Abstract
Visualization of in vivo mRNA localization provides a tool for understanding steps in the mechanism of transport. Here we detail a method of fluorescently labeling mRNA transcripts and microinjecting them into Xenopus laevis oocytes followed with imaging by confocal microscopy. This technique overcomes a significant hurdle of imaging RNA in the frog oocyte while providing a rapid method of visualizing mRNA localization in high resolution.
Keywords: RNA localization, Xenopus, oocyte, RNA transport, confocal microscopy
1. INTRODUCTION
RNA localization is a conserved mechanism of establishing cell polarity in a variety of cell types and organisms. Such spatial regulation of gene expression can define specialized regions of the cell, and prominent examples include germ layer specification during vertebrate development and cytoskeletal rearrangements involved in cell motility (1–3).
Visualization of RNA distribution patterns has provided valuable insights into RNA transport steps and mechanisms. A number of techniques have been developed to visualize subcellular RNA localization, including in situ hybridization with digoxigenin- and fluorescently-labeled probes (4–6), molecular beacons (7), and fluorescent protein tethering (8,9).
RNA localization has been studied extensively in Xenopus laevis oocytes, where RNAs are localized during oogenesis and underlie patterning along the animal-vegetal axis (3–6, 10–18). The Xenopus oocyte offers several significant advantages for studies of RNA transport. First, oocytes are easily obtained through non-lethal surgery. Each surgery can yield thousands of oocytes, making the system amenable to biochemical analyses. Second, oocytes are large in size, easily visible in detail under standard light microscopes, offering facile microinjection of RNA, proteins, DNA, and antibodies, which can be targeted into the nucleus or cytoplasm. Third, isolated oocytes are amenable to culture outside of the frog (19,20). However, one disadvantage is increasing opacity as yolk protein accumulates during oogenesis, complicating imaging approaches. Here we describe a method of visualizing RNA localization in Xenopus oocytes that overcomes this issue while providing striking, high-resolution images of in vivo RNA transport.
2. MATERIALS
2.1 In vitro RNA transcription
DEPC-treated deionized H2O (DEPC-H2O): Add 1–2 drops DEPC (Sigma-Aldrich) per 100 ml deionized H2O. Incubate for 30 minutes at room temperature. Autoclave.
10× Transcription Buffer (10× Tx): 60mM MgCl2, 400 mM Tris-HCl (pH 7.5), 20 mM spermidine-HCl. Store as 1 ml aliquots at −20 °C.
20x cap/NTP mix: 10 mM CTP, 10 mM ATP, 9 mM UTP, 2 mM GTP, 20 mM G(ppp)G Cap Analog (New England Biolabs # S1407L). Store as 25 μl aliquots at −20 °C.
Fluorescent nucleotides: Chromatide Alexa Fluor 488-5-UTP or 546-14-UTP (Invitrogen # C11403 and # C11404, see Note 6).
G-50 solution: Hydrate 5 g Sephadex G-50 beads (Sigma Aldrich) in 100 ml deionized H2O. DEPC-treat as detailed above. Before use, add the following: 0.5 ml 0.2 M EDTA, 1 ml 1 M Tris-HCl pH 8.0, 0.5 ml 20% SDS (all solutions must be RNase-free). Store at 4 °C.
G-50 column: Remove and discard the plunger from a 3 ml syringe (BD Biosciences) and place the barrel of the syringe into a 15 ml conical tube (Corning). Plug the syringe with a small amount of glass wool (a plug about half the size of a penny). Swirl the G-50 solution (see Materials 2.1.5) to resuspend beads. Add 2 ml G-50 solution to the empty column. Spin for 1 minute at 1,000 × g in benchtop centrifuge. Add 200 μl DEPC-H2O to each column. Spin. Repeat wash twice more for a total of three washes. Remove the column to a fresh 15 ml conical tube prior to use.
2.2 Oocyte microinjection
Needles: To make beveled glass needles with an outer diameter of ~0.05 mm, we pull 3.5 inch capillaries (Drummond Scientific item # 3-000-203-G/X) using a Sutter Instrument Co. micropipette puller. Needles are beveled to an angle of 40° using a Narishige Co. EG-4 micropipette grinder.
Microinjection Apparatus: Harvard Apparatus model # PLI-100.
Injection dish: We line a small plastic dish with 1/8th inch thick black foam rubber, cut to fit and secured to the dish with double-sided tape. The white oocytes stand out against the black foam background.
