Summary
The essential Mycobacterium tuberculosis Ser/Thr protein kinase (STPK), PknB, plays a key role in regulating growth and division, but the structural basis of activation has not been defined. Here we provide biochemical and structural evidence that dimerization through the kinase-domain (KD) N-lobe activates PknB by an allosteric mechanism. Promoting KD pairing using a small-molecule dimerizer stimulates the unphosphorylated kinase, and substitutions that disrupt N-lobe pairing decrease phosphorylation activity in vitro and in vivo. Multiple crystal structures of two monomeric PknB KD mutants in complex with nucleotide reveal diverse inactive conformations that contain large active-site distortions that propagate >30 Å from the mutation site. These results define flexible, inactive structures of a monomeric bacterial receptor KD and show how “back-to-back” N-lobe dimerization stabilizes the active KD conformation. This general mechanism of bacterial receptor STPK activation affords insights into the regulation of homologous eukaryotic kinases that form structurally similar dimers.
Introduction
Protein phosphorylation mediated by receptor Ser/Thr protein kinases (STPKs) is widely used to transduce extracellular signals into intracellular responses. In contrast to the regulation of eukaryotic receptor STPKs (Huse and Kuriyan, 2002) the control mechanisms of prokaryotic family members are not well understood. In bacteria, eukaryotic-like STPKs are especially prevalent in pathogens (Cozzone, 2005). Ser/Thr phosphorylation prompts numerous, broad effects--including changes in transcription, metabolic flux, cell growth, cell division, protein localization, and immune defense/pathogenesis (Kang et al., 2008; Park et al., 2008; Sharma et al., 2006b; Thakur and Chakraborti, 2006; Williams, 1999)--each requiring precise regulation of kinases for accurate signal transduction.
In Mycobacterium tuberculosis (Mtb), for example, STPKs have been estimated to phosphorylate several hundred proteins (Greenstein et al., 2005), and over 250 Ser/Thr phosphoproteins have been identified recently (Chao et al., 2010; Prisic et al., 2010). While two of the 11 Mtb STPKs are soluble kinases, nine are predicted transmembrane receptors with an N-terminal “eukaryotic-like” kinase domain (KD) linked through a single transmembrane helix to an extracellular sensor domain (Av-Gay and Everett, 2000). Orthologs of the Mtb transmembrane receptor Protein Kinase B (PknB) are the most widely distributed STPKs in the prokaryotic kingdom (Jones and Dyson, 2006). PknB is essential for Mtb growth (Fernandez et al., 2006; Sassetti et al., 2003), and it phosphorylates diverse substrates, including proteins involved in peptidoglycan synthesis (Parikh et al., 2009), cell division (Dasgupta et al., 2006), the stress response (Park et al., 2008), transcription (Sharma et al., 2006), metabolic control (O’Hare et al., 2008), and other STPKs (Kang et al., 2005; Prisic et al., 2010). A key challenge is to understand how bacterial receptor STPKs such as PknB respond to extracellular signals.
The PknB KD structure, the first of a bacterial STPK, showed that characteristic features of the active conformations of eukaryotic STPKs are conserved in bacterial family members (Ortiz-Lombardia et al., 2003; Young et al., 2003). Like many eukaryotic STPKs, autophosphorylation of a sequence element called the activation loop strongly stimulates PknB kinase activity (Boitel et al., 2003). The KD contains characteristic N- and C-lobes that bracket the ATP binding site. The KD forms a back-to-back dimer through a conserved interface in the N-lobe. A functional role for the N-lobe interface was supported by the finding that the Mtb PknE KD crystallized as a structurally analogous dimer despite divergence of the intersubunit contact residues (Gay et al., 2006). Remarkably, structurally analogous dimers are formed by certain human STPKs including PKR, the cytosolic dsRNA-dependent protein kinase (Dar et al., 2005), and Ire1, which mediates the unfolded protein response (Lee et al., 2008). Dimerization through the N-lobe activates PKR and related kinases (Dey et al., 2005; Dey et al., 2007), as well as the Mtb PknD KD in vitro (Greenstein et al., 2007), suggesting that structurally and functionally similar interfaces regulate these STPKs. The structures of the monomeric forms of these eukaryotic and bacterial KDs, however, have not been reported.
To establish the role of the PknB N-lobe dimer interface and determine the conformational changes associated with dimer formation, we characterized the activities and structures of monomeric PknB mutants. We show here that dimerization of the unphosphorylated PknB KD promotes auto- and trans-phosphorylation in vitro. Mutations that disrupt the N-lobe interface decrease protein phosphorylation in Mycobacterium smegmatis strains expressing full-length Mtb PknB, establishing that N-lobe interactions are required for substrate phosphorylation in vivo. To discover the structural basis of this shared activation mechanism, we determined multiple crystal structures of PknB KD N-lobe interface mutants. Rather than adopting a single conformation, these mutant monomers populate diverse conformations containing hallmarks of “inactive” eukaryotic STPKs, including extensive remodeling of the active site. The unexpected variations in these structures reveal the flexibility of a monomeric bacterial receptor STPK. This work supports a general model for regulation of structurally homologous bacterial and eukaryotic kinases in which a flexible, inactive monomer shifts to the active conformation by dimerization and autophosphorylation.
Results
Although the wild-type PknB KD crystallized as a dimer with intersubunit contacts mediated by conserved residues (Ortiz-Lombardia et al., 2003; Young et al., 2003), the affinity and the function of the dimer have not been established. Equilibrium analytical ultracentrifugation experiments show that the phosphorylated intracellular domain of PknB1-330 (comprising the KD and juxtamembrane linker) has an apparent dissociation constant of ~2.9 mM (Figure S1). Because the interaction is weak, an inducible dimerization system was used to control the association of the KD in vitro and determine the impact of dimerization on phosphoryl transfer activity. We adapted the system previously used to study dimerization of the PknD KD (Greenstein et al., 2007) by constructing C-terminal fusions of PknB1-330 with the FK506-binding protein (FKBP) or the FKBP-rapamycin-binding (FRB) domain of the FKBP-rapamycin-associated protein (Brown et al., 1994; Choi et al., 1996). In this system, the small-molecule rapamycin promotes association of the FKBP and FRB domains, increasing the local concentration of the attached PknB KDs (Clemons, 1999; Crabtree and Schreiber, 1996).
Recombinant PknB1-330-FKBP purified from E. coli contained two predominant species carrying six or seven phosphates, as indicated by mass spectrometry (Table 1). Activity of the phosphorylated PknB1-330-FKBP and PknB1-330-FRB was confirmed by transphosphorylation of myelin basic protein, a non-cognate STPK substrate (Figure 1A). The phosphorylated PknB1-330 fusion proteins displayed similar activities in the absence or presence of rapamycin, and are not further activated by an increase in KD concentration in the 10–50 nM range. Addition of saturating rapamycin to a mixture of phosphorylated PknB1-330-FKBP and PknB1-330-FRB resulted in dimerization detectable by gel shift (Figure S2), but did not alter the phosphorylation of myelin basic protein. These results suggest that dimerization is not required for substrate phosphorylation when the PknB KD is already phosphorylated.
Table 1.
N-lobe dimer interface mutations affect PknB autophosphorylation
| PknB Mutant | Calculated Mass (Da) | Observed Masses (Da)a | PO4 sites |
|---|---|---|---|
| PknB-FKBP | 47,875.0 | 48,399.0 (avg. of 6P & 7P)bc | NDd |
| Arg10Ala-FKBP | 47,789.9 | 48,186 (5P), 48,113 & 48,273 (4P & 6P), 48,045 & 48,376 (3P & 7P) | ND |
| Leu33Asp-FKBP | 47,876.9 | 48,114.9 (3P), 48,033.0 (2P) | T171, T173, T294 |
| Asp76Ala-FKBP | 47,831.0 | 48,067.2 (3P), 47,988.0 (2P) | T171, T173, T294 |
| Asp138Asn-FKBP | 47,874.0 | 47,873.1 (0P) | none |
| Asp138Asn1-308 | 33,478.7 | 33,480.4 (0P) | none |
The species are listed in order of abundance, and the number of phosphates (P) is indicated in parentheses.
