Abstract
The invasion of malignant glioma cells into the surrounding normal brain precludes effective clinical treatment. In this report, we investigated the role of the NH2-terminal FERM domain in the regulation of the promigratory function of Pyk2. We report that the substitution of residues that constitute a small cleft on the surface of the F3 module of the FERM domain do not significantly alter Pyk2 expression but result in the loss of Pyk2 phosphorylation. A monoclonal antibody, designated 12A10, specifically targeting the Pyk2 FERM domain was generated and recognizes an epitope located on the β5C-α1C surface of the F3 module of the FERM domain. Amino acid substitutions in the F3 module that resulted in the loss of Pyk2 phosphorylation also inhibited the binding of 12A10, suggesting that the 12A10 epitope overlaps a site that plays a role in Pyk2 activity. Conjugation of 12A10 to a membrane transport peptide led to intracellular accumulation and inhibition of glioma cell migration in a concentration-dependent manner. A single chain Fv fragment of 12A10 was stable when expressed in the intracellular environment, interacted directly with Pyk2, reduced Pyk2 phosphorylation, and inhibited glioma cell migration in vitro. Stable intracellular expression of the 12A10 scFv significantly extended survival in a glioma xenograft model. Together, these data substantiate a central role for the FERM domain in regulation of Pyk2 activity and identify the F3 module as a novel target to inhibit Pyk2 activity and inhibit glioma progression.
Introduction
Glioblastoma is the most common primary central nervous system tumor and its biology presents significant problems for successful treatment. Chief among these hurdles is the aggressive local invasion of malignant cells from the original tumor. Invasion into the surrounding normal brain renders complete surgical resection impossible. Similarly, chemotherapy and ionizing radiation alone or in combination have produced only a modest increase in median survival due to problems both with the effective targeting of the invading cells and their innate resistance to current chemotherapeutic agents (1). Effective treatment will ultimately require a more thorough understanding of the signaling pathways that drive glioma invasion as well as the identification and specific targeting of the critical signaling effectors.
The nonreceptor tyrosine kinase Pyk2 is highly expressed in the nervous system and uniquely located within cells to transduce information from interactions with the extracellular matrix and soluble mediators through cell surface integrins, receptor tyrosine kinases, and G-protein–coupled receptors into the activation of intracellular signaling pathways that modulate cell growth and migration (2–4). Pyk2 expression is up-regulated in invasive glioma cells relative to cells in the primary tumor core in glioblastoma tumor samples (5) and the phosphorylation of Pyk2 mediates heregulin-stimulated glioma invasion (6). In addition, immunohistochemical analysis of astrocytoma tissue samples of varying pathologic grades indicates Pyk2 expression increases with increasing tumor grade (7). We have previously shown that overexpression of Pyk2 stimulated glioma cell migration (8). Conversely, silencing Pyk2 expression inhibited glioma cell migration in vitro and prolonged survival in an intracranial xenograft model (9, 10). Together, these results support a role for Pyk2 in glioma progression and suggest that Pyk2 inhibition may target glioma invasion and potentially increase efficacy of adjuvant therapies.
Pyk2 contains a number of functional domains including an NH2-terminal FERM domain, a central kinase domain, and two COOH-terminal proline-rich sequences that mediate interactions with proteins containing SH3 domains (11, 12). It is well-appreciated that Pyk2 kinase activity is regulated by increases in intracellular-free calcium (3). However, it is much less well-understood how increased cytoplasmic calcium leads to kinase activation. FERM domains, compact clover-shaped structures composed of three structural modules (designated A, B, and C or F1, F2, and F3 respectively), are typically involved in linking intracellular proteins to the cytoplasmic tails of transmembrane proteins (13). The functional activity of the prototypical FERM domain proteins ezrin, radixin, and moesin is regulated by FERM domain–mediated intramolecular associations (14, 15). Compelling evidence for a similar autoregulatory role of the FERM domain has been described for the closely related focal adhesion kinase FAK. Structural studies have shown that the FAK FERM domain binds directly to the kinase domain inhibiting access to the catalytic cleft preventing phosphorylation of the activation loop (16). Although a similar intramolecular interaction between the Pyk2 FERM domain and the Pyk2 kinase domain has not been shown, experimental results nevertheless support a substantive role for the Pyk2 FERM domain in the regulation of Pyk2 activity (17, 18). Previously, we showed that selected mutations within the Pyk2 FERM domain inhibited Pyk2 phosphorylation and reduced the capacity of Pyk2 to stimulate glioma cell migration (19). In the present study, we show that specific targeting of a cleft on the surface on the F3 module of the Pyk2 FERM domain inhibits glioma cell migration and prolongs survival in a glioma xenograft model. These results further support a regulatory role for the Pyk2 FERM domain and suggest it may represent a novel target to inhibit Pyk2 activity and limit glioma invasion.
Materials and Methods
Antibodies
The anti-FLAG M2 monoclonal antibody (mAb) was from Sigma. The rabbit anti-HA mAb and the polyclonal anti-Pyk2 antibody were from Upstate Biotechnology. The anti-phosphotyrosine mAb pY20 was from BD Biosciences. The anti-Pyk2 mAb OT126 was from United States Biologicals. The horse radish peroxidase–conjugated Fcγ fragment–specific goat anti-mouse IgG and FITC–conjugated anti-mouse were from Jackson ImmunoResearch Laboratories.
Expression Constructs
The construction of the FLAG-epitope tagged wild-type Pyk2 and the HA epitope–tagged Pyk2 FERM domain has been previously described (9). The HA epitope–tagged wild-type FAK has been previously described (8). Pyk2 containing select amino acid substitutions (W104A, Y135C, I308E, D346A, D349A) and the Pyk2 FERM I308E variant have been previously described (19). Additional Pyk2 amino acid substitutions (K42A, R306E, R309A, I348E, Y351A, and R353A) were introduced into FLAG-tagged Pyk2 using the Quickchange site–directed mutagenesis kit (Stratagene). The FAK FERM domain, encoding FAK residues R35-P362, was amplified by PCR and cloned in-frame downstream of a 3× HA epitope in pcDNA3. In the Pyk2 FERM (FAKF3) construct, the Pyk2 FERM F3 module (residues D261-A366) was replaced by the corresponding FAK F3 module (residues D254-P362) by splice overlap extension PCR and cloned in-frame downstream of a 3× HA epitope in pcDNA3. The Pyk2 F3 module sequence encoding amino acid residues D261-A366 was cloned into the inducible expression vector pET28 (Novagen) downstream of a 6× His tag.
Generation of Monoclonal Antibody 12A10
The mouse mAb 12A10 was generated against the F3 module of the Pyk2 FERM domain. The pET28 Pyk2 F3 construct was transformed into Escherichia coli BL21. Bacteria were grown at 30°C to mid-log phase (OD600 = 0.5) and protein expression induced by the addition of isopropyl-l-thio-B-d-galactopyranoside to a final concentration of 0.1 mmol/L. Sixty minutes after induction, bacterial cells were pelleted and frozen at −80°C. Frozen pellets were thawed on ice in CelLytic B-cell lysis reagent (Sigma) containing protease inhibitors. The lysates were clarified by centrifugation and recombinant F3 was purified by batch adsorption on a Ni-NTA resin followed by fast protein liquid chromatography on a Resource Q column (GE Healthcare).
