Abstract
Purpose
Prognosis in renal cell carcinoma (RCC) is dependent on tumor stage at presentation, with significant differences in survival between early and late stage disease. Currently, there are no screening tests or biomarkers identified for the early detection of kidney cancer. Here, we investigate if serum amino acid profiles are a potentially useful biomarker in patients with RCC.
Materials and Methods
The concentrations of 26 different amino acids were determined in serum taken pre-operatively from 189 RCC patients and 104 age and sex matched controls.
Results
Statistically significant changes were observed in patient levels of 15 different amino acids, with 13 being decreased and two being elevated. A logistic regression model utilizing eight amino acids including cysteine, ornithine, histidine, leucine, tyrosine, proline, valine and lysine was created to distinguish cases from controls. A receiver operator curve based on this model had an area under the curve of 0.81. This same model also had predictive value in predicting overall survival and tumor recurrence in RCC patients.
Conclusions
Our findings suggest that serum amino acid levels may be useful as a screening tool for the identification of individuals with RCC and predicting patient outcomes.
Keywords: Kidney Cancer, biomarker, serum, amino acids
Introduction
In the United States, it is estimated that there will be over 50,000 new cases of RCC in 2011, with more that 13,000 deaths from the disease.1 The rate at which RCC is being detected in the population appears to be increasing.2 The primary determinant of prognosis in RCC is stage at presentation. Small tumors confined to the kidney (T1) have 5-year survival rates which exceed 90%, while advanced tumors that have metastasized outside the kidney have rates less than 20%.3 Unfortunately, most individuals with locally confined disease have no obvious symptoms, and therefore over one-third of individuals with the disease are detected only when the cancer is locally advanced or metastatic.4 In fact, most early stage kidney cancer is detected serendipitously, usually when a patient is having an abdominal CT scan for unrelated symptomatology.5 Given the large differences in outcome between early and late stage tumors, serum biomarkers to detect individuals with early stage tumors would be extremely valuable.
The kidney plays a central role in the clearance of nitrogenous substances and in regulating plasma amino acid levels. A key function of normal renal tubule epithelial cells is to reabsorb amino acid from the glomerular filtrate. Since most renal epithelial tumors arise from renal tubule cells, it may be that these tumors exhibit alterations in their ability to reabsorb amino acids from the filtrate, a change which may be reflected in the blood. Furthermore, tumor cells in general are known to require larger than normal amounts of some amino acids in order to grow, perhaps related to the metabolic shift in tumors from respiration to fermentation, the so-called Warburg effect.6 Consistent with this idea is the finding that certain types of tumors have higher than normal requirements for several amino acids including methionine, glutamine, and asparagines.7-9 Finally, the relatively large size of even early stage I (<7cm) or II (7-10+ cm) renal tumors enhances the possibility that they could exert an overall effect on the serum levels of a particular amino acid.
In this manuscript, we have examined the amino acid profiles of a large group of RCC patients (pre-surgery) and compare them to age and sex matched controls. Our findings show that serum amino acids may be a useful biomarker in the identification and risk stratification of individuals with RCC.
Materials and Methods
Patients and Samples
Serum was obtained from RCC patients and controls from the Fox Chase Cancer Center Biosample Repository. The cases were collected by the FCCC Keystone Program in Personalized Kidney Cancer Keystone Therapy between 2004 and 2010. Each RCC patient that is undergoing surgery consented and blood was collected, serum isolated, and stored at −70°C. Control serums come from a variety of sources including FCCC employees, individuals undergoing routine cancer screening, or spouses of patients. A control group was created by randomly matching each of the first 104 cases analyzed by age and sex. The study was approved by the Fox Chase Cancer Center Institutional Review Board.
Amino Acid Analysis
Serum amino acid levels for each sample was quantitated using a Biochrom 30 amino acid analyzer as previously described 10. Each sample was assayed once, as inter-day assay repeatability was previously established by processing 27 different samples on two different days resulting in an average CV for all of the amino acids of 6.7% (range 3.5-14.2%).
