Abstract
Background
L1 retroelements may play a central role in morphogenesis through epigenetic mechanisms involving recruitment of chromatin modifying protein complexes. Retroelements are repressed in terminally differentiated cells, and highly active in embryonic, undifferentiated, and transformed cells. It is not clear if the modulation of differentiation by L1 is a “cause” or “effect”. The aim of this study was to determine if murine embryonic kidney cells of clonal origin (mK4 cells) harbor retrotransposition events upon ectopic expression of L1, and the impact of L1 on embryonic kidney cell differentiation. Given that L1 is reactivated by AHR ligands, we also sought to investigate the effects of benzo(a)pyrene (BaP) and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) on the genetic network of mK4 cells.
Methods
mK4 cells overexpressing human LIRP were assessed for changes in proliferation and expression of molecular markers of cellular differentiation.
Results
L1RP increased proliferation rates and markedly downregulated differentiation programming in mK4 cells. These genetic alterations were recapitulated by exogenous activation of L1 by AHR ligands.
Conclusion
L1 regulates nephrogenesis in vitro via both insertional and non-insertional mechanisms that disrupt mesenchymal to epithelial transition. Thus, a feedback loop involving L1, WT1 and AHR may play a role in regulation of kidney morphogenesis.
Keywords: 5’ untranslated region, Benzo(a)pyrene (BaP), histone acetylation, histone methylation, Long Interspersed Nuclear Element-1 (LINE-1 or L1), nephrogenesis, polycyclic aromatic hydrocarbons (PAHs), Wilms’ Tumor suppressor (WT1)
Introduction
L1RP, an active human retroelement, is 99.9% identical to the consensus L1 sequence (Kimberland et al., 1999), and contains two open reading frames, ORF1 and ORF2 respectively. L1 ORF2 encodes endonuclease (EN) and reverse transcriptase (RT) activities (Feng et al., 1996). The RT function is essential for self-propagation of L1 within the genome (Martin et al., 1998). Inhibition of L1-encoded RT significantly inhibits replication of transformed cell lines and induces differentiation (Sciamanna et al., 2005). Expression of RT-coding genes is generally active in early embryos, germ cells, and tumor tissues, but repressed in terminally differentiated cells (Spadafora, 2004). Thus, L1 may be involved in genetic reprogramming during early embryonic development. This interpretation is consistent with our recent demonstration that reactivation of L1 by genotoxic carcinogens occurs in mK4 mouse embryonic kidney cells (Teneng et al., 2007). The L1 reactivation process involves i) promoter-associated epigenetic changes at the DNA and histone levels, ii) dynamic regulation of protein complexes involving aryl hydrocarbon receptor (AHR), E2F and Rb transcription factors and iii) corepressor and coactivator recruitment including histone deacetylases (HDACs), histone methylases, and DNA methyl transferases (DNMTs), histone and DNA demethylases and histone acetylases (HATs). Further, AHR-mediated L1 reactivation was established in studies showing that AHR null mouse primary cells or human cervical cancer cells (HeLa) transfected with siRNA against AHR exhibit impaired L1 reactivation following BaP challenge. Rescue of the knockdown phenotype through forced overexpression of AHR protein in siRNA-treated cells restored BaP-dependent reexpression of L1, confirming that AHR is in fact involved in L1retroelement reactivation upon carcinogen challenge (Teneng et al., 2007). These events in turn lead to L1 expression and in some instances L1 reinsertion, events that may contribute to regulation of cellular proliferation and differentiation (for a review see (Montoya-Durango and Ramos, 2010)).
Studies of chromatin dynamics in oocytes and early stage embryo development are limited. A comparison of sperm and embryonic stem cells (ESC) showed similar patterns of DNA methylation, with only small differences found at regulatory regions (Albert and Peters, 2009; Edwards, 2006), suggesting that genome-wide demethylation of cytosine is characteristic of pluripotent and germ cell lines (Imamura et al., 2006). In addition to the modulation of DNA methylation, histone covalent modifications are orchestrated early after zygote formation. In the one to two cell embryo, protamines of the paternal genome are substituted by maternal histone H3.3, and later replaced by histones H3.1 and H3.2 (Torres-Padilla et al., 2006), with covalent mono-, di- and tri-methylation of histones H3K4 and H3K27 appearing thereafter (Santos et al., 2005; Torres-Padilla et al., 2006). Conversely, the epigenetic silencing mark H4K20 trimethyl is detected only at the 8-cell stage embryo and maintains significant disparity as compared to histone H3 epigenetic marks (Bhaumik et al., 2007; Biron et al., 2004). Most of these marks however, are already present in the maternal lineage where a nucleosomal status is found upon fertilization (Liu et al., 2004). Thus, the maternal and paternal epigenetic codes are markedly different, with the maternal epigenetic machinery providing chromatin modifiers, such as the polycomb protein (PcG) Ezh2 which modulates H3K27 di-/tri-methylation, and PRC1 complex that controls heterochromatic DNA regions to establish epigenetic chromatic codes during embryogenesis (Bhaumik et al., 2007). A key role for L1 during embryogenesis comes from studies of X-chromosome inactivation (XCI) in female embryonic cells (Chow et al., 2010). L1 elements serve as the building blocks for creation of broad domains of silent chromatin (Pauler et al., 2009) to which actively transcribed genes are recruited and embedded into heterochromatin (Chow et al., 2010), and participate in the synthesis of non-coding RNAs through bidirectional promoter activity (Soifer and Rossi, 2006). A similar mechanism may operate in autosomal DNA domains containing L1 insertions (Chow et al., 2010; Pauler et al., 2009). Human cell lines derived from females subject to random XCI exhibit two different heterochromatic domains characterized by enrichment in histone H3 trimethylated at lysine-9 (H3K9me3) and histone H4 trimethylated at lysine-20 (H4K20me3), respectively (Chadwick and Willard, 2004). Interestingly, we previously identified these marks in the mouse L1 promoter (Montoya-Durango et al., 2009), and found that their expression is dependent on the presence of functional retinoblastoma proteins. Thus, L1 likely plays a role in morphogenesis through epigenetic mechanisms involving L1 positioning within specific domains, generation of non coding RNAs, and recruitment of chromatin modifying protein complexes specialized on nucleosomal histone(s) trimethylation and heterochromatin formation.
Morphogenesis is regulated by genetic and epigenetic networks that control chromatin dynamics and gene expression respectively (Li, 2002), and by structural variations such as genetic imprinting, XCI, and recombination events in cells of the lymphoid lineage. These events are controlled through orchestrated waves of signaling cascades that when compromised disrupt phenotypic expression, or become lethal (De Robertis et al., 2000; Gurdon and Bourillot, 2001). The study of differentiation programming during early development has been facilitated by the mK4 cell model, an established model of mesenchymal-to-epithelial transition during nephrogenesis (Kispert et al., 1998; Valerius et al., 2002). During the nephrogenic transition, mesenchyme differentiates into epithelium and gives rise to the glomerulus and proximal and distal tubules (Almeida and Mandarim-de-Lacerda, 2002; Pohl et al., 2000). As in other organ systems, renal morphogenesis is characterized by changes in spatiotemporal expression of differentiation genes (Patterson and Dressler, 1994; Pohl et al., 2000). Gene array studies using mK4 cells have shown that these cells express epithelial conversion-related genes, including Pax-2, Pax-8, Wnt-4, cadherin-6, collagen IV, and LFB3 (Valerius et al., 2002). As such, mK4 cells represent a “freeze frame” molecular picture of induced metanephric mesenchyme during kidney development.
