Abstract
Background & objectives:
Hepatitis C virus (HCV) induces an immune response of the host, manifested by the formation of anti-HCV antibodies mediated by adaptive and innate immunity. Toll-like receptors (TLRs) play a pivotal role in innate immunity system. This study was aimed to investigate the promoter region polymorphism and expression of TLR3 gene in patients with chronic HCV infection.
Methods:
Patients with chronic HCV infection (N=180) and an equal number of age-sex matched controls were included in the study. Patients positive for HCV-RNA were subjected to analysis of TLR3 polymorphism by direct sequencing of PCR products verified by comparing with the sequences reported in the National Centre for Biotechnology Information (NCBI) database (accession number: NT 022792). Expression of TLR3 gene was analyzed by semiquantitative RT-PCR using housekeeping β-actin gene as the internal control.
Results:
Polymorphisms at position -288G/A and -705A/G were identified. The results were significant in -705 allele (P=0.004) OR 2.79(1.46-5.42) and were associated with high risk of HCV infection. In silico sequence analysis showed the presence of ectropic viral integration site 1 encoded factor, in which G at -705 results in the loss of this site. The -7C/A polymorphism was not seen in our study cohort. The expression of TLR3 was upregulated in chronic HCV patients compared to healthy controls.
Interpretation & conclusions:
Polymorphism in the -705A/G allele at the promoter region of the TLR3 gene may predispose individual to HCV infection. However association of TLR3 expression with polymorphism of TLR3 promoter was not found.
Keywords: HCV, polymorphism, SNP, TLR3
More than 210 million people are affected per year by hepatitis C virus (HCV) worldwide and has been recognized as a major cause of chronic hepatitis, endstage cirrhosis and hepatocellular carcinoma1. HCV is known for extraordinary variability manifested by constant alterations of its phenotype and the formation of so-called quasispecies2, this genetic heterogeneity allows HCV to escape immune pressure and to establish chronic infection. It results in the lack of efficient vaccine against this RNA virus in spite of great efforts of several laboratories worldwide3. Virus induces an immune response of the host manifested by the formation of anti-HCV antibodies and specific cell-mediated immunity. The cell mediated immunity, can be broadly categorized into adaptive and innate immunity. Adaptive immunity is mediated by clonally restricted T and B lymphocytes, which is characterized by features of immunological specificity and memory4. In contrast, innate immunity, being phylogenetically older, is not able to recognize antigens in a specific way, but possesses restricted ability to distinguish ‘self from not self”. The discovery of the importance of the toll-like receptors in host defense against pathogens has led to an increasing interest in innate immunity5. Cells sense viral infection by detecting viral proteins6 and/or nucleic acid, the double stranded RNA (dsRNA), a byproduct of the replicative cycle of many RNA virus in particular6. Viral proteins and dsRNA are recognized through receptors such as the evolutionary conserved toll-like receptors (TLR)7. Of the 10 TLRs identified in human, TLR-3 has been identified to respond to dsRNA a molecular signature of viruses8 that activates nuclear factor-ĸB (NF-ĸB) and the interferon- β (IFN-β) promoter9.
A role for TLR3 in viral infection has been suggested based on the demonstration that TLR3 knockout mice were unable to mount a full response to measles virus (MV) infection10. Genetic variations in the TLRs have been correlated with susceptibility to various disease including sepsis or malignancies11–14 and to vaccination efficiency15. To date, there have been no reports of studies on TLR3 polymorphism in subjects with chronic HCV infection from Indian population. Since host genetic factor with respect to the TLR3 polymorphism and susceptibility to viral infections showed that ethnic diversity may in part reflect the ethnic diversity of host viral susceptibility, population-based association studies are useful for examining genes with a role in common multifactorial diseases that have a strong environment component16.
The aim of the study was to assess genetic variations of the promoter region of TLR3 gene and it expression in chronic HCV infection in patients from northern and north-eastern part of India.
