Abstract
Diterpenes are a structurally diverse class of molecules common in plants, although they are very rarely found in bacteria. We report the identification in Mycobacterium tuberculosis (Mtb) of three diterpenes proposed to promote phagolysosome maturation arrest. MS analysis reveals that these diterpenes are novel compounds not previously identified in other organisms. The diterpene with highest abundance in Mtb has a mass fragmentation pattern identical to edaxadiene, which is produced in vitro from geranylgeranyl diphosphate by the enzymes Rv3377c and Rv3378c [Mann FM et al. (2009) J Am Chem Soc 131, 17526–17527]. A second diterpene found in Mtb has a similar mass spectrum, and is always observed in the same proportion relative to edaxadiene, indicating that it is a side product of the Rv3378c reaction in vivo. We name this second diterpene olefin edaxadiene B. The least abundant of the three diterpenes in Mtb extracts is tuberculosinol, a dephosphorylated side-product of the edaxadiene pathway intermediate produced by Rv3377c [Nakano C et al. (2009) Chembiochem 10, 2060–2071; Nakano C et al. (2005) Chem Commun (Camb) 8, 1016–1018]. A frameshift in Rv3377c in Mtb completely eliminates diterpene production, whereas expression of Rv3377c and Rv3378c in the nonpathogenic M. smegmatis is sufficient to produce edaxadiene and edaxadiene B. These studies define the pathway of edaxadiene and edaxadiene B biosynthesis in vivo. Rv3377c and Rv3378c are unique to Mtb and M. bovis, making them candidates for selective therapeutics and diagnostics.
Keywords: cyclase, edaxadiene, Rv3377c, Rv3378c, tuberculosinol
Introduction
With more than 40 000 members, terpenes (or isoprenoids) are among the most varied class of natural products [1]. These compounds occur throughout nature, ranging from essential metabolites such as sterols and hormones to unique secondary metabolites that serve in defense and communication [2]. In bacteria, several Streptomyces species have been shown to encode diterpene synthases [3–5], although very few diterpenes have been directly isolated from these organisms, and their functions are poorly understood [3,5,6]. Several recent studies have shown that Mycobacterium tuberculosis (Mtb) also contains genes encoding diterpene synthases [7–9]. The demonstration that the Mtb enzymes Rv3377c and Rv3378c produce the unique diterpene edaxadiene from geranylgeranyl diphosphate (GGPP) in vitro showed that these enzymes are functional class II and I diterpene synthases, respectively [7]. Notably, Rv3377c and Rv3378c were identified in a transposon mutagenesis screen to identify Mtb genes involved in phagolysosome fusion arrest [10], suggesting a potential function for mycobacterial diterpenes.
Suggestive of a functionally integrated pathway, Rv3377c and Rv3378c are proximal to each other and several other lipid-metabolizing enzymes within the Mtb genome, including dxs2 (Rv3379c), encoding a synthase for isoprenoid precursors, and idsB (Rv3383c), a predicted GGPP synthase. Rv3377c and Rv3378c, along with the hypothetical protein Rv3376, are co-inherited exclusively in Mtb and M. bovis and are not found in other Mycobacterium spp. These genes have a lower GC content than the rest of the Mtb genome, suggesting that they were acquired recently, apparently just prior to the divergence of Mycobacterium bovis and Mtb from their common ancestor.
Diterpene synthases catalyze the cyclization of GGPP, the universal 20-carbon diterpene precursor, into cyclic products [11]. Diterpene synthases can be mechanistically divided into two groups, both of which use the formation of a highly-reactive carbocation to initiate cyclization [12]. Class I enzymes contain a characteristic DDxxD motif and catalyze diphosphate ionization-initiated reactions, whereas class II enzymes utilize a DxDD motif to catalyze protonation-initiated cyclization reactions [11,13]. Enzymes in both class I and II require a divalent ion for catalysis. Sequence homology to ent-copalyl diphosphate synthases, the occurrence of a conserved active-site motif, as well as the pyrophosphate in the diterpene product indicate that Rv3377c encodes a class II diterpene synthase. Recent studies indicate that Rv3378c is a class I diterpene synthase that converts the Rv3377c intermediate into the diterpene edaxadiene in vitro [7].