Retroelements are transcriptionally repressed in terminally differentiated cells, and highly active in embryonic, undifferentiated, and transformed cells (Packer et al., 1993; Poznanski and Calarco, 1991). Additionally RT activity is high in embryonic and transformed cells and extremely low, or undetectable, in terminally differentiated cells (Deragon et al., 1990; Medstrand and Blomberg, 1993). This inverse correlation between L1 expression and differentiation suggests that expression of retroelements influences differentiation programming. It is not clear if the loss of differentiation associated to L1 expression is a “cause” or an “effect”, nor is it clear whether RT expression and activity are required for modulation of differentiation programming. Thus the aim of this study was to determine if embryonic kidney cells harbor L1 retrotransposition events, and the impact of L1 on embryonic kidney cell differentiation. Given that L1 is reactivated by environmental chemical carcinogens, we also sought to investigate the influence of the environmental hydrocarbons, benzo(a)pyrene (BaP) and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), on the genetic network of mK4 cells. To answer these questions, we transfected mK4 cells with three plasmids; L1RP, an RT mutant counterpart with a single amino acid change at position 105 in the RT active site, or empty plasmid. Expression of differentiation markers was analyzed via real time PCR or immunostaining. Evidence is presented that L1 expression, as well as BaP or TCDD treatments, alter morphogenetic networks and compromise differentiation programming in mK4 cells. We conclude that early kidney morphogenesis in vitro is regulated via L1 epigenetic mechanisms and this regulation is subject to interference by environmental hydrocarbons.
EXPERIMENTAL PROCEDURES
Cell cultures
Murine kidney cells of clonal origin isolated from a mouse carrying the Hoxa 11-SV40 Tag (mK4 cells) (Valerius et al., 2002), were grown in M199 medium, supplemented with 10% FBS and 1% penicillin-streptomycin. Cells were maintained at 37°C and 5% CO2.
Retrotransposition assay
A plasmid based retrotransposition assay that uses a reporter cassette conferring antibiotic resistance to cells harboring retrotransposition events was used (Moran et al., 1996). This plasmid contained a highly active retrotransposon called L1RP, for the disease X-linked retinitis pigmentosa (XLRP) during which an L1 insertion disrupts the RP gene (Schwahn et al., 1998). The plasmid harbors the L1RP inserted into a reporter cassette containing an antisense copy of a selectable marker (neo), a heterologous promoter (P′), and a polyadenylation signal (A′). The neo gene was disrupted by a globin intron in the opposite transcriptional orientation. L1RP was tagged with a neomycin (neo) reporter cassette containing an antisense copy of the antibiotic resistance gene containing a γ-globin intron in the sense orientation. Transcription from either the L1 5’-UTR or a heterologous promoter (P1), splicing of the intron, reverse transcription, and insertion of the cDNA into chromatin was responsible for sustained expression of the neomycin gene which rendered cells resistant to geneticin (G418). Transcripts from the L1 promoter cannot induce expression of the reporter cassette since the intron is in antisense orientation, and lies downstream of the 3’UTR. Individual G418 resistant cells continued growing and formed bonafide clones on culture plates. Stable L1RP expressing cells were trypsinized and counted, and seeded (1 × 106 cells per 10-cm plate) and grown in the presence of G418 (400 μg/mL) until resistant clones were visible. Individual foci were isolated and propagated for DNA isolation and PCR analyses or fixed with formaldehyde and stained with trypan blue (Sigma, St. Louis, MO) and number of foci counted.
Quantitative real time-PCR
Total RNA was extracted using trizol reagent, quantified, DNAse treated and 200-500 ng used for cDNA synthesis. For real time-PCR analyses, the double strand DNA binding dye method was used. Following reverse transcription, real time amplifications were performed using SYBR Green (BIORAD, Redmond, WA). For each reaction, 25 μL of 2x SYBR green was mixed with 10 μM final concentration of forward and reverse primers. One μL of cDNA was added and the final volume aliquoted up to 50 μL with DEPC water. The cycling conditions consisted of an initial denaturation step at 95°C for 3 minutes, and 50 cycles at 95°C for 30 seconds, 55°C for 30 seconds and 72°C for 45 seconds. All experiments were completed in triplicate. (Primers for Mouse L1ORF 1: Fw: 5′CTGGAGAGCAGAAGACCGAAAGG 3′; Rv: 5′ACACACCGAAAATCTAGAC3′; Mouse GAPDH Fw: 5′GCCTTCCGTGTTCCTA CC3′; Rv 5′ AGAGTGGGAGTTGCTGTTG3′; mouse 18S Fw: CGTCTGCCCTATCAACTTTC3′; Rv 5′GCCTGCTGCCTTCCTTGG3′, Mouse E cadherin Fw: 5′CGACCCTGCCTCTGAATCC3′; Rv: 5′CTTTGTTTCTTTGTCCTTCGTTGG3′, mouse Igf1r Fw: 5′TCCCTCAGGCTTCATCC3′; Rv: 5′GAGCAGAAGTCACCGAATC3′, mouse Igf2r Fw: 5′ AGTATGTGAACGGCTCTG3′; Rv: 5′TCTGTGATTGTCTGGATAGG3′, mouse sfrp1 Fw: 5′GCAGTTCTTCGGCTTCTAC3′; Rv: 5′ATGGAGGACACACGGTTG3′, mouse Wnt4 Fw: 5′GTAGCCTTCTCACAGTCCTTTG3′ Rv: 5′GGTACAGCACGCCAGCAC3′).
Transient and stable transfections
Cells were grown to 80% confluence and transfected at a ratio of 1:2 Lipofectamine 2000 (μL) to DNA (μg) without antibiotics. Six hr later, transfection medium was replaced with fresh culture medium, and 48 hr later cells were harvested, RNA extracted and analyzed or else cells were selected for stable integration of plasmids in the presence of hygromycin.
Immunofluorescence
mK4 cells were seeded on glass slides placed in 6-well tissue culture plates. Cells were transiently transfected with L1RP expressing plasmid or empty vector as control. Forty eight hr later cells were fixed with 5% formaldehyde and quenched using 100 mM glycine. Cell permeabilization was accomplished with 0.4% Triton X100. Following incubation with primary antibody targeting L1 ORF1 and FITC coupled secondary antibody, cells were washed and incubated with DAPI for nuclear staining. Slides were washed with 100% ethanol, stained with DAPI and visualized on a fluorescent microscope.
Cell growth assay
mK4 stable transfectants harboring wild type human L1RP, RT mutant or empty vector were counted and equal amounts seeded on 48 well plates in triplicate. Cells were trypsinized and counted daily for up to 6 days.
Soft agar colony formation assay
L1RP transfected cells were seeded on soft agar. Cells were either pretreated with 2-O-tetradecanoylphorbol 13-acetate (TPA), a tumor promoter (Kim et al., 2008) or DMSO. Fresh medium was added every 3 days. Plates were stained two weeks later and visualized under a microscope for colony formation.
