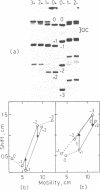
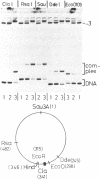
Abstract
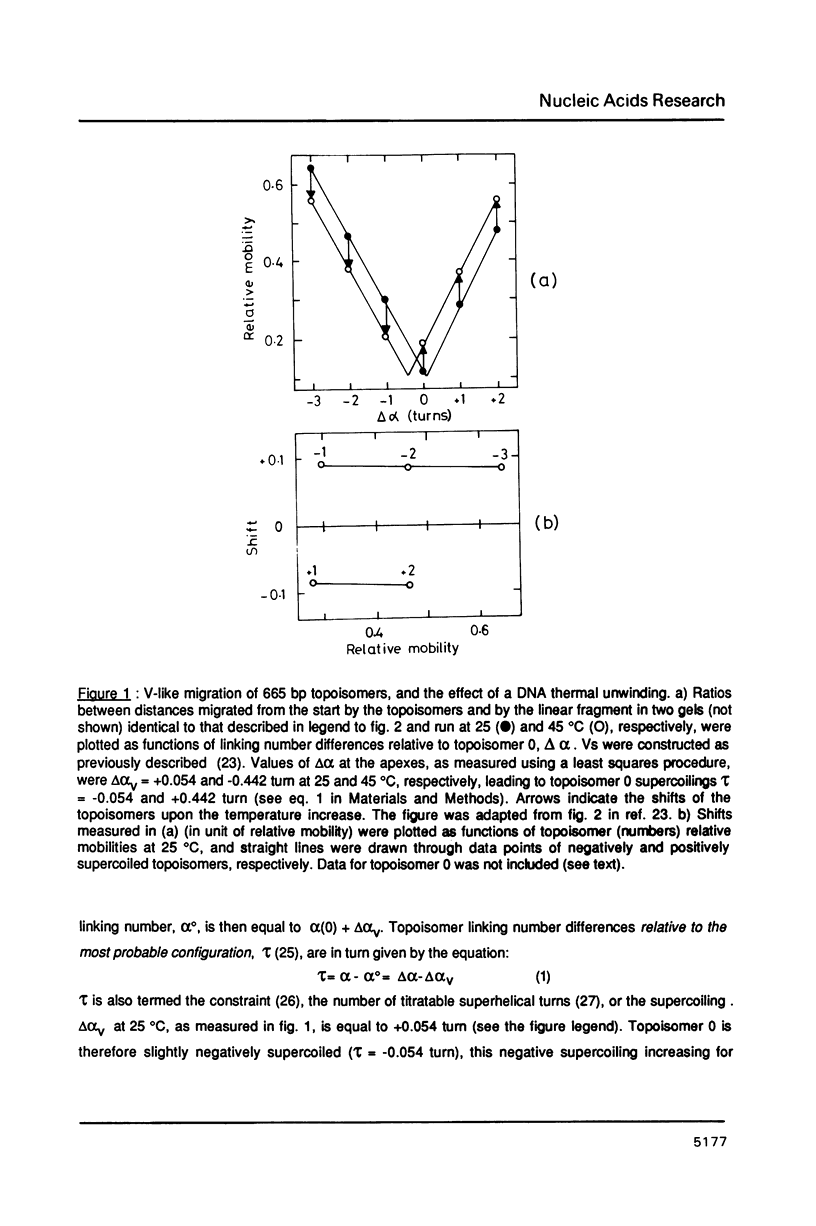
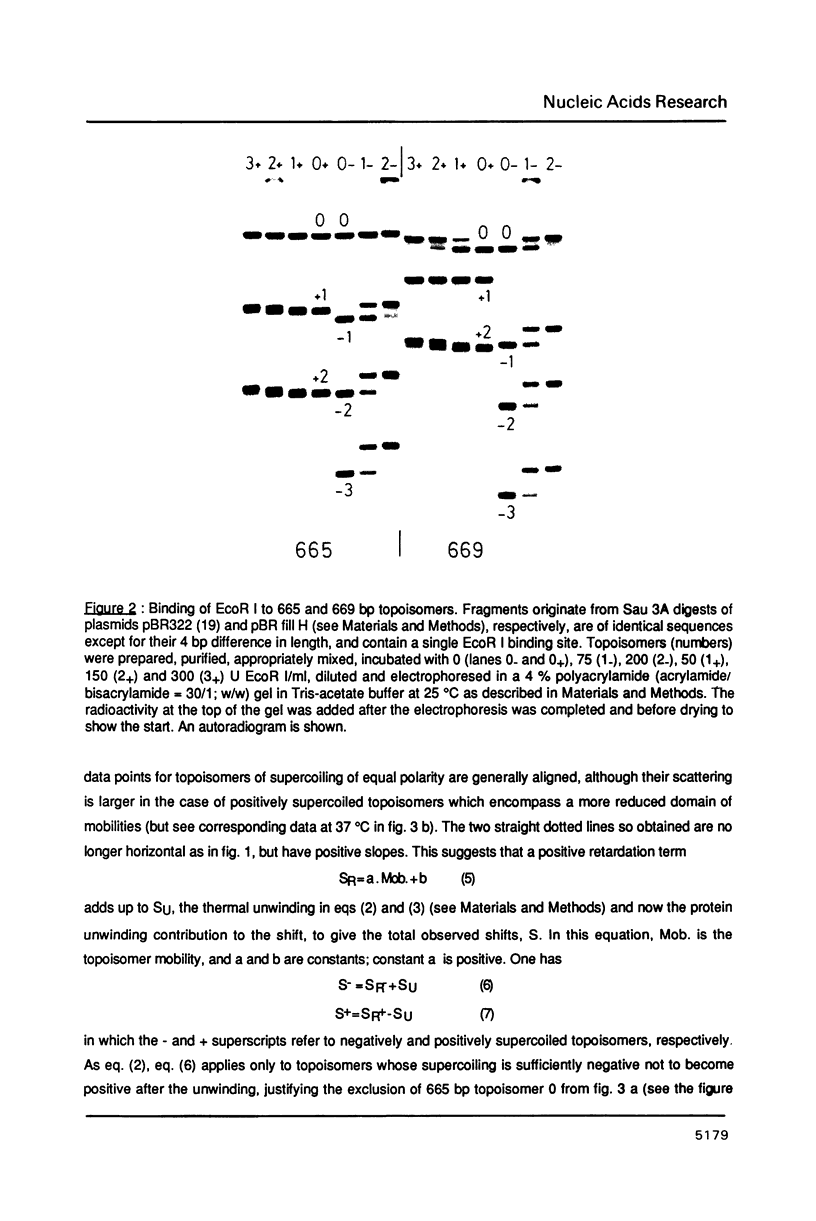
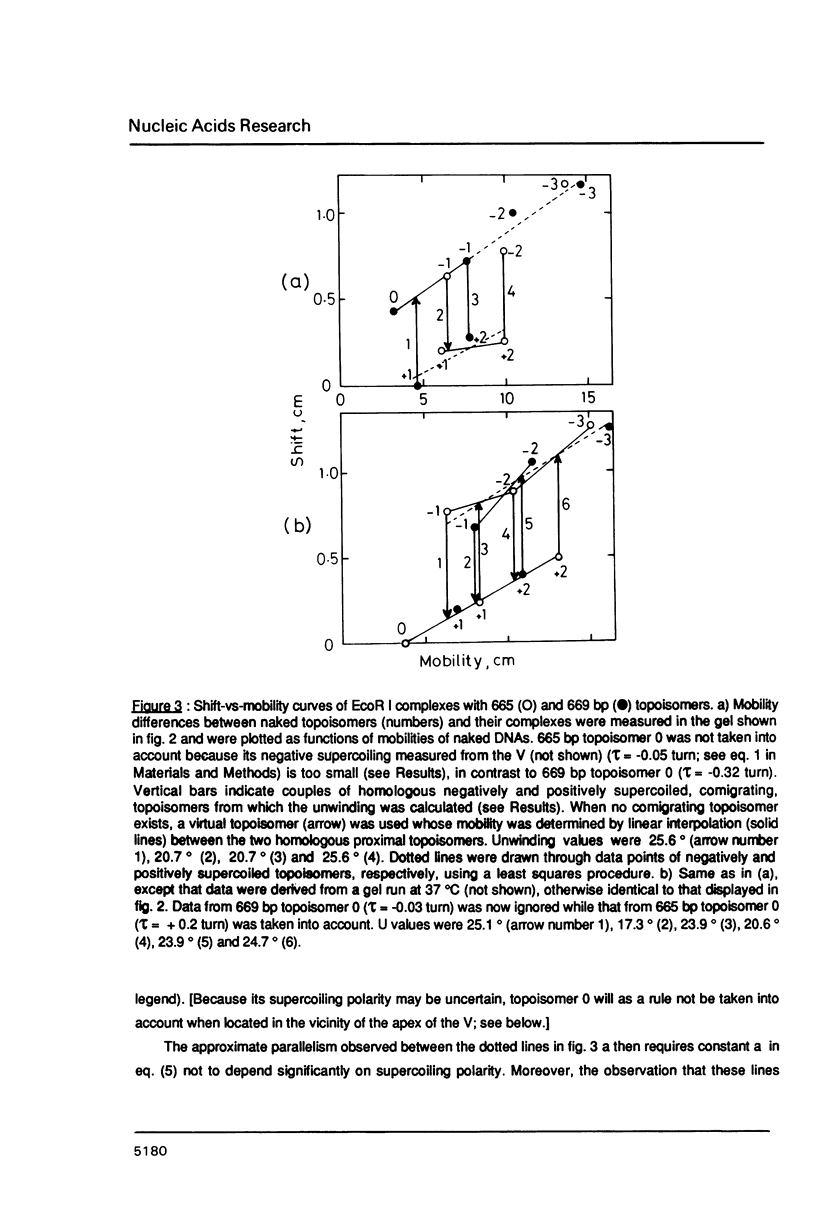
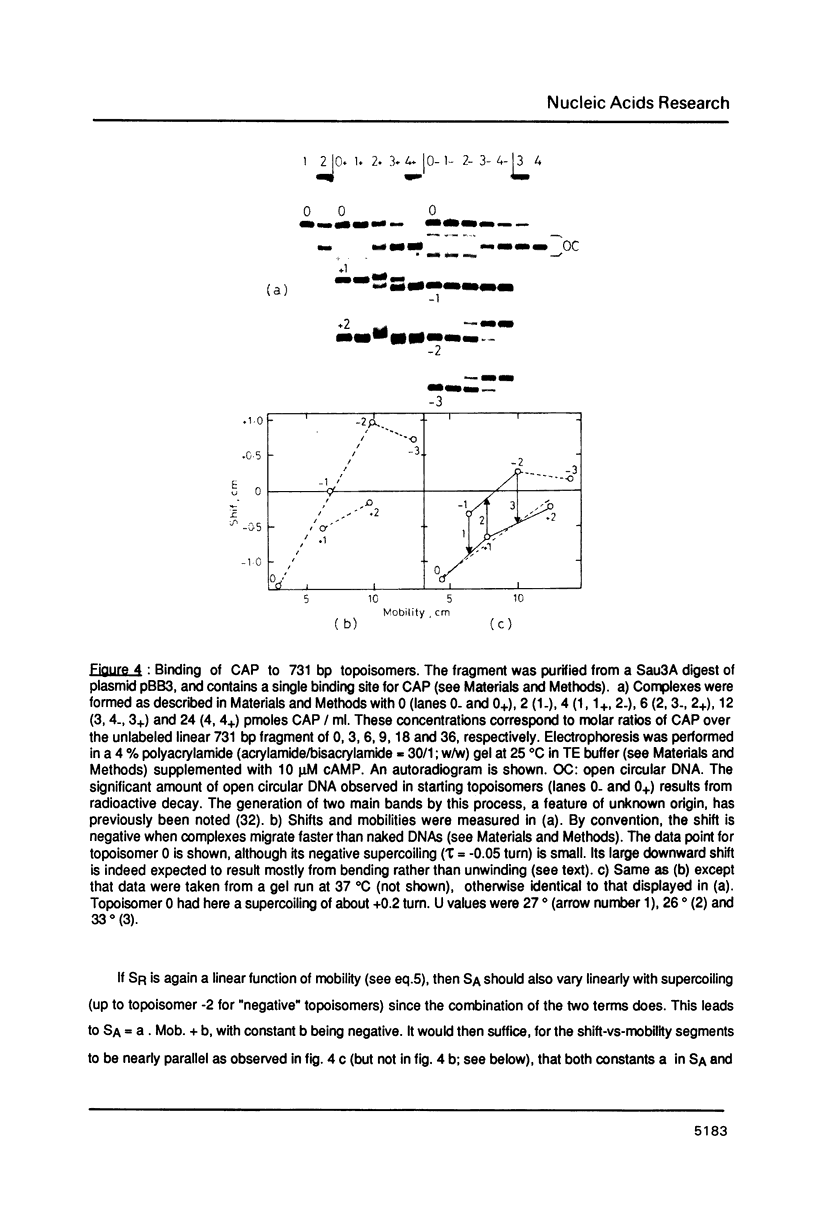
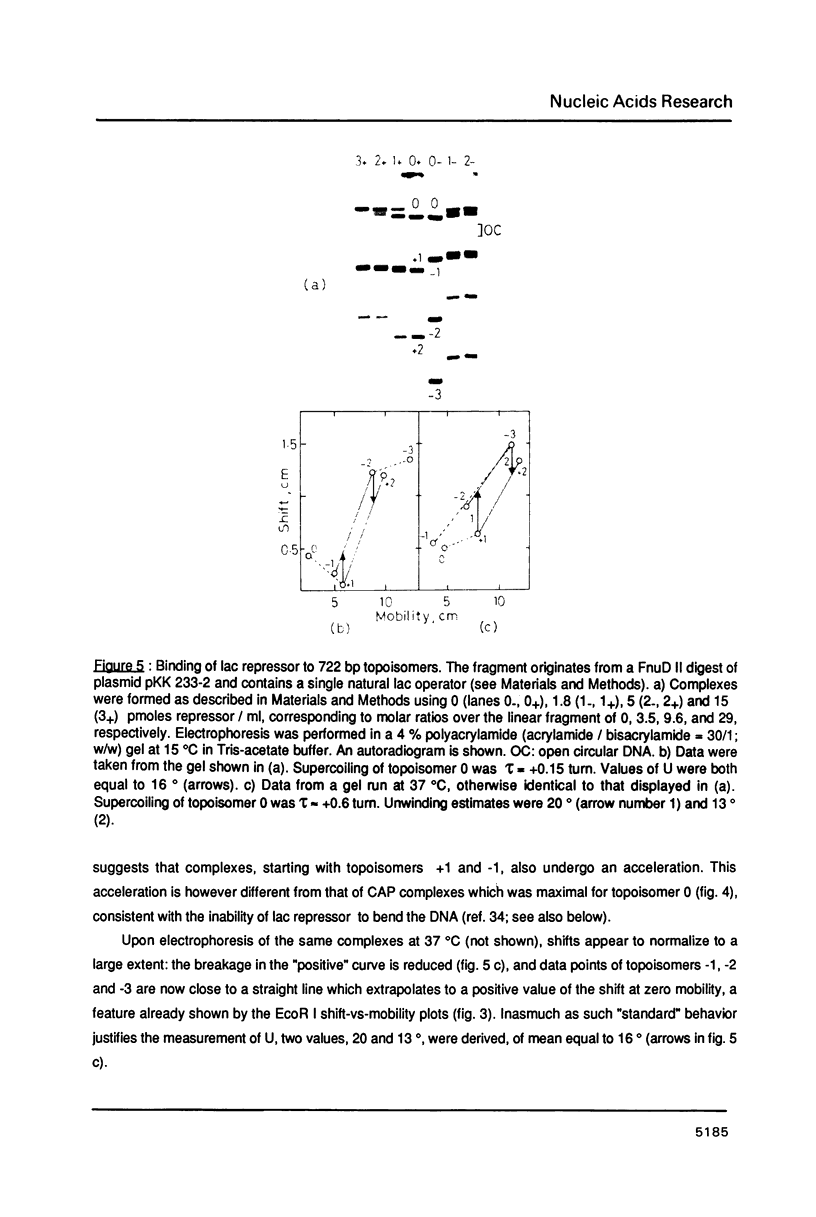
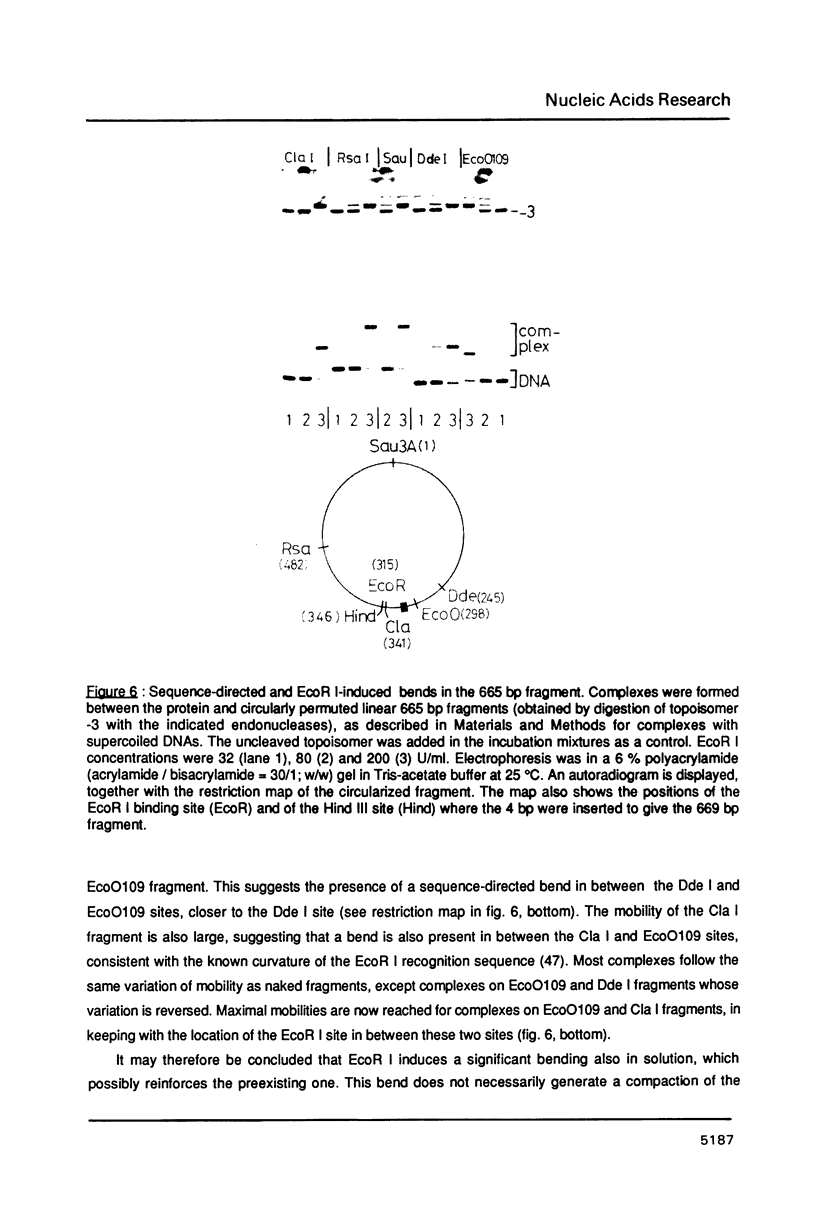
An electrophoretic procedure for the measurement of the helix unwinding induced by a sequence-specific protein is described. The method, which was applied here to EcoR I, CAP and lac repressor, involved the migration of the complexes with positively and negatively supercoiled DNA minicircles carrying a single protein binding site. Mobility shifts of complexes relative to naked DNAs appeared to be a result of i) the unwinding; of ii) an increase in the molecular frictional coefficient, which led to a retardation; of iii) bending, in the particular case of CAP, which induced an acceleration; and of iv) looping, in the case of lac repressor, which also resulted in an acceleration. Under conditions where the migration of the naked topoisomers was V-like (topoisomer mobility showed the same linear increase with both negative and positive supercoilings; Zivanovic et al. (1986) J. Mol. Biol., 192, 645-660), the protein unwinding contribution to mobility was assumed to be identical to that experimentally observed in the case of a thermal unwinding: all negatively supercoiled topoisomers were retarded and all positively supercoiled