Abstract
Notch signaling is essential for proper cardiac development. We recently identified missense variants in the NOTCH1 receptor in patients with diverse left ventricular outflow tract (LVOT) malformations (NOTCH1G661S and NOTCH1A683T) that reduce ligand-induced Notch signaling. Here, we examine the molecular mechanisms that contribute to reduced signaling and perturbed development. We find that NOTCH1A683T exhibits reduced S1 cleavage due to impaired trafficking through the endoplasmic reticulum (ER). This observation is consistent with improper localization of the variant receptor to the ER and decreased presentation at the cell surface. In contrast, the nearby mutation NOTCH1G661S exhibits reduced cell-surface presentation in the absence of overt folding or trafficking defects. To examine the implications of these variants in disease pathogenesis, we investigated their effect on epithelial-to-mesenchymal transition (EMT), a critical process for development of the outflow tract. We find that these LVOT-associated NOTCH1 alleles can contribute to defective EMT in endothelial cell lines through impaired induction of Snail and Hes family members. These data represent the first description of a molecular mechanism underlying NOTCH1 mutations in individuals with LVOT malformations, and have important implications regarding the functional contribution of these alleles to a complex set of developmental defects.
Keywords: Notch signaling, Left ventricular outflow tract, Bicuspid aortic valve, Epithelial to mesenchymal transition (EMT)
1. Introduction
Defects involving the left ventricular outflow tract (LVOT) comprise a clinically significant group of congenital cardiovascular malformations. LVOT malformations, including bicuspid aortic valve (BAV), aortic valve stenosis (AVS), coarctation of the aorta (COA), and hypoplastic left heart syndrome (HLHS), are present in 1 in 1000 live births, and account for a significant portion of infant mortality [1–3]. Although the etiology of LVOT malformations is unclear, both environmental and genetic components play a role in disease pathogenesis. For example, prenatal exposure to solvents or high phenylalanine levels (secondary to maternal phenylketonuria) have been associated with higher incidence of LVOT malformations [4]. In addition, linkage analysis demonstrates a strong genetic component for these malformations. A non-parametric linkage analysis of LVOT malformation shows linkage to three chromosomes with overlapping linkage peaks suggesting a common genetic cause [5]. Mutations in NOTCH1 have been reported in two families with bicuspid aortic valve (BAV) and calcific AVS [6], in sporadic BAV [7,8], and by our group in BAV, AVS, COA, and HLHS [9]. In support of the presumed common genetic pathogenic mechanism, we identified NOTCH1 missense variants in patients across the LVOT phenotypic spectrum and found that these alleles reduce ligand-dependent Notch signaling [9].
Notch signaling is an evolutionarily conserved pathway that regulates cell fates and tissue formation during embryogenesis, including cardiac development [10,11]. The NOTCH1 receptor is synthesized as a large polypeptide with 36 EGF-like repeats in the extracellular domain, three NOTCH/Lin repeats, a transmembrane domain, a transactivating domain, and intracellular domain with six ankyrin repeats to facilitate protein–protein interactions. Mammalian NOTCH1 is synthesized as a single 300-kDa polypeptide in the endoplasmic reticulum (ER) and cleaved by a furin convertase during posttranslational processing in the Golgi complex into p120 and p180 (S1 cleavage, see Fig. 1A). Following cleavage, the two portions of the protein are presented as a functional heterodimer on the cell surface. Ligands of the Delta and Jagged families presented on adjacent cells can interact with the extracellular domain of NOTCH1. This interaction triggers two subsequent cleavages (S2 and S3), resulting in the release of the intracellular domain (NICD). NICD translocates into the nucleus, where it functions in the activation of downstream targets including members of the Hairy-Enhancer of Split (Hes) family of transcription factors [12].
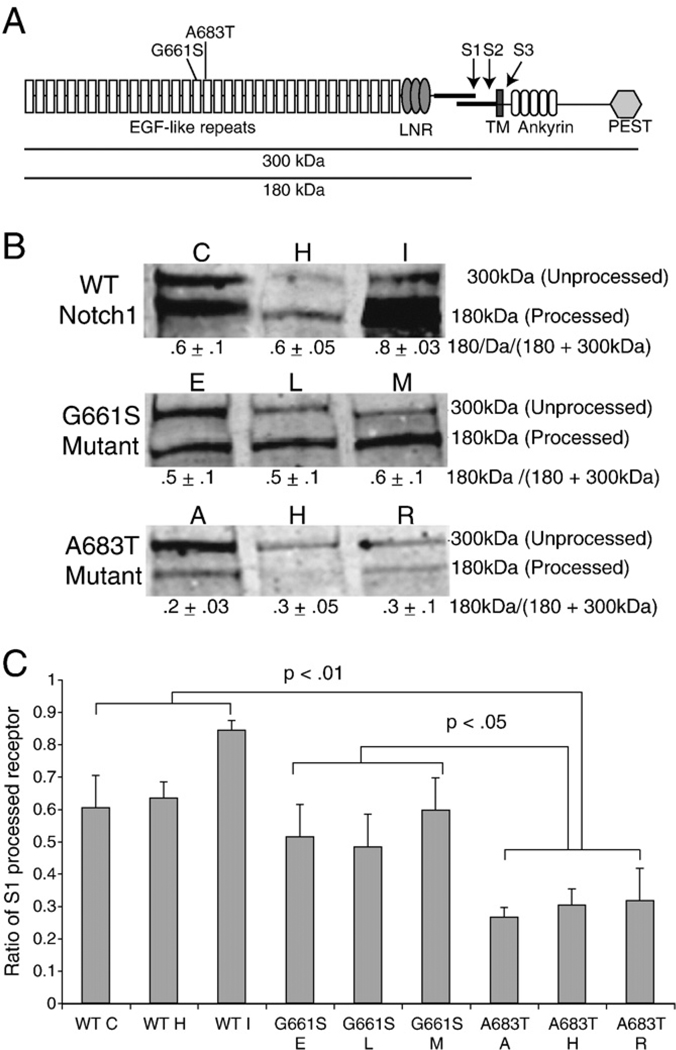
Fig. 1.
Reduced S1-cleavage of NOTCH1A683T receptor. A. Schematic showing the expected NOTCH1 products after typical processing events. The full length NOTCH1 protein (300 kDa) is cleaved at site 1 (S1) before presentation at the plasma membrane as a functional heterodimer (p180 and p120). Interaction with ligand triggers two subsequent cleavages (S2 and S3), releasing the intracellular domain (NICD) to activate downstream transcription factors. The LVOT-associated mutations (G661S, A683T) are located in the seventeenth and eighteenth EGF repeats in the extracellular domain (EC). B. Total protein lysates from three independent NIH3T3 cell lines stably expressing either N-terminally HA-tagged wild-type rNOTCH1 (clones C, H, I), rNOTCH1G661S (clones E, L, M), or NOTCH1A683T (clones A, H, R) receptor were analyzed by western blot using an anti-HA antibody. Note the decreased levels of the 180 kDa band (S1 Processed) in cells expressing the rNOTCH1A683T receptor compared with wild-type rNOTCH1-expressing cells. Results from a representative experiment are shown. C. Band intensities were quantified and the percentage of total receptor that has undergone S1 cleavage was calculated (180 kDa/180+300 kDa). The average ratio of S1-cleaved protein is shown as the mean±SD from three independent experiments. p-values were calculated using ANOVA followed by Bonferroni post hoc.
Recent research supports several important roles for Notch signaling during cardiac development. In the mouse, targeted deletion of Notch1 or its nuclear partner RBPJK/CBF1/Su(H) results in impaired epithelial-to-mesenchymal transition (EMT) during cardiac cushion development, leading to a collapsed endocardium and the absence of cushion cells in the mesenchyme [13]. Combined loss of Notch1 downstream targets, Hey1 and HeyL also causes impaired EMT in mice [14]. EMT occurs in the endocardium around E9.0 to form the cardiac cushions, and is critical for proper outflow tract and atrioventricular canal development. During cardiac EMT, endocardial cells undergo significant changes in gene expression including Notch1-dependent induction of α-SMA, Snail1, and Snail2 [15,16]. In addition to its role in EMT, recent data indicates that Notch signaling plays additional critical roles in both the neural crest and the secondary heart field during development [17, 18].
