Abstract
Despite their compositional complexity, lipidomes comprise a large number of isobaric species that cannot be distinguished by conventional low resolution mass spectrometry and therefore in-depth MS/MS analysis was required for their accurate quantification. Here we argue that the progress in high resolution mass spectrometry is changing the concept of lipidome characterization. Because exact masses of isobaric species belonging to different lipid classes are not necessarily identical, they can now be distinguished and directly quantified in total lipid extracts. By streamlining and simplifying the molecular characterization of lipidomes, high resolution mass spectrometry has developed into a generic tool for cell biology and molecular medicine.
High-resolution mass spectrometry streamlines the shotgun analysis of total lipid extracts. It can distinguish and quantify isobaric lipid species and has potential to become the “gold standard” lipidomics methodology.
As an independent scientific discipline, lipidomics aims at the quantitative characterization of the full lipid complement produced by cells, organisms, or tissues (reviewed in Wenk 2005; Oresic et al. 2008; Dennis 2009). According to different estimates, eukaryotic lipidomes might comprise from 10,000 to 100,000 species originating from a few hundred individual lipid classes (van Meer 2005; van Meer et al. 2008; Yetukuri et al. 2008). Lipidomics relies heavily on mass spectrometry, which helps to recognize individual molecules of varying molecular lipid structures at the low picomole sensitivity (reviewed in Ivanova et al. 2009; Blanksby and Mitchell 2010; Shevchenko and Simons 2010).
Irrespective of a particular method implementation, mass spectrometry characterizes lipid molecules in two major ways. First, it determines their intact masses. Second, by colliding molecular ions of lipid precursors with neutral gas, tandem mass spectrometry dissociates them into several structure-specific fragments (reviewed in Hsu and Turk 2009) such that individual molecular species can be recognized even in total lipid extracts (reviewed in Han and Gross 2005; Niemela et al. 2009). Top-down lipidomics is a strategy that aims at the rapid quantitative characterization of global changes of the lipidome composition and is solely reliant on accurately determined masses of intact lipids (Houjou et al. 2005; Schwudke et al. 2007a). The method operates with the integral abundances of clusters of lipid molecules with exactly the same mass-to-charge ratios (m/z) and does not necessarily identify individual species. In contrast, bottom-up lipidomics strategy aims at the accurate quantification of individual molecular species and relies entirely upon the detection of characteristic fragment ions by tandem mass spectrometry (Ejsing et al. 2006, 2009; Schwudke et al. 2006).
The specificity of both top-down and bottom-up approaches depends on the ability of a mass spectrometer to distinguish the peaks of intact molecular ions or their fragments. It is characterized by the instrument's resolving power (or, synonymously, mass resolution) R, which is usually calculated as a ratio of the ion peak mass to the peak width at the half of its height to the mass of the peak (full width at half maximum [FWHM]) (Gross 2006). To exemplify mass resolution in more “visual” terms, we could assume that two equally abundant peaks having (m/z)1 = R and (m/z)2 = R + 1 are expected to overlap at 50% of their height (this, however, only holds true for some “ideal” device—in reality, mass resolution depends on the ion mass and also mass range of the instruments is limited). Mass resolution depends on the type of mass analyzer employed in a particular mass spectrometer, as well as actual experiment settings: often, it is intentionally compromised for the sake of achieving better sensitivity or spectra acquisition rate (reviewed in Glish and Burinsky 2008; Griffiths and Wang 2009).
Until very recently it was generally assumed that unit mass resolution, under which a mass spectrometer should be able to distinguish peaks separated by 1 Da (like isotopic peaks of a singly charged ion) is sufficient for the vast majority of lipidomics applications. Indeed, molecules of glycerophospolipids—the most abundant component of both eukaryotic and prokaryotic lipidomes—are often isobaric and the same nominal mass (Yergey et al. 1983; Brenton and Godfrey 2010) might represent several individual species of different lipid classes. Direct measurement of intact masses did not allow their unequivocal assignment to individual species or even to lipid classes and it was required to fragment the entire cluster of plausible precursors and detect fragment ions specific for each of these isobaric molecules (Ekroos et al. 2002; Ejsing et al. 2006). Therefore, reaching higher than unit mass resolution by compromising the sensitivity might only result in marginal increase of the overall lipid identification capacity. Also, only a decade ago the mass resolution of over 50,000 was only achievable at FT ICR (Fourier transform ion cyclotron resonance) mass spectrometers featured with superconducting magnets that were expensive and complex in operation and maintenance.
Orbitrap mass analyzers, first introduced in hybrid linear ion trap–Orbitrap tandem mass spectrometers (LTQ Orbitrap) (Makarov et al. 2006a,b), have been changing the paradigm of lipidomics analysis. The mass resolution of 100,000 together with sub-ppm mass accuracy (Olsen et al. 2005) has become routinely achievable in both MS and MS/MS modes. Combining Orbitrap and linear ion trap analyzers within a hybrid tandem instrument enabled efficient MS/MS experiments and increased the dynamic range of ion detection to more than 1000-fold (Scigelova and Makarov 2006). However, high mass resolution comes at the price of reduced spectra acquisition rate. It might take up to 2 sec to acquire a spectrum at 100,000 mass resolution, which is impractical for the analysis by liquid chromatography tandem mass spectrometry (LC-MS/MS) (Olsen et al. 2009). However, acquisition time is less important for shotgun lipidomics, in which total extracts are directly infused into a mass spectrometer. Because the composition of infused analyte does not change in the course of analysis, both MS and MS/MS spectra could be acquired at the minute time scale (Schwudke et al. 2006, 2007b).
