Abstract
The cAMP response element-binding protein (CREB) is a highly conserved transcription factor that integrates signaling through the cAMP-dependent protein kinase A (PKA) in many eukaryotes. PKA plays a critical role in Dictyostelium development but no CREB homologue has been identified in this system. Here we show that Dictyostelium utilizes a CREB-like protein, BzpF, to integrate PKA signaling during late development. bzpF– mutants produce compromised spores, which are extremely unstable and germination defective. Previously, we have found that BzpF binds the canonical CRE motif in vitro. In this paper, we determined the DNA binding specificity of BzpF using protein binding microarray (PBM) and showed that the motif with the highest specificity is a CRE-like sequence. BzpF is necessary to activate the transcription of at least 15 PKA-regulated, late-developmental target genes whose promoters contain BzpF binding motifs. BzpF is sufficient to activate two of these genes. The comparison of RNA sequencing data between wild type and bzpF– mutant revealed that the mutant fails to express 205 genes, many of which encode cellulose-binding and sugar-binding proteins. We propose that BzpF is a CREB-like transcription factor that regulates spore maturation and stability in a PKA-related manner.
Introduction
Cyclic AMP and cAMP-dependent protein kinase A signaling regulate many cellular events in various eukaryotes. In Dictyostelium discoideum, intracellular cAMP plays a central role in development (Saran et al., 2002). PKA is required for early developmental events, including cell aggregation and regulation of gene expression, and for later events, including the transition from slug to fruiting body and the terminal differentiation of spores and stalk cells (Loomis, 1998). The molecular mechanisms that follow PKA activation are largely unknown.
The cAMP response element binding protein, CREB, is a basic-leucine zipper (bZIP) transcription factor and a central downstream target of PKA in many eukaryotes. Dimeric CREB proteins bind the conserved DNA sequence 5′ TGACGTCA 3′, CRE, which is found in the promoters of many cAMP-inducible genes (Montminy et al., 1986), regulating the transcription of target genes in response to increased intercellular cAMP (Johannessen et al., 2004). In mammalian cells, PKA phosphorylates Ser133 in the signal-responsive Kinase-Inducible Domain (KID) of CREB (Gonzalez et al., 1991). CREB phosphorylation leads to interaction with the KIX domain of the co-activators CBP and p300. CREB also has two glutamine-rich domains, which attract the basal transcriptional machinery TFIID (Mengus et al., 2005), and a bZIP domain, which mediates dimerization and DNA binding (Dang et al., 1989; O'Shea et al., 1989).
CREB belongs to the large CREB/ATF family. One of the CREB/ATF subfamilies consists of membrane-bound transcription factors. For example, the liver-specific CREB-like protein CREB3L3/CREB-H binds the canonical CRE, as well as CRE-like sequences, such as the box-B element, and it can suppress hepatoma cell proliferation (Chin et al., 2005; Omori et al., 2001). CREB3L1/OASIS also binds CRE and CRE-like sequences and functions in bone formation (Murakami et al., 2009; Omori et al., 2002). Most members of this subfamily localize to the endoplasmic reticulum (ER) and respond to ER stress (Kondo et al., 2005; Zhang et al., 2006).
The pivotal role of PKA activity in development and in the regulation of gene expression suggested that CREB might have a central function in Dictyostelium development as well, but numerous attempts to isolate the Dictyostelium CREB have failed. In this study, we report that BzpF is a CREB-like transcription factor that functions downstream of PKA during the late stages of Dictyostelium development and regulates sporulation. Loss of BzpF results in severe defects in spore maturation and maintenance.
Material and Methods
Cell culture, strain maintenance, and development
We maintained Dictyostelium discoideum strains, which are described in Table S1, at 22°C in HL5 liquid medium (Sussman, 1987) with the indicated supplements. We collected cells at the mid-log growth phase and washed them to induce development (Shaulsky and Loomis, 1993). We deposited 5×107 cells on a 16 cm2 nitrocellulose filter placed on top of a paper pad soaked in 2 ml of PDF buffer (20.1 mM KCl, 5.3 mM MgCl2, 9.2 mM K2HPO4, 13.2 mM KH2PO4, 0.5 mg/ml streptomycin sulfate, pH 6.4) and incubated them at 22°C in a humid chamber in the dark. When the cells were antibiotic-resistant, we grew them without antibiotics for 24 hours before development.
Plasmid construction and mutant generation
bzpF (DDB_G0279529) is a single-copy gene on the Crick strand of chromosome 3, coordinates 2206682 to 2208577. To knock bzpF out, we first amplified the flanking regions of the gene by PCR using two primer sets; (1) bZIP14NKOA5-sacII: 5′-TCCCGCGGCCCATTTTGTTCTGAATCTCATT-3′ and bZIP14NKOA3-hindIII: 5′-TCAAGCTTTCTTGTTGGCTACCATTGGA-3′ (2) bZIP14NKOB5-kpnI: 5′-TCGGTACCCTCCAATCCCTTCTCCTCAA-3′ and bZIP14NKOB3-sacII: 5′-TCCCGCGGCACCTCACTTGAACGTCGTC-3′, and ligated the products to each other after SacII digestion. After amplifying the ligated fragment with the outside primer set, we cloned the products into the KpnI and HindIII sites of a modified pLPBLP plasmid (Faix et al., 2004) of which the SacII site was removed (pLPBLPΔSacII). The resulting construct is a bzpF-knockout vector with a deletion of most of the bzpF coding-region (+414 to +1654). We linearized the plasmid with SacII and transformed it into AX4 cells (Adachi et al., 1994; Kuspa and Loomis, 1992). We selected the transformants with 10 μg/ml Blasticidin and verified that bzpF was knocked out by PCR across the homologous recombination junctions and by quantitative RT-PCR.
For bzpF overexpression, we cloned the coding region of bzpF from +10 to +1885 under the actin15 promoter in the pDXA-GFP2 vector (Levi et al., 2000), transformed it into bzpF– cells and selected for resistance to 10 μg/ml G418.
Protein Binding Microarrays assay
We purified the GST-fused bZIP region of the BzpF protein (BzpFzip-GST) after expression in E. coli as described (Parikh et al., 2010a). We used the fusion protein to analyze BzpF binding site specificity using two different Protein Binding Microarray (PBM) designs, with oligonucleotide sequences as described (Berger et al., 2008; Lam et al., 2011) and protocols as described (Berger et al., 2006; Lam et al., 2011). In downstream analyses, we used a relaxed E-score threshold (0.4 instead of 0.45), following (Grove et al., 2009), and because all of the 8-mers between 0.4 and 0.45 contained either the core CREB site (ACGT) and/or a full CREB half-site (TGACG/CGTCA), and the same 8-mers scored highly in both experiments. In genomic scans, we eliminated data of 8-mers that did not meet the significance threshold in any of the experiments. We used the first data set to generate the position weighted matrix for BzpF binding and to calculate the binding scores. The results of both analyses are provided in supplementary dataset S1.
Electrophoresis mobility shift assay
We used the fusion protein BzpFzip-GST in an electrophoresis mobility shift assay (EMSA) as described (Parikh et al., 2010a) with the dCRE1-containing oligonucleotide, 5′-AGCTAAAATATGACGTATTAACTTTT-3′ (dCRE1 sequence is indicated by boldface), and the mutated CRE oligonucleotide (mCRE), 5′-AGCTAAAATACACAAAATTAACTTTT-3′, (mutations are underlined). Oligonucleotide labeling, incubation, electrophoresis, and autoradiography were done as described (Parikh et al., 2010a).
RNA preparation and quantitative RT-PCR
We prepared total RNA using Trizol (Invitrogen) and treated the samples with DNaseI to remove residual genomic DNA. We subjected the RNA to reverse transcription using an oligo-dT primer and performed quantitative PCR on an Opticon3 system (MJ Research and Biorad) using gene-specific primer sets (Table S2). We used the Delta-Delta-Ct method to detect relative changes in RNA abundance (Schmittgen et al., 2000; Winer et al., 1999) against the reference gene IG7 (Bloomfield and Pears, 2003).
Spore and stalk purification
We collected spores from AX4 fruiting bodies using a metal loop. We then collected the remaining structures and washed the residual spores through a 40 μm cell strainer (BD falcon) to purify stalks.
Sporulation efficiency, detergent resistance and spore-germination efficiency
We collected all the fruiting bodies from a quarter nitrocellulose filter into 5 ml of 0.1% NP-40 and 1 mM EDTA in 20 mM potassium phosphate pH 6.2 (KK2). For sporulation efficiency, we counted the spores with phase-contrast microscopy. For detergent resistance, we plated spores on SM agar (at least 800 spores for AX4, 2000 for bzpF–) in association with bacteria (Klebsiella pneumoniae) and counted plaques in the bacterial lawn after 3-6 days. For spore-germination efficiency, we collected sori from 2-day old fruiting bodies, resuspended them in HL5 at 2×106 cells/ml and shook them at 200 RPM for 6 hours at 22°C. We then counted the spores and the amoebae with phase-contrast microscopy. Germination efficiency is the ratio between germinated amoebae and the total cell number.
