Abstract
Background & objectives:
Intravenous device (IVD) associated nosocomial blood stream infections due to staphylococci are major cause of morbidity and mortality. The present study was carried out to assess the frequency of staphylococcal IVD associated infections in a paediatric ward of a tertiary case hospital. Prevalence of resistance to commonly used antimicrobials in hospital acquired staphylococcal isolates was also tested.
Methods:
Children admitted in paediatric wards with IVD for more than 48 h were enrolled. Blood, IVD tip at the time of removal, skin swab at the site of insertion of IVD and nasal swab were collected and cultured by standard protocol. All staphylococcal isolates from any source were analyzed for antimicrobial susceptibility by disk diffusion method. Genotyping matching of those staphylococcal isolates was done which were isolated from different sites of the same patient, but were phonotypically similar. Genotype of blood isolate was compared with genotype of isolate from nose/IVD/skin.
Results:
Staphylococcus aureus was the most frequent blood isolate (8.7%) followed by Candida (2.9%), coagulase negative staphylococci (CoNS 2.6%), Pseudomonas spp. (0.4%), Klebsiella spp. (0.3%) and Escherichia coli (0.1%). Isolation of microorganisms from blood was significantly higher in patients whose skin, IVD and nose were colonized by same microorganism (P<0.001). None of the staphylococcal isolate was found to be resistant to glycopeptides (vancomycin and teicoplanin). High penicillin and oxacillin resistance was present in both S. aureus (penicillin resistance; 76.8%, oxacillin resistance; 66.7%) and CoNS (penicillin resistance; 73.3%, oxacillin resistance; 60.0%). Among CoNS biotypes, S. haemolyticus was commonest blood isolate while S. epidermidis was commonest isolate from Skin/nose. Only 33.3 per cent of S. aureus blood stream infections and most of S. epidermidis and S. haemolyticus blood infections were IVD associated.
Interpretation & conclusions:
Staphylococci were the major causative agent of nosocomial blood stream infections. All episodes of septicaemia due to S. epidermidis and S. haemolyticus were IVD associated while only 1/3 of S. aureus septicaemia was IVD associated.
Keywords: Coagulase negative staphylococci, hospital acquired septicaemia, IVD associated staphylococcal septicaemia, Staphylococcus aureus
Nosocomial blood stream infections (BSIs) have been identified as one of the most frequent nosocomial infections in paediatric patients1–3. The risk of nosocomial infections depends on the host characteristics, the number of interventions, invasive procedures, asepsis of techniques, the duration of stay in the hospital and inappropriate use of antimicrobials. Most often the endogenous flora of the patient, which may be altered because of hospitalization, is responsible for nosocomial infections4. Many microorganisms are responsible for nosocomial septicaemia, commonest being Gram-positive cocci primarily staphylococci. These are commensal on human body surfaces and colonize intravenous devices, which become a focus of infection in hospitalized individuals more so in immuno-compromised patients. Colonizing microorganisms may enter into blood stream and cause septicaemia5. The emergence of antibiotic resistant strains of staphylococci is considered a major problem in most hospitals. Large proportions of nosocomial staphylococcal strains are resistant to penicillin. Centres for Disease Control and Prevention reported that the methicillin resistant staphylococcal strains are resistant to several other antibiotics in both large and small hospitals6.
We have noticed that staphylococci were the commonest cause of nosocomial septicaemia in our paediatric ward (unpublished observation). Therefore, the present investigation was planned to study the frequency of staphylococcal intravenous device (IVD) associated infections in children admitted in paediatric ward of a tertiary care hospital in north India. Percentage of resistance to commonly used antimicrobials in our hospital acquired staphylococcal strains was also tested.
Material & Methods
The study was done in Chhatrapati Shahuji Maharaj Medical University (CSMMU), a tertiary care hospital at Lucknow, Uttar Pradesh, from January 2003 to January 2006. All consecutive children admitted in paediatric wards with IVD in peripheral vein for more than 48 h, were followed up and if they developed fever (>99 °F) or had >1 °F rise in existing body temperature, they were enrolled. From each patient, venous blood, IVD tip at the time of removal, skin swab from the site of IVD insertion and nasal swab were collected for culture. Skin swabs were collected from the site of catheter insertion by rolling the wet swab (dipped in sterile saline) and were transported into screw-capped tube. Nasal swab were collected from anterior nasal chamber by rolling the swab and transported into screw-capped tube. Two ml of venous blood was collected from each child and aseptically transported into screw- capped blood culture bottle containing 10 ml brain heart infusion (BHI) broth with 0.05 per cent sodium polyanethol sulphonate (SPS). Tip of each catheter (1.5 inches) was clipped with a sterile scissor and taken into a sterile screw capped vial. All the samples were sent to the Microbiology laboratory of CSMMU without delay for culture. The study protocol was approved by the ethics committee and informed consent was taken from guardian of each patient.