Material & Methods
Subject: A total of 180 patients with chronic HCV (CHC) infection were included in the study between 2005 and 2009 with prior written informed consent. An equal number of age- and sex- matched healthy controls (voluntary blood donors) who had no history of hepatitis, hepatobiliary diseases, negative for anti-HCV and autoimmune hepatitis as well as HCV RNA were also included in the study. The sample size has been estimated assuming prevalence rate of 10 per cent in the study population with a 5 per cent precision and chance level of 5 and 90 per cent power level using Raosoft® (www.raosoft.com). This figure is higher than the prevalence rate of HCV infection among the Indian population of 1.8-2.5 per cent17–19. Patients positive for other hepatotropic virus were not included in the study. Patients attending the medicine out-patient department of Lok Nayak Hospital, New Delhi during the study period were selected consecutively, on the basis of clinical investigation, positive for anti-HCV using 3rd generation ELISA kit confirmed by HCV RNA positivity and received liver biopsies. Liver histology was assessed for scoring the disease activity grade quantitatively in chronic hepatitis patients with prior consent according to the modified version of the Knodell histological activity index (HAI)20. HCV genotyping was done as describe earlier21. Patients who were positive for other hepatotropic virus, HIV, autoimmune hepatitis, alcoholic and drug abuse were excluded. The study was approved by the institutional ethical committee of Maulana Azad Medical College, New Delhi.
TLR3 polymorphism analysis: Genomic DNA was extracted from whole blood standard Proteinase-K digestion and phenol/ chloroform extraction procedure as described earlier22. Promoter region polymorphism of TLR3 was analysed by direct sequencing of PCR products using forward and reverse primer and the variants were verified by comparing with the sequences reported in the National Centre for Biotechnology Information (NCBI) database (gene designed using Primer 3 software version 0.4023 bank accession number: NT 022792) from -811 spanning to exon I using two overlapping pairs of primers designed using Primer 3 software version 0.403. The forward primer PTLR3FWD1 5’CCATGTTTGGCTCTTTCTC 3’ (19 mer) and the reverse primer PTLR3REV1 5’ TTGGATGACTGCTAGCCTTCC 3’ (21-mer) cover a region from -493 to 94 of exon I [as per Human Genome Organization (HUGO) nomenclature] generating an amplicon of 586 bp. The second pair of overlapping primers used was forward primer PTLR3FWD2 5’GCAAACCACTCACCTCCCTA 3’ (20-mer) and PTLR3REV2 5’ CTCATGCGAAGCTGTCAGAA 3’ to amplify a 516 bp product in the promoter region. Polymerase chain reaction (PCR) was performed in a total volume of 25 μl containing 50 ng genomic DNA, 1x Buffer (10 mM Tris, pH 9.0 50mM KCl and 0.01% gelatin), 200μM dNTPs, 10 pmole of each primer (PTLR3FWD1 and PTLR3REV1), 1mM MgCl2 and 0.2U of Taq DNA Polymerase (New England Biolab, England). Following the initial denaturation step (95°C for 5 min), samples were subjected to 30 cycles of PCR consisting of 94°C for 1 min, 65°C for 30 sec; and 72°C for 1 min, followed by a final extension for 10 min at 72°C. The second PCR reaction was carried out with the same master mix concentration using 10 pmol of primer (PTLR3FWD2 and PTLR3REV2) and annealing temperature 55°C for 30 sec. PCR-RFLP analysis for the polymorphic site rs3775296 (exon 2, UTR SNP) was done as described earlier24 followed by sequencing of random samples to confirm them by forward and reverse primer.
In silico analysis of TLR3 promoter region: In silico sequence analysis for DNA-binding domain of the studied promoter region was done using the software MATINSPECTOR v2.2 (genomatix, Munich, Germany; http://transfac.gbf.de).
Sequencing: The cycle sequencing of the amplicon (PCR products) of the promoter region was carried out using ABI Prism Big Dye terminator Cycle sequencing system (PE Applied Biosystem, USA). Variations in the amplified regions were screened with multiple alignments of the sequences based on clustal X output (Rosalind Franklin Centre for genomics Research; http://www.hgmp.mcc.ac.uk).
Total RNA extraction: Total RNA was isolated from peripheral blood mononuclear cells (PBMC) obtained from sixty randomly selected CHC patients and thirty healthy controls (serologically negative for HCV, HBV and HIV antibodies). PBMC were isolated using standard Ficoll-Paque (Amersham Biosciences AB, Uppsala, Sweden) separation in which 1 to 2 × 106 PBMCs were used for total RNA extraction using the RNAeasy mini kit (Qiagen, Hilden, Germany) according to manufacturer's instructions.