Spore staining
We collected spores from 30-hour fruiting bodies into 0.5 ml KK2 buffer. For staining with the fluorescent vital dye propidium iodide (PI; Sigma), we mixed the spores with 2.6 μg/ml propidium iodide and incubated at room temperature for 30 minutes (Levraud et al., 2003). We examined the spores by fluorescence microscopy and by flow cytometry with a BD Biosciences LSRII system equipped with a 488 nm argon laser. In some cases, we monitored GFP signals with the same equipment.
For lectin staining, we used FITC-labeled Arachis hypogaea lectin (Sigma) at a concentration of 40 μg/ml. After treating spores with or without urea (Wang et al., 2003), we incubated the spores with the FITC-lectin for 30 min at room temperature, washed them twice with KK2 buffer and examined the samples with fluorescence microscopy (Fosnaugh et al., 1994).
To stain for cellulose within the spore coat, we used Calcofluor white (Sigma). We incubated the spores with 0.01% (w/v) solution of Calcofluor in KK2 buffer at room temperature for 3-5 minutes. We washed the spores three times with KK2 buffer and examined the samples by fluorescence microscopy.
8-Br-cAMP-induction of sporulation
We developed cells on nitrocellulose filters for 19–20 hours, harvested the structures, dissociated them by repeated pipetting in EDTA-KK2 (Katoh et al., 2004) and washed them thrice with KK2. We induced sporulation as described (Wang et al., 1996) except for using 15 mM 8-Br-cAMP (Sigma) instead of 20 mM.
Induction of gene expression
We developed cells for 19 hours on filters, disassociated and resuspended them as above. We treated the cells with or without 20 mM 8-Br-cAMP for 2 hours at 22°C in Petri dishes. We then collected the cells and prepared RNA for quantitative RT-PCR analysis.
Computational analysis of potential BzpF target genes
We searched the D. discoideum genome for genes that contain at least one dCRE motif in the region between -600 to +150 relative to the ORF. We then calculated the target score according to two criteria: the BzpF-binding preference of the dCRE motifs and co-expression with bzpF. To find targets with high affinity for BzpF, we used the PBM assay binding preference results (Table S3, threshold > 0.4). We obtained the score of each putative target gene by summing the binding preference scores of all the dCRE motifs. The score was downweighted if the dCRE motifs were located in transcribed regions, according to the RNA-seq data. We used published expression data (Parikh et al., 2010b) to search for genes whose expression patterns correlate with that of bzpF during development. Using the Dynamic Time Warp (DTW) method (Sakoe and Chiba, 1978), we aligned the time course data with slightly shifted phases (±1 time point) to compare the time series of the expression levels and then used Pearson's correlation on the DTW-transformed expression profiles to evaluate the correlation with the bzpF expression pattern. To determine significance (p < 0.15), we permuted the BzpF binding preference scores among the 8-mers and repeated the analysis 30 times to obtain a null distribution. We used that distribution to determine the p-value of the experimental data.
RNA Sequencing
We collected RNA from cells at 25 hours of development and processed the samples as described (Parikh et al., 2010b), except that we performed the poly(A) selection twice. We analyzed the RNA-seq data using the data mining software Orange (Curk et al., 2005).
Results
bzpF encodes a CREB-like protein
bzpF was implicated as a CREB gene by transcriptional network analysis (Parikh et al., 2010a). To test this possibility, we analyzed the predicted BzpF protein sequence (631 amino acids) and found a PKA-phosphorylation consensus sites at S438, inside the bZIP region, and glutamine-rich Q-domains at the N-terminal region (Fig. 1A). However, the characteristic KID motif was not readily evident in BzpF. By BLAST search, we found that BzpF is similar to bZIP proteins in Naegleria, to the mammalian CREB3L3 and CREB3L1, and to the Arabidopsis bZIP49 and bZIP17 proteins, which belong to the CREB/ATF subfamily which is involved in ER stress response (Audas et al., 2008; Tajima et al., 2008). Nevertheless, BzpF does not have a putative transmembrane domain, which is characteristic of the CREB/ATF subfamily members (Llarena et al., 2010). We also found that BzpF has a close homolog in Dictyostelium purpureum. Therefore, we propose that BzpF is a Dictyostelid type CREB-like protein.
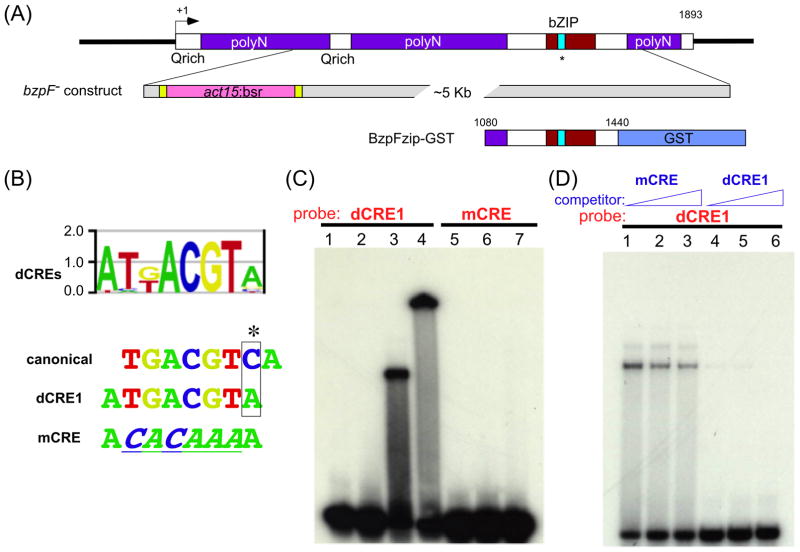
Fig. 1. bzpF is a CREB-like gene.
(A) A schematic representation of bzpF: This single-exon gene is 1893 bp-long with multiple poly-N (purple boxes) and Q-rich coding regions. The bZIP domain (red box) is close to the carboxyl terminus; the cyan colored region inside the bZIP domain indicates a potential CRE binding domain, and the asterisk represents the PKA phosphorylation consensus site. The bzpF– construct: The majority of the coding region (+414 to +1654) is replaced by a Blasticidin S resistance (bsr) cassette (pink), which is flanked by loxP recombination sites (yellow) and a plasmid backbone (grey). The BzpFzip-GST construct: The C-terminal region (+1080 to +1440) including the bZIP domain is fused with GST.
(B) The DNA binding motif of BzpF: An oligonucleotide-chip containing all possible 8 mers was used to measure the BzpF binding preference. The DNA sequences (dCREs) bound by BzpFzip-GST in vitro are summarized in a position weight matrix, which is shown as a sequence logo (Berger et al., 2006). The matrix includes the canonical CRE (canonical) and the sequence with the highest affinity for BzpF (dCRE1). The mutated-CRE oligonucleotide (mCRE) differs by 6 nucleotides from the dCRE1 sequence. The mutated nucleotides are underlined and italicized.
(C) Electrophoresis mobility shift assay: Radioactively labeled dCRE1-containing oligonucleotides (lanes 1-4) and mCRE-containing oligonucleotides (negative control: lanes 5-7) were used as probes. Lane 1 and 5 – no protein added; lane 2 and 6 – a control bZIP protein (DimAzip-GST); lanes 3 and 7 – BzpFzip-GST protein; lane 4 – BzpFzip-GST with anti-GST antibody. (D) Competition EMSA: We added unlabeled nonspecific mCRE (lanes 1-3; 20, 100 and 500 fold excess, respectively) or specific dCRE1 (lanes 4-6; 20, 100 and 500 fold excess, respectively) to the mixture of BzpFzip-GST and the radiolabeled dCRE1.
The D. purpureum bzpF gene model (DPU_G0065610; May 2011) in DictyBase predicted a 249-amino acids protein, 382 amino acids shorter than the D. discoideum BzpF. We assembled RNA-seq reads (Parikh et al., 2010b) and found an unidentified intron 47-bp upstream of the predicted start codon of the DictyBase gene model. In addition, we found three sequencing errors around the predicted stop codon. We predict that the D. purpureum bzpF gene encodes a 531–amino acids protein that has 64.1% identity and 69.9% similarity to the D. discoideum BzpF protein (Fig. S1). Most of the protein sequence, including the bZIP region at the C-terminus, are conserved in both BzpFs except for a poly-Asparagine sequences at the N-terminus of the D. discoideum protein.