Data Analysis
Data analysis was performed using Statistica 9.1 software (StatSoft, Tulsa Oklahoma). Amino acid data was log transformed to ensure normal distribution for parametric tests. For univariate analysis, unpaired two-sided t-tests were used with P<0.05 being deemed significant with no correction for multiple testing. An unpaired test was deemed appropriate as not all the cases were matched with controls. Correlations were determined using Pearson's R and were considered significant if P<0.05. Factor analysis was performed using the principle components extraction method with a maximum of seven factors. A scree plot of the extracted Eigenvalues suggested that only the first three were likely to be significant. For Padj values, a general linear model was used. ANOVA in combination with Tukey's LSD test was used to determine differences between multiple groups. To create a prediction model for cases and controls, stepwise-backward logistic regression was performed using all 26 amino acids as variables. At each step the least predictive variable was removed based on the Wald score. The final model contained only those variables with Wald scores with P<0.05. For differences in survival between two groups, Kaplan Meier's analysis was performed using a log-rank test. For multivariate survival analysis, Cox Proportional Hazard modeling was performed.
Results
Patient and Control Characteristics
Serum was obtained from 189 RCC patients at Fox Chase Cancer Center between the years of 2004 and 2010 before undergoing nephrectomy. The characteristics of the patients are shown on Table 1. The median age of the patients was 58 years old with the majority of the patients being male and white. An appropriate control group was assembled from non-cancer patient samples in the Fox Chase Biosample repository by individually matching for sex, race and age (within two years) for the first 104 patient samples obtained. As expected, no significant differences were found in the distribution of age, sex, race or BMI between the control and patient group as a whole.
Table 1. Characteristics of RCC cases and controls.
| Case (n=189) | Control (n=104) | P value | ||
|---|---|---|---|---|
| Age | Median | 58 | 57 | 0.49 |
|
|
||||
| Range | (25-87) | (36-81) | ||
|
| ||||
| Sex | Male | 129 (68%) | 71 (69%) | 0.93 |
|
|
||||
| Female | 60 (32%) | 32 (31%) | ||
|
| ||||
| BMI | Mean | 29.9 (n=61) | 27.6 (n=97) | 0.09 |
|
| ||||
| White | 167 (88%) | 93 (89%) | 0.97 | |
|
|
||||
| Race | Black | 20 (11%) | 8 (7%) | |
|
|
||||
| Asian | 1 (0.5%) | 1 (0.9%) | ||
|
|
||||
| Unknown | 1 (0.5%) | 2 (1.9%) | ||
|
| ||||
| I | 97 (51%) | |||
|
|
||||
| Stage | II | 24 (13%) | ||
|
|
||||
| III | 29 (15%) | |||
|
|
||||
| IV | 39 (21%) | |||
|
| ||||
| ccRCC | 122 (65%) | |||
|
|
||||
| Type | pRCC | 29 (15%) | ||
|
|
||||
| Other | 38 (21%) | |||
|
| ||||
| Pre-Op GFR (SD) | 80.2 (23.5) | |||
Abbreviations: BMI, Body Mass Index; ccRCC, clear renal cell carcinoma; pRCC, papillary renal cell carcinoma; Other includes adenocarcinoma with mixed subtype (n=15), chromophobe (n=13), cyst associated (n=4), sarcomatoid (n=2), carcinoma (n=1), small cell (n=2), granular cell (n=1). Pre-OP GFR, pre-operative glomerular filtration rate
Amino Acid Analysis
Each serum sample was analyzed for amino acid content using an amino acid analyzer. Twenty-six compounds were quantitated for each sample (Fig. 1). Comparison of patients and controls revealed that 15 of the 26 amino acids showed statistically significant differences in the means between cases and controls (Table 2). Thirteen (taurine, threonine, serine, asparagine, glutamate, glycine, alanine, citrulline, methionine, tyrosine, ornithine, phenylalanine, histidine, and proline) were significantly decreased in RCC patients and two (arginine and cysteine) were elevated. The largest percent differences between the means were observed for histidine and ornithine.
Figure 1.
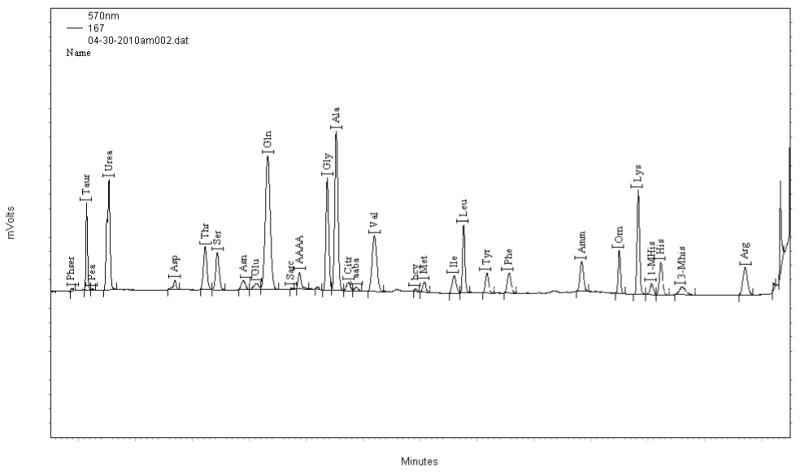
Trace file of human plasma from BioChrom30 amino acid analyzer. X-axis shows the elution time in minutes after injection. Y-axis shows relative absorbance at 570 nm.