The species with molecular mass 48,399.0 (representing an average of 6–7 phosphates) was the most abundant species.
A molecular mass of 48,350.0 (6P) was the lowest molecular weight species observed.
ND - not determined in this study.
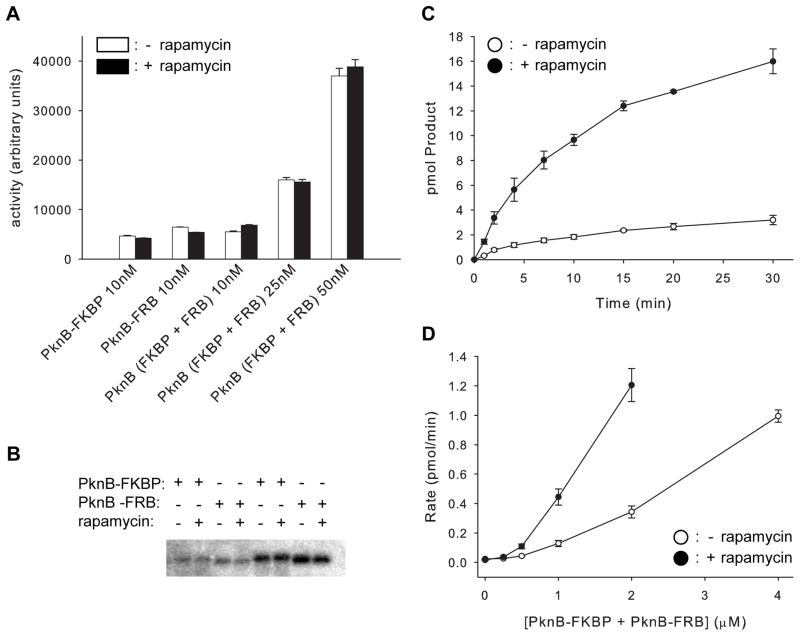
Figure 1. Dimerization regulates PknB activity.
(A) Dimerization does not activate phosphorylated PknB. Myelin basic protein is phosphorylated similarly by phosphorylated PknB1-330-FKBP, PknB1-330-FRB or an equimolar mixture of these fusion proteins (PknB (FKBP + FRB)) in the presence (filled bars) or absence (open bars) of 10 μM rapamycin. Total kinase concentrations are indicated. Error bars show the standard deviation of three independent experiments. See also Figure S2.
(B) Rapamycin does not affect autophosphorylation of isolated, unphosphorylated PknB1-330-FKBP or PknB1-330-FRB. Autoradiogram showing γ-32P incorporation into 4 μM of PknB-FKBP or PknB-FRB incubated for 10 min (first four lanes) or 20 min (last four lanes). The ratios of activities with and without rapamycin are 1.0 +/− 0.1 and 0.99 +/− 0.1, respectively.
(C) Dimerization activates unphosphorylated PknB. Autophosphorylation of unphosphorylated PknB1-330-FKBP and PknB1-330-FRB in the presence (filled circles) or absence (open circles) of rapamycin. Error bars show the standard deviation of three independent experiments.
(D) Kinase concentration dependence of autophosphorylation rates reflects an intermolecular reaction. Autophosphorylation rate is plotted against kinase concentration for the mixture of unphosphorylated PknB1-330-FKBP and PknB1-330-FRB in the presence (filled circles) or absence (open circles) of rapamycin. Error bars show the standard deviation of three independent experiments.
Dimerization activates intermolecular PknB autophosphorylation
To test if dimerization affects the activity of the unphosphorylated KD, PknB1-330-FKBP and PknB1-330-FRB purified from E. coli were dephosphorylated using PstP, the Mtb Ser/Thr protein phosphatase that antagonizes PknB (Boitel et al., 2003). Autophosphorylation of unphosphorylated PknB1-330-FKBP or PknB1-330-FRB individually is not affected by the presence or absence of rapamycin (Figure 1B). When PknB1-330-FKBP and PknB1-330-FRB are dimerized with rapamycin, however, autophosphorylation is stimulated 4.7 +/− 0.1 fold over the rapamycin-free control (Figure 1C), indicating quantitatively that dimerization activates unphosphorylated PknB.
To assess whether autophosphorylation results from intermolecular or intramolecular interactions, we measured the dependence of the reaction rate on enzyme concentration. An intramolecular reaction would display a simple linear dependence of reaction rate on enzyme concentration. The rate of autophosphorylation for the combination of unphosphorylated PknB1-330-FKBP and PknB1-330-FRB, however, exhibits a non-linear dependence on KD concentration in the presence and absence of rapamycin (Figure 1D). This higher-order concentration dependence supports a rate-limiting intermolecular phosphorylation mechanism both for the KD monomer and dimer. This conclusion is consistent with the previous observation that the PknB KD can phosphorylate an inactive KD mutant in a reaction that depends on intermolecular interactions through the G-helix (Mieczkowski et al., 2008).
Dimerization controls phosphorylation through an allosteric mechanism
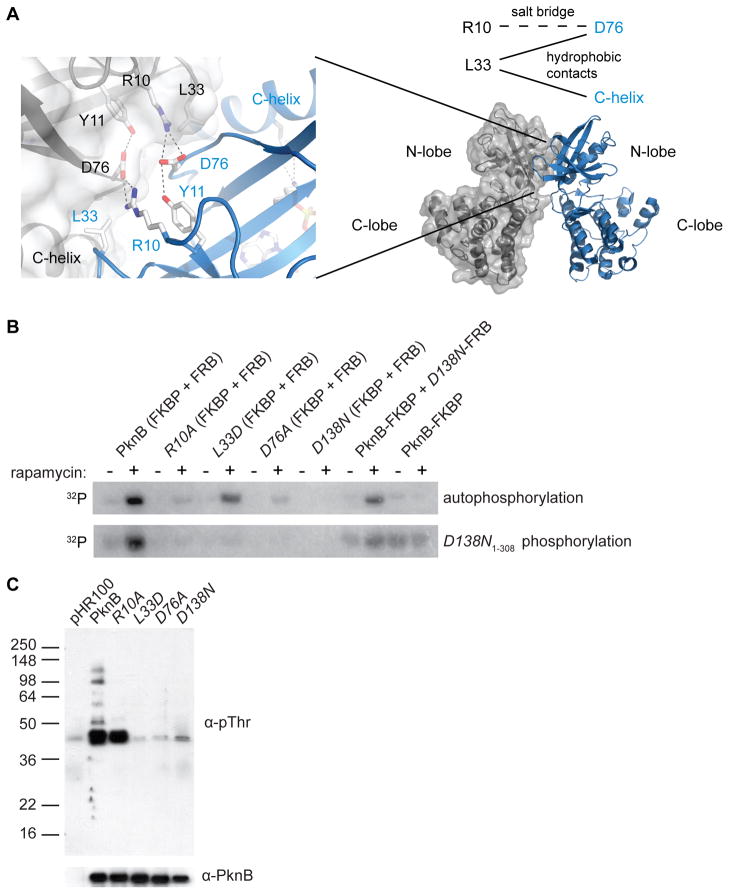
The crystal structure of the PknB KD dimer revealed a number of intermolecular interactions between invariant N-lobe residues, including a salt bridge between Arg10 and Asp76 and hydrophobic contacts between Leu33 and both Asp76 and the C-helix (Figure 2A). These classes of interactions are conserved not only in PknB orthologs, but also in human PKR and closely related kinases (Dey et al., 2005; Dey et al., 2007), suggesting that they are critical for kinase structure and/or function. Substitutions of residues in the predicted PknD KD N-lobe interface decrease kinase activation in vitro (Greenstein et al. 2007). To determine if PknB dimerization activates via allosteric effects that transmit the N-lobe interactions to the active site, we assayed the individual dimer-interface mutants Arg10Ala, Leu33Asp, and Asp76Ala for autophosphorylation activity. Using the dephosphorylated PknB kinase fusions, rapamycin increased autophosphorylation of the Arg10Ala, Leu33Asp, and Asp76Ala mutants only 1.8+/−0.4, 3.1+/−0.3, and 1.5+/−0.03 fold, respectively (Figure 2B). Compared to the 4.7-fold increase of autophosphorylation of the wild-type KD in the presence of rapamycin, these results support the idea that the N-lobe interface allosterically activates the PknB KD.