BzpF binds the canonical CRE in vitro (Parikh et al., 2010a). To find the preferred DNA motif for BzpF, we tested the GST-fused bZIP region of BzpF (BzpFzip-GST, Fig. 1A) against a microarray that contains all possible 8-mer oligonucleotides. We show the result as a position weight matrix in Fig. 1B. We named the sequence, dCRE1, for which BzpFzip-GST had the highest preferences. dCRE1 differs by one nucleotide from the canonical CRE sequence, creating an asymmetric site (Fig. 1B). We confirmed the preference of BzpF to dCRE1 by EMSA. BzpFzip-GST bound the dCRE1-containing oligonucleotide (Fig. 1C, lane 3) but not a mutated oligonucleotide (mCRE; Fig. 1C, lane 7). The shifted band was super-shifted in the presence of an antibody against GST (Fig. 1C, lane 4), suggesting that the binding was indeed mediated by the BzpFzip-GST. We used the GST-fused bZIP region of DimA, which is a well-characterized Dictyostelium bZIP protein, as a control. This bZIP region did not bind the dCRE1-containing oligonucleotide (Fig. 1C, lane 2), indicating the binding specificity of BzpF to dCRE1. We also performed a competition assay by adding unlabeled oligonucleotides to the reaction. Increasing amounts of the mutated oligonucleotide had almost no effect (Fig. 1D, lanes 1-3), whereas the binding was decreased by the addition of unlabeled dCRE1 (Fig. 1D, lanes 4-6). These results suggest that BzpF binds to dCRE1 in a sequence-specific manner, extending the finding that BzpF binds to the canonical CRE (Parikh et al., 2010a).
bzpF is expressed preferentially in spores during late culmination
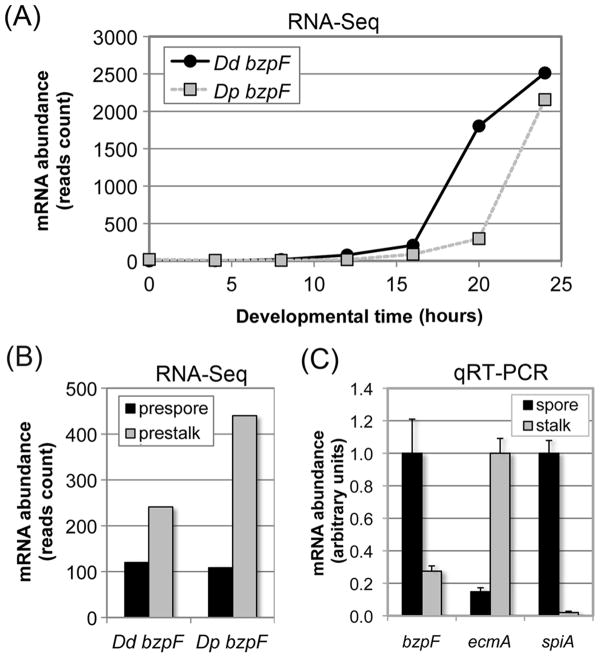
We examined the expression profile of bzpF during development by quantitative RT-PCR and by mining the developmental RNA-seq database (Parikh et al., 2010b). bzpF expression is not detectable at 0-12 hours of development. The mRNA is first detectable at 16 hours and it continues to accumulate through the end of development (Fig. 2A), suggesting a function during terminal differentiation. We also checked the expression of the bzpF ortholog in the related species D. purpureum. We observed a similar pattern of mRNA abundance with a slight temporal delay (Fig. 2A), as expected from the observation that D. purpureum develops more slowly than D. discoideum (Parikh et al., 2010b). The RNA-seq data also suggest that the bzpF mRNA is enriched in prestalk cells during slug migration, about 2-fold in D. discoideum and 4-fold in D. purpureum relative to prespore cells (Fig. 2B). To examine the cell-type specificity in fruiting bodies, where the transcript is at peak abundance, we purified spores and stalk cells and measured bzpF expression. We found that bzpF is expressed in both cell types, but it is enriched about 4 fold in the spores (Fig. 2C). By measuring the mRNA levels of the stalk specific gene ecmA (Harwood et al., 1993) and the spore specific gene spiA (Richardson et al., 1991) we found that the spore sample contains no more than 14.8% stalk RNA and the stalk sample contains no more than 2.0% spore RNA (Fig. 2C).
Fig. 2. bzpF is expressed during late development in D. discoideum and D. purpureum.
(A) The expression profile of bzpF during development: Adapted from the published RNA-sequencing data of D. discoideum (Dd bzpF, black circles) and D. purpureum (Dp bzpF, grey squares) (Parikh et al., 2010b). Time points (hours) are indicated on the x-axis and mRNA abundance (normalized read-counts) on the y-axis.
(B) Preferential expression of bzpF in prestalk cells: Adapted from the published RNA-sequencing data of D. discoideum (Dd bzpF) and D. purpureum (Dp bzpF) (Parikh et al., 2010b). bzpF mRNA is detectable in both prespore cells (black bars) and prestalk cells (gray bars) in slugs. The y-axis indicates mRNA abundance (normalized read counts). (C) Preferential expression of bzpF in spores: We performed quantitative RT-PCR on RNA from spores (black bars) and stalk cells (grey bars) enriched from mature D. discoideum fruiting bodies. The enrichment of the isolated cell types is illustrated with the stalk-specific marker ecmA and the spore-specific marker spiA. The sample with the most abundant mRNA was normalized to 1 unit (y-axis) for each gene. The data are represented as mean + s.e.m of four replicates.
D. discoideum and D. purpureum have diverged approximately 400 million years ago (Sucgang et al., 2011). The similarity between the expression patterns of bzpF in these two organisms (Fig. 2A,B) suggests constant selective surveillance of this trait and an evolutionary conserved function. We propose that BzpF activity is required during late stages of development in both cell types, preferentially in the spores.
bzpF– mutants produce compromised spores
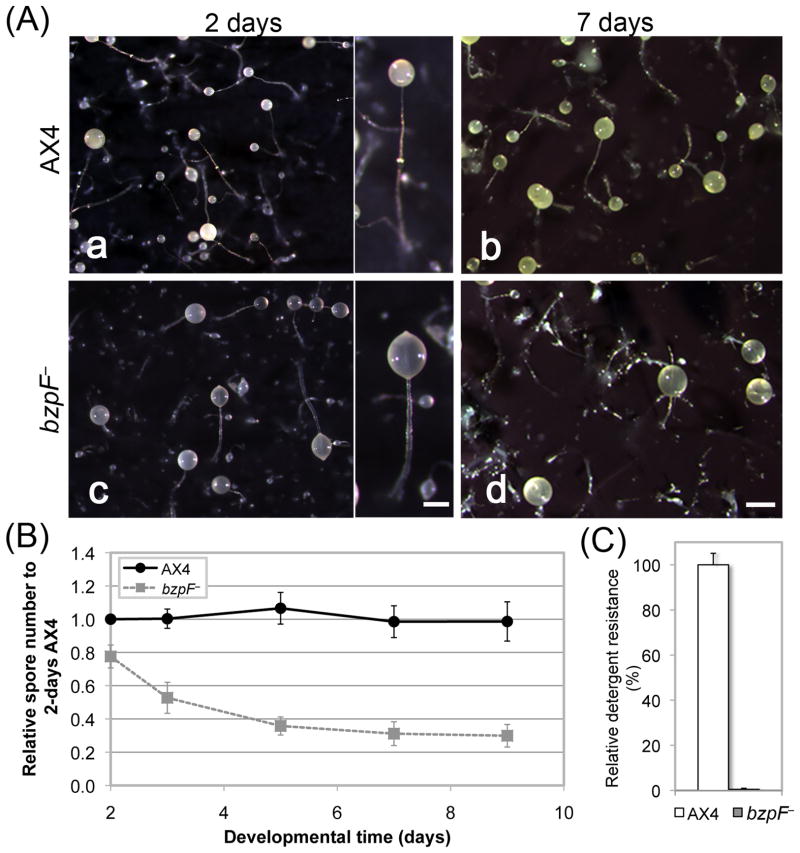
To test the developmental role of bzpF, we generated a mutant by deleting most of the coding region (Fig. 1A). The mutant cells grow normally on bacteria and in HL5 liquid medium. During development on bacterial plates, the mutant exhibits aberrant fruiting-body morphology with thick, irregular stalks and light-colored, translucent sori (data not shown). We also developed axenically grown cells on nitrocellulose filters (Fig. 3A). Under these conditions, the bzpF– mutants are morphologically indistinguishable from the wild type during the first 16 hours. At the end of development the mutants exhibit asynchronous culmination. Fruiting body formation is completed at 28-30 hours and the stalks are short and thick (Fig. 3A a, c). The bzpF– sori appear abnormal – they are more translucent than those of the wild type (Fig. 3A c). Upon prolonged incubation, most of the mutant sori slipped down the stalks and the remaining sori become more translucent, while the AX4 sori become opaque and yellow (Fig. 3A b, d). These observations suggest that the support for sori is weak and that spore development and spore stability are compromised in the bzpF– strain.