Table 2. Amino Acid Mean and t-Test for Cases vs Control.
| Case (n=189) | Control (n=104) | |||||
|---|---|---|---|---|---|---|
| Amino Acid | Mean μM |
SD | Mean μM |
SD | p T-test 2-sided | padjusted |
| Taurine | 159.4 | 52.4 | 174.3 | 58.2 | 0.0319 | .691 |
|
|
||||||
| Aspartate1 | 32.4 | 14.3 | 35.9 | 16.8 | 0.1419 | .717 |
|
|
||||||
| Threonine | 134.7 | 40.1 | 153.6 | 40.4 | 0.0001 | .015 |
|
|
||||||
| Serine | 132.1 | 33.3 | 142.9 | 41.0 | 0.0322 | .691 |
|
|
||||||
| Asparagine | 68.3 | 19.5 | 78.1 | 25.8 | 0.0012 | .205 |
|
|
||||||
| Glutamate | 98.9 | 56.9 | 129.7 | 102.4 | 0.0373 | .743 |
|
|
||||||
| Glutamine | 854.7 | 182.1 | 867.0 | 213.3 | 0.7509 | .190 |
|
|
||||||
| Glycine | 287.9 | 80.5 | 321.1 | 110.9 | 0.0074 | .244 |
|
|
||||||
| Alanine | 451.6 | 122.4 | 527.5 | 163.3 | <0.0001 | .003 |
|
|
||||||
| Citrulline | 34.7 | 12.2 | 38.4 | 9.7 | 0.0040 | .066 |
|
|
||||||
| α-amino butyric acid | 21.3 | 9.3 | 21.0 | 10.7 | 0.5714 | .018 |
|
|
||||||
| Valine | 254.1 | 58.8 | 268.0 | 66.6 | 0.1003 | .219 |
|
|
||||||
| Total Homocysteine | 14.5 | 6.6 | 15.4 | 9.4 | 0.9271 | .060 |
|
|
||||||
| Methionine | 23.7 | 6.5 | 25.7 | 8.0 | 0.0287 | .742 |
|
|
||||||
| Isoleucine | 67.8 | 19.8 | 69.3 | 22.8 | 0.7742 | .005 |
|
|
||||||
| Leucine | 156.5 | 39.0 | 161.6 | 47.0 | 0.4789 | .001 |
|
|
||||||
| Tyrosine | 66.9 | 18.2 | 74.5 | 19.8 | 0.0008 | .105 |
|
|
||||||
| Phenylalanine | 79.0 | 19.5 | 86.5 | 44.8 | 0.1314 | .126 |
|
|
||||||
| Ornithine | 97.8 | 32.4 | 126.3 | 55.2 | <0.0001 | .00001 |
|
|
||||||
| Lysine | 206.1 | 50.7 | 217.4 | 53.7 | 0.0698 | .092 |
|
|
||||||
| 1-methyl-histidine | 19.1 | 13.8 | 18.3 | 10.5 | 0.8477 | .374 |
|
|
||||||
| Histidine | 77.4 | 19.7 | 90.0 | 22.2 | <0.0001 | .00002 |
|
|
||||||
| 3-methyl-histidine2 | 22.9 | 6.1 | 24.0 | 5.8 | 0.0845 | .675 |
|
|
||||||
| Arginine | 98.7 | 31.1 | 84.0 | 33.8 | <0.0001 | .00001 |
|
|
||||||
| Total Cysteine | 401.8 | 98.2 | 374.5 | 87.6 | 0.0172 | <.000001 |
|
|
||||||
| Proline | 214.3 | 83.2 | 230.9 | 63.8 | 0.0373 | .373 |
|
| ||||||
| Factor 1 | 0.131 | 0.934 | -0.237 | 1.074 | 0.0025 | NA |
|
|
||||||
| Factor 2 | -0.070 | 0.864 | 0.128 | 1.203 | 0.1049 | NA |
|
|
||||||
| Factor 3 | 0.032 | 1.019 | -0.058 | 0.967 | 0.461 | NA |
Aspartate co-elutes with reduced glutathione.