Chemical treatments of mK4 cells
Cells were grown in 10-cm plates and allowed to reach 80% confluence. Fresh medium containing DMSO vehicle or 3 μM BaP was added to cells for 9, 15, or 51 hr followed by RNA extraction. Cells were treated for a period of 48 hr with toluene vehicle or 1 nM TCDD. Cells were harvested and processed for quantitative PCR as described above.
Statistical Analysis
ANOVA followed by Tukey’s post hoc test were used to evaluate the statistical significance of the findings.
RESULTS
mK4 Cells harbor stable L1RP retrotransposition events that modulate cellular proliferation
Cells transfected with empty (pGESH), L1RP, or RT 105 mutant plasmids, were grown in the presence of 200 μg/mL hygromycin. The presence of the plasmid encoded hygromycin resistance independent of L1 retrotransposition events. Since transcripts originating from P′ cannot be spliced in the opposite orientation, the neo gene product cannot be synthesized and cells remain sensitive to G418 (Figure 1A). Following hygromycin selection, cells expressing the plasmid were counted and 1×106 cells plated and selected for retrotransposition events with G418 until resistant clones were visible. G418-resistant cells were fixed with formaldehyde and stained with trypan blue (data not shown), or individual clones expanded for genomic DNA analyses. Genomic DNA isolated from six independently expanded clones showed the presence of reintegrated L1RP. PCR analyses confirmed loss of the globin intron, as confirmed by appearance of a 1 kb product and the presence of a spliced and integrated L1 transcript into genomic DNA (Figure 1B). These findings established that embryonic mK4 cells undergo complete cycles of L1 retrotransposition and that expression of L1RP is stable in the mK4 cell genome.
Figure 1. L1RP Retrotransposition in primary cells.
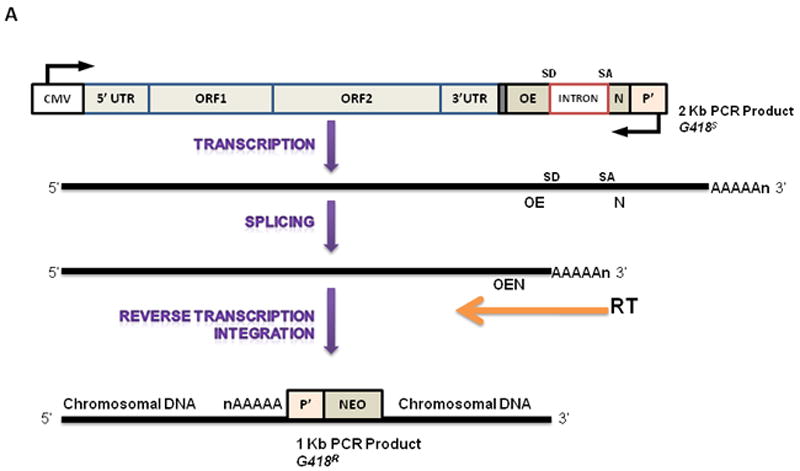
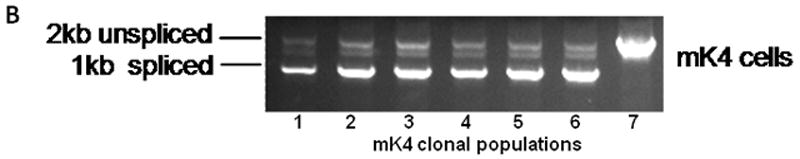
(a) Illustration of synthetic L1RP construct and the mechanism of reinsertion-dependent neomycin resistance in mK4 cells. (b) Six clones from each experiment were isolated and expanded on G418-containing medium. Genomic DNA was extracted and used as the template for PCR. Lanes 1-6, DNA was isolated from G418-resistant clones; lane 7 DNA from original unspliced L1RP plasmid.
Next, the effects of stable L1RP expression on cellular proliferation were examined. Expression of L1 was confirmed by immunostaining and real time PCR (not shown). mK4 cells expressing L1RP, RT mutant or empty plasmid were counted and seeded at equal densities (2.0 ×103 cells/cm2) in triplicate. Expression of L1RP increased mK4 replication rates significantly compared to mutant, or empty plasmid-expressing cells as indicated by differences in cell density (data not shown) and cell counts (Figure 2). We next probed into the possibility that L1 induces features of the transformed phenotype. Transfected cells were pretreated with 2-O-tetradecanoylphorbol 13-acetate (TPA) to act as a tumor promoter (Kim et al., 2008), or left untreated and grown in culture for three weeks. Transformed HeLa cells were used as a control for positive growth. Colony formation was analyzed following fixation of cells with formaldehyde and staining with trypan blue. No colonies were formed by L1RP-expressing mK4 cells or respective controls, but colonies were formed in HeLa cells, indicating the L1 expression does not drive soft agar colony formation in mK4 cells (not shown).
Figure 2. Stable expression of L1RP in mK4 cells accelerates cellular proliferation.
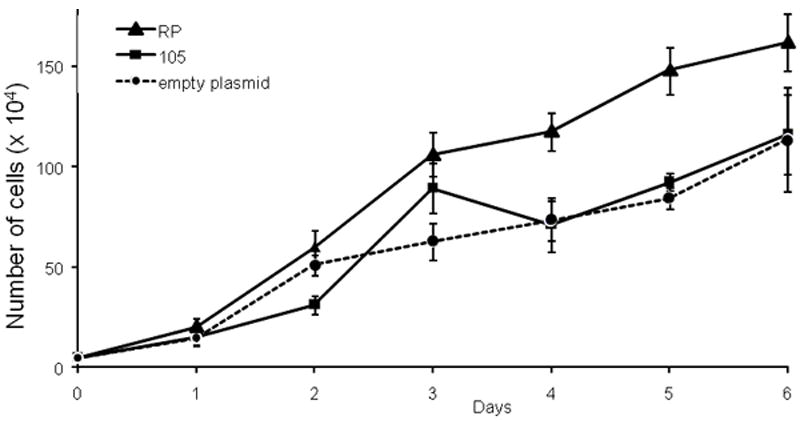
Graphical representation of results from cell counts performed for 6 consecutive days after initial seeding. mK4 cells were transfected with L1RP, L1 reverse transcriptase (RT) mutant (p105), or empty plasmid (pGESH) and selected for hygromycin resistance. After selection, cells were counted and seeded at a density of 2.0 × 103 cells/cm2 in 48 well-plates. Assays were performed in triplicate and data are representative of a typical experimental result.
Transient expression of L1 modulates MET in mK4 cells
The impact of L1RP on mK4 cellular differentiation was examined in mK4 cells transfected with L1RP, mutant or empty plasmid for 48 hr. Expression of L1 was confirmed by immunostaining with antisera against L1 ORF1 protein (Figure 3A), showing that ORF1 signal was mostly cytoplasmic in L1 RP expressing cells and undetectable in cells harboring the empty control plasmid (Figure 3A). Next, RNA was used as template for cDNA synthesis and real time PCR analyses of molecular markers of renal cell differentiation (Igf1r, Igf-2r, Wnt4, sprf-1, E-cadherin and Wt1). L1RP reduced the mRNA levels of several differentiation markers, indicating that L1 expression affects MET transition in mK4 cells (Figure 3B). Immunostaining analyses indicated a marked decrease in WT1 and IGF1R protein levels (Figure 3C). Together, these data indicate that L1 ORF1 accumulates in the cytoplasm and is associated with reduction at the mRNA and protein levels of morphogenetic markers of differentiation.
