Abstract
Common variable immunodeficiency is the most common form of symptomatic primary antibody failure in adults and children. Replacement immunoglobulin is the standard treatment of these patients. By using a differential proteomic approach based on 2D-DIGE, we examined serum samples from normal donors and from matched, naive, and immunoglobulin-treated patients. The results highlighted regulated expression of serum proteins in naive patients. Among the identified proteins, clusterin/ApoJ serum levels were lower in naive patients, compared to normal subjects. This finding was validated in a wider collection of samples from newly enrolled patients. The establishment of a cellular system, based on a human hepatocyte cell line HuH7, allowed to ascertain a potential role in the regulation of CLU gene expression by immunoglobulins.
1. Introduction
Common variable immunodeficiency (CVID), the most frequent symptomatic primary immunodeficiency, is a rare disease, with a prevalence estimated to be between 1 : 25,000 to 1 : 50,000 [1]; unlike many genetic immune defects, the vast majority of patients with CVID are adults between the ages of 20 and 40 years, although many are found outside this age range [2]. CVID is characterized by significantly decreased levels of IgG, IgA, and/or IgM, with poor or absent antibody production, which results in reduction of immune defense [3]. Patients with CVID have an increased susceptibility to infections of the respiratory system and the gastrointestinal tract. These infections can cause irreversible changes of the affected organs, including bronchiectasis, chronic obstructive pulmonary disease, intestinal mucosal atrophy, chronic diarrhea, and protein-wasting enteropathy [2–5]. The clinical course of patients with CVID may also be complicated by a wide spectrum of autoimmune diseases, including systemic immune (e.g., systemic lupus erythematosus or rheumatoid arthritis) or organ-specific disorders (such as thyroiditis, diabetes mellitus type I, vitiligo) [6–14].
The incidence of malignancy appears overall increased in CVID, occurring in up to 15% of patients [15]. Increase in cancer is found most attributable to stomach cancer and non-Hodgkin lymphoma [2, 16–21].
The primary treatment of CVID is replacement of antibody, achieved by either an intravenous or subcutaneous route of Ig, usually in doses of 400 to 600 mg/kg body weight/month [22, 23]. Several gene mutations, associated with approximately 10% of CVID have been recently identified. The affected genes include ICOS (inducible costimulator molecules) [24], CD19 [25, 26], TNFRSF13C (encoding for BAFF-R: B-cell activating factor of the TNF family receptor) [27], and TNFRSF13B, (encoding for TACI: trans-membrane activator and calcium modulating cyclophilin ligand interactor) [28]. All the proteins encoded by these genes are essential for the implementation of proper and effective antibody responses; in fact, they are regulatory molecules variously expressed on B and T lymphocytes, involved at different levels along the route of maturation and activation of B lymphocytes. The complexity of the immune system and its molecular repertoire predicts that additional genetic defects underlying CVID could be identified, which may partly account for the clinical phenotypic heterogeneity of these patients.
One of the most challenging applications of expression proteomics is the quantitative analysis of serum proteins for comparative purposes; availability of serum collections from patients at different steps of disease progression and therapeutic treatments can lead to the discovery of useful biomarkers for diagnosis, evaluation of therapeutic efficacy, and for the clarification of the mechanisms underlying complex diseases [29]. In this paper, we describe the application of 2D-DIGE (two dimension: differential in gel electrophoresis) technology to CVID. On one hand, we compared the serum proteome in a small cohort of newly enrolled, naive patients, to the proteome of healthy individuals, with the aim to detect protein expression differences. Secondly, we compared the serum protein profiles of patients at diagnosis with those occurring one year after intravenous Ig treatment (IVIg), to assess the possible modulation made by the therapy. Clusterin/ApoJ, encoded by the CLU gene, was downregulated in naive patients compared to normal subjects. Serum Clusterin/ApoJ levels were evaluated by western blotting on a wider set of samples, confirming the actual downregulation of this protein in the serum of naive patients. Incubation of the hepatocyte cell line HuH7 with human polyclonal IgG increased the constitutive expression of CLU mRNA.
2. Materials and Methods
2.1. Patients
Patients enrolled for this study were diagnosed and are currently treated at the Center for Diagnosis and Treatment of Adult Primary Immunodeficiency, Division of Clinical Immunology and Allergy of the University of Naples “Federico II”. Diagnosis of CVID was performed according to the diagnostic criteria established by the European Society for Immunodeficiencies (ESID). 15 patients were enrolled (7 males and 8 females), with an average age of 30 at diagnosis. Three naive patients enrolled in the study contributed with serum samples for 2D-DIGE analyses at the time of diagnosis (N1–N3, Table 1) and one year after the beginning of IVIg therapy (I1–I3). Serum samples of 6 naive patients, which were diagnosed with CVID at later times during the collection phase, were characterized by western blot analysis (N4–N9, Table 2). Six additional patients (I4–I9), already receiving IVIg treatment for at least five years, were also tested by western blot analysis. Serum samples from 12 normal donors (C1–C12, 5 male, 7 female; average age 29) were used to generate two pools for 2D-DIGE analysis (P1 and P2, Table 1); six randomly selected samples from the cohort of normal donors were also individually used for western blot analysis. The main clinical features of the patients are reported in Tables 1 and 2.
Table 1.
Clinical and laboratory data of healthy donors (C1–C12) and patients (Pt.1–Pt.3) contributing to serum samples for 2D-DIGE analysis.
| ID | Sex | Serum sample | Age (years) | Total protein level (g/dL) | Ig level (mg/dL) | ERS (mm/1st h) | CRP (mg/dL) | Observed complications | ||
|---|---|---|---|---|---|---|---|---|---|---|
| IgG | IgA | IgM | ||||||||
| C1 | M | 32 | 7 | 1400 | 250 | 150 | 14 | 0.35 | — | |
| C2 | F | 24 | 6.5 | 1100 | 200 | 80 | 14 | 0.33 | — | |
| C3 | M | P1 | 33 | 7 | 1200 | 180 | 85 | 18 | 0.33 | — |
| C4 | M | 29 | 6.8 | 1180 | 300 | 98 | 11 | 0.36 | — | |
| C5 | F | 30 | 6.9 | 1110 | 100 | 100 | 7 | 0.4 | — | |
| C6 | M | 32 | 7.2 | 1300 | 180 | 110 | 7 | 0.35 | — | |
| C7 | F | 27 | 7 | 1280 | 270 | 70 | 4 | 0.30 | — | |
| C8 | F | 31 | 6.6 | 900 | 260 | 90 | 20 | 0.5 | — | |
| C9 | M | P2 | 30 | 6.8 | 980 | 360 | 180 | 2 | 0.5 | — |
| C10 | F | 28 | 7.2 | 1160 | 220 | 150 | 4 | 0.35 | — | |
| C11 | F | 26 | 6.8 | 990 | 230 | 90 | 2 | 0.35 | — | |
| C12 | F | 27 | 7 | 1200 | 250 | 90 | 16 | 0.30 | — | |
| Pt. 1 | M | N1 | 33 | 6.2 | 7 | 6.1 | 10.5 | 9 | <0.31 | R, A |
| I1 | 34 | 7.5 | 400 | 6.1 | 10 | 20 | 0.5 | R, A | ||
| Pt. 2 | F | N2 | 12 | 5.9 | 171 | 6.1 | 4.1 | 7 | 1 | R |
| I2 | 13 | 6.5 | 350 | 6.1 | 6 | 14 | 0.5 | R | ||
| Pt. 3 | M | N3 | 59 | 6.3 | 273 | 13.9 | 38 | 18 | 1.79 | R, GI |
| I3 | 60 | 7 | 380 | 15 | 50 | 24 | 1.4 | R, GI | ||
ESR: erythrocyte sedimentation rate; CRP: C-reactive protein; R: respiratory tract infections; GI: gastrointestinal tract infections; A: autoimmunity.