Culture of the samples: Skin and nasal swabs were inoculated on to 5 per cent sheep blood agar (SBA). All the catheter tips were processed on 5 per cent sheep blood agar by the blood agar roll techniques for semi- quantitative culture by the method of Maki et al7. A colony count of > 15 on SBA was considered significant and further processed. All the inoculated plates were incubated aerobically at 37°C for 24 h; if plates showed no growth after 24 h these were further incubated for 24 h. The blood containing brain heart infusion broth was incubated overnight at 37°C and subcultures were done on 5 per cent sheep blood agar. Inoculated plates were incubated aerobically at 37°C for 24 h. In case first culture was sterile, final subculture was done on appearance of turbidity or on 7th day of incubation, whichever was earlier.
Identification: Smears were prepared from plates showing growth and stained by Gram's stain. Gram-positive cocci in cluster were tested by modified oxidase, catalase and slide and tube coagulase tests. Oxidase negative, catalase positive and coagulase positive strains were Staphylococcus aureus. Oxidase negative, catalase positive and coagulase negative strains were coagulase negative staphylococci (CoNS). CoNS were further biotyped for species identification by using Kloos & Bannerman scheme8.
Antibiotic susceptibility test (AST): AST of all staphylococcal isolates from all the sources was performed as per Clinical Laboratory Standards Institute (CLSI) guidelines9, using Muller-Hinton agar (MHA) (Hi-media, Mumbai, India). The following antibiotic discs, procured from Hi-media (Mumbai) were used; penicillin (10 U), oxacillin (1 μg), vancomycin (30 μg), teikoplanin (30 μg), cefazolin (30 μg), ciprofloxacin (30 μg). A standardized inoculum matching 0.5 McFarland density gradient was swabbed onto the surface of agar plate three times, rotating plate 60 degrees to ensure even distribution. After 5 min of inoculation, discs were applied to plate by sterile forceps and incubated at 37°C for 24 h. A report of resistant, intermediate or sensitive was recorded as per CLSI recommendations9. Standard strain of S. aureus ATCC 29213 was used as a control strain.
Genotyping of isolates: Staphylococcal isolates from those patients whose blood culture was positive for staphylococci were subjected to genotyping10. Genotype of blood isolate was compared with genotype of isolate from nose/IVD/skin. Genotyping by random amplification of polymorphic DNA (RAPD)10 was performed only if blood isolate was phenotypically similar (biotyping, AST pattern) to isolate from any other site (IVD/nose/skin).
Extraction of DNA: Briefly, four to five discrete colonies of Staphylococcus sp. were suspended in 200 μl lysis buffer (Tris 10 mM, EDTA 2 mM, NaCl 0.4 M and Triton X-100 0.5%) and boiled for 20 min. Ten μg of proteinase K was added. The sample was vortexed and incubated at 56°C for 2 h followed by boiling at 100°C for 10 min to inactivate proteinase K. DNA purification was done by addition of equal volume of phenol: chloroform (24:1) followed by chloroform only. Aqueous phase was finally transferred in 2.5 volume of ethanol and 10 μl sodium acetate (0.3 M final conc.) was added. The tubes were kept at 20°C overnight. The sample was centrifuged at 10,000 g for 10 min and the pellet was washed with chilled 70 per cent ethanol. The pellet was allowed to air dry and finally suspended in 25 μl of sterile triple distilled water for RAPD analysis11.
