Abstract
Non-syndromic cleft palate (CP) is a common birth defect with a complex and heterogeneous etiology involving both genetic and environmental risk factors. We conducted a genome wide association study (GWAS) using 550 case-parent trios, ascertained through a CP case collected in an international consortium. Family based association tests of single nucleotide polymorphisms (SNP) and three common maternal exposures (maternal smoking, alcohol consumption and multivitamin supplementation) were used in a combined 2 df test for gene (G) and gene-environment (G×E) interaction simultaneously, plus a separate 1 df test for G×E interaction alone. Conditional logistic regression models were used to estimate effects on risk to exposed and unexposed children. While no SNP achieved genome wide significance when considered alone, markers in several genes attained or approached genome wide significance when G×E interaction was included. Among these, MLLT3 and SMC2 on chromosome 9 showed multiple SNPs resulting in increased risk if the mother consumed alcohol during the peri-conceptual period (3 months prior to conception through the first trimester). TBK1 on chr. 12 and ZNF236 on chr. 18 showed multiple SNPs associated with higher risk of CP in the presence of maternal smoking. Additional evidence of reduced risk due to G×E interaction in the presence of multivitamin supplementation was observed for SNPs in BAALC on chr. 8. These results emphasize the need to consider G×E interaction when searching for genes influencing risk to complex and heterogeneous disorders, such as non-syndromic CP.
INTRODUCTION
Cleft palate (CP) is a common birth defect where both genetic and environmental components contribute to the etiology. CP has a lower birth prevalence compared to cleft lip with/without cleft palate (CL/P): 1/2500 live births vs. 1/700; but CP shows less variability in birth prevalence across populations compared to CL/P. Almost half of all livebirths with CP occur in infants with another congenital anomaly or some identifiable malformation syndrome [Genisca et al., 2009]. The current study focuses on isolated, non-syndromic CP (CP) which shows strong familial aggregation; has documented environmental risk factors; and is etiologically heterogeneous.
Both population based studies and family studies suggest a strong genetic component to the etiology of non-syndromic CP. Risk of CP among first degree relatives of cases is 56 times greater (95%CI=37–85) than the general population in Norway [Sivertsen et al., 2008]; and Grosen et al. (2009) reported a 15-fold higher risk (95%CI=13–17) in Denmark. Twin and family studies of CP also argue for strong genetic control. In a nationwide study of Danish twins, Grosen et al. (2009) reported probandwise concordance rates of 33% for CP among monozygotic twins compared to 7% among dizygotic twins, and the latter was only slightly higher than the 3% recurrence risk seen between full siblings [Grosen et al., 2009]. Despite this evidence for some genetic component, no single gene model can explain the strong familial aggregation of CP [Marazita, 2002]. Part of this difficulty could reflect locus heterogeneity, i.e. several different genes contribute to the etiology of CP. In the absence of external information to separate CP families into more homogeneous subgroups, however, statistical tests on modest sized samples will always have limited power to detect effects of individual causal genes.
Although several genes have been identified for syndromic forms of CP, few have been identified as influencing risk to non-syndromic CP, and none have sufficient evidence to be defined as directly causal. This could reflect the difficulty in amassing sufficiently large samples of cases. Recently, Ghassibe et al. (2010) showed a translocation in a single multiplex CP family with Pierre Robin sequence (which involves CP) included the FAF1 gene on chromosome (chr.) 1p. Subsequent analysis of an intronic marker (rs3827730) in FAF1 showed evidence of linkage and association in a collection of case-parent trios from several populations (mostly of European ancestry). Ghassibe et al. (2010) also showed evidence of decreased expression in blood samples from a small number of CP patients, although the function of FAF1 in craniofacial development remains poorly defined. Li et al. (2009) also reported SNP rs7205289 in the microRNA (miRNA-140) region of chr. 16 showed evidence of association in a sample of 557 non-syndromic oral cleft cases (388 CL/P and 169 CP cases) compared to 306 healthy controls from Western China.
Part of the difficulty in identifying genes controlling risk to non-syndromic CP could also reflect biological interaction between high risk alleles and exposure to environmental risk factors during early development. There are several recognized environmental risk factors for non-syndromic CP, including maternal smoking and alcohol consumption [Little et al., 2004; DeRoo et al., 2008]. Johnson and Little (2008) reviewed the role of multi-vitamin and folate supplementation on risk to oral clefts. A few candidate gene studies have investigated G×E interaction with maternal smoking, although none used a genome wide approach. Starting with Hwang et al. (1995) several CP case-control studies of G×Smoking interaction with an intronic marker in TGFA on chr. 2q gave suggestive evidence, as did some case-parent trio studies [Maestri et al., 1997]. However, meta-analysis of 5 case-control studies did not reveal compelling evidence for G×Smoking interaction [Zeiger et al., 2005]. Recently, evidence of G×E interaction was reported between SNP rs7205289 (in miRNA-140) and maternal passive smoking in an analysis of 162 CP cases and 304 healthy controls [Li et al., 2010]. In this Chinese sample, children whose mother was exposed to passive smoking were 4.7 times more likely to have CP if they carried the high-risk allele.