Figure 3. Ectopic expression of L1RP in mK4 cells inhibits mesenchymal-epithelial transition in mK4 cells.
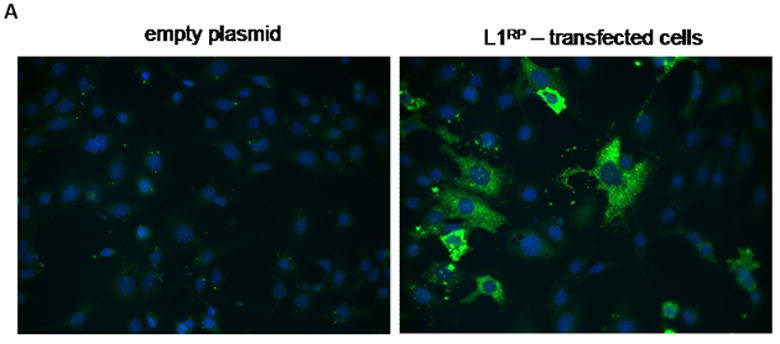
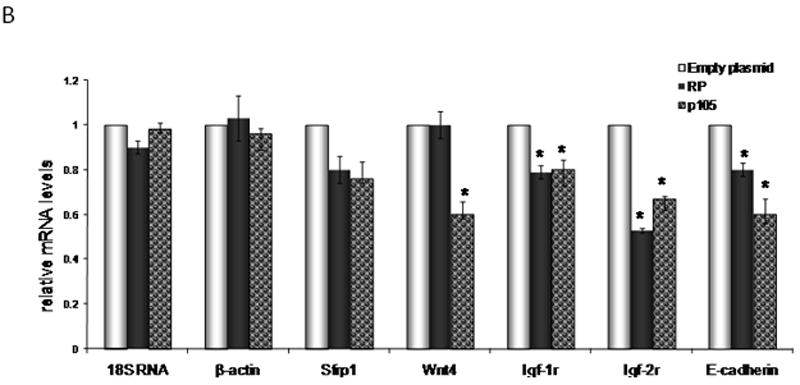
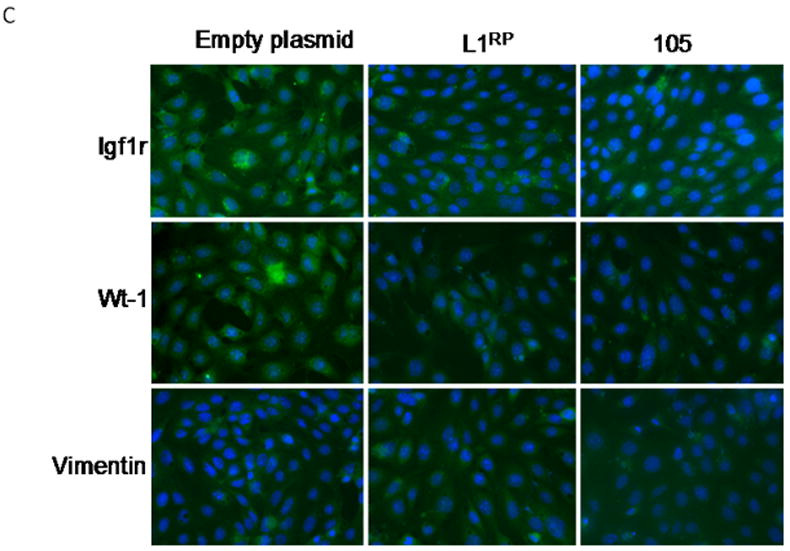
mK4 cells were transiently transfected with L1RP or empty plasmid (pGESH). Forty eight hr after transfection, cells were processed for immunostaining or RNA extraction. (a) Immunofluorescence staining of L1RP using L1 ORF1 antibodies. Cells were transfected on glass slides and immunostained for L1 ORF1. Real time PCR analyses confirmed high expression of ectopic L1RP-specific mRNA in transiently transfected mK4 cells (not shown). (b) mK4 cells were transiently transfected with L1RP, RT mutant (p105) or empty plasmid (pGESH). Forty eight hr later RNA was extracted and processed for real time PCR analyses of mK4 differentiation markers Wnt4, Igfr1, Igf2r and Wt-1. β-actin and 18 S RNA were used as controls. (c) mK4 cells transfected on glass slides as in (a) were immunostained with antibodies targeting IGF1R, WT1 or vimentin (control) protein respectively. Expression of L1RP or RT mutant reduces IGF1R and WT1 protein levels in mK4 cells. Asterisk indicates statistically significant differences in gene expression as compared to empty plasmid (p<0.05).
Early L1 reactivation by AHR ligands alters MET genetic networks in mK4 cells
L1 is rapidly reactivated upon activation of AHR signaling by hydrocarbon ligands of AHR such as BaP and TCDD (Teneng et al., 2007). Intact AHR signaling was confirmed in studies showing a 440-fold increase in Cyp1a1 mRNA levels in BaP-treated mK4 cells compared to vehicle controls (Figure 4A). Time-dependent changes in Sfrp1, Igf1r, Igf2r, Wnt4, and E-cadherin were observed in BaP-treated mK4 cells. While no changes in marker expression were generally seen in BaP-treated cells at 9 or 15 hr (except for increased Sfrp1 at 15 hr), cells treated for 51 hr showed downregulation of mRNA levels for Sfrp1, Igfr1, and Wnt4 (Figure 4B). Next, we examined the expression of WT1 isoforms in mK4 cells challenged with BaP. WT1 mRNA species exhibit combinatorial patterns that include variants resulting from exon 5 splicing (+17 and −17 amino acids), or +KTS and −KTS alternative splice variants that originate from splicing of exon 9. We exposed cells to BaP and collected samples at different time points (Figure 5). The +KTS and −KTS isoforms were upregulated by BaP treatment compared to vehicle controls (Figures 5A and 5B). Transient upregulation of the +17aa variant was seen at 1 and 15 hr, with a return to control levels by 51 hr (Figure 5C). No changes in the expression of the -17 isoform were noted (Figure 5D). Thus, early reactivation of L1 by BaP is associated with marked alterations in renal cell differentiation networks in mK4 cells. To determine if changes in the relative abundance of WT1 splice variants impacted downstream WT1 targets, the expression of Synd1, Pax2, Egfr, and RaRα was examined (Figure 6). The results showed that BaP leads to oscillating patterns in WT1 targets characterized by early downregulation, followed by upregulation of Synd1 and RaRα, and a late robust downregulation of WT1 target gene expression.
Figure 4. BaP downregulates morphogenetic markers of differentiation in mK4 cells.