RAPD analysis: Three different random amplification primers were used (primer 10265’-TACATTCGAGGACCCCTAAGTG-3’, primer 12045’-ATGTAAGCTCCTGGGGATTCAC-3’and primer 10145’-AAGTAAGTGACTGGGGTGAGCG-3’)10. PCR mixture consisted of 10 mm Tris HCl (pH 9.0), 50 mM KCl, 2.5 mM MgCl2, 0.01% gelatin, 100 pico moles of each primer and triton X-100 (0.01%). Deoxyribonucleotide triphosphates (Banglore Genei, India) were used at a final concentration of 0.2 mM. For each reaction, 0.5 U Taq DNA polymerase (Banglore Genei, India) was added. PCR consisted of 40 cycles of consecutive denaturation, annealing and DNA chain extension (1 min at 94°C, 1 min at 25°C, 2 min at 74°C) in a thermer cycler (Techne Progene, USA). Amplicons were analyzed on 1.5 per cent agarose gel electrophoresis and photographed10. Amplified products were analyzed on 1.5 per cent agarose gel electrophoresis. Isolates showing 100 per cent similar bands were considered identical.
Statistical analysis: Statistical analysis was done by Chi-square test for significance of difference in isolation of microorganisms from blood (septicaemia) of the patients whose skin, IVD and nose was colonized by same microorganism. The data were analyzed by using SPSS software ‘version 10’ on windows XP. P<0.05 was considered significant12.
Results
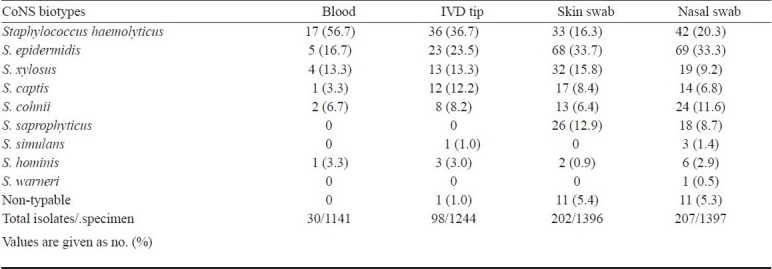
Total samples collected from 1399 hospitalized children were, 1141 venous blood, 1244 IVD, 1396 skin swabs and 1397 nasal swabs. S. aureus was the most frequent isolate; a total of 8.7 per cent venous blood (99/1141), 16.5 per cent nasal swab (231/1397) 13.6 per cent skin swab (190/1396) and 13 per cent catheter tip (162/1244) were positive for S. aureus (Table I). Coagulase negative staphylococci (CoNS) were second commonest bacterial isolate; S. haemolyticus was the most frequent isolate from blood and IVD while S. epidermidis was commonest from skin and nasal swab. The detailed distribution of CoNS species in different samples is shown in Table II. Candida was second most frequent isolate followed by CoNS, Pseudomonas spp., Klebsiella spp., and Escherichia coli (Table I). Significance of difference in isolation of microorganisms from blood (septicaemia) of the patients whose skin, IVD and nose was colonized by same microorganism was studied and it was seen that isolation of microorganisms from blood (septicaemia) was significantly higher in those patients whose skin/ IVD and/or nose was colonized by same microorganism (P<0.001).
Table I.
Isolation rate of microorganisms from hospitalized children with clinical suspicion of hospital acquired septicemia
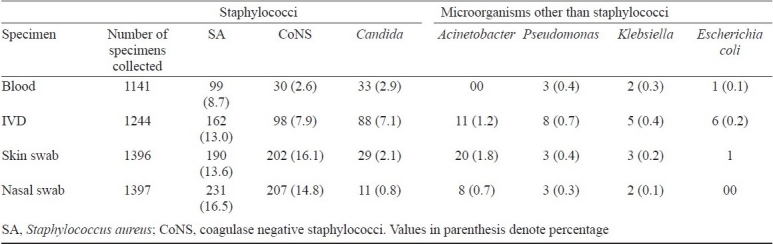
Table II.