Table 2.
Clinical and laboratory data of naive (N4–N9) and IVIg-treated patients (I4–I9) contributing to serum samples for western blot determinations of serum clusterin levels.
| ID | Sex | Age (years) | Total protein level (g/dL) | Ig level (mg/dL) | ESR (mm/1st h) | CRP (mg/dL) | Observed complications | ||
|---|---|---|---|---|---|---|---|---|---|
| IgG | IgA | IgM | |||||||
| N4 | M | 33 | 6.9 | 288 | 6.2 | 7.3 | 10 | <0.31 | R |
| N5 | F | 44 | 5.9 | 235 | 12.2 | 43.5 | 28 | 4.59 | R, GI |
| N6 | F | 30 | 5.6 | 7 | 0 | 4.5 | 27 | 7.54 | R, GI, M |
| N7 | F | 38 | 5.7 | 230 | 12.1 | 43.4 | 26 | 4.50 | R, GI |
| N8 | M | 30 | 6.2 | 69 | <6.2 | <4.5 | 4 | <0.31 | R, GI, A |
| N9 | F | 13 | 6.6 | 253 | 6.2 | 8.2 | 8 | 0.75 | R |
| I4 | M | 13 | 7.4 | 418 | 6.1 | 13.3 | 13 | 0.35 | R, GI |
| I5 | M | 20 | 6.6 | 1000 | 13.9 | 54 | 12 | 0.31 | R, GI |
| I6 | F | 39 | 7.1 | 910 | 5.8 | 15 | 10 | 0.31 | R, GI |
| I7 | F | 19 | 6.4 | 700 | 6.1 | 10.5 | 7 | 0.33 | R |
| I8 | M | 47 | 7.2 | 600 | 63 | 4.1 | 8 | 0.31 | R, GI, A |
| I9 | F | 17 | 6.7 | 481 | 6.9 | 4.1 | 10 | 0.31 | R, GI |
ESR: erythrocyte sedimentation rate; CRP: C-reactive protein; R: respiratory tract infections; GI: gastrointestinal tract infections; A: autoimmunity; M: malignancy.
2.2. 2D-DIGE Analysis
For 2D-DIGE analysis, serum samples were subjected to albumin and immunoglobulin depletion on Q-Proteome Albumin/IgG columns (Qiagen), as suggested by the manufacturer. Protein concentration of the albumin/IgG-depleted sera was determined according to Bradford method (BioRad protein assay, BioRad). Samples were finally precipitated in acetone/methanol (8 : 1, v/v) for 16 h, at −20°C and recovered by centrifugation at 16,000 ×g for 30 min, at 4°C. Proteins were solubilised in 7 M urea, 2 M thiourea, 4% CHAPS, 30 mM Tris-HCl. Protein concentrations were redetermined using the Bradford method (Bio-Rad). Before labelling, the pH of the samples was adjusted to pH 8.5 with HCl solutions. Each labelling reaction was performed with 50 μg of the protein extracts, in a 10 μl volume, in the presence of 400 pmol of Cy2-, Cy3-, or Cy5-dyes (minimal labelling dyes, GE Healthcare). Two pools of 6 serum samples from normal donors were generated from a collection of 12 samples (C1 to C12); the two pools, P1 and P2 (Table 1), were individually labelled with Cy3 and Cy5 dye, respectively. A dye-swapping strategy was used to label serum samples from the three enrolled patients (Pt. 1, Pt. 2, and Pt. 3); accordingly, two sera from naive patients were labelled with Cy3, while the third was labelled with Cy5; in a complementary manner, two sera from IVIg-treated patients were labelled with Cy5, and the third was labelled with Cy3. Four mixtures of the 8 samples (50 μg each) were labelled with Cy2 dye, as the internal standard required by the 2D-DIGE protocol. The labelling reactions were performed in the dark for 30 min, at 0°C and were stopped by addition of 1 mM lysine. The four sample mixtures, including appropriate Cy3- and Cy5-labeled pairs and a Cy2-labeled control, were generated and supplemented with 1% IPG buffer (v/v), pH 3–10 NL (GE Healthcare), 1.4% DeStreak reagent (v/v) (GE Healthcare), and 0.2% DTT (w/v) to a final volume of 450 μl in 7 M urea, 2 M thiourea, and 4% CHAPS. The mixtures (150 μg of total protein content) were used for passive hydration of immobilized pH gradient IPG gel strips (24 cm, pH 3–10 NL) for 16 h, at 20°C. Isoelectric focusing (IEF) was carried out with an IPGphor II apparatus (GE Healthcare) up to 75,000 V/h at 20°C (current limit set to 50 μA per strip). The strips were equilibrated in 6 M urea, 2% SDS, 20% glycerol, and 0.375 M Tris-HCl (pH 8.8), for 15 min, in the dark, in the presence of 0.5% DTT (w/v), and then in the presence of 4.5% iodacetamide (w/v) in the same buffer, for additional 15 min. Equilibrated IPG strips were transferred onto 12% polyacrylamide gels, within low-fluorescence glass plates (ETTAN-DALT 6 system, GE Healthcare). The second-dimension SDS-PAGE was performed using a Peltier-cooled DALT II electrophoresis unit (GE Healthcare) at 1.5 W/gel for 16 h. Gels were scanned with a Typhoon 9400 variable mode imager (GE Healthcare) using appropriate excitation/emission wavelengths for Cy2 (488/520 nm), Cy3 (532/580 nm), and Cy5 (633/670 nm). Images were captured in the Image-Quant software (GE Healthcare) and analyzed using the DeCyder 6.0 software (GE Healthcare). A DeCyder differential in-gel-analysis (DIA) module was used for spot detection and pairwise comparison of each sample (Cy3 and Cy5) to the Cy2 mixed standard present in each gel. The DeCyder biological variation analysis (BVA) module was then used to simultaneously match all of the protein-spot maps from the gels, and to calculate average abundance ratios and statistical parameters across triplicate samples (Student's t-test).