Considering gene-environment (G×E) interaction in the context of a genome-wide association study (GWAS) can be an important step in identifying genes controlling risk to complex and heterogeneous disorders, such as non-syndromic CP. Manning et al. (2011) showed how a variety of statistical tests for G×E interaction can be incorporated into GWAS, even meta-analyses of several studies. We conducted a GWAS to identify genes controlling risk to isolated, non-syndromic oral clefts (including CP) under a case-parent trio design in an international consortium [Beaty et al., 2010]. Here we report on 550 CP trios where tests for G×E interaction with three common maternal exposures (maternal smoking, alcohol consumption and vitamin supplementation) are considered. The genome-wide approach has the advantage of being unbiased in its coverage of the human genome, and the case-parent trio study design has the advantage of minimizing confounding due to population stratification. This provided a unique opportunity to search for genes influencing risk to a common birth defect alone or through interaction with maternal exposures.
METHODS
Case-parent trios
Case-parent trios were drawn from several groups who formed an international consortium to conduct a genome-wide search for genes influencing risk to oral clefts using a case-parent trio design [Beaty et al., 2010]. Table I lists numbers of trios noting the CP proband's gender from each recruitment site and in the total sample. Most cases were ascertained through surgical treatment centers, although population based ascertainment was used in Norway. To minimize potential misclassification of non-syndromic CP, cases were examined by either a clinical geneticist or experienced health care provider to rule out syndromic forms (except in Norway where medical records were reviewed). As often reported, there were more female CP cases (56%) compared to males. Racial ancestry of cases fell into two broad categories: 49% (272 of 550) of CP cases were of European ancestry (including European Americans), 47% (259 of 550) were of Asian ancestry and 3% (19 of 550) were categorized as African ancestry (mostly African Americans) or `other' racial groups (including mixed ancestry).
Table I.
Gender of 550 isolated, non-syndromic cleft palate (CP) cases in the international consortium study by recruitment site
| Sites | Male | Female | Total | |
|---|---|---|---|---|
| Counts | Counts | Counts | % | |
| Denmark | 8 | 5 | 13 | 2% |
| Norway | 52 | 58 | 110 | 20% |
| Iowa | 19 | 22 | 41 | 7% |
| Maryland | 19 | 24 | 43 | 8% |
| Pittsburgh | 7 | 8 | 15 | 3% |
| Utah | 31 | 33 | 64 | 12% |
| Singapore | 22 | 35 | 57 | 10% |
| Taiwan | 29 | 50 | 79 | 14% |
| Shangdong Prov. | 16 | 22 | 38 | 7% |
| Hubei Prov. | 19 | 26 | 45 | 8% |
| Sichuan Prov. | 18 | 22 | 40 | 7% |
| Korea | 4 | 1 | 5 | 1% |
| Total | 244 (44%) | 306 (56%) | 550 (100%) | 100% |
Genotyping
The Center for Inherited Disease Research (CIDR) genotyped DNA samples using Illumina's 610Quad platform and 99.1% passed CIDR quality control (QC) [Beaty et al., 2010]. Genotypes on 589,945 SNPs (99.56% of those attempted) were released and then underwent further QC analysis to set up 4 types of flags for each SNP: 1) unacceptably high rates (>5%) of missing genotype calls, 2) low minor allele frequency (MAF<0.01), 3) unacceptably high rates of Mendelian errors (>5%) between parents and child, and 4) significant deviation (p<10−5) from Hardy Weinberg equilibrium (HWE) among parents within recruitment site or across populations but within European and Asian groups separately. This QC process flagged 14.6% of all SNPs (mostly for low MAF), leaving ~498K SNPs available.
Exposure assessment
Maternal exposure was assessed through a structured interview focused on the peri-conceptual period (3 months prior to conception through the first trimester) because palatal development is completed during weeks 8–9 of gestation (often before the pregnancy is recognized). Three maternal exposures were assessed as simple yes/no responses: personal cigarette smoking by the mother, any alcohol consumption, and any use of multi-vitamin supplements (not limited to folate). Table II presents rates of these maternal exposures in the total sample of mothers and stratifying by European, Asian and African/other ancestry groups. Rates of maternal exposures varied considerably across these groups.
Table II.
Exposure rates for maternal smoking, multivitamin supplementation and alcohol consumption for the total CP group and for three ancestry groups
| Ancestry Group | Maternal smoking | Maternal alcohol consumption | Maternal multivitamin supplementation | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Not exposed | Exposed | Unk. | %E | Not exposed | Exposed | Unk. | %E | Not exposed | Exposed | Unk. | %E | |
| European | 198 | 74 | 0 | 27% | 162 | 109 | 1 | 40% | 88 | 157 | 27 | 58% |
| Asian | 250 | 9 | 0 | 4% | 246 | 10 | 3 | 4% | 197 | 52 | 10 | 20% |
| Afr/Other | 15 | 3 | 1 | 16% | 15 | 3 | 1 | 16% | 4 | 13 | 2 | 68% |
| Total | 463 | 86 | 1 | 16% | 423 | 122 | 5 | 22% | 289 | 222 | 39 | 40% |
Statistical analysis
GWAS
The conventional TDT was used to test individual SNPs in the genome-wide marker panel using PLINK [Purcell et al., 2007] http://pngu.mgh.harvard.edu/purcell/plink/) and PBAT [Lange et al., 2003] (v3.6; http://www.biostat.harvard.edu/~clange/default.htm). Additionally, a total of 14,486 SNPs on the X chromosome were examined using FBAT [Laird et al., 2000] http://www.biostat.harvard.edu/fbat/default.html), which is equivalent to the genotypic TDT under an additive model on independent trios [Laird and Lange, 2006]. Manhattan plots of −log10(p-value) were generated over all autosomal and X-linked markers. Under a Bonferroni correction for ~500K tests, this data set would require a significance level of ≤10−7 for genome-wide significance. Quantile-quantile (QQ) plots were also generated.