It is not known whether Mtb synthesizes diterpenes in vivo and the structures of any such compounds have not been described. To determine whether Mtb produces diterpenes, we examined the functions of Rv3377c and Rv3378c in diterpene synthesis in culture. In the present study, we show that Mtb produces three diterpenes from GGPP, all of which localize to cell membranes. Expression of Rv3377c and Rv3378c in the nonpathogenic species Mycobacterium smegmatis is sufficient to produce significant quantities of edaxadiene, suggesting that these genes constitute a committed biosynthetic pathway. Consistent with the idea that Rv3377c catalyzes the first step in the pathway, a frameshift mutation in Rv3377c in Mtb H37Rv abolishes all diterpene synthesis, and the introduction of Rv3377c on a plasmid complements this phenotype. Rv3378c catalyzes the second step in the pathway, yielding two products in vivo. The results obtained demonstrate that the products of two genes required to arrest phagolysosome fusion form a complete pathway that synthesizes novel diterpenes in Mtb.
Results
In vivo production and localization of Mtb diterpenes
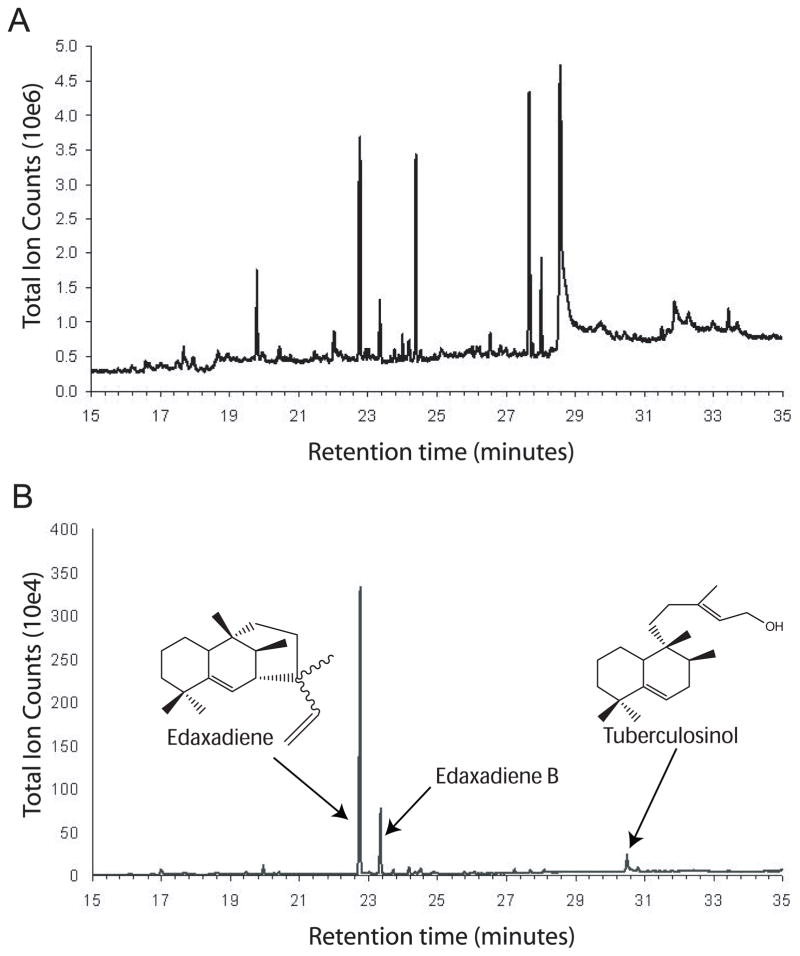
To detect diterpenes in Mtb, we grew Mtb H37Rv to mid-log phase and extracted lipids using chloroform:methanol for analysis by GC/MS. From a GC/MS total-ion chromatogram, we detected several dozen low molecular weight species (Fig. 1A). When the same extract was analyzed for lipid compounds containing ions characteristic of diterpenes that have been generated in vitro using putative Mtb diterpene synthases (ions 80, 189 and 191), we identified three unique species (Fig. 1B). Analyzing the mass spectral fragmentation patterns of the three suspected diterpenes, we found that the most abundant species is identical to edaxadiene (Fig. S1A) and the least abundant diterpene was indistinguishable from tuberculosinol (Fig. S1B). Unexpectedly, we also found an additional analyte with a similar retention time to edaxadiene, and a distinct but related mass fragmentation pattern (Figs 1B and S1C). The presence of a putative molecular ion of 272 (m/z) and a fragment ion of 257 indicate that this compound is a diterpene olefin related to edaxadiene, which we name edaxadiene B. Edaxadiene and edaxadiene B eluted earlier (at 23.2 and 23.8 min, respectively) from the GC column than tuberculosinol (30.3 min) and were present in significantly higher abundance.
Fig. 1.
GC trace of Mtb chloroform:methanol extracts. Lipids from Mtb cells grown to mid-log phase were extracted in chloroform:methanol and analyzed by GC/MS to detect low-molecular weight compounds. (A) Trace showing all species detected by GC/MS. (B) GC trace showing only those species containing ions characteristic of Mtb diterpenes (ions 80, 189 and 191). Chemical structures of diterpenes are adapted from previous studies [7,17] (Fig. S1).