topoisomers were accelerated to the same extent. In contrast, the mobility contribution of the frictional term, as well as those of bending and looping, appeared to vary strongly with the magnitude of the supercoiling, but only weakly with its polarity. As a consequence, these latter contributions may approximately cancel when one is measuring the difference between the shifts observed for two comigrating, negatively and positively supercoiled, topoisomers, allowing the unwinding to be calculated. While estimates obtained for EcoR I, 23 +/- 3 degrees, and CAP, about 29 degrees, were in good agreement with previous measurements using topoisomerase I, the value found for lac repressor, 13 to 16 degrees, was significantly smaller.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amouyal M., Buc H. Topological unwinding of strong and weak promoters by RNA polymerase. A comparison between the lac wild-type and the UV5 sites of Escherichia coli. J Mol Biol. 1987 Jun 20;195(4):795–808. doi: 10.1016/0022-2836(87)90485-2. [DOI] [PubMed] [Google Scholar]
- Bauer W. R. Structure and reactions of closed duplex DNA. Annu Rev Biophys Bioeng. 1978;7:287–313. doi: 10.1146/annurev.bb.07.060178.001443. [DOI] [PubMed] [Google Scholar]
- Carey J. Gel retardation at low pH resolves trp repressor-DNA complexes for quantitative study. Proc Natl Acad Sci U S A. 1988 Feb;85(4):975–979. doi: 10.1073/pnas.85.