Semi-quantitative reverse transcriptase–polymérase chain réaction (RT-PCR): Expression of TLR3 gene at the transcriptional level was analyzed by semi-quantative RT-PCR using housekeeping β-actin gene as the internal control. Total RNA 1μg was used to synthesize cDNA with 0.1 OD of Random hexamer (Fermentas, Lithuania) and 100U of Moloney Murine Leukemia Virus (M-MuLV, MMLV) Reverse Transcriptase (New England Biolabs, England) at 42°C for 60 min. The TLR3 primer specific for exon-exon boundary used was forward 5’TCTGGAAAGGCGCAACC.3’ and reverse 5’CCGTTGGACTCTAATTCAAGAT 3’ to amplify a 276 bp amplicon. Primer specific for β-actin gene used as internal control was forward 5’ GAAGGATTCCTATGTGGGCG 3’ and reverse 5’ TGGTGGTAAAGCTGTAC 3’ to amplify a 465 bp amplicon. The PCR conditions were as follows: denaturation at 94°C for 30 sec, annealing at 55°C for 30 sec, and extension at 72°C for 30 sec. The PCR buffer contained 10mM Tris HCl (pH 10), 2.0 mM MgCl2 and 50 mM KCl with 0.3U Taq polymerase. After 30 cycles an addition extension at 72°C for 10 min was performed. The amplified products were then subjected to densitometric analysis to compare the band intensity with β-actin as the internal control.
Statistical analysis: Differences in the allele and genotype frequencies of the studied gene regions in different groups of with chronic HCV and in age- and sex- matched controls were analysed using χ2 test or Fisher's exact test. Genotype frequencies at each locus were found to deviate significantly from Hardy-Weinberg equilibrium. The odd ratios (ORs) and 95% confidence intervals (CIs) were obtained using unconditional logistic regression analysis. Crude ORs and ORs adjusted for different possible confounders: age (continuous variable), sex, HCV infection (yes or no), HCV RNA status (positive or negative) were calculated. All analyses were performed by the statistical package for social science, version 10 (SPSS, Chicago, IL).
Results
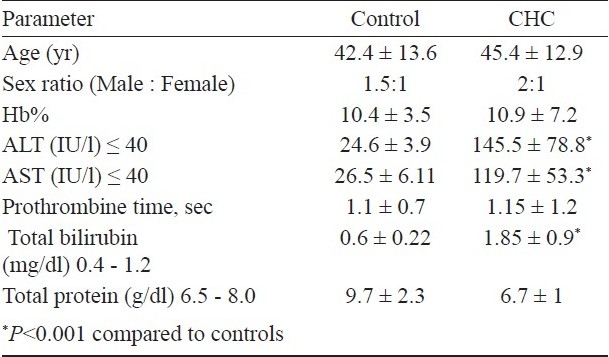
A total of 180 patients with hepatitis C related chronic hepatitis who were positive for HCV RNA (Fig. 1) were studied for the promoter region polymorphism of TLR3. There was no significant difference for age and sex between the cases and controls (Table I). Alanine aminotransferase (ALT), aspartate amino-transferase (AST) and total bilirubin levels were significantly higher in cases (P<0.001) compared to controls. The prothrombin time was on the higher side in chronic hepatitis patients compared to controls. The mean HAI was 7.4 ± 2.2 with prior diagnosis of chronic HCV infection. Of the 180 patients, 84 (46.5%) were infected with genotype 3, followed by 43 (24%) with genotype 1, 28 (15.5%) with genotype 2, and 25 (14%) with genotype 4.
Fig. 1.
