Abstract
Genetically engineered mice are critical experimental models for the study of breast cancer biology. Transgenic mice, employing strong mammary epithelial promoters to drive oncogenes, develop carcinomas with phenotypes corresponding to the molecular pathway activated. Gene-targeted (knockout) mice, in which tumor suppressors are deleted, develop mammary neoplasms with phenotypes primarily including patterns seen in spontaneous mouse mammary tumors, albeit at higher rates. Improved genetic engineering, using inducible gene expression, somatic gene transduction, conditional alleles, and crossbreeding for combined/compound genetic engineering yields precise molecular models with exquisite experimental control and phenotypes with comparative pathologic validity. Mammary gland transplantation technology adds a practical and validated method for assessing biologic behavior of selected mammary tissues. Overall, the many mouse models available are a rich resource for experimental biology with phenocopies of breast cancer subtypes, and a variety of practical advantages. The challenge is matching the model to the experimental question.
Human cancers are genotypically, phenotypically, and behaviorally heterogeneous. However, specific molecular alterations (e.g., Erbb2 amplification, found in 25% of human breast cancers) have been successfully modeled in mice.
There is no perfect model system for studying breast cancer. Breast cancer in women constitutes an array of different diseases, and it is not realistic to consider breast cancer as a single disease. Experimental models are needed to reflect each part of the array. Cell culture lines are the most practical, and a panel of 51 standard cell lines is currently available reflecting most of the intrinsic subtypes of breast cancer (Neve et al. 2006). These cell lines are adapted to growth on two-dimensional (2D) plastic dishes and represent an oversimplified biology out of context. Growth in 3D culture is more contextual, but still simplified (Kenny et al. 2007). Even xenografting of human cell lines in mice is artificial, short-circuiting the precancer stage, the development of a permissive environment, and eliminating the immune system. Mouse models, particularly the genetically defined models offer a step in the direction of biologic reality. Choosing among the mouse models of breast cancer is not a trivial exercise. Literally thousands of specifically engineered mice have been created to study the pathology, molecular biology, natural history, and response to therapy in breast cancer (Borowsky 2007). These successes of mouse models of breast cancer stem from the important foundations in mouse mammary biology. First, the biology of the mouse mammary gland has been studied for many years. The normal development, lactation related changes, and spontaneous tumors in the mouse mammary gland are well characterized. Second, several mammary specific gene promoters have been characterized, permitting ready transgenic engineering to incorporate genes into the mammary glands of mice (Borowsky 2003). Third, the mammary gland of mice is transplantable (Daniel et al. 1968). Nevertheless, several important challenges to the validity (Cardiff et al. 2004) of the mouse models of breast cancer remain. Few of the models are accurate representations of the estrogen receptor (ER) positive human breast cancers. Metastases, which occur in several models, are hematogenous and almost exclusively pulmonary, whereas human breast cancer metastases are characterized by nearly universal lymphatic spread and nodal metastasis that precedes or is synchronous with disseminated hematogenous metastasis. Human breast cancer does metastasize to the lung, but also commonly to the liver, bone, and brain, and these sites are rarely affected in the mouse models without specific designs to adapt the cells for nonpulmonary homing.
Many of the current genetically engineered mouse models of breast cancer have been previously reviewed and categorized (Cardiff et al. 2000; Cardiff 2003). Using a gross oversimplification, the existing models can be grouped into two categories: (1) transgenic models, those with specific oncogene expression, usually through highly active mammary-specific promoter-driven expression; and (2) gene-targeted models, those with an increased susceptibility to mammary tumorigenesis, usually through gene targeting (knockout) of a tumor suppressor. Each of these methods has specific strengths and problems, and new refinements of the engineering seek to solve some of the problems. Methods for introducing defined genetic changes in somatic cells in the mammary gland (rather than germline) have also been developed, perhaps constituting a third category which includes conditional alleles, temporally inducible expression, localized inducible expression, and transplantation. These methods often require a complex combination of genetically engineered components, and as such are covered only briefly here, techniques reviewed in (Heyer et al. 2010). In any event, the work of many labs has resulted in an array of mice modeling specific subsets of human breast cancer. As an introduction to the array, some of the models are tabulated with the corresponding human tumor types in Table 1. This is not a complete list, and is offered with apologies to the many labs and investigators who have created unique and useful mouse models of breast cancer. Some intrinsic subtypes of human cancer have been modeled much more commonly, whereas the ER positive “luminal A” and “luminal B” human cancers are seen much less often in the array of genetically engineered mice.1
Table 1.
Human breast cancer types in order of frequency with corresponding mouse models, phenotypes, and validation comments
| Human breast cancer types | Relative frequency (approx.) | Mouse models | Phenotype and behavior | Validation comments |
|---|---|---|---|---|
| Invasive ductal carcinoma | 75% | |||
| ER positive (luminal A, luminal B) | 1/2 of 75% | Stat1−/−; some Tp53 tumors | Highly ER positive; weak to moderate ER positivity in many | Low penetrance; highly variable; interanimal and intra-animal heterogeneity |
| ERBB2 amplified (some are also ER+) | 1/4 of 75% | Tg(neu); Tg(PymT); Erbb2 knockin | Solid, minimally glandular growth, metastatic | Highly consistent phenotypes. ER is weak or negative. Erbb2 knockin develops endogenous Erbb2 locus amplification in one-half of tumors. |
| “Triple negative” (basal type) | 1/4 of 75% | Some Brca/Tp53 tumors; Tg(Sv40Tag) | Range from basal-like to spindled (below) | Less variability than the Tp53-only model, specificity to a spectrum from basal to spindled suggests a common pathway. |
| Lobular carinoma (most are ER+) | 10% | Ecad/Tp53 knockout | Discohesive round infiltrative cells; metastatic | Morphology highly comparable to human disease except ER-negative and higher grade than typical human invasive lobular carcinoma. |
| Mucinous (colloid) carcinoma | 3% | None | ||
| Tubular carcinoma | 2% | None | ||
| Medullary carcinoma | 1% | None | ||
| Myoepithelioma | Rare | Tg(Wnt); Tg(Fgf8); Tp53 knockouts | Highly variable, inter- and intra-animal heterogeneity | The “Wnt” pathway pathology includes all of the common mammary tumor phenotypes seen spontaneously in mice. |
| Salivary gland type | Rare | Tg(Wnt); Tg(Fgf8); Tp53 knockouts | Highly variable, inter- and intra-animal heterogeneity | |
| Metaplastic spindled cell | Rare | Brca1/Tp53 knockouts; Tg(MMP3) | Range from basal-like (above) to spindled | Epithelial mesenchymal transition in these models with consistent evolution of spindle cell lesions from mammary epithelium. |
| Metaplastic squamous/pilar | Rare | Tg(Wnt); Tg(Fgf8); Tp53 knockouts | Typically a pilomatricoma (hair follical tumor) phenotype | Squamous and pilar differentiation are common in spontaneous and mutagen-induced mouse mammary tumors, whereas squamous differentiation is uncommon in human breast cancer. |
| Secretory carcinoma | Rare | Wap-Cre/EN fusion | Papillary without clear secretory features | Example of a precise human to mouse molecular model without corresponding phenotype association. |
| Precancer and benign diseases | Clinical relevance | Mouse models | Phenotype and behavior | Validation comments |
| Ductal carcinoma in situ | Associated with invasive cancer | MINO transplants; Tp53 transplants; Tg(SV40Tag) | MINOs have an array of dysplasias with progression to Erbb2 tumor phenotypes; Tp53 and Tg(SV40Tag) models have high-grade in situ neoplasias. | MINO and Tp53 transplants fulfill “test-by-transplantation” while the Tg(SV40Tag) areas of in situ disease arise multifocally making it hard to trace the progression of a single lesion. |
| Lobular neoplasia (LCIS) | Increased relative risk | None | Ecad/Tp53 knockout mice do not develop a recognizable in situ lesion, the early tumors are very different from human LCIS. | |
| Hyperplasia | Common, benign | Many | ||
| Intraductal papillomas | Common, sometimes atypical | Tg(Wnt); Tg(Fgf8); Tp53 knockouts | Papillary lesions in mice often become invasive, whereas invasive papillary carcinomas in human breast are rare. | |
Note, in addition to breast cancer subtypes, mouse models of precancer (ductal carcinoma in situ) and nonneoplastic breast diseases are also included.
