Abstract
Asthma is the most common chronic childhood disease in developed nations and its prevalence has increased in the world over the last 25 years. It is a complex disease with both genetic and environmental risk factors. Asthma is caused by multiple interacting genes, some having a protective effect and others contributing to the disease pathogenesis, with each gene having its own tendency to be influenced by the environment. This article reviews the current state of the genetics of asthma in six categories, viz. epidemiology, management, aetiology, family and twin studies, segregation and linkage studies, and candidate genes and single nucleotide polymorphisms (SNPs).
Keywords: Aetiology, asthma, linkage, prevalence, SNPs
Introduction
Asthma is one of the most serious allergic diseases and the most common chronic childhood disease in developed nations1. It has been characterized by increased responsiveness of the tracheobronchial tree to a multiplicity of stimuli2–4, increased infiltration of various inflammatory cells especially eosinophils into the airway, epithelial damage, airway smooth-muscle hypertrophy5, constriction, variable airway obstruction usually associated with inflammation in the conducting airways of the lungs6 and mucous hypersecretion in the bronchiolar walls of the lung7. Asthma is critically dependent on a series of cell adhesion molecule-mediated interactions between vascular endothelium and leukocytes7, leading to symptoms8 and elevation in total serum IgE9. It is manifested physiologically by widespread narrowing of the air passages and clinically by paroxysms of dyspnoea, cough, wheezing and tightness, provoked by one or more triggers such as physical exertion and airway irritants (cold, dry air, smoke, etc.)4,10. It is an episodic disease, with acute exacerbations interspersed with symptom-free periods. Typically, most attacks are short-lived, lasting minutes to hours, and clinically the patient seems to recover completely after an attack. However, there can be a phase in which the patients experience some degree of airway obstruction daily. This phase can be mild, with or without superimposed severe episodes, or can be much more serious, with severe obstruction persisting for days or weeks; the latter condition is known as “acute severe asthma”. In unusual circumstances, acute episodes can cause death4. Asthma exacerbations are characteristically worse at night and can progress to severe airflow obstruction, shortness of breath, and respiratory distress and insufficiency. Rarely, severe sequel such as hypoxic seizures, respiratory failure, and death can occur10.
Here we review the latest information on the genetic basis of asthma which is one of the most intriguing diseases affecting people of all ages, gender, race and ethnicities. Familial and segregation studies have an important role in asthma aetiology and several candidate genes on all the human chromosomes play their roles in initiation and/or inhibition of different pathways of asthma disease.
Epidemiology
Epidemiological studies carried out in different countries indicate the prevalence of respiratory allergy as 15-30 per cent11 and asthma affects in the range of 3.5-20 per cent of the population in any country12. The documented increase in asthma prevalence over the last 25 years is likely due to changes in our environment or lifestyle because changes in our genetic makeup would take more than several generations to occur13. Worldwide, asthma cases are increasing at a rate of 50 per cent every decade, and according to the World Health Organization, by the year 2020, asthma, along with chronic obstructive pulmonary disease (COPD) will become the third leading cause of death. An estimated 300 million people in the world currently have asthma and there may be an additional 100 million persons with asthma by 202514.
Unlike in the case of most other diseases, the prevalence of asthma is the highest in developed countries such as the United States, the United Kingdom, Australia, New Zealand and North-west Europe8, and the least in Macau1,14,19 (Table I). About half of the people with asthma develop it before age 10, and most develop it before the age of 30. Among younger children, asthma develops twice as frequently in boys than in girls, however, after puberty it is more common in girls. The prevalence of asthma is higher in urban areas than in rural1,20. Poverty and malnutrition exacerbate asthma in children, leading to compromised lung function.
Table I.
Prevalence of asthma in different countries
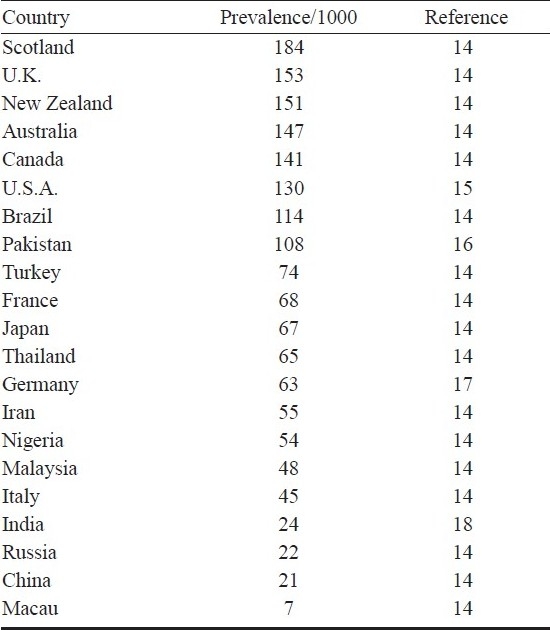
It has been reported that India has approximately 15-20 million asthmatics and 10 to 15 per cent of Indian children between the ages of 5 and 11 yr show symptoms of asthma. In India, there is a median prevalence of about 2.4 per cent in adults of over 15 yr of age21. In one of the largest epidemiological multi-centric studies on the prevalence of asthma in Indian adults using a uniform, validated and standardized methodology, a prevalence of 1.69-3.47 per cent was observed22. Female gender, increasing age, family history of asthma, history suggestive of atopy, lower socio-economic status and urban residence were significantly associated with asthma22. In a study in Mumbai, the prevalence of asthma in adults was 3.5 and 17 per cent when broad definitions including asymptomatic bronchial hyper-responsiveness were used23. In rural children in Delhi, parental smoking, paracetamol intake, current exposure to cat, exposure to traffic pollution were found to be significantly associated with current wheezing24 whereas in children aged 4-15 yr in Chandigarh, a prevalence of 7 per cent was observed25. India accounts for a third of the world's asthma patients20.
Management
Asthma can be suspected in a patient based on history, patterns of symptoms and physical examination. The gold standard for the diagnosis of asthma remains spirometry demonstrating > 12 per cent and >200 ml improvement in FEV1 after bronchodilation26. Bronchoprovocation tests can be used to confirm bronchial hyper-responsiveness. Pulse oximetry and arterial blood gas analysis are used to assess the severity of acute asthma attack and chest X-ray is used to rule out asthma mimickers and related complications such as pneumothorax. Asthma is classified according to the persistence of symptoms and their severity26. Asthma treatment includes environmental control and medication. Quick relief medications are intended to open up the airways to improve breathing during an acute exacerbation. Long-term medications are taken even when no symptoms are present, to minimize lung inflammation. Once an individual has been diagnosed with asthma, a physician will develop an asthma action plan to help the patient to monitor the condition. Although the worldwide market for asthma medication is currently worth of US$5.5 billion a year to the pharmaceutical industry1,27,28, there is no cure for asthma and only the symptoms can be controlled.