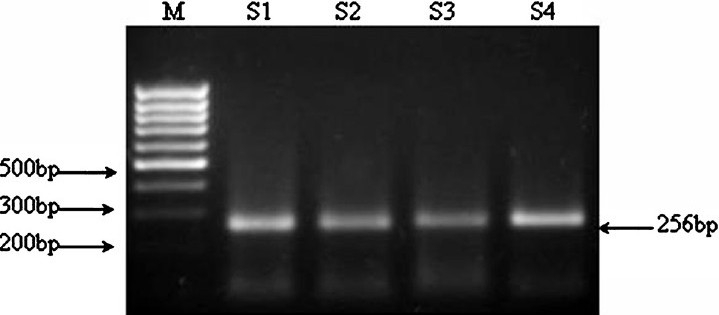
Representative photograph of HCV RNA +ve product (5’UTR-region), amplified from patients of chronic hepatitis.
Table I.
Demographic and aetiological characteristics of chronic hepatitis C (CHC) cases and control
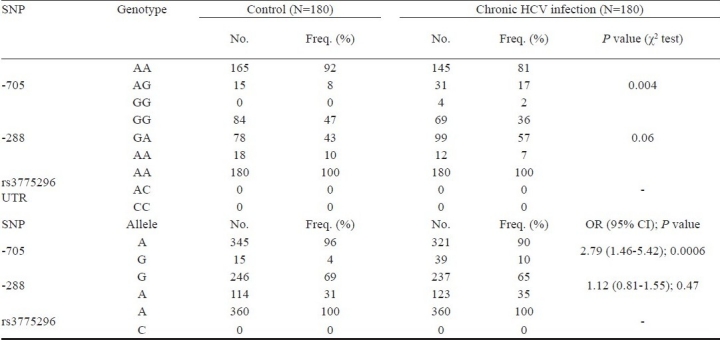
Analysis of the TLR3 promoter region SNP with HCV infection: The -7C/A (rs3775296) polymorphism was not found in our study. The screening of variants in the promoter region of the TLR3 gene resulted in identification of polymorphism at position – 288 G to A and –705 A to G (Fig. 2a, 2b) by forward primer and confirmed by reverse primer (as per HUGO nomenclature). The results were significant in -705 allele (P<0.05) and were associated with predisposition of HCV infection OR 2.79 (95% CI 1.46-5.42). In -288 allele the result was not significant (P=0.47) which indicated moderate risk for HCV infection (OR= 1.21; CI = 0.81 - 1.55). The -7C/A (rs3775296) polymorphism was not seen (Table II).
Fig. 2a.
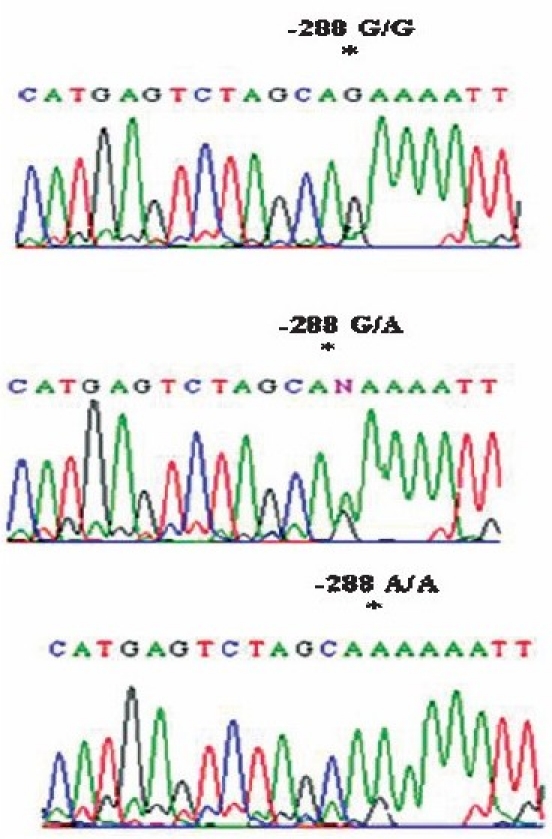
Partial chromatograms representing (a) the homozygous G/G, A/A genotype and (b) the heterozygous G/A genotype at position-288 in the TLR3 promoter region depicting G/A variation.
Fig. 2b.
