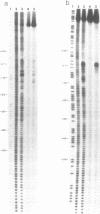
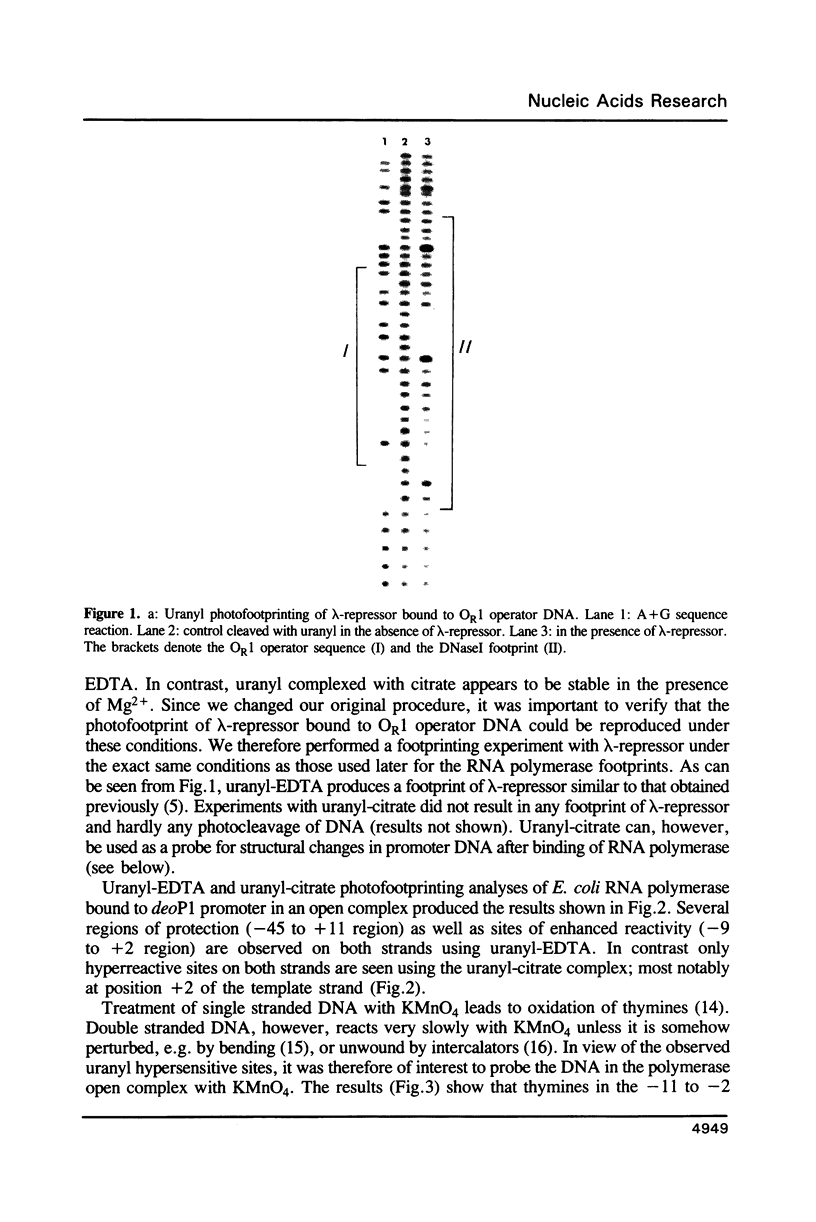
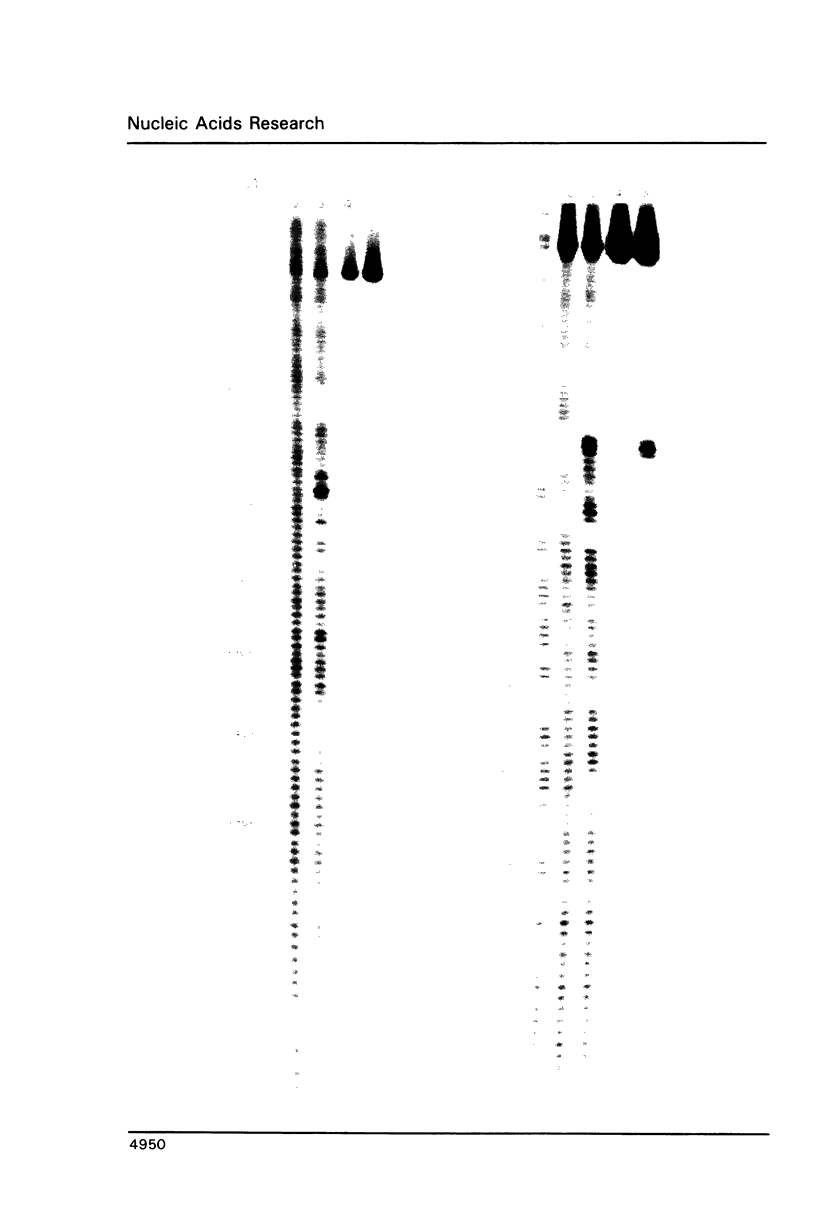
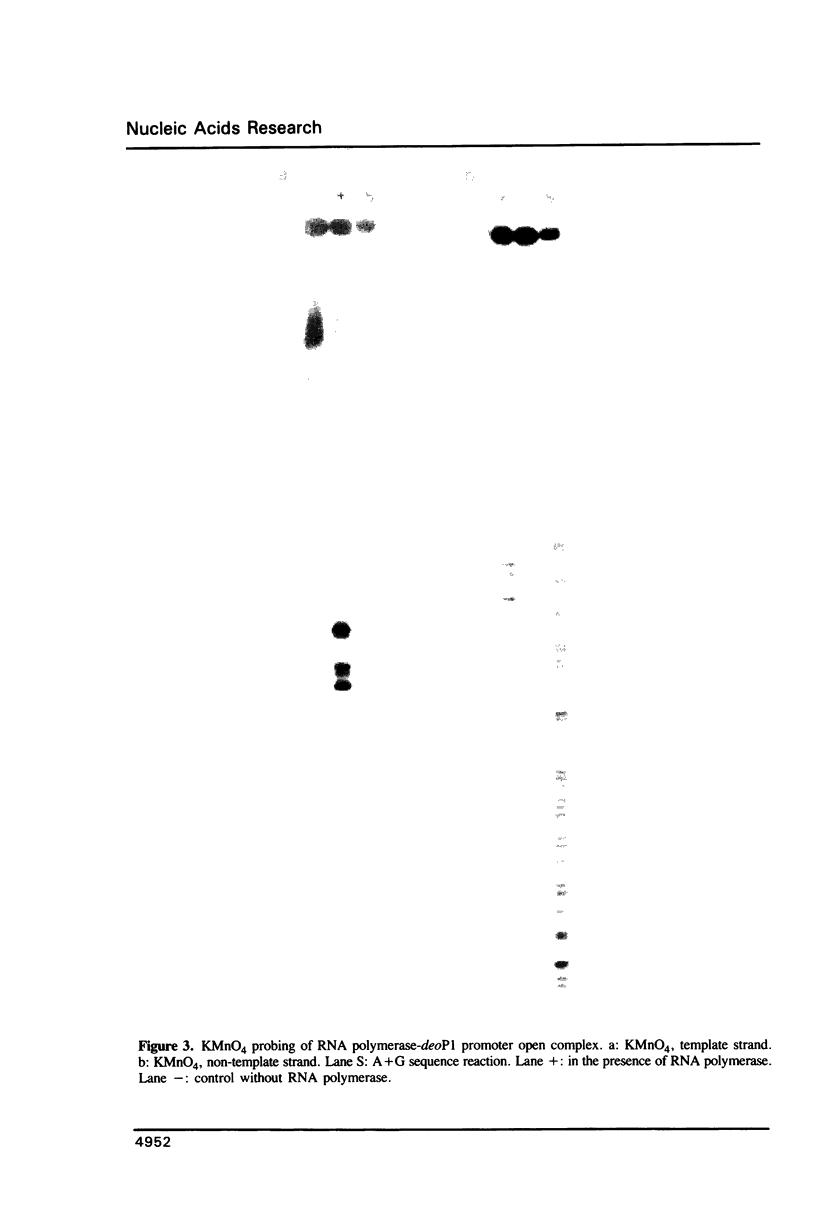
Abstract
Employing a newly developed uranyl photofootprinting technique (Nielsen et al. (1988) FEBS Lett. 235, 122), we have analyzed the structure of the E. coli RNA polymerase deoP1 promoter open complex. The results show strong polymerase DNA backbone contacts in the -40, -10, and most notably in the +10 region. These results suggest that unwinding of the -12 to +3 region of the promoter in the open complex is mediated through polymerase DNA backbone contacts on both sides of this region. The pattern of bases that are hyperreactive towards KMnO4 or uranyl within the -12 to +3 region furthermore argues against a model in which this region is simply unwound and/or single stranded. The results indicate specific protein contacts and/or a fixed DNA conformation within the -12 to +3 region.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amouyal M., Buc H. Topological unwinding of strong and weak promoters by RNA polymerase. A comparison between the lac wild-type and the UV5 sites of Escherichia coli. J Mol Biol. 1987 Jun 20;195(4):795–808. doi: 10.1016/0022-2836(87)90485-2. [DOI] [PubMed] [Google Scholar]
- Borowiec J. A., Zhang L., Sasse-Dwight S., Gralla J. D. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J Mol Biol. 1987 Jul 5;196(1):101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- Burgess R. R., Jendrisak J. J. A procedure for the rapid, large-scall purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975 Oct 21;14(21):4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- Burrows H. Shortening of Right Humerus and Left Thigh due to Unobserved Fractures in Early Life (Infantile Scurvy?). Proc R Soc Med. 1910;3(SECT):165–165. doi: 10.1177/003591571000301582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenchick A., Beabealashvilli R., Mirzabekov A. Topography of interaction of Escherichia coli RNA polymerase subunits with lac UV5 promoter. FEBS Lett. 1981 Jun 1;128(1):46–50. doi: 10.1016/0014-5793(81)81076-9. [DOI] [PubMed] [Google Scholar]