GxE interaction
To test for GxE interaction, we followed a strategy proposed first by Lange et al. (2003) for family studies and later by Kraft et al. (2007) for case-control studies. The PBAT package was used to compute a combined score test with 2 df test for G and GxE interaction together, and the separate 1 df test for GxE interaction alone. This provides the opportunity to detect genes where the GxE interaction enhances G effects, as well as detecting deviation from predicted effects of G and E (the classical definition of statistical interaction). Autosomal markers were used in the initial screen with tests for SNP effects under an additive model ignoring all exposures, followed by the 2 df test for joint effects of G and GxE interaction and the 1 df test for GxE interaction alone. Any marker with a −log10(p-value) for either the 1 df test or the 2 df test >6 (i.e. p-value≤10−6) was selected for further analysis.
All genes identified as close to genome wide significant in this PBAT analysis were further examined with a genotypic TDT using conditional logistic regression models. In these models, the observed genotype of the case is compared to genotypes of 3 possible “pseudo-sib” controls in each trio. A conventional conditional logistic regression model specifies the log-odds of being the observed case in the i-th trio as: logit[P(casei)]=β0+βG(Gi)+ βGxE(Gi*Ei) where G=0, 1, or 2 reflects the number of high risk alleles in the case/”pseudo-sib” control, and E=0/1 for unexposed and exposed trios, respectively. The regression coefficients (βG and βGxE, respectively) represent effects of G and GxE interaction on risk. Exposure specific odds ratio of being a CP case for carriers of a high risk allele were calculated; i.e. for unexposed carriers, this becomes OR(CP|G no E)=exp(βG), and for exposed carriers, this is OR(CP|G and E)=exp(βG+βGE). For any gene with several SNPs showing evidence of possible GxE interaction, these estimated OR were plotted along with p-values from the 1 df test from the likelihood ratio test (LRT) for either βGE alone or the 2 df combined test of βG=βGE=0.
RESULTS
A conventional search for marginal gene (G) effects influencing risk to CP in the total sample of 550 trios showed no markers achieved significance at a strict genome-wide level (p≤10−7). Supporting Figure 1 presents a Manhattan plot for all autosomal and X-linked SNPs. There were 7 SNPs with asymptotic p~10−6, and two of these mapped to the DSC3 gene on chr. 18, which contained several additional SNPs giving p<0.001. Four other genes (TPP2, HTR1B, BCL6 and MEST) had a single SNP with p~10−6, plus at least one other SNP yielding p<0.001. Thus, while the initial GWAS failed to yield evidence of individual genes controlling risk to CP in these 550 case-parent trios (see QQ plot in Supporting Figure 2), several SNPs did yield suggestive evidence.
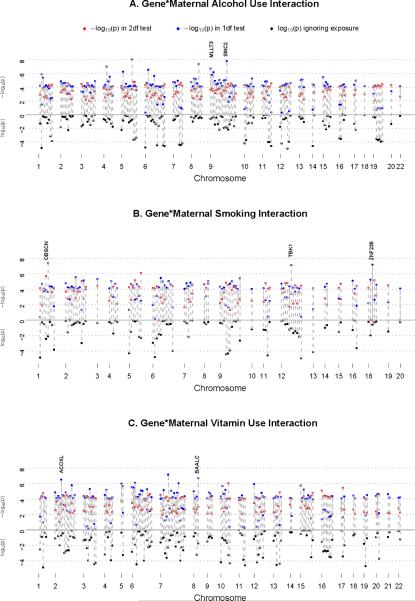
A genome-wide screen for GxE interaction was carried out using PBAT under the strategy described above, where both the 2 df test for gene (G) and gene-environment (GxE) interaction and the 1 df test for (GxE) interaction alone were examined. This screening process revealed several markers achieving genome-wide significance, especially for the 1 df test of GxE interaction (Supporting Figure 3). To further investigate this evidence, Figure 1 presents “double Manhattan plots” to summarize evidence for G and GxE interaction effects for each of the three maternal exposures (Figure 1A for GxAlcohol; 1B for GxSmoking and 1C for GxVitamin). In these plots, the bottom half shows the log10(p-value) for the conventional family based test of SNP effects ignoring exposure (where more significant results fall farther below the mid-line). In the top half, the −log10(p-values) are presented for each autosomal SNP from both the 2 df test of G and GxE interaction together (red dots) and the 1 df test for GxE interaction alone (blue dots). Only SNPs yielding asymptotic p<0.0001 in either of these two tests were included to minimize clutter. Dashed lines connect p-values from the marginal test ignoring exposures (below mid-line) to those models considering GxE interaction in either the 1 df test or the 2 df test (above mid-line). Here the very strongest signals for G effects ignoring exposures were omitted (i.e. p-value<0.00001 in the conventional TDT) to highlight those SNPs showing evidence of GxE interaction, which favors identifying genes suggesting GxE interaction. We focused on SNPs in genes yielding p<10−6 in one or another test for GxE interaction for further analysis. As seen in Figure 1A, 8 markers gave p<10−6 in the 1 df test for GxAlcohol interaction among autosomal SNPs, including 3 in MLLT3 on chr. 9q22.