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann S., McLaughlin L. W. DNA curvature in native and modified EcoRI recognition sites and possible influence upon the endonuclease cleavage reaction. J Mol Biol. 1988 Aug 20;202(4):823–834. doi: 10.1016/0022-2836(88)90561-x. [DOI] [PubMed] [Google Scholar]
- Diekmann S., Wang J. C. On the sequence determinants and flexibility of the kinetoplast DNA fragment with abnormal gel electrophoretic mobilities. J Mol Biol. 1985 Nov 5;186(1):1–11. doi: 10.1016/0022-2836(85)90251-7. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Ebright R. H., Kolb A., Buc H., Kunkel T. A., Krakow J. S., Beckwith J. Role of glutamic acid-181 in DNA-sequence recognition by the catabolite gene activator protein (CAP) of Escherichia coli: altered DNA-sequence-recognition properties of [Val181]CAP and [Leu181]CAP. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6083–6087. doi: 10.1073/pnas.84.17.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick C. A., Grable J., Melia M., Samudzi C., Jen-Jacobson L., Wang B. C., Greene P., Boyer H. W., Rosenberg J. M. Kinked DNA in crystalline complex with EcoRI endonuclease. Nature. 1984 May 24;309(5966):327–331. doi: 10.1038/309327a0. [DOI] [PubMed] [Google Scholar]
- Gartenberg M. R., Crothers D. M. DNA sequence determinants of CAP-induced bending and protein binding affinity. Nature. 1988 Jun 30;333(6176):824–829. doi: 10.1038/333824a0. [DOI] [PubMed] [Google Scholar]