Distribution of coagulase negative staphylococci (CoNS) species in different samples collected from children with clinical suspicion of hospital acquired septicaemia
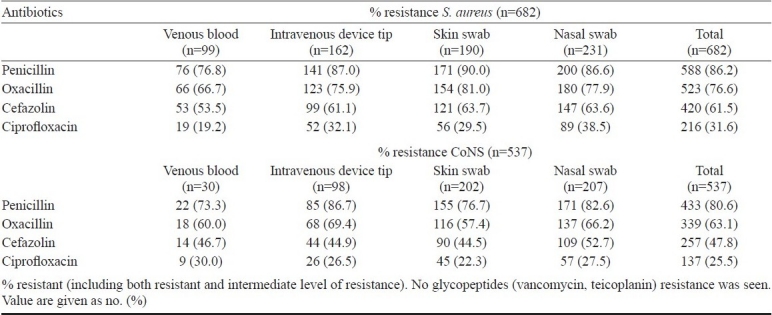
Antimicrobial resistance (AMR) pattern: Antimicrobial sensitivity testing of S. aureus (n=682) and CoNS (n=537) isolates was done. None of the isolate was found to be resistant to glycopeptides (vancomycin and teicoplanin). Penicillin resistance was high in both S. aureus 86.2 per cent (588/682) and CoNS 80.6 per cent (433/537). A total of 76.7 per cent of S. aureus isolates were resistance to oxacillin, 61.5 per cent to cefazolin and 31.6 per cent to ciprofloxacin (Table III). Among CoNS isolates, 63.1 per cent were resistant to oxacillin, 47.8 per cent to cefazolin and 25.5 per cent to ciprofloxacin.
Table III.
Antimicrobial resistance pattern of S. aureus and CoNS isolated from different clinical samples from children with clinical suspicion of hospital acquired septicaemia
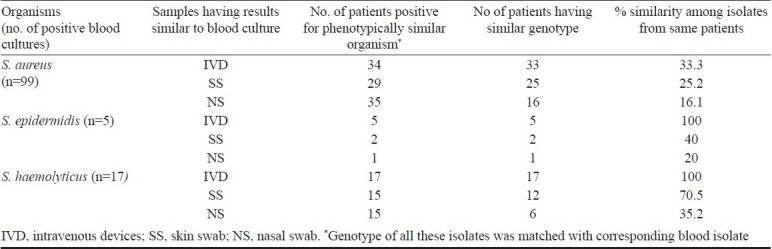
RAPD analysis: RAPD assay was performed to analyze the similarity among the staphylococcal isolates from different specimens of the same patient. Staphylococcal isolates (S. aureus, S. epidermidis and S. haemolyticus) from those subjects whose blood cultures were positive for phenotypically similar staphylococci (similar biotype and AMR pattern) were studied by RAPD. Of the 99 patients whose blood culture was positive for S. aureus, 34 were also having IVD culture positive for phenotypically similar S. aureus, of which 33 blood and IVD isolates were also genetically similar; showing that 33.3 per cent episodes of S. aureus blood stream infections are IVD associated. Twenty nine patients were also having skin swabs positive for phenotypically similar S. aureus, of which 25 were also genetically similar to blood isolates. Thirty four patients were having nasal swabs positive for phenotypically similar S. aureus, of which 16 strains were genotypically identical to blood isolate (Table IV). Five patients had S. epidermidis isolated from their blood. All of them had genotypically similar isolate from IVD while only 2 had same isolate from skin and only one had same isolate from nose. Of the 17 patients whose blood culture was positive for S. haemolyticus, all had genotypically similar isolate from IVD. Only 12 of them had same isolate from skin and only 6 of them had same isolate from nose (Table IV).
Table IV.
RAPD analysis result of common isolates from children with clinical suspicion of hospital acquired septicaemia
Discussion
Nosocomial blood stream infections (BSI) are one of the most serious and potentially life threatening infectious diseases in paediatric patients. Early diagnosis and therapy are essential for the prevention of morbidity and mortality13. The accurate prediction of likely pathogens and antimicrobial resistance pattern is crucial for successful therapy. In the present study, S. aureus was found to be the most frequent blood isolate followed by Candida, CoNS, Pseudomonas spp., Klebsiella spp. and E. coli. Among CoNS, S. haemolyticus was commonest blood isolate followed by S. epidermidis. Berner et al14, retrospectively analyzed 1037 bacteraemic episodes in children in a German tertiary care center during a 10 years period and noted that Gram-positive bacteria accounted for two third of all bacteraemic episodes in paediatric patients. In another study13 131 episodes of blood stream infection in a paediatric ICU in the UK in a 3 year period were studied. They found that Gram-positive and Gram-negative bacteria accounted for 63 and 31 per cent respectively, 6 per cent were yeast. US medical centers published the data on blood stream infection in 1991 and 1997 focusing on paediatric patients14. The key findings were that significant increase in the overall blood stream infection rate was caused by each of the following pathogens groups: CoNS, S. aureus, enterococci and Candida species. In contrast, the blood stream infection rate caused by Gram-negative bacilli remained stable during the decade. They reported that the greatest increase in blood stream infection rates was observed in coagulase negative staphylococci followed by Candida spp. S. aureus colonization varied from 8.0 to 71.2 per cent. Candida isolation reported from various specimens varies from 3.2 to 26.9 per cent15,16. Studies done during 1986 to 2002 reported that the ratio of S. aureus to CoNS was around 1:117,18 and majority of nosocomial CoNS isolates were obtained from blood samples, catheters and wounds indicating that CoNS have increasingly been recognized as important agents of nosocomial infection. S. epidermidis and S. haemolyticus were the most responsible CoNS species for septicaemia in paediatric patients19, while in another study S. epidermidis was reported most frequent blood isolate followed by S. haemolyticus20.