Figure 2. The PknB N-lobe interface is required for efficient autophosphorylation and substrate phosphorylation.
(A) Invariant residues Arg10, Leu33, and Asp76 form intermolecular contacts between PknB monomers (grey and blue (left)) (PDB code: 1mru). Hydrogen bonds are indicated by dotted lines. Reciprocal interactions include the Arg10-Asp76 ion pair and Leu33 hydrophobic contacts with Asp76 and the C-helix (top right).
(B) Mutations in the PknB N-lobe dimer interface impair autophosphorylation and phosphorylation of inactive PknB in vitro. Autoradiogram showing autophosphorylation of unphosphorylated PknB1-330 fusions (top) and transphosphorylation of inactive Asp138Asn PknB1-308 KD (bottom). Dimerization with the inactive Asp138Asn mutant (D138N-FRB; Lanes 11 & 12) activates the paired active subunit (PknB-FKBP), indicating that the dimer interface plays an allosteric role.
(C) Mutations in the PknB N-lobe dimer interface impair substrate phosphorylation in vivo. Western blots of M. smegmatis strains overexpressing the indicated full-length Mtb PknB variant or vector alone (pHR100) were probed using anti-phosphoThr (α-pThr; top) or anti-PknB (α-PknB; bottom) antibodies.
Supporting this idea, we found using mass spectrometry that the dimer-interface mutations also reduced autophosphorylation of the KD fusions expressed in E. coli. While the predominant wild-type PknB1-330-FKBP species carried six or seven phosphates, the predominant Arg10Ala1-330-FKBP species carried five phosphates and the Leu33Asp1-330-FKBP and Asp76Ala1-330-FKBP species carried three or two phosphates (Table 1). Further MS/MS analysis of trypsin digested PknB fusions showed that the Leu33Asp1-330-FKBP and Asp76Ala1-330-FKBP variants were phosphorylated at the positions in the activation loop known to be essential for activity, Thr171 and Thr173 (Boitel et al., 2003), as well as Thr294 in the juxtamembrane linker.
The idea that PknB KD is activated via an allosteric mechanism through the kinase N-lobe predicts that N-lobe interactions will increase autophosphorylation even if one monomer within the dimer is catalytically inactive (Greenstein et al., 2007). To test this prediction, we inactivated the PknB KD by replacing the catalytic Asp138 with Asn (Table 1). When Asp138Asn1-330-FKBP and Asp138Asn1-330-FRB are combined, no autophosphorylation is detected either in the presence or absence of rapamycin (Figure 2B). However, in the presence of active, unphosphorylated PknB1-330-FKBP, inactive Asp138Asn1-330-FRB stimulates the autophosphorylation activity by 3.7 +/− 0.5 fold. Thus, N-lobe pairing of catalytically active and inactive subunits stimulates kinase autophosphorylation.
N-lobe interface mutations decrease substrate phosphorylation in vitro and in vivo
To test the effects of the interface mutations on substrate phosphorylation, we quantified the phosphorylation of a shorter, catalytically inactive PknB construct (Asp138Asn1-308), which is separated easily from the longer, active PknB constructs. The unphosphorylated wild-type PknB1-330 fusions phosphorylated Asp138Asn1-308, and this activity was stimulated by rapamycin (Figure 2B, bottom left). In contrast, the N-lobe interface mutations in the KD fusions effectively eliminated phosphorylation of Asp138Asn1-308 to yields of phosphorylated products comparable to those observed using the inactive Asp138Asn1-330 PknB fusions (Figure 2B). Partial activity was observed when a functional dimer interface was restored using an active (PknB1-330-FKBP) and inactive (Asp138Asn1-330-FRB) subunit. These data demonstrate quantitatively that in vitro substrate phosphorylation by PknB depends on N-lobe dimerization.
To determine the role of the N-lobe interface in vivo, we overexpressed variants of full-length Mtb PknB in the nonpathogenic mycobacterium, M. smegmatis. When phosphorylation was measured by Western blotting with anti-phosphoThr antibodies, several cellular substrates were detected (Figure 2C). These phosphorylation levels were reduced upon expression of the catalytically inactive Asp138Asn mutant, the dimer interface mutants Arg10Ala, Leu33Asp, and Asp76Ala, or vector alone. Western blotting using an anti-PknB antibody showed that the levels of PknB variants were similar in the overexpression strains. These data suggest that the N-lobe dimer interface promotes substrate phosphorylation and mediates activation of Mtb PknB in vivo.
Dimer-interface mutants adopt novel N-lobe conformations
The dependence on N-lobe dimerization for allosteric PknB activation suggests that the conformation of the monomeric kinase should be inactive. To test this idea and to explore the structural relationship between dimerization and activity, we determined the crystal structures of two of the dimer-interface mutants. Because the PknB1-330 construct used for biochemical studies was more difficult to crystallize, mutations were introduced into PknB1-308 for structural studies (Young et al., 2003). Unexpectedly, the Leu33Asp variant in complex with the ATP analog, ATPγS, yielded multiple distinct crystal forms, which grew under identical or nearly identical conditions (Table S1). These crystals formed similar lattices with comparable intermolecular interactions. Here we describe four PknB dimer-interface mutant structures: three structures of Leu33Asp PknB1-308 bound to ATPγS·Mn2+ at 2.9 Å, 2.0 Å, and 1.6 Å resolution (space groups P212121, P21, and P212121, respectively) and one structure of Asp76Ala PknB1-308 bound to ATPγS·Mn2+ at 2.4 Å resolution (space group P212121). X-ray data-collection and refinement statistics are presented in Table 2.
Table 2.
Data collection and refinement statistics for Mn:ATPγS-bound PknB KD mutant structures. Highest resolution shell is shown in parentheses. See also Table S1
| Leu33Asp-1 | Leu33Asp-2 | Leu33Asp-3 | Asp76Ala | |
|---|---|---|---|---|
| Data Collection
| ||||
| Space Group | P21 | P212121 | P212121 | P212121 |
| Unit cell dimensions | ||||
| a, b, c (Å) | 80.93, 51.54, 141.41 | 39.34, 51.60, 155.13 | 40.19, 50.63, 132.74 | 40.46, 51.08, 134.75 |
| Resolution (Å) | 2.0 | 1.6 | 2.9 | |
| Wavelength (Å) | 1.116 | 1.116 | 1.893 | 1.116 |
| Rsym | 0.097 (0.424) | 0.034 (0.315) | 0.087 (0.407) | 0.070 (0.42) |
| I/μI | 11.95 (2.25) | 18.2 (3.4) | 11.2 (3.1) | 17.7 (2.5) |
| Completeness (%) | 100 (100) | 97.9 (88.2) | 100 (99.9) | 97.0 (96.5) |
| Solvent Content (%) | 37.5 | 43.9 | 34.6 | 38.6 |
|
| ||||
| Refinement
| ||||
| Rwork/Rfree | 0.201/0.258 | 0.155/0.186 | 0.210/0.278 | 0.215/0.288 |
| RMS bonds (Å) | 0.015 | 0.013 | 0.011 | 0.006 |
| RMS angles (º) | 1.58 | 1.52 | 1.0 | 1.1 |
| Non-hydrogen atoms | 9124 | 2593 | 2058 | |
| Solvent molecules | 907 | 352 | 50 | 15 |
| Metals | 2 Mn2+ | 1 Mn2+ | 1 Mn2+ | 1 Mn2+ |
| Ordered protein residues/monomer | 267 | 274 | 269 | 266 |
| Other molecules | Tris, SO4 | |||
Contains four monomers in the asymmetric unit; number of metals and ordered protein residues refers to the values for a single monomer.