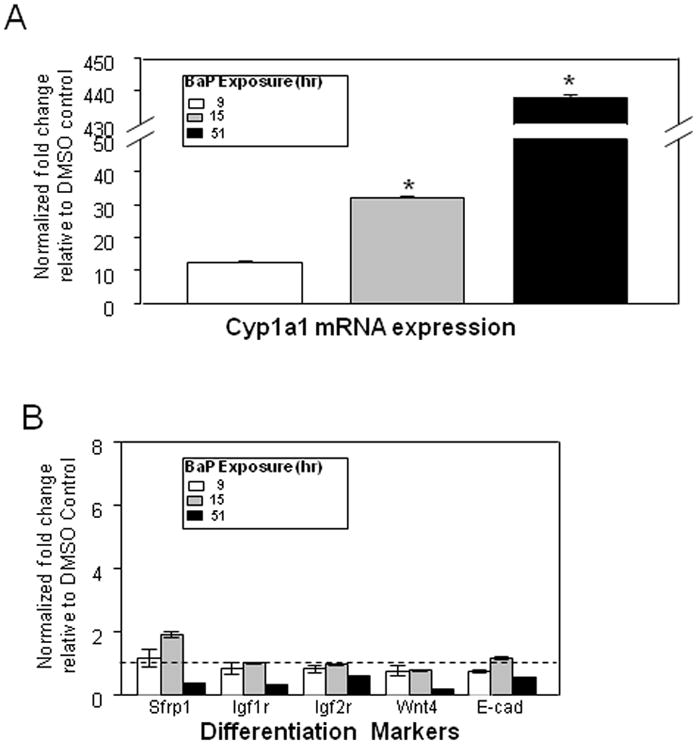
mK4 cells were grown in 10-cm plates and allowed to reach 80% confluence. Cells were treated with DMSO vehicle or 3 μM BaP for 48 hr. RNA was harvested 9, 15, or 51 hr after BaP challenge and cDNA used to analyze changes in expression of mK4-associated developmental markers. (a) qPCR results for time-dependent variations in Cyp1a1 mRNA expression after BaP challenge. (b) qPCR results for time-dependent variation of mK4 differentiation markers Sfrp1, Igf1r, Igf2r, Wnt4, and E-cadherin. Results were normalized to DMSO vehicle control. Bars represent the standard error of the mean for fold changes. All experiments were performed independently at least three times. Asterisk indicates statistically significant differences in gene expression as compared to empty plasmid (p<0.05).
Figure 5. Environmental hydrocarbons alter the expression of WT1 variants in mK4 cells.
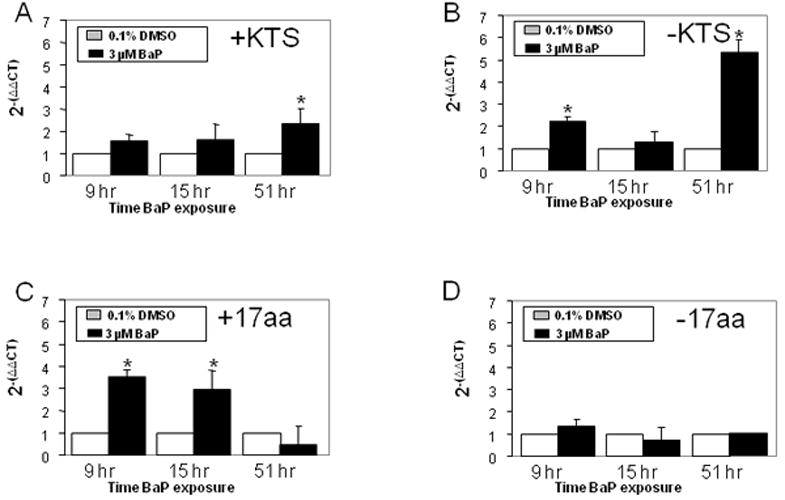
mK4 cells were grown in 10-cm plates and allowed to reach 80% confluence. Next, cells were treated with DMSO vehicle or 3 μM BaP for 9, 15, or 51 hr. RNA was harvested at each time point and cDNA used to analyze changes in expression of WT1 variants. Normalized changes in gene expression after qPCR are shown for (a) +KTS; (b) −KTS; (c) +17 aa; and (d) − 17aa. Bars represent the standard error of the mean. Results for triplicate assays performed independently at least three times. Asterisk indicates statistically significant differences in gene expression as compared to empty plasmid (p<0.05).
Figure 6. BaP alters the expression of WT1 targets in mK4 cells.
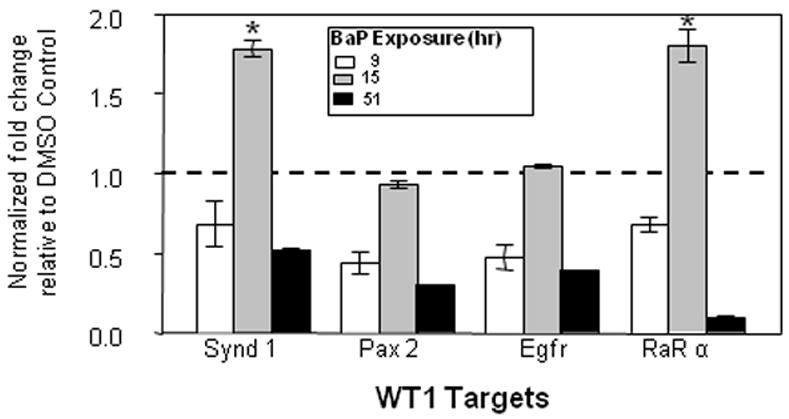
mK4 cells were grown in 10-cm plates and allowed to reach 80% confluence. Next, cells were treated with DMSO vehicle or 3 μM BaP for 9, 15, or 51 hr. RNA was harvested at each time point and cDNA used to analyze changes in expression of the WT1 targets Synd1, Pax2, Egfr, and RARα. Normalized changes in gene expression after qPCR are shown. Bars represent the standard error of the mean. Results for triplicate assays performed independently at least three times. Asterisk indicates statistically significant differences in gene expression as compared to empty plasmid (p<0.05).
Lastly, the cellular responses to 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD, dioxin) were examined in this cellular model to determine if modulation of differentiation networks by BaP was dependent upon its mutagenic actions at the DNA level. As seen with BaP, TCDD decreased Sfrp1, Igfr1, Igfr2 and Wnt4 expression levels (Figure 7A), and induced overexpression of +KTS, -KTS, and +17 variants, while the -17 aa variant decreased by half compared to vehicle control (Figure 7B). In keeping with these relationships, TCDD decreased all downstream WT1 targets examined (Figure 7C). These findings indicate that the signaling pathways and cellular responses to AHR ligands are reproducible and likely independent of DNA damage.
Figure 7. TCDD dysregulates morphogenetic and WT1 networks in mK4 cells.