In this study we examine the impact of LVOT-associated NOTCH1 mutations on Notch processing and induction of EMT. Two previously identified missense NOTCH1 variants that reduce JAGGED1 dependent Notch signaling are observed across a wide spectrum of LVOT phenotypes. One variant (NOTCH1G661S) was present in patients with AVS, CoA, and HLHS, as well as in a patient with bicuspid aortic valve (BAV), reinforcing the idea of a common pathogenic mechanism. Interestingly, although these mutations are found at highly conserved sites within the NOTCH1 protein, alignment with other EGF repeats suggests that they might be well-tolerated substitutions. Indeed, all of these variants were also observed in an unaffected parent, so they can be tolerated in some developmental conditions. Given these conflicting findings, we examined the functional effects of these mutations on NOTCH1 protein maturation, trafficking and function, as well as their effects on the induction of EMT in endothelial cell lines, to determine how these missense variants might contribute to human disease.
2. Materials and methods
2.1. Cell culture and plasmids
NIH3T3 cells were cultured as previously described [9]. The HMEC-1 microvascular endothelial cell line was provided by the Centers for Disease Control and Prevention (Atlanta, Ga) and cultured as previously described [19]. Rat cDNAs encoding N-terminally HA-tagged rat NOTCH1, mutant NOTCH1G661S, and mutant NOTCH1A683T were described previously [9]. GenBank Accession numbers are NM_017617.3 for human NOTCH1 and NM_001105721.1 for rat NOTCH1. Protein numbering reflects the initiation codon as codon 1. To generate stable NOTCH1 cell lines, 4×104 NIH3T3 cells were plated in 24 well plates and transfected using Lipofectamine 2000 with 0.8 µg HA-tagged wild-type or mutant NOTCH1 expression vectors. Stable lines were generated by expanding individual colonies after culturing the cells for 15 days after transfection in the presence of G418 (0.6 mg/ml). NOTCH1 expression levels were determined by western blot. Cell lines exhibiting a range of NOTCH1 expression levels were expanded for use in further studies. Cell lines exhibiting similar levels of wild-type or mutant NOTCH1 were maintained to facilitate comparison between cell lines.
2.2. Western blot
1.5 × 105 NIH3T3 cells stably expressing NOTCH1, mutant NOTCH1G661S, or mutant NOTCH1A683T were plated in 6 well plates. After 24 h cells were lysed, run on a 6% SDS-PAGE gel, transferred to nitrocellulose, and NOTCH1 protein was detected with a mouse anti-HA antibody (HA-7, 1:1000, Sigma-Aldrich) and Alexafluor anti-mouse 688 secondary antibody (1:20,000, Invitrogen). Band intensity was quantified using Li-Cor Odyssey 2.1 software. All exposures were in the linear range, as determined by this software. p300 and p180 bands were quantified, and the % of total protein cleaved was calculated as p180/(p180+p300). Each experiment was performed in triplicate and statistical analysis was performed (one way ANOVA, followed by Bonferroni post hoc). Western blots after co-culture assays were performed essentially as above, using the following primary antibodies: SNAIL 1(Cell Signaling Technologies #3895 1:1000), SNAIL 2 (Cell Signaling Technologies #9585 1:500), Hey2 (Millipore #AB15632 1:1000), HEYL (Millipore #MAB10094, 1:1000) and α-Tubulin (Sigma-Aldrich T5168 1:1000). In some cases, western blots were developed using ECL plus, following the manufacturers recommendations.
2.3. Cell surface biotinylation
1.5 × 105 NIH3T3 cells stably expressing NOTCH1, mutant NOTCH1G661S, or mutant NOTCH1A683T were plated in 6 well plates. After 24 h, cells were incubated with 0.5 mg/ml sulfo-NHS-biotin (Pierce) for 2 h at 4 °C. After washing, cell lysates were prepared in RIPA buffer. Ten percent of each sample (35 µl) was removed and stored at −80 °C. The remaining lysate (315 µl) was incubated with 40 µl of streptavidin agarose resin (Thermo Scientific) overnight at 4 °C. Avidin-bound biotinylated samples (50 µl) and their respective total lysates (35 µl) were analyzed by western blot as described above. The amount of NOTCH1 at the cell surface was quantified as biotinylated receptor (B)/total amount of expressed receptor (T) and expressed as the relative ratio of biotinylated NOTCH1. Each experiment was performed in triplicate and statistical analysis was performed (one way ANOVA, followed by Bonferroni post hoc).
2.4. Immunofluorescent protein localization
2×104 NIH 3 T3 cells stably expressing NOTCH1, mutant NOTCH1G661S, or mutant NOTCH1A683T were plated on acid treated coverslips in 24 well plates. The following primary antibodies were used: mouse (m) α-HA (1:1000, Sigma), rabbit (Rb) α-HA (1:1500, Abcam), Rbα-EEA1 (1:500,Abcam),Rbα-Rab7 (1:100,Cell Signaling),r α-calnexin (1:500, Abcam), m α-GM130 (1:100), and m α-smooth muscle actin (1:200, Sigma). Alexa secondary antibodies (Molecular Probes) were used at a dilution 1:1000: goat (gt) α-m 488 and gt α-Rb 594. For quantification, 100 cells were counted for each experiment on each cell line, and three independent experiments were performed. The samples were blinded before quantification.
2.5. EMT induction by co-culture
2×105 HMEC-1 cells were plated in a 6-well dish and transfected 24 h later using Lipofectin (Invitrogen) with 2 µg of NOTCH1, mutant NOTCH1G661S, ormutant NOTCH1A683T expression vectors.In a separate 6-well plate, 2×105 HMEC-1 cells were transfected with 2 µg of pEF-BOS (vector), or pBOS-JAG1. 12 h post-transfection, HMEC-vector or HMEC-JAG1 was co-cultured with HMECs expressing wild-type or mutant NOTCH1 for 24 h. Total RNA was isolated for RT-PCR. Alternatively, 2×104 HMEC-1 cells were plated on coverslips and transfected 24 h later using Lipofectin (Invitrogen) with 1 µg of NOTCH1, mutant NOTCH1G661S, or mutant NOTCH1A683T expression vectors. After co-culture with control HMEC-1rJAG1-expressingHMEC-1 cells, cells were examined by immunofluorescence.
3. Results
3.1. S1-cleavage of the NOTCH1A683T receptor is reduced
The NOTCH1G661S and NOTCH1A683T variants cause impaired ligand-dependent Notch1 activation and are associated with LVOT malformations [9].To determine the molecular cause of the impaired signaling, we examined whether these mutations affect NOTCH1 receptor processing. The NOTCH1 receptor is synthesized as a single 300-kDa polypeptide in the endoplasmic reticulum and cleaved (S1) by a furin convertase during posttranslational processing in the Golgi complex into p120 and p180 before its presentation at the cell surface. S1 cleavage has generally been believed to be required for CBF1 dependent Notch signaling, although more recent data suggests that there is not an absolute requirement for S1 cleavage in NOTCH1 [20,21]. The reduction in ligand-induced signaling when S1 cleavage is inhibited suggests that variants that reduce cleavage may exhibit reduced ligand-dependent signaling. S1 cleavage analysis in NIH3T3 cells transiently overexpressing wild-type or variant NOTCH1 suggested that S1 cleavage was reduced in NOTCH1A683T [9]. However, the high protein expression levels found in transient transfection can impair protein processing and trafficking, thus we confirmed and extended these observations in cell lines stably expressing varying amounts of NOTCH1 protein. Independent, stable cell lines were produced expressing NOTCH1WT, NOTCH1G661S or NOTCH1A683T. Western blot analysis was used to select independent cell lines expressing varying amounts of NOTCH1 receptor to control for effects of protein expression levels.