Five Balb/C mice were each given 40 µg of purified F3 in RIBI adjuvant (Sigma) via i.p. injection followed by two booster administrations at days 14 and 28. The immunoreactivity of the sera following the second booster injection against the purified F3 immunogen was compared with the immunoreactivity of mouse prebleed sera by ELISA assay. Three mice with substantial anti-F3 serum titers were splenectomized and the B-lymphocytes fused with the myeloma cell line P3X63-Ag8.653 (ATCC-1580) to generate hybridomas. Subcloning of the hybridomas was done by limiting dilution. Hybridoma supernatants were screened for immunoreactivity with the full-length Pyk2 FERM domain (residues R39-A366) isolated from cell lysates of 293 cells transfected with HA epitope–tagged Pyk2 FERM by a capture sandwich ELISA. The binding of hybridoma supernatant was detected by incubation with a 1:10,000 dilution of a horse radish peroxidase–conjugated Fcγ fragment–specific goat anti-mouse IgG. The wells were then developed with 10 µmol/L O-phenylene diamine and were read at 490 nmol/L. Six hybridomas that exhibited significant immunoreactivity with the wild-type Pyk2 FERM domain were subsequently screened against the Pyk2 FERM variant I308E (19). One hybridoma, designated 12A10, bound to the wild-type Pyk2 FERM domain but failed to bind to Pyk2 FERM I308E. The 12A10 hybridoma was expanded and IgG purified from culture media using Protein G Sepharose column chromatography. The 12A10 hybridoma was isotyped and found to be IgG1.
Synthesis of 12A10–Membrane Transport Sequence Peptide Conjugate
Membrane transport sequence (MTS) peptide conjugated 12A10 mAb was graciously provided by InNexus Biotechnology, Inc., and was generated as previously described (20). The MTS peptide (KGEGAAVLLPVLLAAPG) was obtained from Genemed Synthesis. Purified 12A10 mAb was dialyzed against PBS (pH 6.0) buffer, oxidized by adding 1/10 volume of 200 mmol/L NaIO4, and incubated at 4°C for 30 min in the dark. The reaction was terminated by adding glycerol to a final concentration of 30 mmol/L and the sample dialyzed at 4°C for 1 h against PBS buffer (pH 6.0). The MTS peptide (50-fold molar excess) was added and the sample was incubated at 37°C for 1 h. The resulting antibody-peptide conjugate was dialyzed against PBS (pH 7.4) and stored at 4°C.
Cloning of 12A10 VH and VL Genes and scFv Construction
To generate the 12A10 scFv, total RNA from 12A10 hybridoma cells was isolated using the Trizol reagent. The first strand cDNA was synthesized with a kit from BD Biosciences. The cDNAs for the VH and VL domains were amplified by PCR using Taq polymerase and degenerate primers as described (21). The PCR products for VH and VL were ligated into the pCR2.1 TA cloning vector (Invitrogen) and sequenced. The 12A10 scFv was generated by joining the VH and VL sequences together by splice overlap PCR using oligonucleotides primers that encoded a (Gly4Ser)3 linker between the COOH terminus of the VH and the NH2 terminus of the VL. The resulting PCR fragment was ligated upstream of a HA epitope in expression vector pcDNA3 (Invitrogen) and designated pc12A10 scFv. For stable transduction of glioma cell lines, a fragment containing the HA epitope–tagged 12A10 scFv was excised from pc12A10 scFv and ligated into the lentiviral plasmid vector pWPXL (Addgene). The 12A10 scFv was expressed by the EF1-α promoter as part of a bicistronic transcript encoding DsRed using the encephalomyocarditis virus 5′ internal ribosome entry site (22).
Lentiviral Transduction
Recombinant lentiviruses were produced by transient transfection of 293T cells with 20 µg of the appropriate lentiviral transfer vector construct, 15 µg of psPAX2 packaging plasmid, and 5 µg of pMD2G-VSVG envelope vector by calcium phosphate precipitation. Recombinant lentivirus containing supernatants were harvested 48 h after transfection. For lentiviral transduction, medium containing recombinant 12A10 scFv lentiviruses was added to subconfluent cultures of cells. Control cells were transduced with pWPXL expressing GFP. Forty-eight hours after infection, cells were harvested and GFP- or RFP-positive cells were collected by mass sorting on a fluorescence-activated cell sorting Vantage flow cytometer (BD Biosciences).
Immunoblotting and Immunoprecipitation
Cells were washed in cold PBS, lysed by addition of 1 mL IPB buffer [137 mmol/L NaCl, 20 mmol/L Tris (pH 7.5), 1% NP40, and 10% glycerol containing protease and phosphatase inhibitors] and incubated on ice for 30 min. Lysates were clarified by centrifugation at 16,000 × g for 10 min at 4°C. Protein content of the lysate was determined using the BCA assay (Sigma). Immunoblotting and immunoprecipitation of cleared lysates was done as described (19). Detection was done with horseradish peroxidase–conjugated secondary antibodies and enhanced chemiluminescence (Perkin-Elmer Life Sciences) or by IR detection with the Odyssey Infrared Imaging System (LI-COR Biosciences). Quantitation of band intensity was done with the Odyssey application software v3.0.
Cell Culture and Transfection
The human glioblastoma cell line SF767 and the 293T packaging cells were routinely passaged in DMEM containing 10% fetal bovine serum, 1% nonessential amino acids, 2 mmol/L glutamine, 100 units/mL penicillin, and 10 µg/mL streptomycin. For transfection, subconfluent cultures were transfected with the Effectene reagent (Qiagen) as previously described (19). The primary GBM line 8 (GBM8) was established directly from a patient surgical sample and maintained as a subcutaneous flank xenograft through serial passaging in immune deficient mice (23). GBM8 flank tumor xenografts were harvested, mechanically disaggregated, and grown in short term culture (5–7 d) in DMEM containing 2.5% fetal bovine serum, 1% nonessential amino acids, 2 mmol/L glutamine, 100 units/mL penicillin, and 10 µg/mL streptomycin for lentiviral transduction before intracranial implantation.
Generation of Intracranial Xenograft Tumors
Female athymic nude mice (age 4–5 wk) were randomized into groups of 8 that received either wild-type control–transduced cells or the matching cell type transduced with 12A10 scFv. Power analysis indicated that a sample size of 8 animals for each group will have 80% power to detect a probability of 0.90 that the time till onset of a moribund state in one group is less than the time until onset of a moribund state in another group using a Wicoxon (Mann-Whitney) rank sum test with a 0.05 two-sided significance level. Animals were anesthetized and cells (7 × 105 in 10 µL) were injected into the right basal ganglia using a small animal stereotaxic frame (TSE Systems). Following injection, the craniotomy was filled with bone wax and the skin closed with silk sutures. Mice were weighed daily and observed for the onset of neurologic symptoms or until moribund. When reaching the study end-point, animals were euthanized and formalin-perfused brains were harvested for tissue analysis. The orthotopic xenograft tumor model was done with a protocol approved by the Mayo Institutional Animal Care and Use Committee.
Radial Migration Assay
A monolayer radial migration assay was used as previously described (24). Linear migration from the initial seeded area was determined for at least 10 replicate samples for each infected construct. Specific migration rates were calculated by normalizing the measurements to nonspecific migration on bovine serum albumin. The absolute migration and migratory rates were calculated and group means determined.
Cell Cycle Progression Assay
Cell cycle analysis was done as previously described (9). The percentage of cells in S-phase was calculated using the Modfit Program (Verity Software House, Inc.).
Statistics
Survival of mice with xenografts was determined by Kaplan-Meier analysis using GraphPad Prism 4.0 software (GraphPad, Inc.). Differences between survival curves were compared with the log-rank test. Significance was set at criterion level P value of <0.01.