MAMMARY-SPECIFIC TRANSGENIC ANIMALS
The overexpression of an oncogene under the control of one of the mammary-specific promoter sequences in the mouse mammary gland results in multifocal (nearly diffuse) expression throughout the luminal epithelium of all of the mammary glands of the genetically engineered mouse. The commonly used promoters, including the mouse mammary tumor virus long terminal repeat (MMTV-LTR), the whey acidic protein (WAP), and the β lactoglobin (BLG) promoter sequence are all highly active in luminal epithelial cells, all drive high levels of transcription, and all have some hormonal dependence (hormone responsive elements and glucocorticoid responsive elements) creating a potential for “artificial” hormone and corticoid sensitivity through direct modulation of the oncogene expression. This is regarded as “artificial” because this mechanism is not involved in human breast cancer; that is, oncogene expression in human breast cancer is not known to depend directly on transcriptional activation via hormonal signaling.
To avoid targeting only luminal epithelium some groups have used a keratin 14 promoter sequence (Derksen et al. 2006). Keratin 14 (K14) is expressed in mammary basal cells, often coexpressed in mature myoepithelial cells but not in mature luminal epithelium. It is clear that these basal cells can differentiate into luminal cells, whereupon they down-regulate K14 expression. K14 promoters used to drive oncogene expression may therefore result in a bias away from luminal differentiation (or alternatively selection of transformed cells independent of the oncogene). K14 is also expressed in other organs such as skin, salivary glands, and pancreas resulting in a compromise of mammary specificity. Nonetheless, this strategy permits targeting of a different potential cell of origin in cancer, helping to address questions of the requirements for cell of origin susceptibility.
The use of the tetracycline inducible promoter sequences (utilizing a mammary-specific promoter to drive the tet trans-activator and a second tet responsive element driven promoter) substantially reduces the hormone dependence because the promoter activity is regulated by the presence of tetracycline/doxycycline as the major rate limiter (Gunther et al. 2002; Moody et al. 2002). Still, the MMTV-LTR used to drive the trans-activator is not equally active in all mammary cell types. Nonetheless, the benefits of exogenous control of oncogene expression which can be driven at high levels, even with changes in estrogen and glucocorticoid signaling, is critical to a wide variety of study designs.
As a result of the diffuse gland-wide epithelial expression driven by strong mammary promoters, multifocal tumorigenesis is the rule in these animals. This is unlike human cancer, which arises in a single focus, or, less often, in just a few separate foci. The earliest tumors in these mice are typically seen in the proximal glands (nearest the nipples) likely because this is the oldest part of the gland, which develops over the first 6–12 weeks of life by extending and branching outward from the nipple to fill the mammary fat pads. Multiple synchronous and metachronous tumors arise, therefore, depending primarily on the duration of oncogene expression in a particular area of the mammary tree. The tumor phenotypes are directly related to the oncogene such that “signature” phenotypes are now recognized for a handful of molecular activation “pathways”(Rosner et al. 2002; Du et al. 2006). In many instances, individual genes within a given pathway have been shown to result in tumors with identical phenotypes. This has provided a functional proof, in fact, that the molecular models are accurate and fulfills an argument like Koch's postulate. If an oncogene, in this case, rather than a microorganism, is isolated from a diseased patient, and can be readministered to an animal reproducing the disease, the agent (oncogene) is considered the cause of the disease.
These animals, created at a time of emerging understanding of the molecular pathways involved in human cancers, have shown that the tumor phenotypes which emerge are directly related to the molecular pathway affected by the transgene. More simply, the genotype creates or causes the phenotype of the tumor, or conversely, the phenotype is a readout of the genotype. This concept has been characterized by the Robert Cardiff laboratory, where the histopathology for most of the available genetically engineered mouse models of mammary cancer have been reviewed. Cardiff has dubbed the concept “pathway pathology,” denoting the relationship between the molecular biologic pathway and the resulting histomorphologic pathology. In particular, he has shown that genetic engineering of the Wnt pathway, the Erbb2 pathway, and also Myc and Ras activation have highly characteristic “signature” phenotypes. Wnt pathway tumors are myoepithelial, papillary, or pilocytic/squamous phenotypes that resemble human salivary and skin adnexal tumors more closely than breast cancers (Rosner et al. 2002).
GENE-TARGETED KNOCKOUT MODELS
In contrast to the transgenic models, tumor suppressor knockout mice typically develop tumors after longer latency and with much lower multifocality. In the standard germline knockout mice, tumor susceptibility is not limited to the mammary gland, and in general these animals are most strongly prone to lymphomas and sarcomas. Nevertheless, mammary epithelial neoplasms do occur providing a model mimicking both hereditary cancer syndromes and the somatic mutation or loss of tumor suppressors.
Even within a genetically identical tumor suppressor knockout colony, heterogeneity of tumors is the rule. It appears that somatic molecular changes to activate additional molecular pathways, in addition to the genetic engineering, are required in tumor development with varying latencies and phenotypes. The most common phenotypes are those in the Wnt pathway, which are also the most commonly seen spontaneous tumors in mice, and are identical to the tumors seen in insertional mutagenesis studies. Wnt and the Wnt pathway member Fgf3 were, in fact, originally cloned as mouse tumor related genes, based on mouse mammary tumor virus insertion site analysis in viral-induced tumors. Of considerable recent interest, combined tumor suppressor knockout mice employing more complex Cre recombinase “conditional” alleles may have consistent cancer phenotypes which compare closely to more common human breast cancer phenotypes (Jonkers and Berns 2002; Borowsky 2003). One of these models, for example, knocks out Tp53 and Ecad, resulting in a cancer with striking similarity to invasive lobular carcinoma, the most common “special type” breast cancer encountered in women (Derksen et al. 2006). Of note, this phenotype had never been seen spontaneously in viral mutagenesis, chemical mutagenesis, or even arising in the Tp53 knockout by itself. A more complete description of these Ecad mice is included below.