Current asthma management is aimed at reducing airways inflammation by using daily “controller” anti-inflammatory medications, minimizing proinflammatory environmental exposures, and controlling co-morbid conditions that can worsen asthma. Less inflammation typically leads to better asthma control, including less need for “quick-reliever” asthma medication (i.e., beta-agonist bronchodilators) and fewer exacerbations10.
Aetiology
Studies of family history, twins, familial aggregation and segregation studies in asthma have convincingly shown that the disease has a strong genetic component20. A heterogeneous condition of asthma may predominate in different geographic locations, and is strongly influenced by environmental factors that may differ among populations and at different ages. However, it is likely that the risk of developing asthma is greatest when both genetic and environmental risk factors are present simultaneously13.
The inheritance of asthma and allergy does not follow the classical Mendelian patterns of inheritance6. However, rarely monogenic cases of atopic disease have been documented and the majority of atopic asthma is likely to be the result of numerous interacting genetic and environmental factors1. It is a commonly believed that asthma is caused by multiple interacting genes, some having a protective effect and others contributing to the disease pathogenesis, with each gene having its own tendency to be influenced by the environment.
Family and twin studies
Familial aggregation of asthma was probably first described by Sennertus in 165027. Two large studies were performed the inheritance of atopy, one in 191628 and the other in 192429. In the first and second studies 48.4 and 58.4 per cent of family history cases respectively were reported and autosomal dominant inheritance of atopy was suggested. Schwartz30 reported that the prevalence rates of asthma in the 1,634 relatives of the 161 asthmatic subjects was 6.6 per cent, but, in the 1,790 relatives of the control group, only 1 per cent. Sibbald et al31 reported that the overall prevalence of asthma in the first degree relatives of asthmatics was 13 per cent and in the relatives of controls only 4 per cent. This indicates that there is a considerable genetic component in the pathogenesis of asthma.
A large twin study reported in 1971 was a questionnaire--based study of 6,996 twin pairs from the Swedish Twin Registry. In this study the monozygotic (MZ) concordance for self-reported asthma was 19 per cent and dizygotic (DZ) concordance was 4.8 per cent. Another large Finnish study investigated 13,888 twin pairs and showed a concordance rate of 0.13 for MZ twins and 0.7 for DZ twins, and under the multifactorial threshold model, the heritability of asthma combining the sexes was 36 per cent6. Duffy et al32 in a questionnaire-based study of 3,808 Australian twin pairs showed a correlation of self-reported asthma of 0.65 among MZ twins and 0.24 among DZ twins. The heritability was 60 per cent for females and 75 per cent for males. Harris et al33 studied 5,864 Norwegian twins in a study on health and development in Oslo. The proband-wise concordance for asthma was 0.45 for MZ twins and 0.25 for DZ twins. A population-based twin family study in 16 yr old Finnish twins and their parents34 presented combined twin/family data on the inheritance of asthma. The heritability of asthma was approximately 79 per cent, whereas 21 per cent was due to unique environmental factors34. Huovinen et al35 have studied 262 Finnish twin pairs, and reported that in addition to allergic diseases, educational level and physical activity were associated with adult onset asthma. Nystad et al36 have studied 3334 pairs of Norwegian twins aged 18-35 yr and established that the phenotypic correlation between disease and symptom was 0.67 for asthma and wheezing.
Twin studies have generally shown that concordance rates for asthma are significantly higher in MZ twins than in DZ twins, whether reared apart or together. Broad-sense heritability estimates derived from twin studies range from 36 to 75 per cent14. Twin studies have revealed a 0.74 concordance between monozygotic twins and a 0.35 concordance between dizygotic twins, implicating a genetic contribution to asthma development10.
Segregation and linkage analysis
Segregation analysis can provide insight into the genetics of a trait, e.g. the number of genes involved and the genetic model: dominant or recessive, polygenic, and those with environmental effects. Using this type of analysis, the heritability, mode of inheritance, penetrance and frequency of a trait are being estimated and also indicated the involvement of major genes6.
A large study performed by the European Community Respiratory Health Survey Group analyzed the pooled data from 13,963 families (consisting of 75,392 randomly selected individuals) using complex seg-regation analysis. The results of this study showed further evidence of genetic regulation of asthma and a model with a two-allele gene with codominant inheritance fitted the data best, assuming a major gene has to be involved in the pathogenesis of asthma, but the penetrance of such a gene is low37. Jenkins et al38 presented a segregation analysis of 7,394 families in which 15.9 per cent of the index individuals had asthma. A segregation analysis of physician-diagnosed asthma in 3,369 randomly selected individuals from 906 nuclear families done by Holberg et al39 in Tucson, AR, USA, showed evidence of a polygenic or an oligogenic model with some evidence of a recessive gene, explaining only part of the segregation.
Many segregation analyses of total serum IgE-concen-tration in asthma have been studied and most of these studies conclude that IgE levels are highly heritable. Several studies have shown a strong association between atopy and bronchial hyper-responsiveness40–42. The complexity of the immunological network involved in the pathogenesis of asthma, atopy, its related traits and the existence of different asthma phenotypes are consistent that different genes may be involved in the pathogenesis of asthma6.
Ober et al43 conducted a genome-wide screen in the Hutterites, a religious isolate of European ancestry, to identify genes that influence asthma and asthma-associated phenotypes. A primary sample of 361 individuals and a replication sample of 292 individuals were evaluated by a genome-wide screen using 292 autosomal and three X-Y pseudoautosomal markers. Using the semi-parametric likelihood ratio, χ2 test and the transmission-disequilibrium test, 12 markers in 10 regions were identified that showed possible linkage to asthma or an associated phenotype. They showed markers in four regions (5q23-31, 12q15-24.1, 19q13 and 21q21) with possible linkage in both the primary and replication samples and have also shown linkage to asthma phenotypes in other samples; two adjacent markers in one additional region (3p24.2-22) showing possible linkage were reported for the first time in the Hutterites. Recently, Pillai et al44 have identified five major quantitative asthma phenotypes.
Kleeberger and Peden45 have studied different environmental factors (physical, chemical, nutritional, behavioral, etc.) in isolation which have been shown to affect asthma and related phenotypes but their interaction effects have been missed46.
Bouzigon et al47 reported that polymorphisms in 17q21 confer higher risk in early onset asthma and the risk increases further when there is exposure to environmental tobacco smoke in early life. This region contains four genes all of which could have potential role in asthma pathogenesis46,47. Teerlink et al48 revealed genome-wide significant evidence of linkage to region 5q13 and suggestive evidence for linkage to region 6p21. Both the 5q13 and 6p21 regions were previously identified as regions of interest in other genome-wide scans for asthma-related phenotypes48.
Table II provides the chromosome regions involved in causing asthma identified by linkage analysis. More than 100 loci on 22 autosomes, X and Y chromosomes have been linked to asthma8,10,49,50. Chromosome 12 appears to harbour maximum susceptible genes for asthma than any other chromosome. Interestingly, only one locus has been established on each of chromosomes 3, 15, 18 and 22. Of these loci associated with asthma, some had very strong association.
Table II.