Figure 1.
Double Manhattan plots for SNP effects ignoring maternal exposures (lower half) and considering G and G×E interaction for three maternal exposures. Blue dots represent –log10(p-value) from 1 df test of G×E interaction alone; red dots represent – log10(p-value) from 2 df test of G and G×E interaction. Only autosomal SNPs showing p<0.0001 for one of these two tests are displayed. Dashed lines connect SNP showing this level of significance in one test considering G×E interaction with their corresponding p-value when interaction is ignored. Panel A: Results for GxMaternal Alcohol Consumption interaction; Panel B: Results for GxMaternal Smoking interaction; Panel C: Results for GxMaternal Vitamin Use interaction.
Table III lists all genes (including 3 pseudo-genes and 1 open reading frame) showing evidence of GxE interaction for any of the three maternal exposures at p≤10−6, along with a total count of SNPs mapping to this gene and the count of additional SNPs yielding p<0.01 in either the 1 df test for GxE alone or the 2 df test for G and GxE interaction together. We dropped 5 genes with one SNP each yielding p<10−6 in either test for GxE interaction but ≤3 additional SNPs showing p<10−2 (AGXT2, HMP19, PRDM14, BTN2A and ETV6). While we examined all genes listed in Table III (including pseudo-genes), here we focus on recognized genes showing evidence of GxE interaction (noted in bold in Table III and labeled in Figure 1).
Table III.
Genes yielding p-values <10−6 in either the 2 df test for G and GxE or the 1 df test for GxE with one or more maternal exposures in genome wide screen using PBAT on 550 CP case-parent trios. Total counts of SNPs mapping to genes and numbers of SNPs yielding p<0.01 in either test
| Gene (Chr) | # SNPs p<0.01 | Total # SNPs | Exp. | Gene (Chr) | # SNPs p<0.01 | Total # SNPs | Exp. | ||
|---|---|---|---|---|---|---|---|---|---|
| in 2 df test | in 1 df test | in 2 df test | in 1 df test | ||||||
| LOC645762 (4) | 2 | 8 | 70 | Alc | ZNF236 (18) | 3 | 8 | 39 | Smk |
| c6orf105 (6) | 11 | 18 | 126 | Alc | ACOXL (2) | 8 | 10 | 82 | Vit |
| MLLT3 (9) | 4 | 8 | 144 | Alc | LOC391828 (5) | 3 | 4 | 52 | Vit |
| SMC2 (9) | 5 | 5 | 141 | Alc | LOC392027 (7) | 6 | 6 | 214 | Vit |
| OBSCN (1) | 10 | 10 | 27 | Smk | BAALC (8) | 3 | 7 | 61 | Vit |
| TBK1 (12) | 6 | 4 | 18 | Smk | |||||
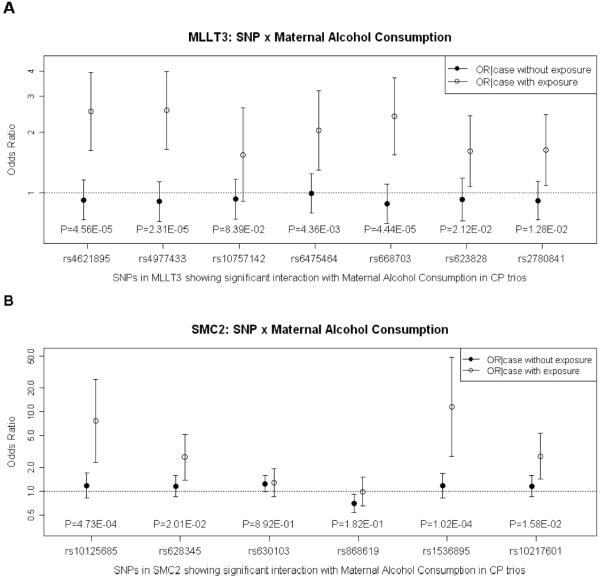
When considering maternal alcohol exposure, MLLT3 and SMC2 on chr. 9 yielded evidence of GxAlcohol interaction. A total of 144 SNPs mapped to MLLT3, and 3 of these gave strong evidence of GxAlcohol interaction in the 1 df test (rs4621895, p=1.9*10−7; rs668703, p=6.6*10−7; and rs4977433, p=1.7*10−6). A cluster of 7 adjacent SNPs showed evidence of G and GxE interaction, even though none were significant when exposure to maternal alcohol was ignored (see column 3 in Supporting Table I). Six of these 7 SNPs yielded nominal significance (p<0.05) in both the 1 df test for GxE interaction alone and in the 2 df test for combined effects of G and GxE interaction (last 2 columns of Supporting Table I). Pairwise linkage disequilibrium (LD) as measured by r2 in Asian and European parents separately could not account for this pattern alone (Supporting Figure 4).