To determine the cellular localization of these diterpenes, we prepared cell wall, cell membrane and cytosol fractions from γ-irradiated H37Rv cells and culture filtrate from live Mtb cultures. Lipids were extracted in either hexane (for cell wall) or chloroform:methanol (for culture filtrate, cytosol and cell membrane) and analyzed using GC/MS. Edaxadiene, edaxadiene B and tuberculosinol were detected in the cell membrane fraction. No diterpenes were detectable in the cytosol; the culture filtrate and cell membrane contained only trace amounts of edaxadiene (Table 1).
Table 1.
Localization and relative abundance of Mtb diterpenes. γ-irradiated Mtb cells (Colorado State University) were separated into cell wall, cell membrane and cytosol fractions, and lipids were extracted using hexane (for cell wall) or chloroform:methanol (for cytosol and cell membrane). Culture filtrate prepared from live Mtb cultures was lyophilized, and lipids were extracted in chloroform:methanol. Tuberculosinol, edaxadiene and edaxadiene B were detected by GC/MS and the relative abundances are indicated.
| Fraction | Tuberculosinol | Edaxadiene | Edaxadiene B |
|---|---|---|---|
| Cell membrane | + | +++ | ++ |
| Cell wall | − | + | − |
| Cytosol | − | − | − |
| Culture filtrate | − | + | − |
Rv3377c is necessary for diterpene synthesis in vivo
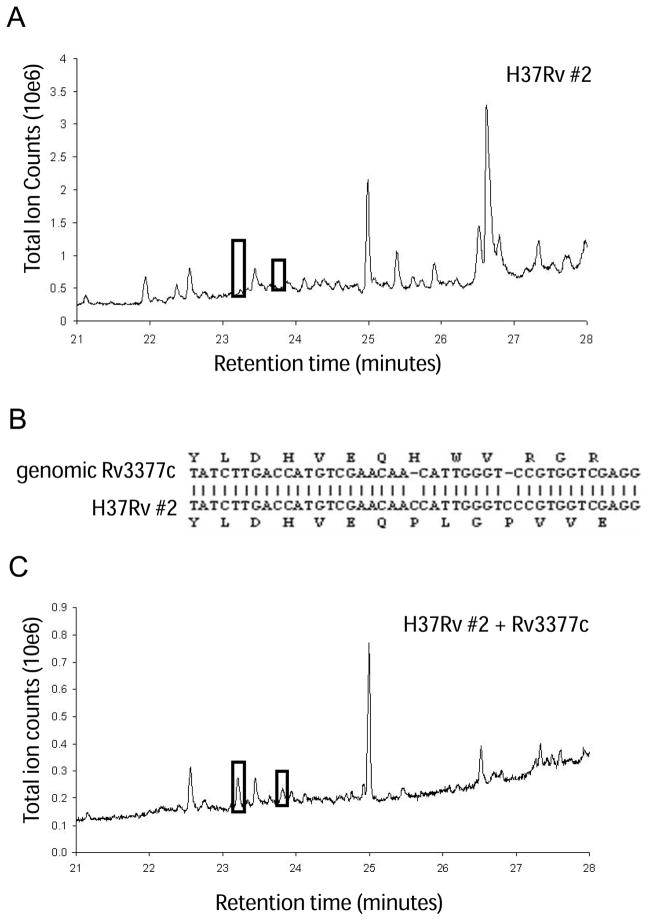
To define the biosynthetic pathway for diterpene production in Mtb, we compared diterpene production in several common laboratory Mtb strains. Chloroform:methanol extracts from five different Mtb isolates all showed the presence of the three diterpenes (Table S1). Surprisingly, one stock of H37Rv (#2) produced neither tuberculosinol, edaxadiene, nor edaxadiene B (Fig. 2A and Table S1). H37Rv #1 and #2 were obtained as separate glycerol stocks from the same source (M. Schelle, University of California, Berkeley, CA, USA) as the other Mtb strains analyzed, and they were grown under the same conditions. H37Rv #2, however, was subcultured in liquid medium for approximately 9 months longer than H37Rv #1. To determine how diterpene production is disrupted in H37RV #2, we isolated mRNA and used it as a template for RT-PCR. Rv3377c transcripts were detected in the mutant strain (data not shown), although sequencing of the cDNA revealed two single-base insertions in Rv3377c (Fig. 2B). These two insertions disrupt the reading frame just upstream of the sequence encoding the catalytic-site motif and presumably eliminate production of the active enzyme. Notably, complementing this mutant H37Rv strain with a plasmid containing Rv3377c under the control of an inducible promoter restored the production of edaxadiene and edaxadiene B (Fig. 2C). These results indicate that the reaction catalyzed by Rv3377c is the first committed step in diterpene production in Mtb, and that Rv3377c is required to produce all three Mtb diterpenes.