Results
The Monoclonal Antibody 12A10 Binds to a Functional Site in the Pyk2 FERM Domain
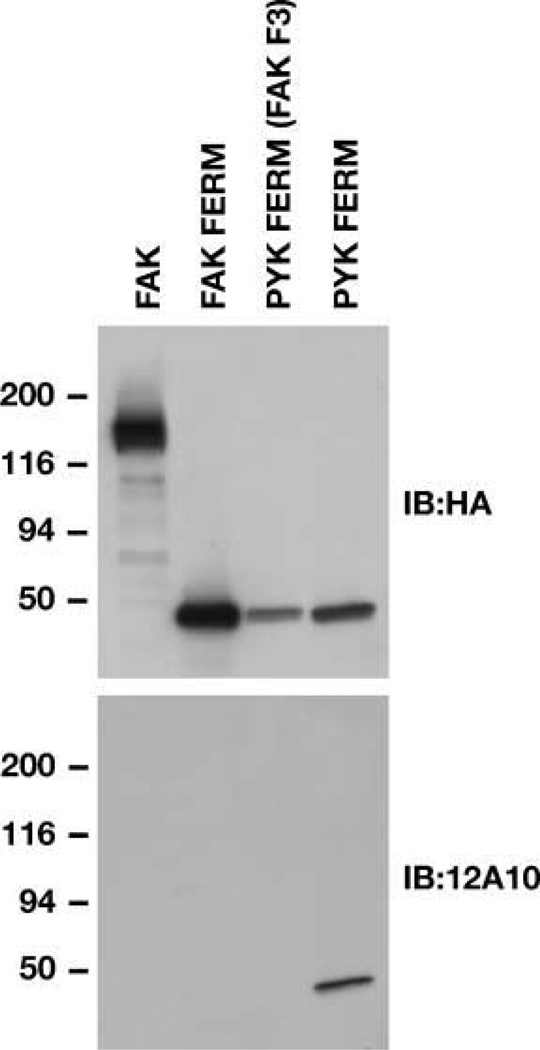
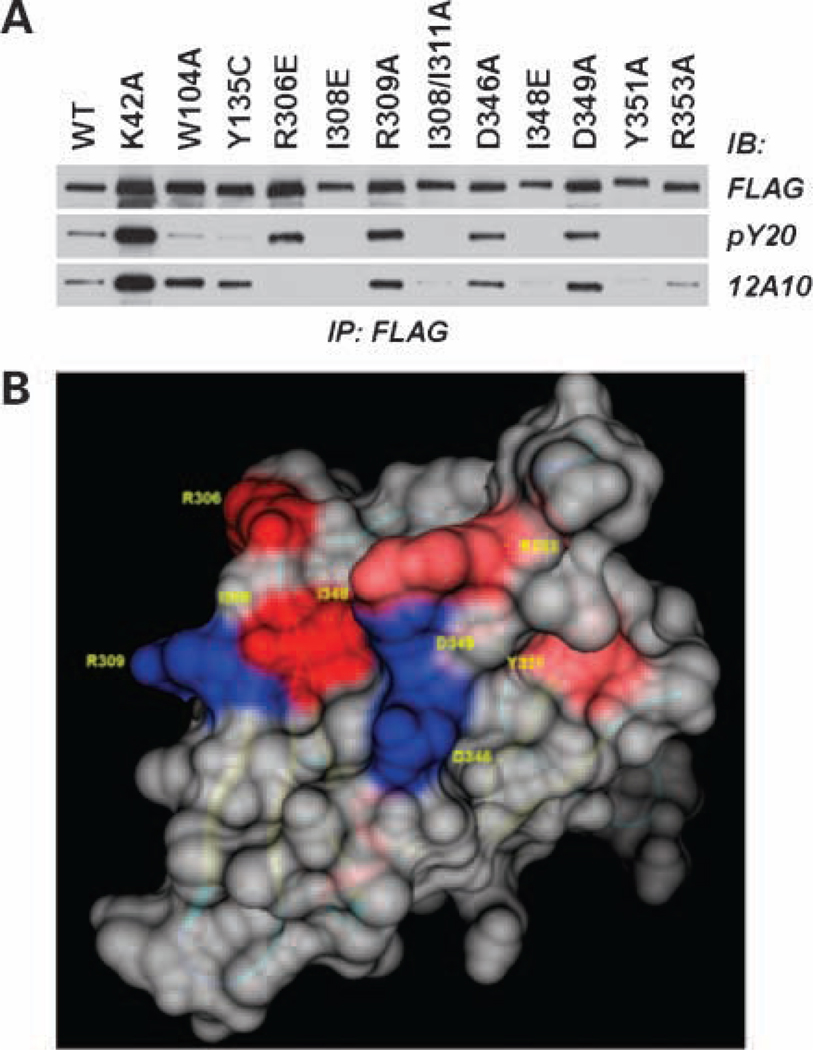
Previously, we showed that the Pyk2 FERM domain played an important role in the promigratory effect of Pyk2 in glioma cells (19). Notably, substitution of I308 in strand β5C of the F3 module of the Pyk2 FERM domain inhibited Pyk2 phosphorylation. In addition, the I308E substitution blocked the inhibitory activity of the autonomously expressed Pyk2 FERM domain. To further investigate the role of the Pyk2 FERM domain in regulating the promigratory activity of Pyk2, we generated a mAb targeting the F3 module of Pyk2 using recombinant F3 domain as immunogen. The mAb designated 12A10 reacted specifically with the F3 module of the Pyk2 FERM domain (Fig. 1). 12A10 did not react with full-length FAK, the FAK FERM domain, or a chimeric Pyk2 FERM domain containing the F1 and F2 modules of the Pyk2 FERM domain and the F3 module of the FAK FERM domain. Several residues in the Pyk2 FERM F3 module were selected for site directed mutagenesis based on a three-dimensional model of the Pyk2 FERM domain (19) and available ligand-bound FERM domain crystal structures (25–27) indicating the importance of a shallow groove formed by residues from strand β5C and helix α1C on the surface of the F3 module in ligand binding. Cells transfected with FLAG-tagged Pyk2 or Pyk2 variants were lysed and immunoprecipitated with anti-FLAG antibodies. The effect of each of the substitutions on Pyk2 phosphorylation and 12A10 binding was examined by immunoblotting the immunoprecipitates with the anti-phospho tyrosine antibody pY20 or antibody 12A10 (Fig. 2A). Although slight variations in expression were observed, none of the substitutions significantly reduced Pyk2 expression relative to expression of Pyk2 wild-type as indicated by blotting the immunoprecipitates with anti-FLAG antibody. However, the substitutions had variable effects on Pyk2 phosphorylation. Consistent with previous results (19), the I308E substitution abrogated Pyk2 phosphorylation. The I308E substitution also resulted in the loss of mAb 12A10 binding. Similarly, the I348E, Y351A, and R353A substitutions also resulted in a loss of Pyk2 phosphorylation and either abolished (I348E, Y351A) or significantly reduced (R353A) mAb 12A10 binding. In contrast, the R309A, D346A, or D349A substitutions did not alter Pyk2 phosphorylation or inhibit the binding of mAb 12A10. The R306E substitution did not inhibit Pyk2 phosphorylation, but this substitution resulted in the loss of mAb 12A10 binding. By comparison, differential effects on 12A10 binding and phosphorylation were observed following substitution of residues highly conserved among FERM domains and previously linked to JAK3 kinase function (28). Pyk2 residue W104 is located in sheet β4 of F1 and occupies the base of the cleft separating subdomains F1 and F3. Residue Y135 is the first residue of the linker segment connecting subdomains F1 and F2. The W104A and Y135C substitutions significantly reduced Pyk2 phosphorylation but did not affect mAb12A10 binding. Substitution of K38 to alanine in the F1 subdomain of the FAK FERM resulted in increased phosphorylation of FAK when expressed in mammalian cells (29). Substitution of the corresponding residue in Pyk2, K42, resulted in an increase in Pyk2 phosphorylation but did not affect the binding of mAb 12A10. These results indicate that the epitope of the 12A10 mAb maps to the β5C-α1C surface of the F3 module of the Pyk2 FERM domain.
Figure 1.
The mAb 12A10 reacts specifically with Pyk2 FERM domain. Lysates of 293 cells transfected with the indicated 3× HA epitope–tagged constructs were analyzed by blotting with anti-HA antibody. Blots were stripped and reprobed with mAb 12A10.
Figure 2.
The mAb 12A10 epitope maps to a surface on the F3 module of the Pyk2 FERM domain. SF767 cells were transfected with FLAG-tagged Pyk2 wild-type (WT) or the indicated FLAG-tagged Pyk2 variant. Pyk2 was immunoprecipitated (IP) with anti-FLAG mAb and immunoblotted (IB) with anti-phosphotyrosine mAb pY20 or mAb 12A10. Antiphosphotyrosine blot was stripped and reprobed with anti-FLAG. B, space fill model of the Pyk2 FERM F3 module highlighting the β5C-α1C surface. Substitution of residues colored blue did not affect mAb 12A10 binding, whereas substitution of residues shaded red abolished or reduced mAb 12A10 binding.