Conditional logistic regression models were used to estimate the odds ratio of having CP given the infant carried one risk allele in the absence of exposure [OR(CP|G no E)] and in its presence [OR(CP|G and E)], along with their 95%CI. Estimated OR(CP|G no E) and OR(CP|C and E) for these 7 SNPs in MLLT3 are presented in Figure 2A along with p-values from a LRT for the 1 df test in the conditional logistic regression framework. Here, an additive model was used and the apparent `high-risk allele' became the target allele (which was the minor allele for rs4621895, rs4977433, rs648703 and rs2780841, but the major allele for rs10757142 and rs6475464 -- see Supporting Table I). Estimated OR(CP|G and E) and their 95%CI for a heterozygous child exposed to maternal alcohol consumption were distinctly higher (open circles) compared to a similar unexposed child (solid circles).
Figure 2.
Estimated OR(CP|G no E) and OR(CP|G and E) for maternal alcohol consumption from logistic regression on 550 case-parent trios. P-values from 1 df LRT for G×E interaction are shown along the X axis. Panel A: Estimated OR(CP|G no E) and OR(CP|G and E) for 7 SNPs in MLLT3. Panel B: Estimated OR(CP|G no E) and OR(CP|G and E) for 7 SNPs in SMC2.
Although none 141 SNPs mapping to SMC2 on chr. 9q31.1 were significant at the p<0.0001 level when maternal exposures were ignored, some did achieve nominal significance. When maternal alcohol consumption was considered, SNP rs1536895 yielded p=1.53*10−8 in the 1 df test for GxE interaction from PBAT and an adjacent SNP approached genome wide significance (rs10125685, p=9.83*10−6). These SNPs identified a region spanning 11kb (and including 6 SNPs) where evidence of GxAlcohol interaction was apparent. When conditional logistic regression models were used to estimate exposure specific ORs, all 6 of these SNPs suggested modest G effects ignoring exposure (see column 3 of Supporting Table II for p-values from the LRT), and 5 of these 6 were also significant in the 2 df test for combined G and GxAlcohol interaction. Figure 2B shows estimated OR(CP|G no E) and OR(CP|G and E), plus their 95%CI, for these 6 SNPs under an additive model. For 4 separate SNPs, the putative high risk allele was associated with a doubling of risk when the fetus was exposed to maternal alcohol consumption, although some confidence intervals were quite wide.
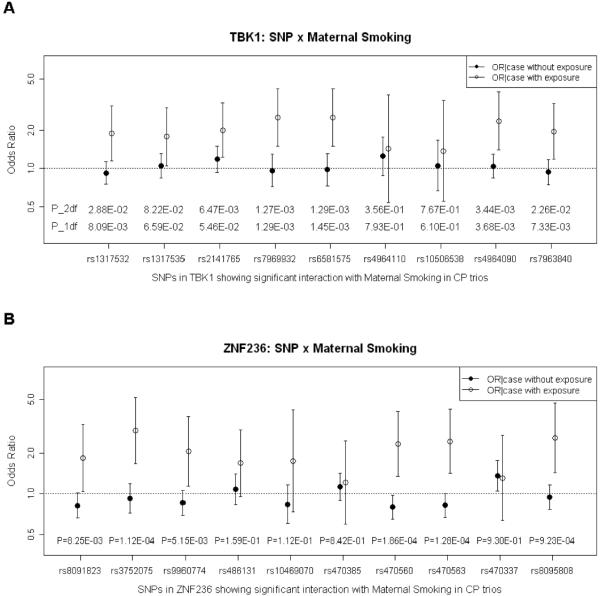
Eighteen SNPs mapped to TBK1 on chromosome 12q14.2, but only one was nominally significant when exposure to maternal smoking was ignored (rs2141765; p=0.0095). However, 4 SNPs were significant at p<0.01 in the 2 df test for G and G×Smoking interaction, and 6 were significant at this level in the 1 df test for G×Smoking interaction alone (including rs7969932 with p=7.86*10−8), forming a cluster of 9 SNPs spanning 30 kb. In a conditional logistic regression model, 6 of these 9 SNPs were nominally significant in the 2 df test for combined effects of G and G×E interaction, and 5 were significant in the 1 df test for G×E interaction alone (Figure 3A which shows p-values for both the 1 df and 2 df tests; also Supporting Table III).
Figure 3.
Estimated OR(CP|G no E) and OR(CP|G and E) considering G×E interaction with maternal smoking in logistic regression on 550 case-parent trios. Panel A: Estimated OR(CP|G no E) and OR(CP|G and E) for 9 SNPs in TBK1. P-values from 2 df LRT for G and G×E interaction plus that from 1 df LRT for G×E interaction are shown along the X axis. Panel B: Estimated OR(CP|G no E) and OR(CP|G and E) for 10 SNPs in ZNF236. P-values from 1 df LRT for G×E interaction are shown along the X axis.