In our study none of the staphylococcal isolates was found to be resistant to glycopeptides (vancomycin and teicoplanin) while high penicillin and oxacillin resistance was present in both S. aureus and CoNS. Silvia et al21 have also reported reduced susceptibility to glycopeptides in 5.4 per cent of CoNS, isolated from blood samples of critically ill haematology patients. Reduced susceptibility to teicoplanin in blood CoNS isolates was also shown22. Resistance to methicillin among S. aureus increased from 1.5 per cent in 1986 to 31.2 per cent in 200218. This also implies an increase in resistance to other antimicrobials such as macrolides, lincosamides, aminoglycosides, and quinolones. All S. aureus isolates have remained uniformly susceptible to glycopeptides and novel antimicrobials (linezolid, and quinupristin/dalfopristin)18.
With regard to CoNS, the increase in resistance to methicillin was even greater, reaching to 61.3%. In most cases, this was associated with resistance to multiple antimicrobials18. Antimicrobial resistance pattern of staphylococci varied from study to study. High resistance to penicillin (80 - 90.6% in CNS & 78.6 - 88.9% in S. aureus), oxacillin (20.8 - 98.4% in CNS & 24.5 - 87.2% in S. aureus), cefazolin (2.7 - 80% in CNS & 1.1 - 23.7% in S. aureus) and ciprofloxacin (11 - 66% in CNS 13.1 - 25% in S. aureus) was reported2,23–25. In the present study, cefazolin resistance was slightly lower than oxacillin resistance. Clinically all oxacillin resistant strains are resistant to all betalactams and should not be treated with penicillins or cephalosporins. All strains showing discrepancy in results were tested by us for detection of mecA gene and other phenotypic methods28. It was noted that oxacillin disc diffusion test is less specific than mec A detection method. Except occasional reports of vancomycin resistant staphylococci, maximum number of studies had reported 0 per cent prevalence of vancomycin resistant staphylococci23,26,27. Difference in (i) prescription practices in the hospitals28, (ii) geographical locations2,29,3, (iii) methods of AMR testing29,30, (iv) sample size24,31, (v) types of clinical samples are different reasons for these variations3,31.
It was found that 1/3 of S. aureus blood isolates and 100 per cent of CoNS blood isolates were identical to IVD isolate .Gemme et al32 reported that in young infants 65 per cent of CoNS blood isolates were identical to catheter while Nataro et al33 reported that 42 per cent of the staphylococcal blood isolated from hospitalized infants were genotypically identical to catheter isolates. Another study reported that 64 per cent paediatric patients with CoNS in their blood had same bacteria present on their catheter tips4. Similarity of skin and nose isolates to that of blood isolates was not that frequent, varying between 16-70 per cent in our study. This implies that the source of at least some of the blood isolates in hospitalized paediatric patients is not endogenous.
In conclusion, staphylococci were found to be the major causative agent of nosocomial blood stream infections in paediatric population. Penicillin and oxacillin resistance was high in these isolates. Only 33.3 per cent of S. aureus blood infections and majority of CoNS (S. haemolyticus and S. epidermidis) blood infections were IVD associated.
Acknowledgments
Authors acknowledge the Indian Council of Medical Research, New Delhi, for financial support.