Fig. 3. bzpF– cells exhibit morphological defects during development.
(A) Morphology: We developed cells for 2 and 7 days on dark filters and photographed the multicellualr structures of AX4 (a and b, respectively) and the bzpF– mutant (c and d, respectively). Bar = 0.2 mm in the magnified image, 0.5 mm in d. (B) Spore stability: We counted spores (AX4 - circles, solid line; bzpF– - squares, dashed line) between 2 - 9 days of incubation on filters (x-axis) as a fraction of the value observed for AX4 at 2 days (y-axis). The data are the means ± s.e.m of four replicates. (C) Detergent resistance: We incubated AX4 spores (white bar) and bzpF– spores (grey bar) in 0.1% NP-40 and plated them on nutrient agar in association with bacteria. The number of plaques formed on the bacterial lawn relative to the number of spores plated (%) is the detergent resistance (y-axis). The data are the means + s.e.m of three replicates.
We compared the spore stability of the bzpF– mutant to that of AX4 after several days of development. The spore number of the bzpF– mutant is only slightly lower than that of AX4 at 2 days, but it is markedly decreased after 3-5 days, suggesting spore instability (Fig. 3B). We also examined the detergent resistance of the spores and found that the bzpF– spores are much more sensitive than their AX4 counterparts (Fig. 3C), further suggesting spore instability. We note that the declines in spore count and in detergent resistance were not due to germination within the sori because we have not found hatched amoebae and empty spore shells in the mutant sori.
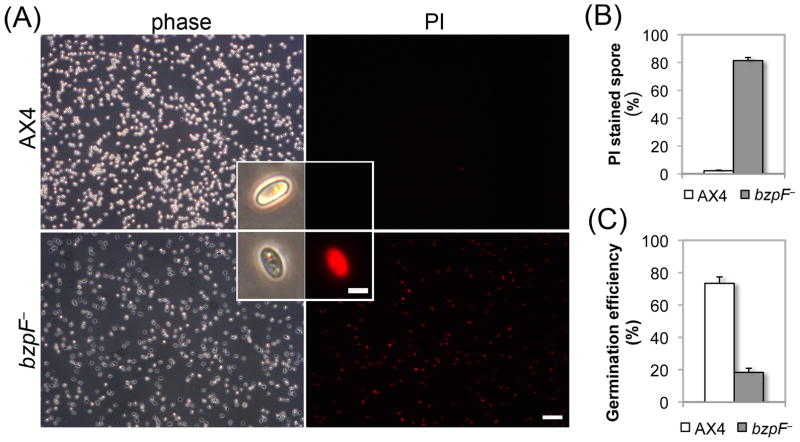
To test the potential cause of the spore instability, we examined whether the spore coat of the bzpF– spores is compromised. The Dictyostelium spore coat is composed of three layers; the central mid layer, which includes galactose-rich polysaccharides and cellulose, is sandwiched between two protein-rich layers (West, 2003). A compromised outer layer could result in a porous, less protective spore coat (Fosnaugh et al., 1994) that can be detected by staining of mid layer components with a fluorescently-labeled lectin, which recognizes polysaccharides, and with calcofluor, which stains cellulose. The bzpF– spores were stained slightly stronger than AX4 spores with both FITC-labeled lectins and with calcofluor (Fig. S2A and B). Stronger FITC-labeled lectin staining could also be caused by altered glycosylation level (Wang et al., 2003). We therefore treated the spores with urea permeabilization, which strips the external layer barrier proteins before the FITC-lectin staining. This experiment ruled out the possibility of altered N-glycosylation because the staining level of AX4 and bzpF– spores were nearly identical after urea treatment (Fig. S2). Therefore, the stronger staining with FITC-lectin and calcofluor (Fig. S2A and B) indicates that the bzpF– spore coat outer layer might be slightly compromised, possibly due to the loss of some urea-soluble components from the spore coat. The bzpF– spores also have visible defects. Compared to the birefringent wild-type spores, most of the bzpF– spores appear dark under phase-contrast microscopy (Fig. 4A). Viable spores can exclude propidium iodide (PI) (Cornillon et al., 1994; Yoshino et al., 2007) so we stained the spores with PI to monitor spore integrity. The difference between bzpF– and AX4 is dramatic (Fig. 4A). The AX4 spores are almost completely free of PI staining (1.7% positive, Fig. 4B), whereas most of the bzpF– spores are stained (78.1% positive, Fig. 4B). In addition, almost all of the stained spores are phase-dark (Fig. 4A), suggesting that the membrane of the bzpF– spores is compromised.
Fig. 4. bzpF– spores are compromised.
(A) PI staining of spores. Phase-contrast (phase) and fluorescence (PI) images of each field of AX4 and bzpF– spores, as indicated. Magnified spores are shown as inserts. Bars = 40 μm (low magnification) and 5 μm (inserts). (B) Quantitation of PI staining: We counted the PI-stained spores of AX4 (white bar) and bzpF– (grey bar) by flow cytometry and the fraction (%) of PI positive-spores is shown (y-axis). Data are the means + s.e.m of five replicates. (C) Germination: We incubated AX4 (white bar) and bzpF– spores in nutrient medium. The number of germinated amoebae is shown as a fraction (%) of the total spores (y-axis). The data are the means + s.e.m of three replicates.
Finally, we examined spore-germination by incubating the spores in liquid medium. The fraction of hatched amoebae in the bzpF– population (18.4%, Fig. 4C) is lower than in AX4 (73.4%, Fig. 4C), consistent with the fraction of PI-negative, phase-bright spores in these strains.
Overall, the bzpF– spores are compromised, most likely due to defects in the cell membrane inside the spore coat. We postulate that these defects lead to spore instability within the sori, resulting in reduced viability, detergent sensitivity, and poor germination.
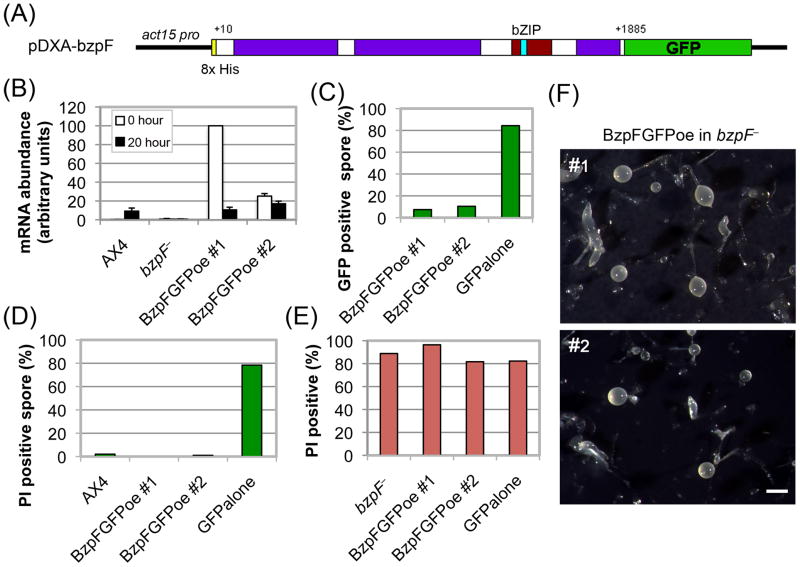
Ectopic expression of bzpF
We fused the bzpF coding region with the GFP coding region and cloned the fusion gene under the ubiquitous actin15 promoter (Fig. 5A). We transformed the bzpF– strain with the expression vector and observed high but variable levels of bzpF mRNA in the transformants during growth (0 hours) and during late development (20 hours) (Fig. 5B). Western-blot analysis revealed small amounts of the full-size BzpF-GFP fusion protein and high amounts of apparent degradation products (data not shown). We then examined the spores of the transformants by flow cytometry and found that only about 10% are GFP positive (Fig. 5C). In contrast, bzpF– cells transformed with a GFP vector alone show 85% GFP-positive spores (Fig. 5C). These results suggest that the BzpF-fusion protein may be unstable upon ectopic expression. Another possibility is the potential expression of a truncated BzpF peptide (1-137 amino acids) that may be produced as a consequence of the deletion in bzpF–. This potential peptide would lack the bZIP domain but it would carry a short conserved region (1-56 amino acids, see Fig. S1) that may have a dominant negative effect over the transgenic BzpF.
Fig. 5. Ectopic expression of bzpF complements the compromised spore phenotype.