12A10 Inhibits Glioma Cell Migration
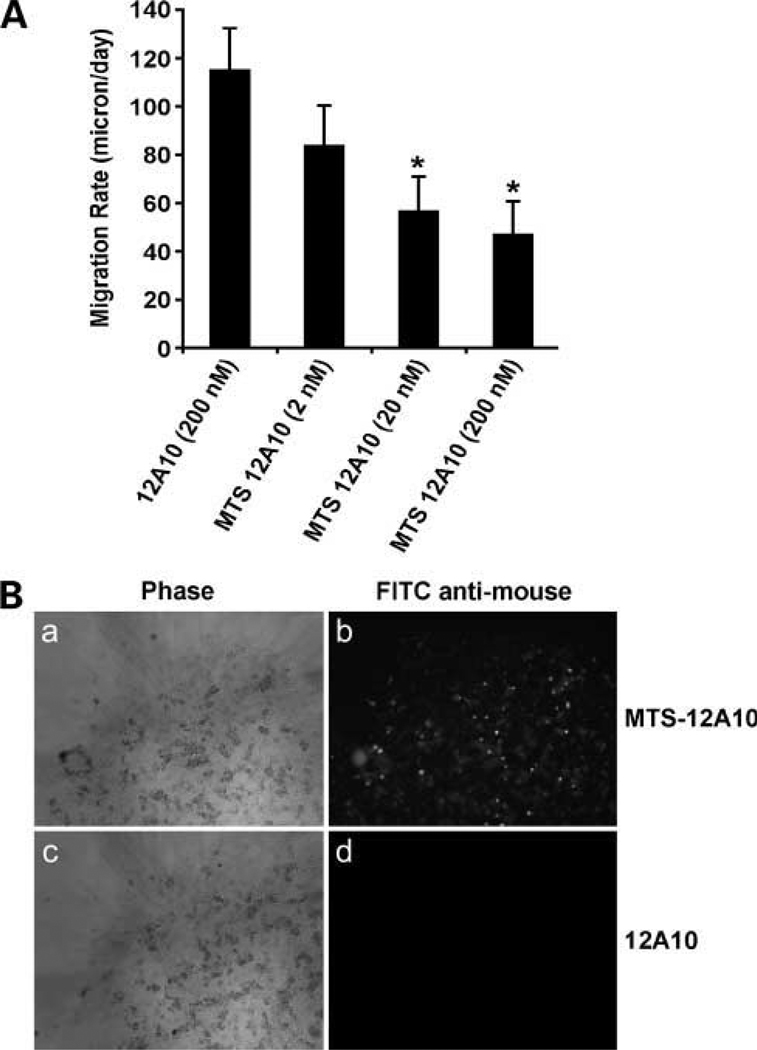
The binding of 12A10 to the F3 module of the Pyk2 FERM was inhibited by F3 amino acid substitutions that inhibited Pyk2 phosphorylation. These results suggested that the binding of mAb 12A10 might interfere with Pyk2 function. To test this possibility, mAb 12A10 was conjugated to a MTS peptide (20, 30) that enabled intracellular localization of 12A10 antibody. SF767 cells were incubated with unconjugated mAb 12A10 or varying concentrations of MTS-conjugated mAb 12A10 for 16 hours and the effect on cell migration examined with a radial migration assay (Fig. 3A). Unconjugated mAb 12A10 did not inhibit glioma cell migration. MTS-conjugated 12A10 mAb inhibited glioma cell migration in a concentration-dependent manner with 50% inhibition at 200 nmol/L. Inhibition was dependent on internalization as cells treated with MTS-conjugated 12A10 were positive for intracellular murine IgG by immunofluorescence, whereas cells treated with unconjugated 12A10 were negative for intracellular immunofluorescence (Fig. 3B).
Figure 3.
MTS-conjugated 12A10 mAb inhibits glioma cell migration. A, SF767 cells were treated with unconjugated 12A10 or MTS-conjugated 12A10 at the indicated concentration for 16 h, and the effect on migration rate was determined by radial migration assay (*, P < 0.05). B, after a migration period of 24 h, cells were fixed, permeabilized, and incubated with a FITC-tagged anti-mouse IgG. A and B, SF767 cells treated with MTS-12A10. C and D, SF767 cells treated with unconjugated 12A10. Phase contrast images are in A and C. fluorescent images are in B and D.
Construction and Characterization of 12A10 scFv
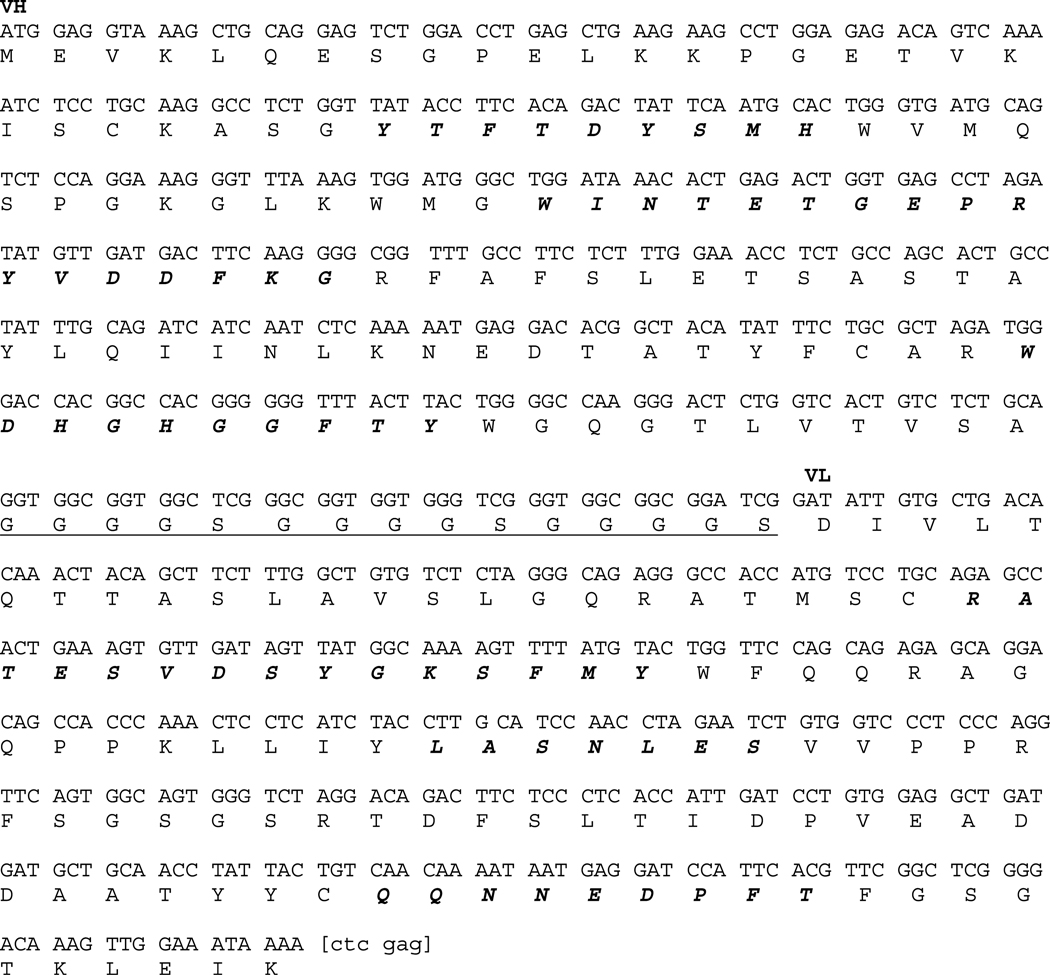
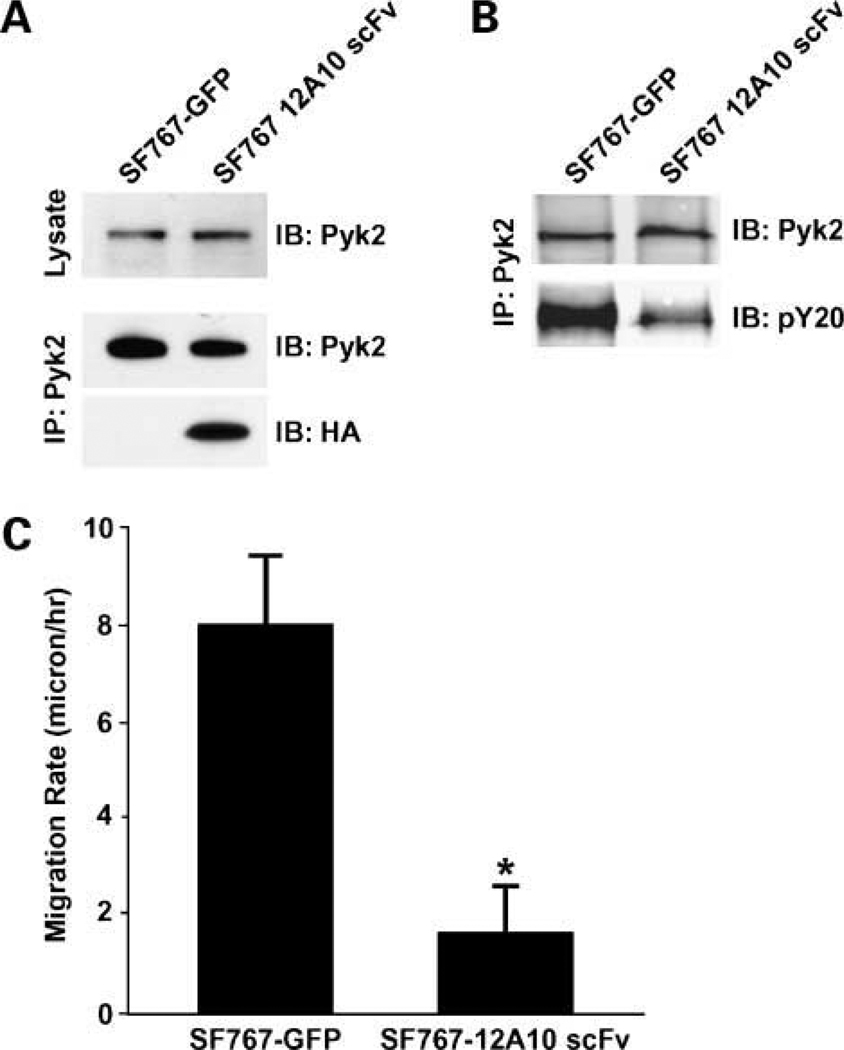
To evaluate the potential of specifically targeting the F3 module of the Pyk2 FERM domain, we developed a 12A10 single chain antibody. The 12A10 scFv fragment was generated by joining fragments encoding the VH and VL with a (G4S)3 linker. The nucleotide and deduced amino acid sequence of the 12A10 scFv is given in Fig. 4. SF767-GFP control cells or SF767 cells stably expressing 12A10 scFv were generated by lentiviral transduction and collected by mass sorting on a flow cytometer. Intracellular expression of the 12A10 scFv did not alter cell growth as cell cycle analysis indicated that the percentage of SF767-12A10 scFv in S-phase was not different than that of the control SF767 cells (31.41 ± 0.28 versus 32.46 ± 1.49 respectively; P = 0.38). In addition, immunoblotting of whole cell lysates indicated that expression of the 12A10 scFv did not alter endogenous Pyk2 expression (Fig. 5A). The intracellular expression and function of scFvs is often unpredictable due to problems often associated with correct folding in the reducing intracellular environment (31). To determine whether the 12A10 scFv retained its capacity to interact with Pyk2 intracellularly, we did immunoprecipitation experiments. Control SF767 cells or SF767-12A10 scFv cells were lysed, the endogenous Pyk2 immunoprecipitated with anti-Pyk2 antibodies, and the immunoprecipitates probed for the presence of the 12A10 scFv. As shown in Fig. 5A, 12A10 scFv was coimmunoprecitated with Pyk2 indicating that the 12A10 scFv retained its capacity to bind to Pyk2 in the intracellular environment. To determine the effect of 12A10 scFv expression on Pyk2 activity, Pyk2 was immunoprecipitated from SF767 GFP