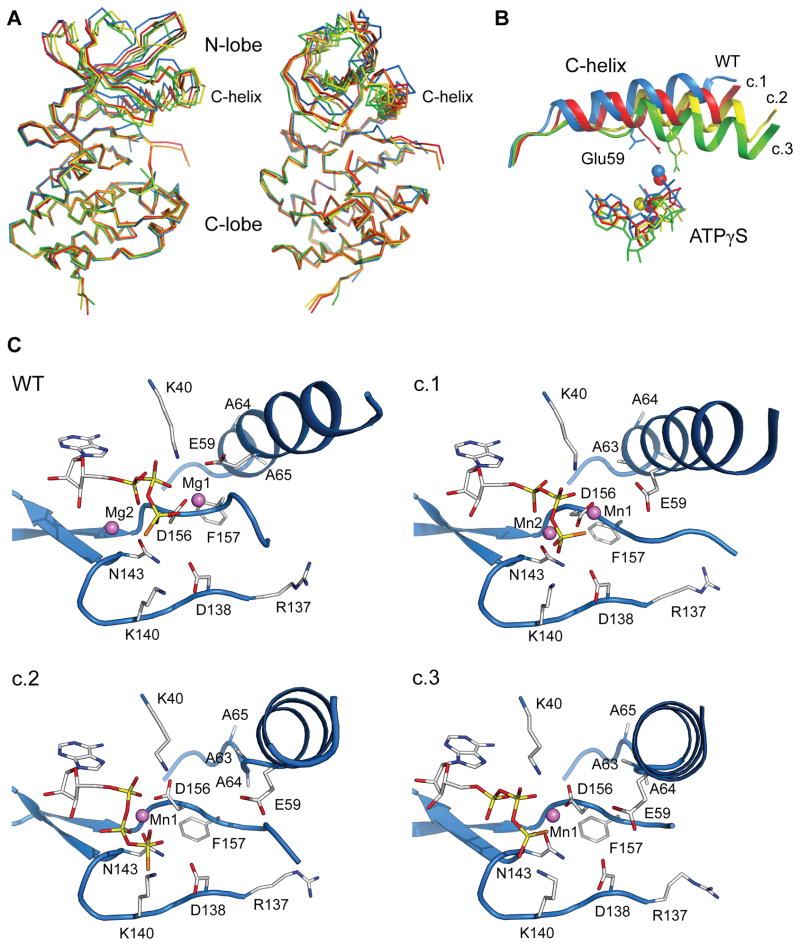
The structures clustered into three distinct conformations, distinguished primarily by differences in the N-lobe (Figure 3A). Conformation 1 (most similar to wild-type PknB) was found in two monomers of Leu33Asp with 2 Mn2+ ions and the Asp76Ala variant with a single Mn2+ ion coordinating the nucleotide. These Leu33Asp KD structures occurred in the monoclinic crystal form that contained four molecules representing two similar conformations in the asymmetric unit. The two conformations, although restrained by non-crystallographic symmetry throughout most of refinement, differed by a slight rigid-body rotation of the N-lobe (chains A and B, Cα root-mean-square-deviation (rmsd) vs. wild-type PknB: 1.01 Å, and chains C and D, Cα rmsd vs. wild-type PknB: 1.13 Å). The Asp76Ala KD structure (Cα rmsd vs. wild-type PknB: 1.08 Å) is very similar to chains A and B of the Leu33Asp KD in conformation 1 (Figure S3). Conformation 2 (more structurally distinct from wild-type PknB; Cα rmsd: 1.76 Å) and conformation 3 (most dissimilar from wild-type PknB; Cα rmsd: 2.15 Å) were observed for the Leu33Asp KD mutant bound to a single Mn2+. Conformations 1 and 2 were reproduced in multiple Leu33Asp KD crystals containing either manganese or magnesium and ATPγS or AMP-PNP (data not shown).
Figure 3. Dimer interface mutations cause conformational changes in the PknB N-lobe and active site.
(A) The monomeric Leu33Asp PknB KD shows large structural changes in the N-lobe compared to wild-type PknB. The dimeric wild-type PknB KD (blue, one monomer shown) and four distinct structures of the Leu33Asp KD (conformation 1 chain A: red; conformation 1 chain C: orange; conformation 2: yellow; conformation 3: green) are superimposed using the Cα atoms of C-lobe residues 100–155 and 185–285. Nucleotide and metals are omitted for clarity. See also Figure S3.
(B) The C-helix shifts downward toward the C-lobe and inward toward the active site in the Leu33Asp PknB KD structures (conformation 1 (c.1): red; conformation 2 (c.2): yellow; conformation 3 (c.3): green) compared to wild-type PknB (blue). The C-terminal turn of the C-helix unwinds in the Leu33Asp KD conformations 2 and 3, the Lys40-Glu59 ion pair is broken and the C-helix N-terminus (right; Ser51) shifts up to 11 Å. In concert, the bound nucleotide shifts away from the active conformation.
(C) Conformational changes occur in the active site of the Leu33Asp PknB KD compared to wild-type PknB. Each panel shows identical views of Leu33Asp conformations 1, 2, and 3 (c.1, c.2, and c.3) when the C-lobes are superimposed. C-helix shifts break the Lys40-Glu59 ion pair and couple directly to the DFG motif through contacts of Phe157 with C-helix residues Ala63 and Ala64. These multiple conformations suggest that the monomeric PknB KD is more flexible than the dimer.
In all the structures of these KD variants, PknB was monomeric, with no crystal contacts at the N-lobe interface containing the substitution. The structures show a small domain closure relative to the wild-type PknB KD and large shifts within the N-lobe. Similar to most eukaryotic protein kinases, the primary hinge for domain movement is in the linker between the N- and C-lobe, corresponding to PknB residues 93–99. In each conformation, backbone torsion-angle changes in this region account for most of the domain shifts relative to the wild-type PknB KD.
Dimer-interface mutations cause C-helix and active site changes
The structural changes in the N-lobe altered the conformation of residues 51–65, comprising the regulatory C-helix (Figure 3B). The C-terminus of this helix in wild-type PknB forms part of the N-lobe dimer interface, and intersubunit interactions to the C-helix are lost in the monomeric mutants. The C-helix carries key residues of the active site and is thought to contact protein substrates. In eukaryotic kinases, regulatory signals often toggle the C-helix between characteristic active and inactive positions (Huse and Kuriyan, 2002). In PknB KD conformation 1, the C-helix rotates toward the C-lobe and into the active site, with a displacement of ~4 Å at the helix N-terminus (Ser51) relative to the wild-type PknB KD dimer. Conformation 2 exhibits ~9 Å displacement of Ser51 compared to wild-type PknB due to an unwinding of the last turn of the helix (alanine residues 63–65). Ala64 in this segment makes direct hydrophobic contacts with Leu33 of the partner monomer in the WT dimer. The largest C-helix movement was observed in conformation 3, with a similar unwinding of the last turn of the helix and an ~11 Å translational shift of Ser51.