- Goulet I., Zivanovic Y., Prunell A. Helical repeat of DNA in solution. The V curve method. Nucleic Acids Res. 1987 Apr 10;15(7):2803–2821. doi: 10.1093/nar/15.7.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet I., Zivanovic Y., Prunell A., Revet B. Chromatin reconstitution on small DNA rings. I. J Mol Biol. 1988 Mar 20;200(2):253–266. doi: 10.1016/0022-2836(88)90238-0. [DOI] [PubMed] [Google Scholar]
- Hanas J. S., Bogenhagen D. F., Wu C. W. DNA unwinding ability of Xenopus transcription factor A. Nucleic Acids Res. 1984 Jan 25;12(2):1265–1276. doi: 10.1093/nar/12.2.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley J. L., Gregori T. J. Cloning multiple copies of a DNA segment. Gene. 1981 May;13(4):347–353. doi: 10.1016/0378-1119(81)90014-7. [DOI] [PubMed] [Google Scholar]
- Hsieh W. T., Wells R. D. Influence of negative supercoiling and of the proximity of left-handed Z-DNA on the Escherichia coli lactose repressor-operator interaction. J Biol Chem. 1987 Oct 25;262(30):14576–14582. [PubMed] [Google Scholar]
- Hsieh W. T., Whitson P. A., Matthews K. S., Wells R. D. Influence of sequence and distance between two operators on interaction with the lac repressor. J Biol Chem. 1987 Oct 25;262(30):14583–14591. [PubMed] [Google Scholar]
- Kim R., Kim S. H. Direct measurement of DNA unwinding angle in specific interaction between lac operator and repressor. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):451–454. doi: 10.1101/sqb.1983.047.01.053. [DOI] [PubMed] [Google Scholar]