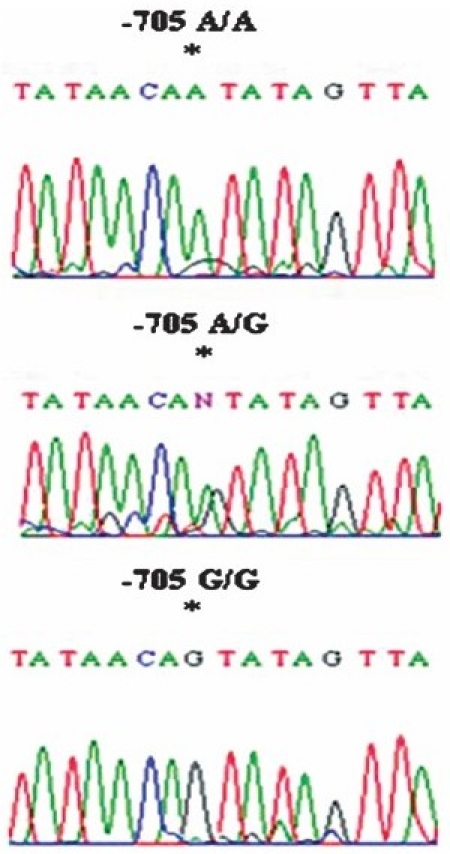
Partial chromatograms representing the homozygous G/G, A/A genotype and heterozygous G/A genotype at position-705 in the TLR3 promoter region depicting G/A variation.
Table II.
Genotypic and allele status for the -705 A/G, -288 G/A, rs3775296, polymorphism of TLR3 in patients with chronic hepatitis C (CHC) and healthy controls of Indian origin
In silico analysis of promoter region: In silico sequence analysis for DNA-binding domain of the promoter region showed the presence of ectropic viral integration site 1 encoded factor “ataacaATATagttata” with a MATINSPECTOR core similarity of 0.750 and an affinity of 0.835, in which G at -705 resulted in the loss of this site. The STATs binding site “TTCTTGGAA” and the interferon-stimulated response element (ISRE) were located at upstream of -40 bp and at around + 2 bp of the TLR3 promoter region respectively. Both the STATs binding site and the ISRE were conserved in all the patients.
Expression profile of TLR3 by semiquantitative RT-PCR correlated with the promoter region polymorphism of TLR3: The expression of TLR3 at the transcriptional level was determined by semiquantitative RT-PCR using housekeeping β-actin as the internal control by densitometric analysis (Fig. 3). The expression of TLR3 was upregulated in chronic HCV patients compared with controls (Fig. 4a). Analysis of the TLR3 expression profile for each samples showed that there were no association with regards to TLR3 SNPs variants of -705A/G (Fig. 4b), -288G/A (Fig. 4c), rs3775296 (Fig. 4d) and in HCV related liver disease.
Fig.3.
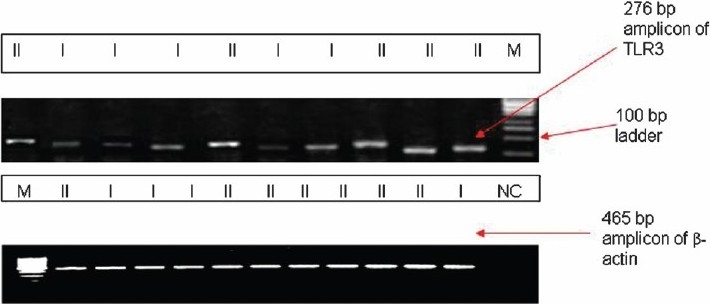
Representative photograph of semi-quantitative RT-PCR for TLR3 compared to β-actin as the internal control.
Fig. 4a–4d.
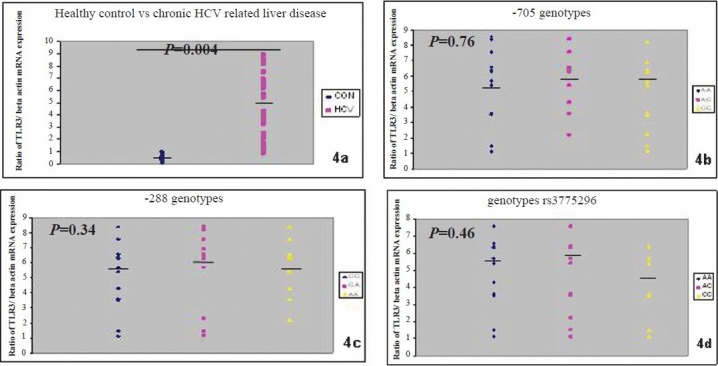
TLR3 mRNA expression in PBMC in relation to β-actin as control from patients with chronic hepatitis C and normal control (4a) with regard to SNPs of -705A/G (4b), -288 G/A (4c), rs3775296 (4d). Medians and P values are given.