Tryptophan co-elutes with 3-methylhistidine.
Since so many of amino acid levels were altered, we decided to examine how the levels of different amino acids were correlated with each other in the entire dataset (Supplemental Fig. 1). With the exception of arginine, we found that there was a statistically significant positive correlation between most of the different amino acid pairs, with the strength of the correlation varying depending on the pairs examined. The strongest correlations were between leucine, isoleucine, and valine (R=0.85-0.89), while the mean correlation co-efficient (R) between different amino acids excluding arginine was 0.39.
To explore these correlations in more depth, we performed Factor analysis using principle component extraction. We found that a single primary factor could explain 45% of the overall variance in amino acid levels, and the first three factors together could explain 62.6% of the variance. However, when the calculated factor scores for each case and control were examined, only the primary factor was shown to be significantly different between cases and controls (Table 2). No correlation was observed between this primary factor and re-operative glomerular filtration rates (GFR) in patients, indicating that this factor was not related to decreased kidney function.
Because of the significant correlation between different amino acids and the strength of the primary factor, we suspected that some of the significant differences observed in univariate t-tests might be due to this underlying “general” correlation. To control for this, we also determined the significance value in which each amino acid was adjusted for this factor (Table 2, padjusted). When adjusted in this way, nine amino acids including threonine, alanine, α-aminobutyrate, isoleucine, leucine, ornithine, histidine, arginine and cysteine still showed significant differences between cases and controls.
Logistic Regression Model
We next created a logistic regression model by performing a backward-stepwise logistic regression procedure to identify which of the twenty-six amino acids had significant predictive value (P<0.05) with regards to a sample being either a case or control. The final model contained eight different amino acids (cysteine, ornithine, histidine, leucine, tyrosine. proline, valine, and lysine) and the receiver-operator curve (ROC) for this model gave an AUC 0.81 (Supplemental Table 2, Fig. 2). Because the number of potential predictor variables in the model was relatively large compared to the total number of samples, we were somewhat concerned about the model over-fitting the data.11 To address this possibility, we performed 10-fold cross validation on the sample set. This procedure involves using 90% of the data set as the analysis group (used to build the model) and 10% as the validation group in ten different unique iterations. Performing this procedure using the eight amino acids identified above to make the model, we found using ROC analysis that the mean AUC for the analysis group vs. the validation group was not significantly different (0.81 vs. 0.79, p=0.28, Supplemental Table 3). This result suggests that the model is not over-fitting the data to a significant degree.
Figure 2.
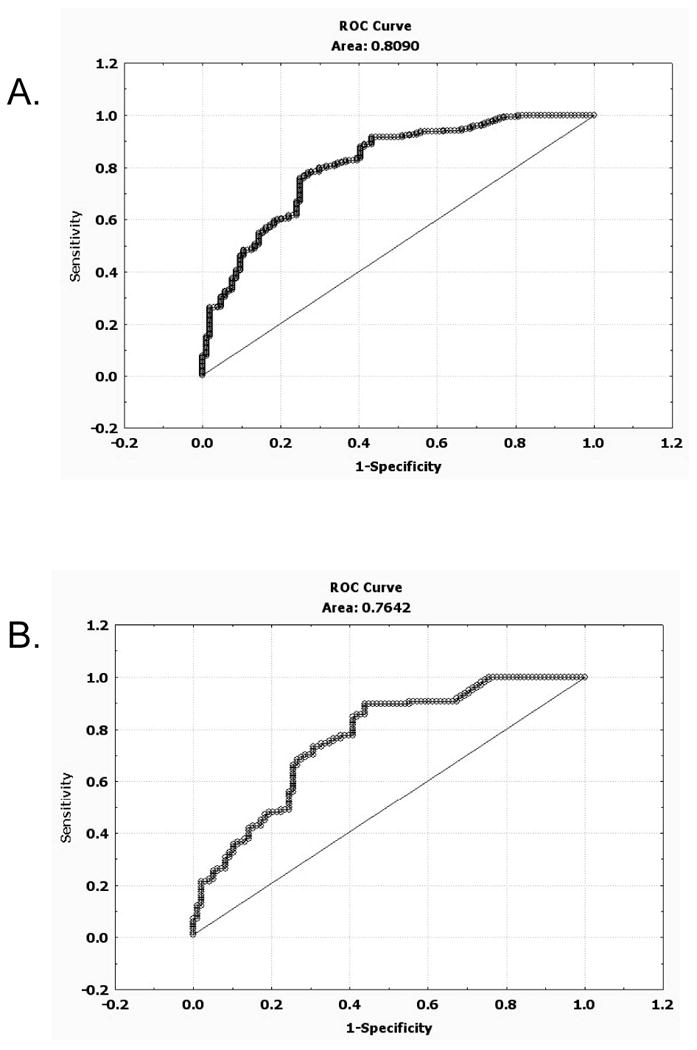
ROC curves for logistic regression model. A) ROC for logistic regression model presented in Table 3. Samples include all patients (n=189) and all controls (n=104) B) ROC for only early stage patients (n=121) and all controls (n=104).