- Gamper H. B., Hearst J. E. A topological model for transcription based on unwinding angle analysis of E. coli RNA polymerase binary, initiation and ternary complexes. Cell. 1982 May;29(1):81–90. doi: 10.1016/0092-8674(82)90092-7. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heumann H., Lederer H., Baer G., May R. P., Kjems J. K., Crespi H. L. Spatial arrangement of DNA-dependent RNA polymerase of Escherichia coli and DNA in the specific complex. A neutron small angle scattering study. J Mol Biol. 1988 May 5;201(1):115–125. doi: 10.1016/0022-2836(88)90443-3. [DOI] [PubMed] [Google Scholar]
- Heumann H., Ricchetti M., Werel W. DNA-dependent RNA polymerase of Escherichia coli induces bending or an increased flexibility of DNA by specific complex formation. EMBO J. 1988 Dec 20;7(13):4379–4381. doi: 10.1002/j.1460-2075.1988.tb03336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen C., Buchardt O., Henriksen U., Nielsen P. E. Photocleavage of DNA and photofootprinting of E. coli RNA polymerase bound to promoter DNA by azido-9-acridinylamines. Nucleic Acids Res. 1988 Jul 11;16(13):5755–5770. doi: 10.1093/nar/16.13.5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen C., Jensen K. F., Nielsen P. E. A specific and efficient photoreaction between E. coli RNA polymerase and T+1 in the lacUV5 or deoP1 promoter. Nucleic Acids Res. 1988 Oct 25;16(20):9545–9555. doi: 10.1093/nar/16.20.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen C., Nielsen P. E. Detection of intercalation-induced changes in DNA structure by reaction with diethyl pyrocarbonate or potassium permanganate. Evidence against the induction of Hoogsteen base pairing by echinomycin. FEBS Lett. 1988 Apr 11;231(1):172–176. doi: 10.1016/0014-5793(88)80725-7. [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Pabo C. O., Sauer R. T. Bacteriophage lambda repressor and cro protein: interactions with operator DNA. Methods Enzymol. 1980;65(1):839–856. doi: 10.1016/s0076-6879(80)65078-2. [DOI] [PubMed] [Google Scholar]