ZNF236 on chromosome 18q22–q23 encompassed 39 SNPs, one of which yielded evidence of influencing risk when exposures were ignored (rs470337, p=0.015). However, rs372075 gave p=6.75*10−8 in the 1 df test for G×E interaction and rs470563 gave p=6.91*10−6 in the 2 df test. A block of 10 SNPs (spanning 57 kb) was examined using conditional logistic regression models, and again rs470337 was significant when maternal smoking was ignored (p-value=0.016; see Supporting Table IV). However, when maternal smoking was considered, 7 of these 10 SNPs were significant in the 2 df test for G and G×E interaction combined (Supporting Table IV). These SNPs showed no evidence of influencing risk for unexposed infants, i.e. the 95%CI of the OR(CP|G no E) always overlapped the null hypothesis value of one. Figure 4B shows estimated OR(CP|G and E) were distinctly higher for 6 of these SNPs.
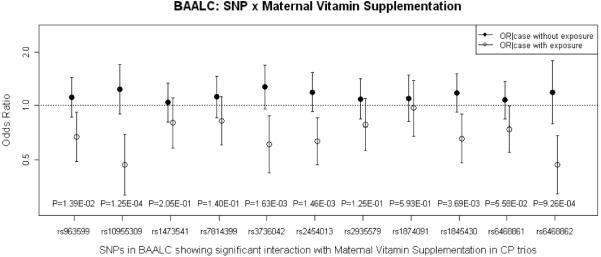
Figure 4.
Estimated OR(CP|G no E) and OR(CP|G and E) for 11 SNPs in BAALC considering G×E interaction with maternal vitamin supplementation in logistic regression on 550 CP case-parent trios. P-values from 1 df LRT for G×E interaction are shown along the X axis.
Among the 61 SNPs mapping to BAALC on chr. 8q22.3, a block of 11 SNPs (spanning 34kb) yielded one SNP with significant evidence in the 1 df test for G×E interaction (rs6468862; p=2.03*10−7), plus 4 additional SNPs yielding nominal significance from PBAT. Only rs6468862 showed strong evidence of influencing risk in conditional logistic regression when maternal exposures were ignored (see column 3 of Supporting Table V). However, when G×E interaction was included in the model, 6 SNPs became significant in either the 1 or 2 df test (Supporting Table V), and the estimated OR(CP|G no E) and OR(CP|G and E) were distinct (see Figure 4).
Among the genes in Table III, OBSCN on chr. 1q42.13 and ACOXL on chr. 2q13 deserve additional mention. The 27 SNPs mapping to OBSCN included 22 SNPs spanning 114 kb, 18 of which were nominally significant for G effects ignoring maternal exposures. When G×Smoking interaction was included, 17 of these 22 showed significant evidence in the 2 df test for G and G×Smoking interaction combined. Thus, OBSCN may represent “quantitative interaction” where exposure to maternal smoking enhances G effects. Among all the genes considered, only OBSCN showed any evidence of heterogeneity between trios of Asian and European ancestry in formal tests of heterogeneity considering G×Smoking interaction (data not shown). Because exposure to smoking is much lower among Asian mothers, however, it is difficult to confirm the absence of G×Smoking interaction in this group. Supporting Figure 5 shows estimated OR(CP|G no E) and OR(CP|G and E) for these 22 SNPs. A total of 126 SNPs mapped to the ACOXL gene, and these included a block of 24 adjacent SNPs (spanning 64 kb) of which 11 were significant in the 1 df test for G×Vitamin interaction (including rs7602030, p=3.13*10−7). When conditional logistic regression models were used to estimate OR(CP|G no E) and OR(CP|G and E), 10 SNPs in ACOXL showed significantly lower risk to the child if the mother used multivitamin supplements (see Supporting Figure 6).
Supporting Figures 7–10 illustrate estimated effects as OR(CP|G no E) and OR(CP|G and E) for 3 pseudo-genes (LOC645762 on chr. 4; LOC391828 on chr. 5; and LOC392027 on chr. 7) and for the open reading frame c6orf105 on chr. 6. Each of these putative genes met the criteria used to select genes giving evidence of G×E interaction, but 2 of these 3 pseudo-genes (LOC645762 and LOC391828) have been dropped from the latest version of the human genome (Build 37) and the inferred status of the remaining pseudo-genes makes it difficult to assess their true relevance.
DISCUSSION
Our initial GWAS of 550 CP case-parent trios failed to show any markers achieving genome-wide significance after Bonferroni correction (Supporting Figures 1 and 2), although DSC3 (desmocollin 3; MIM ID *600271) on chr. 18q12.1 showed several SNPs giving suggestive evidence. When markers were screened using family based association tests considering both G and G×E interaction, however, several markers yielded p-values at or near genome-wide significance (Supporting Figure 3). Several genes had SNPs showing evidence attaining or approaching genome-wide significance plus blocks of adjacent SNPs yielding additional statistical evidence (Table III). We estimated OR(CP|G no E) and OR(CP|G and E) for blocks of adjacent SNPs to illustrate how common maternal exposures can alter effects of markers on risk to nonsyndromic CP.