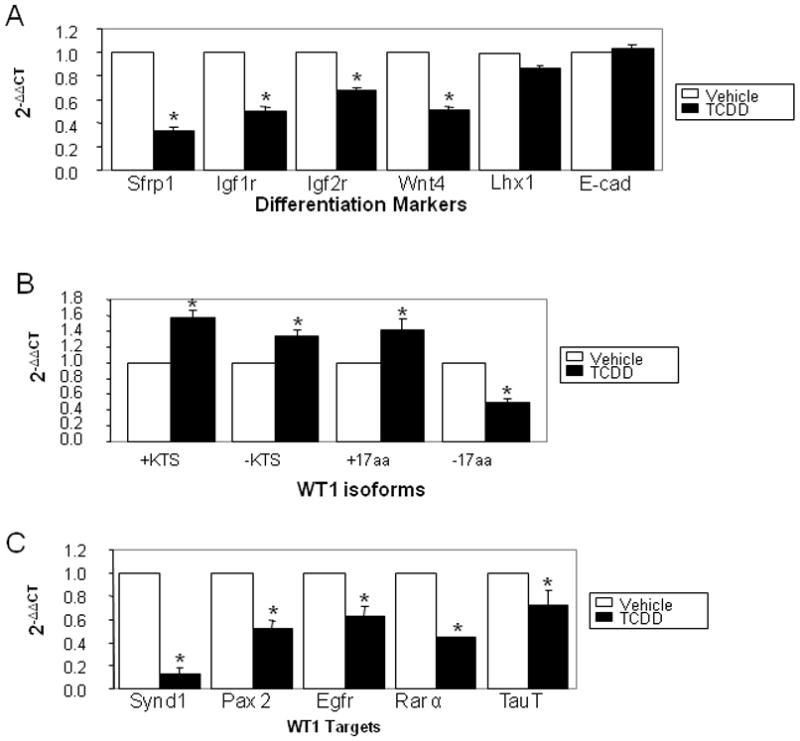
mK4 cells were treated with toluene vehicle or 1 nM TCDD. RNA was harvested 48 hr after TCDD challenge and cDNA used to analyze changes in expression of mK4-associated developmental markers, WT1 variants, or WT1 target genes respectively. (a) qPCR results show variations in expression of mK4 differentiation markers Synd1, Pax2, Egfr, and RARα following TCDD challenge. (b) qPCR results for TCDD-induced changes in WT1 variants +KTS, -KTS, +17aa, -17aa. (c) qPCR results for variation in WT1 network targets Synd1, Pax2, Egfr, RARα, and TauT. Bars represent the standard error of the mean. Results for triplicate assays performed independently at least three times. Asterisk indicates statistically significant differences in gene expression as compared to empty plasmid (p<0.05).
DISCUSSION
The sheer magnitude of L1 in the mammalian genome has long fueled the hypothesis that over the course of evolution, L1 insertions structurally modify genes to regulate gene expression via mutational and/or epigenetic mechanisms. The actions of L1 are also believed to involve non-insertional mechanisms mediated via transcription of coding and non-coding L1 RNAs, or overexpression of RT within the cell. We show here for the first time that L1 regulates embryonic kidney mK4 cell proliferation and differentiation via insertional and non-insertional mechanisms. mK4 cells were found to harbor L1 retrotransposition events, and wild type L1RP increased proliferation rates in mK4 cells compared to RT mutant, or empty plasmid-transfected cells, and mediated a molecular switch toward lesser differentiated phenotypes. Thus, retrotransposition may be an ongoing process throughout the developmental period of non-germ cells which functions to regulate genome plasticity and to define cellular fates. Interestingly, the functionality of L1 in mK4 cells also involved insertion-independent mechanisms, as changes in cellular differentiation were observed in cells expressing mutant RT constructs deficient in retrotransposition activity. The extent to which endogenous activities influence non-insertional patterns of L1 function is not yet clear, thus definitive answers about the role of insertion independent mechanisms await further analyses.
The regulatory function of L1 on development of kidney structures likely involves epigenetic remodeling of chromatin. This interpretation is consistent with our previous demonstration that corepressors are recruited to L1 promoter in the presence of the E2F/Rb complex (Montoya-Durango et al., 2009), and reports that L1 influences neuronal development and differentiation via genetic silencing mechanisms (Muotri et al., 2005). Thus, early activation of L1 might be required for the maintenance of undifferentiated phenotypes during early embryogenesis, followed by induction of differentiation through epigenetic L1 silencing. The involvement of AHR (aryl hydrocarbon receptor) in the regulation of L1 epigenetics also needs to be considered. AHR is a key regulator of WT1 signaling in the developing kidney (Falahatpisheh and Ramos, 2003), and loss of AHR protein in vivo correlates with downregulation of renal cell differentiation markers and WT1 expression in metanephric kidneys (Ramos et al., 2006). Importantly, we have shown that L1 is regulated at the transcriptional level by AHR protein, and that both AHR null cells and knock-down of the AhR by siRNA fail to respond to L1 induction by hydrocarbons (Teneng et al., 2007). Together, these data support a scenario in which activation of AHR by environmental hydrocarbons alters L1 epigenetic silencing via removal of pRb from the L1 promoter, followed by disassembly of heterochromatin-associated protein complexes in a genome-wide scale from DNA regions where L1 is present.
Of note was the finding that L1 modulated expression WT1, a master regulator of kidney and urogenital developmental programming (Falahatpisheh and Ramos, 2003). WT1 functions as a transcriptional regulator of many genes involved in renal cell differentiation, including Bmp7, Pax2, Egfr and Sall1 (Hartwig et al., 2010). In addition, WT1 binds RNA to control genetic networks of differentiation via non-transcriptional mechanisms (Bor et al., 2006; Gessler et al., 1992). Thus, modulation of genetic and epigenetic programs in embryonic kidney cells via ectopic L1 may be directly linked to the downregulation of WT1 protein, and the consequent induction of lesser differentiated phenotypes. This interpretation is consistent with the ability of hydrocarbon ligands of AHR to coordinately modulate L1 and WT1 in embryonic kidney cells, implicating L1 and WT1 in a cellular feedback loop in which L1 reactivation mediates marked downregulation of morphogenetic markers of renal cell development. Interestingly, L1-mediated shifts toward undifferentiated phenotypes did not compromise anchorage-dependent growth, suggesting that L1 does not induce acquisition of neoplastic phenotypes.
A previous study tracking embryonic L1 events in transgenic mice using an L1-EGFP construct whose expression requires a cycle of transcription, reverse transcription, and integration into a transcriptionally-permissive genomic region has shown that retrotransposition occurred exclusively in mouse testes (Prak et al., 2003). No somatic retrotransposition was detected, suggesting low frequency retrotransposition, consistent with low levels of L1 RNA in organs other than the testes. Given genetic similarities and linkages between gonadal and urogenital development, it is likely that L1 functions as a master regulator of early nephrogenesis via regulatory functions at the level of chromatin through heterochromatin formation and possibly RNA interference.
We have reported previously that hydrocarbon challenge leads to proteasomal degradation of AHR protein in vivo, and delayed renal cell differentiation associated with altered WT1 splice variants (Falahatpisheh and Ramos, 2003; Ramos et al., 2006; Teneng et al., 2011). Thus, it is likely that during development, hydrocarbons activating L1 retroelements through dysregulation of AHR and WT1 networks elicit phenotypic and genotypic changes that negatively impact embryogenesis and predispose to later-onset adult disease. This hypothesis is consistent with the notion that genomic changes during critical developmental periods defines susceptibility to disease and set the stage for adult-onset disease (Barker, 1995; Beaulac-Baillargeon and Desrosiers, 1987). The impact on genetic regulation in the embryo is likely not restricted to the developing kidney and may in fact contribute to morphogenetic deficits in other tissues where AHR and/or WT1 play important roles (for review see (Ramos and Nanez, 2009) and references herein). For instance, AHR -/- mice present defects in vascular structures, extramedular hematopoiesis, reduced liver weight, cardiac hypertrophy, and hypertension (Fernandez-Salguero et al., 1995; Lahvis et al., 2000; Lahvis et al., 2005; Lund et al., 2003; Lund et al., 2006; Schmidt et al., 1996). In humans, fetal exposure to environmental hydrocarbons as seen in pregnant women who smoke may disrupt morphogenesis pathways regulated by L1 retroelements and predispose to diverse pathologies during adulthood. This suggestion is consistent with the occurrence of low birth weights and tissue hypoplasia seen in newborns exposed to tobacco smoke constituents during early developmental periods (Barker, 1995; Nelson et al., 1999a; Nelson et al., 1999b).