Erbb2 MODELS
Erbb2 amplification is seen in 25% of human breast cancers. Erbb2 is normally expressed on the surface of both mouse and human luminal mammary epithelial cells, but the expression is quantitatively increased as a result of gene copy amplification. Because the mechanism appears to simply involve increased expression, mouse models employing highly active promoters rather than amplification provide a valid surrogate molecular model of the human disease. The William Muller laboratory was discontent to study only one method of Erbb2 over expression in the target mouse mammary epithelium, and have produced a series of engineered mice including the Erbb2/ERBB heterodimer mimic, Polyomavirus middle t antigen (PyV-mT), several Erbb2 mutations, and most recently, an activation mutant Erbb2 targeted to the native locus. This last model is designed to express an activated form of the Erbb2 molecule at normal levels, under normal promoter regulation. Remarkably, one result of this manipulation is that there is locus amplification of Erbb2 in some of the resulting tumors. The details of the variety of Erbb2 manipulations were recently reviewed (Ursini-Siegel et al. 2007) and can be found in Lee and Muller (2010).
Included in the category of Erbb2 models, is one of the most commonly used transgenic mammary carcinoma models, the FVB/n Tg(MMTV-PyVmT) mouse. This mouse employs a surrogate molecule that mimics the effect of Erbb2 heterodimers (i.e., Erbb2/Erbb3 dimers) but in a ligand-independent and dimerization-independent manner. Otherwise, the downstream activation is very similar with activation of PI3K, Shc, Src, and also PP2a (Dilworth 2002). This model is therefore molecularly relevant to human cancers, at least those with Erbb2 amplification. In addition, this model has important practical advantages. Tumorigenesis is 100% penetrant, and tumors have a consistent latency occurring by 20 weeks of age in all cases. In fact, small nascent tumors can be detected microscopically much earlier, sometimes as early as 4 weeks of age. Tumors appear to arise from in situ lesions within a normally developed ductal tree. Multiple foci of tumor occur in all of the mammary glands, and the earliest tumors arise in the region of the proximal (nearer the nipple) gland. These animals develop pulmonary metastases at very high rates.
The comparative pathology of these Erbb2 models with human cancers includes several important considerations. First, as mentioned above, these mice develop tumors with a signature phenotype. The phenotype is characterized by solid growth with minimal but evident residual glandular lumen formation, and a peripheral and central zone of larger and more flattened cells (respectively) representing tumoral differentiation. In contrast, human Erbb2 amplified breast cancers are highly heterogeneous, with sclerosing and solid and high and low grade all occurring in human populations. The human amplified tumors are more likely to be high grade with solid and trabecular growth patterns, but essentially all phenotypes are possible. The mouse tumors are consistently estrogen receptor weak expressers. As discussed below, the comparative pathology of ER expression in mouse versus human is a distinct issue, and this is also true in the Errb2 models. Human Erbb2 amplified breast cancers are most often ER negative, but may also be positive, even strongly and diffusely positive. In contrast, the mouse tumors are either negative or weakly and focally positive, but have been shown to be at least partially estrogen responsive (Namba et al. 2005).
NONGERMLINE GENETICALLY ENGINEERED MOUSE MODELS
A number of approaches for generating chimeric mice bearing tumor prone genetics in a subset or minority of cells have been developed (Heyer et al. 2010). Introduction of transgenes via viral infection in vivo has been employed (Bartek et al. 1991) and transgenic mammary expression (driven by the MMTV-LTR) of a viral receptive sequence (“RCAS”) has been used to increase the efficiency of this approach (Du et al. 2006). This has the added benefit of reducing the foci of initiation, more closely modeling human cancer, but has the disadvantage of relatively low efficiency and technical difficulty usually requiring nipple cannulation and duct lavage with the viral preparation.
None of the nongermline approaches is better developed or as reproducible in utility as mammary transplantation. Transplantation of the mouse mammary gland was pioneered in the DeOme lab and has been used to study the biology of specific mammary tissues for many years (Medina and DeOme 1970a,b; Medina et al. 1970; Cardiff et al. 2002). The developing mammary gland can be cleared from the inguinal (#4) fat pad by removing the proximal gland at three weeks of age. This is performed in mice syngeneic to the expected transplanted tissue or in immune incompetent nude (nu/nu) or SCID (Rag−/−) mice. Normal mammary gland fragments can be transplanted in this way, and show normal branching morphogenesis, filling the remaining stromal fat pad. After serial transplantation, normal mammary tissues senesce after several generations (Daniel and Young 1971). The history and experimental details involving the development of these techniques is provided in the articles by Medina (2010) and Cardiff and Kenney (2011).
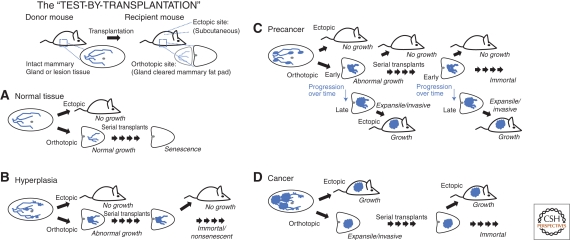
Transplantation of normal, hyperplastic, putative neoplastic or preneoplastic lesions, or selected cells allows evaluation of the specific biologic potential with respect to (1) immortalization, (2) phenotype/morphology, (3) proliferative rates, (4) malignant potential, and (5) metastatic potential. This evaluation has been called a “test-by-transplantation,” in which experimental criteria define the lesion rather than associative morphology,2 as illustrated in Figure 1. The test may be too stringent in some respects, because some locally invasive tumors arising in the fat pad may not be able to grow ectopically without further adaptations, but the important point to notice is the role of experimental criteria–based definitions that can be used in mice. Meanwhile human disease potential must be assessed by odds ratios after best efforts at curative treatment (i.e., one cannot study untreated tumors in people). The use of mammary gland transplantation reduces the complexity encountered in the native transgenic models in which it is impossible3 to trace independent lineages from the multiple emerging foci of neoplasia seen in these models. Isolation of individual gland segments, foci of hyperplasia, cysts, or other lesions can be accomplished in this way. Also notable, in mice bearing tumor suppressor knockouts such as the Tp53 knockout mouse, transplantation of the mammary gland permits evaluation of the effect which would otherwise be severely time limited because of the other malignancies (chiefly lymphomas) which occur in the knockout animals. In contrast, transplantation of Tp53 null mammary epithelium from a younger knockout animal into a Tp53 wild-type (“normal” syngeneic) mouse precleared fat pad allows the development and progression to malignancy to be evaluated over the lifespan of the mouse.
Figure 1.
The test by transplantation. Transplantation in orthotopic or ectopic sites provides an experimental definition for normal (A), hyperplastic (B), precancer (C), or cancer (D) in mammary epithelial tissues. Fresh tissue fragments can be harvested from a “donor” mouse and transplanted into a “recipient” mouse, either in the orthotopic site—a gland-cleared mammary fat pad—where the tissue proximal to the lymph node in the inguinal mammary stroma is removed at 3 weeks of age; or, in an ectopic site such as subcutaneous stroma. (A) Normal tissue transplanted into the orthotopic site yields a normal gland outgrowth, but does not grow ectopically. After sequential “serial” transplant generations, the normal tissue will senesce, eventually resulting in no outgrowth. (B) Hyperplasias, in contrast, will not senesce after multiple serial transplant generations, but they still will not grow ectopically. (C) Precancer tissues are defined by immortal growth in serial transplantation, but are also defined by a progression to a lesion that will grow ectopically. (D) Cancer tissues, meanwhile, are consistently able to grow in either the orthotopic or ectopic site.