Western blot analysis of cell lines stably expressing wild-type or variant (G661S, A683T) NOTCH1 reveals the presence of full length, unprocessed NOTCH1 as a band of 300 kDa and S1 processed NOTCH1 as a band of 180 kDa in all cell lines (Fig. 1B). As suggested by previous work, we found that S1-processing is consistently reduced for NOTCH1A683T; the ratio of total protein processed (p180/(p180+ p300)) averaged .66±.1 for the three wild-type NOTCH1 lines, but only .29±.03 (p<.01) for the NOTCH1 A683T cell lines. In contrast, the ratio of total processed protein averaged .52±.03 for the NOTCH1G661S cell lines, indicating a slight but statistically insignificant reduction in S1 processing compared to wild-type (Fig. 1B,C). These data confirm and extend our previous findings, confirming that alterations in NOTCH1A683T S1-processing are a result of the missense mutation and are not caused by protein overexpression.
3.2. Cell surface presentation of NOTCH1G661S and NOTCH1A683T is reduced
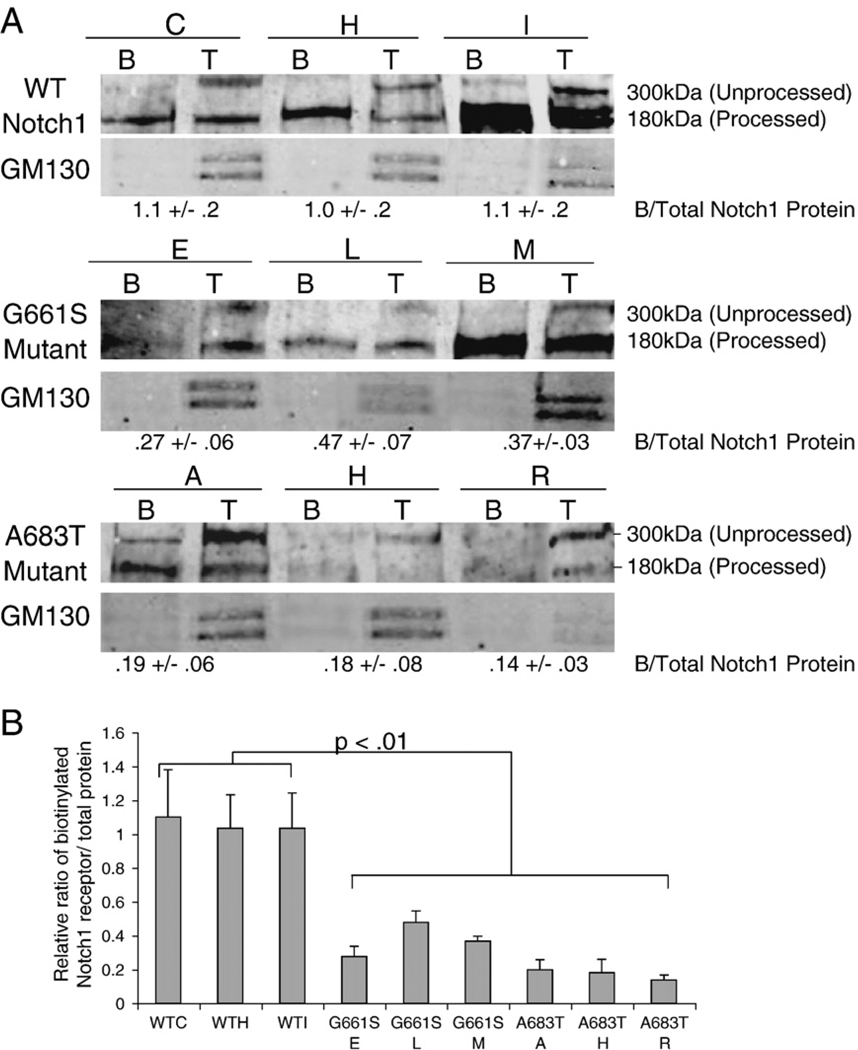
Given the decrease in ligand-induced signaling of NOTCH1G661S and NOTCH1A683T mutants [9], and the reduction of S1-cleaved NOTCH1A683T, the amount of mutant receptor present at the plasma membrane may be altered compared to wild-type. To address this hypothesis, we performed cell-surface biotinylation experiments to directly assess receptor presentation on the cell surface. In cell lines stably expressing wild-type NOTCH1 receptor, the majority of biotinylated receptors corresponded to the 180 kDa form (Fig. 2A), indicating that under our conditions only the S1 cleaved form efficiently reaches the cell surface. Likewise, the majority of biotinylated NOTCH1G661S and NOTCH1A683T receptors corresponded to the 180 kDa form with significant amounts of p300 at the cell surface seen in only one NOTCH1A683T line (Fig. 2A). Interestingly, the relative amounts of receptor on the cell surface were significantly reduced in NOTCH1G661S and NOTCH1A683T cell lines compared to lines expressing wild-type receptor. The relative ratio of biotinylated receptor (expressed on the cell surface) to total amount of receptor protein was 1.05±.04 for the three wild-type lines, whereas it was 0.38±.1 (p<.01) for NOTCH1G661S and 0.17±.03 (p<.01) for NOTCH1A683T expressing cell lines (Fig. 2A,B). Thus, NOTCH1 receptors with mutations at either G661 or A683 are found at reduced levels on the cell surface compared to wild-type receptors. In the NOTCH1A683T mutation, this reduced cell-surface presentation may be a secondary effect of reduced S1 cleavage. Further, since S1 cleavage of NOTCH1G661S appears to be normal in our analyses, the molecular mechanism underlying the reduced cell-surface expression may be distinct for each variant.
Fig. 2.
Reduced amounts of NOTCH1G661S and NOTCH1A683T are presented at the plasma membrane. A. Proteins expressed on the plasma membrane of NIH3T3 stable cell lines expressing either rNOTCH1WT, rNOTCH1G661S, or rNOTCH1A683T receptor were labeled by biotinylation. Ten percent of the total protein extract was removed (T) after which the remaining lysate was subjected to streptavidin pull down (B), to isolate cell-surface proteins. Western blot analysis of total protein lysate (T) and cell-surface proteins (B) was performed using an anti-HA antibody. The 180 kDa band present in all biotinylated fractions indicates that only the S1 cleaved form of rNOTCH1 reaches the plasma membrane efficiently. Note the relative decrease in biotinylated receptor in cell lines expressing rNOTCH1G661S or rNOTCH1A683T mutants compared to cell lines expressing wild-type NOTCH1. anti-GM130 antibody was utilized to assess fraction purity. Representative data are shown. B. Band intensities were quantified and the ratio of biotinylated NOTCH1 to total NOTCH1 was calculated. Results are graphed as mean±SD from three independent experiments. p-values were calculated using ANOVA followed by Bonferroini post hoc.
3.3. NOTCH1A683T receptor protein accumulates in the endoplasmic reticulum
The decrease in S1 cleavage and the reduced levels of NOTCH1A683T present at the cell surface compared to wild-type could both result from impaired intracellular trafficking. For instance, it is possible that the observed reduction in S1 cleavage of NOTCH1A683T could be a consequence of either impaired S1 cleavage in the Golgi, or of reduced transport of the receptor from the endoplasmic reticulum (ER) to the Golgi. To test these hypotheses, we analyzed NOTCH1 intracellular localization by immunofluorescence in cells stably expressing wild-type or mutant receptors. In most cells expressing wild-type NOTCH1, the receptor was found localized to small, cytoplasmic vesicle-like structures, and present at the cell surface, similar to previous descriptions [22]. Similar localization was observed in cell lines expressing NOTCH1G661S. In contrast, the majority of cells expressing the variant NOTCH1A683T receptor exhibited protein localization in the perinuclear region with a corresponding reduction in protein at the cell surface and in vesicle-like bodies. To quantify the mislocalization of NOTCH1A683T, 100 cells from each cell line were counted, and classified as having protein largely in perinuclear regions (see, for example Fig. 3A. panels i, l) or having protein throughout the cell (Fig. 3A, panels a and j). Three independent experiments were performed for each cell line, demonstrating a significant increase in the number of cells with perinuclear protein staining for cells expressing NOTCH1A683T compared to cells expressing NOTCH1G661S or wild-type NOTCH1 (Fig. 3B).