For preparative protein separations, 600 μg of unlabeled protein samples from albumin/IgG depleted, pooled serum samples from normal donors and the three CVID patients were used to passively hydrate 24 cm strips for first dimension (pH 3–10 NL IPG strips, GE Healthcare). The first and second dimension runs were carried out as previously described for the analytical separations. After 2-DE, gels were fixed and stained with SyproRuby fluorescent stain (invitrogen). After spot matching with the master gel from the analytical step in the BVA module of DeCyder software, a pick list was generated for spot picking by a robotic picker (Ettan spot picker, GE Healthcare).
2.3. Protein Identification and Bioinformatic Analysis
Isolated spots were minced and washed with water. Proteins were reduced, S-alkylated, and in-gel digested with trypsin, as previously reported [30]. Digest samples were desalted and concentrated on microC18 ZipTips (Millipore Corp., Bedford, MA) using acetonitrile as eluent before MALDI-TOF-MS analysis. Peptide mixtures were loaded on the MALDI target together with α-cyano-4-hydroxycinnamic acid as matrix, using the dried droplet technique. Samples were analyzed with a Voyager-DE PRO spectrometer (Applied Biosystems, Framingham MA, USA). Spectra were acquired in reflectron mode; internal mass calibration was performed with peptides deriving from trypsin autoproteolysis. ProFound software package was used to identify spots from independent nonredundant human sequence databases by peptide mass fingerprint experiments [31]. Candidates with ProFound Est'd Z scores >1.8 (corresponding to P < 0.05 for a significant identification) were further evaluated by the comparison with molecular mass and pI experimental values obtained from 2-DE. The occurrence of protein mixtures was excluded by sequential searches for additional protein components using unmatched peptide masses. Protein identification was confirmed by performing PSD fragment ion spectral analysis of the most abundant mass signal within each MALDI-TOF-MS spectrum, as previously reported [32].
Gene ontology classification of the identified proteins was performed through the web-accessible DAVID (v 6.7) annotation system (http://david.abcc.ncifcrf.gov/home.jsp) [33, 34]. Briefly, the identified proteins were converted into RefSeq-protein identifiers through the DAVID Gene ID conversion tool; the new list was then submitted to functional annotation clustering.
2.4. Western Blotting Analysis
Western blot analysis was used to validate differential expression data obtained by proteomic analysis. Equal volumes of diluted serum samples, corresponding to 0.5 μL of whole serum from 6 normal donors, 6 newly enrolled naive, CVID patients, and 6 inpatients undergoing IVIg replacement therapy since >5 years were separated on 10% polyacrylamide gels by SDS-PAGE and then blotted on PVDF membranes (GE Healthcare). Filters were blocked in PBS containing 5% nonfat dry milk and incubated with a 1 : 250 dilution of the polyclonal, anticlusterin primary antibody H-330 (Santa Cruz Biotechnology), for 2 h. Anti-rabbit IgG horseradish peroxidase conjugated was used as secondary antibody; bands were visualised by ECL kit (GE Healthcare) on a Chemi Doc XRS apparatus (BioRad). Filters were finally probed with the mouse monoclonal anti-ApoH antibody 1D2 (Santa Cruz Biotechnology). Chemiluminescent signals were quantitated through the Quantity One 1-D Analysis Software (BioRad); background signals were subtracted from each value. The net chemiluminescence value due to anticlusterin antibody from each sample was normalized to the corresponding value of ApoH signal. The normal serum sample with highest normalized clusterin expression was arbitrary set to 1, and the normalized clusterin expression of all remaining samples was referred to this value. The t-test method was used for statistical evaluations of relative clusterin levels among the control, naive, and IVIg-treated groups.
2.5. Cell Cultures and Real-Time PCR Analysis
HuH7 cells were cultured in Dulbecco Modified Eagle's Medium supplemented with 10% foetal bovine serum, 1% penicillin/streptomycin, 2 mM L-glutamine (complete medium) at 37°C, under 5% CO2. Cultures were gradually adapted to serum-free medium (Opti-MEM, Invitrogen) by sequential passages in media with increasing concentration of Opti-MEM (1 : 1, 1 : 2, 1 : 3 complete : serum-free medium, resp.), then to 100% serum-free medium. Cells were finally cultured for at least three additional passages in serum-free medium, then evaluated for proliferation by standard growth curves. Adapted HuH7 cells were transferred to 6-well plates and grown in Opti-MEM, then treated with 0.5% (w/v) bovine serum albumin (Sigma-Aldrich) or human polyclonal IgG (Ig VENA, Kedrion Biopharmaceuticals) for 4 h. Total RNA was extracted from triplicate cultures from each treatment with Trizol reagent (Invitrogen). For reverse transcription of DNase-treated total RNA samples (1 μg per sample), the SuperScript VILO cDNA synthesis kit (Invitrogen) was used, with pdN6 primers. Quantitative real-time PCR was performed on a 7500 Real-Time PCR System (Applied Biosystems) using 10 μL of EXPRESS SYBR GreenER qPCR SuperMix with Premixed ROX (Invitrogen), 10 μM each of forward and reverse primers and 10 μL of diluted cDNA (1 : 200). The oligonucleotide primers were designed by the Primer Express software (Applied Biosystems); their sequences were the following: CLU forward, 5′-CGCTGACCGAGGCGTG-3′; CLU reverse, 5′-CCACTCTCCCAGGTCAGCAG-3′; APOH forward, 5′-TGCTATTGCAGGACGGACCT-3′; APOH reverse, 5′-GGCTCATAGAATGTTTTTAACGGG-3′; ACTB forward, 5′-CGTGCTGCTGACCGAGG-3′, ACTB reverse, 5′-GAAGGTCTCAAACATGATCT-3′. For calculation of relative mRNA expression, ACTB (β-actin) transcripts were used as the reference mRNA; data analysis was carried out according to the ΔΔCt method [35, 36].