Reorganization of the C-helix in the dimer-interface mutants results in rearrangements of active-site residues that disrupt key features that mediate catalytic activity. The conserved Lys40-Glu59 ion pair, a hallmark of active STPKs (Huse and Kuriyan, 2002; Johnson et al., 1996), was broken in all of the dimer-interface mutant structures (Figure 3C). Lys40, which coordinates the α- and β-phosphates of the nucleotide, is stabilized in the active conformation by the close interaction with Glu59. The Glu59 carboxylate rotates ~45º away from Lys40 in Leu33Asp conformation 1 and ~90º away in conformations 2 and 3 compared to wild-type PknB (Figure 3B). As a result, the conformation of Lys40 is variable in the mutant structures, allowing the nucleotide phosphates to bind in a nonproductive orientation in conformation 2.
Coincident with shifts in the C-helix, the conserved DFG motif (Asp156-Gly158) adopts inactive conformations. Asp156, which coordinates the primary metal ion (Mg1) and positions the nucleotide β- and γ-phosphates, shifts from gauche- in the wild-type PknB KD to gauche+ in conformations 2 and 3 (Figure 3C). Phe157 shifts away from the “active” conformation observed in the wild-type PknB KD dimer and a flip in the Phe157 main-chain carbonyl group accompanies these changes in the side-chain position. In the monomer structures, the C-helix shifts into the position that Gly158 occupies in the active KD conformation.
The structures of the monomeric mutants also reveal changes in metal binding in the active site. In the wild-type PknB KD structure, the inner metal (Mg1) is bound by the β- and γphosphates of ATPγS and the carboxyl group of Asp156, while the outer metal (Mg2) binds the α- and γ-phosphates and the oxygen Oδ1 of Asn143 (Young et al., 2003). In the N-lobe-interface mutants, the second manganese ion (Mn2) is present only in Leu33Asp conformation 1 (Figure 3C). In this conformation, however, Mn2 is significantly shifted toward the interior of the active site, positioning it only ~3.8 Å away from Mn1. Subtle shifts in residue positions preserve the coordination of Mn2. In conformations 2 and 3 of the Leu33Asp PknB KD, the unwinding of the last turn of the C-helix results in larger changes in the active-site residues (Figure 3C). This reorganization appears to be incompatible with binding both Mn2+ ions. The N-lobe and nucleotide conformation tilt relative to the C-lobe and bring Asn143 “beneath” the nucleotide where it coordinates Mn1. This shift partially frees Asp156, which adopts a distinct rotamer with only one carboxyl oxygen coordinating Mn1. These shifts in comparison to the wild-type PknB KD eliminate the metal coordination common to active STPK conformations.
The activation loop, which forms a substrate-binding platform flanked by the DFG and APE motifs (SPE in PknB), also shows large differences between wild-type PknB and the monomeric mutants (Figure 3C). The ordered portion of the wild-type PknB activation loop (residues 159–163) extends away from the active site under the C-helix similar to the conformations observed in active eukaryotic kinases. In the Leu33Asp and Asp76Ala PknB KD structures, however, the activation loop extends past the end of the C-helix in a different, relatively straight conformation. The dimer-interface mutants show similar disorder in the activation loop compared to the wild-type PknB KD, with no interpretable electron density for residues 164–178, 164–177 or 165–178 (conformation 1), 161–178 (conformation 2) or 160–178 (conformation 3). This partial ordering contrasts with many inactive eukaryotic kinases, where the activation loop assumes a folded, protease-resistant conformation that blocks access to the ATP binding site (Huse and Kuriyan, 2002; Nolen et al., 2004).
Discussion
Protein interactions regulate STPKs by diverse mechanisms that control the populations of the active and inactive conformations of the KD. Here we characterized the role of the conserved, back-to-back, N-lobe dimer interface identified in the crystal structure of the Mtb PknB KD (Ortiz-Lombardia et al., 2003; Young et al., 2003). Our studies reveal that dimerization of the unphosphorylated PknB KD through the N-lobe interface activates the kinase (autophosphorylation increases ~5-fold in vitro) via a distinctive allosteric mechanism. An intermolecular interaction initiates autophosphorylation, as judged by the concentration dependence of the reaction and the ability of the KD to phosphorylate inactive subunits. The N-lobe dimer positions the PknB active sites away from each other, blocking access of the phosphorylation sites on one monomer to the active site of the other. This geometry, together with the ability of a catalytically inactive subunit to stimulate a wild-type partner, suggests that the dimer phosphorylates other dimers or monomers and the N-lobe contacts stabilize the active conformation of the PknB KD.
PknB N-lobe dimerization stimulates autophosphorylation not only in vitro--our data go beyond previous studies and suggest that this interface mediates PknB activation in vivo. The overexpressed, full-length Mtb PknB stimulates phosphorylation of endogenous proteins in M. smegmatis, and single substitutions in the N-lobe dimer interface that impede KD dimerization reduce or abolish this activity. Although the cellular protein phosphorylation stimulated by the expressed PknB can be direct or indirect, these results directly link N-lobe dimerization to PknB activation in mycobacteria.
Earlier work implicates a distinct C-lobe interface involving the PknB G-helix in trans-autophosphorylation of the activation loop (Mieczkowski et al., 2008). Taken together, these studies suggest that two interfaces--the N-lobe contacts that activate the KD and G-helix contacts in the C-lobe that mediate substrate recognition--contribute to autophosphorylation.
Inactive monomeric bacterial STPK structure
Previous structures of the PknB and PknE KD dimers left open the question of the conformational effects of the N-lobe intersubunit interactions. Our crystal structures of PknB KD dimer-interface mutants reveal the monomeric conformations of a bacterial STPK and explain the allosteric effects of dimerization. Unlike a switch between unique active and inactive conformations, the structural heterogeneity of the monomeric variants in multiple crystal forms obtained under similar conditions indicates that the PknB KD monomers are more flexible than the active dimer. The loss of direct intersubunit contacts destabilizes the last turn of the C-helix, which provides the mechanism of communication between the dimer interface and the active site. The release of the C-terminal anchor of the C-helix propagates and telescopes small structural changes (1.5–3 Å) at the mutation sites to changes of up to 11 Å at the C-helix N-terminus, >30 Å from the Leu33 substitution. These shifts at the active site rupture critical interactions required for substrate binding and catalysis. Thus, the structural polymorphism of the monomers reveals that the N-lobe dimer interface selects the active conformation by stabilizing particular productive features while maintaining flexibility in segments that move during turnover.
Comparison to inactive conformations of eukaryotic kinases
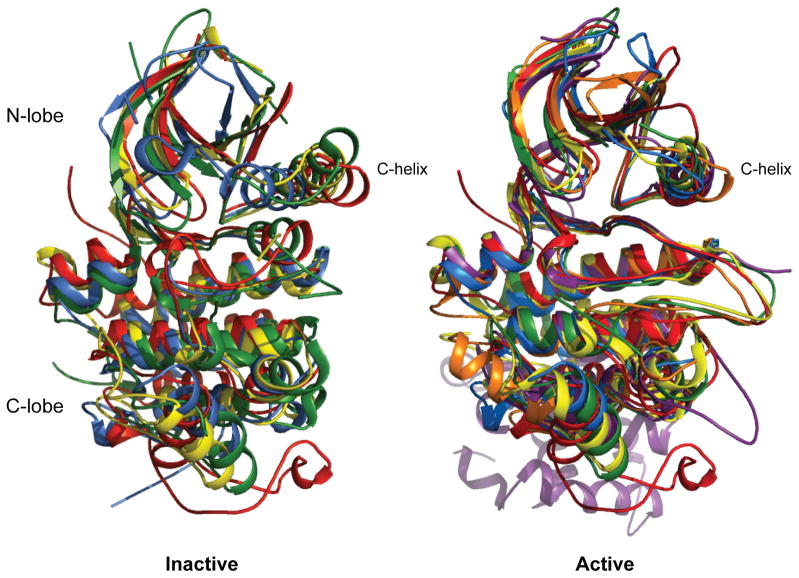
The activation mechanism of PknB represents an unprecedented conformational transition in the kinase family. In the PknB KD monomer mutant structures, the C-helix moves toward the active site, a shift opposite to the direction seen in most inactive eukaryotic kinases (Figure 4). In the inactive, nucleotide-bound structures of CDK2 (Brown et al., 1999), Src (Cowan-Jacob et al., 2005), and EGFR (Zhang et al., 2006), for example, the C-helix is swung back and away from the active site, increasing the distance between the essential lysine-glutamate ion pair. This inactive conformation is often maintained by other parts of the structure, such as direct interactions of the unphosphorylated activation loop with the C-helix that block helix movement. By contrast, in conformation 1 of the Leu33Asp and Asp76Ala PknB KDs, the orientation of the C-helix relative to the C-lobe differs by ~30º compared to the CDK2 and Src inactive states, and the tilting of the C-helix moves Glu59 away from a productive conformation. In conformations 2 and 3, the unwinding of the last turn of the C-helix also couples to distinctive shifts in this helix and the DFG motif. On the other hand, the active conformations of wild-type PknB (Young et al., 2003) and the eukaryotic kinases CDK2 (Russo et al., 1996), Src (Sicheri et al., 1997), EGFR (Zhang et al., 2006), PKR (Dar et al., 2005) and Ire1 (Lee et al., 2008) contain similar orientations of the C-helix and N-lobe (Figure 4). Thus, release of the PknB N-lobe intersubunit contacts increases the flexibility of the KD, which populates distinctive inactive conformations.