With recent evidence for mammary “stem” cells, and mammary cancer “stem” cells, transplantation into cleared fat pad has been an important functional test for the potential of individual cells (Alvi et al. 2003). In many examples, dissociated cells are injected in serial dilution studies, but in some examples, a single cell has reportedly been transplanted giving rise to a complete ductal tree (Shackleton et al. 2006). This work is reviewed in detail in the article by Visvader and Smith (2011).
THE MINO MODEL
In order to overcome the problem of multifocality inherent in transgenic oncogene expression models, serial transplantation of precancerous mammary epithelia derived from the FVB/n Tg(MMTV-PyVmT) mouse, and resulted in a consistent progression model (Maglione et al. 2001, 2004; Namba et al. 2004). This model, called mammary intraepithelial neoplasia outgrowth (MINO) has become an important model of the human precancer lesion, ductal carcinoma in situ (DCIS). Each of the six MINO lines meets the test-by-transplantation criteria: (a) grows in gland-cleared fat pad (orthotopic); (b) does not grow in the subcutis (ectopic); (c) does not senesce over many generations of transplantation; and (d) consistently transforms to a phenotype characterized by an ability to grow in the subcutis (ectopic). The time or latency to transformation is consistent within a given MINO line over multiple transplant generations, though different lines have different latencies. Gene expression analysis and hierarchical clustering shows that a MINO transplant and the transformed lesion arising within it are more closely related than any two MINO lines or any two transformed tumors (Maglione et al. 2004; Namba et al. 2004). This is exactly analogous to findings in human DCIS and paired invasive cancers (Sgroi et al. 1999), providing a molecular validity of the model even in the context of tumor (and pretumor) heterogeneity. This model has now been used in a series of preclinical trials and chemoprevention studies capitalizing on the high reproducibility/consistency of the model as well as the ease of setting up a simultaneous cohort of animals through transplantation (Abbey et al. 2004, 2006; Namba et al. 2005, 2006).
TUMOR SUPPRESSOR KNOCKOUTS
A number of mouse models of tumor suppressor loss, either as germline knockouts, or as somatic “conditional” knockouts in specific cell subsets have been developed. The most commonly studied is the Tp53 knockout mouse (Medina et al. 2002). Several additional tumor suppressors have also been targeted in the study of mammary cancer progression. Rb has been targeted, but most often, the combination of Tp53 and Rb is targeted using the SV40 large T antigen as a transgene (Maroulakou et al. 1994). Because SV40 large T functions primarily to inactivate these two tumor suppressors, this transgenic mouse is therefore included here with the tumor suppressor knockouts. Additional tumor suppressors targeted include the Bloom's syndrome gene Blm1, the familial breast cancer genes Brca1, Brca2, and E-Cadherin.
Tp53 KNOCKOUTS
Three approaches have been taken in order to study Tp53 knockout absent the confounding factors of nonmammary tumors. The nonmammary tumors encountered most commonly in these germline (total body) Tp53 knockout models are (in order of incidence rates) thymic lymphomas, other lymphoma, hemangiosarcoma, teratoma, osteosarcoma, and Hardarian gland tumors (Cressman et al. 1999; Kuperwasser et al. 2000). Mammary tumors occur in these mice at ∼1%–2%. This is a significant practical limitation, but avoidable using one of three approaches. (1) Tp53 null mammary epithelium transplanted into syngeneic or immune incompetent and Tp53 wild type recipient mice using gland-cleared mammary fat pad transplantation. The approach uses the same mammary transplantation techniques described above and, similarly, permits the evaluation of the growth and phenotype of an isolated segment of mammary gland. When Tp53 mammary epithelium is transplanted, a wide variety of latencies and phenotypes were seen and selective serial retransplantation of several desirable phenotypes has been employed in order to perpetuate these as independent models (Jerry et al. 2000; Kuperwasser et al. 2000; Medina et al. 2002, 2005). (2) Cre recombinase conditional Tp53 allele (loxP flanked exons via “knock-in” gene targeting) crossed with a mammary-specific Cre expressing transgene. These mice will have somatic cell conversion to a null allele of either a heterozygous or homozygous conditional allele. The result is a loss of Tp53 in most, but not all of the mammary epithelial cells occurring after gland development (Lin et al. 2004). (3) Transgene expression in the mammary epithelium of either SV40 large T antigen (which binds to and inactivates Tp53 and Rb) (Maroulakou et al. 1994) or a mutant form of the Tp53 protein with dominant-negative activity such as the R270H mutation or R172H mutation (Li et al. 1994, 1998; Wijnhoven et al. 2005). All three of these approaches have been employed in the study of mammary cancer.
Regardless of the approach, dominant-negative transgene, null epithelium transplantation, or conditional allele, the phenotypes of Tp53 null mammary tumors are similar or dissimilar depending on perspective. They are all characterized by a high level of heterogeneity. Tumor phenotypes range from the well differentiated papillary, pilar, and myoepethelioma patterns often seen as spontaneous lesions in mice to higher grade lesions with solid and spindled growth patterns and often with high levels of nuclear atypia (Nguyen et al. 2011). There is strong evidence that Tp53 mutation leads to genetic instability, chromosomal abnormalities, and aneuploidy (Donehower et al. 1995; Jerry et al. 2000; Kuperwasser et al. 2000). Conceptually, this might be expected. If the major tumor suppressor effect of Tp53 is checkpoint control for DNA damage, the expected effect of loss of this control would depend on the frequency and array of DNA damage within a developing cancer. If the DNA damage is random, as would be the case in spontaneous tumor studies but also in chemical carcinogen (DMBA) or irradiation experiments, the driving tumor genotype would also be random. Randomness is reduced only because faster growing (higher grade) tumors will evolve more quickly once they are initiated. The prediction would be a shift toward a more poorly differentiated (higher grade) tumor, and this is what is observed experimentally.
Because of the vast array of tumor types arising in this Tp53 background, a number of more useful phenotypes have been featured in reports in the literature, and in some cases maintained as serial transplants for further study (Kuperwasser et al. 2000; Medina et al. 2002; Lin et al. 2004). In one study, premalignant lesions with ER positivity and ER dependence were serially transplanted permitting a detailed study of the time course of progression and estrogen dependence (Medina et al. 2003).
BRCA1 KNOCKOUTS
The molecular biology of Brca1 and Brca2 and the mouse models of mutations of these genes have been reviewed recently (Moynahan 2002; Evers and Jonkers 2006). Both genes are DNA repair partners, and human cancers with mutations are characterized by rapid and extensive chromosomal aberrations.
Brca1 homozygous null alleles in the germline are embryonic lethal (Moynahan 2002). Mice heterozygous for Brca1 loss show no increase in tumor development over the background rate of spontaneous tumors (Cressman et al. 1999; McCarthy et al. 2007). The addition of Tp53 knockout to the Brca1 knockout does increase the rate of mammary tumors, though the overall tumor rate (tumors of all types) remains the same (Cressman et al. 1999). Through the use of a Cre conditional Brca1 allele, a combined homozygous somatic mammary-specific loss of Brca1 with heterozygous loss of p53 resulted tumor phenotypes constituting an interesting spectrum. Tumors were solid in growth and highly proliferative with anaplastic nuclei. These carcinomas were predominantly ER and PR negative and all were ErbB2 negative. They frequently displayed basal keratins (cytokeratins 5/6 and 14) and also developed both squamous and spindle cell metaplasia (McCarthy et al. 2007). The tumor types have been assessed by gene expression profile, and this confirms that an array of intrinsic subtypes are encountered (Wright et al. 2008). Although the spindle and squamous metaplastic carcinomas are rare in humans, they do occur. Meanwhile, the basal keratin-expressing, ER/PR/Erbb2-negative phenotype is an important poor prognosis group in human populations. The role of estrogen receptor and progesterone receptor in the initiation of tumors in these mice has been documented (Poole et al. 2006; Li et al. 2007; Jones et al. 2008). The importance of hormone signaling is surprising, as the tumors themselves are estrogen and progesterone receptor negative.