High mass resolution of modern instruments offers robust practical solutions for both top-down and bottom-up shotgun lipidomics because isobaric precursor ions and fragments can now be distinguished and independently quantified (Schwudke et al. 2007a). This enhances both the specificity and throughput of top-down shotgun screens and enables accurate structural characterization of individual molecular species in tandem mass spectrometric experiments.
IDENTIFYING LIPIDS BY ACCURATE MASSES OF INTACT PRECURSORS
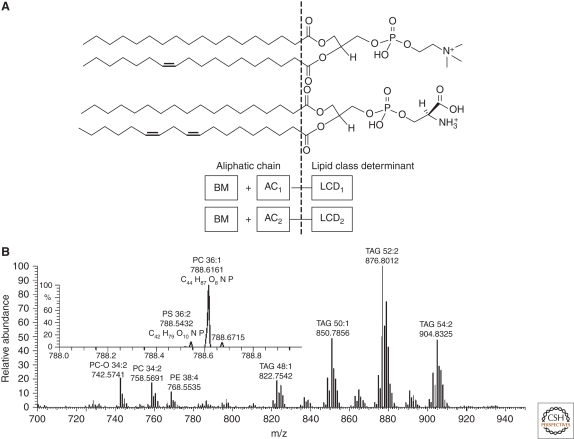
Why might species of different lipid classes have isobaric masses? Let us first consider glycerophospholipids (GPL) as a representative example. For clarity, we assume that the chemical structure of GPL molecules is composed of two elements, each of which may independently vary: a head group together with glycerol backbone with two ester or ester and ether moieties attached (here we will term it as a lipid class determinant, or LCD) and an aliphatic complement (AC), comprising methylene (-CH2-) and methylidene (-CH = CH-) groups (Fig. 1).
Figure 1.
Mass differences between LCDs could be balanced by altering the complementary AC so that masses of certain species from different lipid classes might be isobaric, although their exact masses will still be different. For example, the mass difference between the LCDs of phosphatidylcholine (PC) and phosphatidylserine (PS) species is 1.9429 Da. Apparently, masses of PC and PS species could be isobaric if they have the same number of carbon atoms in their fatty acid moieties and the PS molecule has one double bond more compared to the PC. Although they have the same nominal masses (i.e., they are isobaric), their exact masses will differ by 0.0726 Da and it should be possible to distinguish their peaks in high resolution mass spectra (Fig. 1).
How could we determine whether the exact masses are unique for species of a given lipid class, or if they overlap with masses of species from other classes? With no compositional constraints applied, major lipid classes can be clustered into several groups (Table 1). Exact masses of lipids from these groups do not overlap and their peaks in MS spectra could be attributed unequivocally, no matter what fatty acid or fatty alcohol moieties they comprise. Each group differs by the unique number of N, O, and P atoms in their molecular cations or anions. Some groups consist of more than one lipid class, implying that species of different lipid classes could have the same exact masses. For example, it might not be possible to judge solely from the accurately measured masses whether a peak belongs to the lipid of PC or phosphatidylethanolamine (PE) class because they are within the same group. At the same time, if a peak in the high resolution spectrum matches the mass of a sphingomyelin (SM) species, its attribution is unequivocal. We underscore that Table 1 presents the composition of heteroatoms of molecular ions, rather than neutral lipid molecules. Several lipid classes (such as, for example, triacylglycerols [TAG]) are barely detectable in the protonated form [M + H]+, but produce abundant adducts with cations of alkaline metals or ammonium [M + NH4]+ (McAnoy et al. 2005; Schwudke et al. 2006; Hsu and Turk 2010). Note that grouping of lipid classes differs between positive and negative modes, because some lipids are detectable as adducts, whereas others are not. For example, in negative ion mode PCs produce abundant adducts with acetate or formiate anions (Ekroos et al. 2003), whereas PEs are detected as deprotonated anions. Because of the different heteroatom composition of their anionic forms, they fell into different groups and their peaks could be unequivocally distinguished.
Table 1.