(A) The BzpF ectopic expression construct: We fused most of the bzpF coding region (+10 to +1885) with GFP and expressed it in the pDXA vector under the actin15 promoter in bzpF– mutants. (B) Quantitation of bzpF expression: We measured the abundance of bzpF mRNA in AX4, bzpF–, and two independent ectopic expression transformants (BzpFGFPoe) by qRT-PCR in vegetative cells (0 hour, white bars) and in developing cells (20 hour, black bars). The data (arbitrary units, y-axis) are the means and standard errors of five replicates. (C) Partially penetrant expression of BzpF-GFP: We determined the fraction of GFP positive spores (%, y-axis) by flow cytometry of the ectopic expression strains and of a control strain expressing GFP alone (x-axis). (D) Complementation of the compromised spore phenotype in spores that express bzpF-GFP: We stained spores of the indicated strains (x-axis) with PI and used flow cytometry to determine the proportion (%, y-axis) of PI-positive (compromised) spores among the wild type (AX4) and among the GFP-positive spores of the transgenic strains. (E) Lack of complementation in spores that do not express BzpF-GFP: We stained spores of the indicated strains (x-axis) with PI and used flow cytometry to determine the proportion (%, y-axis) of PI-positive (compromised) spores among the parental (bzpF–) and among the GFP-negative spores of the transgenic strains. (F) The fruiting body morphology of the bzpFoe strains: Filter development of two bzpF-overexpressor strains (#1 and #2) showing morphologies that are nearly identical to the bzpF– phenotypes. Bar = 0.5 mm.
The availability of strains that produce 10% BzpF-GFP-positive spores allowed us to test whether ectopic expression of the BzpF-GFP fusion protein could complement some of the bzpF– phenotypes. To test the compromised spore phenotype, we stained the spores with PI and used flow cytometry to simultaneously count GFP-positive and PI-positive spores. Almost none of the BzpF-GFP-expressing spores were stained with PI, similar to the level observed in AX4, whereas most of the bzpF– spores expressing GFP alone were stained with PI (Fig. 5D). Among the BzpF-GFP-negative spores in the same populations, almost all are stained with PI (Fig. 5E). These results suggest that ectopic expression of the BzpF-GFP fusion protein is sufficient to complement the bzpF– mutation with respect to the spore instability phenotype. Expression of GFP alone is not a confounding factor and the partial penetrance of the ectopic expression system allows us to suggest that the complementation, and therefore the function of BzpF, are cell autonomous.
We also developed the transformed cells on nitrocellulose filters to test the effect of the ectopically-expressed fusion protein on morphogenesis. During the first 16 hours, the transformed cells develop normally, as do the bzpF– cells. Later in development the BzpF-GFP transformants exhibit asynchronous culmination and aberrant fruiting bodies (Fig. 5F), also identical to the bzpF– phenotypes (Fig. 3A) and different from AX4, suggesting that the overexpression vector is not capable of complementing the bzpF– mutation with respect to developmental morphology. This failure to complement is probably due to the low proportion of cells (10%) that express the full-length fusion protein in the developing mass and to the cell-autonomous nature of the BzpF function.
PKA-C activation does not suppress the bzpF– phenotype
Activation of PKA can suppress many developmental defects in Dictyostelium, indicating that PKA-C functions downstream of the mutated gene responsible for the defect (Loomis, 1998; Richardson and Loomis, 1992; Wang and Kuspa, 1997). Previous analyses implicated BzpF as a member of the PKA-pathway and suggested that it functions downstream of PKA-C (Parikh et al., 2010a), but our analysis shows that BzpF lacks several characteristic CREB domains so it might not function downstream of PKA. It was therefore of interest to test the relationship between PKA-C activation and bzpF function. We postulated that if BzpF functions downstream of PKA-C, then activation of PKA-C would not suppress the bzpF– phenotype. 8-Br-cAMP is a membrane-permeable analogue of cAMP that activates PKA by dissociating the PKA-R regulatory subunit from the PKA-C catalytic subunit (Kay, 1989; Maeda, 1988). We developed AX4 and bzpF– cells on filters for 19-20 hours, disaggregated the multicellular structures, incubated the cells with or without 15 mM 8-Br-cAMP, and measured the proportion of spores that have formed. In the control experiment, 75.2% of the treated AX4 cells sporulate in response to 8-Br-cAMP whereas bzpF– achieves 46.5% sporulation, which is significantly lower (Student's t-test, p=0.02) (Fig. 6A). This finding suggests that activation of PKA-C is not sufficient to fully induce sporulation in the absence of BzpF. Neither strain produces a significant number of spores without 8-Br-cAMP (Fig. 6A).
Fig. 6. Activating PKA fails to rescue the sporulation defect of the bzpF– mutant.
We developed AX4 cells (white bars) and bzpF– cells (grey bars), disaggregated them, and incubated them with and without 8-Br-cAMP as indicated. (A) Sporulation efficiency (y-axis) was calculated as the proportion (%) of cells that became spores. The data are the means + s.e.m of 3 replicates. (B) We tested the germination efficiency of 8-Br-cAMP-induced spores by treatment with 0.1% NP-40. The data are presented as the fraction (%) of detergent-resistant spores relative to the total AX4 spores. The data are the means + s.e.m of 4 replicates.
We also tested the detergent resistance of the induced spores. The bzpF– spores have very low detergent resistance (Fig. 6B), similar to the level observed after filter development (Fig. 3D), whereas the AX4 spores are detergent resistant (Fig. 6B). These findings indicate that activation of PKA-C does not suppress the defects of the bzpF– strain. We suggest that PKA-C is not downstream of BzpF in a pathway that regulates sporulation, which is consistent with the idea that BzpF functions downstream of PKA-C or that the two proteins function in parallel pathways.
The transcriptional targets of BzpF
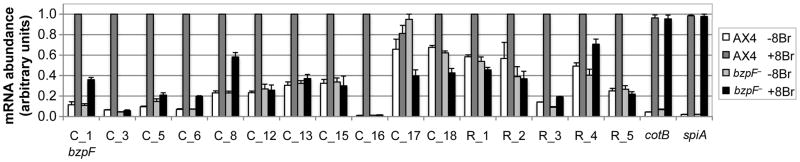
If BzpF is a transcription factor, it should be both necessary and sufficient for the expression of genes that contain its binding motifs in their promoters. We searched the Dictyostelium genome (Eichinger et al., 2005) for genes that carry any one of the dCRE motifs (Table S3) in their potential promoter regions and found 1197 candidates. We narrowed the list down using computational and experimental approaches. First, we postulated that the expression of BzpF-target genes would be changed after bzpF is expressed, so we searched the Dictyostelium wild type transcriptome (Parikh et al., 2010b) for genes whose developmental expression correlates well with that of bzpF. Second, we calculated the score of each putative target gene following two criteria: the BzpF-binding preference from the PBM assay results and the co-expression with bzpF. We found 18 genes whose promoter regions include dCRE motifs with high binding affinities and whose expression patterns were positively correlated (Pearson's r > 0.8) with bzpF during development (Table S4). There is no gene with negative correlation (r < –0.8) with bzpF. To find additional potential target genes of BzpF, we also performed RNA-seq on late-stage AX4 and bzpF– samples (Dataset S2) and found 13 additional genes that have at least one dCRE motif (with a lower stringency than the above criteria) and show reduced expression in bzpF– (Table S5). We also checked whether the other 18 genes exhibit differential expression in the mutant strain and found that 11 were indeed expressed at a lower level in bzpF– (Table S4).
Next, we examined whether the expression of those genes is induced by activation of PKA-C in a BzpF-dependent manner. We chose 11 genes, including bzpF, from the first approach (Table S4, bold font) and 5 genes from the second approach (Table S5, bold font) and performed quantitative RT-PCR on 8-Br-cAMP-treated samples. Expression of 15 of the 16 genes is inducible by 8-Br-cAMP in AX4 (Fig. 7) and all 15 are less abundant in the bzpF– mutant. The prespore genes, cotB and spiA, which are known to be regulated by PKA, are induced to almost the same level in both strains, suggesting that the PKA-pathway in bzpF– can be activated by 8-Br-cAMP. These data indicate that BzpF is necessary for the transcriptional activation of several terminal differentiation genes that are also regulated by the PKA pathway. These 15 genes are potential direct targets of BzpF.
Fig. 7. Candidate BzpF-target genes are regulated by PKA.
We developed AX4 and bzpF– cells and treated them with or without 8-Br-cAMP as indicated. We then tested the abundance of the candidate target transcripts by quantitative RT-PCR. The gene identities are indicated on the x-axis (Tables S4 and S5). The data are the means + s.e.m of 3-5 replicates, normalized to the maximal abundance of each gene.