Discussion
Being a persistently infecting virus, HCV was forced to evolve sophisticated escape strategies for both the innate and the adaptive immune system. Patients in the acute phase of HCV infection were much more likely to respond to therapy that is designed to eradicate the virus than the patients who progress to chronicity24. The 0.3kb TLR-3-promoter region was responsible for virus-enhanced promoter activity. In human, TLR3 appears to be involved in both maintenance of promoter integrality and promoter specific virus responsive element, besides the interferon-stimulated response element (ISRE) at -0.1 kb, the Sp l binding site at 0.2 kb are also responsible for both virus-mediated upregulation and basal expression of TLR10. There were several transcriptional starting sites for human TLR325 a characteristic feature of GC-rich sequence-driven gene transcription16. Polymorphisms within the exon-4 region of TLR-3 gene have been shown to be associated with type-1 diabetes26. However, the impact of rare mutations on disease susceptibility could only be traced by large sequencing approaches. This study aimed to asses the role of a promoter region polymorphism in TLR3 gene that may mediate a link between exposure to a putative environmental precipitant and the adaptive immune system. Polymorphism of TLR3 at the -705 promoter region (A > G) may predispose an individual to HCV infection as observed in all patients under investigation compared to normal controls. The STATs binding site “TTCTTGGAA” and the ISRE are located at upstream of -40 bp and at around + 2 bp of the TLR3 promoter region respectively, which is in contrast to that described earlier10,27. Both the STATs binding site and the ISRE were found to be conserved in all the cases suggesting that the IFN-β promoter activation through TLR3 is conserved in Indian population. In silico sequence analysis for DNA-binding domain of the studied promoter region showed the presence of ectropic viral integration site 1 encoded factor “ataacaATATagttata” with a MATINSPECTOR core similarity of 0.750 and an affinity of 0.835, in which G at -705 resulted in the loss of this site. The findings need to be confirmed in a larger population and in vitro analysis which is the main limitation of the study.
In this study, the SNPs at -288 G/A, rs 3775296 within the promoter region were not associated with HCV infection, similar to that shown by other28,29. The (C/A) SNP at position -7 (rs3775296) in the TLR3 promoter region in Chinese population had frequency of 23 per cent29 whereas, this polymorphism was not found in our study and it has been detected at a prevalence rate of 11.1 per cent in Japanese, 4.4 per cent in Chinese, 3.3 per cent in sub Saharan African and 1.7 per cent in European as per HapMapn project30. Two new polymorphisms of TLR3 at the promoter region the -705 (A > G) and -288 (G > A) were observed by direct sequencing in which the allele G of the SNPs -705 A/G increased the risk for HCV infection.
Since the newly found SNP at -705 A/G was located in the promoter region within 1 kb upstream of exon 1 in the TLR3 gene in close proximity to the regulatory regions, it can be postulated that it might modulate or influence the promoter activity of TLR3. SNPs within the promoter region of TLR3 might affect transcriptional activity but we have not found any association with the expression level of TLR mRNA in PBMC which indicates that SNPs in the promoter region have least role to play with respect to the expression of TLR3 gene.
The TLR3 expression on peripheral blood monocytes was significantly upregulated in chronic HCV infection compared to control in our study irrespective of the HCV genotype and even at an early histological stage Tanabe et al10 demonstrated that MV strains with IFN- β inducible properties up-regulate TLR3 expression following their infection. Increased TLR3 expression level resulted in enhanced IFN- β secretion in response to polyI: C, this is consistent with the implicated function of TLR-3 and also suggests the existence of positive feedback regulation of IFN α/β signaling in response to virus infection31. Expression analysis of TLR3 gene at PBMC did not show any association with the SNPs located at rs3775296 in our study, whereas Aksar et al29 have reported that TLR3 gene expression was related to hepatic TLR3 gene expression but not PBMC. Contrary to their role in provoking the host defense mechanism, increased expressions of TLR3 in severe liver disease promote a suppressive effect to the adaptive immune response in chronic HCV infection. Molecular signals through TLR3 ligands such as HCV dsRNA in its replicative forms enhance the receptor expression and responsiveness. HCV appears to be a ‘double hit’ that could mark the difference between pathogenesis and tolerance induced by HCV signals as overactivation of TLRs triggers TLR-signaling-inhibitory processes, instead of inducing the expression of immune and pro-inflammatory genes32.