Major lipid classes grouped according to the number of heteroatoms
| Lipid class | Positive modea | Negative modea | N | O | P |
|---|---|---|---|---|---|
| DAG | – | [DAG + Ac]− | 0 | 7 | 0 |
| TAG | – | [TAG + Ac]− | 0 | 8 | 0 |
| PA | – | [PA − H]− | 0 | 8 | 1 |
| CholFA | [Chol-FA + NH4]+ | – | 1 | 2 | 0 |
| Cer | [Cer + H]+ | [Cer − H]− | 1 | 3 | 0 |
| DAG | [DAG + NH4]+ | – | 1 | 5 | 0 |
| TAG | [TAG + NH4]+ | – | 1 | 6 | 0 |
| PE-O | [PE-O + H]+ | [PE-O − H]− | 1 | 7 | 1 |
| LPE | [LPE + H]+ | [LPE − H]− | 1 | 7 | 1 |
| PC-O | [PC-O + H]+ | – | 1 | 7 | 1 |
| LPC | [LPC + H]+ | – | 1 | 7 | 1 |
| HexCer | [HexCer + H]+ | [HexCer − H]− | 1 | 8 | 0 |
| PE | [PE + H]+ | [PE − H]− | 1 | 8 | 1 |
| PC | [PC + H]+ | – | 1 | 8 | 1 |
| PA | [PA + NH4]+ | – | 1 | 8 | 1 |
| PC-O | – | [PC-O + Ac]− | 1 | 9 | 1 |
| LPC | – | [LPC + Ac]− | 1 | 9 | 1 |
| PG | – | [PG − H]− | 0 | 10 | 1 |
| PS | [PS + H]+ | [PS − H]− | 1 | 10 | 1 |
| PG | [PG + NH4]+ | – | 1 | 10 | 1 |
| PC | – | [PC + Ac]− | 1 | 10 | 1 |
| PI | – | [PI − H]− | 0 | 13 | 1 |
| PI | [PI + NH4]+ | – | 1 | 13 | 1 |
| SM | [SM + H]+ | – | 2 | 6 | 1 |
| CerPE | [CerPE + H]+ | [CerPE − H]− | 2 | 6 | 1 |
| SM | – | [SM + Ac]− | 2 | 8 | 1 |
Abbreviations: DAG, diacylglycerols; PA, phosphatidic acid; Chol-FA, cholesterol esters of fatty acids; FA, fatty acids; Cer, ceramides; HexCer, hexosylceramides; PE-O, PE; PC-O, 1-alkyl-2-acyl PC; LPE, lyso-PE; LPC, lyso-PC; PI, phosphatidylionositols; PG, phosphatidylglycerols; CerPE, ceramide PE; Ac, acetate. Other abbreviations are spelled out in the text.
aMolecular ions commonly detectable in the spectra of total lipid extracts using 7.5 mm ammonium acetate in isopropanol/methanol/chloroform 4:2:1 (v/v/v) solvent.
Would even higher mass resolution further increase the specificity of top-down lipidomics? A mass difference of 0.009 Da spaces the second isotopic peak of a lipid species and the monoisotopic peak of the species from the same lipid class that lacks one double bond. Computational modeling suggests that although these peaks are not completely resolved even at 100,000 mass resolution, they still could be recognized as individual signals by a conventional spectra processing software. Within the entire space of lipid compositions there are no isobaric pairs with closer exact masses and therefore we speculate that higher mass resolution (e.g., exceeding 500,000) might only result in the marginal improvement of lipid identification.
We underscore that the grouping of lipid classes in Table 1 presumed that no restrictions on the length and number of double bonds in their fatty acid/fatty alcohol moieties were applied. However, some of these moieties might not occur because of the stringent specificity of lipid biosynthesis pathways. Therefore, applying some broad, yet reasonable compositional restrictions (such as limiting the number of double bonds or length of fatty acid/fatty alcohol moieties, restricting fatty acids to those having even number of carbon atoms, etc.) dramatically increases the specificity of mass assignment.
Therefore we conclude that the acquisition of high mass resolution spectra provides sufficiently specific means for taking a rapid informative “snapshot” of the lipidome composition, even though the exact molecular assignment of certain peaks might remain ambiguous.
HIGH-RESOLUTION MASS SPECTROMETRY FOR TOP-DOWN LIPIDOMICS SCREENS
In top-down shotgun lipidomics the identification and quantification of lipid species relies on accurate masses and abundances of their molecular ions, respectively. Top-down analysis is a high-throughput screening tool that rapidly reveals global changes within the entire lipidome. Although these screens could be accompanied by MS/MS experiments, these would only target a small number of selected precursors. Therefore, for top-down screens high mass resolution is of paramount importance because their specificity and accuracy depend on how many individual species (rather than unresolved clusters of different precursors with close masses) could be distinguished in MS spectra of total lipid extracts.
A typical top-down lipidomics screen encompasses up to 100 individual samples, from each of which five to 10 MS spectra are acquired. LipidXplorer software preprocesses the entire dataset such that individual spectra are organized in a single flat file database that is further interrogated by user-defined queries written in the molecular fragmentation query language (MFQL) (Herzog et al. 2011). In each MS spectrum the lipid class-specific MFQL query checks if plausible precursor masses match the elemental composition expected for the corresponding molecular species. If required, optional search criteria, such as the odd or even number of carbon atoms in the fatty acid moieties, expected number of double bonds, etc., are applied. These criteria could also be bundled by logical Boolean operations (AND, OR, NOT) (Schwudke et al. 2007b) helping to resolve ambiguous peaks assignments. In this way, the interpretation does not rely on a preconceived knowledge of the lipid composition of analyzed samples and is not associated with any particular mass spectrometry platform. However, the confidence of lipid identification and the number of unequivocally recognized lipid classes strongly depend on the mass resolution and mass accuracy of the employed instrument.