MBSH buffer: 88mM NaCl, 1 mM KCl, 2.4 mM NaHCO3, 0.82 mM MgSO4, 0.33 mM Ca(NO3)2, 0.41 mM CaCl2, 10mM HEPES (pH 7.6).
2.3 Oocyte culture
Collagenase Solution: 3 mg/ml collagenase (Sigma-Aldrich # C0130), 0.1 M KPO3+ (pH 7.4).
24 well plates (Sigma-Aldrich #CLS 3527).
Antibiotic Stocks: Nystatin (10,000 U/ml, Gibco, store in 1 ml aliquots at −20 °C). Penicillin/Streptomycin (10,000 U/ml, 10 mg/ml, Gibco, store in 100 μl aliquots at −20 °C). Gentamicin (10 mg/ml, Gibco, store at 4 °C).
Incomplete Oocyte Culture Medium (In-OCM): 50% L-15 medium (Sigma-Aldrich), 15 mM HEPES (pH 7.6), 1 mg/ml insulin (Sigma-Aldrich). We typically make a 50 ml stock, which can be stored at 4 °C for up to two months.
Complete Oocyte Culture Medium (OCM): 980 μl In-OCM, 5 μl nystatin stock, 10 μl gentamicin stock, 5 μl penicillin/streptomycin stock. Sterilize using a 0.22 μm syringe filter (Millipore # SLGP033RS). Make fresh, do not store.
2.4 Oocyte fixation and immunofluorescence
Glass vials for fixation (Fisher # 03-339-26B).
10x MEM Stock: 1 M MOPS (pH 7.4), 20 mM EGTA, 10 mM MgSO4. Store at room temperature.
MEMFA: 1 ml 10x MEM, 1 ml 37% formaldehyde, 8 ml deionized H2O. Prepare fresh for each use.
Methanol: Anhydrous methanol (Alfa Aesar # 41467), must be fresh.
Proteinase K Solution: 0.1 M Tris-HCl pH 7.5, 10 mM EDTA, 50 μg/ml Proteinase K (Sigma-Aldrich). Prepare fresh for each use.
PBT: PBS (pH 7.4), 0.2 % BSA, 0.1 % Triton X-100 (Roche).
Alexa Fluor 633 - coupled secondary antibodies (Invitrogen # A21070 [goat anti-rabbit IgG] or # A21050 [goat anti-mouse IgG], see Note 6).
2.5 Confocal Microscopy
Murray’s Clearing Medium: 2 parts benzyl benzoate (MD Biomedicals), 1 part benzylalcohol (Fisher). Store at room temperature.
Imaging dishes: FluoroDish (WPI Inc. # FD3510-100).
3. METHODS
3.1 Preparation of fluorescently labeled RNA by in vitro transcription
Linearize plasmid DNA containing the relevant sequence and upstream promoter sites for transcription by T7, SP6, or T3 RNA polymerase.
Resuspend DNA at 1 μg/μl with DEPC-H2O.
-
Add the following reagents, in order, to a sterile 1.5 ml tube:
2 μl 10× Tx buffer
1 μl 20x cap/NTP mix
11 μl DEPC-H2O
1 μl 0.2 M DTT
1 μl RNasin (Promega)
1 μl linearized DNA template (see Methods 3.1.1)
1 μl 1 mM Alexa Fluor 546-14-UTP
1 μl [α-32P]UTP (1 μCi/μl, Perkin Elmer)
1 μl RNA Polymerase (Promega)
Mix reagents gently and centrifuge briefly (10 sec. in a microcentrifuge).
Incubate for 2-4 hours at 37° C, covering the tube with aluminum foil to prevent photobleaching of fluorophore.
Add 1 μl 1 mg/ml RNase-free DNase (Promega) to degrade template DNA.
Incubate 15 minutes at 37° C.
Add 79 μl 20 mM EDTA (pH 8.0) to stop the reaction.
Remove 1 μl to a separate tube, labeled as “input” and save.
Spin reaction through a 1 ml G50 column, which will retain unincorporated nucleotides while excluding full length mRNA.
-
Concentrate the RNA by ethanol precipitation:
Add 250 μl 100% ethanol, 10 μl 7M ammonium acetate, 1 μl carrier RNA (yeast tRNA at 10 μg/μl) or 1 μl glycogen (20 mg/ml).
Mix well and freeze until solid on dry ice or at −80°C for ~30 minutes.
Spin at maximum speed in microcentrifuge for 15 minutes. Remove the supernatant. Wash the pellet with 150 μl of cold 75% ethanol.