Model Performance on Tumor Grade and Type
We next examined how the model performed relative to pathologic tumor stage. First we examined the model likelihood values (model score) for each of the samples relative to their tumor grade (Fig. 3A). Early stage tumors (stage 1 and stage 2) have slightly lower model scores than late stage tumors (stage 3 and stage 4), but are still significantly elevated relative to the control samples. ROC analysis on only stage I and stage 2 samples compared to controls gives an AUC of 0.76, only slightly lower than the total data set (Fig. 2B). We also examined how the model performed on different histological subtypes of kidney cancer (Fig. 3B). The mean value was not significantly different among clear cell, papillary, and a mixture of other types of kidney tumors.
Figure 3.
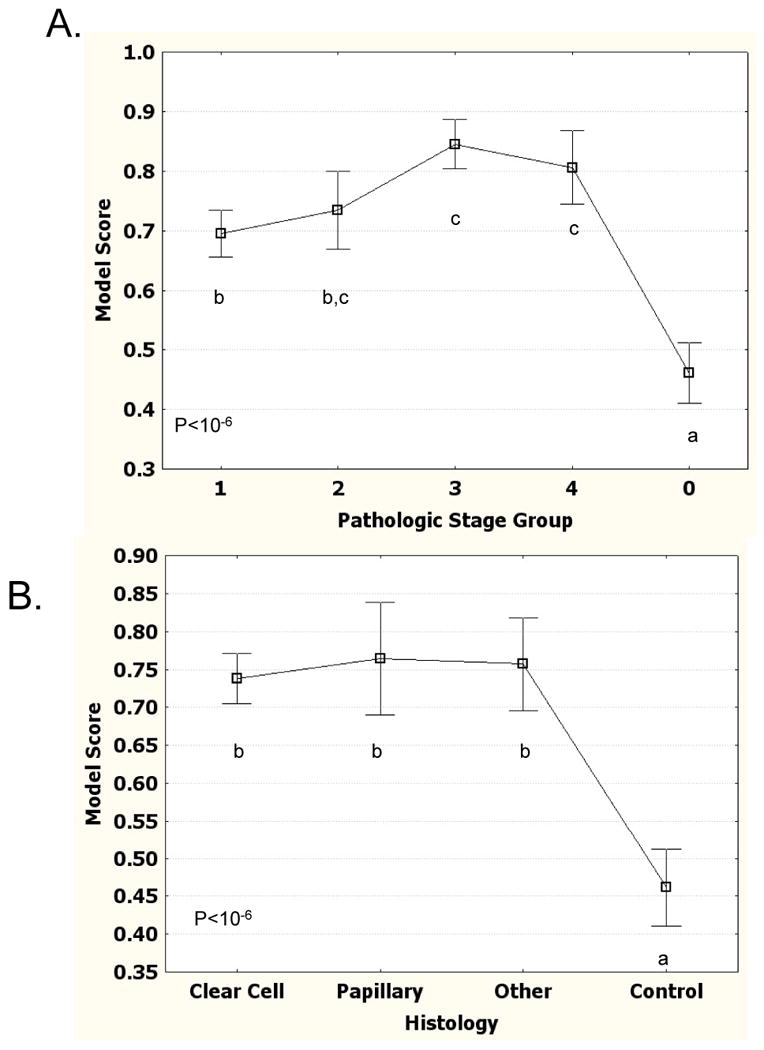
Patient Logistic Regression Model Scores stratified by tumor grade and type. A) Logistic Regression Model score stratified by tumor grade. Mean score for each grade is shown. Error bars show 95% confidence interval of mean. Stage 0 are control samples. Letters show significant differences (P<0.05) based on Tukey HSD test. Points lacking same letter are different. B) Logistic regression model score stratified by tumor type.