3. Results and Discussion
3.1. Identification of Serum Proteins Differentially Expressed in CVID
To evaluate distinctive signatures of serum proteins in CVID, we analyzed three available naive patients (Pt. 1–3, Table 1). The corresponding serum samples, taken at the time of diagnosis, were used in 2D-DIGE experiments and compared to pooled serum samples of normal donors (P1 and P2). Serum samples of the three patients were also taken after one year of therapy with IVIg, to evaluate the effects of replacement therapy on serum protein expression. Table 1 summarizes the clinical and laboratory data of the three patients at diagnosis (N1, N2, and N3) and following one year from replacement therapy with intravenous immunoglobulins (I1, I2, and I3). To obtain partial enrichment of serum proteins, all the samples were subjected to affinity chromatography for depletion of albumin and immunoglobulins. The 2D-DIGE analysis on the depleted samples allowed to identify statistically significant variations between the different conditions. Figure 1 shows the image of the gel, selected as the “Master gel”, for spot matching among the 4 analytical gels (run for the quantitative analysis) Figure 1(a), and the preparative gel (run for spot picking and identification) Figure 1(b). Namely, 27 protein spots, reported in Figure 1(a), showed significant relative expression ratios in at least one out of the three possible comparisons (i.e., naive versus normal donors, naive versus IVIg treatment, IVIg treatment versus normal donors, Table 3). To identify the differentially expressed proteins, the spots of interest were excised from the preparative gel (Figure 1(b)), digested with trypsin and subjected to MALDI-TOF-MS analysis. Table 3 shows the details of each protein spot identification. The occurrence of multiple close spots identified as the same protein species, occurring with the horizontal train's aspect typical of glycosylated serum proteins, led to the final recognition of 9 differentially expressed polypeptide entries (Table 3).
Figure 1.
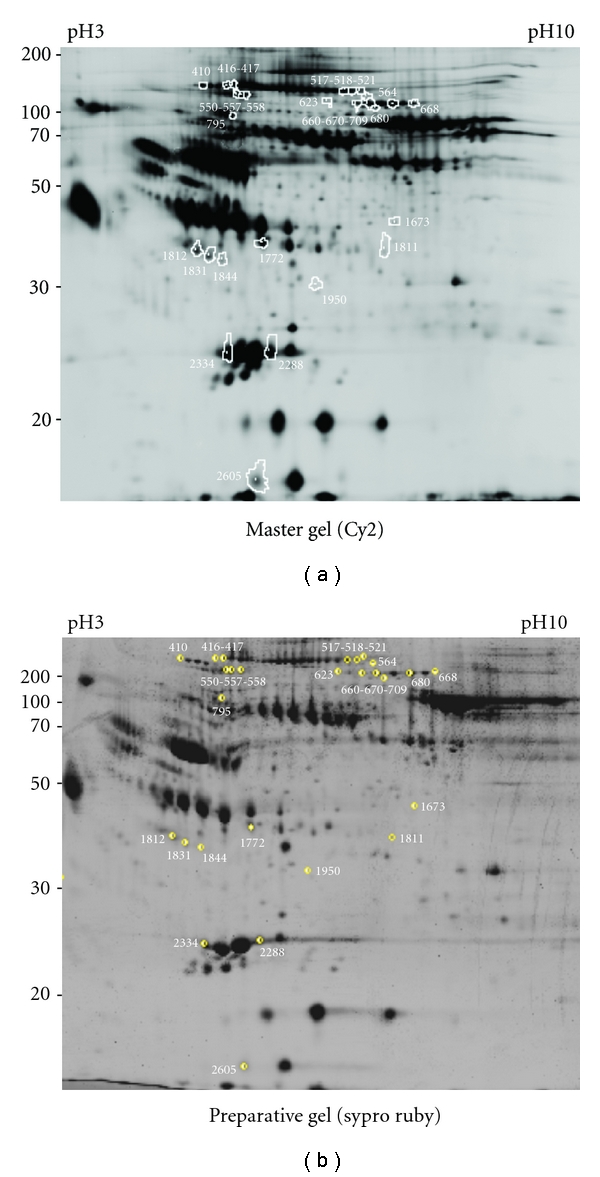
2-DE map of differentially expressed proteins in CVID. The figure shows the position on the master (a) and on the preparative (b) gels of the 27 deregulated spots, as revealed by the comparison of serum samples from normal and CVID patients. In (a), the spots are surrounded by a white border; matched spots within the preparative gel (b) are highlighted by a yellow circle, also indicating the picking surface for the robotic spot picker. The representative image reported in (a) shows to the Cy2-labelled proteins on the scanned master gel; protein spots in (b) were visualized by Sypro Ruby fluorescent staining.
Table 3.
Relative expression and MALDI-TOF-MS-based identification of deregulated spots, as revealed by 2D-DIGE analysis of serum samples.
| Naive/Normal donors | Naive/IVIg donors | IVIg/Normal donors | Protein identification | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spot no. |
Av. Ratio | t-test | Av. Ratio | t-test | Av. Ratio | t-test | Protein name | Gene name | Accession number | Matched peptides | Sequence coverage | Est'd Z score | MW (theor) | pI (theor) |
| 410 | 4.51 | 0.013 | 1.35 | 0.30 | 3.33 | 0.0018 | ||||||||
| 416 | 2.02 | 0.21 | 1.25 | 0.67 | 1.62 | 0.031 | Alpha-1-antitrypsin | SERPINA1 | P01009 | 10 | 28% | 2.34 | 44324 | 5.37 |
| 417 | 1.89 | 0.047 | 1.21 | 0.33 | 1.56 | 0.0081 | ||||||||
| 518 | 4.92 | 0.12 | 1.31 | 0.91 | 3.77 | 0.0051 | ||||||||
| 517 | 5.49 | 0.086 | 1.41 | 0.74 | 3.89 | 0.013 | Alpha-2-macroglobulin | A2M | P01023 | 17 | 17% | 2.32 | 160796 | 5.95 |
| 521 | 5.94 | 0.087 | 1.49 | 0.71 | 4.00 | 0.0082 | ||||||||
| 550 | 2.20 | 0.010 | 1.10 | 0.58 | 1.99 | 0.042 | ||||||||
| 557 | 2.47 | 0.0072 | 1.02 | 0.83 | 2.42 | 0.053 | Ceruloplasmin | CP | P00450 | 12 | 25% | 2.34 | 120085 | 5.41 |
| 558 | 2.52 | 0.0027 | −1.00 | 0.90 | 2.53 | 0.042 | ||||||||
| 564 | 7.48 | 0.11 | 1.04 | 0.78 | 7.16 | 0.013 | Alpha-2-macroglobulin | A2M | P01023 | 9 | 8% | 1.96 | 160796 | 5.95 |
| 623 | 4.27 | 0.048 | 1.07 | 0.66 | 3.99 | 0.29 | Alpha-2-macroglobulin | A2M | P01023 | 12 | 10% | 2.33 | 160796 | 5.95 |
| 666 | 7.08 | 0.0047 | 1.05 | 0.72 | 6.72 | 0.036 | ||||||||
| 668 | 2.75 | 0.043 | 1.21 | 0.57 | 2.27 | 0.035 | ||||||||
| 670 | 5.72 | 0.010 | 1.23 | 0.55 | 4.63 | 0.023 | Complement factor B | CFB | P00751 | 20 | 32% | 2.38 | 83000 | 6.66 |
| 680 | 6.20 | 0.013 | 1.32 | 0.50 | 4.71 | 0.046 | ||||||||
| 709 | 5.22 | 0.065 | 2.79 | 0.10 | 1.87 | 0.014 | ||||||||
| 795 | −1.96 | 0.022 | −1.42 | 0.061 | −1.38 | 0.033 | Afamin | AFM | P43652 | 8 | 13% | 2.25 | 66577 | 5.58 |
| 1673 | 2.61 | 0.12 | −1.41 | 0.44 | 3.68 | 0.039 | Not identified | |||||||
| 1772 | 2.53 | 0.016 | 1.30 | 0.17 | 1.94 | 0.025 | Haptoglobin | HP | P00738 | 12 | 32% | 2.35 | 43349 | 6.13 |