Asthma related genes and their location*

Candidate genes and SNPs
Table II shows the list of common candidate genes in asthma with their locations derived from a large number of single nucleotide polymorphisms (SNPs) studies. The following are some of the extensively studied candidate genes and SNPs associated with asthma, with special reference to studies in the Indian population:
1. A Disintegrin and Metalloprotease33 (ADAM 33): This is a member of the “A Disintegrin and Metalloprotease” (ADAM) family proteins with diverse functions that reflect the complex domain structure of these molecules79. This gene has been identified by positional cloning and localized on to chromosome 20p13 as a susceptibility gene for asthma43. This is the most extensively studied and highly polymorphic gene with 14119 bps, 22 exons and 21 introns. Case-control and family-based association studies have confirmed a link between ADAM33 and asthma. Its restricted expression to mesenchymal cells as well as its association with bronchial hyper-responsiveness and accelerated decline in lung functions over time strongly point to its involvement in the structural airway components of asthma. Extensive alternative splicing, expression during branching morphogenesis in the developing foetus, impaired lung function in childhood, the production of a soluble form linked to chronic asthma, and tight epigenetic regulation indicate a level of complexity in the way ADAM33 influences the disease phenotype. ADAM33 function includes activation, proteolysis, adhesion, fusion, and intracellular signaling. The crystal structure of the catalytic domain of ADAM33 has been resolved around the nonselective matrix metalloproteinase inhibitor (marimastat) in addition to the zinc binding site77. Angela et al80 supported the hypothesis that ADAM33 polymorphisms influence lung function in early life and epithelial-mesenchymal dysfunction in the airways may predispose individuals toward asthma, being present in early childhood before asthma becomes clinically expressed. ADAM33 contains over 55 SNPs, some of which play an important role in asthma and related traits. Polymorphisms in the ADAM33 are associated with an accelerated decline in forced expiratory volume in the first second (FEV1) in the spirometery of general population and these are not only risk factors for the development of asthma, but also for COPD. Thus, polymorphisms in ADAM33 constitute important risk factors for the development of respiratory diseases in a large subset of the general population78. Bijanzadeh et al81 reported that there are not significant association between T1 SNP of the ADAM33 and asthma in an Indian population.
2. Interleukin-4 (IL-4): This is located on chromosome 5 at position q31 with 32675 bps, 10 exons and 9 introns. IL-4 is a cytokine secreted by helper T cell type 2 (TH-2 cells) that stimulates the production of IgE and induces eosinophil-mediated attacks against allergens82. Chiang et al83 established that polymorphism in the promoter of the IL-4 is associated with asthma and is a disease modifier in terms of the severity of airway hyper-responsiveness (AHR). A total of 16 polymorphisms were identified in the IL4, of which one in the promoter (C-589T) and other on the 5’ untranslated region (C-33T) of the IL4 have been identified that influence total serum IgE levels and bronchial hyper-responsiveness84. Nagarkati et al85 indicated that the promoter of the IL4 gene is invariant in Indian population and Bijanzadeh et al81 reported that there are no significant association between this SNP of the IL-4 and asthma in an Indian population.
3. β-chain of the high-affinity receptor for IgE (FcεRIβ): This is localized on chromosome 11q13 with a length of 8.74 kb. This is responsible for immediate reactions and also is found on the surface of mast cells, basophils, eosinophils and Langerhan's cells. The binding of allergen to the receptor-bound IgE leads to degranulation of the cell and the synthesis and release of cytokines (IL-4), and activated inflammatory cells. The β-chain is not essential for FcεRI function, but it stabilizes the surface expression of the receptor and acts as an amplifying element within it1,6,16. A G/A polymorphism in intron 2, a (CA)n repeat polymorphism in intron 5, and a C/T polymorphism in 3’-UTR were established as significant association with asthma86. A promoter-dependent mechanism with altered transcriptional regulation of Fc and epsi; RIβ may be involved for its association with asthma87.
4. PDH finger protein 11 (PHF11): This is localized on chromosome 13q14 and contains 10 exons, 9 introns and with 32973 bps. PHF11 has 17 SNPs associated with asthma59. Public databases identified an alternative first exon with multiple overlapping variants that produce alternative start methionines for protein translation70.
5. IL-4 receptor-α ( IL-4Rα): IL-4 uses the α-chain of the IL-4 Receptor (IL-4Rα) as a part of the respective receptor systems. This gene is located on chromosome 16 and represents an ideal candidate gene for atopy susceptibility because of its pivotal role in IL-4 signaling and its key role in allergic inflammation by promoting IgE production and Th2 cell development72,88.
6. G-protein-coupled receptor for asthma (GPRA): This is localized on chromosome 7p with 7 SNPs. A hierarchical genotyping design was used to identify this gene. The data implied that this gene is involved in the pathogenesis of atopy and asthma and may have application in other inflammatory diseases64.
7. Dipeptidyl-peptidase 10 (DPP10): This is localized on chromosome 2q14-2q32 and shares features with members of the S9B family of DPP serine proteases, which includes DPP4, a widely expressed enzyme that plays a central role in chemokine processing as part of the innate immune system. The locus displays a complex pattern of transcript splicing, with eight alternate first exons; four of which strongly associated with asthma87.
8. Interferon gamma (IFNG): This locus is localized on 12q21 and established as a candidate gene for asthma on the basis of its role in pathophysiology and positive linkage demonstrated in some populations69.
9. Inducible nitric oxide synthase (iNOS): This gene is localized in the CC chemokine cluster region on chromosome 17q11.2-q12 and a linkage has been observed to asthma and atopy. iNOS is expressed predominantly from inflammatory cells such as T cells and macrophages and the resultant nitric oxide that is produced causes mucus hypersecretion, upregulation of Th2 and downregulates Th1 responses49,89,90.
10. Inositol polyphosphate 4 phosphatase type I (INPP4A): The gene for INPP4A lies in the region 2q11.2 and an association to atopic asthma has been demonstrated. INPP4A is a magnesium independent phosphatase which negatively regulates PI3K-Akt signaling important for various pathophysiological pathways in asthma58.
11. CD 14: The gene for CD 14 receptor is located on chromosome 5q31.1 and this lipopolysaccharide receptor for endotoxin modulates the Th1-Th2 responses during early childhood. An association between C-159T functional polymorphism and asthma has been demonstrated49,50,91.
12. TNF- α and TNF-β: The genes for TNF- 0α and TNF – β have been localized within the MHC region on chromosome 6p21 and are major pro-inflammatory cytokines important in the pathogenesis of asthma. An association with polymorphisms has been associated with asthma in both atopic and non-atopic subjects and with elevated total serum IgE levels49,50,92,93.
13. Clara cell secretory protein (CC16): The genes for Clara cell secretory protein is localized on the chromosome 11q13 and encodes a 16kDa protein secreted from the Clara cells in the respiratory system. It is an important anti-inflammatory molecule limiting the synthesis of leucotrienes and prostaglandins and inhibits chemotaxis of inflammatory cells. An association with asthma has been demonstrated in both family-based and case-control studies50,94.