The top-down lipidomics screen of human blood plasma samples collected from 70 male patients with various manifestations of metabolic syndrome (obesity, insulin resistance, hypertension) serves here as a representative example (Graessler et al. 2009). Internal standards were spiked into plasma samples prior to one-step lipid extraction by methyl-tert-butyl ether (MTBE) (Matyash et al. 2008). It was previously shown that MTBE extracts lipids as efficiently as “gold standard” protocol of Bligh and Dyer (1959) or Folch et al. (1957). However, MTBE phase into which lipids are partitioned, is lighter than water and forms an upper layer in the two-phase system. It is convenient that nonextractable residual material (proteins, oligosaccharides, etc.) is collected in the water phase at the bottom of the test tube after gentle centrifuging. Therefore, it is easy to recover a clean lipid extract that is also free from “floating” solid particles disturbing or even completely blocking electrospray from micrometer-size nozzles (Kameoka et al. 2001).
Total lipid extracts were directly infused into a LTQ Orbitrap XL mass spectrometer and survey mass spectra were acquired within less than 3 min time at the target mass resolution of 100,000. High mass resolution, better than 5 ppm mass accuracy and practical compositional constraints identified lipid species directly solely relying on their accurately determined m/z (Fig. 2).
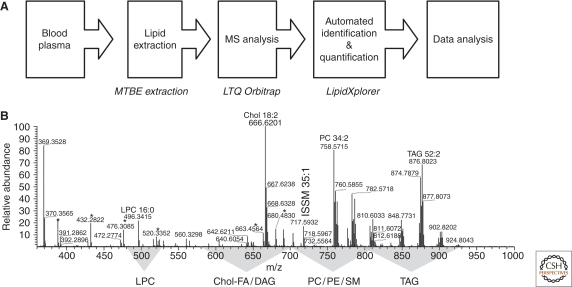
Figure 2.
Top-down lipidomics screen of human blood plasma extracts. (A) A workflow of a top-down lipidomics screen encompassing blood plasma samples collected from 70 patients. In total, 151 high-resolution spectra were acquired. (B) Representative MS spectrum of a total blood plasma extract acquired at the LTQ Orbitrap XL mass spectrometer. Here only the most abundant species are annotated; m/z ranges in which peaks of the species of major lipid class were located, are highlighted. Most abundant peaks of chemical noise are designated with asterisks. IS, internal standard spiked into samples prior to lipid extraction.
Within this screen high mass resolution alone distinguished peaks of isobaric species of several classes: ether lipids and ester lipids comprising fatty acids with odd number of carbon atoms; cholesterol esters and DAG; PC-O and TAG. Monoisotopic peaks of SM species were also distinguished from first isotope peaks of PCs. Altogether, 28 lipid species were resolved within the isobaric clusters. Unequivocal assignment of elemental compositions to peak masses allowed us to disregard background peaks. Indeed, from approximately 2500 peaks detected in each survey MS spectrum, only approximately 100 (<4%) represented monoisotopic peaks of bona fide lipid species.
To distinguish the species from lipid classes that might have the same exact masses, we imposed several compositional constraints. If the accurate mass (and, hence, unequivocally determined elemental composition) could be assigned to the species with odd and even number of carbon atoms in their fatty acid moieties, the software attributed the peak to the latter species. For example, for the peak with m/z 744.5538 detected in positive ion mode the software suggested the sum composition of C41H79O8N1P1 that could be interpreted as PE 36:2 or PC 33:2. The latter would imply that one of the fatty acid moieties must be odd and therefore the peak was recognized as the PE molecule. Of course, such compositional constraints should only be applied in the organism-specific manner: in particular, odd numbered fatty acid moieties are relatively rare in mammalian lipid species, yet are commonly found in major lipids of Caenorhabditis elegans (Entchev et al. 2008). Although top-down screens might rely on tentative peak assignments, the molecular identity of peaks showing interesting phenotype-specific abundance changes could be unequivocally established in follow-up MS/MS experiments.
Limited use of “slow” MS/MS, as compared to “fast” MS, is a major time-saving factor. For accurate quantification, MS/MS spectra of minor lipid species are usually acquired for 5–30 sec. Considering that more than 250 precursors are usually detectable in a survey MS spectrum, it might take 30–90 min to compete the analysis, instead of 3 min used in MS-based bottom-up screening. Acquiring MS/MS spectra from all detectable precursors would have reduced the analysis throughput by 10-fold, whereas most of the acquisition time would be spent on fragmenting precursors with unaffected abundances.
Altogether, within the constraints imposed on elemental compositions and detection reproducibility of lipid peaks, we were able to recognize and quantify 95 lipids from 10 major classes: 5 lipids belonging to Chol-FA, 4 to DAG, 5 to LPC, 13 to PC and 13 to PC-O, 8 to PE and 5 to PE-O, 10 to SM, 31 to TAGs, and free cholesterol. Statistical analysis of the variation of lipid abundances suggested that in male hypertensive patients the content of PE-O and PC-O species was decreased specifically and independently of other common manifestations of metabolic syndrome, such as obesity and insulin resistance. The molecular compositions of eight most affected PC-O and PE-O precursors were established in subsequent MS/MS experiments.