Fig. 2.
Frameshift mutations in Rv3377c disrupt diterpene production in Mtb. The strain H37Rv #2 does not produce any diterpenes. (A) GC trace from H37Rv #2. Black boxes indicate where edaxadiene and edaxadiene B are expected to elute. (B) Sequencing of Rv3377c from H37Rv #2 shows two single-base insertions in the gene, upstream of the active site. (C) GC trace of H37Rv #2 complemented with a plasmid expressing Rv3377c. Black boxes indicate the retention times of edaxadiene and edaxadiene B.
Reconstitution of the edaxadiene pathway in M. smegmatis
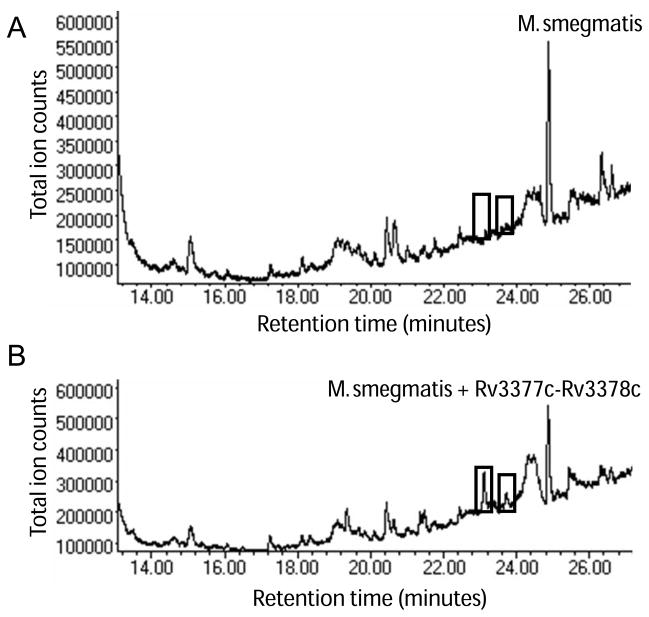
To determine the full biosynthetic pathway of these diterpenes, we tested whether Rv3377c and Rv3378c are sufficient to produce all three diterpenes in M. smegmatis, a nonpathogenic mycobacterium species. M. smegmatis lacks orthologs of Rv3377c and Rv3378c, and GC/MS analysis of lipids extracted from wild-type M. smegmatis showed no evidence of diterpenes (Fig. 3A). We transformed wild-type M. smegmatis mc2115 with a vector harboring an inducible promoter driving both Rv3377c and Rv3378c. When transformed with vector alone, or when expression was not induced, M. smegmatis did not produce detectable levels of diterpenes (data not shown). When both Rv3377c and Rv3378c were expressed in M. smegmatis mc2115, edaxadiene and edaxadiene B were produced, whereas tuberculosinol was below detection levels (Fig. 3B). The diterpenes produced by Rv3377c and Rv3378c in M. smegmatis and those found in Mtb have identical GC retention times and mass spectra. These results support a working model of diterpene biosynthesis in Mtb whereby Rv3377c and Rv3378c act in sequence to convert GGPP to edaxadiene and the previously unidentified product, edaxadiene B.
Fig. 3.
Engineering diterpene production in M. smegmatis. (A) GC trace of wild-type M. smegmatis mc2115, which lacks orthologs of Rv3377c and Rv3378c. Boxes indicate the elution times of edaxadiene and edaxadiene B. (B) GC trace of M. smegmatis overexpressing Rv3377c and Rv3378c. Boxes show edaxadiene and edaxadiene B.
Discussion
Lipids, including pthiocerol dimycocerate and sulpholipid-1, are critical mediators of Mtb virulence. Consistent with this role, the genes responsible for diterpene synthesis, Rv3377c and Rv3378c, were identified in a genetic screen for factors involved in initial fusion arrest events [10]. In the present study, we demonstrate for the first time that Mtb grown in broth culture produces membrane-associated diterpenes implicated in blocking phagolysosome fusion (Fig. 1 and Table 1). Thus, the proteins necessary for diterpene production are functional under normal growth conditions, even when the bacteria are outside the host macrophage. Observing diterpenes under laboratory culture conditions implies that they are typically present in the Mtb membrane, which appropriately situates the diterpenes to interact with the host cell upon contact. Inhibition of phagolysosome fusion is a fast process, taking place within 4 min of the bacteria engaging the host cell [14]. This time frame is too short for Mtb to synthesize new protein or the lipid factors necessary for arrest. The factors involved in early fusion arrest must therefore be present and active before the bacteria enters the host cell, and the diterpenes detected in Mtb membranes are well positioned to assist in this process.