While none of the genes listed in Table III are recognized candidates for oral clefts, further investigation is warranted. MLLT3 (myeloid/lymphoid or mixed-lineage leukemia) is a large gene on 9p22 often involved in translocations; although the region of signal represented in Figure 2A contains only intronic SNPs. The SMC2 (structural maintenance of chromosomes 2) gene on 9q31.1 plays a role in mitotic chromosome condensation and DNA repair, and SNPs represented in Figure 2B are upstream of coding sequences. TBK1 (tank-binding kinase 1 on chr. 12q14) is a key signaling molecule in the NFκB pathway, however, the region of signal shown in Figure 3A lies outside the gene. ZNF236 gene on chr. 18q22–q23 has two alternatively spliced transcripts and is widely expressed; although the region represented in Figure 3B includes mostly intronic SNPs. The BAALC (brain and acute leukemia gene cytoplasmic) gene on chr. 8q22.3 is largely expressed in neural tissues and has several isoforms. The region of signal represented in Figure 4 lies partially outside this gene, encompassing part of the hypothetical protein coding region FLJ10489. The OBSCN gene (obscurin) on chr 1q42.13 containing several immunoglobulin-like domains, and is located close to WNT3A, recently reported to be associated with non-syndromic CL/P in a Chinese sample [Yao et al., 2010]. It is important to remember location of a peak signal in a GWAS may lie some distance from any etiologic variant, so these genes showing peak signals may not be functional.
Our GWAS results argue the genetic etiology of non-syndromic CP is distinct from CL/P because none of the genes identified in our analysis of non-syndromic CL/P trios were identified here (Beaty et al., 2010). This difference cannot be attributed to the smaller numbers of CP trios alone, because analysis of 547 CL trios showed clear support for regions such as chr. 8q24 and genes such as IRF6. In this study of CP trios, we only achieved genome-wide significance when G×E interaction was explicitly considered, thus it remains prudent to separate CP from CL/P.
Statistical vs. biological interaction
Biological interaction is said to exist when both a gene (G) and an environmental factor (E) contribute in the etiology of a disease, and E alters the impact of G on risk (increasing or decreasing risk among carriers of G). Statistical interaction, however, is defined as a detectable deviation from predicted joint effects of G and E under a specific model (typically an additive or a multiplicative model). Thus, statistical interaction may or may not correspond to biological interaction.
Several patterns of statistical interaction are possible [Kraft and Hunter, 2005], including “pure interaction” where neither the gene (G) nor the environmental exposure (E) has any detectable effect alone, but when both are present there is a strong impact on risk. More typical is “quantitative interaction” where the effect of one exposure is enhanced by the presence of the second, e.g. G may have a modest effect in the absence of E but a greater effect in its presence. The converse is termed “qualitative interaction”, where the G effects are reversed in the presence of exposure E. These are crude categories and if dose effects are strong, one could envision a transition point where extreme levels of exposure would blur the lines between categories. Power to detect statistical interaction in observational studies is determined by the true type of interaction (pure, quantitative, etc.), but more importantly by the allele frequencies at the unobserved G controlling risk and by the prevalence of exposure to the environmental risk factor E. Ignoring G×E interaction when it exists could easily lead to overlooking the role of genes influencing risk. Without confirmatory data from independent samples, it is difficult to conclude with certainty the suggestive evidence for G×E interaction seen here is important to the etiology of CP. However, replication studies will require adequate numbers of CP cases to provide sufficient statistical power to detect G×E interaction, and this study is the largest collection of CP case-parent trios reported to date. Nonetheless, the present analysis does clearly show ignoring potential G×E interaction will overlook genes controlling risk to a common birth defect of complex and heterogeneous etiology.
Supplementary Material
ACKNOWLEDGEMENTS
We sincerely thank all of the families at each recruitment site for participating in this international study, and we gratefully acknowledge the invaluable assistance of clinical, field and laboratory staff who contributed to making this work possible. We are particularly grateful to K. Durda and J. L'Heureux of the University of Iowa for assistance with samples and phenotype data: L. Henkle for technical assistance; J. Resick, C. Brandon, and K. Bardi of the University of Pittsburgh for assistance with recruitment; W. Carricato, K. Deeley, and J. Ruff of the University of Pittsburgh for sample handling. We also thank Dr. Christoph Lange for providing guidance in using the PBAT software. Funding to support data collection, genotyping and analysis came from several sources, some to individual investigators (R01-DE-014581, R01-DE-016148, P50-DE-016215, R21-DE-016930, R01-DE-09886, R37-DE-08559, U01-DE-020057, R37-DE-0-8559, U01-DE-020057, R01-DE-012472, R01-DE-0106877) and some to the cleft consortium itself (U01-DE-018993); part of this research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences. This project was part of the Gene, Environment Association Studies (GENEVA) Consortium funded by the National Human Genome Research Institute (NHGRI) to enhance communication and collaboration among researchers conducting genome-wide studies of complex diseases. Our group benefited greatly from the work and efforts of the entire consortium, especially the Coordinating Center (directed by B. Weir and C. Laurie of the University of Washington; U01-HG004446) in data cleaning and preparation for submission to the Database for Genotypes and Phenotypes (dbGaP). We also acknowledge the leadership of T. Manolio of NHGRI and E. Harris of National Institute of Dental and Craniofacial Research (NIDCR). Genotyping services were provided by the Center for Inherited Disease Research (CIDR).