and SF767-12A10 scFv cells, and the immunoprecipitates blotted with the anti-phospho tyrosine antibody pY20. There was a 57% reduction in Pyk2 phosphorylation in the SF767-12A10 scFv cells relative to the control cells (Fig. 5B). That a comparable amount of Pyk2 was present in the immunoprecipitates was verified by reprobing the blots with and anti-Pyk2 antibody. Next, we tested the effect of 12A10 scFv expression on glioma cell migration. Intracellular expression of the 12A10 scFv significantly inhibited the migration of SF767 12A10 scFv relative to the migration of control SF767 cells transduced with vector alone (Fig. 5C).
Figure 4.
Nucleotide and deduced amino acid sequence of the 12A10 scFv. VH and VL sequences are joined by a (G4S)3 linker peptide (underlined). CDR regions are indicated in italics. An Xho restriction site, enclosed in brackets on the 3′ end, is used to join a 3× HA epitope tag.
Figure 5.
The 12A10 scFv reacts intracellularly with Pyk2, reduces Pyk2 phosphorylation, and inhibits glioma cell migration. A, whole cell lysates of SF767-GFP control cells or SF767-12A10 scFv cells were immunoblotted with anti-Pyk2 mAb (top). Lysates of SF767 control cells or SF767-12A10 scFv cells were immunoprecipitated with the polyclonal anti-Pyk2 antibody and the precipitates blotted with the anti-Pyk2 mAb (middle). The blots were stripped and reprobed with the anti-HA antibody to show that the 12A10 scFv was associated with endogenous Pyk2 (bottom). B, lysates of SF767-GFP control cells or SF767-12A10 scFv cells were immunoprecipitated with polyclonal anti-Pyk2 antibody and the precipitates immunoblotted with the anti-phosphotyrosine antibody pY20. Blots were stripped and reprobed with anti-Pyk2 antibody to verify equal amounts of Pyk2 in the immunoprecipitate. C, radial migration assay of SF767-GFP and SF767-12A10 scFv cells on 10 µg/mL laminin. *, P < 0.05.
Expression of 12A10 scFv Extends Survival of Orthotopic Xenograft Mice
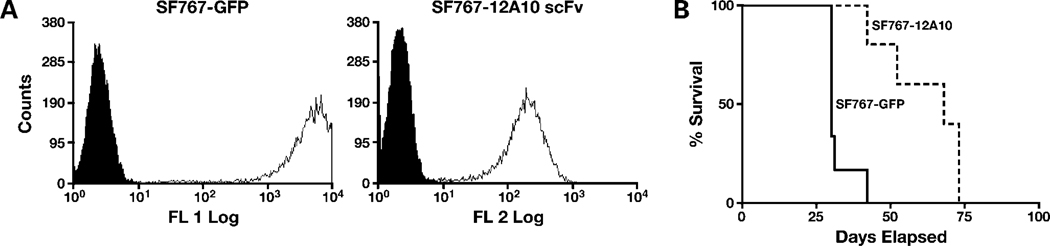
To examine the effect of targeting the Pyk2 FERM domain on tumor progression in vivo, SF767 glioma cells with stable expression of the 12A10 scFv were generated by transduction with a lentiviral construct encoding the 12A10 scFv and red fluorescent protein. SF767 control cells were transduced with the same lentiviral vector expressing only GFP. Transduced cells were mass sorted on a flow cytometer (Fig. 6A), and positive cells were intracranially implanted into nude mice. Mice with xenografts established with control SF767 cells survived a mean of 30 days (Fig. 6A). In contrast, the mean survival duration for the mice with xenografts established with SF767 cells expressing the 12A10 scFv was 68 days, which was significantly longer than the control group (P = 0.0014). One mouse developed an unrelated abdominal distention due to an intestinal obstruction and was euthanized on day 34. Two mice developed neurologic symptoms consistent with those related with tumor burden and were sacrificed at days 42 and 68. The remaining mice remained healthy without demonstrating any neurologic symptoms requiring euthanasia and were sacrificed on day 73. Brains obtained from mice were paraffin embedded, sectioned, and stained for gross inspection for tumor. Brain slices from the 42-day SF767 12A10 scFv survivor mouse had observable tumor cells, whereas the remaining 73-day survival SF767 12A10 scFv mice had no gross observable tumor burden (data not shown).