Spin for 3 minutes at maximum speed in microcentrifuge. Remove supernatant and allow the pellet to dry for 3 minutes, at 37°C.
Resuspend RNA in 11 μl DEPC-H2O. Remove 1 μl to a separate tube, labeled as “incorporated”.
Determine the percent incorporation (see Note 1) and bring the RNA to a final concentration of 50 nM in DEPC-H2O (see Note 2 for troubleshooting RNA yield).
The RNA should be frozen in 5 μl aliquots, for single use (to avoid freeze/thaw cycles), and can be stored for several months at −80°C.
3.2 Oocyte microinjection
RNA preparation: Thaw an aliquot of RNA (50 nM), and denature the RNA at 70° C for 3–5 minutes. Spin for 10 minutes at maximum speed in microcentrifuge to remove particulates, remove to a fresh tube and keep on ice.
Oocyte preparation: Surgically remove oocytes from albino Xenopus laevis females (Nasco) and defolliculate by incubation in Collagenase Solution for 15 minutes, gently shaking (~200 rpm) at 18 C. Check to ensure ovary has released the defolliculated oocytes (see Note 4). Wash the oocytes three times with MBSH buffer. Manually sort stage III/IV oocytes (21), which are 250–400 μm in diameter.
-
Oocyte microinjection:
Calibrate needle with DEPC-H2O to deliver 2 nl per injection.
Load RNA into needle.
Place sorted oocytes in injection dish in MBSH buffer.
Carefully inject each oocyte with 2 nl of RNA at 50 nM.
Expel RNA, rinse needle with DEPC-H2O, and load the next RNA for injection (see Note 3).
3.3 Oocyte culture
Place injected oocytes in a well of a sterile 24 well plate.
Remove buffer and replace with 400 μl OCM per well.
Incubate oocytes at 18° C for time points ranging between 8 and 48 hours.
After culture, remove any dead oocytes; we routinely observe >90% survival.
3.4 Oocyte fixation and immunofluorescence
Place cultured oocytes in glass vials and rinse with MBSH.
-
If you are co-imaging RNA and protein distribution, skip to Methods 3.4.3. If you are imaging RNA alone, fix oocytes as follows:
Remove MBSH and replace with 1 ml MEMFA (see Note 5).
Rock for 20 minutes at room temperature.
Wash oocytes once with 1 ml MBSH. Skip to Methods 3.4.4.
-
For immunofluorescence, treat the oocytes with Proteinase K, followed by fixation and antibody incubation, as follows:
Remove MBSH and add Proteinase K solution. Incubate for three minutes at room temperature.
Remove Proteinase K solution and wash twice with 1 ml MEMFA (see Note 5).
Rock vials for one hour at room temperature in 1 ml MEMFA.
Remove MEMFA and add 1 ml PBT, rock at room temperature for 15 minutes. Repeat PBT wash twice, for a total of three washes.
Replace PBT with 500 μl of fresh PBT plus 2% BSA and 2% goat serum and rock for two hours at room temperature.
Replace solution with 500 μl PBT plus 2% BSA, 2% goat serum, and the appropriate dilution of primary antibody. (You may use 250 μl, if antibody is limiting.) Rock vials overnight at 4 °C.
Replace the primary antibody solution with 500 μl PBT and rock at room temperature for 1.5 hours. Wash twice more with PBT, for a total of three washes.
Replace PBT with 500 μl PBT plus 2% BSA, 2% goat serum, and the appropriate dilution of the secondary antibody. Rock vials overnight at 4 °C.
Replace solution with 500 μl PBT and rock at room temperature for 1.5 hours. Wash twice more with PBT, for a total of three washes.
-
Dehydrate oocytes as follows:
Remove half of the volume of buffer (MBSH or PBT), and replace with an equal volume of anhydrous methanol.
Remove half of the buffer/methanol solution and replace with an equal volume of anhydrous methanol. Repeat.
Remove all of the solution, replace with anhydrous methanol.
Wash once with anhydrous methanol.
Oocytes can be stored in methanol at −20° C until ready to image.
3.4 Imaging of RNA and protein distribution by confocal microscopy
Place Murray’s Clearing Medium into a Fluorodish.
Transfer oocytes from glass vials to an imaging dish, taking care to transfer as little methanol as possible.
Wait for several minutes for the oocytes to become optically clear. The oocytes should also sink to the bottom of the imaging dish. If not, gently tap the oocytes with forceps to break surface tension.