Surprisingly, we identify three lipid species with mass spectral fragmentation patterns characteristic of diterpenes (Fig. 1B). Previous in vitro biochemical studies suggested that only a single diterpene, edaxadiene, would be present in Mtb [8,9]. The most abundant diterpene that we could detect in both Mtb membranes and whole-cell extracts shows a mass and fragmentation pattern identical to edaxadiene. The abundance of edaxadiene in Mtb extracts suggests that it is the major pathway product and that it is unlikely that there are subsequent modifications in vivo.
However, Mtb also produces another diterpene with a distinct GC retention time but similar mass fragmentation pattern to edaxadiene (Figs S1B,C). This diterpene has the same putative molecular ion (272) as edaxadiene, as well as a 257 ion characteristic of diterpene olefins (M-CH3), and is likely a minor product of Rv3378c not detected in vitro. We propose naming this side product edaxadiene B. Because of its low abundance and the difficulty in purifying sufficient quantities, the molecular structure of edaxadiene B remains unresolved. The production of more than one end product by a class I diterpene synthase is not uncommon in plants [13,15]. Several well-characterized plant enzymes, such as levopimaradiene/abietadiene synthase, produce multiple diterpenes [16]. In most cases, only one of the plant diterpenes is biologically active. It remains to be determined which form of edaxadiene is active in vivo.
The third diterpene that we detected in Mtb has a mass spectral fragmentation pattern identical to that of tuberculosinol, a dephosphorylated intermediate in the edaxadiene pathway. In the presence of Rv3377c alone in vitro, GGPP is converted into the intermediate halimadienyl diphosphate, which can be detected as tuberculosinol after dephosphorylation [8,9,17]. The presence of tuberculosinol in the Mtb membrane implies the existence of an endogenous phosphatase that can convert a small fraction of the halimadienyl diphosphate into tuberculosinol. This may act as a detoxifying strategy because high levels of prenyl phosphates are toxic to bacteria and are converted to less toxic alcohols by endogenous phosphatases [18]. The identity of the protein responsible for this dephosphorylation reaction remains unknown.
We investigated whether the three diterpenes identified are produced by common Mtb strains. All of the Mtb strains that we surveyed except one produce the three diterpenes with approximately the same abundance (Table S1). The exception is H37Rv #2, which does not produce any diterpenes (Fig. 2A and Table S1). This H37Rv strain, which was grown in liquid culture for approximately 9 months longer than the other H37Rv strain analyzed, has two single-base insertions in Rv3377c that disrupt the reading frame upstream of the active site (Fig. 2B). Without active Rv3377c, none of the three diterpenes are produced. When a plasmid-encoded copy of Rv3377c is introduced into this diterpene mutant strain and protein expression is induced, edaxadiene and edaxadiene B are generated (Fig. 2C). These results confirm that Rv3377c performs the first committed step in the production of all three diterpenes in Mtb.
The related, nonpathogenic species M. smegmatis does not contain genes homologous to Rv3377c or Rv3378c and does not produce detectable levels of diterpenes in vivo as shown by GC/MS (Fig. 3A). However, when both Rv3377c and Rv3378c are expressed in M. smegmatis from a plasmid-based inducible promoter, edaxadiene and edaxadiene B are produced (Fig. 3B), indicating these are the only two proteins necessary for diterpene production in Mycobacterium. This two-step pathway from the substrate GGPP to final diterpene products, with no further modifications, is shorter than a typical diterpene pathway in plants or other microbes. An abbreviated pathway may reflect that Rv3377c and Rv3378c, as well as the adjacent gene Rv3376, were likely acquired recently through horizontal gene transfer. If recent acquisition is indeed the case and only these three genes were transferred, then it is likely that additional enzymes, such as P450 cytochromes, dehydrogenases or oxidoreductases that typically modify plant diterpenes, are not involved in diterpene synthesis in Mtb.