14. Uteroglobin related protein1(UGRP1): The genes for uteroglobin related protein 1 is localized on the chromosome 5q32 and encodes for a secretory protein in the airways with anti-inflammatory activity. Studies evaluating polymorphisms in URGP1 have demonstrated both association61 and lack of association with asthma62.
15. Transforming growth factor beta 1(TGF β1): The genes encoding for TGF β1 is localized on the chromosome 6q11-q2. TGF β1 is an important protein with both pro-inflammatory and anti-inflammatory properties. An association with asthma has been demonstrated and both increased protection and increased risk is seen with different haplotypes of the TGF β1 gene63.
16. Signal transducer and activator of transcription 6 (STAT6): The gene for STAT6 is located on chromosome 12q13. It is a member of the STAT family of transcription factors which plays a central role in IL-4 mediated biological responses. An association with asthma has been demonstrated in the Indian population49,95.
17. Mast cell chymase (CMA1): The gene for mast cell chymase is located on chromosome 14q11.2 and encodes for a serine protease expressed in mast cells and is important for inflammation and airway remodeling. An association has been observed with asthma and increased total IgE96.
18. N-acetyltransferase 2 (NAT2): The gene for NAT2 is located on chromosome 8p22 and is responsible for N-acetylation and influence susceptibility to atopic disorders. An association with asthma, increased total IgE and eosinophilia has been observed in the Indian population49,97,98.
19. Late cornified envelope like proline-rich1(LELP1): The gene for LELP1 is located in chromosome 1q21 and encompasses a small proline rich protein gene cluster and has been associated with atopy54.
20. Eotaxin(SCYA11): The gene for eotaxin is located on chromosome 17q21.1-q21.2 and encodes for the chemokine that is a specific attractant for eosinophils and has been implicated in asthma76.
21. Acid mammalian chitinase (CHIA): The gene for CHIA is localized on 1q13.1-21.3 and is important as an effector response for IL-13, shifts the inflammation towards Th2 and act as a chemo-attractant for inflammatory cell and has been associated with asthma53.
22. Interleukin-10(IL-10): The gene for IL-10 is located on chromosome 1q31-q32. IL-10 is an anti-inflammatory cytokine primarily produced by monocytes and macrophages and plays a key role in asthma56.
23. Interleukin -21 (IL-21): The gene for IL-21 is located on chromosome 4q26-q27 and encodes for a multifunctional cytokine which is produced by activated CD4+ T cells and affects growth and survival of numerous immune cells. It is important in asthma as it also regulates IgE production and has been implicated in asthma60.
24. Chemokine receptor 2 (CCR2): The gene for CCR 2 is localized on chromosome 3p21.31 and encodes for members of a large family of G protein-coupled receptors and plays an important role in asthma pathogenesis and has been implicated in the Indian population99.
These are the most common genes studied worldwide. The association of asthma with the remaining genes is less established and their study is restricted to a limited population.
Some of these genes may also be involved with other phenotypes such as helminthic infections (FcεRIβ and IL-4)1,41, COPD, cardiovascular diseases, congenital thrombotic thrombocytopenia (TTP), Crohn's disease (ADAM33)77,82, renal cell carcinoma, blood malignancies (PHF11)59, tuberculosis (TB), hyperparathyroidism, prostate cancer, insulin dependent diabetes mellitus (IDDM), leprosy and chronic hepatitis B infection (vitamin D receptor)70.
Conclusion and future prospects
Asthma is one of the most serious and intriguing allergic diseases. Asthma aggregates within families and is a complex multifactorial disease with the involvement of environment and genetic components. Our preliminary pedigree analysis revealed that autosomal recessive pattern of inheritance was prominent in asthma; parental consanguinity100 and serum intracellular cell adhesion molecule-1 (ICAM-1)101 was significantly associated with asthma, whereas the ABO blood system102, IL-4 and ADAM33 specific gene variants81, and serum E-selectin101 were not associated with asthma. More than 100 loci have been reported to be associated with asthma and there are also indications that mutation in a major gene can cause asthma. Due to an increasing number of current studies being done in genetics of asthma, there is an increasing list of inducer and inhibitor candidate genes for asthma. There are more than 100 candidate genes in every chromosome which are identified to have an association with asthma and the strength of association of these SNPs with asthma varies in different parts of the world. More studies are needed to determine the exact function of these genes, gene-gene interactions and the gene-environment interactions which are undoubtedly complex and remain elusive for the time being even with whole genome-wide association studies.
Further studies on asthma with the genomics data and tools, to map, identify the specific gene/s, and phenotype specific SNPs will help to unravel the pathways involved in asthma aetiology and employ pharmacogenomics to design better drugs for an individualized treatment plan. Thus with a fruitful interaction among researchers involved in pathophysiology, epidemiology, clinical research and genetics of asthma, this century holds promise for a better understanding of the pathology, diagnosis, prevention, treatment and management of asthma.
References
- 1.Palmer LJ, Cookson WOCM. Genomic approaches to understanding asthma. Genome Res. 2000;10:1280–7. doi: 10.1101/gr.143400. [DOI] [PubMed] [Google Scholar]
- 2.Lee YC, Cheon KT, Lee HB, Kim W, Rhee YK, Kim DS. Gene polymorphisms of endothelial nitric oxide synthase and angiotensin-converting enzyme in patient with asthma. Allergy. 2000;55:959–63. doi: 10.1034/j.1398-9995.2000.00724.x. [DOI] [PubMed] [Google Scholar]