Image oocytes using an inverted confocal microscope. The entire field of oocytes can first be imaged using a 10× objective, which facilitates scoring localization for entire batches of oocytes. For high-resolution imaging of individual oocytes, a 20× or 63× objective should be used. (See also, Note 8.)
Examples of RNAs visualized using this protocol are shown in Figure 1. An example of RNA and protein co-localization is shown in Figure 2.
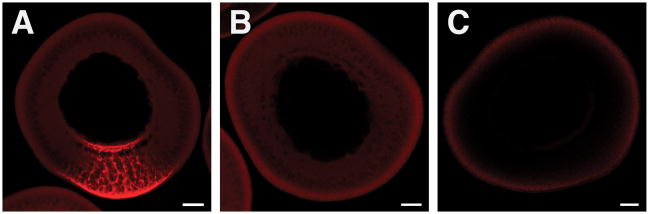
Figure 1.
Vegetal RNA Localization in Xenopus Oocytes. Stage III oocytes were injected with Alexa-546-labeled VLE RNA (A) or Alexa-546-labeled vector control RNA (B) and cultured to allow localization. Control oocytes were uninjected (C). The VLE (Vg1 Localization Element; 11) directs RNA localization to the vegetal cortex, bottom, while the vector control RNA (of similar length, synthesized from pSP73) is uniformly distributed throughout the oocyte cytoplasm. Scale bar = 50 μm.
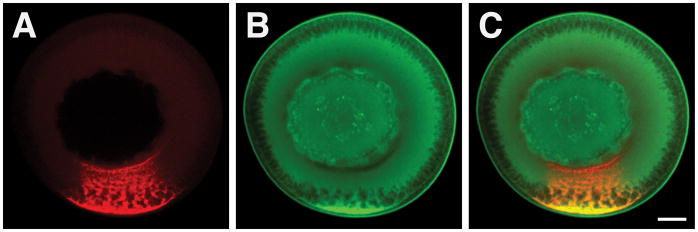
Figure 2.
RNA and Protein Colocalization in Xenopus Oocytes. Stage III oocytes were injected with Alexa-546-labeled VLE RNA and cultured to allow localization of the injected RNA. Immunofluorescence was subsequently performed using antibodies directed against PTB/hnRNP I, an RNA binding protein that associates with VLE RNA and is required for VLE RNA localization (6). Vegetal localization of VLE RNA is shown in the red channel (A), and PTB distribution is visualized in the green channel (B). Co-localization of VLE RNA and PTB protein is evident in the overlay of the red and green channels, and is shown in yellow (C). Scale bar = 50 μm.
Acknowledgments
We would like to thank Mowry lab members past and present, who helped to develop these methods. This work was supported by NIH grant # R01GM071049 to KLM.
Footnotes
Problems with low RNA yield could be due to one or more of the following issues:
- Low template DNA concentration: Quantify template concentration to ensure addition of 1 μg of linear DNA per transcription reaction.
- Non-linear template DNA: Run a sample of template on an agarose gel to confirm complete linearization of plasmid DNA.
- Impurity of template DNA: After linearization of the template DNA, treat with proteinase K, followed by phenol extraction to ensure removal of any contaminating protein. After ethanol precipitation, wash the DNA pellet with 75% ethanol to remove residual salts.
- Loss of RNA during precipitation: Be careful to completely freeze precipitation reaction as described in Methods 3.3.11.b. Do not phenol extract the RNA.
- RNase contamination: Verify that all enzymes are RNase-free; DEPC-treat all solutions as described in Materials 2.1.1.
The steps below can help to prevent problems with needle clogging during microinjection:
- Always centrifuge the RNA and carefully place on ice before loading into the needle. Centrifugation removes large particulate matter that quickly clogs the needle.
- Ensure slight outward pressure in the needle. This will prevent viscous cytoplasm from entering the needle upon piercing the oocyte.
- Keep the tip of the needle immersed in liquid whenever possible. If you have to take the needle out of liquid, quickly expel a drop to cover the tip. RNA can dry rapidly and clog the needle.
Problems with oocyte viability generally stem from extended collagenase treatment during oocyte isolation or non-sterile conditions during oocyte culture. The following precautions are necessary:
- Careful collagenase treatment is essential when isolating oocytes. Ovary should be cut with scissors before placing in collagenase solution. Defoliculation is complete when the majority of ovary chunks have completely released individual oocytes. Incubation should be closely monitored especially when using a new batch of collagenase for the first time; lots can vary greatly in activity. Some lots may take up to 25 minutes to reduce ovary to defolliculated oocytes. However, other lots may complete this task in half the time. Buffer pH is also extremely critical; if defolliculation takes an abnormally long time the buffer pH should be checked.