Figure 6.
12A10 scFv expression increases survival in an intracranial xenograft model. Control SF767 cells expressing GFP or SF767 cells expressing 12A10 scFv along with dsRed were generated by lentiviral transduction. A, fluorescence-activated cell sorting histograms of mass-sorted populations with cell number on the ordinate and fluorescence intensity of the abcissa. B, survival curves of athymic nude mice with intracranial xenografts of SF767-GFP or SF767-12A10 scFv cells. Survival curves show a significant survival benefit for the mice with SF767 12A10 scFv xenografts (P = 0.0014).
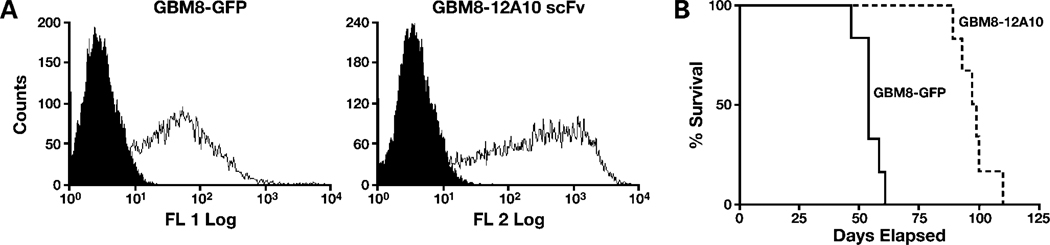
To substantiate the results obtained with the SF767 glioma cell line, we examined the effect of intracellular expression of the 12A10 scFv on survival of mice with intracranial xenografts established with a primary glioblastoma xenograft cell line GBM8. GBM8 is from a panel of serially propagated GBM xenografts shown to maintain the morphologic and molecular characteristics of the corresponding patient tumor (32, 33). Control GBM8 cells and GBM8-12A10 scFv were established by lentiviral transduction, and transduced cells were mass sorted on a flow cytometer (Fig. 7A). Consistent with the results obtained with the SF767 cell line, mice with GBM8-12A10 scFv xenografts survived a mean of 98 days (Fig. 7B), which was significantly longer than control transduced GBM8 cells that survived a mean of 54 days (P = 0.0005).
Figure 7.
12A10 scFv expression increases survival of mice with primary GBM cell xenografts. Control GBM8 cells expressing GFP or GBM8 cells expressing 12A10 scFv along with dsRed were generated by lentiviral transduction. A, fluorescence-activated cell sorting histograms of mass-sorted populations. B, survival curves of athymic nude mice with intracranial xenografts of GBM8-GFP or GBM8-12A10 scFv cells. Mice with GBM8-12A10 xenografts exhibited a significant survival benefit relative to mice with control GBM8 xenografts (P = 0.0005).
Discussion
We have previously reported that the NH2-terminal FERM domain of Pyk2 played a critical role in regulating the Pyk2-mediated stimulation of glioma migration (19). In the current study, we investigated the functional importance of the F3 module of the FERM domain in the regulation of Pyk2 activity. The major findings of this report are (a) a mAb, 12A10, was generated that recognizes an epitope located on the β5C-α1C surface of the F3 module of the FERM domain and overlaps a site associated with Pyk2 activity; (b) conjugation of 12A10 to a membrane transport peptide led to intracellular accumulation and inhibition of glioma cell migration; (c) a scFv fragment of 12A10 was stable when expressed in the intracellular environment, interacted directly with Pyk2, reduced Pyk2 phosphorylation, and inhibited glioma cell migration; and (c) stable expression of 12A10 scFv significantly extended survival in a glioma xenograft model. Together, these data substantiate a central role for the FERM domain in regulation of Pyk2 activity and identify the F3 module as a potential novel target to inhibit Pyk2 activity and inhibit glioma progression.
Pyk2 function is modulated in large part by the actions of several discrete protein modules. A central kinase domain mediates the transphosphorylation of Pyk2 itself (34), leading to the recruitment of Src and subsequent phosphorylation of additional Pyk2 tyrosine residues. These phosphorylated tyrosine residues enable interaction of Pyk2 with a variety of SH2 containing adapter and signaling molecules. Pyk2 also contains several proline-rich domains and an NH2-terminal FERM domain. Two COOH-terminal proline-rich sequences and a third proline-rich motif juxtaposed between the FERM domain and kinase domain provide docking sites for SH3 containing proteins instrumental in effective signal transduction. FERM domains were first described in the ezrin/radixin/moesin family of proteins that function as conformationally regulated cross-linkers of cortical actin filaments and the plasma membrane (35). The NH2-terminal FERM domain of the FAK tyrosine kinase has been of considerable interest recently as a potential autoregulatory domain. The crystal structure of a FAK fragment containing the FERM and kinase domains revealed that the FAK FERM domain–regulated FAK activity by directly interacting with the kinase domain and blocking access to the catalytic cleft (16). Displacement of the FERM domain would therefore be required for FAK activation. The mechanism for displacement of the FERM domain remains to be defined, but it has been proposed that displacement may result from the binding of an activating protein to the FERM domain (16). The FERM domain of Pyk2 shares significant identity with the FAK FERM domain and has also been previously implicated in regulation of function (36). However, we and others (17) have been unable to observe an interaction between the Pyk2 FERM domain and the kinase domain, suggesting that the mechanism of regulation of Pyk2 activity by the Pyk2 FERM domain is different that that proposed for FAK. Indeed, Kohno et al. (18) have recently suggested that the Pyk2 FERM domain regulates Pyk2 activity domain by mediating a Ca2+/calmodulin dependent Pyk2 homodimer formation and resultant transphosphorylation. It was shown that expression of an autonomous Py2 FERM domain inhibited Pyk2 phosphorylation likely through the formation of a Pyk2 FERM-Pyk2 heterodimer that blocked transphosphorylation. They reported that Ca2+/calmodulin bound to α-helix 2 in the F2 module, a site that is distinct from the F3 module surface described in the current study. Interestingly, a second Ca2+/calmodulin binding site was very recently reported within the catalytic domain of Pyk2 (37). The relationship between these two sites and the regulation of Pyk2 activity remains to be determined.
We have previously shown that expression of an autonomous Pyk2 FERM domain specifically inhibited Pyk2 phosphorylation and the capacity of Pyk2 to stimulate glioma cell migration (19). One explanation for this result is that the expression of an autonomous FERM domain could inhibit Pyk2 activity by blocking the formation of the Pyk2 homodimer (18). An alternative possibility is that expression of an autonomous FERM domain could competitively inhibit FERM domain–mediated protein-protein interactions that regulate Pyk2 activity. Previous results (19) and results of the current study suggest that these interactions may be mediated by the F3 module of the FERM domain. Specifically, substitution of residues on the β5C-α1C face of the F3 module resulted in the loss of Pyk2 phosphorylation and the inhibitory activity of the exogenously expressed FERM domain. The F3 module adopts the fold of a phosphotyrosine binding and pleckstrin-homology domain (38). Notably, the cleft formed between β5C and α1C on the surface of the F3 module has been shown to serve as a recognition site both for phosphorylated peptides, which contain an NPXY motif (38–40) as well as nonphosphorylated peptide sequences (25–27) indicating multiple modes of ligand recognition. We tested the potential importance of the β5C-α1C cleft in mediating interactions critical to regulation of Pyk2 activity by raising a mAb to this surface based on a hypothesis that it could potentially interfere with protein-protein interactions mediated by this cleft. The mAb 12A10 reacted specifically with the Pyk2 FERM domain and did not react with the FAK FERM domain despite the significant sequence identity between these two domains. The epitope of 12A10 mapped to the F3 module as the binding of 12A10 antibody was inhibited by mutation of residues that contribute to the β5C-α1C surface of F3. Moreover, there was a strong correlation between those mutations that resulted in loss of Pyk2 phosphorylation and those mutations that abrogated 12A10 binding, suggesting that the 12A10 epitope overlaps a site that was congruent with Pyk2 activity. The functional significance of this site was corroborated by studies investigating the role of this site in mediating the promigratory effect of Pyk2 on glioma migration. Treatment of glioma cells with 12A10 conjugated to a membrane transport peptide significantly inhibited glioma cell migration in vitro. Inhibition was dependent on intracellular accumulation as unconjugated 12A10 had no effect on migration. Moreover, expression of a 12A10 scFv in the cytoplasm maintained its ability to interact specifically with Pyk2, reduced Pyk2 phosphorylation, inhibited glioma cell migration in vitro, and significantly increased survival of mice with intracranial xenografts, suggesting that antibody binding to this site interfered with important functional protein interactions. Together, these results support a critical role for the F3 module surface recognized by 12A10 in the promigratory function of Pyk2 in glioma.