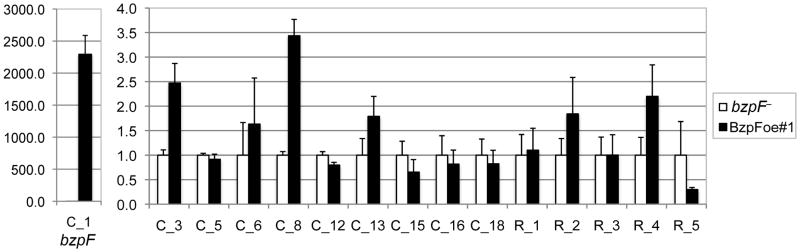
We also tested whether BzpF is sufficient for the transcriptional activation of the putative target genes. Since these target genes are not expressed in vegetative cells, we measured mRNA abundance in vegetative cells that express BzpF ectopically. We prepared RNA from vegetatively growing bzpF– and a BzpF-GFP transformant. Using quantitative RT-PCR we found an increase in the expression of 2 putative target genes in the BzpF-overexpressing strain relative to bzpF– (Fig. 8), suggesting that BzpF is both necessary and sufficient for the expression of these two genes.
Fig. 8. BzpF is sufficient for expression of its target genes.
We used quantitative RT-PCR to test the abundance of putative target transcripts in vegetative bzpF– cells (white bars) and bzpF-overexpressor cells with the ectopic BzpF-GFP expression vector (black bars). The gene identities are indicated on the x-axis (Tables S4 and S5). The data are the means + s.e.m of 3-9 replicates, normalized to the abundance of each gene in bzpF–.
The results shown in Figs 7 and 8 supported the idea that BzpF is a transcription factor because it is both necessary and sufficient for the transcriptional activation of its target genes.
Biological processes regulated by BzpF
Comparing the transcriptomes of the bzpF– mutant and the wild type revealed 205 genes that are under-expressed in bzpF– (log2(AX4/bzpF–)>1, AX4 raw read count>100). Only 24 of the genes contain dCRE in their upstream regions, so we suspect that most of these transcriptional changes are secondary consequences of the mutation. The genes encode 13 cellulose-binding domain-containing proteins, 4 spore germination proteins, and 3 GATA transcription factors (Table S6). We also checked the differential expression of cell-type marker genes (Parikh et al., 2010b) between the wild type strain AX4 and the bzpF– mutants. We found that 19 out of 633 prespore genes and 22 out of 800 prestalk genes exhibit differential expression (Table S7). In bzpF–, the expression levels of cotA, B and C, which are prespore markers, are not significantly different from those in AX4 (cotA, B and C log2(AX4/bzpF–)= 0.58, 0.05, 0.12, respectively). The expression level of cotE, which encodes a spore coat protein, is greatly decreased in the bzpF– strain (log2(AX4/bzpF–)= 1.72) (Table S6 and S7). The expression of ecmA, which a prestalk markers, exhibited almost no difference (log2(AX4/bzpF–)= 0.21). On the other hand, the expression level of another prestalk marker, ecmB, in the bzpF– strain is less than half of the wild type level (log2(AX4/bzpF–)= 1.03). These data suggest that BzpF might be involved in both cell-type differentiation processes – stalk differentiation and late spore maturation through direct or indirect transcriptional regulation of developmental genes. A Gene Ontology (GO) enrichment analysis of the 205 genes is shown in Table 1. These Genes are enriched in the GO categories “lipid biosynthesis” and “fatty acid biosynthesis”. The most differentially expressed genes (log2(AX4/bzpF–)>3) are enriched in “cellulose binding” and “sugar binding” (Table 1).
Table 1. Gene Ontology analysis of transcripts that are less abundant in bzpF–
| Log2(AX4/bzpF–) | matched (%)/input genes | GO term | Category | P-value | Enrichment |
|---|---|---|---|---|---|
| >1 | 107(52.2%)/205 | lipid biosynthetic process | Biological process | 0.0582 | 4.54 |
| fatty acid biosynthetic | Biological process | 0.0582 | 6.70 | ||
| >3 | 26(40.6%)/64 | cellulose binding | Molecular function | 0.0174 | 30.49 |
| sugar binding | Molecular function | 0.0174 | 27.58 | ||
| extracellular region | Cellular component | 0.0066 | 13.16 |
Discussion
Our studies show that BzpF is a CREB-like transcription factor, which functions downstream of PKA-pathway and regulates spore maturation and stability during late Dictyostelium development. BzpF binds the canonical CRE motif as well as the related dCRE motif.
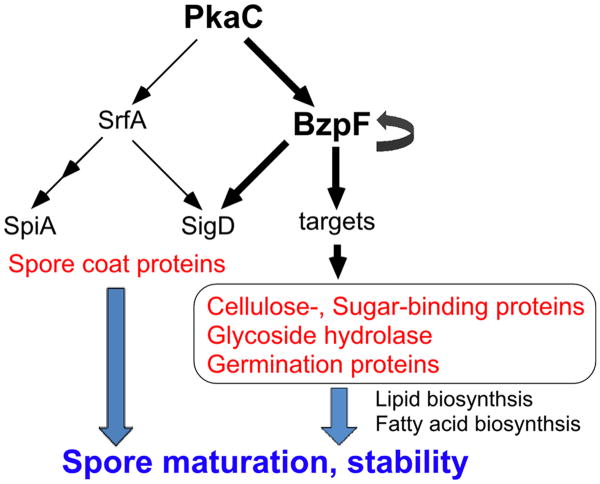
Dictyostelium spores have a thick spore coat layer, which contains proteins and polysaccharides. The spore coat proteins and the galactose-rich polysaccharides first accumulate in prespore vesicles during early-mid development and are secreted by exocytosis under the control of PKA at the beginning of sporulation. Subsequently, cellulose is synthesized and extruded from the cells to form the mid layer, which is sandwiched by protein-rich outer and inner layers. The cellulose layer interacts with the coat glycoproteins to form a protein-impermeability barrier (West, 2003; West et al., 2009). Spore integrity could be compromised at many levels, including defects in the cellulose layer and the galactose rich polysaccharides layer, glycosylation defects, and inner membrane defects. The loss of BzpF causes severe spore defects that are manifested as spore instability and low germination efficiency. Genes that are down regulated in the mutant are involved in lipid and fatty acid biosynthesis as well as cellulose and sugar-binding, although most of these genes are probably not regulated directly by BzpF. These findings suggest that BzpF affects membrane biosynthesis and spore coat formation through indirect transcriptional regulation of cellulose and sugar binding proteins, which in turn promote maturation of spores and maintenance of their dormancy (Fig. 9). Some of the BzpF targets we have identified are members of the GATA transcription factor gene family, so it is possible that BzpF regulates a sporulation-specific cascade of transcriptional events.
Fig. 9. A proposed model of the PKA-BzpF pathway.
PKA regulates several transcription factors including BzpF and SrfA, which are involved in terminal differentiation. Activation of PKA can drive the activation of BzpF, which positively regulates its own expression. BzpF expression is required for spore maturation and stability, probably through direct and indirect transcriptional activation of genes that encode spore coat proteins, cellulose- and sugar-binding proteins, Glycoside hydrolases and germination proteins, as well as lipid and fatty acid biosynthesis.
Another essential factor of sporulation is the MADS-box transcription factor SrfA (Escalante and Sastre, 1998, 2002). Loss of this gene results in spores that are round and phase-dark. These spores have reduced calcofluor staining and reduced expression of spiA, which encodes a late spore coat protein. PKA activity leads to srfA expression during culmination and SrfA regulates spiA expression indirectly (Fig. 9). Unlike srfA– spores, bzpF– spores are oval and well-stained with calcofluor, even though they are phase-dark. Moreover, the expression of cotB, spiA and srfA in bzpF– is almost indistinguishable from the wild type, suggesting that BzpF and SrfA have largely parallel roles in the regulation of sporulation (Fig. 9). Nevertheless, there might be some overlap between the two pathways because the expression of six SrfA-induced genes (sigL-1, rnpA, sigK, sigM, sigD and sigI) is significantly lower in bzpF– than in AX4 and three of the genes contain dCRE motifs in their promoter regions (Fig. 9, Table S6). In many cases, the expression levels of developmental genes might be regulated by the coordination of multiple transcription factors.
In Dictyostelium development, PKA plays pivotal roles in the control of transcriptional regulation during multiple processes, including aggregation, prespore/prestlak differentiation and spore maturation. BzpF acts downstream of PKA, but, it is not expressed at aggregation stage. Other transcription factors, including other bZIP proteins such as bzpD and bzpH, which are expressed at the early stages, may transduce the PKA signals to regulate developmental gene expression. The expression of bzpF is initially detected at 16 hrs in both cell types. It is enriched in prestalk cells during slug migration and then accumulates in spores during culmination. Consistent with this observation, the bzpF– mutant exhibits two stalk-related phenotypes in addition to the severe spore defects: thick stalks and weak support of the lower cup, which results in slippage of the sori down the stalk. Also, some of the target genes of BzpF are preferentially expressed in prestalk cells (Table S6, S7). The prestalk marker genes aslA-1 and ecmB, which are normally expressed in the upper and lower cups during culmination, are down-regulated in bzpF– (Table S7), suggesting that they might be indirect targets of BzpF and their down-regulation may be related to the weak lower cup support phenotype. We propose that BzpF regulates two developmental processes – early prestalk differentiation and late spore maturation – raising the possibility that BzpF interacts with different components in those cell types. The fact that the 19 Dictyostelium bZIP proteins can form various homo- and hetero-dimers (Deppmann et al., 2006; Huang et al., 2006) suggests that BzpF may dimerize with other bZIP proteins in the different cell types and such hetero-dimer complexes may have specific roles and targets.