At the DNA and chromatin levels, L1 retrotransposition events may lead to genetic rearrangements and mutations, as well as epigenetic silencing of otherwise transcriptionally active chromatin regions by forming nucleation sites for DNA and nucleosomal hypermethylation and heterochromatin formation, as is the case for the X-chromosome (Chow et al., 2010). Given that the L1 promoter is bidirectional (Yang and Kazazian, 2006), reactivation under stress conditions may also impact the expression of neighboring eukaryotic genes that otherwise would remain silenced, thus leading to dedifferentiation of tissues and disease. New genomic tools allowing for deep sequencing of the human genome will shed light into whether L1 orchestrates expression of chromatin regions where gene clusters associated with development become L1 targets.
Acknowledgments
FUNDING: This work was supported in part by National Institutes of Health (NIH, USA) [Grant 5R01 ES004849 and ES014443 to K.S.R.]
Footnotes
COMPETING INTEREST: The authors declare no potential competing interests.
References
- Albert M, Peters AH. Genetic and epigenetic control of early mouse development. Curr Opin Genet Dev. 2009;19(2):113–121. doi: 10.1016/j.gde.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Almeida JR, Mandarim-de-Lacerda CA. Quantitative study of the comma-shaped body, S-shaped body and vascularized glomerulus in the second and third human gestational trimesters. Early Human Development. 2002;69(1-2):1. doi: 10.1016/s0378-3782(02)00021-x. [DOI] [PubMed] [Google Scholar]
- Barker DJ. The fetal and infant origins of disease. Eur J Clin Invest. 1995;25(7):457–463. doi: 10.1111/j.1365-2362.1995.tb01730.x. [DOI] [PubMed] [Google Scholar]
- Beaulac-Baillargeon L, Desrosiers C. Caffeine-cigarette interaction on fetal growth. Am J Obstet Gynecol. 1987;157(5):1236–1240. doi: 10.1016/s0002-9378(87)80301-0. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14(11):1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- Biron VL, McManus KJ, Hu N, Hendzel MJ, Underhill DA. Distinct dynamics and distribution of histone methyl-lysine derivatives in mouse development. Dev Biol. 2004;276(2):337–351. doi: 10.1016/j.ydbio.2004.08.038. [DOI] [PubMed] [Google Scholar]
- Bor YC, Swartz J, Morrison A, Rekosh D, Ladomery M, Hammarskjold ML. The Wilms’ tumor 1 (WT1) gene (+KTS isoform) functions with a CTE to enhance translation from an unspliced RNA with a retained intron. Genes Dev. 2006;20(12):1597–1608. doi: 10.1101/gad.1402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc Natl Acad Sci U S A. 2004;101(50):17450–17455. doi: 10.1073/pnas.0408021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E, Greally JM, Voinnet O, Heard E. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141(6):956–969. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- De Robertis EM, Larrain J, Oelgeschlager M, Wessely O. The establishment of Spemann’s organizer and patterning of the vertebrate embryo. Nat Rev Genet. 2000;1(3):171–181. doi: 10.1038/35042039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deragon JM, Sinnett D, Labuda D. Reverse transcriptase activity from human embryonal carcinoma cells NTera2D1. EMBO J. 1990;9(10):3363–3368. doi: 10.1002/j.1460-2075.1990.tb07537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RG. Genetics, epigenetics and gene silencing in differentiating mammalian embryos. Reprod Biomed Online. 2006;13(5):732–753. doi: 10.1016/s1472-6483(10)60665-7. [DOI] [PubMed] [Google Scholar]
- Falahatpisheh MH, Ramos KS. Ligand-activated Ahr signaling leads to disruption of nephrogenesis and altered Wilms’ tumor suppressor mRNA splicing. Oncogene. 2003;22(14):2160–2171. doi: 10.1038/sj.onc.1206238. [DOI] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87(5):905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Fernandez-Salguero P, Pineau T, Hilbert DM, McPhail T, Lee SS, Kimura S, Nebert DW, Rudikoff S, Ward JM, Gonzalez FJ. Immune system impairment and hepatic fibrosis in mice lacking the dioxin-binding Ah receptor. Science. 1995;268(5211):722–726. doi: 10.1126/science.7732381. [DOI] [PubMed] [Google Scholar]
- Gessler M, Konig A, Bruns GA. The genomic organization and expression of the WT1 gene. Genomics. 1992;12(4):807–813. doi: 10.1016/0888-7543(92)90313-h. [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Bourillot PY. Morphogen gradient interpretation. Nature. 2001;413(6858):797–803. doi: 10.1038/35101500. [DOI] [PubMed] [Google Scholar]
- Hartwig S, Ho J, Pandey P, Macisaac K, Taglienti M, Xiang M, Alterovitz G, Ramoni M, Fraenkel E, Kreidberg JA. Genomic characterization of Wilms’ tumor suppressor 1 targets in nephron progenitor cells during kidney development. Development. 2010;137(7):1189–1203. doi: 10.1242/dev.045732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura M, Miura K, Iwabuchi K, Ichisaka T, Nakagawa M, Lee J, Kanatsu-Shinohara M, Shinohara T, Yamanaka S. Transcriptional repression and DNA hypermethylation of a small set of ES cell marker genes in male germline stem cells. BMC Dev Biol. 2006;6:34. doi: 10.1186/1471-213X-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HG, Lee KW, Cho YY, Kang NJ, Oh SM, Bode AM, Dong Z. Mitogen- and stress-activated kinase 1-mediated histone H3 phosphorylation is crucial for cell transformation. Cancer Res. 2008;68(7):2538–2547. doi: 10.1158/0008-5472.CAN-07-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberland ML, Divoky V, Prchal J, Schwahn U, Berger W, Kazazian HH., Jr Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Human molecular genetics. 1999;8(8):1557–1560. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- Kispert A, Vainio S, McMahon A. Wnt-4 is a mesenchymal signal for epithelial transformation of metanephric mesenchyme in the developing kidney. Development. 1998;125(21):4225–4234. doi: 10.1242/dev.125.21.4225. [DOI] [PubMed] [Google Scholar]
- Lahvis GP, Lindell SL, Thomas RS, McCuskey RS, Murphy C, Glover E, Bentz M, Southard J, Bradfield CA. Portosystemic shunting and persistent fetal vascular structures in aryl hydrocarbon receptor-deficient mice. Proc Natl Acad Sci U S A. 2000;97(19):10442–10447. doi: 10.1073/pnas.190256997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahvis GP, Pyzalski RW, Glover E, Pitot HC, McElwee MK, Bradfield CA. The aryl hydrocarbon receptor is required for developmental closure of the ductus venosus in the neonatal mouse. Mol Pharmacol. 2005;67(3):714–720. doi: 10.1124/mol.104.008888. [DOI] [PubMed] [Google Scholar]
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3(9):662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- Liu H, Kim JM, Aoki F. Regulation of histone H3 lysine 9 methylation in oocytes and early pre-implantation embryos. Development. 2004;131(10):2269–2280. doi: 10.1242/dev.01116. [DOI] [PubMed] [Google Scholar]