HIGH RESOLUTION MS/MS FOR THE CHARACTERIZATION OF INDIVIDUAL LIPID SPECIES
Lipid fragmentation pathways might look simple, compared to, for example, peptides. Indeed, lipid molecules contain only a few heteroatoms, whose bonds with carbon atoms are cleaved by low-energy collision-induced dissociation (Hsu and Turk 2009). Therefore, it is a common notion that mass resolution might be unimportant for lipid MS/MS because only a few fragments are produced.
In our experience, high resolution MS/MS contributes to the accurate characterization of molecular species in two ways. First, precise masses of fragment ions reveal important details of precursor structures. Second, completely resolved peaks of isobaric fragments enable accurate assignment of intensities to the characteristic reporter ions and simplify the quantification of molecular species. Below we substantiate this notion with a few representative case studies.
Accurate Interpretation of Spectra of Polyunsaturated Species
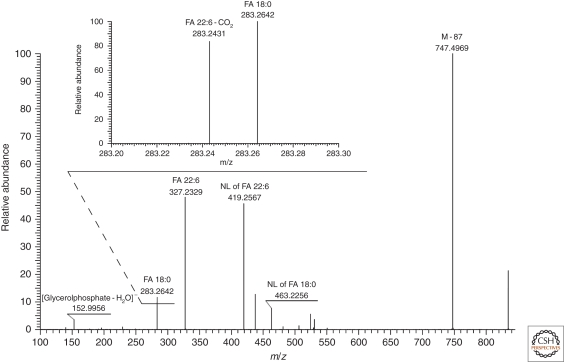
One problem alleviated by high resolution MS/MS is the quantification of glycerophospholipids comprising moieties of polyunsaturated fatty acids (PUFA). Following collisional activation, acyl anions of PUFA moieties could further lose CO2 (reviewed in Griffiths 2003). In this way, the acyl anion of docosahexaenoic (22:6) FA produces a fragment with m/z 283.2420, which is isobaric to the acyl anion of abundant stearic (18:0) FA (m/z 283.2632). Their peaks overlap in low resolution MS/MS spectra and therefore the quantification of individual species relies on a correction factor determined in the series of MS/MS experiments with lipid standards containing 22:6 moieties performed under the fixed collision energy offset (Ejsing et al. 2006). However, in high resolution Fourier transform (FT) MS/MS spectra peaks of [18:0]− and [22:6-CO2]− anions were completely resolved (Fig. 3) and their individual abundances were accounted for directly.
Figure 3.
FT MS/MS spectrum of PS 18:0/22:6 acquired in negative mode with the target mass resolution of 30,000 acquired on a LTQ Orbitrap XL instrument. Peaks of the acyl anion of FA 18:0 and the product of CO2 loss from the acyl anion of FA 22:6 were fully resolved.
FT MS/MS also supported the identification of rare species with unusual fatty acid moieties. For example, in shotgun analysis of rat retina extracts we detected a large number of peaks whose exact masses pointed to uncommon PE and PC species comprising very long chain PUFA moieties with more than 26 carbon atoms and more than four double bonds. Abundant peaks of acyl anions along with corresponding products of CO2 losses were observed in FT MS/MS spectra (Fig. 4). Characteristic fragment masses determined with the low ppm accuracy supported unequivocal identification of these very uncommon species.
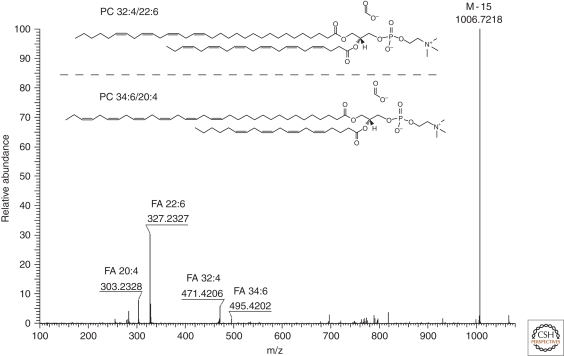
Figure 4.
FT MS/MS spectrum of a low abundant precursor ion with m/z 1066.7490 (0.8 ppm) detected by shotgun analysis of a total lipid extract of rat retina in negative ion mode. Spectrum interpretation revealed that this precursor ion represents two very long chain polyunsaturated species of PC 32:4/22:6 and PC 34:6/20:4 ([M + HCOO]−). As judged from the relative intensity of acyl anion fragments, FA 32:4 and 34:6 are at the sn − 1 position, whereas FA 22:6 and FA 20:4 are at the sn − 2 position of the glycerol backbone of the corresponding PC species. Masses of fragment ions were determined with the low ppm accuracy: [M-15]- fragment at m/z 1006.7218 (−5.2 ppm), acyl anion of FA 34:6 at m/z 495.4202 (−1.1 ppm), acyl anion of FA 32:4 at m/z 471.4206 (−0.3 ppm), acyl anion of FA 22:6 at m/z 327.2327 (−0.8 ppm), acyl anion of FA 20:4 at m/z 303.2328 (−0.5 ppm), and the product of neutral loss of CO2 from the acyl anion of FA 22:6 at m/z 283.2431 (−0.1 ppm).