References
- 1.Piette A, Verschraegen G. Role of coagulase-negative staphylococci in human disease. Vet Microbiol. 2009;134:45–54. doi: 10.1016/j.vetmic.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Altiparmak MR, Güngör K, Pamuk GE, Pamuk ON, Ozgenç R, Oztürk R. Temporary Catheter infections in hemodialysis patients: results from a single center in Turkey. Acta Clin Belg. 2003;58:345–9. doi: 10.1179/acb.2003.58.6.003. [DOI] [PubMed] [Google Scholar]
- 3.Leibovitch I, Lai TF, Senarath L, Hsuan J, Selva D. Infectious keratitis in South Australia: Emerging resistance to cephazolin. Eur J Opthalmol. 2005;15:23–6. [PubMed] [Google Scholar]
- 4.Lodha R, Natchu UC, Nanda M, Kabra SK. Nosocomial infections in pediatric intensive care units. Indian J Pediatr. 2001;68:1063–70. doi: 10.1007/BF02722358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jain A, Agarwal A. Biofilm production, a marker of pathogenic potential of colonizing and commensal staphylococci. J Microbiol Methods. 2009;76:88–92. doi: 10.1016/j.mimet.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 6.CDC National Nosocomial Infections Surveillance (NNIS) System Report, Data Summary from January 1990-May 1999, issued June 1999. Am J Infect Control. 1999;27:520–32. doi: 10.1016/s0196-6553(99)70031-3. [DOI] [PubMed] [Google Scholar]
- 7.Maki DG, Weise CE, Sarafin HW. A semi quantitative culture method for identifying intravenous-catheter-related infection. N Engl J Med. 1977;296:1305–9. doi: 10.1056/NEJM197706092962301. [DOI] [PubMed] [Google Scholar]
- 8.Kloos WE, Bannerman YL. Staphylococcus and micrococcus. In: Murry RP, Baron EJ, Pfaller MA, Tenover MC, Yolken RH, editors. Manual of clinical microbiology. 7th ed. Washington DC: American Society of Microbiology; 1999. pp. 262–82. [Google Scholar]
- 9.15th Informational Supplement testing M100-S15. Wayne (PA): CLSI; 2005. Clinical Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- 10.Burnie JP, Naderi-Nasab M, Loudon KW, Matthews RC. An epidemiological study of blood culture isolates of coagulase negative staphylococci demonstrating hospital acquired infection. J Clin Microbiol. 1997;35:1746–50. doi: 10.1128/jcm.35.7.1746-1750.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaffe RI, Lane JD, Albury SV, Niemeyer DM. Rapid extraction from and direct identification of clinical samples of methicillin resistant staphylococci using the PCR. J Clin Microbiol. 2000;38:3407–12. doi: 10.1128/jcm.38.9.3407-3412.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenwood PE, Nikulin MS. A guide to chi-squared testing. New York: Wiley; 1996. [Google Scholar]
- 13.Gray J, Gossain S, Morris K. Three year survey of bacteraemia and fungemia in a pediatric intensive care unit. Pediatr Infect Dis J. 2001;20:416–21. doi: 10.1097/00006454-200104000-00009. [DOI] [PubMed] [Google Scholar]
- 14.Berner R, Sauter S, Duffner U, Brandis M, Niemeyer CM. Bacteremic episodes in pediatric oncologic patients, especially caused by the Streptococcus viridans group. Clin Padiatr. 1998;210:256–60. doi: 10.1055/s-2008-1043888. [DOI] [PubMed] [Google Scholar]
- 15.Kuehnert MJ, Webb RM, Jochimsen EM. Staphylococcus aureus blood stream infections among patients undergoing electroconvulsive therapy traced to breaks in infection control and possible extrinsic contamination by propofol. Anesth Analg. 1997;85:420–5. doi: 10.1097/00000539-199708000-00031. [DOI] [PubMed] [Google Scholar]
- 16.Glowacki M, Quraishi ZA, Zakhireh B. Risk factors of nosocomial bacteremia associated with pulmonary artery catheters in a critical care unit. J Am Osteopath Assoc. 1990;90:509–14. [PubMed] [Google Scholar]
- 17.Chlebicki MP, Teo EK. Review of peripherally inserted central catheters in the Singapore acute-care hospital. Singapore Med J. 2003;44:531–5. [PubMed] [Google Scholar]