- OCM quality must be maintained to successfully culture oocytes. In-OCM should be a bright red/pink color. Any other color indicates that the pH has deviated from normal and the In-OCM should be discarded. Store In-OCM no longer than two months at 4 °C. We always make complete OCM fresh immediately before use to culture oocytes. Some antibiotics lose their efficacy after repeated freeze/thaw cycles, resulting in bacterial or fungal contamination. We avoid this issue by aliquoting the anti-microbial stock solutions in small volumes, as described in Materials 2.3.3.
- Successfully cultured oocytes maintain their spherical shape and do not stick to the bottom of the plate well during culture. Microinjection is a stressful procedure for oocytes, approximately 10% of oocytes will not survive even under the best conditions. Greater than 30% oocyte death after culture should be a warning sign that oocytes are not healthy and the experiment may be jeopardized.
The 10x MEM stock is good for several months as long as it remains colorless; store at room temperature wrapped in aluminum foil. 20 minutes is the minimum time for fixation; however, we can also fix for several hours without negatively affecting oocyte quality.
Autofluorescence of yolk proteins in the oocyte places limitations on the choice of fluorescent nucleotides and secondary antibodies. If possible, fluorescent nucleotides and secondary antibodies giving emission at longer wavelengths (e.g., 546 nm, 633 nm) should be used, as autofluorescence is significant at shorter wavelengths (e.g., 488 nm).
Problems with signal detection can occur, and may arise from a number of sources:
- No RNA signal/no localization: Adequate controls are critical to dissect issues with RNA signal. Negative controls include both uninjected oocytes (see Fig. 1C), and injection of a non-localizing control RNA (such as β-globin, see Fib. 1B), which should be uniformly distributed in the oocyte cytoplasm. Additionally, a positive control RNA that is known to exhibit localization (see Fig. 1C) is also critical. If both the non-localizing RNA and the positive control show no fluorescence increase over background, there may be problems in synthesis of fluorescently labeled RNA. If, however, fluorescence of the positive control RNA is evident throughout the cytoplasm, the oocyte quality may be insufficient to support localization, and the experiment must be repeated.
- No immunofluorescence (IF) signal: Antibody selection is crucial. The optimal antibody to use is one previously shown to work for IF in Xenopus oocytes or tissues. However, many antibodies have been shown to work for IF in other systems, but have not been tested in Xenopus. We have had success using such antibodies; however, when using an antibody that has not been tested for IF in any system, a series of careful controls must be used to ensure useful data. One essential negative control is the secondary only control, which is treated in an identically to the experiment oocytes but without incubation with primary antibody. A useful positive control when testing unknown antibodies is a proven antibody that works for IF in Xenopus oocytes. Finally, the blocking solution should use serum from the animal that the secondary was made; we routinely use goat serum and goat secondary antibodies.
- Uncleared oocytes: Problems with oocyte clearing can result in residual opacity and inability to visualize signal deep into the oocyte. Often the nucleus cannot be visualized at all. This can be avoided by carefully dehydrating the oocytes into anhydrous methanol during fixation. Fresh anhydrous methanol and multiple washes are required for dehydration, as even trace amounts of water will prevent clearing. Also, when transferring oocytes from methanol to Murray’s Clearing Medium, great care should be taken to minimize the amount of methanol transferred with the single drop containing the oocytes. This ensures that the Murray’s Clear penetrates the oocytes without being diluted with methanol.
Additional imaging tips: We generally image with a fairly open pinhole (>1 Airy Unit), as fluorescence intensity can be quite weak. However, results may vary from batch to batch of ooytes. Additionally, some autofluorescent subcellular structures may be visualized in the 488 nm (green channel), which can be useful for orienting oocytes along the animal/vegetal axis.