Based on sequence analysis, BzpF is neither a canonical CREB nor a classical CREB3 type protein. Our results suggest that BzpF may function downstream of PKA-C in D. discoideum although we have no conclusive evidence that it is directly regulated by PKA-C through phosphorylation. Future studies will be needed to determine whether PKA regulates BzpF and how BzpF regulates the transcription of each target gene we found here. Nevertheless, BzpF is a CREB-like transcription factors because it binds dCRE1, a motif only 1-bp different from the mammalian CRE, and it functions in concert with PKA-C to regulate terminal differentiation in Dictyostelium.
Supplementary Material
Highlights.
Dictyostelium bZIP protein BzpF binds to CRE-like sequence with high specificity.
bzpF is expressed preferentially in spores during late culmination stage.
Loss of BzpF results in severe defects in spore maturation and stability.
We found 15 PKA-regulated, late-developmental genes as potential BzpF direct targets.
Many under-expressed genes in bzpF– encode cellulose- and sugar-binding proteins.
Acknowledgments
We thank A. Parikh for assistance with the RNA-seq data analysis, I. Mann, E. Chan, and H. van Bakel for technical and computational support, and A. Kuspa for useful discussions. We are grateful to the Dicty stock center for vectors. This work was supported by grants from the National Institute of Child Health and Human Development (P01 HD39691), the Slovenian Research Agency (P2-0209, J2-2197, L2-1112, Z7-3665) and the Canadian Institutes of Health Research (MOP-77721).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi H, Hasebe T, Yoshinaga K, Ohta T, Sutoh K. Isolation of Dictyostelium discoideum cytokinesis mutants by restriction enzyme-mediated integration of the blasticidin S resistance marker. Biochem Biophys Res Commun. 1994;205:1808–1814. doi: 10.1006/bbrc.1994.2880. [DOI] [PubMed] [Google Scholar]
- Audas TE, Li Y, Liang G, Lu R. A novel protein, Luman/CREB3 recruitment factor, inhibits Luman activation of the unfolded protein response. Mol Cell Biol. 2008;28:3952–3966. doi: 10.1128/MCB.01439-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, Khalid F, Zhang W, Newburger D, Jaeger SA, Morris QD, Bulyk ML, Hughes TR. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, 3rd, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield G, Pears C. Superoxide signalling required for multicellular development of Dictyostelium. J Cell Sci. 2003;116:3387–3397. doi: 10.1242/jcs.00649. [DOI] [PubMed] [Google Scholar]
- Chin KT, Zhou HJ, Wong CM, Lee JM, Chan CP, Qiang BQ, Yuan JG, Ng IO, Jin DY. The liver-enriched transcription factor CREB-H is a growth suppressor protein underexpressed in hepatocellular carcinoma. Nucleic Acids Res. 2005;33:1859–1873. doi: 10.1093/nar/gki332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornillon S, Foa C, Davoust J, Buonavista N, Gross JD, Golstein P. Programmed cell death in Dictyostelium. J Cell Sci. 1994;107(Pt 10):2691–2704. doi: 10.1242/jcs.107.10.2691. [DOI] [PubMed] [Google Scholar]
- Curk T, Demsar J, Xu Q, Leban G, Petrovic U, Bratko I, Shaulsky G, Zupan B. Microarray data mining with visual programming. Bioinformatics. 2005;21:396–398. doi: 10.1093/bioinformatics/bth474. [DOI] [PubMed] [Google Scholar]
- Dang CV, McGuire M, Buckmire M, Lee WM. Involvement of the ‘leucine zipper’ region in the oligomerization and transforming activity of human c-myc protein. Nature. 1989;337:664–666. doi: 10.1038/337664a0. [DOI] [PubMed] [Google Scholar]
- Deppmann CD, Alvania RS, Taparowsky EJ. Cross-species annotation of basic leucine zipper factor interactions: Insight into the evolution of closed interaction networks. Mol Biol Evol. 2006;23:1480–1492. doi: 10.1093/molbev/msl022. [DOI] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, Tunggal B, Kummerfeld S, Madera M, Konfortov BA, Rivero F, Bankier AT, Lehmann R, Hamlin N, Davies R, Gaudet P, Fey P, Pilcher K, Chen G, Saunders D, Sodergren E, Davis P, Kerhornou A, Nie X, Hall N, Anjard C, Hemphill L, Bason N, Farbrother P, Desany B, Just E, Morio T, Rost R, Churcher C, Cooper J, Haydock S, van Driessche N, Cronin A, Goodhead I, Muzny D, Mourier T, Pain A, Lu M, Harper D, Lindsay R, Hauser H, James K, Quiles M, Madan Babu M, Saito T, Buchrieser C, Wardroper A, Felder M, Thangavelu M, Johnson D, Knights A, Loulseged H, Mungall K, Oliver K, Price C, Quail MA, Urushihara H, Hernandez J, Rabbinowitsch E, Steffen D, Sanders M, Ma J, Kohara Y, Sharp S, Simmonds M, Spiegler S, Tivey A, Sugano S, White B, Walker D, Woodward J, Winckler T, Tanaka Y, Shaulsky G, Schleicher M, Weinstock G, Rosenthal A, Cox EC, Chisholm RL, Gibbs R, Loomis WF, Platzer M, Kay RR, Williams J, Dear PH, Noegel AA, Barrell B, Kuspa A. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante R, Sastre L. A Serum Response Factor homolog is required for spore differentiation in Dictyostelium. Development. 1998;125:3801–3808. doi: 10.1242/dev.125.19.3801. [DOI] [PubMed] [Google Scholar]
- Escalante R, Sastre L. Regulated expression of the MADS-box transcription factor SrfA mediates activation of gene expression by protein kinase A during Dictyostelium sporulation. Mech Dev. 2002;117:201–208. doi: 10.1016/s0925-4773(02)00203-4. [DOI] [PubMed] [Google Scholar]
- Faix J, Kreppel L, Shaulsky G, Schleicher M, Kimmel AR. A rapid and efficient method to generate multiple gene disruptions in Dictyostelium discoideum using a single selectable marker and the Cre-loxP system. Nucleic Acids Res. 2004;32:e143. doi: 10.1093/nar/gnh136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosnaugh K, Fuller D, Loomis WF. Structural roles of the spore coat proteins in Dictyostelium discoideum. Dev Biol. 1994;166:823–825. doi: 10.1006/dbio.1994.1362. [DOI] [PubMed] [Google Scholar]
- Gonzalez GA, Menzel P, Leonard J, Fischer WH, Montminy MR. Characterization of motifs which are critical for activity of the cyclic AMP-responsive transcription factor CREB. Mol Cell Biol. 1991;11:1306–1312. doi: 10.1128/mcb.11.3.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove CA, De Masi F, Barrasa MI, Newburger DE, Alkema MJ, Bulyk ML, Walhout AJ. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 2009;138:314–327. doi: 10.1016/j.cell.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood AJ, Early A, Williams JG. A repressor controls the timing and spatial localisation of stalk cell-specific gene expression in Dictyostelium. Development. 1993;118:1041–1048. doi: 10.1242/dev.118.4.1041. [DOI] [PubMed] [Google Scholar]
- Huang E, Blagg SL, Keller T, Katoh M, Shaulsky G, Thompson CR. bZIP transcription factor interactions regulate DIF responses in Dictyostelium. Development. 2006;133:449–458. doi: 10.1242/dev.02240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen M, Delghandi MP, Moens U. What turns CREB on? Cell Signal. 2004;16:1211–1227. doi: 10.1016/j.cellsig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Katoh M, Shaw C, Xu Q, Van Driessche N, Morio T, Kuwayama H, Obara S, Urushihara H, Tanaka Y, Shaulsky G. An orderly retreat: Dedifferentiation is a regulated process. Proc Natl Acad Sci U S A. 2004;101:7005–7010. doi: 10.1073/pnas.0306983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay RR. Evidence that elevated intracellular cyclic AMP triggers spore maturation in Dictyostelium. Development. 1989;105:753–759. [Google Scholar]
- Kondo S, Murakami T, Tatsumi K, Ogata M, Kanemoto S, Otori K, Iseki K, Wanaka A, Imaizumi K. OASIS, a CREB/ATF-family member, modulates UPR signalling in astrocytes. Nat Cell Biol. 2005;7:186–194. doi: 10.1038/ncb1213. [DOI] [PubMed] [Google Scholar]