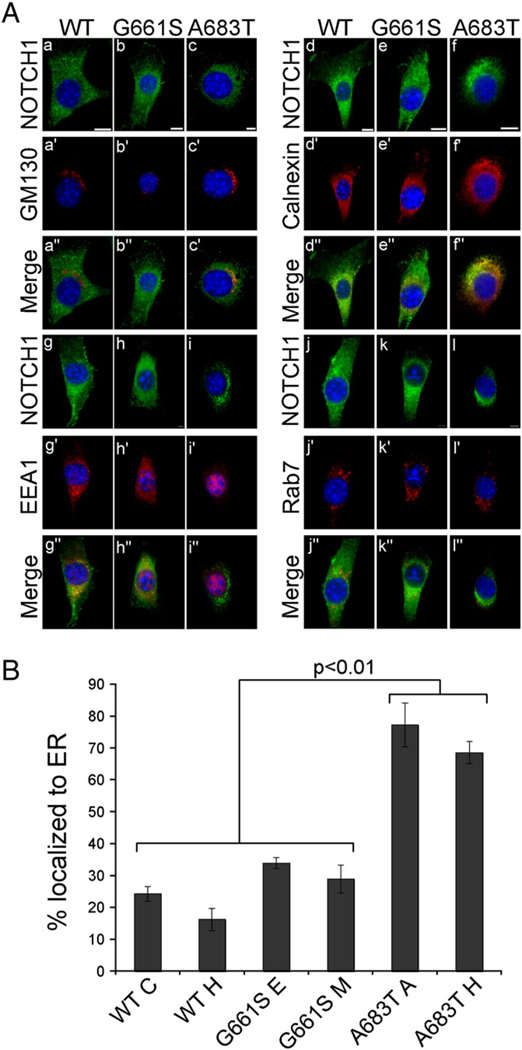
Fig. 3.
The LVOT-associated mutant NOTCH1A683T exhibits improper intracellular trafficking and localization to the endoplasmic reticulum (ER). A. The subcellular location of rNOTCH1 receptors in stable NIH 3T3 cells was assayed by immunofluorescence. Costaining of NOTCH1 with the Golgi-marker GM130 (a–c) did not reveal co-localization in cells expressing rNOTCH1WT (a–a”), rNOTCH1G661S (b–b”), or rNOTCH1A683T (c–c”). Costaining of Notch1 with ER-marker calnexin (d–f) revealed strong co-localization of the mutant rNOTCH1A683T protein with calnexin in the majority of cells observed. Costaining of NOTCH1 with early endosome marker EEA1 (g–i) revealed some co-localization although no significant differences were observed. Similarly, some co-localization with late endosome marker Rab7 was observed, although the degree of co-localization was similar among all cell lines (j–l). Scale bars are 10 µm. B. Two stable lines expressing wild-type or mutant NOTCH1 were co-stained for ER-marker calnexin and NOTCH1 (anti-HA) as above. The co-localization of NOTCH1 and calnexin was assessed in 100 cells, and the percent of cells exhibiting NOTCH1 localization in the ER is shown. In cells expressing rNOTCH1A683T, co-staining reveals a significant increase in localization of mutant protein to the ER (p<.001). In addition, ER localization is slightly lower in cell line WT H compared to lines expressing NOTCH1G661S (p <.05). % ER localization is calculated from three independent experiments. Values were analyzed by ANOVA followed by Bonferroni post hoc.
To further define the intracellular localization of the variant NOTCH1A683T protein, we performed immunofluorescence with markers for Golgi (GM130), ER (Calnexin), early endosome (EEA1), and late endosome (Rab7). In the majority (73%) of cells expressing NOTCH1A683T, mutant protein co-localized with an ER marker, calnexin (Fig. 3A: f”; B), whereas only 20% of cells expressing wild-type NOTCH1 exhibited co-localization with calnexin (Fig. 3A: d”; B). These findings indicate that trafficking of the mutant NOTCH1A683T receptor from the ER to the Golgi is impaired, leading to less protein present in the Golgi for S1 cleavage, and ultimately less protein presented at the cell surface. This suggests that the A683T mutation may impair proper protein folding, leading to the retention of the protein in the ER.
In contrast to our findings for NOTCH1A683T protein, the localization pattern in the majority (68%) of cells expressing mutant NOTCH1G661S was similar to that seen in wild-type cells, although the cell-surface expression was sometimes reduced (Fig. 3A: compare, for example, panel a with panel b), consistent with the biotinylation data reported above. We did not observe co-localization with Golgi markers in wild-type NOTCH1, NOTCH1G661S, or NOTCHA683T (Fig. 3A:a–c), reflecting that NOTCH1 protein is only transiently present in the Golgi. Both wild-type NOTCH1 and NOTCH1G661S exhibited some co-localization with EEA1 and Rab7, although no significant differences were observed (Fig. 3A: g–i, j–l), indicating that endocytic processing of the mutant NOTCH1G661S receptor is not demonstrably different than that of wild-type NOTCH1 receptor. These findings reinforce the suggestion that distinct mechanisms underlie the reduction in ligand-induced Notch signaling observed in the NOTCH1A683T and the NOTCH1G661S variants.
3.4. NOTCH1G661S and NOTCH1A683T mutations cause reduced expression of Notch-responsive EMT markers
These findings, together with our previous report [9] demonstrate that the A683T and G661S variants in NOTCH1 variably affect protein folding, processing and cell-surface presentation, resulting in a partial loss of ligand-induced Notch signaling. It remains to be demonstrated that this reduction in signaling has an overt effect on developmental processes that require Notch function. To address this question, we examined the effect of missense variants in the NOTCH1 receptor on the pathway's function in EMT. During cardiac EMT, endocardial cells that overlie the atrioventricular (AV) canal and outflow tract lose apical–basal polarity, invade the cardiac jelly, and form the endocardial cushions. EMT in the outflow tract is an essential process for proper development of the aortic valve [23]. Recent work has shown that Snail2 is directly upregulated by Notch in endothelial cells and that expression of both Snail1 and Snail2 is required for cardiac cushion EMT in vivo [15].