Recent work to determine the cell of origin for Brca1 mouse tumors has shown a surprising and critical association. By directly comparing the keratin 14 driven cre conditional to the BLG cre conditional Brca1 knockouts a distinct difference in tumor phenotypes was shown. BLG targeting of the luminal progenitors and conversely K14 targeting of basal cells was confirmed with cell surface phenotype sorting, and resulted in the luminal progenitor cells giving rise to the “basal” subtype cancers with the greatest similarity to human breast cancers. Meanwhile the basal cells give rise to phenotypes uncommon in human cancers. (Molyneux et al. 2010). In parallel work on human tumors the luminal progenitor also emerges as the likeliest cell of origin (Lim et al. 2009). The mouse experiments suggest that there may be a plasticity in cancer cells which includes “dedifferentiation” or the ability for cancer cells to go “in reverse” in the differentiation hierarchy/ontogeny. In other words, the relationship between the cell of origin and tumor phenotype may be more complicated than once suspected. Further evidence for dedifferentiation and even redifferentiation toward alternate lineages requires detailed cell progeny tracking and orthotopic transplantation.
BRCA2 KNOCKOUTS
Similar to Brca1, Brca2 germline homozygous null mice are embryonic lethal and somatic loss of Brca2 does not result in increased mammary tumor incidence by itself. Using a keratin 14 promoter to drive Cre expression and combined with Tp53 heterozygous or homozygous loss, the incidence of mammary (and also skin) tumors was dramatically increased (Jonkers et al. 2001). Mammary tumor phenotypes in these mice were heterogeneous, but the more basaloid appearance was described as being the most common. This is a distinct contrast with human Brca2 related tumors, which are usually ER positive (luminal phenotype) without Ck5 expression and without Erbb2 amplification (Bane et al. 2007).
ECAD KNOCKOUTS AND LOBULAR CARCINOMA
Another of the models employing the Cre conditional knockout alleles in combination is a combined epithelial cell–specific knockout of E-cadherin and Tp53 (Derksen et al. 2006). E-cadherin loss of expression is associated with the most common “special type” of breast cancer in humans, invasive lobular carcinoma (ILC), and some have advocated a diagnosis based on this molecular criterion. The term “special type” in breast diagnostics implies that these tumors have both (a) a characteristic phenotype, and (b) special behavioral or prognostic characteristics. In the case of ILC, the phenotype consists of discohesive cells with small nuclei, and glassy cytoplasm often containing intracytoplasmic mucin. ILC is almost always estrogen receptor positive, and carries a better prognosis than nonspecial type mammary carcinomas (invasive ductal carcinomas). Metastases in ILC are less frequent and may have long latency.
This model of E-cadherin and TP53 loss in the mouse mammary epithelium results in invasive mammary carcinomas with several critical features in common with ILC (Alvarez et al. 2006). Perhaps the most striking is the histologic phenotype, characterized by discohesive cells with the same kind of “single file” and targetoid to diffuse invasive architecture seen in the human phenotype. The real success of the model, however, is the elucidation of the role of E-cadherin loss in the initiation of ILC. Hitherto, in the human disease, it was largely unknown if the E-cadherin loss helped to initiate the disease, if it helped to drive the special behavioral characteristics, or if it was only a marker of these special properties but unrelated mechanistically. Here, the mouse model proves that the E-cadherin loss can contribute to the initiation of an invasive carcinoma with a human ILC like phenotype, and constitutes a new “signature” phenotype, which is a phenocopy of the human tumor. The model is not absolutely faithful to the human phenotype in several respects, however. First, the mouse tumors are estrogen receptor negative, whereas human tumors are almost always positive (perhaps 100% if strict diagnostic criteria are used). Second, and potentially related to the first, the mouse tumors tend to be more densely cellular and have higher proliferation rates. Individual cell cytology may be more pleomorphic. Human tumors with these features are excluded from the “special type” category and imply the same prognosis and behavior as nonspecial type mammary carcinomas (invasive ductal carcinoma). The findings in this mouse model show that a molecular classification of tumors with E-cadherin loss defines a group of tumors including ILC as a sub-set. The mouse model permits the study of the progression in this molecular/phenotypic “pathway pathology” representing a major advance. Already, the mouse model has helped to address the controversy of “lobular carcinoma in situ” (LCIS) as a potential precursor to ILC. Despite cytologic similarities in the human, epidemiologic evidence implies that LCIS does not progress to ILC and is therefore regarded as a “risk marker” lesion rather than a “pre-invasive” cancer (Tchou and Morrow 2003). The mouse model appears to confirm this, because the earliest lesions do not expand or repopulate the existing gland tree, as seen in LCIS but instead become invasive very early in progression (Derksen et al. 2006). Another critical observation is seen in a follow-up paper, in which the cre recombinase is driven by the wap promoter instead of K14 (Derksen et al. 2011). The tumor phenotype is identical (still a “pleomorphic” lobular carcinoma morphology) suggesting that the genetics are dominant over the cell of origin, at least in this specific context. Also extremely interesting is the widespread (human-like) metastasis pattern with lymph node, liver, intestinal, and bone involvement as well as lung (Derksen et al. 2011).
TUMOR MICROENVIRONMENT IN MOUSE MODELS
The mouse mammary fat pad is a specialized stroma for mammary epithelial growth. In contrast to the human breast, it has a very low ratio of collagenous or fibrous stroma to adipose tissue. Although scant and hard to visualize, an organized collagenous stroma structures the mouse fat pad prior to gland development. Along with gland development, a periductal stroma develops, similar to human breast tissue. Mouse models have been employed to show roles for myoepithelium and stroma in limiting epithelial cancer progression (Hu et al. 2008; Bissell and Hines 2011) In some cases, the tumor associated stroma can promote cancer development of otherwise normal (i.e., nonengineered) epithelia and can even reprogram uncommitted progenitors from nonmammary origins to become a part of the mammary cancer (Boulanger et al. 2007; Booth et al. 2008). Mouse models, unlike human cell xenografts in mice, permit study of the role of inflammation. Interestingly, roles for both immunoprotection against cancer as well as immune cell promotion of progression and metastases have been shown (DeNardo et al. 2009). This area is covered in much greater detail in the article by Coussens and Pollard (2011). In one provocative series of experiments, it was shown that macrophages contribute to metastasis in the PyMT transgenic mouse by associating with tumor cells and escorting them away from the tumor (Wyckoff et al. 2007; Kedrin et al. 2008). The details of this experiment are discussed in the article by Condeelis and Weissleder (2010).