| 1811 | −2.32 | 0.031 | −1.55 | 0.19 | −1.50 | 0.19 | Ficolin-3 | FCN3 | O75636 | 7 | 19% | 1.95 | 30353 | 6.22 |
| 1812 | −2.52 | 0.043 | −1.42 | 0.27 | −1.78 | 0.084 | ||||||||
| 1831 | −2.75 | 0.047 | −1.53 | 0.22 | −1.79 | 0.091 | Clusterin | CLU | P10909 | 6 | 11% | 1.89 | 50062 | 5.89 |
| 1844 | −2.54 | 0.047 | −1.32 | 0.40 | −1.91 | 0.081 | ||||||||
| 1950 | −2.41 | 0.045 | −1.87 | 0.15 | −1.29 | 0.41 | Not identified | |||||||
| 2288 | −4.62 | 0.0042 | −1.72 | 0.19 | −2.68 | 0.053 | Apolipoprotein A-I | APOA1 | P02647 | 11 | 31% | 2.24 | 28078 | 5.27 |
| 2334 | −4.99 | 0.012 | −1.83 | 0.41 | −2.73 | 0.14 | Apolipoprotein A-I | APOA1 | P02647 | 14 | 43% | 2.36 | 28078 | 5.27 |
| 2605 | −3.46 | 0.013 | −1.22 | 0.43 | −2.84 | 0.011 | Not identified | |||||||
3.2. Serum Protein Expression in CVID
Among the identified proteins, gene ontology classification highlighted common participation of the genes encoding alpha-2-macroglobulin, clusterin, complement factor B, ficolin-3, α-1 antitrypsin to inflammatory responses (GOTERM_BP_ALL, “acute inflammatory response”, P = 1.5E − 7), with genes coding for α1-antitrypsin, ceruloplasmin, complement factor B, and haptoglobin being also described as involved in acute-phase response (SP_PIR_KEYWORDS, “acute phase”, P = 1.7E − 7). Most of the spots corresponding to these proteins were upregulated in naive patients, compared to healthy controls (Table 3). Moreover, a quantitative comparison of their relative levels showed less pronounced up-regulation of these proteins in IVIg-treated patients versus normal donors, thus suggesting a mild reduction in the inflammatory status of patients after one-year IVIg therapy. The relative expression ratios of these inflammatory biomarkers were not statistically significant in the comparison of naive versus IVIg-treated patients (Table 3). Collectively, these data indicate that in naive CVID patients there is a smouldering inflammation, which is attenuated, but not abolished, after one-year therapy.
3.3. Validation Analysis of Clusterin Expression in CVID
Three different protein spots (1812, 1831 and 1844), identified as clusterin/ApoJ, were downrepresented in the serum of naive patients, compared to healthy donors (Table 3). The clusterin/ApoJ proteins, encoded by the CLU gene, are produced as a variety of intracellular and secreted isoforms, which are associated with systemic and cellular functions [37–39]. Clusterin/ApoJ protein function was originally associated with inhibition of complement cascade, inflammation, and autoimmunity; its involvement in apoptosis and DNA repair allows also to predict a role of CLU gene in human cancer [37–39]. In the liver, the secreted form of clusterin/ApoJ, sCLU, is produced following alcohol-induced liver injuries [40] and following LPS, tumour necrosis factor, or interleukin-1 stimulation [41]. Interestingly, clusterin expression is downregulated in rheumatoid arthritis [42]. In two out of the three patients evaluated by 2D-DIGE, IVIg treatment resulted in a normalization of the relative abundance of these spots (1812, 1831, 1844), after one-year therapy (Figure 2(a)).
Figure 2.
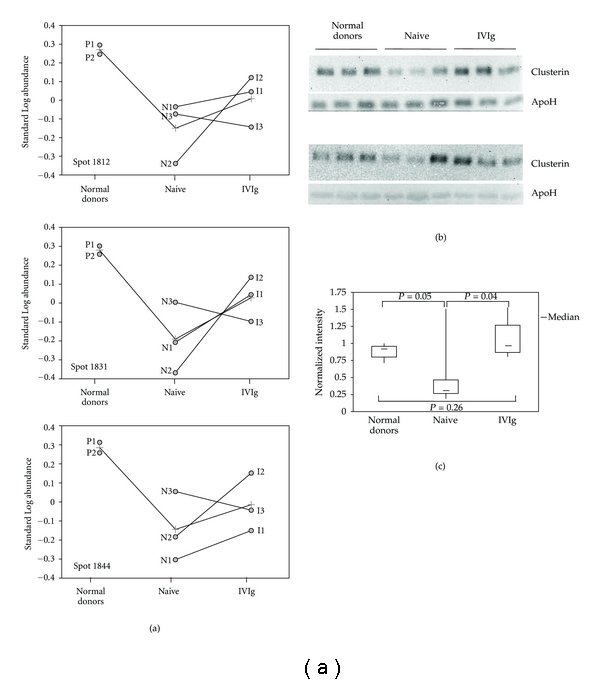
Clusterin expression in CVID. (a) The panel shows the graphic output of DeCyder software (BVA module) for spots 1812, 1831 and 1844, further identified as clusterin. The line connecting normal donors (pools P1 and P2), naive patients (N1–N3), and IVIg-treated patients (T1–T3) in each of the panels represents average levels of the corresponding protein spot. The lines connecting naive samples to the corresponding IVIg-treated samples denote the change in protein expression after one-year substitutive therapy for each of the three patients. (b) Western blot analysis of clusterin in serum samples from normal donors, naive, and IVIg-treated patients. The experiment was performed on individual serum samples from normal donors and newly enrolled, naive and IVIg-treated patients. Each gel was loaded with serum samples from three healthy controls, three naive patients, and three IVIg-treated individuals for >5 years. The filters were first incubated with clusterin antibodies, then with an apolipoprotein H antibody for individual normalization of protein contents. (c) The normal serum with highest normalized expression of clusterin was set to 1, for relative analysis of normalized clusterin expression in the samples loaded on the western blot of Figure 2(b). The data were then used to build the shown box plot. The bottom and top of each box represent the 25th and 75th percentile, respectively; the thick band inside each box shows the 50th percentile (the median). The ends of the whiskers represent the minimum and maximum values of each group of data. The brackets indicate the statistical comparisons between groups with the corresponding P values.