Serum Amino Acid Profiles and Outcome
In patient samples, model score was examined in relation to both time to recurrence in patients in which the tumor was entirely removed by surgery, and overall survival time. For this analysis we divided the patients into two groups, those with model scores above and below the median (0.79). We found patients with lower regression scores had significantly fewer recurrences (log-rank test, p=0.003, Fig. 4A). In addition, patients with lower logistic regression scores had significantly increased overall survival compared to those with higher scores (p=0.006, log-rank test; Fig. 4B). However, since the above median group also had a significantly higher percentage of stage 3 and stage 4 tumors compared to the below median group (53.7% vs. 18.8%), this suggested that stage might be driving these findings. In an attempt to control for stage effects, we performed Cox Multivariate hazard analysis using stage and model score as covariates (Supplemental Table 4). Although the Hazard ratio for the model score is still greater than 1 in both recurrence-free and overall survival, the P value is no longer significant (p=0.11 and p=0.12, respectively).
Figure 4.
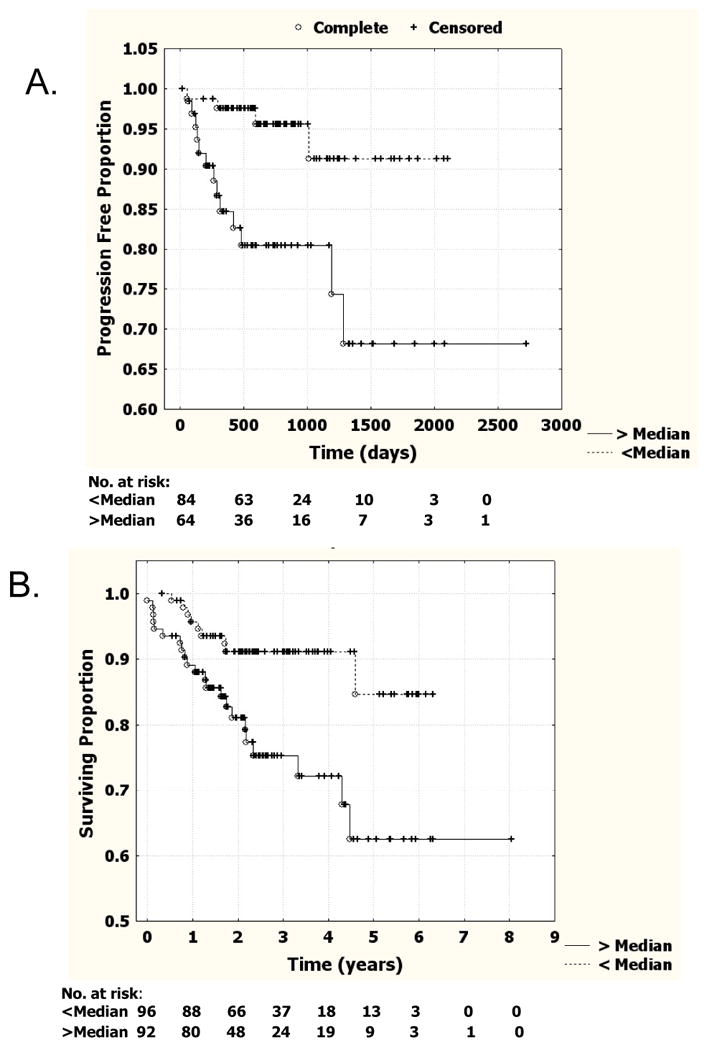
Survival curves stratified by logistic regression model score. A) Cancer recurrence for all RCC patients deemed tumor free after surgery (n=147) stratified by logistic regression score either being above or below the median for all patients (0.79). Tick marks indicate individuals that were censored. B) Overall survival for all RCC patients deemed tumor free after surgery (n=147) stratified by logistic regression score either being above or below the median for all patients.