SUMMARY
The combination of advanced techniques for genetically engineering the mouse mammary gland, and transplantation techniques for isolating and maintaining individual mammary lesions with specific characteristics has resulted in a new level of mammary cancer modeling. Pathologic validation remains critical in evaluating these new models (Cardiff et al. 2004). Newer molecular analyses, gene expression profiles, and sequencing are included, and can be used to quickly compare tumors within and between models and human tumor samples.4 Validation is the process of delineating the attributes of an experimental system that accurately match the attributes of human disease. A guide and vocabulary for defining these attributes in mouse models of mammary cancer has been offered (Cardiff 2001).
Even when precisely defined molecular changes are engineered to match the human disease, important differences between the model and the disease are inevitable, and need to be characterized and considered in experimental design whenever one is using the model. There has been great success in modeling specific molecular alterations, and many of these have yielded characteristic and consistent phenotypes. Models of Erbb2 amplification are perhaps the most well represented, and include the MINO model of DCIS, which offers a practical and consistent model of precancer progression. Other models such as the Tp53 knockouts recapitulate the genetic instability common in human breast cancer. The phenotypes that result are heterogeneous, and some of these reflect common human breast cancer phenotypes, but some do not.
These differences between mouse models and human disease leave new challenges for improving the models, but also reflect the difficulty inherent in modeling human cancers. Human cancers are genotypically, phenotypically, and behaviorally heterogeneous, with multiple recognized types and potential overlaps of these types adding to the complexity. A perfect model should be molecularly defined and phenotypically consistent. Inevitably this means that the model reflects only a small proportion of human cancers, and implies a need for hundreds of distinct models. Alternatively, a given model might give rise to a complex array of genotypes and phenotypes, but this limits the practical and statistical utility by removing consistency. Ultimately, the model must serve the needs of the experiment. The precise questions of the experiment and the expected endpoints to be measured may be relevant and realistic in one model, but not in another. The future of mouse models of breast cancer will see further development of specifically engineered models with reproducible well validated attributes. These models will be freely available and carefully archived, just as eukaryotic cell lines are today (Xu et al. 2003). The availability of the models and the validation data will make easy selection for specific experimental questions possible.
ACKNOWLEDGMENTS
This work was supported in part by grants K26 RR024037 (A.D.B.) and U01 CA14582 (PI: Cardiff). I thank Robert Cardiff, Robert Munn, Ed Hubbard, and Jeff Rosen for critical commentary.
Stat1−/− mice on the 129S6/SvEv background develop mammary tumors with low penetrance, but are uniquely estrogen receptor–positive, and therefore are a part of the table although the complete description has not yet been published (SR Chan and R Schrieber, pers. comm., 2009).
The history of the original development of the “test by transplantation” is provided by Cardiff and Kenney (2011).
Theoretically, new imaging reporters with combinatorial recombination of multiple fluorescent proteins might be used to color code individual initiation foci, but this has not been applied to mammary tumorigenesis studies, to my knowledge (as in Livet et al. 2007).
Several of the common mouse models discussed here have been compared in cross-species gene expression analysis–based hierarchical clustering (Herschkowitz et al. 2007). These data were interpreted to show that some models have “signature” genomic profiles similar to the “signature” phenotypes described by Cardiff and similar subsequent analyses. Other models, including p53 mutations and also the chemical carcinogen DMBA-induced tumors, have much greater heterogeneity spanning different human tumor subtypes. Gene expression subtyping is not yet a part of the routine clinical assessment of patient cancers, and interpretation of the gene expression data out of context is probably an oversimplification of the complex tumor biology. As such, it is just one of many measurements of tumor attributes for comparison and validation.
Editors: Mina J. Bissell, Kornelia Polyak, and Jeffrey M. Rosen
Additional Perspectives on The Mammary Gland as an Experimental Model available at www.cshperspectives.org
REFERENCES
- Abbey CK, Borowsky AD, Gregg JP, Cardiff RD, Cherry SR 2006. Preclinical imaging of mammary intraepithelial neoplasia with positron emission tomography. J Mammary Gland Biol Neoplasia 11: 137–149 [DOI] [PubMed] [Google Scholar]
- Abbey CK, Borowsky AD, McGoldrick ET, Gregg JP, Maglione JE, Cardiff RD, Cherry SR 2004. In vivo positron-emission tomography imaging of progression and transformation in a mouse model of mammary neoplasia. Proc Natl Acad Sci 101: 11438–11443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez JV, Perez D, Chodosh LA 2006. mILC-ing the mouse mammary gland: A model for invasive lobular carcinoma. Cancer Cell 10: 347–349 [DOI] [PubMed] [Google Scholar]
- Alvi AJ, Clayton H, Joshi C, Enver T, Ashworth A, Vivanco MM, Dale TC, Smalley MJ 2003. Functional and molecular characterisation of mammary side population cells. Breast Cancer Res 5: R1–R8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bane AL, Beck JC, Bleiweiss I, Buys SS, Catalano E, Daly MB, Giles G, Godwin AK, Hibshoosh H, Hopper JL, et al. 2007. BRCA2 mutation–associated breast cancers exhibit a distinguishing phenotype based on morphology and molecular profiles from tissue microarrays. Am J Surg Pathol 31: 121–128 [DOI] [PubMed] [Google Scholar]
- Bartek J, Bartkova J, Kyprianou N, Lalani EN, Staskova Z, Shearer M, Chang S, Taylor-Papadimitriou J 1991. Efficient immortalization of luminal epithelial cells from human mammary gland by introduction of simian virus 40 large tumor antigen with a recombinant retrovirus. Proc Natl Acad Sci 88: 3520–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell MJ, Hines WC 2011. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med 17: 320–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth BW, Mack DL, Androutsellis-Theotokis A, McKay RD, Boulanger CA, Smith GH 2008. The mammary microenvironment alters the differentiation repertoire of neural stem cells. Proc Natl Acad Sci 105: 14891–14896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowsky AD 2003. Genetically engineering a mouse. Comp Med 53: 249–250 [PubMed] [Google Scholar]
- Borowsky A 2007. Special considerations in mouse models of breast cancer. Breast Dis 28: 29–38 [DOI] [PubMed] [Google Scholar]