In conclusion, presence of two previously unreported SNPs at -705 and -288 promoter region of TLR3 has been identified. The TLR3 SNP of -705 was associated with chronic HCV infection. This study suggests a possible means whereby environmental agents may interact with the innate and adaptive immune systems in the pathogenesis of the disease. Larger studies in various populations are required to explore further the link between the innate and adaptive immune systems in relation to HCV infection, in an attempt to identify additional susceptibility loci and indications of the initiator of the disease.
References
- 1.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–67. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 2.Martell M, Esteban JI, Quer J, Genescà J, Weiner A, Esteban R, et al. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–9. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stoll-Keller F, Barth H, Fafi-Kremer S, Zeisel MB, Baumert TF. Development of hepatitis C virus vaccines: challenges and progress. Expert Rev Vaccines. 2009;8:333–45. doi: 10.1586/14760584.8.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janeway CA., Jr How the immune system protects the host from infection. Microbes Infect. 2001;3:1167–71. doi: 10.1016/s1286-4579(01)01477-0. [DOI] [PubMed] [Google Scholar]
- 5.Rock FL, Hardiman G, Timans JC, Kastelein RA, Bazan JF. A family of human receptors structurally related to Drosophila Toll. Proc Natl Acad Sci USA. 1998;95:588–93. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooke GS, Hill AV. Genetics of susceptibility to human infectious disease. Nat Rev Genet. 2001;2:967–77. doi: 10.1038/35103577. [DOI] [PubMed] [Google Scholar]
- 7.Miettinen M, Sareneva T, Julkunen I, Matikainen S. IFNs activate toll-like receptor gene expression in viral infections. Genes Immun. 2001;2:349–55. doi: 10.1038/sj.gene.6363791. [DOI] [PubMed] [Google Scholar]
- 8.Vercammen E, Staal J, Beyaert R. Sensing of viral infection and activation of innate immunity by toll-like receptor 3. Clin Microbiol Rev. 2008;21:13–25. doi: 10.1128/CMR.00022-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhiman N, Ovsyannikova IG, Vierkant RA, Ryan JE, Pankratz VS, Jacobson RM, et al. Associations between SNPs in toll-like receptors and related intracellular signaling molecules and immune responses to measles vaccine: preliminary results. Vaccine. 2008;26:1731–6. doi: 10.1016/j.vaccine.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanabe M, Kurita-Taniguchi M, Takeuchi K, Takeda M, Ayata M, Oyura H, et al. Mechanism of up-regulation of human Toll-like receptor receptor 3 secondary to infection of measles virus-attenuated strains. Biochem Biophys Res Commun. 2003;311:39–48. doi: 10.1016/j.bbrc.2003.09.159. [DOI] [PubMed] [Google Scholar]
- 11.Tabeta K, Georgel P, Janssen E, Du X, Hoebe K, Crozat K, et al. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc Natl Acad Sci USA. 2004;101:3516–21. doi: 10.1073/pnas.0400525101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooke GS, Hill AV. Genetics of susceptibility to human infectious disease. Nat Rev Genet. 2001;2:967–77. doi: 10.1038/35103577. [DOI] [PubMed] [Google Scholar]
- 13.El-Omar EM, Ng MT, Hold GL. Polymorphisms in Toll-like receptor genes and risk of cancer. Oncogene. 2008;27:244–52. doi: 10.1038/sj.onc.1210912. [DOI] [PubMed] [Google Scholar]