- Jordan S. R., Pabo C. O. Structure of the lambda complex at 2.5 A resolution: details of the repressor-operator interactions. Science. 1988 Nov 11;242(4880):893–899. doi: 10.1126/science.3187530. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K., Buc H., Spassky A., Wang J. C. Mapping of single-stranded regions in duplex DNA at the sequence level: single-strand-specific cytosine methylation in RNA polymerase-promoter complexes. Proc Natl Acad Sci U S A. 1983 May;80(9):2544–2548. doi: 10.1073/pnas.80.9.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo A., Sakaguchi N., Melchers F. Organization of the murine Ig-related lambda 5 gene transcribed selectively in pre-B lymphocytes. EMBO J. 1987 Jan;6(1):103–107. doi: 10.1002/j.1460-2075.1987.tb04725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefèvre J. F., Lane A. N., Jardetzky O. A description of conformational transitions in the Pribnow box of the trp promoter of Escherichia coli. Biochemistry. 1988 Feb 23;27(4):1086–1094. doi: 10.1021/bi00404a002. [DOI] [PubMed] [Google Scholar]
- Nielsen P. E., Jeppesen C., Buchardt O. Uranyl salts as photochemical agents for cleavage of DNA and probing of protein-DNA contacts. FEBS Lett. 1988 Aug 1;235(1-2):122–124. doi: 10.1016/0014-5793(88)81245-6. [DOI] [PubMed] [Google Scholar]
- O'Halloran T. V., Frantz B., Shin M. K., Ralston D. M., Wright J. G. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell. 1989 Jan 13;56(1):119–129. doi: 10.1016/0092-8674(89)90990-2. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Lewis M. The operator-binding domain of lambda repressor: structure and DNA recognition. Nature. 1982 Jul 29;298(5873):443–447. doi: 10.1038/298443a0. [DOI] [PubMed] [Google Scholar]
- Ramstein J., Lavery R. Energetic coupling between DNA bending and base pair opening. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7231–7235. doi: 10.1073/pnas.85.19.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznikoff W. S., Siegele D. A., Cowing D. W., Gross C. A. The regulation of transcription initiation in bacteria. Annu Rev Genet. 1985;19:355–387. doi: 10.1146/annurev.ge.19.120185.002035. [DOI] [PubMed] [Google Scholar]