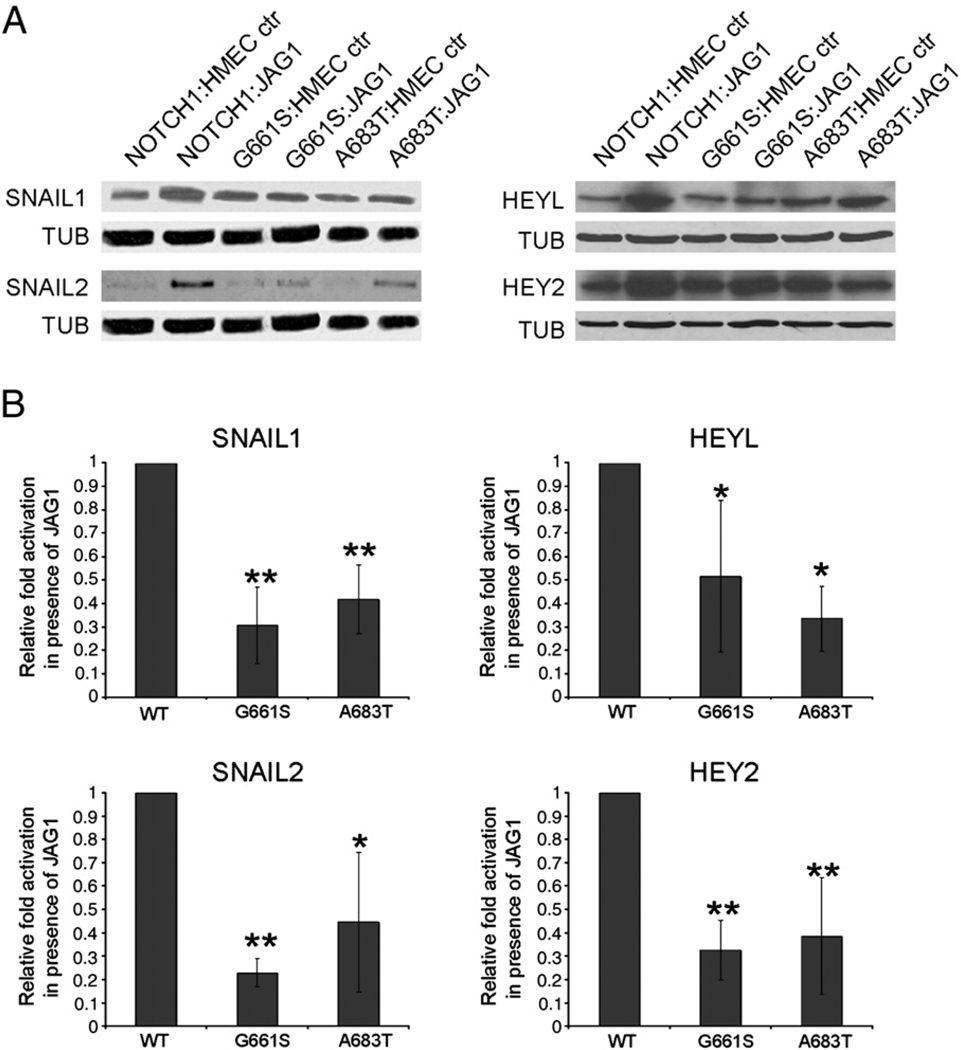
To examine the effects of LVOT-associated NOTCH1 variants on EMT we took advantage of the recent finding that co-culture of HMEC-1 cells (an immortalized human microvascular endothelial cell line) with HMEC-1 cells expressing JAGGED1 can induce EMT in signal-receiving cells, as demonstrated by induction of α-SMA, a direct Notch target [24] and phenotypic changes indicative of mesenchymal transition [16]. We utilized western blot analysis to examine the relative expression of SNAIL1 and SNAIL2 after HMEC cells expressing NOTCH1WT or variant NOTCH1 (NOTCH1G661S, NOTCH1A683T) were co-cultured with HMEC cells expressing JAGGED1. A robust induction of SNAIL1 and SNAIL2 was observed when HMECs expressing wild-type NOTCH1 were co-cultured with HMEC-JAGGED1 cells (Fig. 4A). In contrast, when HMEC cells expressing either variant receptor were co-cultured with HMEC-JAGGED1, we observed reduced SNAIL1 induction (69% reduction for NOTCH1G661S and 58% reduction for NOTCH1A683T) compared to that seen in NOTCH1WT co-cultured with HMEC-JAGGED1. Both variants also exhibit significant reductions in JAGGED1-dependent induction of SNAIL2 (77% reduction for NOTCH1G661S and 55 % reduction for NOTCH1A683T) (Fig. 4B). In addition, HEYL and HEY2 represent direct Notch targets that play important roles in cardiac EMT [14]. As observed for SNAIL1 and SNAIL2, we find that JAGGED1-responsive induction of both HEYL and HEY2 during EMT is significantly reduced in cells expressing either NOTCH1G661S or NOTCH1A683T compared to cells expressing wild-type NOTCH1 (Fig. 4). These data indicate that two LVOT-associated missense alleles, NOTCH1G661S and NOTCH1A683T, have an impaired ability to induce the expression of Notch-responsive markers of EMT.
Fig. 4.
Reduced JAGGED1 dependent Notch1 activation of SNAIL1 and SNAIL2 in LVOT-associated mutants. A. HMECs expressing WT or mutant rNOTCH1 were co-cultured with HMEC cells transfected with a control vector (HMEC-ctr) or a JAGGED1 expression vector (JAG1) for 24 h. Western blot analysis was performed using antibodies specific for SNAIL1, SNAIL2, HEY2, HEYL, or Tubulin. Representative results are shown indicating that these markers are upregulated when signal-receiving cells expressing wild-type NOTCH1 receptor are co-cultured with cells expressing JAGGED1, while activation of expression is reduced when signal-receiving cells express NOTCH1A683T or NOTCH1G661S. B. The induction of, SNAIL1, SNAIL2, HEYL and HEY2 by JAGGED1 was compared in HMEC cells expressing wild-type NOTCH1, NOTCH1G661S or NOTCH1A683T. Band intensities were quantified and normalized to the value for tubulin. Fold activation by JAGGED-expressing cells was calculated (level when co-cultured with JAGGED1 expressing cells/level when co-cultured with HMEC control cells), and the value for fold activation in cells expressing wild-type NOTCH1 was set to one for comparison across experiments (actual fold increases were 6.1±1.2 for SNAIL1 and 8.0±4.3 for SNAIL2, 3.2±1.1 for HEYL, and 2.5±1.2 for HEY2). We observe an overall 50–80% reduction in the expression levels of SNAIL1, SNAIL2, HEYL and HEY2 in cells expressing missense variants of NOTCH1 compared to cells expressing wild-type NOTCH1. Each experiment was performed at least three times in duplicate, and statistical analysis was performed (**=p<.01, *=p<.05 by ANOVA, Dunnett post hoc).
3.5. NOTCH1G661S and NOTCH1A683T mutants exhibit impaired induction of EMT
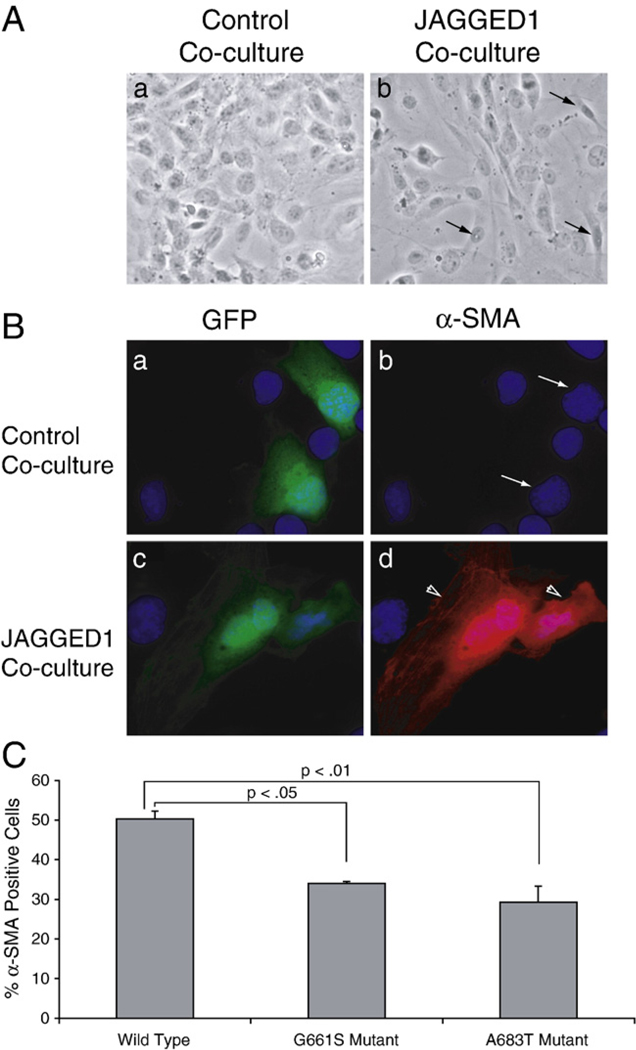
In order to determine whether a partial reduction in Notch signaling can affect the efficiency of the cellular transition of endothelial to mesenchymal cell types, we performed co-culture assays as described above followed by phase-contrast microscopy and immunofluorescence. Epithelial cells appear as a unicellular layer, with uniformly spaced cell–cell junctions and apical–basal polarity. In contrast, mesenchymal cells exhibit a lack of intracellular adhesion, with irregularly shaped, elongated cells. Mesenchymal cells also exhibit a distinct leading edge polarity indicative of their dynamic migratory capability. HMEC-1 cells were co-transfected with expression vectors for wild-type or mutant NOTCH1, along with a GFP expression vector to mark transfected cells. Co-culture of HMECs expressing exogenous NOTCH1 receptor with Jagged1-expressing cells caused morphological changes in some signal-receiving cells consistent with EMT (Fig. 5A). To examine the relative efficiency of EMT in cells expressing wild-type NOTCH1 compared to cells expressing missense variants of NOTCH1, cells were stained with an anti-α-SMA antibody and examined by immunofluorescence. α-SMA expression and cell morphology were observed in GFP positive signal-receiving cells (which are presumed to express exogenous wild-type or mutant NOTCH1). Co-culture of HMECs expressing wild-type NOTCH1 with JAGGED1-expressing cells induced EMT in ∼50% of GFP positive cells as determined by loss of apical–basal polarity, loss of distinct intracellular junctions, and expression of EMT marker, α-SMA (Fig. 5B,C). In contrast, co-culture of HMECs expressing NOTCH1G661S or NOTCH1A683T variant receptors with JAGGED1-expressing cells induced EMT in only 34% and 29% of GFP positive cells respectively (Fig. 5B,C p<0.05). These results indicate that a partial reduction in Notch1 signaling, which leads to reduced expression of target genes can result in reduced efficiency of EMT in endothelial cells.