- Kuspa A, Loomis WF. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc Natl Acad Sci U S A. 1992;89:8803–8807. doi: 10.1073/pnas.89.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam KN, van Bakel H, Cote AG, van der Ven A, Hughes TR. Sequence specificity is obtained from the majority of modular C2H2 zinc-finger arrays. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkq1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi S, Polyakov M, Egelhoff TT. Green fluorescent protein and epitope tag fusion vectors for Dictyostelium discoideum. Plasmid. 2000;44:231–238. doi: 10.1006/plas.2000.1487. [DOI] [PubMed] [Google Scholar]
- Levraud JP, Adam M, Luciani MF, de Chastellier C, Blanton RL, Golstein P. Dictyostelium cell death: early emergence and demise of highly polarized paddle cells. J Cell Biol. 2003;160:1105–1114. doi: 10.1083/jcb.200212104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llarena M, Bailey D, Curtis H, O'Hare P. Different mechanisms of recognition and ER retention by transmembrane transcription factors CREB-H and ATF6. Traffic. 2010;11:48–69. doi: 10.1111/j.1600-0854.2009.00997.x. [DOI] [PubMed] [Google Scholar]
- Loomis WF. Role of PKA in the timing of developmental events in Dictyostelium cells. Microbiol Mol Biol Rev. 1998;62:684–694. doi: 10.1128/mmbr.62.3.684-694.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda M. Dual Effects of cAMP on the Stability of Prespore Vesicles and 8-bromo CAMP-enhanced Maturation of Spore and Stalk Cells of Dictyostelium discoideum. Develop Growth & Differ. 1988;30:573–587. doi: 10.1111/j.1440-169X.1988.00573.x. [DOI] [PubMed] [Google Scholar]
- Mengus G, Fadloun A, Kobi D, Thibault C, Perletti L, Michel I, Davidson I. TAF4 inactivation in embryonic fibroblasts activates TGF beta signalling and autocrine growth. EMBO J. 2005;24:2753–2767. doi: 10.1038/sj.emboj.7600748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montminy MR, Sevarino KA, Wagner JA, Mandel G, Goodman RH. Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc Natl Acad Sci U S A. 1986;83:6682–6686. doi: 10.1073/pnas.83.18.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Saito A, Hino S, Kondo S, Kanemoto S, Chihara K, Sekiya H, Tsumagari K, Ochiai K, Yoshinaga K, Saitoh M, Nishimura R, Yoneda T, Kou I, Furuichi T, Ikegawa S, Ikawa M, Okabe M, Wanaka A, Imaizumi K. Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat Cell Biol. 2009;11:1205–1211. doi: 10.1038/ncb1963. [DOI] [PubMed] [Google Scholar]
- O'Shea EK, Rutkowski R, Kim PS. Evidence that the leucine zipper is a coiled coil. Science. 1989;243:538–542. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
- Omori Y, Imai J, Suzuki Y, Watanabe S, Tanigami A, Sugano S. OASIS is a transcriptional activator of CREB/ATF family with a transmembrane domain. Biochem Biophys Res Commun. 2002;293:470–477. doi: 10.1016/S0006-291X(02)00253-X. [DOI] [PubMed] [Google Scholar]
- Omori Y, Imai J, Watanabe M, Komatsu T, Suzuki Y, Kataoka K, Watanabe S, Tanigami A, Sugano S. CREB-H: a novel mammalian transcription factor belonging to the CREB/ATF family and functioning via the box-B element with a liver-specific expression. Nucleic Acids Res. 2001;29:2154–2162. doi: 10.1093/nar/29.10.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh A, Huang E, Dinh C, Zupan B, Kuspa A, Subramanian D, Shaulsky G. New components of the Dictyostelium PKA pathway revealed by Bayesian analysis of expression data. BMC Bioinformatics. 2010a;11:163. doi: 10.1186/1471-2105-11-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh A, Miranda ER, Katoh-Kurasawa M, Fuller D, Rot G, Zagar L, Curk T, Sucgang R, Chen R, Zupan B, Loomis WF, Kuspa A, Shaulsky G. Conserved developmental transcriptomes in evolutionarily divergent species. Genome Biol. 2010b;11:R35. doi: 10.1186/gb-2010-11-3-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson DL, Hong CB, Loomis WF. A prespore gene, Dd31, expressed during culmination of Dictyostelium discoideum. Dev Biol. 1991;144:269–280. doi: 10.1016/0012-1606(91)90421-x. [DOI] [PubMed] [Google Scholar]
- Richardson DL, Loomis WF. Disruption of the sporulation-specific gene spiA in Dictyostelium discoideum leads to spore instability. Genes Dev. 1992;6:1058–1070. doi: 10.1101/gad.6.6.1058. [DOI] [PubMed] [Google Scholar]
- Sakoe H, Chiba S. Dynamic programming algorithm optimization for spoken word recognition. IEEE Transactions on Acoustics, Speech, and Signal Processing. 1978;26:43–49. [Google Scholar]
- Saran S, Meima ME, Alvarez-Curto E, Weening KE, Rozen DE, Schaap P. cAMP signaling in Dictyostelium. Complexity of cAMP synthesis, degradation and detection. J Muscle Res Cell Motil. 2002;23:793–802. doi: 10.1023/a:1024483829878. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Zakrajsek BA, Mills AG, Gorn V, Singer MJ, Reed MW. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal Biochem. 2000;285:194–204. doi: 10.1006/abio.2000.4753. [DOI] [PubMed] [Google Scholar]
- Shaulsky G, Loomis WF. Cell type regulation in response to expression of ricin A in Dictyostelium. Dev Biol. 1993;160:85–98. doi: 10.1006/dbio.1993.1288. [DOI] [PubMed] [Google Scholar]
- Sucgang R, Kuo A, Tian X, Salerno W, Parikh A, Feasley CL, Dalin E, Tu H, Huang E, Barry K, Lindquist E, Shapiro H, Bruce D, Schmutz J, Salamov A, Fey P, Gaudet P, Anjard C, Mohan MB, Basu S, Bushmanova Y, van der Wel H, Katoh-Kurasawa M, Dinh C, Coutinho PM, Saito T, Elias M, Schaap P, Kay RR, Henrissat B, Eichinger L, Rivero F, Putnam NH, West CM, Loomis WF, Chisholm R, Shaulsky G, Strassmann JE, Queller DC, Kuspa A, Grigoriev I. Comparative genomics of the social amoebae Dictyostelium discoideum and Dictyostelium purpureum. Genome Biol. 2011;12:R20. doi: 10.1186/gb-2011-12-2-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman M. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol. 1987;28:9–29. doi: 10.1016/s0091-679x(08)61635-0. [DOI] [PubMed] [Google Scholar]
- Tajima H, Iwata Y, Iwano M, Takayama S, Koizumi N. Identification of an Arabidopsis transmembrane bZIP transcription factor involved in the endoplasmic reticulum stress response. Biochem Biophys Res Commun. 2008;374:242–247. doi: 10.1016/j.bbrc.2008.07.021. [DOI] [PubMed] [Google Scholar]
- Wang B, Kuspa A. Dictyostelium development in the absence of cAMP. Science. 1997;277:251–254. doi: 10.1126/science.277.5323.251. [DOI] [PubMed] [Google Scholar]
- Wang N, Shaulsky G, Escalante R, Loomis WF. A two-component histidine kinase gene that functions in Dictyostelium development. EMBO J. 1996;15:3890–3898. [PMC free article] [PubMed] [Google Scholar]
- Wang F, Metcalf T, van der Wel H, West CM. Initiation of mucin-type O-glycosylation in dictyostelium is homologous to the corresponding step in animals and is important for spore coat function. J Biol Chem. 2003;278:51395–51407. doi: 10.1074/jbc.M308756200. [DOI] [PubMed] [Google Scholar]
- West CM. Comparative analysis of spore coat formation, structure, and function in Dictyostelium. Int Rev Cytol. 2003;222:237–293. doi: 10.1016/s0074-7696(02)22016-1. [DOI] [PubMed] [Google Scholar]
- West CM, Nguyen P, van der Wel H, Metcalf T, Sweeney KR, Blader IJ, Erdos GW. Dependence of stress resistance on a spore coat heteropolysaccharide in Dictyostelium. Eukaryot Cell. 2009;8:27–36. doi: 10.1128/EC.00398-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- Yoshino R, Morio T, Yamada Y, Kuwayama H, Sameshima M, Tanaka Y, Sesaki H, Iijima M. Regulation of ammonia homeostasis by the ammonium transporter AmtA in Dictyostelium discoideum. Eukaryot Cell. 2007;6:2419–2428. doi: 10.1128/EC.00204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, Kaufman RJ. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124:587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.