Footnotes
Web Resources: Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
REFERENCES
- Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, et al. A genome wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet. 2010;42:525–29. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRoo LA, Wilcox AJ, Drevon CA, Lie RT. First-trimester maternal alcohol consumption and the risk of infant oral clefts in Norway: a population-based case-control study. Am J Epidemiol. 2008;168:638–46. doi: 10.1093/aje/kwn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genisca AE, Frías JL, Broussard CS, Honein MA, Lammer EJ, Moore CA, et al. National Birth Defects Prevention Study. Orofacial clefts in the National Birth Defects Prevention Study, 1997–2004. Am J Med Genet A. 2009;149A:1149–58. doi: 10.1002/ajmg.a.32854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassibe M, Desmyter L, Langenberg F, Claes O, Boute B, et al. Am Hum Genet. 2010. FAF1, the first gene associated with cleft palate. in press. [Google Scholar]
- Grosen D, Chevrier C, Skytthe A, Bille C, Molsted K, et al. A cohort study of recurrence patterns among more than 54,000 relatives of oral cleft cases in Denmark: Support for the multifactorial threshold model of inheritance. J Med Genet. 2009;47:162–68. doi: 10.1136/jmg.2009.069385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosen D, Bille C, Petersen I, Skytthe A, Hjelmborg J, et al. Risk of oral clefts in twins. Epidemiol. 2011;22:313–319. doi: 10.1097/EDE.0b013e3182125f9c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SJ, Beaty TH, Panny SR, Street NA, Joseph JM, et al. Association study of transforming growth factor alpha (TGFα) TaqI polymorphism and oral clefts: Indication of gene-environment interaction in a population based sample of infants with birth defects. Am J Epidemiol. 1995;141:629–36. doi: 10.1093/oxfordjournals.aje.a117478. [DOI] [PubMed] [Google Scholar]
- Johnson CY, Little J. Folate intake, markers of folate status and oral clefts: is the evidence converging? Intl J Epidemiol. 2008;37:1041–1058. doi: 10.1093/ije/dyn098. [DOI] [PubMed] [Google Scholar]
- Kraft P, Hunter D. Integrating epidemiology and genetic association: the challenge of gene-environment interaction. Phil Trans Soc B. 2005;360:1609–16. doi: 10.1098/rstb.2005.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft P, Yen YC, Stram DO, Morrison J, Gauderman WJ. Exploiting gene-environment interaction to detect genetic associations. Hum Hered. 2007;63:111–19. doi: 10.1159/000099183. [DOI] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19:S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Laird NM, Lange C. Family-based designs in the age of large-scale gene-association studies. Nat Rev Genet. 2006;7:385–94. doi: 10.1038/nrg1839. [DOI] [PubMed] [Google Scholar]
- Lange C, Silverman EK, Xu X, Weiss ST, Laird NM. A multivariate family based association test using generalized estimating equations: FBAT-GEE. Biostatistics. 2003;4:195–206. doi: 10.1093/biostatistics/4.2.195. [DOI] [PubMed] [Google Scholar]
- Li L, Meng T, Jia Z, Zhu G, Shi B. Single nucleotide polymorphism associated with nonsyndromic cleft palate influences the processing of miR-140. Am J Med Genet A. 2009;152A:856–62. doi: 10.1002/ajmg.a.33236. [DOI] [PubMed] [Google Scholar]
- Li L, Zhu G, Meng T, Wu J, Shi B. Mother passive smoking, a genetic polymorphism in pre-miR-140 and non-syndromic cleft palate: a biological and epidemiological study of gene-environment interaction. 2010. in press. [Google Scholar]
- Little J, Cardy A, Munger RG. Tobacco smoking and oral clefts: a meta-analysis. Bull World Health Organ. 2004;82:213–18. [PMC free article] [PubMed] [Google Scholar]
- Manning AK, LaValley M, Liu CT, Rice K, An P, Liu Y, Miljkovic I, Rasmussen-Torvik L, Harris TB, Province MA, Borecki IB, Florez JC, Meigs JB, Cupples LA, Dupuis J. Meta-analysis of gene-environment interaction: Joint estimation of SNP and SNPxEnvironment regression coefficients. Genet Epidemiol. 2011;35:11–18. doi: 10.1002/gepi.20546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestri NE, Beaty TH, Hetmanski JB, Smith A, McIntosh I, et al. Application of transmission disequilibrium tests to nonsyndromic oral clefts: Including candidate genes and environmental exposures in the models. Am J Med Genet. 1997;73:337–44. doi: 10.1002/(sici)1096-8628(19971219)73:3<337::aid-ajmg21>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Marazita ML. Segregation Analysis. In: Wyszynski D, editor. Cleft Lip and Palate: From Origin to Treatment. Oxford University Press; Oxford: 2002. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivertsen A, Wilcox AJ, Skjaerven R, Vindense HA, Abyholm F, et al. Familial risk of oral clefts by morphological type and severity: population based cohort study of first degree relatives. BMJ. 2008;336:432–34. doi: 10.1136/bmj.39458.563611.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T, Yang L, Li PQ, Wu H, Xie HB, et al. Association of Wnt3A gene variants with non-syndromic cleft lip with or without cleft palate in Chinese population. Arch Oral Biol. 2010;56:73–78. doi: 10.1016/j.archoralbio.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Zeiger JS, Beaty TH, Liang KY. Oral clefts, maternal smoking and TGFA: Meta-analysis of gene-environment interaction. Cleft Palate-Craniofac J. 2005;42:58–63. doi: 10.1597/02-128.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.