To validate the data originating from 2D-DIGE, we used a different approach, based on western blot analysis of additional, naive, and IVIg-treated CVID patients. In this analysis, we included 6 sera from newly diagnosed CVID patients (naive group), 6 sera from normal donors, and 6 sera from patients undergoing replacement therapy with intravenous immunoglobulins for >5 years (IVIg group). Clinical and laboratory data of the patients are summarised in Table 2. Each gel was loaded with 3 randomized samples from each of the three groups. A clusterin antibody was used to detect clusterin by western blot analysis, while an apolipoprotein H antibody was used for normalization of clusterin levels in each sample. Figure 2(b) shows that clusterin levels in normal subjects were fairly constant, whereas in naive patients, the majority of samples exhibited low levels of clusterin, with the exception of a single naive sample, showing high clusterin expression. The IVIg group was finally characterized by good expression of clusterin in serum. These data were analyzed by a statistical software, and subjected to t-test analysis to evaluate whether significant differences were occurring among the three groups. As shown in Figure 2(c), the analysis confirmed a narrow range for clusterin expression in the serum of normal donors; clusterin expression in the naive group showed a strong enrichment in low-expressing serum samples, highlighting a significant statistical value, compared to controls (P = 0.05), despite the single highly expressing sample. In the IVIg group, a narrower distribution of samples and lack of samples with low clusterin expression can be observed, revealing a statistically relevant difference with respect to the naive group (P = 0.04); finally, the IVIg versus normal donor comparison (P = 0.26) does not show significant differences. These results confirmed the downregulation of clusterin expression in naive patients; in addition, they confirm the trend, observed following 2D-DIGE analysis, in restoring clusterin expression in IVIg-treated patients. One of the difficulties encountered in the study had to face a slow recruitment rate of naive patients, which is an obvious consequence of the low prevalence of CVID. Thus, further studies on larger collections of serum samples from matched, naive, and IVIg-treated patients will be required for a complete evaluation of clusterin expression and correlation analyses with disease progression and therapeutic efficacy. Since CVID actually spans over different primary immunodeficiencies, systematic proteomic approaches on a wide array of affected patients may give relevant contributions for the assessment of the expression signatures characterizing such heterogeneous disorders. The availability of large datasets of differential expression for a panel of serum proteins, including clusterin, may indeed contribute to the accurate stratification of individual patients, according to the peculiar features of the primary immunodeficiency affecting them.
3.4. Human IgGs Increase CLU Gene Expression in Human Hepatocyte Cell Line HuH7
Data reported above suggested that the differentially expressed proteins in serum from CVID patients are derived from peripheral organs, mostly from liver. It seemed therefore of interest to develop a model system for in vitro analysis, based on human hepatocytes, to clarify the potential role of immunoglobulins in modulating the expression of the genes, whose products were altered in CVID. To this aim, we firstly adapted cultures of the human hepatocyte line HuH7, to grow under conditions lacking serum proteins, by gradual deprivation from bovine serum. The presence of growth factors necessary for cell survival and proliferation in culture was insured by the gradual adaptation of these cells to low-protein medium, with a defined composition of trophic factors (Opti-MEM). After stabilization under novel culture conditions, we did not observe alterations in the proliferation of adapted cultures (data not shown). Adapted HuH7 cultures were then treated with bovine serum albumin or human IgG, at protein concentrations comparable to those of Ig present in the circulating blood (0.5% w/v). The relative expression of mRNAs for clusterin was evaluated by quantitative real-time PCR experiments. Apolipoprotein H transcript levels were measured as an additional target in real-time PCR experiments, being this gene unregulated in CVID. As shown in Figure 3, the levels of CLU transcripts were increased (about 40%) in cells treated with immunoglobulins, compared to untreated cells (P = 0.0001) or those treated with BSA (P = 0.0003). By contrast, norelevant changes in mRNA expression were observed for APOH transcripts. These findings suggest that human immunoglobulins exert a modulating activity on the expression of CLU gene in vitro. This is in agreement with what is observed in CVID patients, in which lower expression of clusterin correlates to the naive status (low IgG), while a trend towards normalized levels of circulating clusterin was associated with increased levels of circulating IgG in patients subjected to replacement therapy.
Figure 3.
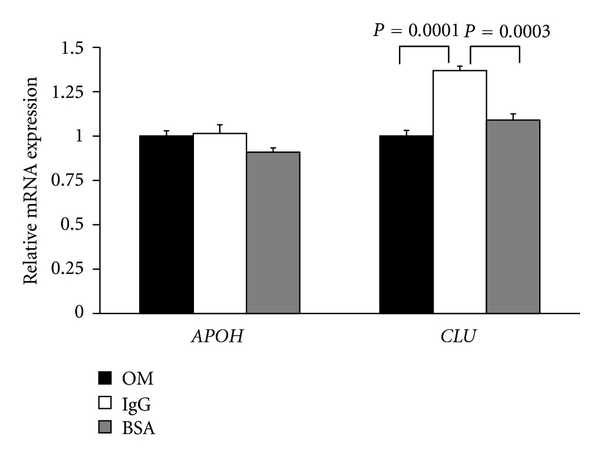
Real-time PCR analysis of clusterin transcript expression in HuH7 cells treated with immunoglobulins. The chart shows the expression of apolipoprotein H (APOH) or clusterin (CLU) transcripts in HuH7 cells adapted to culture in Opti-MEM (OM: black bars), or treated with immunoglobulins (Ig: white bars) or bovine serum albumin (BSA: gray bars). The bars indicate the average expression levels and the observed standard deviations. The expression levels of CLU or APOH transcripts were normalized to β-actin mRNA levels.
The cohort of subjects enrolled in this study was entirely made of CVID patients receiving IgG administration by intravenous route. However, IgG replacement therapy can also be carried out via subcutaneous route of administration to guarantee the quality of life of patients; in fact, home subcutaneous IgG therapy can be considered a valid option for many of them. Subcutaneous administration of IgG is also a successful approach in most patients with previous adverse reactions to IVIg (23). Further studies on larger cohorts of patients subjected to IgG treatments, both by intravenous and subcutaneous administrations are needed to clarify whether clusterin can be considered a biomarker of efficacy for IgG treatments in CVID.