Intracellular molecular targeting with endogenously expressed antibody-based biologics (intrabodies) or cell-penetrating whole antibody or scFvs (transbodies) represents an intriguing area of therapeutic discovery with an increasing number successful applications (41–43). Intracellular expression of scFvs can inhibit cellular processes through a variety of different mechanisms (41). Utilizing peptide tags that encode intracellular trafficking signals, scFv intrabodies have been used to inhibit intracellular transport of cell surface receptors or to misdirect localization of the targeted protein (44–46). Intrabodies interacting with signaling effectors have been used to inhibit their activity or target them for degradation thereby inhibiting downstream signaling events (47, 48). In addition to inhibiting protein-protein interactions, intracellular scFvs can function to block protein-DNA interactions. Thus, a nucleus directed anti–cyclin-E scFv inhibited its function and growth of a breast cancer cell line (49). Our current results show that direct intracellular expression of the 12A10 scFv or MTS conjugation of 12A10 mAb resulted in modulation of Pyk2-mediated glioma cell phenotype and as such suggests a potential role of either approach for therapeutic application.
In summary, these data substantiate an important role for the FERM domain in regulation of the promigratory activity of Pyk2 in glioma cells. Previously, we have shown that mutations within the FERM domain, particularly those within the F3 module, significantly inhibited Pyk2 activity and this inhibition correlated with loss of stimulation of migration. The results of the current study show that direct targeting of this surface by a mAb inhibited glioma migration and significantly improved survival in a xenograft model of glioma. These observations support the therapeutic potential of the development of small molecule inhibitors targeting this site in the Pyk2 FERM domain as a novel approach for specific inhibition of Pyk2 activity and the reduction of glioma invasion.
Acknowledgments
We thank Tammy Brehm-Gibson for assistance with generation of the 12A10 mAb, Natalie Meurice for assistance with modeling of the F3 domain, and Nikki Boruff and Dan Riggs for assistance with the graphics.
Grant support: NIH grants CA103956 and CA108961 (J.C. Loftus).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Girault JA, Costa A, Derkinderen P, Studler JM, Toutant M. FAK and PYK2/CAKβ in the nervous system: a link between neuronal activity, plasticity and survival? Trends Neurosci. 1999;22:257–263. doi: 10.1016/s0166-2236(98)01358-7. [DOI] [PubMed] [Google Scholar]
- 3.Avraham H, Park SY, Schinkmann K, Avraham S. RAFTK/Pyk2-mediated cellular signalling. Cell Signal. 2000;12:123–133. doi: 10.1016/s0898-6568(99)00076-5. [DOI] [PubMed] [Google Scholar]
- 4.Watson JM, Harding TW, Golubovskaya V, et al. Inhibition of the calcium-dependent tyrosine kinase (CADTK) blocks monocyte spreading and motility. J Biol Chem. 2001;276:3536–3542. doi: 10.1074/jbc.M006916200. [DOI] [PubMed] [Google Scholar]
- 5.Hoelzinger DB, Mariani L, Weis J, et al. Gene expression profile of glioblastoma multiforme invasive phenotype points to new therapeutic targets. Neoplasia. 2005;7:7–16. doi: 10.1593/neo.04535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Horst EH, Weber I, Ullrich A. Tyrosine phosphorylation of PYK2 mediates heregulin-induced glioma invasion: novel heregulin/HER3-stimulated signaling pathway in glioma. Int J Cancer. 2005;113:689–698. doi: 10.1002/ijc.20643. [DOI] [PubMed] [Google Scholar]
- 7.Gutenberg A, Bruck W, Buchfelder M, Ludwig HC. Expression of tyrosine kinases FAK and Pyk2 in 331 human astrocytomas. Acta Neuropathol (Berl) 2004;108:224–230. doi: 10.1007/s00401-004-0886-3. [DOI] [PubMed] [Google Scholar]
- 8.Lipinski CA, Tran NL, Bay C, et al. Differential role of proline-rich tyrosine kinase 2 and focal adhesion kinase in determining glioblastoma migration and proliferation. Mol Cancer Res. 2003;1:323–332. [PubMed] [Google Scholar]
- 9.Lipinski CA, Tran NL, Menashi E, et al. The tyrosine kinase Pyk2 promotes migration and invasion of glioma cells. Neoplasia. 2005;7:435–445. doi: 10.1593/neo.04712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipinski CA, Tran NL, Viso C, et al. Extended survival of Pyk2 or FAK deficient orthotopic glioma xenografts. J Neuro-oncol. 2008;90:181–189. doi: 10.1007/s11060-008-9656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Avraham S, London R, Fu Y, et al. Identification and characterization of a novel related adhesion focal tyrosine kinase (RAFTK) from megakaryocytes and brain. J Biol Chem. 1995;270:27742–27751. doi: 10.1074/jbc.270.46.27742. [DOI] [PubMed] [Google Scholar]
- 12.Sasaki H, Nagura K, Ishino M, Tobioka H, Kotani K, Sasaki T. Cloning and characterization of cell adhesion kinase β, a novel protein-tyrosine kinase of the focal adhesion kinase subfamily. J Biol Chem. 1995;270:21206–21219. doi: 10.1074/jbc.270.36.21206. [DOI] [PubMed] [Google Scholar]
- 13.Mangeat P, Roy C, Martin M. ERM proteins in cell adhesion and membrane dynamics. Trends Cell Biol. 1999;9:187–192. doi: 10.1016/s0962-8924(99)01544-5. [DOI] [PubMed] [Google Scholar]
- 14.Edwards SD, Keep NH. The 2.7 A crystal structure of the activated FERM domain of moesin: an analysis of structural changes on activation. Biochemistry. 2001;40:7061–7068. doi: 10.1021/bi010419h. [DOI] [PubMed] [Google Scholar]
- 15.Pearson MA, Reczek D, Bretscher A, Karplus PA. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell. 2000;101:259–270. doi: 10.1016/s0092-8674(00)80836-3. [DOI] [PubMed] [Google Scholar]
- 16.Lietha D, Cai X, Ceccarelli DF, Li Y, Schaller MD, Eck MJ. Structural basis for the autoinhibition of focal adhesion kinase. Cell. 2007;129:1177–1187. doi: 10.1016/j.cell.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunty JM, Gabarra-Niecko V, King ML, Ceccarelli DF, Eck MJ, Schaller MD. FERM domain interaction promotes FAK signaling. Mol Cell Biol. 2004;24:5353–5368. doi: 10.1128/MCB.24.12.5353-5368.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohno T, Matsuda E, Sasaki H, Sasaki T. Protein-tyrosine kinase CAKβ/PYK2 is activated by binding Ca2+/calmodulin to FERM F2 α2 helix and thus forming its dimer. Biochem J. 2008;410:513–523. doi: 10.1042/BJ20070665. [DOI] [PubMed] [Google Scholar]
- 19.Lipinski CA, Tran NL, Dooley A, et al. Critical role of the FERM domain in Pyk2 stimulated glioma cell migration. Biochem Biophys Res Commun. 2006;349:939–947. doi: 10.1016/j.bbrc.2006.08.134. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Y, Lou D, Burkett J, Kohler H. Chemical engineering of cell penetrating antibodies. J Immunol Methods. 2001;254:137–145. doi: 10.1016/s0022-1759(01)00410-0. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z, Raifu M, Howard M, et al. Universal PCR amplification of mouse immunoglobulin gene variable regions: the design of degenerate primers and an assessment of the effect of DNA polymerase 3′ to 5′ exo-nuclease activity. J Immunol Methods. 2000;233:167–177. doi: 10.1016/s0022-1759(99)00184-2. [DOI] [PubMed] [Google Scholar]