High Resolution MS/MS Distinguishes Structurally Related Isobaric Species
In similar way, high resolution MS/MS simplified structural assignments in complex mixtures of isobaric species that are commonly encountered in the analysis of TAG. TAG are major energy storage and free fatty acids resource (reviewed in Gross and Han 2007). It is therefore not surprising that their molecular composition is highly diverse: in higher eukaryotes (Schwudke et al. 2006; Entchev et al. 2008) and mammals (McAnoy et al. 2005; Bartz et al. 2007) each isobaric cluster of precursors sharing the same number of carbon atoms and double bonds might represent five to 10 individual molecules differing by their fatty acid composition. In mammals TAG often include fatty acids with odd number of carbon atoms and branched fatty acids acquired through the food chain (Quehenberger et al. 2010). TAG might also include species with oxidized fatty acids and also recently identified alkyldiacylglycerols (Bartz et al. 2007).
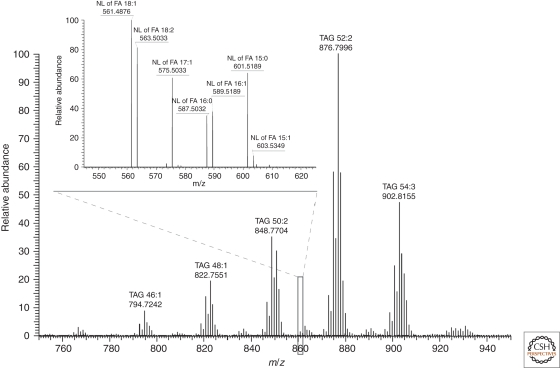
As an example, let us consider the analysis of molecular composition of precursor ion with m/z 860.7703 (Fig. 5). Note that, within 1.5 Da precursor isolation window, several species are typically coisolated for MS/MS. Some of them might be obscured by background and remained unrecognized in survey MS spectra, yet their fragments will be detectable in MS/MS spectra because of much reduced chemical noise. In this example, the accurate mass of isolated peak (m/z 860.7703) pointed to TAG 51:3 (mass difference of 0.2 ppm) having an odd numbered fatty acid moiety, whereas a putative isobaric ether lipid TAG–O 52:3 (expected m/z 860.8066; 42.1 ppm) was a less likely candidate. In FT MS/MS spectra fragments were clearly assigned to neutral losses of ammonia and fatty acid moieties of 15:1 (m/z 603.5349; −0.4 ppm); 15:0 (m/z 601.5189; −0.2 ppm); 16:1 (m/z 589.5189; −0.2 ppm); 16:0 (m/z 587.5032; −0.3 ppm); 17:1 (m/z 575.5033; −0.2 ppm); 18:2 (m/z 563.5033; −0.2 ppm); and 18:1 (m/z 561.4876; −0.2 ppm). Considering fragment intensities, TAG 15:0 / 18:1 / 18:2; TAG16:0 / 17:1 / 18:2; and TAG 16:1 / 17:1 / 18:1 are major molecular species among the cofragmented isobaric precursors.
Figure 5.
Part of the FT MS spectrum of a total extract of human adipose tissue acquired in positive ion mode at a LTQ Orbitrap XL mass spectrometer. TAG peaks were detected as ammonium adducts [M + NH4]+ and are annotated by m/z and total number of carbon atoms and double bonds. Following MS/MS fragmentation, TAG precursors underwent neutral losses of fatty acid moieties and ammonium. Inset shows a part of FT MS/MS spectrum acquired from precursor(s) having m/z 860.7703. Peaks of neutral loss fragments are designated with their m/z, the acronym NL XX:Y stands for the product of neutral loss of the acid moiety having XX carbon atoms and Y double bonds. Target mass resolution defined at m/z 400 was 100,000 and 30,000 in MS and MS/MS modes, respectively.
We further inspected the MS/MS spectrum seeking evidences whether putative TAG–O 52:3 was also present among minor cofragmented precursors. However, m/z of expected fragments of FA neutral losses from putative TAG–O 52:3 would have been 60 ppm off of m/z of observed fragments. Despite very low level of chemical noise, fragment peaks corresponding to a putative ether TAG species were undetectable. Both MS and MS/MS spectra suggested that the fragmented precursor was indeed TAG 51:3 that contained the odd numbered fatty acids 15:1, 15:0, and 17:1 and there was no evidence of the presence of alternative isobaric ether species. We note that in low resolution MS/MS spectra the presence of odd numbered fatty acid moieties (that, according to a common notion, should not be produced by mammals) could have been a frequent source of confusing structural assignments.
CONCLUDING REMARKS
In this article, we intended to show how the advent of novel high resolution mass spectrometers is changing the basic concept of lipidome characterization. For a long period of time the importance of achieving high mass resolution has been recognized, yet not accorded the attention it really deserves. Part of the problem was that applying high resolution machines to routine lipidomics analysis was technically challenging and required the level of expertise only available in a few dedicated mass spectrometric laboratories. This changed considerably with the introduction of the family of Orbitrap tandem mass spectrometers and routinely achieving the mass resolution of 100,000 both in MS and MS/MS.
The demand for higher mass resolution in lipidomics has one simple rationale: unique sum compositions can be attributed to fully resolved peaks of isobaric precursors or fragments. As shown above, this dramatically simplified the interpretation of MS and MS/MS spectra, which, in turn, streamlined qualitative and quantitative characterization of complex eukaryotic lipidomes. However, even a very high mass resolution does not distinguish molecules with identical masses and does not circumvent other well known analytical limitations, such as dynamic range, signal-to-noise, ion statistics, etc.