- Boulanger CA, Mack DL, Booth BW, Smith GH 2007. Interaction with the mammary microenvironment redirects spermatogenic cell fate in vivo. Proc Natl Acad Sci 104: 3871–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardiff RD 2001. Validity of mouse mammary tumour models for human breast cancer: Comparative pathology. Microsc Res Tech 52: 224–230 [DOI] [PubMed] [Google Scholar]
- Cardiff RD 2003. Mouse models of human breast cancer. Comp Med 53: 250–253 [PubMed] [Google Scholar]
- Cardiff RD, Kenney N 2011. A compendium of the mouse mammary tumor biologist: From the initial observations in the house mouse to the development of genetically engineered mice. Cold Spring Harb Perspect Biol 3: a003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardiff RD, Anver MR, Gusterson BA, Hennighausen L, Jensen RA, Merino MJ, Rehm S, Russo J, Tavassoli FA, Wakefield LM, et al. 2000. The mammary pathology of genetically engineered mice: The consensus report and recommendations from the Annapolis meeting. Oncogene 19: 968–988 [DOI] [PubMed] [Google Scholar]
- Cardiff RD, Bern HA, Faulkin LJ, Daniel CW, Smith GH, Young LJ, Medina D, Gardner MB, Wellings SR, Shyamala G, et al. 2002. Contributions of mouse biology to breast cancer research. Comp Med 52: 12–31 [PubMed] [Google Scholar]
- Cardiff RD, Rosner A, Hogarth MA, Galvez JJ, Borowsky AD, Gregg JP 2004. Validation: The new challenge for pathology. Toxicol Pathol 32 (Suppl. 1): 31–39 [DOI] [PubMed] [Google Scholar]
- Condeelis J, Weissleder R 2010. In vivo imaging in cancer. Cold Spring Harb Perspect Biol 2: a003848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Pollard JW 2011. Leukocytes in mammary development and cancer. Cold Spring Harb Perspect Biol 3: a003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cressman VL, Backlund DC, Hicks EM, Gowen LC, Godfrey V, Koller BH 1999. Mammary tumor formation in p53- and BRCA1-deficient mice. Cell Growth Differ 10: 1–10 [PubMed] [Google Scholar]
- Daniel CW, Young LJ 1971. Influence of cell division on an aging process. Life span of mouse mammary epithelium during serial propagation in vivo. Exp Cell Res 65: 27–32 [DOI] [PubMed] [Google Scholar]
- Daniel CW, De Ome KB, Young JT, Blair PB, Faulkin LJ Jr 1968. The in vivo life span of normal and preneoplastic mouse mammary glands: A serial transplantation study. Proc Natl Acad Sci 61: 53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, Coussens LM 2009. CD4+ T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 16: 91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derksen PW, Braumuller TM, van der Burg E, Hornsveld M, Mesman E, Wesseling J, Krimpenfort P, Jonkers J 2011. Mammary-specific inactivation of E-cadherin and p53 impairs functional gland development and leads to pleomorphic invasive lobular carcinoma in mice. Dis Model Mech [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derksen PW, Liu X, Saridin F, van der Gulden H, Zevenhoven J, Evers B, van Beijnum JR, Griffioen AW, Vink J, Krimpenfort P, et al. 2006. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell 10: 437–449 [DOI] [PubMed] [Google Scholar]
- Dilworth SM 2002. Polyoma virus middle T antigen and its role in identifying cancer-related molecules. Nat Rev Cancer 2: 951–956 [DOI] [PubMed] [Google Scholar]
- Donehower LA, Godley LA, Aldaz CM, Pyle R, Shi YP, Pinkel D, Gray J, Bradley A, Medina D, Varmus HE 1995. Deficiency of p53 accelerates mammary tumorigenesis in Wnt-1 transgenic mice and promotes chromosomal instability. Genes Dev 9: 882–895 [DOI] [PubMed] [Google Scholar]
- Du Z, Podsypanina K, Huang S, McGrath A, Toneff MJ, Bogoslovskaia E, Zhang X, Moraes RC, Fluck M, Allred DC, et al. 2006. Introduction of oncogenes into mammary glands in vivo with an avian retroviral vector initiates and promotes carcinogenesis in mouse models. Proc Natl Acad Sci 103: 17396–17401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers B, Jonkers J 2006. Mouse models of BRCA1 and BRCA2 deficiency: Past lessons, current understanding and future prospects. Oncogene 25: 5885–5897 [DOI] [PubMed] [Google Scholar]
- Gunther EJ, Belka GK, Wertheim GB, Wang J, Hartman JL, Boxer RB, Chodosh LA 2002. A novel doxycycline-inducible system for the transgenic analysis of mammary gland biology. FASEB J 16: 283–292 [DOI] [PubMed] [Google Scholar]
- Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, et al. 2007. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 8: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer J, Kwong LN, Lowe SW, Chin L 2010. Non-germline genetically engineered mouse models for translational cancer research. Nat Rev Cancer 10: 470–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Yao J, Carroll DK, Weremowicz S, Chen H, Carrasco D, Richardson A, Violette S, Nikolskaya T, Nikolsky Y, et al. 2008. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell 13: 394–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerry DJ, Kittrell FS, Kuperwasser C, Laucirica R, Dickinson ES, Bonilla PJ, Butel JS, Medina D 2000. A mammary-specific model demonstrates the role of the p53 tumor suppressor gene in tumor development. Oncogene 19: 1052–1058 [DOI] [PubMed] [Google Scholar]
- Jones LP, Tilli MT, Assefnia S, Torre K, Halama ED, Parrish A, Rosen EM, Furth PA 2008. Activation of estrogen signaling pathways collaborates with loss of Brca1 to promote development of ERα-negative and ERα-positive mammary preneoplasia and cancer. Oncogene 27: 794–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers J, Berns A 2002. Conditional mouse models of sporadic cancer. Nat Rev Cancer 2: 251–265 [DOI] [PubMed] [Google Scholar]
- Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A 2001. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet 29: 418–425 [DOI] [PubMed] [Google Scholar]
- Kedrin D, Gligorijevic B, Wyckoff J, Verkhusha VV, Condeelis J, Segall JE, van Rheenen J 2008. Intravital imaging of metastatic behavior through a mammary imaging window. Nat Methods 5: 1019–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Petersen OW, et al. 2007. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol 1: 84–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperwasser C, Hurlbut GD, Kittrell FS, Dickinson ES, Laucirica R, Medina D, Naber SP, Jerry DJ 2000. Development of spontaneous mammary tumors in BALB/c p53 heterozygous mice. A model for Li–Fraumeni syndrome. Am J Pathol 157: 2151–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EYHP, Muller WJ 2010. Oncogenes and tumor suppressor genes. Cold Spring Harb Perspect Biol 2: a003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Greenberg N, Stephens LC, Meyn R, Medina D, Rosen JM 1994. Preferential overexpression of a 172Arg–>Leu mutant p53 in the mammary gland of transgenic mice results in altered lobuloalveolar development. Cell Growth Differ 5: 711–721 [PubMed] [Google Scholar]
- Li B, Murphy KL, Laucirica R, Kittrell F, Medina D, Rosen JM 1998. A transgenic mouse model for mammary carcinogenesis. Oncogene 16: 997–1007 [DOI] [PubMed] [Google Scholar]
- Li W, Xiao C, Vonderhaar BK, Deng CX 2007. A role of estrogen/ERα signaling in BRCA1-associated tissue-specific tumor formation. Oncogene 26: 7204–7212 [DOI] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, et al. 2009. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med 15: 907–913 [DOI] [PubMed] [Google Scholar]