- 3.Aron Y, Busson M, Polla BS, Lockhart A, Swierczewski E, Favatier F. Analysis of hsp70 gene polymorphism in allergic asthma. Allergy. 1999;54:165–70. doi: 10.1034/j.1398-9995.1999.00859.x. [DOI] [PubMed] [Google Scholar]
- 4.McFadden JER. Asthma. In: Fauci AS, Braunwald E, Kasper DL, Hauser SL, Longo DL, Jameson JL, editors. Harrison's principles of internal medicine. New York: McGrav-Hill; 2005. pp. 1508–16. [Google Scholar]
- 5.Xin X, Zhian F, Binyan W, Changzhong C, Wenwei G, Yongtang J, et al. A genomewide search for quantitative-trait loci underlying asthma. Am J Hum Genet. 2001;69:1271–7. doi: 10.1086/324650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Los H, Koppelman GH, Postma DS. The importance of genetic influences in asthma. Eur Respir J. 1999;14:1210–27. doi: 10.1183/09031936.99.14512109. [DOI] [PubMed] [Google Scholar]
- 7.Lin SJ, Chang LY, Yan DC, Huang YJ, Lin TJ, Lin TY. Decreased intercellular adhesion molecule-1 (CD54) and L-selectin (CD62L) expression on peripheral blood natural killer cells in asthmatic children with acute exacerbation. Allergy. 2003;58:67–71. doi: 10.1034/j.1398-9995.2003.t01-1-23697.x. [DOI] [PubMed] [Google Scholar]
- 8.Haagerup A, Bjerke T, Schiotz PO, Binderup HG, Dahl R, Kruse TA. Asthma and atopy-a total genome scan for susceptibility genes. Allergy. 2002;57:680–6. doi: 10.1034/j.1398-9995.2002.23523.x. [DOI] [PubMed] [Google Scholar]
- 9.Eden E, Mitchel D, Mehlman B, Khouli H, Nejat M, Grieco MH, et al. Atopy, asthma, and emphysema in patients with severe a-1-antitrypysin deficiency. Am J Respir Crit Care Med. 1997;156:68–74. doi: 10.1164/ajrccm.156.1.9508014. [DOI] [PubMed] [Google Scholar]
- 10.Liu AH, Spahn JD, Leung DYM. Nelson textbook of pediatrics. New Delhi: Elsevier; 2004. Childhood asthma; pp. 760–74. [Google Scholar]
- 11.Singh AB, Kumar P. Aeroallergens in clinical practice of allergy in India.An overview. Ann Agric Environ Med. 2003;10:131–6. [PubMed] [Google Scholar]
- 12.12 Lundback B. Epidemiology of rhinitis and asthma. Clin Exp Allergy. 1998;2(Suppl 2):3–10. [PubMed] [Google Scholar]
- 13.Yeatts K, Sly P, Shore S, Weiss S, Martinez F, Geller A, et al. A brief targeted review of susceptibility factors, environmental exposures, asthma incidence, and recommendations for future asthma. Environ Health Perspect. 2006;114:634–40. doi: 10.1289/ehp.8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masoli M, Fabian D, Holt S, Beasley R. Global burden of asthma. Southmpton: Medical Research Institute of New Zealand and University of Southampton; 2004. World map of prevalence of clinical asthma; pp. 12–5. [Google Scholar]
- 15.Bloom B, Cohen RA. Summary Health Statistics for U.S. Children: National Health Interview Survey, 2006. Vital Health Stat. 2006;10:42–6. [PubMed] [Google Scholar]
- 16.Shahzad K, Akhtar S, Mahmud S. Prevalence and determinants of asthma in adult male leather tannery workers in Karachi, Pakistan: A cross sectional study. BMC Public Health. 2006;6:292–9. doi: 10.1186/1471-2458-6-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stock S, Redaelli M, Luengen M, Wendland G, Civello D, Lauterbach KW. Asthma: prevalence and cost of illness. Eur Respir J. 2005;25:47–53. doi: 10.1183/09031936.04.00116203. [DOI] [PubMed] [Google Scholar]
- 18.The Collaborative Study on the Genetics of Asthma (CSGA). A genome-wide search for asthma susceptibility loci in ethnically diverse populations. Nat Genet. 1997;15:389–92. doi: 10.1038/ng0497-389. [DOI] [PubMed] [Google Scholar]
- 19.Palmer LJ, Cookson WOCM. Using single nucleotide polymorphisms as a means to understanding the pathophysiology of asthma. Respir Res. 2001;2:102–12. doi: 10.1186/rr45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cookson WOCM. Asthma genetics. Chest. 2002;121:7S–13S. doi: 10.1378/chest.121.3_suppl.7s-a. [DOI] [PubMed] [Google Scholar]
- 21.Jindal SK. Asthma control in the first decade of 21 century. Indian J Med Res. 2007;125:604–7. [PubMed] [Google Scholar]
- 22.Aggarwal AN, Chaudhry K, Chhabra SK, D’Souza GA, Gupta D, Jindal SK, et al. Prevalence and risk factors for bronchial asthma in Indian adults: A multicentre study. Indian J Chest Dis Allied Sci. 2006;48:13–22. [PubMed] [Google Scholar]
- 23.Chowgule RV, Shetye VM, Parmar JR, Bhosale AM, Khandagale MR, Phalnitkar SV, et al. Prevalence of respiratory symptoms, bronchial hyperreactivity, and asthma in a megacity: results of the European Community Respiratory Health Survey in Mumbai (Bombay) Am J Respir Crit Care Med. 1998;158:547–54. doi: 10.1164/ajrccm.158.2.9708064. [DOI] [PubMed] [Google Scholar]
- 24.Sharma SK, Banga A. Prevalence and risk factors for wheezing in children from rural areas of north India. Allergy Asthma Proc. 2007;28:647–53. doi: 10.2500/aap.2007.28.3059. [DOI] [PubMed] [Google Scholar]
- 25.Jaiswal A. Pets can cause asthma. Chandigarh Tribune. 2008. May 1-2, Available from: http://www.tribuneindia.com/2008/20080507/cth1.htm .
- 26.Bateman ED, Hurd SS, Barnes PJ, Bousquet J, Drazen JM, Fitz Geralde M, et al. Global strategy for asthma management and prevention: GINA executive summary. Eur Respir J. 2008;31:143–78. doi: 10.1183/09031936.00138707. [DOI] [PubMed] [Google Scholar]
- 27.Wiener AS, Zieve I, Fries JH. The inheritance of allergic disease. Ann Eugenics. 1936;7:141–62. [Google Scholar]
- 28.Cooke RA, Vander VA. Human sensitization. J Immunol. 1916;1:201–305. [Google Scholar]
- 29.Spain WC, Cooke RA. Studies in specific hypersensitiveness.The familial occurrence of hayfever and bronchial asthma. J Immunol. 1924;9:521–69. [Google Scholar]
- 30.Schwartz M. Heredity in bronchial asthma. Acta Allergol. 1952;5(Suppl II):1–19. [PubMed] [Google Scholar]
- 31.Sibbald B, Horn ME, Brain EA, Gregg I. Genetic factors in childhood asthma. Thorax. 1980;35:671–4. doi: 10.1136/thx.35.9.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duffy DL, Martin NG, Battistutta D, Hopper JL, Mat-hews JD. Genetics of asthma and hay fever in Australian twins. Am Rev Respir Dis. 1990;142:1351–8. doi: 10.1164/ajrccm/142.6_Pt_1.1351. [DOI] [PubMed] [Google Scholar]