Figure 4. Unique conformational changes inactivate PknB compared to eukaryotic kinases.
The C-helix of monomeric, inactive PknB shifts in the opposite direction as eukaryotic kinases. Superposition of inactive (left) and active (right) kinase conformations of PknB (blue; Leu33Asp conformation 1 and 1mru), CDK2 (red; 1b38 and 1jst), Src (yellow; 1y57 and 1ad5), EGFR (green; 2gs7 and 2gs6), PKR (orange; no inactive structure and 2a19), and Ire1 (violet; no inactive structure and 2rio).
PknB regulation model
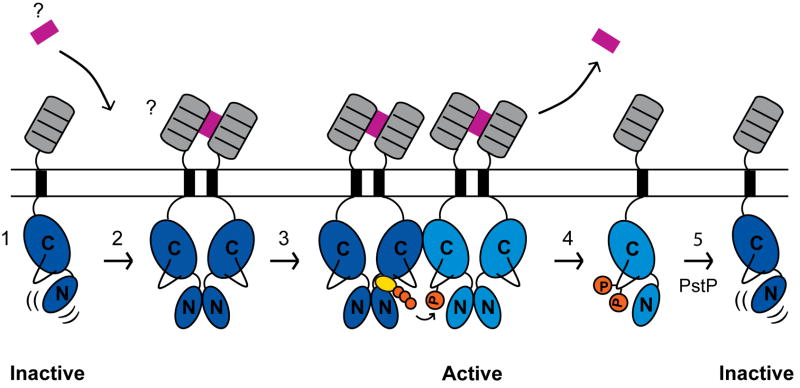
Although no structures of the Mtb PknD KD have been reported, the finding that N-lobe dimerization also activates the PknD KD in vitro (Greenstein et al., 2007) suggests that back-to-back pairing may regulate multiple mycobacterial receptor STPKs. Our quantitative measurements showing that dimerization helps activate the unphosphorylated KD supports a stepwise model for regulation of PknB and PknD (Figure 5). The unphosphorylated KD monomer represents the inactive form. Dimerization promoted, for example, by ligand binding to the extracellular domain, rigidifies the KD dimer N-lobe and stabilizes the active kinase conformation. This activation facilitates intermolecular autophosphorylation, which produces a fully active kinase domain that can phosphorylate substrates. When the extracellular dimerization signal dissipates, the phosphorylated kinase remains active. The PstP phosphatase antagonizes activation by dephosphorylating the KD (Boitel et al., 2003).
Figure 5. Model for PknB regulation.
(1) The unphosphorylated PknB monomer (kinase domain: dark blue) is the inactive species. (2) Dimerization, possibly mediated by ligand binding (violet) to the extracellular PASTA domains (gray), activates the intracellular kinase domains, triggering intermolecular autophosphorylation (kinase domain: cyan) and subsequent transphosphorylation of substrates. (4) Upon dimer dissociation, phosphorylated kinase monomers amplify the signal and remain active until (5) dephosphorylation by the Mtb Ser/Thr protein phosphatase, PstP, regenerates the inactive, unphosphorylated monomer.
We hypothesize that ligand binding to the PknB extracellular sensor domain promotes dimerization. The eukaryotic kinases PKR and Ire1, which form similar KD dimers but have unrelated sensor-domain architectures, are both activated by oligomerization promoted by ligand binding. The self-association of the isolated PknB and PknE KDs is very weak (low mM range (Gay et al., 2006)), and the isolated, extracellular sensor domains of PknB, PknD, and PknE kinases are all monomers at typical protein concentrations in solution (data not shown and (Barthe et al., 2010; Good et al., 2004)), suggesting that neither the receptor KDs nor the sensor domains dimerize without added ligands. Although the ligand that initiates PknB activation is unknown, the sensor domain contains PASTA domains thought to bind unlinked peptidoglycan fragments (Shah et al., 2008).
Implications for other STPKs
This regulatory mechanism shared by Mtb PknB and PknD (Greenstein et al., 2007) has several important and versatile features. The activity of the phosphorylated monomer can amplify external signals and enable phosphorylation to continue after the dimerization signal has dissipated. In addition, the intermolecular character of KD phosphorylation supports the idea that STPKs phosphorylate and activate other kinases in the cell. The finding that PknB can phosphorylate PknA exemplifies this kind of kinase network (Kang et al., 2005). This suggests that crosstalk between Mtb STPKs may form specific signaling networks with multiple combinations of inputs and phosphorylation outputs. As a result, responses to specific ligands could be tuned by the combination of ligand affinity, intermolecular binding, and the geometry of kinase binding sites in the signaling molecule.
The nine receptor kinases in Mtb cluster into three sequence clades, each containing three members (Greenstein et al., 2007). The N-lobe interface sequences are conserved within each clade, raising the possibility that kinases within a clade could form functional allosteric heterodimers. This idea has not been tested, however, and potential ligands that could bring together different STPKs remain to be defined.
Especially in light of the similarities to the structurally divergent PknD (Greenstein et al. 2007) and our demonstration that PknB dimer interface mutants fail to mediate protein phosphorylation in M. smegmatis, the general features of the PknB regulation mechanism are likely to be conserved in homologous bacterial receptor STPKs. The PknB orthologs in B. subtilis, PrkC, and S. pneumonia, StkP, are predicted to have conserved N-lobe surfaces similar to PknB. Although the structures of these bacterial STPKs have not been reported, these proteins form dimers in vivo, involving the extracellular PASTA domains and the transmembrane domain (Madec et al., 2002; Pallova et al., 2007). Importantly, ligand-promoted dimerization of an engineered P. aeruginosa receptor kinase, PpkA, fused through the extracellular C-terminus to a FKBP variant activates the kinase in vivo (Mougous et al., 2007). These biochemical and microbiological studies provide support for a general role for N-lobe dimerization in activating bacterial receptor STPKs.
The flexibility of the PknB monomer structures may represent a broader mechanism for regulation applicable to structurally similar eukaryotic protein kinases. Mtb PknB, human PKR, and human Ire1 form N-lobe dimers with striking similarities. Like PknB, dimerization through the N-lobe interface activates human PKR (Dey et al., 2005). Monomeric, inactive PKR is susceptible to proteolysis of the activation loop (Anderson and Cole, 2008), suggesting that the inactive conformation of the PKR and PknB activation loops are similarly disordered and positioned away from the active site. The functional and structural similarities between PknB and PKR support the hypothesis that the inactive PKR monomer will have structural similarities to monomeric, inactive PknB. Although the Ire1 and PknB KD dimers stabilize particular features of the KD structures (Lee et al., 2008; Young et al., 2003), the functions of these dimers appear to be somewhat different. Rather than activating the KD, as in PknB, the Ire1 dimer acts as a scaffold to promote assembly of a larger oligomer that enhances the activity of the appended ribonuclease domains on cognate substrates (Korennykh et al., 2009). Distinct from Ire1, the phosphorylated PknB KD is not further activated by an increase in dimer concentration, supporting the idea that PknB does not function as a higher-order oligomer. While bacterial STPKs may form complexes with other proteins (Mougous et al., 2007), the available data suggest that activated PknB functions as either a monomer or dimer.