The ability of immunoglobulins to affect CLU gene expression may account on potential mechanisms based on Fc receptors to transduce signals upon binding to their ligands. In addition to canonical Fcγ receptors expressed by immune cells, several organs (including placenta, intestine, kidney, lung, and liver) actually express the FCRN gene encoding for the neonatal Fc receptor, FcRn [43]. Besides its involvement in the transfer of maternal IgG to the foetus or neonate, FcRn acts as a homeostatic receptor for serum stabilization of IgG in adults. Recently, an inverse correlation between FCRN mRNA expression in peripheral blood mononuclear cells and the extent of bronchiectasis in CVID has been demonstrated [44]. These authors highlighted a faster decline in serum IgG concentration during the 2nd week after IVIg infusion in patients with lower FCRN mRNA level [44]. Whether proper regulation of gene expression by circulating IgG may be affected in CVID, and whether altered function of FcRn receptor may contribute to the disease may be an emerging scenario in CVID.
4. Conclusions
A selective pattern of protein expression is represented in serum of naive patients affected by common variable immunodeficiency. Among the differentially expressed proteins, Clusterin/ApoJ may constitute a potential biomarker in CVID, since it is downregulated in the serum of most CVID patients, and its levels are close to those showed by normal subjects, in patients undergoing IVIg replacement therapy. These results prompted us to set-up and analyze a cellular model, based on the human hepatocyte cell line HuH7 adapted to serum-free conditions, to evaluate whether IgG stimulation may contribute to CLU gene regulation. Our results suggest that CLU gene transcripts are upregulated by IgG. These results allow to hypothesize that a signal transduction mechanism, based on the neonatal Fc immunoglobulin receptor, may be involved in the observed regulations on CLU gene and its products.
Acknowledgments
This work has been supported by Funds from Ministero dell'Università e della Ricerca Scientifica (PS 126-Ind), from Regione Campania (L.R. n. 5, Annualità 2007) (to N. Zambrano), from Regione Campania (Rete di Spettrometria di Massa, RESMAC) (to A. Scaloni) and from Associazione Immunodeficienze Primitive (www.aip-it.org/) (to G. Spadaro). Authors thank T. Russo and the Centro Regionale di Competenza GEAR (Genomics for Applied Research, Regione Campania) for granting access to the 2D-DIGE facility, and T. Russo and A. Usiello for critical reading.
Abbreviations
- CVID:
Common variable immunodeficiency
- DIGE:
Differential in-gel electrophoresis
- IVIg:
Intravenous immunoglobulin
- PCR:
Polymerase chain reaction.
References
- 1.Rosen FS, Eibl M, Roifman C, et al. Primary immunodeficiency diseases. Clinical and Experimental Immunology, Supplement. 1999;118(1):1–28. [Google Scholar]
- 2.Cunningham-Rundles C, Bodian C. Common variable immunodeficiency: clinical and immunological features of 248 patients. Clinical Immunology. 1999;92(1):34–48. doi: 10.1006/clim.1999.4725. [DOI] [PubMed] [Google Scholar]
- 3.Park MA, Li JT, Hagan JB, Maddox DE, Abraham RS. Common variable immunodeficiency: a new look at an old disease. The Lancet. 2008;372(9637):489–502. doi: 10.1016/S0140-6736(08)61199-X. [DOI] [PubMed] [Google Scholar]
- 4.Deane S, Selmi C, Naguwa SM, Teuber SS, Gershwin ME. Common variable immunodeficiency: etiological and treatment issues. International Archives of Allergy and Immunology. 2009;150(4):311–324. doi: 10.1159/000226232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ardeniz O, Başoğlu OK, Günşar F, et al. Clinical and immunological analysis of 23 adult patients with common variable immunodeficiency. Journal of Investigational Allergology and Clinical Immunology. 2010;20(3):222–236. [PubMed] [Google Scholar]
- 6.Sleasman JW. The association between immunodeficiency and the development of autoimmune disease. Advances in Dental Research. 1996;10(1):57–61. doi: 10.1177/08959374960100011101. [DOI] [PubMed] [Google Scholar]
- 7.Strober W, Chua K. Common variable immunodeficiency. Clinical Reviews in Allergy and Immunology. 2000;19(2):157–181. doi: 10.1385/CRIAI:19:2:157. [DOI] [PubMed] [Google Scholar]
- 8.Di Renzo M, Pasqui AL, Auteri A. Common variable immunodeficiency: a review. Clinical and Experimental Medicine. 2004;3(4):211–217. doi: 10.1007/s10238-004-0027-2. [DOI] [PubMed] [Google Scholar]
- 9.Michel M, Chanet V, Galicier L, et al. Autoimmune thrombocytopenic purpura and common variable immunodeficiency: analysis of 21 cases and review of the literature. Medicine. 2004;83(4):254–263. doi: 10.1097/01.md.0000133624.65946.40. [DOI] [PubMed] [Google Scholar]
- 10.Brandt D, Gershwin ME. Common variable immune deficiency and autoimmunity. Autoimmunity Reviews. 2006;5(7):465–470. doi: 10.1016/j.autrev.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 11.Cunningham-Rundles C. Autoimmune manifestations in common variable immunodeficiency. Journal of Clinical Immunology. 2008;28(supplement 1):S42–S45. doi: 10.1007/s10875-008-9182-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramyar A, Aghamohammadi A, Moazzami K, et al. Presence of idiopathic thrombocytopenic purpura and autoimmune hemolytic anemia in the patients with common variable immunodeficiency. Iranian Journal of Allergy, Asthma and Immunology. 2008;7(3):169–175. [PubMed] [Google Scholar]
- 13.Agarwal S, Cunningham-Rundles C. Autoimmunity in common variable immunodeficiency. Current Allergy and Asthma Reports. 2009;9(5):347–352. doi: 10.1007/s11882-009-0051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arumugakani G, Wood PMD, Carter CRD. Frequency of treg cells is reduced in CVID patients with autoimmunity and splenomegaly and is associated with expanded CD21lo B lymphocytes. Journal of Clinical Immunology. 2010;30(2):292–300. doi: 10.1007/s10875-009-9351-3. [DOI] [PubMed] [Google Scholar]
- 15.Cunningham-Rundles C. How I treat common variable immune deficiency. Blood. 2010;116(1):7–15. doi: 10.1182/blood-2010-01-254417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinlen LJ, Webster AD, Bird AG, et al. Prospective study of cancer in patients with hypogammaglobulinemia. The Lancet. 1985;1:263–266. doi: 10.1016/s0140-6736(85)91037-2. [DOI] [PubMed] [Google Scholar]