- Kim R., Modrich P., Kim S. H. 'Interactive' recognition in EcoRI restriction enzyme-DNA complex. Nucleic Acids Res. 1984 Oct 11;12(19):7285–7292. doi: 10.1093/nar/12.19.7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb A., Buc H. Is DNA unwound by the cyclic AMP receptor protein? Nucleic Acids Res. 1982 Jan 22;10(2):473–485. doi: 10.1093/nar/10.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb A., Spassky A., Chapon C., Blazy B., Buc H. On the different binding affinities of CRP at the lac, gal and malT promoter regions. Nucleic Acids Res. 1983 Nov 25;11(22):7833–7852. doi: 10.1093/nar/11.22.7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer H., Amouyal M., Nordheim A., Müller-Hill B. DNA supercoiling changes the spacing requirement of two lac operators for DNA loop formation with lac repressor. EMBO J. 1988 Feb;7(2):547–556. doi: 10.1002/j.1460-2075.1988.tb02844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laundon C. H., Griffith J. D. Curved helix segments can uniquely orient the topology of supertwisted DNA. Cell. 1988 Feb 26;52(4):545–549. doi: 10.1016/0092-8674(88)90467-9. [DOI] [PubMed] [Google Scholar]
- Le Bret M. Monte Carlo computation of the supercoiling energy, the sedimentation constant, and the radius of gyration of unknotted and knotted circular DNA. Biopolymers. 1980 Mar;19(3):619–637. doi: 10.1002/bip.1980.360190312. [DOI] [PubMed] [Google Scholar]