To our knowledge, Mycobacterium is only the second bacterial genus reported to produce diterpenes in vivo. A small number of other diterpenes, including terpentecin, have been isolated from several Streptomyces species [3–6]. The biological roles diterpenes play in bacteria are poorly understood because only the antibiotic function of terpentecin has been determined. The identification of edaxadiene and edaxadiene B as the endpoints of the biosynthetic pathway in vivo sets the stage for determining the biological functions and mechanism of action of the Mtb diterpenes. The diterpenes may create bacterial membrane microdomains that mediate signaling or, alternatively, they may transfer to host membranes to target host components. Discovering the biological target for the bioactive diterpene will further elucidate the pathways regulated by Mtb lipids during infection.
Experimental procedures
Amplification and cloning
A vector for expression of Rv3377c and Rv3378c from M. tuberculosis H37Rv (Colorado State University) was created using the Gateway System (Invitrogen, Carlsbad, CA, USA). A 2400 bp product was amplified by PCR containing primers 3378F (5′-CTGGTGCCACGCGGTTCTCATATGAACTTGGTTAGCGAA-3′) and 3377R (5′-GAAAGCTGGGTGAAGCTTTCATTGGTTACTCTCATC-3′), 5% dimethylsulfoxide and TurboCX polymerase (Stratagene, La Jolla, CA, USA). The PCR product was purified and used as a template for a second amplification using primers GatewayTHF and GatewayR. Recombinants of this second PCR product in the vector pDONR207 were obtained using BP clonase (Invitrogen) and were confirmed by DNA sequencing. Rv3377c/Rv3378c was moved from pDONR207 using LR clonase (Invitrogen) into destination vector pTETGW (a gift from C. Sassetti, Univerisity of Massachusetts, Worcester, MA, USA), a Mycobacterium expression vector containing Gateway cloning sites, a Mycobacterium origin of replication and a gene encoding hygromycin resistance. The LR reactions were transformed into Escherichia coli DH5α and confirmed by DNA sequencing.
M. smegmatis and Mtb strains and growth conditions
Mtb strains were a kind gift from M. Schelle (University of California, Berkeley). M. smegmatis mc2115 and Mtb strains were grown in Middlebrook 7H9 broth (Difco, Franklin Lakes, NJ, USA) supplemented with 10% Middlebrook OADC Enrichment and 0.05% Tween 80. Strains harboring an Rv3377c expression plasmid, an Rv3377c/Rv3378c expression plasmid or vector only were grown in medium also containing 50μg·mL−1 hygromycin B, and expression of Rv3377c or Rv3377c/Rv3378c proteins was induced with 50 ng·mL−1 anhydrotetracycline (Atc; Sigma, St Louis, MO, USA). Strains were grown at 37 °C with shaking at 220 r.p.m.
Preparation of Mtb membranes and cytosol
γ-irradiated Mtb cells were obtained from the Colorado State University TB Vaccine Testing and Research Materials Contract. Irradiated Mtb H37Rv cells were resuspended in NaCl/Pi containing the protease inhibitors leupeptin (5μg·mL−1) and 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (1 mM ), sonicated on ice and centrifuged at 1500 g for 10 min. The supernatant was saved, and the cell pellet was resuspended, sonicated and centrifuged a second time. The two supernatants were combined and centrifuged at 26 000 g for 1.5 h to pellet the cell wall. The remaining supernatant was centrifuged at 100 000 g for 4 h to separate the cell membranes from the cytosol. The membrane fraction was resuspended in NaCl/Pi containing 1% n-octyl-β-D-glucopyranoside and 1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride overnight at 4°C. Lipids were extracted from the cell membranes and cytosol in 5 mL of 1:1 chloroform:methanol by vortexing for 10 min at maximum speed and centrifugation at 3200 g for 10 min after partition with water. The organic phase was used for GC/MS analysis.
Preparation of Mtb culture filtrate
A 100 mL culture of Mtb H37Rv was grown until an D600 of 0.8 was reached in 7H9 medium supplemented with 10% OADC and 0.05% Tween-80. The cells were removed by centrifugation at 3200 g for 15 min. The supernatant (culture filtrate) was sterilized by passing it through a 0.22μm filter twice, before removal from the biosafety level three facility. The culture filtrate was lyophilized to dryness and lipids were extracted in 8 mL of 1:1 chloroform:methanol with vortexing for 10 min. A water partition separated the phases, and samples were centrifuged at 3000 g for 10 min. The lower organic phase was used for GC/MS analysis.
Lipid extraction from cell pellets
The cell pellet from a 50 mL culture of Mtb or M. smegmatis was resuspended in 4 mL of 1:1 chloroform:methanol and transferred to a glass vial. The cell suspension was vortexed for 10 min. One milliliter of ddH2O was added and the samples were vortexed for an additional 1 min. Cell suspensions were centrifuged at 3000 g for 10 min. The lower, organic phase was removed and used for GC analysis.