Fig. 5.
Cell lines expressing LVOT-associated NOTCH1 variants exhibit defective JAGGED1-induced EMT. A. Phase-contrast micrographs of HMECs expressing wild-type NOTCH1 co-cultured with HMEC cells containing empty vector (a) or JAGGED1 expression vector (b). When NOTCH1 expressing cells are co-cultured with JAGGED1 expressing cells, they undergo a morphological transformation characterized by loss of intercellular junctions, loss of cellular monolayer, formation of filopodia, and irregular cell shape (arrows). B. HMECs co-transfected with a NOTCH1 expression vector and a GFP expression vector were co-cultured with control HMECs (a and b) or HMECs expressing JAGGED1 (c and d). Transfected cells (as identified by GFP expression (a, c)) were examined for α-SMA staining (b and d). Cells underwent EMT only when NOTCH1 expressing HMECs were co-cultured with HMECs expressing JAGGED1 (compare arrows in b to arrowheads in d). C. The efficiency of EMT induction by JAGGED1 was compared between cells expressing wild-type NOTCH1 and cells expressing NOTCH1G661S or NOTCH1A683T. The percentage of transfected cells (as identified by GFP expression) that had also undergone EMT (as defined by morphology and α-SMA expression) was quantified. The percentages reflect the average efficiency of EMT for100 observed cells in three independent experiments. The efficiency of EMT is significantly reduced in cells that express mutant forms of the NOTCH1 receptor (p<0.05, ANOVA followed by Dunnet post hoc).
4. Discussion
4.1. Distinct molecular mechanisms may underlie the reduction in signaling through different mutant Notch1 receptors
We have previously reported that missense variants in the human NOTCH1 gene identified in patients with LVOT defects are correlated with a reduction in ligand-induced CBF1-dependent Notch signaling [9]. Here we examine the molecular mechanism(s) that contribute to this reduction in signaling. The examined variants (NOTCH1G661S and NOTCH1A683T) are found in neighboring EGF repeats (repeats 17 and 18), but our findings indicate that the underlying molecular cause of the reduction in signaling of these mutant receptors may be distinct.
We find that NOTCH1A683T receptors exhibit both reduced S1 cleavage and a reduction in the amount of receptor found at the cell surface (Fig. 1 and Fig. 2). Our data further indicate that mutant NOTCH1A683T localizes to the ER in the majority of cells, in contrast to the usual NOTCH1 protein localization in small vesicle-like bodies, and at the cell surface. These data may indicate that the A683T mutation, which replaces a hydrophobic, nonpolar amino acid with a hydrophilic polar amino acid, interferes with proper protein folding, resulting in the retention of the misfolded protein in the ER. This retention, and the presumed subsequent degradation of misfolded protein would result in a reduced S1 cleavage, reduced cell-surface presentation, and a subsequent reduction in ligand-induced signaling. There exists some precedent that missense mutations in EGF repeats can cause protein misfolding. For instance, the G274S mutation in JAGGED1 identified in patients with tetralogy of Fallot impairs folding of the EGF repeat [25]. It may also be interesting to note that EGF repeat 18 is a reported substrate for fucosylation by the enzyme POFUT1 [26]. In Drosophila, Ofut1 protein possesses an essential, chaperone function that is required for stable, cell-surface expression of NOTCH [27]. In contrast, however, in mammalian cells POFUT1 is not required for cell-surface expression, although fucosylation of Notch receptors is required for optimal ligand binding [28].
In contrast to our findings for NOTCH1A683T, we find that the NOTCH1G661S variant does not overtly affect either folding or S1 processing. However, like NOTCH1A683T, NOTCH1G661S is found at reduced amounts at the cell surface (Fig. 1 and Fig. 2). We presume that the reduced amount of receptor at the cell surface contributes to the observed reduction in ligand-induced signaling, but the mechanisms for this change in protein trafficking are not clear. G661 is found in EGF repeat 17, which has been reported to be modified by addition of an O-linked glucose [26]. In Drosophila, loss of the enzyme that catalyzes this modification causes a temperature sensitive Notch phenotype, and accumulation of the receptor on the cell surface [29]. The functions of EGF glucosylation in mammalian cells are poorly understood [30]. It is also possible that this amino acid change interferes with other modifications of the receptor during intracellular processing. For instance, in mammalian cells, the E3 ubiquitin ligases Cbl and Itch/AIP4 have been shown to modify Notch but the biological importance of these alterations are currently not known [31,32]. Some studies have shown that NOTCH1 cell-surface expression is negatively regulated by the addition of ubiquitin signals during intracellular trafficking [33,34]. Since ubiquitination is often linked to the endocytic pathway, it is possible that improper modifications to the mutant receptor lead to alterations in endocytic trafficking that are not noticeable by immunofluorescence, but which nonetheless affect receptor presentation at the cell surface. These results suggest that diverse molecular mechanisms can perturb Notch1 signaling during cardiac development.
4.2. Reductions in ligand-induced Notch signaling can affect cell fate during EMT
The NOTCH1 variants identified in patients with LVOT malformations are predicted to reduce, but not abolish, Notch signaling. This implies that at least some of the processes during cardiac development that require Notch signaling will have dosage sensitive outcomes. We examined the function of the NOTCH1A683T and NOTCH1G661S variants during JAGGED1-induced EMT to determine whether the partial reductions we described in Notch signaling could influence cell fate decisions. EMT in the outflow tract cushions is necessary for proper formation of the aortic and pulmonary valves. In response to signals from the adjacent myocardium, the endocardium undergoes Notch-dependent EMT via upregulation of the Snail family of transcription factors [13,16,35,36]. Mice with loss-of-function mutations in the Notch pathway display collapsed endocardium, lack mesenchymal cushion cells, and exhibit defective invasion of endocardial cells into the cardiac jelly, indicative of impaired EMT in the outflow tract [13,14]. We find that endothelial cells that express mutant forms of the NOTCH1 receptor protein exhibit reduced JAGGED1-dependent expression of critical EMT inducers Snail1, Snail2, and α-SMA, as well as reduced expression of the canonical Notch targets HEY2 and HEYL (Fig. 4). This reduction in the expression levels of critical mediators of EMT leads to an actual reduced efficiency in JAGGED1-induced EMT compared to cells expressing wild-type NOTCH1 receptors (Fig. 5). Thus our data support the hypothesis that relatively small reductions in NOTCH1 signaling can adversely affect key processes in cardiac development. These findings support the proposal that reduced Notch signaling and the resulting reduction in the efficiency of EMT could serve as a unifying event whereby Notch mutations contribute to diverse LVOT malformations.
4.3. The Notch pathway plays multiple roles during outflow tract development
Although impaired EMT in the outflow tract may provide a unifying pathogenesis for diverse LVOT malformations, there are several additional cardiac developmental processes that may be sensitive to reductions in Notch1 signaling. Dosage sensitivity in any of these developmental decisions could also contribute to cardiac defects in patients carrying Notch1 variant proteins. The outflow tract is generated by interactions among many different cell types, including cardiomyocytes, neural crest derived cells, the secondary heart field, and endothelial cells. Neural crest derived cells give rise to the smooth muscle layer of the aorta and pulmonary artery and contribute to the semilunar valve leaflets. Notch signaling is essential for the differentiation of cardiac neural crest progenitors into smooth muscle cells in vivo, as blocking Notch signaling in the derivatives of the cardiac neural crest results in stenosis of the vessels and aortic arch defects resembling that seen in Alagille syndrome [17]. The smooth muscle layer is believed to be critical for maintaining the structure of the arteries, indicating that defects in Notch signaling resulting from LVOT-associated NOTCH1 alleles in neural crest derived cells could contribute to CoA, which results from a narrowing of the aorta. In addition, recent work has demonstrated that altering Notch signaling levels in cardiomyocytes can result in cardiac defects [37].