- 22.Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pandita A, Aldape KD, Zadeh G, Guha A, James CD. Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes Chromosomes Cancer. 2004;39:29–36. doi: 10.1002/gcc.10300. [DOI] [PubMed] [Google Scholar]
- 24.Giese A, Rief MD, Loo MA, Berens ME. Determinants of human astrocytoma migration. Cancer Res. 1994;54:3897–3904. [PubMed] [Google Scholar]
- 25.Hamada K, Shimizu T, Yonemura S, Tsukita S, Hakoshima T. Structural basis of adhesion-molecule recognition by ERM proteins revealed by the crystal structure of the radixin-ICAM-2 complex. EMBO J. 2003;22:502–514. doi: 10.1093/emboj/cdg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Pereda JM, Wegener K, Santelli E, et al. Structural bases for phosphatidylinositol phosphate kinase type I-γ binding to talin at focal adhesions. J Biol Chem. 2005;280:8381–8386. doi: 10.1074/jbc.M413180200. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Alvarez B, de Pereda JM, Calderwood DA, et al. Structural determinants of integrin recognition by talin. Mol Cell. 2003;11:49–58. doi: 10.1016/s1097-2765(02)00823-7. [DOI] [PubMed] [Google Scholar]
- 28.Zhou YJ, Chen M, Cusack NA, et al. Unexpected effects of FERM domain mutations on catalytic activity of Jak3: structural implication for Janus kinases. Mol Cell. 2001;8:959–969. doi: 10.1016/s1097-2765(01)00398-7. [DOI] [PubMed] [Google Scholar]
- 29.Cohen LA, Guan J-L. Residues within the first subdomain of the FERM-like domain in focal adhesion kinase are important in its regulation. J Biol Chem. 2005;280:8197–8207. doi: 10.1074/jbc.M412021200. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y, Brown TL, Kohler H, Müller S. MTS-conjugated-antiactive caspase 3 antibodies inhibit actinomycin D-induced apoptosis. Apoptosis. 2003;8:631–637. doi: 10.1023/A:1026139627930. [DOI] [PubMed] [Google Scholar]
- 31.Cattaneo A, Biocca S. The selection of intracellular antibodies. Trends Biotechnol. 1999;17:115–121. doi: 10.1016/s0167-7799(98)01268-2. [DOI] [PubMed] [Google Scholar]
- 32.Sarkaria JN, Yang L, Grogan PT, et al. Identification of molecular characteristics correlated with glioblastoma sensitivity to EGFR kinase inhibition through use of an intracranial xenograft test panel. Mol Cancer Ther. 2007;6:1167–1174. doi: 10.1158/1535-7163.MCT-06-0691. [DOI] [PubMed] [Google Scholar]
- 33.Giannini C, Sarkaria JN, Saito A, et al. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro-oncol. 2005;7:164–176. doi: 10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park SY, Avraham HK, Avraham S. RAFTK/Pyk2 activation is mediated by trans-acting autophosphorylation in a Src-independent manner. J Biol Chem. 2004;279:33315–33322. doi: 10.1074/jbc.M313527200. [DOI] [PubMed] [Google Scholar]
- 35.Tsukita S, Yonemura S. Cortical actin organization: lessons from ERM (ezrin/radixin/moesin) proteins. J Biol Chem. 1999;274:34507–34510. doi: 10.1074/jbc.274.49.34507. [DOI] [PubMed] [Google Scholar]
- 36.Dunty JM, Schaller MD. The N termini of focal adhesion kinase family members regulate substrate phosphorylation, localization, and cell morphology. J Biol Chem. 2002;277:45644–45654. doi: 10.1074/jbc.M201779200. [DOI] [PubMed] [Google Scholar]
- 37.Xie J, Allen KH, Marguet A, et al. Analysis of the calcium-dependent regulation of proline-rich tyrosine kinase 2 by gonadotropin-releasing hormone. Mol Endocrinol. 2008;22:2322–2335. doi: 10.1210/me.2008-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eck MJ, Dhe-Paganon S, Trub T, Nolte RT, Shoelson SE. Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell. 1996;85:695–-705. doi: 10.1016/s0092-8674(00)81236-2. [DOI] [PubMed] [Google Scholar]
- 39.Li S-C, Zwahlen C, Vincent SJF, et al. Structure of a Numb PTB domain-peptide complex suggests a basis for diverse binding specificity. Nat Struct Mol Biol. 1998;5:1075–1083. doi: 10.1038/4185. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Z, Lee C-H, Mandiyan V, et al. Sequence-specific recognition of the internalization motif of the Alzheimer's amyloid precursor protein by the X11 PTB domain. EMBO J. 1997;16:6141–6150. doi: 10.1093/emboj/16.20.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams BR, Zhenping Z. Intrabody-based approaches to cancer therapy: status and prospects. Curr Med Chem. 2006;13:1473–1480. doi: 10.2174/092986706776872899. [DOI] [PubMed] [Google Scholar]
- 42.Cao T, Heng BC. Intracellular antibodies (intrabodies) versus RNA interference for therapeutic applications. Ann Clin Lab Sci. 2005;35:227–229. [PubMed] [Google Scholar]
- 43.Manikandan J, Pushparaj PN, Melendez AJ. Protein i: interference at protein level by intrabodies. Front Biosci. 2007;12:1344–1352. doi: 10.2741/2152. [DOI] [PubMed] [Google Scholar]
- 44.Boldicke T, Weber H, Mueller PP, Barleon B, Bernal M. Novel highly efficient intrabody mediates complete inhibition of cell surface expression of the human vascular endothelial growth factor receptor-2 (VEGFR-2/KDR) J Immunol Methods. 2005;300:146–159. doi: 10.1016/j.jim.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 45.Wheeler YY, Kute TE, Willingham MC, Chen S-Y, Sane DC. Intrabody-based strategies for inhibition of vascular endothelial growth factor receptor-2: effects on apoptosis, cell growth, and angiogenesis. FASEB J. 2003:1733–1735. doi: 10.1096/fj.02-0942fje. [DOI] [PubMed] [Google Scholar]
- 46.Zhu Q, Zeng C, Huhalov A, et al. Extended half-life and elevated steady-state level of a single-chain Fv intrabody are critical for specific intracellular retargeting of its antigen, caspase-7. J Immunol Methods. 1999;231:207–222. doi: 10.1016/s0022-1759(99)00158-1. [DOI] [PubMed] [Google Scholar]
- 47.Shin I, Edl J, Biswas S, Lin PC, Mernaugh R, Arteaga CL. Proapoptotic activity of cell-permeable anti-Akt single-chain antibodies. Cancer Res. 2005;65:2815–2824. doi: 10.1158/0008-5472.CAN-04-2898. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka T, Rabbitts TH. Intrabodies based on intracellular capture frameworks that bind the RAS protein with high affinity and impair oncogenic transformation. EMBO J. 2003;22:1025–1035. doi: 10.1093/emboj/cdg106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strube RW, Chen S-Y. Characterization of anti-cyclin E single-chain Fv antibodies and intrabodies in breast cancer cells: enhanced intracellular stability of novel sFv-Fc intrabodies. J Immunol Methods. 2002;263:149–167. doi: 10.1016/s0022-1759(02)00035-2. [DOI] [PubMed] [Google Scholar]