GC/MS
Chloroform extracts were transferred to 2 mL glass vials, containing glass inserts for smaller volumes as appropriate, and run directly on a gas chromatograph-mass spectrometer (GC model 6890, MS model 5973 inert; Agilent Technologies Inc., Santa Clara, CA, USA). An aliquot of the sample (1 μL) was injected into a Cyclosil-B column (Agilent Technologies Inc.) operating at a helium flow rate of 1 mL·min−1. The oven temperature was held at 100 °C for 2 min after injection, and was then ramped to 250 °C at 5 °C·min−1, and finally held at 250 °C for 2 min. The MS ion source was held at 230 °C throughout, with the quadrupole at 150 °C. Full mass spectra were generated for metabolite identification by scanning within the m/z range of 45–350.
RNA extraction from Mtb
Mtb H37Rv (30 mL) was grown until an D600 of 0.8 was reached. Cells were harvested by centrifugation, and the cell pellet was frozen on dry ice and resuspended in 1 mL of Trizol reagent (Invitrogen). The suspension was transferred to a 2 mL screw cap tube containing 0.5 mL of 0.1 mm glass beads, and cells were disrupted using three 30 s pulses in a bead beater. After 5 min of inversion at room temperature, samples were centrifuged at 13000 g for 1 min. The Trizol solution was transferred to a tube containing 500μL of heavy phase lock gel (5 Prime 3 Prime, Inc., Boulder, CO, USA) and 300μL of chloroform was added followed by rapid inversion for 2 min and centrifugation at 13 000 g for 5 min. The aqueous layer was transferred to a tube containing 270μL of isopropanol and mixed with 270μL of 0.8 M sodium citrate/1.2 M NaCl prior to removal from the biosafety level three facility.
RNA was precipitated overnight at −20 °C and then centrifuged at 13 000 g for 10 min at 4 °C. The pellet was washed once with 70% ethanol in diethylpyrocarbonate-treated water, centrifuged for 5 min as above and air-dried. After resuspension of RNA pellets in 90μL of diethylpyrocarbonate-treated water, 10μL of 10×DNase I buffer and 4 units of DNase I (Invitrogen) were added and samples were incubated for 30 min at room temperature. Final purification of RNA was carried out using an RNeasy column in accordance with the manufacturer’s instructions (Qiagen, Valencia, CA, USA).
cDNA synthesis and RT-PCR
cDNA was synthesized from 3 μg of purified RNA using random primers (Invitrogen) and Thermo-X reverse transcriptase (Invitrogen). This cDNA was used as a template for PCR with primers complementary to sequences near the 5′ end of Rv3377c [RT3377F (5′-TGTCGAAGTTTGGACG-3′) and RT3377R (5′-CTTGGCAATCCACAAC-3′)], creating a product of approximately 450 bp. Primers [SigAF (5′-ACATGGTCGAGGTGAT-3′) and SigAR (5′-ATGGTCTTGGACTCGAT-3′)] complementary to sequences in the SigA gene were used as a positive control. To confirm the sequence of Rv3377c from the H37Rv strain, the complete ORFwas amplified from cDNA using primers 3377F (5′-CTGGTGCCACGCGGTTCTCATATGGAGACTTTCAGGACT-3′) and 3377R (5′-GAAAGCTGGGTGAAGCTTTCATTGGTTACTCTCATC-3′) using PFU Turbo CX polymerase (Invitrogen). The 1500 bp product was visualized on a 1% agarose gel, gel purified and sequenced directly.
Supplementary Material
Fig. S1. Mass spectrum fragmentation patterns of Mtb diterpenes.
Table S1. Diterpenes detected in laboratory strains of Mtb.
Acknowledgments
We thank C. Sassetti (Univerisity of Massachusetts) for the gift of pTETGW; M. Schelle (University of California, Berkeley) for Mtb strains; and the TB Vaccine Testing and Research Materials Facility at Colorado State University for providing the γ-irradiated H37Rv cells and Mtb genomic DNA. This work was supported by NIH grant P01AI68135 to T.A.