Notch signaling in the second heart field is also important for proper outflow tract development. The second heart field is a group of mesoderm derived cells that migrate from the anterior pharynx into the heart and gives rise to the myocardium of the outflow tract, right ventricle, interventricular septum, part of the atria, and smooth muscle at the bottom of the outflow vessels [38–43]. Recent work demonstrates that JAGGED1-dependent Notch signaling in the second heart field regulates the migration of neighboring neural crest cells, and EMT in the endocardial cushions [18]. Together, these data indicate that Notch plays multiple roles in tissue–tissue interactions during outflow tract development, representing additional mechanisms where reductions in Notch signaling could perturb outflow tract development. It is not unreasonable to suggest that as we have seen during EMT, reductions in Notch signaling due to missense mutations such as G661S or A683T may affect the efficiency of other cell fate decisions that are critical to outflow tract development.
4.4. Reductions in Notch signaling may sensitize outflow tract development to other genetic and environmental insults
It is important to remember that the NOTCH1 variants identified in patients with LVOT malformations are not sufficient to cause cardiac defects. In all cases, the identical variant was found in an unaffected parent [9]. One attractive explanation for this finding would be that these missense variants, which lead to reductions, but not loss, of Notch signaling, act to sensitize outflow tract development to other genetic or environmental insults. This hypothesis is especially intriguing in light of the fact that cells expressing mutant NOTCH1 receptors exhibit a reduction in the efficiency of EMT, but not a block to the process. In the context of embryonic development, this partial reduction might lead to a developmental window during which other genetic or environmental insults could synergize with the reduction in EMT to perturb development of the outflow tract in some individuals. In other cases, in the absence of additional perturbations, the level of EMT driven by these mutant receptors might be sufficient to support normal cardiac development, explaining the presence of unaffected carriers of the variants. Some precedent exists for this idea. Snail2-deficient mouse embryos exhibit defective EMT at E9.5, which might be considered equivalent to the reductions in EMT efficiency we observe in endothelial cells expressing variant NOTCH1 receptors. However, in Snail2 deficient embryos, EMT recovers by E10.5 due to an increase in Snail1 expression [15]. It is attractive to hypothesize that this transient deficiency during early outflow tract development could sensitize the tissue to other environmental or genetic insults. The severity of these additional insults may determine whether a specific NOTCH1 variant leads to a simple bicuspid aortic valve or severe hypoplastic left heart syndrome. Intriguingly, recent findings from the Stanley lab indicate that reductions in Notch signaling may act stochastically, or in combination with genetic background to produce phenotypic outcomes of varying severity. Specifically, they find that the combination of the Notch112f hypomorphic allele with a Notch1 null allele produces embryos of the genotype Notch112f/− with a wide variation in phenotype [44]. Similarly, individuals carrying a single copy of a NOTCH1 missense mutation may be at heightened risk for cardiac malformations, dependent on genetic background or environmental effects.
This study elucidates the molecular mechanisms by which missense mutations in the NOTCH1 receptor can contribute to LVOT malformations. We find that two LVOT-associated NOTCH1 alleles reduce Jagged-1 dependent Notch1 signaling by altering protein folding and processing and/or protein trafficking. Expression of variant NOTCH1 receptors in endothelial cells results in defective EMT as well as reduced Notch-dependent induction of Snail family genes. Given the results of our study and recent work of others in the field, we propose that slight reductions in NOTCH1 dosage can affect proper induction of endocardial EMT, and that this change predisposes patients to develop varying malformations of the outflow tract when additional critical pathways are perturbed.
Acknowledgements
This work was supported in part by funds from the Ohio State University Department of Molecular Genetics (SEC), and by predoctoral training fellowship Award Number 0815465D from the American Heart Association (MFR).
References
- 1.Pradat P, Francannet C, Harris JA, Robert E. The epidemiology of cardiovascular defects, part I: a study based on data from three large registries of congenital malformations. Pediatr. Cardiol. 2003;24:195–221. doi: 10.1007/s00246-002-9401-6. [DOI] [PubMed] [Google Scholar]
- 2.McBride KL, Marengo L, Canfield M, Langlois P, Fixler D, Belmont JW. Epidemiology of noncomplex left ventricular outflow tract obstruction malformations (aortic valve stenosis, coarctation of the aorta, hypoplastic left heart syndrome) in Texas, 1999–2001. Birth Defects Res. A Clin. Mol. Teratol. 2005;73:555–561. doi: 10.1002/bdra.20169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferencz C, Loffredo CA, Corea-Vilasenor A, Wilson PD. Genetic and Environmental Risk Factors of Major Cardiovascular Malformations. ARmonk: Futura Publishing Co. Inc; 1997. [Google Scholar]
- 4.Jenkins KJ, Correa A, Feinstein JA, Botto L, Britt AE, Daniels SR, Elixson M, Warnes CA, Webb C, Young AHACoCDit. Noninherited risk factors and congenital cardiovascular defects: current knowledge: a scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115:2995–3014. doi: 10.1161/CIRCULATIONAHA.106.183216. [DOI] [PubMed] [Google Scholar]
- 5.McBride KL, Zender GA, Fitzgerald-Butt SM, Koehler D, Menesses-Diaz A, Fernbach S, Lee K, Towbin JA, Leal S, Belmont JW. Linkage analysis of left ventricular outflow tract malformations (aortic valve stenosis, coarctation of the aorta, and hypoplastic left heart syndrome) Eur. J. Hum. Genet. 2009;17:811–819. doi: 10.1038/ejhg.2008.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 7.Mohamed SA, Aherrahrou Z, Liptau H, Erasmi AW, Hagemann C, Wrobel S, Borzym K, Schunkert H, Sievers HH, Erdmann J. Novel missense mutations (p.T596M and p.P1797H) in NOTCH1 in patients with bicuspid aortic valve. Biochem. Biophys. Res. Commun. 2006;345:1460–1465. doi: 10.1016/j.bbrc.2006.05.046. [DOI] [PubMed] [Google Scholar]
- 8.McKellar SH, Tester DJ, Yagubyan M, Majumdar R, Ackerman MJ, Sundt TMr. Novel NOTCH1 mutations in patients with bicuspid aortic valve disease and thoracic aortic aneurysms. J. Thorac. Cardiovasc. Surg. 2007;134:290–296. doi: 10.1016/j.jtcvs.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 9.McBride KL, Riley MF, Zender GA, Fitzgerald-Butt SM, Towbin JA, Belmont JW, Cole SE. NOTCH1 mutations in individuals with left ventricular outflow tract malformations reduce ligand-induced signaling. Hum. Mol. Genet. 2008;17:2886–2893. doi: 10.1093/hmg/ddn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.High FA, Epstein JA. The multifaceted role of Notch in cardiac development and disease. Nat. Rev. Genet. 2008;9:49–61. doi: 10.1038/nrg2279. [DOI] [PubMed] [Google Scholar]
- 11.de la Pompa JL. Notch signaling in cardiac development and disease. Pediatr. Cardiol. 2009;30:643–650. doi: 10.1007/s00246-008-9368-z. [DOI] [PubMed] [Google Scholar]
- 12.Nichols JT, Weinmaster G. Notch signaling—constantly on the move. Traffic. 2007;8:959–969. doi: 10.1111/j.1600-0854.2007.00592.x. [DOI] [PubMed] [Google Scholar]