Acquiring a quality data set of high resolution mass spectra is only a part of the lipidomics pipeline. Importantly, spectra interpretation algorithms and software should support resolution-dependent interpretations. In the simplest scenario, the same signal could be recognized as an occasional spike and removed from a low resolution mass spectrum, yet it should be treated as a bona fide peak in a high resolution spectrum. Note that there is no clear cut numerical threshold that distinguishes high and low resolution spectra, it is more that the actual mass resolution should be used as an obligatory peak attribute in any spectra interpretation routine. There is an urgent need in developing such algorithms and software. As shown above, a practical solution could be to apply flexible data processing and interpretation rules, rather than building instrument- and project-specific spectra database resources.
We envision that in forthcoming years high resolution mass spectrometry will gradually develop into a “gold standard” lipidomics methodology and efforts should be made to adapt biochemical and analytical routines and software to fulfill its potential.
ACKNOWLEDGMENTS
We are grateful for our colleagues in MPI of Molecular Cell Biology and Genetics, Dresden for useful discussions and experimental support and to Kathy Eisenhofer for critical reading of the manuscript. A sample of rat retina was kindly provided by Prof. Vadim Arshavsky, Duke University Eye Center. D.S. is supported by Wellcome Trust/DBT India Alliance and is a recipient of NCBS–Merck & Co International Investigator Award. Work in A.S.'s laboratory is supported by a TRR 83 grant from Deutsche Forsungsgemeinschaft and the Virtual Liver Network program from Bundesministerium f. Bildung u. Forschung.
Footnotes
Editor: Kai Simons
Additional Perspectives on The Biology of Lipids available at www.cshperspectives.org
REFERENCES
- Bartz R, Li WH, Venables B, Zehmer JK, Roth MR, Welti R, Anderson RG, Liu P, Chapman KD 2007. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res 48: 837–847 [DOI] [PubMed] [Google Scholar]
- Blanksby SJ, Mitchell TW 2010. Advances in mass spectrometry for lipidomics. Annu Rev Anal Chem (Palo Alto Calif) 3: 433–465 [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ 1959. A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Brenton AG, Godfrey AR 2010. Accurate mass measurement: Terminology and treatment of data. J Am Soc Mass Spectrom 21: 1821–1835 [DOI] [PubMed] [Google Scholar]
- Dennis EA 2009. Lipidomics joins the omics evolution. Proc Natl Acad Sci 106: 2089–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejsing CS, Duchoslav E, Sampaio J, Simons K, Bonner R, Thiele C, Ekroos K, Shevchenko A 2006. Automated identification and quantification of glycerophospholipid molecular species by multiple precursor ion scanning. Anal Chem 78: 6202–6214 [DOI] [PubMed] [Google Scholar]
- Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A 2009. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci 106: 2136–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekroos K, Chernushevich IV, Simons K, Shevchenko A 2002. Quantitative profiling of phospholipids by multiple precursor ion scanning on a hybrid quadrupole time-of-flight mass spectrometer. Anal Chem 74: 941–949 [DOI] [PubMed] [Google Scholar]
- Ekroos K, Ejsing CS, Bahr U, Karas M, Simons K, Shevchenko A 2003. Charting molecular composition of phosphatidylcholines by fatty acid scanning and ion trap MS3 fragmentation. J Lipid Res 44: 2181–2192 [DOI] [PubMed] [Google Scholar]
- Entchev EV, Schwudke D, Zagoriy V, Matyash V, Bogdanova A, Habermann B, Zhu L, Shevchenko A, Kurzchalia TV 2008. LET-767 is required for the production of branched chain and long chain fatty acids in Caenorhabditis elegans. J Biol Chem 283: 17550–17560 [DOI] [PubMed] [Google Scholar]
- Folch JM, Lees M, Sloane-Stanley GH 1957. A simple method for the isolation and purification of total lipids from animal tissue. J Biol Chem 226: 497–509 [PubMed] [Google Scholar]
- Glish GL, Burinsky DJ 2008. Hybrid mass spectrometers for tandem mass spectrometry. J Am Soc Mass Spectrom 19: 161–172 [DOI] [PubMed] [Google Scholar]
- Graessler J, Schwudke D, Schwarz PE, Herzog R, Shevchenko A, Bornstein SR 2009. Top-down lipidomics reveals ether lipid deficiency in blood plasma of hypertensive patients. PLoS One 4: e6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths WJ 2003. Tandem mass spectrometry in the study of fatty acids, bile acids, and steroids. Mass Spectrom Rev 22: 81–152 [DOI] [PubMed] [Google Scholar]
- Griffiths WJ, Wang Y 2009. Mass spectrometry: From proteomics to metabolomics and lipidomics. Chem Soc Rev 38: 1882–1896 [DOI] [PubMed] [Google Scholar]
- Gross JH 2006. Mass spectrometry: A textbook. Springer, New York [Google Scholar]
- Gross RW, Han X 2007. Lipidomics in diabetes and the metabolic syndrome. Methods Enzymol 433: 73–90 [DOI] [PubMed] [Google Scholar]
- Han X, Gross RW 2005. Shotgun lipidomics: Electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev 24: 367–412 [DOI] [PubMed] [Google Scholar]