- 33.Harris JR, Magnus P, Samuelsen SO, Tambs K. No evidence for effects of family environment on asthma.A retrospective study of Norwegian twins. Am J Respir Crit Care Med. 1997;156:43–9. doi: 10.1164/ajrccm.156.1.9609094. [DOI] [PubMed] [Google Scholar]
- 34.Laitinen T, Rasanen M, Kaprio J, Koskenvuo M, Laitinen LA. Importance of genetic factors in adolescent asthma. Am J Respir Crit Care Med. 1998;157:1073–8. doi: 10.1164/ajrccm.157.4.9704041. [DOI] [PubMed] [Google Scholar]
- 35.Huovinen E, Kaprio J, Laitinen LA, Koskenvuo M. Social predictors of adult asthma: a co-twin case-control study. Thorax. 2001;56:234–6. doi: 10.1136/thorax.56.3.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nystad W, Roysamb E, Magnus P, Tambs K, Harris JR. A comparison of genetic and environmental variance structures for asthma, hay fever and eczema with symptoms of the same diseases: a study of Norwegian twins. Int J Epidemiol. 2005;34:1302–9. doi: 10.1093/ije/dyi061. [DOI] [PubMed] [Google Scholar]
- 37.The European Community Respiratory Health Survey Group. Genes for asthma? An analysis of the European Community Respiratory Health Survey. Am J Respir Crit Care Med. 1997;156:1773–80. doi: 10.1164/ajrccm.156.6.9611068. [DOI] [PubMed] [Google Scholar]
- 38.Jenkins MA, Hopper JL, Giles GG. Regressive logistic modeling of familial aggregation for asthma in 7,394 population-based nuclear families. Genet Epidemiol. 1997;14:317–22. doi: 10.1002/(SICI)1098-2272(1997)14:3<317::AID-GEPI9>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 39.Holberg CJ, Elston RC, Halonen M, Wright AL, Taussing LM, Morgan WJ, et al. Segregation analysis of physician-diagnosed asthma in Hispanic and Non-Hispanic white families.A recessive component? Am J Respir Crit Care Med. 1996;154:144–50. doi: 10.1164/ajrccm.154.1.8680670. [DOI] [PubMed] [Google Scholar]
- 40.Palmer LJ, Cookson WOCM, James AL, Musk AW, Burton PR. Gibbs sampling-based segregation analysis of asthma-associated quantitative traits in a population-based sample of nuclear families. Genet Epidemiol. 2001;20:356–72. doi: 10.1002/gepi.6. [DOI] [PubMed] [Google Scholar]
- 41.Sheikh A. Itch, sneeze and wheeze: the genetics of atopic allergy. J R Soc Med. 2002;95:14–7. doi: 10.1258/jrsm.95.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patel AC, Morton JD, Kim EY, Alevy Y, Swanson S, Tucker J, et al. Genetic segregation of airway disease traits despite redundancy of chloride channel calcium-activated (CLCA) family members. Physiol Genomics. 2006;25:1–27. doi: 10.1152/physiolgenomics.00321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ober C, Cox NJ, Abney M, Rienzo AD, Lander ES, Changyaleket B, et al. Genome-wide search for asthma susceptibility loci in a founder population. Hum Mol Genet. 1998;7:1393–8. doi: 10.1093/hmg/7.9.1393. [DOI] [PubMed] [Google Scholar]
- 44.Pillai SG, Tang Y, Oord EV, Klotsman M, Barnes K, Carlsen K, et al. Factor analysis in the Genetics of Asthma International Network family study identifies five major quantitative asthma phenotypes. Clin Exp Allergy. 2008;38:421–9. doi: 10.1111/j.1365-2222.2007.02918.x. [DOI] [PubMed] [Google Scholar]
- 45.Kleeberger SR, Peden D. Gene-environment interactions in asthma and other respiratory diseases. Annu Rev Med. 2005;56:383–400. doi: 10.1146/annurev.med.56.062904.144908. [DOI] [PubMed] [Google Scholar]
- 46.Kumar A, Ghosh B. Genetics of asthma: a molecular biologist perspective. Clin Mol Allergy. 2009;7:7–16. doi: 10.1186/1476-7961-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bouzigon E, Corda E, Aschard H, Dizier MH, Boland A, Bousquet J, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med. 2008;359:1985–94. doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 48.Teerlink CC, Camp NJ, Bansal A, Crapo R, Hughes D, Kort E, et al. Significant evidence for linkage to chromosome 5q13 in a genome-wide scan for asthma in an extended pedigree resource. Eur J Hum Genet. 2009;17:636–43. doi: 10.1038/ejhg.2008.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malerba G, Pignatti PF. A review of asthma genetics: gene expression studies and recent candidates. J Appl Genet. 2005;46:93–104. [PubMed] [Google Scholar]
- 50.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nature Rev Immunol. 2008;8:169–82. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 51.Kamada F, Suzuki Y, Shao C, Tamari M, Hasegawa K, Hirota T, et al. Association of the hCLCA1 gene with childhood and adult asthma. Genes Immunity. 2004;5:540–7. doi: 10.1038/sj.gene.6364124. [DOI] [PubMed] [Google Scholar]
- 52.Manuel ARF, Gorman LO, Souef PL, Burton PR. Robust estimation of experimentwise P values applied to a genome scan of multiple asthma traits identifies a new region on significant linkage on chromosome 20q13. Am J Hum Genet. 2005;77:1075–85. doi: 10.1086/497997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chatterjee R, Batra J, Das S, Sharma SK, Ghosh B. Genetic association of acidic mammalian chitinase with atopic asthma and serum total IgE levels. J Allergy Clin Immunol. 2008;122:202–8. doi: 10.1016/j.jaci.2008.04.030. [DOI] [PubMed] [Google Scholar]
- 54.Sharma M, Mehla K, Batra J, Ghosh B. Association of a chromosome 1q21 locus in close proximity to a late cornified envelope-like proline-rich 1 (LELP1) gene with total serum IgE levels. J Hum Genet. 2007;52:378–83. doi: 10.1007/s10038-007-0118-5. [DOI] [PubMed] [Google Scholar]
- 55.Hakonarson H, Bjornsdottir US, Halapi E, Palsson S, Adalsteinsdottir E, Gislason D, et al. A major susceptibility gene for asthma maps to chromosome 14q24. Am J Hum Genet. 2002;71:483–91. doi: 10.1086/342205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chatterjee R, Batra J, Kumar A, Mabalirajan U, Nahid S, Niphadkar PV, et al. Interleukin-10 promoter polymorphisms and atopic asthma in north Indians. Clin Exp Allergy. 2005;35:914–9. doi: 10.1111/j.1365-2222.2005.02273.x. [DOI] [PubMed] [Google Scholar]