- Lin SC, Lee KF, Nikitin AY, Hilsenbeck SG, Cardiff RD, Li A, Kang KW, Frank SA, Lee WH, Lee EY 2004. Somatic mutation of p53 leads to estrogen receptor α-positive and -negative mouse mammary tumors with high frequency of metastasis. Cancer Res 64: 3525–3532 [DOI] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW 2007. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450: 56–62 [DOI] [PubMed] [Google Scholar]
- Maglione JE, McGoldrick ET, Young LJ, Namba R, Gregg JP, Liu L, Moghanaki D, Ellies LG, Borowsky AD, Cardiff RD, et al. 2001. Transgenic Polyoma middle-T mice model premalignant mammary disease. Cancer Res 6: 8298–8305 [PubMed] [Google Scholar]
- Maglione JE, McGoldrick ET, Young LJ, Namba R, Gregg JP, Liu L, Moghanaki D, Ellies LG, Borowsky AD, Cardiff RD, et al. 2004. Polyomavirus middle T-induced mammary intraepithelial neoplasia outgrowths: Single origin, divergent evolution, and multiple outcomes. Mol Cancer Ther 3: 941–953 [PubMed] [Google Scholar]
- Maroulakou IG, Anver M, Garrett L, Green JE 1994. Prostate and mammary adenocarcinoma in transgenic mice carrying a rat C3(1) simian virus 40 large tumor antigen fusion gene. Proc Natl Acad Sci 91: 11236–11240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy A, Savage K, Gabriel A, Naceur C, Reis-Filho JS, Ashworth A 2007. A mouse model of basal-like breast carcinoma with metaplastic elements. J Pathol 211: 389–398 [DOI] [PubMed] [Google Scholar]
- Medina D 2010. Of mice and women: A short history of mouse mammary cancer research with an emphasis on the paradigms inspired by the transplantation method. Cold Spring Harb Perspect Biol 2: a004523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina D, DeOme KB 1970a. Carcinogen-induced mammary tumors from preneoplastic nodule outgrowths in BALB-c mice. Cancer Res 30: 1055–1059 [PubMed] [Google Scholar]
- Medina D, DeOme KB 1970b. Effects of various oncogenic agents on tumor-producing capabilities of series D BALB-c mammary nodule outgrowth lines. J Natl Cancer Inst 45: 353–363 [PubMed] [Google Scholar]
- Medina D, DeOme KB, Young L 1970. Tumor-producing capabilities of hyperplastic alveolar nodules in virgin and hormone-stimulated BALB/c f. C3H and C3Hf mice. J Natl Cancer Inst 44: 167–174 [PubMed] [Google Scholar]
- Medina D, Kittrell FS, Hill J, Shepard A, Thordarson G, Brown P 2005. Tamoxifen inhibition of estrogen receptor-α-negative mouse mammary tumorigenesis. Cancer Res 65: 3493–3496 [DOI] [PubMed] [Google Scholar]
- Medina D, Kittrell FS, Shepard A, Contreras A, Rosen JM, Lydon J 2003. Hormone dependence in premalignant mammary progression. Cancer Res 63: 1067–1072 [PubMed] [Google Scholar]
- Medina D, Kittrell FS, Shepard A, Stephens LC, Jiang C, Lu J, Allred DC, McCarthy M, Ullrich RL 2002. Biological and genetic properties of the p53 null preneoplastic mammary epithelium. FASEB J 16: 881–883 [DOI] [PubMed] [Google Scholar]
- Molyneux G, Geyer FC, Magnay FA, McCarthy A, Kendrick H, Natrajan R, Mackay A, Grigoriadis A, Tutt A, Ashworth A et al. 2010. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell 7: 403–417 [DOI] [PubMed] [Google Scholar]
- Moody SE, Sarkisian CJ, Hahn KT, Gunther EJ, Pickup S, Dugan KD, Innocent N, Cardiff RD, Schnall MD, Chodosh LA 2002. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell 2: 451–461 [DOI] [PubMed] [Google Scholar]
- Moynahan ME 2002. The cancer connection: BRCA1 and BRCA2 tumor suppression in mice and humans. Oncogene 21: 8994–9007 [DOI] [PubMed] [Google Scholar]
- Namba R, Maglione JE, et al. 2004. Molecular characterization of the transition to malignancy in a genetically engineered mouse-based model of ductal carcinoma in situ. Mol Cancer Res 2: 453–463 [PubMed] [Google Scholar]
- Namba R, Maglione JE, Young LJ, Borowsky AD, Cardiff RD, MacLeod CL, Gregg JP 2006. Rapamycin inhibits growth of premalignant and malignant mammary lesions in a mouse model of ductal carcinoma in situ. Clin Cancer Res 12: 2613–2621 [DOI] [PubMed] [Google Scholar]
- Namba R, Young LJ, Maglione JE, McGoldrick ET, Liu S, Wurz GT, DeGregorio MW, Borowsky AD, MacLeod CL, Cardiff RD, et al. 2005. Selective estrogen receptor modulators inhibit growth and progression of premalignant lesions in a mouse model of ductal carcinoma in situ. Breast Cancer Res 7: R881–R889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10: 515–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DH, Oketch-Rabah HA, Illa-Bochacal I, Geyer FC, Reis-Filho JS, Mao J-H, Ravani SA, Zavadil J, Borowsky AD, Jerry DJ, et al. 2011. Radiation acts on the microenvironment to affect breast carcinogenesis by distinct mechanisms that decrease cancer latency and affect tumor type. Cancer Cell 19: 640–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AJ, Li Y, Kim Y, Lin SC, Lee WH, Lee EY 2006. Prevention of Brca1-mediated mammary tumorigenesis in mice by a progesterone antagonist. Science 314: 1467–1470 [DOI] [PubMed] [Google Scholar]
- Rosner A, Miyoshi K, Landesman-Bollag E, Xu X, Seldin DC, Moser AR, MacLeod CL, Shyamala G, Gillgrass AE, Cardiff RD 2002. Pathway pathology: Histological differences between ErbB/Ras and Wnt pathway transgenic mammary tumors. Am J Pathol 161: 1087–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgroi DC, Teng S, Robinson G, LeVangie R, Hudson JR Jr, Elkahloun AG 1999. In vivo gene expression profile analysis of human breast cancer progression. Cancer Res 59: 5656–5661 [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE 2006. Generation of a functional mammary gland from a single stem cell. Nature 439: 84–88 [DOI] [PubMed] [Google Scholar]
- Tchou J, Morrow M 2003. Overview of clinical risk assessment. In Managing breast cancer risk (ed. Morrow M, Jordan VC), pp. 3–25 Decker, Hamilton, ON [Google Scholar]
- Ursini-Siegel J, Schade B, Cardiff RD, Muller WJ 2007. Insights from transgenic mouse models of ERBB2-induced breast cancer. Nat Rev Cancer 7: 389–397 [DOI] [PubMed] [Google Scholar]
- Visvader JE, Smith GH 2011. Murine mammary epithelial stem cells: Discovery, function, and current status. Cold Spring Harb Perspect Biol 3: a004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnhoven SW, Zwart E, Speksnijder EN, Beems RB, Olive KP, Tuveson DA, Jonkers J, Schaap MM, van den Berg J, Jacks T, et al. 2005. Mice expressing a mammary gland-specific R270H mutation in the p53 tumor suppressor gene mimic human breast cancer development. Cancer Res 65: 816–8173 [DOI] [PubMed] [Google Scholar]
- Wright MH, Robles AI, Herschkowitz JI, Hollingshead MG, Anver MR, Perou CM, Varticovski L 2008. Molecular analysis reveals heterogeneity of mouse mammary tumors conditionally mutant for Brca1. Mol Cancer 7: 29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, Segall JE, Pollard JW, Condeelis J 2007. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res 67: 2649–2656 [DOI] [PubMed] [Google Scholar]
- Xu F, Sahni H, Settnek S, Gupta A, Phillips J, Zhang D, Beasley J, De Coronado S, Wagner U, Rosso K, et al. 2003. Mouse models of human cancer web-based resources. AMIA Annu Symp Proc 2003: 1056. [PMC free article] [PubMed] [Google Scholar]