Discussion
In the work described here we have examined serum amino acid profiles in a large series of renal cell carcinoma patients compared to age and sex matched controls. We found statistically significant differences in the concentrations of 15 of the 26 amino acids that were quantitated. Because serum amino acid levels tended to be correlated overall, we performed factor analysis to identify if a single underlying factor might be involved in the large number of differences observed between cases and controls. We found that a single underlying factor could account for up to 45% of the variance in amino acid levels, and that this factor was significantly different between cases and controls. A possible biological explanation for this factor would be that kidney tumors might be affecting the reabsorption of amino acids by affecting overall renal function. However, two observations argue against this explanation. First, an analysis of GFR rates in the patient samples show no overall correlation between kidney function and this factor, and second, at least nine amino acids exhibited differences between cases and controls even when adjusted for this underlying factor.
An alternative hypothesis is that the generally lower levels of serum amino acids may be a reflection of the increased usage of amino acids by the tumor for biosynthetic processes. Consistent with this idea, patients with head and neck cancer have also have reduced serum levels of many of the same amino acids we identified as decreased in this study.12 It has been proposed that weight loss in cancer patients may be responsible for this decrease in amino acid levels, but it should be noted in our study there was no difference in BMI between cases and controls.
Our data suggest that serum amino acid profiling may have potential clinical uses in the detection and risk stratification of RCC. We identified a logistic regression model in which a combination of eight amino acids could be used to distinguish cases from controls. ROC analysis of this model indicates that the AUC is 0.81, in a range similar to that used in other cancer screening tests such as Paps smears (0.70) and PSA tests (0.68).13, 14 An important feature of the test is that it was possible to identify early stage tumors with only slightly less efficiency as late stage tumors (AUC 0.76). While the presumed low specificity would likely preclude clinical utility as a general screening test at the current time, it is possible that additional research may refine the clinical utility of such as test (i.e. screening at-risk populations) making this more attractive in the future. A key unanswered question is whether changes in an individual's amino acid profile might be a more sensitive indicator of RCC rather than just absolute levels. Previous work has shown that there is significantly more inter-individual variability than intra-individual variability with regards to plasma amino acid levels.15 Thus by following an individual over a period of time, it might be possible to increase the receiver-operator characteristics of the assay. In addition, it might be possible to find other serum based markers which when included with the amino acid profile might make the assay more specific and powerful.
We also found that our logistic regression model had prognostic utility with regards to predicting cancer recurrence and overall patient survival. Patients with logistic regression model scores above the mean had significantly increased likelihood of cancer recurrence and shorter survival than those with lower scores. Some of this difference appeared to be due to the fact that higher stage cancers tended to have higher model scores. However, the study was insufficiently powered to know with certainty whether the model score may have prognostic utility independent of tumor stage. Another limitation of the study is the lack of detailed information on the control samples.
Conclusions
In summary, we have shown for the first time that alterations in the serum amino acid profiles are characteristic of individuals with both early and late stage RCC. In addition, serum amino acid profiles also had predictive value in predicting overall survival and tumor recurrence in RCC patients. Our studies suggest that serum amino acid sampling may have potential as a blood based screening test for the detection and prognostication of RCC.
Supplementary Material
Supplemental Table 1. Pearson Product Moment Correlations. In the entire data set (293 samples) the Pearson correlation R was determined for each pair of amino acids. The average correlation coefficient for the entire set of amino acids is shown at the bottom of each column.
Supplemental Table 2. Logistic Regression Model parameters.
Supplemental Table 3. 10-fold cross-validation testing. Results from cross validation studies are shown. For each run, one-tenth of the data was withheld and a logistic regression model using cysteine, ornithine, histidine, leucine, tyrosine. proline, valine, and lysine was created. ROC curves were then created for the analysis set and the test set. A different tenth was withheld for each run.
Supplemental Table 4. Cox Proportional Hazards Models. Cox proportional hazards models are shown for both time to failure and overall survival. Model score was used as both a single variable and in a model combined with pathologic stage. P value, Hazard Ratio, and Confidence intervals are shown.
Acknowledgments
Support for this work was provided by funds from the FCCC Keystone Program in Personalized Kidney Cancer Therapy, NIH core grant (CA06927), and an appropriation from the Commonwealth of Pennsylvania to the Fox Chase Cancer Center. We acknowledge the contribution of Jo-ellen Weaver and the Fox Chase Cancer Center Biosample Repository facility, and Debra Kister and Michelle Collins and the Kidney Cancer Database. We also acknowledge Robert B. Kruger for motivating this project.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009 [Google Scholar]
- 2.Hock LM, Lynch J, Balaji KC. Increasing incidence of all stages of kidney cancer in the last 2 decades in the United States: an analysis of surveillance, epidemiology and end results program data. J Urol. 2002;167:57. [PubMed] [Google Scholar]
- 3.Zisman A, Pantuck AJ, Wieder J, et al. Risk group assessment and clinical outcome algorithm to predict the natural history of patients with surgically resected renal cell carcinoma. J Clin Oncol. 2002;20:4559. doi: 10.1200/JCO.2002.05.111. [DOI] [PubMed] [Google Scholar]
- 4.NCI Surveillance, Epidemiology and End Results (SEER) Program. 2010 www.seer.cancer.gov.