Abbreviations
- GGPP
geranylgeranyl diphosphate
- Mtb
Mycobacterium tuberculosis
References
- 1.Buckingham J. Dictionary of Natural Products. 1. Chapman and Hall/CRC; London: 2004. [Google Scholar]
- 2.Lange BM, Rujan T, Martin W, Croteau R. Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc Natl Acad Sci USA. 2000;97:13172–13177. doi: 10.1073/pnas.240454797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tamamura T, Sawa T, Isshiki K, Masuda T, Homma Y, Inuma H, Naganawa H, Hamada M, Takeuchi T, Umezawa H. Isolation and characterization of terpentecin, a new antitumor antibiotic. J Antibiot (Tokyo) 1985;38:1664–1669. doi: 10.7164/antibiotics.38.1664. [DOI] [PubMed] [Google Scholar]
- 4.Dairi T, Hamano Y, Kuzuyama T, Itoh N, Furihata K, Seto H. Eubacterial diterpene cyclase genes essential for production of the isoprenoid antibiotic terpentecin. J Bacteriol. 2001;183:6085–6094. doi: 10.1128/JB.183.20.6085-6094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamano Y, Kuzuyama T, Itoh N, Furihata K, Seto H, Dairi T. Functional analysis of eubacterial diterpene cyclases responsible for biosynthesis of a diterpene antibiotic, terpentecin. J Biol Chem. 2002;277:37098–37104. doi: 10.1074/jbc.M206382200. [DOI] [PubMed] [Google Scholar]
- 6.Motohashi K, Ueno R, Sue M, Furihata K, Matsumoto T, Dairi T, Omura S, Seto H. Studies on terpenoids produced by actinomycetes: oxaloterpins A, B, C, D, and E, diterpenes from Streptomyces sp. KO-3988. J Nat Prod. 2007;70:1712–1717. doi: 10.1021/np070326m. [DOI] [PubMed] [Google Scholar]
- 7.Mann FM, Xu M, Chen X, Fulton DB, Russell DG, Peters RJ. Edaxadiene: a new bioactive diterpene from Mycobacterium tuberculosis. J Am Chem Soc. 2009;131:17526–17527. doi: 10.1021/ja9019287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakano C, Hoshino T. Characterization of the Rv3377c gene product, a type-B diterpene cyclase, from the Mycobacterium tuberculosis H37 genome. Chembiochem. 2009;10:2060–2071. doi: 10.1002/cbic.200900248. [DOI] [PubMed] [Google Scholar]
- 9.Mann FM, Prisic S, Hu H, Xu M, Coates RM, Peters RJ. Characterization and inhibition of a class II diterpene cyclase from Mycobacterium tuberculosis: implications for tuberculosis. J Biol Chem. 2009;284:23574–23579. doi: 10.1074/jbc.M109.023788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pethe K, Swenson DL, Alonso S, Anderson J, Wang C, Russell DG. Isolation of Mycobacterium tuberculosis mutants defective in the arrest of phagosome maturation. Proc Natl Acad Sci USA. 2004;101:13642–13647. doi: 10.1073/pnas.0401657101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christianson DW. Structural biology and chemistry of the terpenoid cyclases. Chem Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 12.Wendt KU, Schulz GE. Isoprenoid biosynthesis: manifold chemistry catalyzed by similar enzymes. Structure. 1998;6:127–133. doi: 10.1016/s0969-2126(98)00015-x. [DOI] [PubMed] [Google Scholar]
- 13.Christianson DW. Unearthing the roots of the terpenome. Curr Opin Chem Biol. 2008;12:141–150. doi: 10.1016/j.cbpa.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russell DG. Mycobacterium tuberculosis: here today, and here tomorrow. Nat Rev Mol Cell Biol. 2001;2:569–577. doi: 10.1038/35085034. [DOI] [PubMed] [Google Scholar]
- 15.Keeling CI, Weisshaar S, Lin RP, Bohlmann J. Functional plasticity of paralogous diterpene synthases involved in conifer defense. Proc Natl Acad Sci USA. 2008;105:1085–1090. doi: 10.1073/pnas.0709466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peters RJ, Flory JE, Jetter R, Ravn MM, Lee HJ, Coates RM, Croteau RB. Abietadiene synthase from grand fir (Abies grandis): characterization and mechanism of action of the ‘pseudomature’ recombinant enzyme. Biochemistry. 2000;39:15592–15602. doi: 10.1021/bi001997l. [DOI] [PubMed] [Google Scholar]
- 17.Nakano C, Okamura T, Sato T, Dairi T, Hoshino T. Mycobacterium tuberculosis H37Rv 3377c encodes the diterpene cyclase for producing the halimane skeleton. Chem Commun (Camb) 2005;8:1016–1018. doi: 10.1039/b415346d. [DOI] [PubMed] [Google Scholar]
- 18.Withers ST, Gottlieb SS, Lieu B, Newman JD, Keasling JD. Identification of isopentenol biosynthetic genes from Bacillus subtilis by a screening method based on isoprenoid precursor toxicity. Appl Environ Microbiol. 2007;73:6277–6283. doi: 10.1128/AEM.00861-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Mass spectrum fragmentation patterns of Mtb diterpenes.
Table S1. Diterpenes detected in laboratory strains of Mtb.