- Lund AK, Goens MB, Kanagy NL, Walker MK. Cardiac hypertrophy in aryl hydrocarbon receptor null mice is correlated with elevated angiotensin II, endothelin-1, and mean arterial blood pressure. Toxicol Appl Pharmacol. 2003;193(2):177–187. doi: 10.1016/j.taap.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Lund AK, Goens MB, Nunez BA, Walker MK. Characterizing the role of endothelin-1 in the progression of cardiac hypertrophy in aryl hydrocarbon receptor (AhR) null mice. Toxicol Appl Pharmacol. 2006;212(2):127–135. doi: 10.1016/j.taap.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Martin SL, Li J, Epperson LE, Lieberman B. Functional reverse transcriptases encoded by A-type mouse LINE-1: defining the minimal domain by deletion analysis. Gene. 1998;215(1):69–75. doi: 10.1016/s0378-1119(98)00252-2. [DOI] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403(6769):501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- Medstrand P, Blomberg J. Characterization of novel reverse transcriptase encoding human endogenous retroviral sequences similar to type A and type B retroviruses: differential transcription in normal human tissues. J Virol. 1993;67(11):6778–6787. doi: 10.1128/jvi.67.11.6778-6787.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya-Durango DE, Liu Y, Teneng I, Kalbfleisch T, Lacy ME, Steffen MC, Ramos KS. Epigenetic control of mammalian LINE-1 retrotransposon by retinoblastoma proteins. Mutat Res. 2009;665(1-2):20–28. doi: 10.1016/j.mrfmmm.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya-Durango DE, Ramos KS. L1 retrotransposon and retinoblastoma: molecular linkages between epigenetics and cancer. Curr Mol Med. 2010;10(5):511–521. doi: 10.2174/156652410791608234. [DOI] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87(5):917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435(7044):903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Nelson E, Goubet-Wiemers C, Guo Y, Jodscheit K. Maternal passive smoking during pregnancy and foetal developmental toxicity. Part 2: histological changes. Hum Exp Toxicol. 1999a;18(4):257–264. doi: 10.1191/096032799678840011. [DOI] [PubMed] [Google Scholar]
- Nelson E, Jodscheit K, Guo Y. Maternal passive smoking during pregnancy and fetal developmental toxicity. Part 1: gross morphological effects. Hum Exp Toxicol. 1999b;18(4):252–256. doi: 10.1191/096032799678840002. [DOI] [PubMed] [Google Scholar]
- Packer AI, Manova K, Bachvarova RF. A discrete LINE-1 transcript in mouse blastocysts. Dev Biol. 1993;157(1):281–283. doi: 10.1006/dbio.1993.1133. [DOI] [PubMed] [Google Scholar]
- Patterson LT, Dressler GR. The regulation of kidney development: new insights from an old model. Curr Opin Genet Dev. 1994;4(5):696–702. doi: 10.1016/0959-437x(94)90136-q. [DOI] [PubMed] [Google Scholar]
- Pauler FM, Sloane MA, Huang R, Regha K, Koerner MV, Tamir I, Sommer A, Aszodi A, Jenuwein T, Barlow DP. H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res. 2009;19(2):221–233. doi: 10.1101/gr.080861.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl M, Stuart RO, Sakurai H, Nigam SK. Branching Morphogenesis During Kidney Development. Annual Review of Physiology. 2000;62(1):595–620. doi: 10.1146/annurev.physiol.62.1.595. [DOI] [PubMed] [Google Scholar]
- Poznanski AA, Calarco PG. The expression of intracisternal A particle genes in the preimplantation mouse embryo. Dev Biol. 1991;143(2):271–281. doi: 10.1016/0012-1606(91)90077-g. [DOI] [PubMed] [Google Scholar]
- Prak ET, Dodson AW, Farkash EA, Kazazian HH., Jr Tracking an embryonic L1 retrotransposition event. Proc Natl Acad Sci U S A. 2003;100(4):1832–1837. doi: 10.1073/pnas.0337627100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos KS, Hadi Falahatpisheh M, Nanez A, He Q. Modulation of biological regulatory networks during nephrogenesis. Drug Metab Rev. 2006;38(4):677–683. doi: 10.1080/03602530600959532. [DOI] [PubMed] [Google Scholar]
- Ramos KS, Nanez A. Genetic regulatory networks of nephrogenesis: deregulation of WT1 splicing by benzo(a)pyrene. Birth Defects Res C Embryo Today. 2009;87(2):192–197. doi: 10.1002/bdrc.20148. [DOI] [PubMed] [Google Scholar]
- Santos F, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol. 2005;280(1):225–236. doi: 10.1016/j.ydbio.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Schmidt JV, Su GH, Reddy JK, Simon MC, Bradfield CA. Characterization of a murine Ahr null allele: involvement of the Ah receptor in hepatic growth and development. Proc Natl Acad Sci U S A. 1996;93(13):6731–6736. doi: 10.1073/pnas.93.13.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwahn U, Lenzner S, Dong J, Feil S, Hinzmann B, van Duijnhoven G, Kirschner R, Hemberger M, Bergen AA, Rosenberg T, Pinckers AJ, Fundele R, Rosenthal A, Cremers FP, Ropers HH, Berger W. Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nature genetics. 1998;19(4):327–332. doi: 10.1038/1214. [DOI] [PubMed] [Google Scholar]
- Sciamanna I, Landriscina M, Pittoggi C, Quirino M, Mearelli C, Beraldi R, Mattei E, Serafino A, Cassano A, Sinibaldi-Vallebona P, Garaci E, Barone C, Spadafora C. Inhibition of endogenous reverse transcriptase antagonizes human tumor growth. Oncogene. 2005;24(24):3923–3931. doi: 10.1038/sj.onc.1208562. [DOI] [PubMed] [Google Scholar]
- Soifer HS, Rossi JJ. Small interfering RNAs to the rescue: blocking L1 retrotransposition. Nat Struct Mol Biol. 2006;13(9):758–759. doi: 10.1038/nsmb0906-758. [DOI] [PubMed] [Google Scholar]
- Spadafora C. Endogenous reverse transcriptase: a mediator of cell proliferation and differentiation. Cytogenetic and genome research. 2004;105(2-4):346–350. doi: 10.1159/000078207. [DOI] [PubMed] [Google Scholar]
- Teneng I, Montoya-Durango DE, Quertermous JL, Lacy ME, Ramos KS. Reactivation of L1 retrotransposon by benzo(a)pyrene involves complex genetic and epigenetic regulation. Epigenetics. 2011;6(3) doi: 10.4161/epi.6.3.14282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teneng I, Stribinskis V, Ramos KS. Context-specific regulation of LINE-1. Genes Cells. 2007;12(10):1101–1110. doi: 10.1111/j.1365-2443.2007.01117.x. [DOI] [PubMed] [Google Scholar]
- Torres-Padilla ME, Bannister AJ, Hurd PJ, Kouzarides T, Zernicka-Goetz M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int J Dev Biol. 2006;50(5):455–461. doi: 10.1387/ijdb.052073mt. [DOI] [PubMed] [Google Scholar]
- Valerius MT, Patterson LT, Witte DP, Potter SS. Microarray analysis of novel cell lines representing two stages of metanephric mesenchyme differentiation. Mechanisms of Development. 2002;112(1-2):219–232. doi: 10.1016/s0925-4773(02)00008-4. [DOI] [PubMed] [Google Scholar]
- Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13(9):763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]