- 5.Cindolo L, Chiodini P, Gallo C, et al. Validation by calibration of the UCLA integrated staging system prognostic model for nonmetastatic renal cell carcinoma after nephrectomy. Cancer. 2008;113:65. doi: 10.1002/cncr.23517. [DOI] [PubMed] [Google Scholar]
- 6.Unwin RD, Craven RA, Harnden P, et al. Proteomic changes in renal cancer and co-ordinate demonstration of both the glycolytic and mitochondrial aspects of the Warburg effect. Proteomics. 2003;3:1620. doi: 10.1002/pmic.200300464. [DOI] [PubMed] [Google Scholar]
- 7.Stern PH, Wallace CD, Hoffman RM. Altered methionine metabolism occurs in all members of a set of diverse human tumor cell lines. J Cell Physiol. 1984;119:29. doi: 10.1002/jcp.1041190106. [DOI] [PubMed] [Google Scholar]
- 8.Yuneva M. Finding an “Achilles' heel” of cancer: the role of glucose and glutamine metabolism in the survival of transformed cells. Cell Cycle. 2008;7:2083. doi: 10.4161/cc.7.14.6256. [DOI] [PubMed] [Google Scholar]
- 9.Rytting M. Peg-asparaginase for acute lymphoblastic leukemia. Expert Opin Biol Ther. 2010;10:833. doi: 10.1517/14712591003769808. [DOI] [PubMed] [Google Scholar]
- 10.Wang L, Jhee KH, Hua X, et al. Modulation of cystathionine beta-synthase level regulates total serum homocysteine in mice. Circ Res. 2004;94:1318. doi: 10.1161/01.RES.0000129182.46440.4a. [DOI] [PubMed] [Google Scholar]
- 11.Concato J, Feinstein AR, Holford TR. The risk of determining risk with multivariable models. Ann Intern Med. 1993;118:201. doi: 10.7326/0003-4819-118-3-199302010-00009. [DOI] [PubMed] [Google Scholar]
- 12.Scioscia KA, Snyderman CH, Wagner R. Altered serum amino acid profiles in head and neck cancer. Nutr Cancer. 1998;30:144. doi: 10.1080/01635589809514654. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell MF, Cantor SB, Brookner C, et al. Screening for squamous intraepithelial lesions with fluorescence spectroscopy. Obstet Gynecol. 1999;94:889. doi: 10.1016/s0029-7844(99)00408-1. [DOI] [PubMed] [Google Scholar]
- 14.Thompson IM, Ankerst DP, Chi C, et al. Operating characteristics of prostate-specific antigen in men with an initial PSA level of 3.0 ng/ml or lower. JAMA. 2005;294:66. doi: 10.1001/jama.294.1.66. [DOI] [PubMed] [Google Scholar]
- 15.Scriver CR, Gregory DM, Sovetts D, et al. Normal plasma free amino acid values in adults: the influence of some common physiological variables. Metabolism. 1985;34:868. doi: 10.1016/0026-0495(85)90112-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Pearson Product Moment Correlations. In the entire data set (293 samples) the Pearson correlation R was determined for each pair of amino acids. The average correlation coefficient for the entire set of amino acids is shown at the bottom of each column.
Supplemental Table 2. Logistic Regression Model parameters.
Supplemental Table 3. 10-fold cross-validation testing. Results from cross validation studies are shown. For each run, one-tenth of the data was withheld and a logistic regression model using cysteine, ornithine, histidine, leucine, tyrosine. proline, valine, and lysine was created. ROC curves were then created for the analysis set and the test set. A different tenth was withheld for each run.
Supplemental Table 4. Cox Proportional Hazards Models. Cox proportional hazards models are shown for both time to failure and overall survival. Model score was used as both a single variable and in a model combined with pathologic stage. P value, Hazard Ratio, and Confidence intervals are shown.