- 57.Postma1 DS, Koppelman GH. Genetics of asthma: Where are we and where do we go? Proc Am Thorac Soc. 2009;6:283–7. doi: 10.1513/pats.200806-047RM. [DOI] [PubMed] [Google Scholar]
- 58.Sharma M, Batra J, Mabalirajan U, Sharma S, Nagarkatti R, Aich J, et al. A genetic variation in inositol polyphosphate 4 phosphatase a enhances susceptibility to asthma. Am J Respir Crit Care Med. 2008;177:712–9. doi: 10.1164/rccm.200705-781OC. [DOI] [PubMed] [Google Scholar]
- 59.Weiss ST, Raby BA. Asthma genetics 2003. Hum Mol Genet. 2004;13:83–9. doi: 10.1093/hmg/ddh080. [DOI] [PubMed] [Google Scholar]
- 60.Chatterjee R, Batra J, Ghosh B. A common exonic variant of interleukin21 confers susceptibility to atopic asthma. Int Arch Allergy Immunol. 2009;148:137–46. doi: 10.1159/000155744. [DOI] [PubMed] [Google Scholar]
- 61.Inoue K, Wang X, Saito J, Tanino Y, Ishida T, Iwaki D, et al. Plasma UGRP1 levels associate with promoter G-112A polymorphism and the severity of asthma. Allergol Int. 2008;57:57–64. doi: 10.2332/allergolint.O-07-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Batra J, Pramod VN, Sharma SK, Ghosh B. Uteroglobin-related protein 1(UGRP1) gene polymorphisms and atopic asthma in the Indian population. Int Arch Allergy Immunol. 2005;136:1–6. doi: 10.1159/000082578. [DOI] [PubMed] [Google Scholar]
- 63.Nagpal K, Sharma S, Rao CB, Nahid S, Niphadkar PV, Sharma SK, et al. TGF beta1 haplotypes and asthma in Indian populations. Allergy Clin Immunol. 2005;115:527–33. doi: 10.1016/j.jaci.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 64.Malerba G, Lindgren CM, Xumerle L, Kiviluoma P, Trabetti E, Laitinen T, et al. Chromosome 7p linkage and GPR154 gene association in Italian families with allergic asthma. Clin Exp Allergy. 2007;37:83–9. doi: 10.1111/j.1365-2222.2006.02615.x. [DOI] [PubMed] [Google Scholar]
- 65.Shyur S, Wang J, Lin CG, Hsiao Y, Liou Y, Wu Y, et al. The polymorphisms of protein-tyrosine phosphatase receptor-type delta gene and its association with pediatric asthma in the Taiwanese population. Eur J Hum Genet. 2008;16:1283–8. doi: 10.1038/ejhg.2008.79. [DOI] [PubMed] [Google Scholar]
- 66.Adachi T, Hanaka S, Masuda T, Yoshihara H, Nagase H, Ohta K. Transduction of phosphatase and tensin homolog deleted on chromosome 10 into eosinophils attenuates survival, chemotaxis, and airway inflammation. J Immunol. 2007;8:105–11. doi: 10.4049/jimmunol.179.12.8105. [DOI] [PubMed] [Google Scholar]
- 67.Sharma S, Ghosh B. Association of an intragenic microsatellite marker in the CC16 gene with asthma in the Indian population. J Hum Genet. 2004;49:677–83. doi: 10.1007/s10038-004-0206-8. [DOI] [PubMed] [Google Scholar]
- 68.Tianpen C, Lin W, Jianmin W, Lihua H, Jungang X. Polymorphisms of IL-4, IL-4Rα, and AICDA genes in adult allergic. J Huazhang U Sci Med Sci. 2003;23:134–7. doi: 10.1007/BF02859936. [DOI] [PubMed] [Google Scholar]
- 69.Nagarkatti R, Rao CB, Rishi JP, Chetiwal R, Shandilya V, Vijayan V, et al. Association of IFNG gene polymorphism with asthma in the Indian population. J Allergy Clin Immunol. 2002;110:410–2. doi: 10.1067/mai.2002.127859. [DOI] [PubMed] [Google Scholar]
- 70.Zhang Y, Leaves NI, Anderson GG, Ponting CP, Broxholme J, Holt R, et al. Positional cloning of a quantitative trait locus on chromosome 13q14 that influences immunoglobulin E levels and asthma. Nat Genet. 2003;34:181–6. doi: 10.1038/ng1166. [DOI] [PubMed] [Google Scholar]
- 71.Haagerup A, Børglum AD, Binderup HG, Kruse TA. Fine-scale mapping of type I allergy candidate loci suggests central susceptibility genes on chromosomes 3q, 4q and Xp. Allergy. 2004;59:88–94. doi: 10.1111/j.1398-9995.2004.00294.x. [DOI] [PubMed] [Google Scholar]
- 72.Risma KA, Wang N, Andrews RP, Cunningham CM, Ericksen MB, Bernstein JA, et al. V75R576 IL-4 receptor is associated with allergic asthma and enhanced il0 -4 receptor function. J Immunol. 2002;169:1604–10. doi: 10.4049/jimmunol.169.3.1604. [DOI] [PubMed] [Google Scholar]
- 73.Ivanov VP, Solodilova MA, Polonikov AV, Khoroshaya IV, Kozhuhov MA, Panfilov VI. Association analysis of C242T and A640G polymorphisms in the gene for p22phox subunit of NADPH oxidase with the risk of bronchial asthma: A pilot study. Russ J Genet. 2008;44:601–8. [PubMed] [Google Scholar]
- 74.Pinto LA, Stein RT, Kabesch M. Impact of genetics in childhood asthma. J Pediatr (Rio J) 2008;84(4 Suppl):S68–75. doi: 10.2223/JPED.1781. [DOI] [PubMed] [Google Scholar]
- 75.Wjst M, Lichtner P, Meitinger T, Grimbacher B. STAT3 single-nucleotide polymorphisms and STAT3 mutations associated with hyper-IgE syndrome are not responsible for increased serum IgE serum levels in asthma families. Eur J Hum Genet. 2009;17:352–6. doi: 10.1038/ejhg.2008.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Batra J, Rajpoot R, Ahluwalia J, Devarapu SK, Sharma SK, Dinda AK, et al. A hexanucleotide repeat upstream of eotaxin gene promoter is associated with asthma, serum total IgE and plasma eotaxin levels. J Med Genet. 2007;44:397–403. doi: 10.1136/jmg.2006.046607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stephen TH, Yang Y, Haitchi HM, Powell RM, Holloway JW, Yoshisue H, et al. The genetics of asthma, ADAM33 as an example of a susceptibility gene. Proc Am Thor Soc. 2006;3:440–3. doi: 10.1513/pats.200603-026AW. [DOI] [PubMed] [Google Scholar]
- 78.Cleo CD, Postma DS, Vonk JM, Bruinenberg M, Schouten JP, Boezen HM. A disintegrin and metalloprotease 33 polymorphisms and lung function decline in the general population. Am J Respir Crit Care Med. 2005;172:329–33. doi: 10.1164/rccm.200411-1486OC. [DOI] [PubMed] [Google Scholar]
- 79.Holgate ST, Davies DE, Murphy G, Powell RM, Holloway JW. ADAM 33: just another asthma gene or a breakthrough in understanding the origins of bronchial hyperresponsiveness? Thorax. 2003;58:466–9. doi: 10.1136/thorax.58.6.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Angela S, Nikolas M, Francine J, Julie AC, Lesley AL, Stephen TH, et al. Polymorphisms in A Disintegrin and Metalloprotease 33 (ADAM33) predict impaired early-life lung function. Am J Respir Crit Care Med. 2005;172:55–60. doi: 10.1164/rccm.200412-1708OC. [DOI] [PubMed] [Google Scholar]