- 17.Filipovich AH, Mathur A, Kamat D, Kersey JH, Shapiro RS. Lymphoproliferative disorders and other tumors complicating immunodeficiencies. Immunodeficiency. 1994;5(2):91–112. [PubMed] [Google Scholar]
- 18.Mellemkjær L, Hammarström L, Andersen V, et al. Cancer risk among patients with IgA deficiency or common variable immunodeficiency and their relatives: a combined Danish and Swedish study. Clinical and Experimental Immunology. 2002;130(3):495–500. doi: 10.1046/j.1365-2249.2002.02004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gompels MM, Hodges E, Lock RJ, et al. Lymphoproliferative disease in antibody deficiency: a multicentre study. Clinical and Experimental Immunology. 2003;134:314–321. doi: 10.1046/j.1365-2249.2003.02253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quinti I, Soresina A, Spadaro G, et al. Long-term follow-up and outcome of a large cohort of patients with common variable immunodeficiency. Journal of Clinical Immunology. 2007;27(3):308–316. doi: 10.1007/s10875-007-9075-1. [DOI] [PubMed] [Google Scholar]
- 21.Yap YL, So JBY. Gastric adenocarcinoma occurring in a young patient with common variable immunodeficiency syndrome. Singapore Medical Journal. 2009;50(6):e201–e203. [PubMed] [Google Scholar]
- 22.Orange JS, Hossny EM, Weiler CR, et al. Use of intravenous immunoglobulin in human disease: a review of evidence by members of the Primary Immunodeficiency Committee of the American Academy of Allergy, Asthma and Immunology. Journal of Allergy and Clinical Immunology. 2006;117(4):S525–S553. doi: 10.1016/j.jaci.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 23.Quinti I, Soresina A, Agostini C, et al. Prospective study on CVID patients with adverse reactions to intravenous or subcutaneous IgG administration. Journal of Clinical Immunology. 2008;28(3):263–267. doi: 10.1007/s10875-007-9169-9. [DOI] [PubMed] [Google Scholar]
- 24.Grimbacher B, Hutloff A, Schlesier M, et al. Homozygous loss of ICOS is associated with adult-onset common variable immunodeficiency. Nature Immunology. 2003;4(3):261–268. doi: 10.1038/ni902. [DOI] [PubMed] [Google Scholar]
- 25.Van Zelm MC, Reisli I, van der Burg M, et al. An antibody-deficiency syndrome due to mutations in the CD19 gene. The New England Journal of Medicine. 2006;354(18):1901–1912. doi: 10.1056/NEJMoa051568. [DOI] [PubMed] [Google Scholar]
- 26.Kanegane H, Agematsu K, Futatani T, et al. Novel mutations in a Japanese patient with CD19 deficiency. Genes and Immunity. 2007;8(8):663–670. doi: 10.1038/sj.gene.6364431. [DOI] [PubMed] [Google Scholar]
- 27.Warnatz K, Salzer U, Rizzi M, et al. B-cell activating factor receptor deficiency is associated with an adult-onset antibody deficiency syndrome in humans. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(33):13945–13950. doi: 10.1073/pnas.0903543106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salzer U, Bacchelli C, Buckridge S, et al. Relevance of biallelic versus monoallelic TNFRSF13B mutations in distinguishing disease-causing from risk-increasing TNFRSF13B variants in antibody deficiency syndromes. Blood. 2009;113(9):1967–1976. doi: 10.1182/blood-2008-02-141937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Molecular and Cellular Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 30.Caratù G, Allegra D, Bimonte M, et al. Identification of the ligands of protein interaction domains through a functional approach. Molecular and Cellular Proteomics. 2007;6(2):333–345. doi: 10.1074/mcp.M600289-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Zhang W, Chait BT. ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Analytical Chemistry. 2000;72(11):2482–2489. doi: 10.1021/ac991363o. [DOI] [PubMed] [Google Scholar]
- 32.Bernardini G, Renzone G, Comanducci M, et al. Proteome analysis of Neisseria meningitidis serogroup A. Proteomics. 2004;4(10):2893–2926. doi: 10.1002/pmic.200400946. [DOI] [PubMed] [Google Scholar]
- 33.Dennis G, Jr., Sherman BT, Hosack DA, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biology. 2003;4(5, article P3) [PubMed] [Google Scholar]
- 34.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Yuan JS, Reed A, Chen F, Stewart CN. Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7, article 85 doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Falgarone G, Chiocchia G. Clusterin: a multifacet protein at the crossroad of inflammation and autoimmunity. Advances in Cancer Research. 2009;104:139–170. doi: 10.1016/S0065-230X(09)04008-1. [DOI] [PubMed] [Google Scholar]
- 38.Klock G, Baiersdörfer M, Koch-Brandt C. Cell protective functions of secretory Clusterin (sCLU) Advances in Cancer Research. 2009;104:115–138. doi: 10.1016/S0065-230X(09)04007-X. [DOI] [PubMed] [Google Scholar]
- 39.Wyatt A, Yerbury J, Poon S, Dabbs R, Wilson M. The chaperone action of Clusterin and its putative role in quality control of extracellular protein folding. Advances in Cancer Research. 2009;104:89–114. doi: 10.1016/S0065-230X(09)04006-8. [DOI] [PubMed] [Google Scholar]
- 40.Bykov I, Junnikkala S, Pekna M, Lindros KO, Meri S. Effect of chronic ethanol consumption on the expression of complement components and acute-phase proteins in liver. Clinical Immunology. 2007;124(2):213–220. doi: 10.1016/j.clim.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 41.Hardardottir I, Kunitake ST, Moser AH, et al. Endotoxin and cytokines increase hepatic messenger RNA levels and serum concentrations of apolipoprotein J (clusterin) in Syrian hamsters. The Journal of Clinical Investigation. 1994;94(3):1304–1309. doi: 10.1172/JCI117449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Devauchelle V, Essabbani A, De Pinieux G, et al. Characterization and functional consequences of underexpression of clusterin in rheumatoid arthritis. Journal of Immunology. 2006;177(9):6471–6479. doi: 10.4049/jimmunol.177.9.6471. [DOI] [PubMed] [Google Scholar]
- 43.Roopenian DC, Akilesh S. FcRn: the neonatal Fc receptor comes of age. Nature Reviews Immunology. 2007;7(9):715–725. doi: 10.1038/nri2155. [DOI] [PubMed] [Google Scholar]
- 44.Freiberger T, Grodecká L, Ravčuková B, et al. Association of FcRn expression with lung abnormalities and IVIG catabolism in patients with common variable immunodeficiency. Clinical Immunology. 2010;136(3):419–425. doi: 10.1016/j.clim.2010.05.006. [DOI] [PubMed] [Google Scholar]


