [PubMed] [Google Scholar]
- Hwang J, Mehrani T, Millar SE, Morasso MI. Dlx3 is a crucial regulator of hair follicle differentiation and cycling. Development. 2008;135:3149–59. doi: 10.1242/dev.022202. Epub 2008 Aug 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs AM, Nicol SM, Hislop RG, Jaffray EG, Hay RT, Fuller-Pace FV. SUMO modification of the DEAD box protein p68 modulates its transcriptional activity and promotes its interaction with HDAC1. Oncogene. 2007;26:5866–76. doi: 10.1038/sj.onc.1210387. [DOI] [PubMed] [Google Scholar]
- Li T, Santockyte R, Shen RF, Tekle E, Wang G, Yang DC, Chock PB. Expression of SUMO-2/3 induced senescence through p53- and pRB-mediated pathways. J Biol Chem. 2006;281:36221–7. doi: 10.1074/jbc.M608236200. [DOI] [PubMed] [Google Scholar]
- Morasso MI, Grinberg A, Robinson G, Sargent TD, Mahon KA. Placental failure in mice lacking the homeobox gene Dlx3. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:162–7. doi: 10.1073/pnas.96.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morasso MI, Radoja N. Dlx genes, p63, and ectodermal dysplasias. Birth Defects Research. Part C, Embryo Today: Reviews. 2005;75:163–71. doi: 10.1002/bdrc.20047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Kanei-Ishii C, Nomura T, Ishii S. TRAF7 sequesters c-Myb to the cytoplasm by stimulating its sumoylation. Mol Biol Cell. 2005;16:5433–44. doi: 10.1091/mbc.E05-08-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Berger M, Lehembre F, Seeler JS, Haupt Y, Dejean A. c-Jun and p53 activity is modulated by SUMO-1 modification. J Biol Chem. 2000;275:13321–9. doi: 10.1074/jbc.275.18.13321. [DOI] [PubMed] [Google Scholar]
- Ouyang J, Valin A, Gill G. Regulation of transcription factor activity by SUMO modification. Methods Mol Biol. 2009;497:141–52. doi: 10.1007/978-1-59745-566-4_9. [DOI] [PubMed] [Google Scholar]
- Pichler A, Melchior F. Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport. Traffic. 2002;3:381–7. doi: 10.1034/j.1600-0854.2002.30601.x. [DOI] [PubMed] [Google Scholar]
- Poukka H, Karvonen U, Janne OA, Palvimo JJ. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1) Proc Natl Acad Sci U S A. 2000;97:14145–50. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JA, Bowden DW, Wright JT, Pettenati MJ, Hart TC. Identification of a mutation in DLX3 associated with tricho-dento-osseous (TDO) syndrome. Human Molecular Genetics. 1998;7:563–9. doi: 10.1093/hmg/7.3.563. [DOI] [PubMed] [Google Scholar]
- Robinson GW, Mahon KA. Differential and overlapping expression domains of Dlx-2 and Dlx-3 suggest distinct roles for Distal-less homeobox genes in craniofacial development. Mechanisms of Development. 1994;48:199–215. doi: 10.1016/0925-4773(94)90060-4. [DOI] [PubMed] [Google Scholar]
- Seeler JS, Dejean A. SUMO: of branched proteins and nuclear bodies. Oncogene. 2001;20:7243–9. doi: 10.1038/sj.onc.1204758. [DOI] [PubMed] [Google Scholar]
- Tateishi Y, Ariyoshi M, Igarashi R, Hara H, Mizuguchi K, Seto A, Nakai A, Kokubo T, Tochio H, Shirakawa M. Molecular basis for SUMOylation-dependent regulation of DNA binding activity of heat shock factor 2. J Biol Chem. 2009;284:2435–47. doi: 10.1074/jbc.M806392200. [DOI] [PubMed] [Google Scholar]
- Tsuruzoe S, Ishihara K, Uchimura Y, Watanabe S, Sekita Y, Aoto T, Saitoh H, Yuasa Y, Niwa H, Kawasuji M, Baba H, Nakao M. Inhibition of DNA binding of Sox2 by the SUMO conjugation. Biochem Biophys Res Commun. 2006;351:920–6. doi: 10.1016/j.bbrc.2006.10.130. [DOI] [PubMed] [Google Scholar]
- Wang J, Li A, Wang Z, Feng X, Olson EN, Schwartz RJ. Myocardin sumoylation transactivates cardiogenic genes in pluripotent 10T1/2 fibroblasts. Mol Cell Biol. 2007;27:622–32. doi: 10.1128/MCB.01160-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang H, Iyer D, Feng XH, Schwartz RJ. Regulation of cardiac specific nkx2.5 gene activity by small ubiquitin-like modifier. J Biol Chem. 2008;283:23235–43. doi: 10.1074/jbc.M709748200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson VG, Rangasamy D. Intracellular targeting of proteins by sumoylation. Exp Cell Res. 2001;271:57–65. doi: 10.1006/excr.2001.5366. [DOI] [PubMed] [Google Scholar]
- Yang SH, Jaffray E, Senthinathan B, Hay RT, Sharrocks AD. SUMO and transcriptional repression: dynamic interactions between the MAP kinase and SUMO pathways. Cell Cycle. 2003;2:528–30. doi: 10.4161/cc.2.6.597. [DOI] [PubMed] [Google Scholar]
- Zhang H, Hu G, Wang H, Sciavolino P, Iler N, Shen MM, Abate-Shen C. Heterodimerization of Msx and Dlx homeoproteins results in functional antagonism. Molecular & Cellular Biology. 1997;17:2920–32. doi: 10.1128/mcb.17.5.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]