- Rubin C. M., Schmid C. W. Pyrimidine-specific chemical reactions useful for DNA sequencing. Nucleic Acids Res. 1980 Oct 24;8(20):4613–4619. doi: 10.1093/nar/8.20.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. B., Gamper H., Hearst J. E. The effects of covalent additions of a psoralen on transcription by E. coli RNA polymerase. Nucleic Acids Res. 1987 Sep 11;15(17):6843–6854. doi: 10.1093/nar/15.17.6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. B., Gamper H., Van Houten B., Hearst J. E. Interaction of Escherichia coli RNA polymerase with DNA in an elongation complex arrested at a specific psoralen crosslink site. J Mol Biol. 1988 Jan 20;199(2):277–293. doi: 10.1016/0022-2836(88)90314-2. [DOI] [PubMed] [Google Scholar]
- Siebenlist U. RNA polymerase unwinds an 11-base pair segment of a phage T7 promoter. Nature. 1979 Jun 14;279(5714):651–652. doi: 10.1038/279651a0. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Simpson R. B. The molecular topography of RNA polymerase-promoter interaction. Cell. 1979 Oct;18(2):277–285. doi: 10.1016/0092-8674(79)90047-3. [DOI] [PubMed] [Google Scholar]
- Spassky A., Kirkegaard K., Buc H. Changes in the DNA structure of the lac UV5 promoter during formation of an open complex with Escherichia coli RNA polymerase. Biochemistry. 1985 May 21;24(11):2723–2731. doi: 10.1021/bi00332a019. [DOI] [PubMed] [Google Scholar]
- Travers A. A. Structure and function of E. coli promoter DNA. CRC Crit Rev Biochem. 1987;22(3):181–219. doi: 10.3109/10409238709101483. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Hansen P., Aiba H., Schümperli D. The structure of tandem regulatory regions in the deo operon of Escherichia coli K12. EMBO J. 1982;1(3):317–322. doi: 10.1002/j.1460-2075.1982.tb01167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen W. P., Jeppesen C., Nielsen P. E. Psoralen photofootprinting of protein-binding sites on DNA. FEBS Lett. 1988 Feb 29;229(1):73–76. doi: 10.1016/0014-5793(88)80800-7. [DOI] [PubMed] [Google Scholar]