- 14.Fukata M, Abreu MT. Role of Toll-like receptors in gastrointestinal malignancies. Oncogene. 2008;27:234–43. doi: 10.1038/sj.onc.1210908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhiman N, Ovsyannikova IG, Vierkant RA, Ryan JE, Pankratz VS, Jacobson RM, et al. Associations between SNPs in toll-like receptors and related intracellular signaling molecules and immune responses to measles vaccine: preliminary results. Vaccine. 2008;26:1731–6. doi: 10.1016/j.vaccine.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neil R. Searching for genes in complex diseases: lessons from systemic lupus erythematosus. J Clin Invest. 2000;105:1503–6. doi: 10.1172/JCI10266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arankalle VA, Chobe P, Banerjee K. HCV in Pune. J Assoc Physicianss India. 1992;40:562–6. [PubMed] [Google Scholar]
- 18.Thyagarajan SP, Mohan KVK, Mohanvalli B. In: Transfusion associated hepatitis: diagnosis, treatment and prevention. Sarin SK, Hess G, editors. New Delhi: CBS Publishers and Distributors; 1998. pp. 27–233. [Google Scholar]
- 19.Chandra M, Khaja MN, Farees N, Poduri CD, Hussain MM, Aejaz Habeeb M, et al. Prevalence, risk factors and genotype distribution of HCV and HBV infection in the tribal population: a community based study in south India. Trop Gastroenterol. 2003;24:193–5. [PubMed] [Google Scholar]
- 20.Ishak K, Baptistta A, Bianchi I, Callea F, De Groote J, Gudat H, et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22:696–9. doi: 10.1016/0168-8278(95)80226-6. [DOI] [PubMed] [Google Scholar]
- 21.Chinchai T, Labout J, Noppornpanth S, Theamboonlers A, Haagmans BL, Osterhaus AD, et al. Comparative study of different methods to genotype hepatitis C virus type 6 variants. J Virol Methods. 2003;109:195–201. doi: 10.1016/s0166-0934(03)00071-5. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. NY, USA: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 1990. p. 73. [Google Scholar]
- 23. http://frodo.wi,mit.edu/primer3/
- 24.Nomura H, Sou S, Tanimoto H, Nagahama T, Kimura Y, Hayashi J, et al. Short-term interferon-alfa therapy for acute hepatitis C: a randomized controlled trial. Hepatology. 2004;39:1213–9. doi: 10.1002/hep.20196. [DOI] [PubMed] [Google Scholar]
- 25.Gongora C, Degols G, Espert L, Hua TD, Mechti N. A unique ISRE, in the TATA-less human Isg20 promoter, confers IRF-1-mediated responsiveness to both interferon type I and type II. Nucleic Acids Res. 2000;28:2333–41. doi: 10.1093/nar/28.12.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pirie FJ, Pegoraro R, Motala AA, Rauff S, Rom L, Govender T, et al. Toll-like receptor 3 gene polymorphisms in South African Blacks with type 1 diabetes. Tissue Antigens. 2005;66:125–30. doi: 10.1111/j.1399-0039.2005.00454.x. [DOI] [PubMed] [Google Scholar]
- 27.Heinz S, Haehnel V, Karaghiosoff M, Schwarzfischer L, Muller M, Krause SW, et al. Species-specific regulation of Toll-like receptor 3 genes in men and mice. J Biol Chem. 2003;24:21502–9. doi: 10.1074/jbc.M301476200. [DOI] [PubMed] [Google Scholar]
- 28.Askar E, Bregadze R, Mertens J, Schweyer S, Rosenberger A, Ramadori G, et al. TLR3 gene polymorphisms and liver disease manifestations in chronic hepatitis C. J Med Virol. 2009;81:1204–11. doi: 10.1002/jmv.21491. [DOI] [PubMed] [Google Scholar]
- 29.Cheng PL, Eng HL, Chou MH, You HL, Lin TM. Genetic polymorphisms of viral infection-associated Toll-like receptors in Chinese population. Transl Res. 2007;150:311–8. doi: 10.1016/j.trsl.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 30. http://hapmap.icbi.nlm.nih.gov/
- 31.Seki E, Brenner DA. Toll-like receptors and adaptor molecules in liver disease: update. Hepatology. 2008;48:322–35. doi: 10.1002/hep.22306. [DOI] [PubMed] [Google Scholar]
- 32.Liew FY, Xu D, Brint EK, O’Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–58. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]