- Liu-Johnson H. N., Gartenberg M. R., Crothers D. M. The DNA binding domain and bending angle of E. coli CAP protein. Cell. 1986 Dec 26;47(6):995–1005. doi: 10.1016/0092-8674(86)90814-7. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McClarin J. A., Frederick C. A., Wang B. C., Greene P., Boyer H. W., Grable J., Rosenberg J. M. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986 Dec 19;234(4783):1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- Newman A. K., Rubin R. A., Kim S. H., Modrich P. DNA sequences of structural genes for Eco RI DNA restriction and modification enzymes. J Biol Chem. 1981 Mar 10;256(5):2131–2139. [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D., Reiness G., Zubay G. Purification and DNA-binding properties of the catabolite gene activator protein. Proc Natl Acad Sci U S A. 1971 Jun;68(6):1222–1225. doi: 10.1073/pnas.68.6.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg J. M., Khallai O. B., Kopka M. L., Dickerson R. E., Riggs A. D. Lac repressor purification without inactivation of DNA binding activity. Nucleic Acids Res. 1977 Mar;4(3):567–572. doi: 10.1093/nar/4.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin R. A., Modrich P. EcoRI methylase. Physical and catalytic properties of the homogeneous enzyme. J Biol Chem. 1977 Oct 25;252(20):7265–7272. [PubMed] [Google Scholar]
- Saucier J. M., Wang J. C. Angular alteration of the DNA helix by E. coli RNA polymerase. Nat New Biol. 1972 Oct 11;239(93):167–170. doi: 10.1038/newbio239167a0. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Thoma F., Brubaker J. M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985 Oct;42(3):799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C., Barkley M. D., Bourgeois S. Measurements of unwinding of lac operator by repressor. Nature. 1974 Sep 20;251(5472):247–249. doi: 10.1038/251247a0. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Peck L. J., Becherer K. DNA supercoiling and its effects on DNA structure and function. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):85–91. doi: 10.1101/sqb.1983.047.01.011. [DOI] [PubMed] [Google Scholar]
- Warwicker J., Engelman B. P., Steitz T. A. Electrostatic calculations and model-building suggest that DNA bound to CAP is sharply bent. Proteins. 1987;2(4):283–289. doi: 10.1002/prot.340020404. [DOI] [PubMed] [Google Scholar]
- Whitson P. A., Hsieh W. T., Wells R. D., Matthews K. S. Influence of supercoiling and sequence context on operator DNA binding with lac repressor. J Biol Chem. 1987 Oct 25;262(30):14592–14599. [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zivanovic Y., Goulet I., Prunell A. Properties of supercoiled DNA in gel electrophoresis. The V-like dependence of mobility on topological constraint. DNA-matrix interactions. J Mol Biol. 1986 Dec 5;192(3):645–660. doi: 10.1016/0022-2836(86)90282-2. [DOI] [PubMed] [Google Scholar]
- Zubay G. The isolation and properties of CAP, the catabolite gene activator. Methods Enzymol. 1980;65(1):856–877. doi: 10.1016/s0076-6879(80)65079-4. [DOI] [PubMed] [Google Scholar]