- 81.Bijanzadeh M, Ramachandra NB, Mahesh PA, Savitha MR, Kumar P, Manjunath BS, et al. Association of IL-4 and ADAM33 gene polymorphisms with asthma in an Indian population. Lung. 2010;188:415–22. doi: 10.1007/s00408-010-9247-2. [DOI] [PubMed] [Google Scholar]
- 82.DeGuzman B, Duck W, Myrick M, Proctor S, Sashti N, Weber MB. Emory: University School of Public Health; 2001. Atopic Asthma and Interleukin-4; pp. 2–4. [Google Scholar]
- 83.Chiang CH, Tang YC, Lin MW, Chung MT. Association between the IL-4 promoter polymorphisms and asthma or severity of hyperresponsiveness in Taiwanese. Respirology. 2007;12:42–8. doi: 10.1111/j.1440-1843.2006.00960.x. [DOI] [PubMed] [Google Scholar]
- 84.Kabesch M, Tzotcheva I, Carr D, Höfler C, Weiland SK, Fritzsch C, et al. A complete screening of the IL4 gene, novel polymorphisms and their association with asthma and IgE in childhood. J Allergy Clin Immunol. 2003;112:893–8. doi: 10.1016/j.jaci.2003.08.033. [DOI] [PubMed] [Google Scholar]
- 85.Nagarkatti R, Kumar R, Sharma SK, Ghosh B. Association of IL4 gene polymorphisms with asthma in north Indians. Int Arch Allergy Immunol. 2004;134:206–12. doi: 10.1159/000078767. [DOI] [PubMed] [Google Scholar]
- 86.Sharma S, Nagarkatti R, Rao CB, Niphadkar PV, Vijayan V, Sharma SK, et al. A_16_C haplotype in the FceRIb gene confers a higher risk for atopic asthma in the Indian population. Clin Genet. 2004;66:417–25. doi: 10.1111/j.1399-0004.2004.00333.x. [DOI] [PubMed] [Google Scholar]
- 87.Sharma S, Ghosh B. Promoter polymorphism in the MS4A2 gene and asthma in the Indian population. Int Arch Allergy Immunol. 2009;149:208–18. doi: 10.1159/000199716. [DOI] [PubMed] [Google Scholar]
- 88.Nagarkatti R, Ghosh B. Identification of single-nucleotide and repeat polymorphisms in two candidate genes, interleukin 4 receptor (IL4RA) and signal transducer and activator of transcription protein 6 (STAT6), for Th2-mediated diseases. J Hum Genet. 2002;47:684–7. doi: 10.1007/s100380200105. [DOI] [PubMed] [Google Scholar]
- 89.Batra J, Singh TP, Mabalirajan U, Sinha A, Prasad R, Ghosh B. Association of inducible nitric oxide synthase with asthma severity, total serum immunoglobulin E and blood eosinophil levels. Thorax. 2007;62:16–22. doi: 10.1136/thx.2006.057935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dizier MH, Besse-Schmittler C, Guilloud-Bataille M, Annesi-Maesano I, Boussaha M, Bousquet J, et al. Genome screen for asthma and related phenotypes in the French EGEA study. Am J Respir Crit Care Med. 2000;162:1812–8. doi: 10.1164/ajrccm.162.5.2002113. [DOI] [PubMed] [Google Scholar]
- 91.Sharma M, Batra J, Mabalirajan U, Goswami S, Ganguly D, Lahkar B, et al. Suggestive evidence of association of C-159T functional polymorphism of the CD14 gene with atopic asthma in northern and northwestern Indian populations. Immunogenetics. 2004;56:544–7. doi: 10.1007/s00251-004-0721-y. [DOI] [PubMed] [Google Scholar]
- 92.Shin HD, Park BL, Kim LH, Jung JH, Wang HJ, Kim YJ, et al. Association of tumor necrosis factor polymorphisms with asthma and serum total IgE. Hum Mol Genet. 2004;13:397–403. doi: 10.1093/hmg/ddh036. [DOI] [PubMed] [Google Scholar]
- 93.Sharma S, Sharma A, Kumar S, Sharma SK, Ghosh B. Association of TNF haplotypes with asthma, serum IgE levels, and correlation with serum TNF-alpha levels. Am J Respir Cell Mol Biol. 2006;35:488–95. doi: 10.1165/rcmb.2006-0084OC. [DOI] [PubMed] [Google Scholar]
- 94.Sharma S, Ghosh B. Association of an intragenic microsatellite marker in the CC16 gene with asthma in the Indian population. J Hum Genet. 2004;49:677–83. doi: 10.1007/s10038-004-0206-8. [DOI] [PubMed] [Google Scholar]
- 95.Nagarkatti R, Rao CB, Vijayan V, Sharma SK, Ghosh B. Signal transducer and activator of transcription 6 haplotypes and asthma in the Indian population. Am J Respir Cell Mol Biol. 2004;31:317–21. doi: 10.1165/rcmb.2003-0128OC. [DOI] [PubMed] [Google Scholar]
- 96.Sharma S, Rajan UM, Kumar A, Soni A, Ghosh B. A novel (TG)n(GA)m repeat polymorphism 254 bp downstream of the mast cell chymase (CMA1) gene is associated with atopic asthma and total serum IgE levels. J Hum Genet. 2005;50:276–82. doi: 10.1007/s10038-005-0252-x. [DOI] [PubMed] [Google Scholar]
- 97.Batra J, Sharma SK, Ghosh B. Arylamine N-acetyltransferase gene polymorphisms: markers for atopic asthma, serum IgE and blood eosinophil counts. Pharmacogenomics. 2006;7:673–82. doi: 10.2217/14622416.7.5.673. [DOI] [PubMed] [Google Scholar]
- 98.Batra J, Ghosh B. N-acetyltransferases as markers for asthma and allergic/atopic disorders. Curr Drug Metab. 2008;9:546–53. doi: 10.2174/138920008784892074. [DOI] [PubMed] [Google Scholar]
- 99.Batra J, Ghosh B. Genetic contribution of chemokine receptor 2 (CCR2) polymorphisms towards increased serum total IgE levels in Indian asthmatics. Genomics. 2009;94:161–8. doi: 10.1016/j.ygeno.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 100.Bijanzadeh M, Mahesh PA, Savitha MR, Kumar P, Jayaraj BS, Ramachandra NB. Inheritance patterns, consanguinity & risk for asthma. Indian J Med Res. 2010;132:48–55. [PubMed] [Google Scholar]
- 101.Bijanzadeh M, Ramachandra NB, Mahesh PA, Savitha MR, Vijayakumar GS, Kumar P, et al. Soluble intercellular adhesion molecule-1 and E-selectin in patients with asthma exacerbation. Lung. 2009;187:315–20. doi: 10.1007/s00408-009-9171-5. [DOI] [PubMed] [Google Scholar]
- 102.Bijanzadeh M, Ramachandra NB, Mahesh PA, Savitha MR, Manjunath BS, Jayaraj BS. Lack of association between asthma and ABO blood group. Lung. 2009;187:389–92. doi: 10.1007/s00408-009-9175-1. [DOI] [PubMed] [Google Scholar]
- 103.Poon AH, Laprise C, Lemire M, Montpetit A, Sinnett D, Schurr E, et al. Association of vitamin D receptor genetic variants with susceptibility to asthma and atopy. Am J Respir Crit Care Med. 2004;170:967–73. doi: 10.1164/rccm.200403-412OC. [DOI] [PubMed] [Google Scholar]