Overall, our work reveals the structural ensemble of the inactive monomer of a bacterial STPK and provides insights into unanticipated mechanisms of protein kinase regulation. This window into the structural and functional implications of STPK dimerization sheds light on key control mechanisms that may open new avenues for therapeutic intervention in tuberculosis.
Experimental Procedures
Cloning, expression and purification
Expression vectors for Mtb PknB1-330, PstP, and PknB1-308 were described previously (Grundner et al., 2005; Pullen et al., 2004; Young et al., 2003). Expression vectors for the PknB-FKBP and -FRB fusions were generated as described (Greenstein et al., 2007), except the fusion constructs were introduced into a modified pET-28b vector encoding an N-terminal His tag and tobacco etch virus (TEV) cleavage site. Mutations were introduced using the QuikChange method (Stratagene). PknB1-308, PknB1-330, PknB fusion proteins, PknB variants and PstP were expressed and purified as described (Greenstein et al., 2007; Grundner et al., 2005; Pullen et al., 2004; Young et al., 2003). His tags of the PknB fusions and PknB1-308 variants were cleaved by incubation with TEV and thrombin, respectively, and removed with a second Chelating Sepharose HP column (GE Healthcare). The PknB1-308 Asp138NAsn, Leu33Asp, and Asp76Ala variants were purified further by gel-exclusion chromatography on a HiLoad 26/60 Superdex 75 column (GE Healthcare) and by ion exchange chromatography with a HiTrap Q HP column (GE Healthcare) using an elution gradient of 75–500 mM NaCl. The PknB fusions were dialyzed into 75–150 mM NaCl, 50 mM Hepes pH 7.5, and 0.5 mM TCEP. Other proteins were dialyzed into 75 mM NaCl, 20 mM Tris pH 7.5, and 0.5 mM TCEP.
In vitro kinase assays
To test the effect of dimerization on phosphorylation, phosphorylated KDs purified from E. coli, PknB-FKBP or PknB-FRB fusions (10 nM each) or the combination of the two (10, 25 or 50 nM total) were incubated in 20 μL of 20 mM Pipes pH 7.5, 75 mM NaCl, 1 mM DTT, 2 mM MnCl2, 54 μM myelin basic protein, and 10 μM rapamycin in ethanol or the equivalent volume of ethanol at 4 ºC for 2 h. Reactions were initiated by adding ATP to 50 μM supplemented with 5 μCi γ32P-ATP. Reactions were quenched after 30 min at room temperature using SDS loading buffer supplemented with 0.2 M DTT and 0.1 M EDTA. Samples were separated on 4–12% NuPAGE Bis-Tris gels by SDS-PAGE, dried, and exposed to a Molecular Dynamics Typhoon 8600 PhosphoImager. 32P incorporation was quantified with ImageQuant 5.2.
To dephosphorylate the PknB fusion proteins, a mixture of 1:10 (w/w) His6-PstP:KD was incubated at 25 °C in 2 mM MnCl2 in 50 mM Hepes pH 8.0 and 1 mM TCEP. PstP was removed with Ni-Sepharose resin. The complete removal of phosphoryl groups was confirmed by Western blotting using anti-pThr antibody (Zymed Laboratories) or mass spectrometry.
To detect autophosphorylation, dephosphorylated PknB-FKBP and PknB-FRB or mutants (2 μM total kinase or amount indicated) were incubated with 50 mM NaCl, 50mM Hepes (pH 7.5), 0.5 mM TCEP, 2 mM MnCl2 and 10 μM rapamycin (Sigma-Aldrich) in ethanol or an equivalent volume of ethanol for 25 min at room temperature. 100 μM ATP supplemented with 5 μCi of γ32P-ATP was added to initiate the 20-μL reactions. After the indicated times or 6 min for the rate measurements, reactions were quenched with SDS loading buffer and analyzed as described above for the phosphorylated kinase assays. Rapamycin-induced changes in activity were calculated using time points of 2, 4 and 8 min. To detect transphosphorylation, PknB-FKBP and PknB-FRB or mutants (1 μM total kinase) were incubated with 20 μM Asp138Asn PknB1-308. Rapamycin-induced changes in activity were calculated at 10, 20 and 40 min.
M. smegmatis strains, media and growth conditions
The full-length Mtb PknB gene was amplified from H37Rv genomic DNA (CSU TB Vaccine Testing and Materials Contract) and cloned into pJSC232 (Jeffery Cox, UCSF). PknB variants were cloned into the mycobacterial expression plasmid pHR100 (Jeffery Cox, UCSF). Plasmids were electroporated into M. smegmatis mc2155 and verified by DNA sequencing. M. smegmatis cultures were grown at 37 ºC in 7H9 Middlebrook medium (Difco) supplemented with 0.5 % [w/v] dextrose, 0.5 % [v/v] glycerol, and 0.05 % [w/v] Tween-80 or on Middlebrook 7H10-agar (Difco) plates with the same supplements. Plasmids were maintained by growth in medium containing kanamycin (20 μg/mL).
Phosphorylation and protein detection in M. smegmatis strains
Protein phosphorylation and the levels of PknB variants were determined by immunoblot analysis. M. smegmatis cells harboring pHR100-PknB, pHR100-PknB R10A, pHR100-PknB Leu33Asp, pHR100-PknB Asp76Ala, pHR100-PknB Asp138Asn, or pHR100 vector alone were grown to mid-log phase (OD600=0.8) and diluted to OD600=0.02 in 25 mL medium containing 0.2 % [w/v] acetamide. After one doubling time, cells were harvested by centrifugation, resuspended in extraction buffer (1% SDS, 20 mM EDTA, 50 mM HEPES pH 7.5, and 1 mM AEBSF), and bead beaten for 2 min at 4 ºC. 1 μg of protein from each strain was separated by a 12% Tris-glycine gel (Invitrogen) and analyzed by Western blotting using rabbit anti-pThr antibodies (Zymed Laboratories) or rabbit anti-PknB KD antibodies (Pacific Immunology).
Mass spectrometry
Analysis of kinase-domain phosphorylation states by mass spectrometry was performed as described (Young et al., 2003).
Crystallization, data collection, processing and crystallographic refinement
Leu33Asp and Asp76Ala PknB1-308 variants were crystallized at 4 °C by vapor diffusion from 10 mg/mL protein, 2 mM ATPγS, and 2 mM MnCl2 mixed 1:1 with precipitant. Table S1 shows the crystallization conditions and cryoprotectants for the PknB mutants. Before data collection, crystals were soaked for 1–15 minutes in the mother liquor plus 10–16% glycerol (or 30% PEG3350 for Asp76Ala) and 2 mM MnCl2, and frozen in liquid nitrogen.
X-ray diffraction data were collected at the ALS beamline 8.3.1 at 100 K and 11,111 eV or 6551 eV. Data were processed using the Elves automation program (Holton and Alber, 2004). Structures were solved by molecular replacement using the ATPγS-bound wild-type PknB KD (Young et al., 2003) as the search model in EPMR or Molrep (Kissinger et al., 1999; Vagin and Teplyakov, 2000). Rigid-body refinement with REFMAC (Murshudov et al., 1997) and/or simulated annealing with CNS (Brunger et al., 1998) were used prior to manual rebuilding. High-resolution structures were rebuilt using ARP/wARP (Perrakis et al., 1999). Models were further improved using O and Coot (Emsley and Cowtan, 2004; Jones et al., 1991) and refined using REFMAC and phenix.refine (Adams et al., 2010), with separate TLS parameters for the N and C lobes (Winn et al., 2001). For crystals containing Mn2+, the number and positions of the metals were verified using anomalous difference electron density maps calculated with data collected above the Mn2+ absorption edge.