References
- 1.Shav-Tal Y, Singer RH. RNA Localization. J Cell Sci. 2005;118:4077–4081. doi: 10.1242/jcs.02543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.St Johnston D. Moving messages: the intracellular localization of mRNAs. Nat Rev Mol Cell Biol. 2005;6:363–75. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- 3.King ML, Messitt TJ, Mowry KL. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell. 2005;97:19–33. doi: 10.1042/BC20040067. [DOI] [PubMed] [Google Scholar]
- 4.Forristall C, Pondel M, Chen L, King ML. Patterns of localization and cytoskeletal association of two vegetally localized RNAs, Vg1 and Xcat-2. Development. 1995;121:201–208. doi: 10.1242/dev.121.1.201. [DOI] [PubMed] [Google Scholar]
- 5.Kloc M, Etkin LD. Two distinct pathways for the localization of RNAs at the vegetal cortex in Xenopus oocytes. Development. 1995;121:287–97. doi: 10.1242/dev.121.2.287. [DOI] [PubMed] [Google Scholar]
- 6.Cote CA, Gautreau D, Denegre JM, Kress TL, Terry NA, Mowry KL. A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol Cell. 1999;4:431–7. doi: 10.1016/s1097-2765(00)80345-7. [DOI] [PubMed] [Google Scholar]
- 7.Bratu DP, Cha BJ, Mhlanga MM, Kramer FR, Tyagi S. Visualizing the distribution and transport of mRNAs in living cells. Proc Natl Acad Sci USA. 2003;100:13308–13313. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertrand EC, Pascal S, Matthias S, Shailesh M, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 9.Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol. 2003;15:1159–68. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- 10.Yisraeli JK, Sokol S, Melton DA. A two-step model for the localization of maternal mRNA in Xenopus oocytes: involvement of microtubules and microfilaments in the translocation and anchoring of Vg1 mRNA. Development. 1990;108:289–98. doi: 10.1242/dev.108.2.289. [DOI] [PubMed] [Google Scholar]
- 11.Mowry KL, Melton DA. Vegetal messenger RNA localization directed by a 340-nt RNA sequence element in Xenopus oocytes. Science. 1992;21:991–4. doi: 10.1126/science.1546297. [DOI] [PubMed] [Google Scholar]
- 12.Deshler JO, Highett MI, Schnapp BJ. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 1997;276:1128–31. doi: 10.1126/science.276.5315.1128. [DOI] [PubMed] [Google Scholar]
- 13.Havin L, Git A, Elisha Z, Oberman F, Yaniv K, Schwartz SP, Standart N, Yisraeli JK. RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev. 1998;12:1593–8. doi: 10.1101/gad.12.11.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Houston DW, King ML, Payne C, Wylie C, Heasman J. The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell. 1998;94:515–24. doi: 10.1016/s0092-8674(00)81592-5. [DOI] [PubMed] [Google Scholar]
- 15.Houston DW, King ML. A critical role for Xdazl, a germ plasm-localized RNA, in the differentiation of primordial germ cells in Xenopus. Development. 2000;127:447–56. doi: 10.1242/dev.127.3.447. [DOI] [PubMed] [Google Scholar]
- 16.Yoon YJ, Mowry KL. Xenopus Staufen is a component of a ribonucleoprotein complex containing Vg1 RNA and kinesin. Development. 2004;131:3035–45. doi: 10.1242/dev.01170. [DOI] [PubMed] [Google Scholar]
- 17.Czaplinski K, Köcher T, Schelder M, Segref A, Wilm M, Mattaj IW. Identification of 40LoVe, a Xenopus hnRNP D family protein involved in localizing a TGF-beta-related mRNA during oogenesis. Dev Cell. 2005;8:505–15. doi: 10.1016/j.devcel.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 18.Messitt TJ, Gagnon JA, Kreiling JA, Pratt CA, Yoon YJ, Mowry KL. Multiple kinesin motors coordinate cytoplasmic RNA transport on a subpopulation of microtubules in Xenopus oocytes. Dev Cell. 2008;15:426–36. doi: 10.1016/j.devcel.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallace RA, Misulovin Z, Wiley HS. Growth of anuran oocytes in serum-supplemented medium. Reprod Nutr Dev. 1980;20:699–708. doi: 10.1051/rnd:19800412. [DOI] [PubMed] [Google Scholar]
- 20.Yisraeli JK, Melton DA. The material mRNA Vg1 is correctly localized following injection into Xenopus oocytes. Nature. 1988;336:592–5. doi: 10.1038/336592a0. [DOI] [PubMed] [Google Scholar]
- 21.Dumont JN. Oogenesis in Xenopus laevis (Daudin). I. Stages of oocyte development in laboratory maintained animals. J Morphol. 1972;136:153–179. doi: 10.1002/jmor.1051360203. [DOI] [PubMed] [Google Scholar]