- Herzog R, Schwudke D, Schuhmann K, Sampaio JL, Bornstein SR, Schroeder M, Shevchenko A 2011. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol 12: R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houjou T, Yamatani K, Imagawa M, Shimizu T, Taguchi R 2005. A shotgun tandem mass spectrometric analysis of phospholipids with normal-phase and/or reverse-phase liquid chromatography/electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom 19: 654–666 [DOI] [PubMed] [Google Scholar]
- Hsu FF, Turk J 2009. Electrospray ionization with low-energy collisionally activated dissociation tandem mass spectrometry of glycerophospholipids: Mechanisms of fragmentation and structural characterization. J Chromatogr B Analyt Technol Biomed Life Sci 877: 2673–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu FF, Turk J 2010. Electrospray ionization multiple-stage linear ion-trap mass spectrometry for structural elucidation of triacylglycerols: assignment of fatty acyl groups on the glycerol backbone and location of double bonds. J Am Soc Mass Spectrom 21: 657–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova PT, Milne SB, Myers DS, Brown HA 2009. Lipidomics: a mass spectrometry based systems level analysis of cellular lipids. Curr Opin Chem Biol 13: 526–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameoka J, Craighead HG, Zhang H, Henion J 2001. A polymeric microfluidic chip for CE/MS determination of small molecules. Anal Chem 73: 1935–1941 [DOI] [PubMed] [Google Scholar]
- Makarov A, Denisov E, Kholomeev A, Balschun W, Lange O, Strupat K, Horning S 2006a. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal Chem 78: 2113–2120 [DOI] [PubMed] [Google Scholar]
- Makarov A, Denisov E, Lange O, Horning S 2006b. Dynamic range of mass accuracy in LTQ Orbitrap hybrid mass spectrometer. J Am Soc Mass Spectrom 17: 977–982 [DOI] [PubMed] [Google Scholar]
- Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D 2008. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res 49: 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAnoy AM, Wu CC, Murphy RC 2005. Direct qualitative analysis of triacylglycerols by electrospray mass spectrometry using a linear ion trap. J Am Soc Mass Spectrom 16: 1498–1509 [DOI] [PubMed] [Google Scholar]
- Niemela PS, Castillo S, Sysi-Aho M, Oresic M 2009. Bioinformatics and computational methods for lipidomics. J Chromatogr B Analyt Technol Biomed Life Sci 877: 2855–2862 [DOI] [PubMed] [Google Scholar]
- Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M 2005. Parts per million mass accuracy on an orbitrap mass spectrometer via lock-mass injection into a C-trap. Mol Cell Proteomics 4: 2010–2021 [DOI] [PubMed] [Google Scholar]
- Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, Lange O, Remes P, Taylor D, Splendore M, et al. 2009. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol Cell Proteomics 8: 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oresic M, Hanninen VA, Vidal-Puig A 2008. Lipidomics: A new window to biomedical frontiers. Trends Biotechnol 26: 647–652 [DOI] [PubMed] [Google Scholar]
- Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, Bandyopadhyay S, Jones KN, Kelly S, Shaner RL, et al. 2010. Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res 51: 3299–3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwudke D, Hannich JT, Surendranath V, Grimard V, Moehring T, Burton L, Kurzchalia T, Shevchenko A 2007a. Top-down lipidomic screens by multivariate analysis of high-resolution survey mass spectra. Anal Chem 79: 4083–4093 [DOI] [PubMed] [Google Scholar]
- Schwudke D, Liebisch G, Herzog R, Schmitz G, Shevchenko A 2007b. Shotgun lipidomics by tandem mass spectrometry under data-dependent acquisition control. Methods Enzymol 433: 175–191 [DOI] [PubMed] [Google Scholar]
- Schwudke D, Oegema J, Burton L, Entchev E, Hannich JT, Ejsing CS, Kurzchalia T, Shevchenko A 2006. Lipid profiling by multiple precursor and neutral loss scanning driven by the data-dependent acquisition. Anal Chem 78: 585–595 [DOI] [PubMed] [Google Scholar]
- Scigelova M, Makarov A 2006. Orbitrap mass analyzer—Overview and applications in proteomics. Proteomics 6: 16–21 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Simons K 2010. Lipidomics: Coming to grips with lipid diversity. Nat Rev Mol Cell Biol 11: 593–598 [DOI] [PubMed] [Google Scholar]
- van Meer G 2005. Cellular lipidomics. Embo J 24: 3159–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW 2008. Membrane lipids: Where they are and how they behave. Nat Rev Mol Cell Biol 9: 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenk MR 2005. The emerging field of lipidomics. Nat Rev Drug Discov 4: 594–610 [DOI] [PubMed] [Google Scholar]
- Yergey J, Heller D, Hansen G, Cotter RJ, Fenselau C 1983. Isotopic distributions in mass spectra of large molecules. Anal Chem 55: 353–356 [Google Scholar]
- Yetukuri L, Ekroos K, Vidal-Puig A, Oresic M 2008. Informatics and computational strategies for the study of lipids. Mol Biosyst 4: 121–127 [DOI] [PubMed] [Google Scholar]