Supplementary Material
Highlights.
Dimerization allosterically activates M. tuberculosis Ser/Thr protein kinase PknB.
N-lobe dimer interface mutations block PknB-dependent phosphorylation in vivo.
N-lobe interface mutants adopt multiple, monomeric, inactive conformations.
N-lobe pairing affords a general mechanism of activation for homologous kinases.
Acknowledgments
We thank Holly Ramage and Jeffrey Cox for the mycobacterial expression plasmids pJSC232 and pHR100, and Jessica Seeliger for advice on growth of M. smegmatis. We are indebted to Christoph Grundner for experimental advice and critical reading of the manuscript. Terry Lang provided invaluable editorial guidance. We thank John Kuriyan for helpful discussions, and especially James Holton, George Meigs, and Jane Tanamachi at Beamline 8.3.1 at Lawrence Berkeley National Laboratory for crystallographic assistance. We appreciate the support of the TB Structural Genomics Consortium. This work was supported by NIH grant R01 GM70962 to T.A.
Footnotes
Accession numbers
Coordinates and structure factors for the models of the PknB Leu33Asp (conformations 1, 2, and 3) and Asp76Ala variant KDs have been assigned Protein Data Bank accession numbers 3ORI, 3ORK, 3ORL and 3ORM, respectively. We also deposited coordinates and structure factors for three PknB Leu33Asp conformations (3ORO, 3ORP and 3ORT) obtained in the absence of bound divalent ions in crystals grown from high concentrations of the chelator, citrate. Although these metal-free structures may not be significantly populated in vivo, they confirm the flexibility of the Leu33Asp mutant KD.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E, Cole JL. Domain stabilities in protein kinase R (PKR): evidence for weak interdomain interactions. Biochemistry. 2008;47:4887–4897. doi: 10.1021/bi702211j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Av-Gay Y, Everett M. The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol. 2000;8:238–244. doi: 10.1016/s0966-842x(00)01734-0. [DOI] [PubMed] [Google Scholar]
- Barthe P, Mukamolova GV, Roumestand C, Cohen-Gonsaud M. The structure of PknB extracellular PASTA domain from mycobacterium tuberculosis suggests a ligand-dependent kinase activation. Structure. 2010;18:606–615. doi: 10.1016/j.str.2010.02.013. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cozzone AJ. Role of protein phosphorylation on serine/threonine and tyrosine in the virulence of bacterial pathogens. J Mol Microbiol Biotechnol. 2005;9:198–213. doi: 10.1159/000089648. [DOI] [PubMed] [Google Scholar]
- Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- Dasgupta A, Datta P, Kundu M, Basu J. The serine/threonine kinase PknB of Mycobacterium tuberculosis phosphorylates PBPA, a penicillin-binding protein required for cell division. Microbiology. 2006;152:493–504. doi: 10.1099/mic.0.28630-0. [DOI] [PubMed] [Google Scholar]
- Dey M, Cao C, Dar AC, Tamura T, Ozato K, Sicheri F, Dever TE. Mechanistic link between PKR dimerization, autophosphorylation, and eIF2alpha substrate recognition. Cell. 2005;122:901–913. doi: 10.1016/j.cell.2005.06.041. [DOI] [PubMed] [Google Scholar]
- Gay LM, Ng HL, Alber T. A conserved dimer and global conformational changes in the structure of apo-PknE Ser/Thr protein kinase from Mycobacterium tuberculosis. J Mol Biol. 2006;360:409–420. doi: 10.1016/j.jmb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Good MC, Greenstein AE, Young TA, Ng HL, Alber T. Sensor domain of the Mycobacterium tuberculosis receptor Ser/Thr protein kinase, PknD, forms a highly symmetric beta propeller. J Mol Biol. 2004;339:459–469. doi: 10.1016/j.jmb.2004.03.063. [DOI] [PubMed] [Google Scholar]
- Greenstein AE, Echols N, Lombana TN, King DS, Alber T. Allosteric activation by dimerization of the PknD receptor Ser/Thr protein kinase from Mycobacterium tuberculosis. J Biol Chem. 2007;282:11427–11435. doi: 10.1074/jbc.M610193200. [DOI] [PubMed] [Google Scholar]
- Greenstein AE, Grundner C, Echols N, Gay LM, Lombana TN, Miecskowski CA, Pullen KE, Sung PY, Alber T. Structure/function studies of Ser/Thr and Tyr protein phosphorylation in Mycobacterium tuberculosis. J Mol Microbiol Biotechnol. 2005;9:167–181. doi: 10.1159/000089645. [DOI] [PubMed] [Google Scholar]
- Holton J, Alber T. Automated protein crystal structure determination using ELVES. Proc Natl Acad Sci U S A. 2004;101:1537–1542. doi: 10.1073/pnas.0306241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- Kang CM, Abbott DW, Park ST, Dascher CC, Cantley LC, Husson RN. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 2005;19:1692–1704. doi: 10.1101/gad.1311105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski C, Iavarone AT, Alber T. Auto-activation mechanism of the Mycobacterium tuberculosis PknB receptor Ser/Thr kinase. Embo J. 2008;27:3186–3197. doi: 10.1038/emboj.2008.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol. 2007;9:797–803. doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- O’Hare HM, Duran R, Cervenansky C, Bellinzoni M, Wehenkel AM, Pritsch O, Obal G, Baumgartner J, Vialaret J, Johnsson K, Alzari PM. Regulation of glutamate metabolism by protein kinases in mycobacteria. Molecular microbiology. 2008;70:1408–1423. doi: 10.1111/j.1365-2958.2008.06489.x. [DOI] [PubMed] [Google Scholar]
- Ortiz-Lombardia M, Pompeo F, Boitel B, Alzari PM. Crystal structure of the catalytic domain of the PknB serine/threonine kinase from Mycobacterium tuberculosis. J Biol Chem. 2003;278:13094–13100. doi: 10.1074/jbc.M300660200. [DOI] [PubMed] [Google Scholar]
- Parikh A, Verma SK, Khan S, Prakash B, Nandicoori VK. PknB-mediated phosphorylation of a novel substrate, N-acetylglucosamine-1-phosphate uridyltransferase, modulates its acetyltransferase activity. J Mol Biol. 2009;386:451–464. doi: 10.1016/j.jmb.2008.12.031. [DOI] [PubMed] [Google Scholar]
- Park ST, Kang CM, Husson RN. Regulation of the SigH stress response regulon by an essential protein kinase in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2008;105:13105–13110. doi: 10.1073/pnas.0801143105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- Russo AA, Jeffrey PD, Pavletich NP. Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat Struct Biol. 1996;3:696–700. doi: 10.1038/nsb0896-696. [DOI] [PubMed] [Google Scholar]
- Shah IM, Laaberki HM, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Gupta M, Krupa A, Srinivasan N, Singh Y. EmbR, a regulatory protein with ATPase activity, is a substrate of multiple serine/threonine kinases and phosphatase in Mycobacterium tuberculosis. Febs J. 2006;273:2711–2721. doi: 10.1111/j.1742-4658.2006.05289.x. [DOI] [PubMed] [Google Scholar]
- Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- Young TA, Delagoutte B, Endrizzi JA, Falick AM, Alber T. Structure of Mycobacterium tuberculosis PknB supports a universal activation mechanism for Ser/Thr protein kinases. Nat Struct Biol. 2003;10:168–174. doi: 10.1038/nsb897. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.