- 13.Timmerman LA, Grego-Bessa J, Raya A, Bertrán E, Pérez-Pomares JM, Díez J, Aranda S, Palomo S, McCormick F, Izpisúa-Belmonte JC, de la Pompa JL. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18:99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer A, Steidl C, Wagner TU, Lang E, Jakob PM, Friedl P, Knobeloch KP, Gessler M. Combined loss of Hey1 and HeyL causes congenital heart defects because of impaired epithelial to mesenchymal transition. Circ. Res. 2007;100:856–863. doi: 10.1161/01.RES.0000260913.95642.3b. [DOI] [PubMed] [Google Scholar]
- 15.Niessen K, Fu Y, Chang L, Hoodless PA, McFadden D, Karsan A. Slug is a direct Notch target required for initiation of cardiac cushion cellularization. J. Cell Biol. 2008;182:315–325. doi: 10.1083/jcb.200710067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noseda M, McLean G, Niessen K, Chang L, Pollet I, Montpetit R, Shahidi R, Dorovini-Zis K, Li L, Beckstead B, Durand RE, Hoodless PA, Karsan A. Notch activation results in phenotypic and functional changes consistent with endothelial-to-mesenchymal transformation. Circ. Res. 2004;94:910–917. doi: 10.1161/01.RES.0000124300.76171.C9. [DOI] [PubMed] [Google Scholar]
- 17.High FA, Zhang M, Proweller A, Tu L, Parmacek MS, Pear WS, Epstein JA. An essential role for Notch in neural crest during cardiovascular development and smooth muscle differentiation. J. Clin. Invest. 2007;117:353–363. doi: 10.1172/JCI30070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.High FA, Jain R, Stoller JZ, Antonucci NB, Lu MM, Loomes KM, Kaestner KH, Pear WS, Epstein JA. Murine Jagged1/Notch signaling in the second heart field orchestrates Fgf8 expression and tissue–tissue interactions during outflow tract development. J. Clin. Invest. 2009;119:1986–1996. doi: 10.1172/JCI38922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leong KG, Hu X, Li L, Noseda M, Larrivée B, Hull C, Hood L, Wong F, Karsan A. Activated Notch4 inhibits angiogenesis: role of beta 1-integrin activation. Mol. Cell. Biol. 2002;22:2830–2841. doi: 10.1128/MCB.22.8.2830-2841.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bush G, diSibio G, Miyamoto A, Denault JB, Leduc R, Weinmaster G. Ligand-induced signaling in the absence of furin processing of Notch1. Dev. Biol. 2001;229:494–502. doi: 10.1006/dbio.2000.9992. [DOI] [PubMed] [Google Scholar]
- 21.Gordon WR, Vardar-Ulu D, L'Heureux S, Ashworth T, Malecki MJ, Sanchez-Irizarry C, McArthur DG, Histen G, Mitchell JL, Aster JC, Blacklow SC. Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PLoS One. 2009;4:e6613. doi: 10.1371/journal.pone.0006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flasza M, Nguyen Huu NS, Mazaleyrat S, Clémence S, Villemant C, Clarke R, Baron M. Regulation of the nuclear localization of the human Nedd4-related WWP1 protein by Notch. Mol. Membr. Biol. 2006;23:269–276. doi: 10.1080/09687860600665010. [DOI] [PubMed] [Google Scholar]
- 23.Grego-Bessa J, Diez J, Timmerman L, de la Pompa JL. Notch and epithelial- mesenchyme transition in development and tumor progression: another turn of the screw. Cell Cycle. 2004;3:718–721. [PubMed] [Google Scholar]
- 24.Noseda M, Fu Y, Niessen K, Wong F, Chang L, McLean G, Karsan A. Smooth Muscle alpha-actin is a direct target of Notch/CSL. Circ. Res. 2006;98:1468–1470. doi: 10.1161/01.RES.0000229683.81357.26. [DOI] [PubMed] [Google Scholar]
- 25.Guarnaccia C, Dhir S, Pintar A, Pongor S. The tetralogy of Fallot-associated G274D mutation impairs folding of the second epidermal growth factor repeat in Jagged-1. FEBS J. 2009;276:6247–6257. doi: 10.1111/j.1742-4658.2009.07333.x. [DOI] [PubMed] [Google Scholar]
- 26.Moloney DJ, Shair LH, Lu FM, Xia J, Locke R, Matta KL, Haltiwanger RS. Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. J. Biol. Chem. 2000;275:9604–9611. doi: 10.1074/jbc.275.13.9604. [DOI] [PubMed] [Google Scholar]
- 27.Okajima T, Reddy B, Matsuda T, Irvine KD. Contributions of chaperone and glycosyltransferase activities of O-fucosyltransferase 1 to Notch signaling. BMC Biol. 2008;6:1. doi: 10.1186/1741-7007-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stahl M, Uemura K, Ge C, Shi S, Tashima Y, Stanley P. Roles of Pofut1 and O-fucose in mammalian Notch signaling. J. Biol. Chem. 2008;283:13638–13651. doi: 10.1074/jbc.M802027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, Pan H, Haltiwanger RS, Bellen HJ. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell. 2008;132:247–258. doi: 10.1016/j.cell.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luther KB, Haltiwanger RS. Role of unusual O-glycans in intercellular signaling. Int. J. Biochem. Cell Biol. 2009;41:1011–1024. doi: 10.1016/j.biocel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jehn BM, Bielke W. c-Cbl binding and ubiquitin-dependent lysosomal degradation of membrane-associated Notch1. J. Biol. Chem. 2002;277:8033–8040. doi: 10.1074/jbc.M108552200. [DOI] [PubMed] [Google Scholar]
- 32.Qiu L, Joazeiro C, Fang N, Wang HY, Elly C, Altman Y, Fang D, Hunter T, Liu YC. Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J. Biol. Chem. 2000;275:35734–35737. doi: 10.1074/jbc.M007300200. [DOI] [PubMed] [Google Scholar]
- 33.Wilkin MB, Carbery AM, Fostier M, Aslam H, Mazaleyrat SL, Higgs J, Myat A, Evans DA, Cornell M, Baron M. Regulation of notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Curr. Biol. 2004;14:2237–2244. doi: 10.1016/j.cub.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 34.Sakata T, Sakaguchi H, Tsuda L, Higashitani A, Aigaki T, Matsuno K, Hayashi S. Drosophila Nedd4 regulates endocytosis of notch and suppresses its ligand-independent activation. Curr. Biol. 2004;14:2228–2236. doi: 10.1016/j.cub.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 35.Eisenberg LM, Markwald RR. Molecular regulation of atrioventricular valvulo-septal morphogenesis. Circ. Res. 1995;77:1–6. doi: 10.1161/01.res.77.1.1. [DOI] [PubMed] [Google Scholar]
- 36.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 37.Kratsios P, Catela C, Salimova E, Huth M, Berno V, Rosenthal N, Mourkioti F. Distinct roles for cell-autonomous Notch signaling in cardiomyocytes of the embryonic and adult heart. Circ. Res. 2010;106:559–572. doi: 10.1161/CIRCRESAHA.109.203034. [DOI] [PubMed] [Google Scholar]
- 38.Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev. Biol. 2005;287:134–145. doi: 10.1016/j.ydbio.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 39.Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev. Cell. 2001;1:435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- 40.Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, Kirby ML. Conotruncal myocardium arises from a secondary heart field. Development. 2001;128:3179–3188. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- 41.Mjaatvedt CH, Nakaoka T, Moreno-Rodriguez R, Norris RA, Kern MJ, Eisenberg CA, Turner D, Markwald RR. The outflow tract of the heart is recruited from a novel heart-forming field. Dev. Biol. 2001;238:97–109. doi: 10.1006/dbio.2001.0409. [DOI] [PubMed] [Google Scholar]
- 42.Zaffran S, Kelly RG, Meilhac SM, Buckingham ME, Brown NA. Right ventricular myocardium derives from the anterior heart field. Circ. Res. 2004;95:261–268. doi: 10.1161/01.RES.0000136815.73623.BE. [DOI] [PubMed] [Google Scholar]
- 43.Waldo KL, Hutson MR, Ward CC, Zdanowicz M, Stadt HA, Kumiski D, Abu-Issa R, Kirby ML. Secondary heart field contributes myocardium and smooth muscle to the arterial pole of the developing heart. Dev. Biol. 2005;281:78–90. doi: 10.1016/j.ydbio.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 44.Ge C, Stanley P. Effects of varying Notch1 signal strength on embryogenesis and vasculogenesis in compound mutant heterozygotes. BMC Dev. Biol. 2010;10:36. doi: 10.1186/1471-213X-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]