- 18.Cuevas O, Cercenado E, Vindel A, Guinea J, Sanchez-Conde M, Sanchez-Somolinos M, et al. Evolution of the antimicrobial resistance of Staphylococcus spp. in Spain: Five Nationwide Prevalence Studies 1986 to 2002. Antimicrob Chemother. 1986;48:4240–5. doi: 10.1128/AAC.48.11.4240-4245.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jog SM, Patole SK. Staphylococcus warneri septicemia in preterm neonates-A reminder. Indian Pediatr. 2002;39:309–10. [PubMed] [Google Scholar]
- 20.Mohan U, Jindal N, Aggarwal P. Species distribution and antibiotic sensitivity pattern of coagulase negative staphylococcal isolates from various clinical specimens. Indian J Med Microbiol. 2002;20:45–6. [PubMed] [Google Scholar]
- 21.Silvia N, Carla F, Marco F, Alberto B, Piero TG, Silvia M, et al. Characterization of coagulase-negative staphylococcal isolates from blood with reduced susceptibility to glycopeptides and therapeutic options. BMC Infect Dis. 2009;9:83–6. doi: 10.1186/1471-2334-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Del’ Alamo L, Cereda RF, Tosin I, Miranda EA, Sader HS. Antimicrobial susceptibility of coagulase-negative staphylococci and characterization of isolates with reduced susceptibility to glycopeptides. Diagn Microbiol Infect Dis. 1999;34:185–91. doi: 10.1016/s0732-8893(99)00034-6. [DOI] [PubMed] [Google Scholar]
- 23.Agvvald-Ohman C, Lund B, Edlund C. Multiresistant coagulase négative staphylococci disseminate frequently between intubated patients in a multidisplinary intensive care unit. Crit Care. 2004;8:R42–7. doi: 10.1186/cc2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mokuolu AO, Jiya N, Adesiyun OO. Neonatal septicaemia in Ilorin: bacterial pathogens and antibiotic sensitivity pattern. Afr J Med Sci. 2002;31:127–30. [PubMed] [Google Scholar]
- 25.Sechi LA, Pinna A, Pusceddu C, Fadda G, Carta C, Zanetti S. Molecular characterization and antibiotic susceptibilities of ocular isolates of Staphylococcus epidermidis. J Clin Microbiol. 1999;37:3031–3. doi: 10.1128/jcm.37.9.3031-3033.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenover FC, Biddle JW, Lancaster MV. Increasing resistance to vancomycin and other glycopeptides in Staphylococcus aureus. Emerg Infect Dis. 2001;7:327–32. doi: 10.3201/eid0702.010237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ojha N, Deodhar L. Antimicrobial susceptibility pattern of nosocomial pathogens. Bombay Hosp J. 1997;39:49–51. [Google Scholar]
- 28.Jain A, Agarwal A, Verma RK. Cefoxitin disc diffusion test for detection of meticillin-resistant staphylococci. J Med Microbiol. 2008;57:957–61. doi: 10.1099/jmm.0.47152-0. [DOI] [PubMed] [Google Scholar]
- 29.Oteo J, Baquero F, Vindel A, Campos J. Spanish members of the European Antimicrobial Resistance Surveillance System. Antibiotic resistance in 3113 blood isolates of Staphylococcus aureus in 40 Spanish hospitals participating in the European Antimicrobial Resistance Surveillance System (2001-2002) J Antimicrob chemother. 2004;53:1033–8. doi: 10.1093/jac/dkh214. [DOI] [PubMed] [Google Scholar]
- 30.Jain A, Agarwal A, Verma RK. Cefoxitin disc diffusion test for detection of meticillin-resistant staphylococci. J Med Microbiol. 2008;57:957–61. doi: 10.1099/jmm.0.47152-0. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds R, Potz N, Colman M, Williams A, Livermore D, MacGowan A. BSAC extended working party on Bacteraemia Resistance Surveillance.Antimicrobial susceptibility of the pathogens of bacteraemia in the UK and Ireland 2001-2002: the BSCA bacteraemia Resistance Surveillance Program. J Antimicrob Chemother. 2004;53:1018–32. doi: 10.1093/jac/dkh232. [DOI] [PubMed] [Google Scholar]
- 32.St Gemme JW, Bell LM, Baumgart S, D’Angio CT, Harris MC. Distinguishing sepsis from blood culture contamination in young infants with blod cultures growing coagulase-negative staphylococci. Pediatrics. 1990;86:157–62. [PubMed] [Google Scholar]
- 33.Nataro JP, Concoran L, Zirin S. Prospective analysis of coagulase-negative staphylococcal infection in hopitalized infants. J Pediatr. 1994;125:798–804. doi: 10.1016/s0022-3476(94)70081-8. [DOI] [PubMed] [Google Scholar]


