Abstract
Current data suggest that DNA–peptide crosslinks are formed in cellular DNA as likely intermediates in the repair of DNA–protein crosslinks. In addition, a number of naturally occurring peptides are known to efficiently conjugate with DNA, particularly through the formation of Schiff-base complexes at aldehydic DNA adducts and abasic DNA sites. Since the potential role of DNA–peptide crosslinks in promoting mutagenesis is not well elucidated, here we report on the mutagenic properties of Schiff-base-mediated DNA–peptide crosslinks in mammalian cells. Site-specific DNA–peptide crosslinks were generated by covalently trapping a lysine-tryptophan-lysine-lysine peptide to the N6 position of deoxyadenosine (dA) or the N2 position of deoxyguanosine (dG) via the aldehydic forms of acrolein-derived DNA adducts (γ-hydroxypropano-dA or γ-hydroxypropano-dG, respectively). In order to evaluate potential of DNA–peptide crosslinks to promote mutagenesis, we inserted the modified oligodeoxynucleotides into a single-stranded pMS2 shuttle vector, replicated these vectors in simian kidney (COS-7) cells, and tested the progeny DNAs for mutations. Mutagenic analyses revealed that at the site of modification, the γ-hydroxypropano-dA-mediated crosslink induced mutations at only ~ 0.4%. In contrast, replication bypass of the γ-hydroxypropano-dG-mediated crosslink resulted in mutations at the site of modification at an overall frequency of ~ 8.4%. Among the types of mutations observed, single base substitutions were most common, with a prevalence of G to T transversions. Interestingly, while covalent attachment of lysine-tryptophan-lysine-lysine at γ-hydroxypropano-dG caused an increase in mutation frequencies relative to γ-hydroxypropano-dG, similar modification of γ-hydroxypropano-dA resulted in decreased levels of mutations. Thus, certain DNA–peptide crosslinks can be mutagenic, and their potential to cause mutations depends on the site of peptide attachment. We propose that in order to avoid error-prone replication, proteolytic degradation of proteins covalently attached to DNA and subsequent steps of DNA repair should be tightly coordinated.
1. Introduction
The chemical properties of DNA render it susceptible to a variety of base and backbone modifications that occur either spontaneously or via exposure to environmental toxicants. Failure to repair such DNA lesions in a timely and error-free manner can lead to a variety of deleterious consequences, including but not limited to damage fixation, mutagenesis, alteration of gene expression and signaling pathways, cellular transformation, or apoptosis. Although a subset of the myriad of potential DNA lesions has been extensively investigated (e.g., cyclobutane pyrimidine dimers), the cellular response to an entire class of adducts, DNA–protein and DNA–peptide crosslinks, has received only modest attention. This is in spite of the current cumulative knowledge suggesting the existence of various DNA–peptide crosslinks in cellular DNA. Stable complexes between DNA and glutathione have been detected in cultured cells following chromate exposure [1,2] and a number of synthetic and naturally occurring peptides have been shown to efficiently conjugate with DNA in vitro [3–10]. In addition, DNA–peptide crosslinks are likely to be formed as intermediate structures during biological processing of DNA–protein crosslinks. It has been hypothesized that proteolytic digestion of proteins that have become covalently attached to DNA takes place as an early step in repair of these lesions [11]. In support of this proposed mechanism, proteasome-mediated degradation of the crosslinked proteins has been shown for topoisomerase I and topoisomerase II covalent complexes [12,13] and the active removal of formaldehyde-induced DNA–protein crosslinks in human cells was inhibited upon treatment with lactacystin, a specific proteasome inhibitor [11].
Although DNA–peptide crosslinks are expected to be deleterious DNA lesions, very little is known about their possible genotoxic effects and cellular processing. In particular, the mutagenic properties of these lesions are described poorly, with the only report on a related subject clearly indicating that certain DNA–peptide crosslinks can cause mutations [2]. Specifically, in human fibroblasts, replication of plasmid DNA adducted with glutathione–chromium–DNA crosslinks, resulted in single base substitutions, predominantly at G:C base pairs [2].
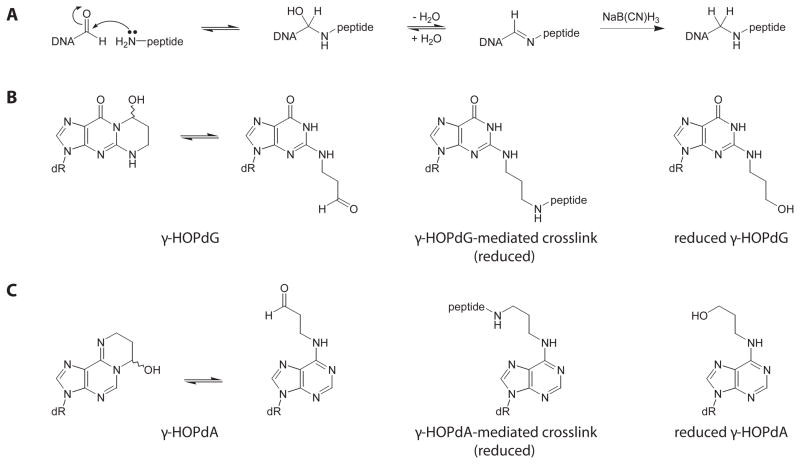
Investigations on the biological properties of DNA–peptide crosslinks are significantly complicated by at least two factors. First, these lesions are extremely diverse, such that they differ in size, amino acid composition, nature of chemical linkage, and sites of the peptide attachment within DNA. Second, it remains a technical challenge to generate site-specific DNA–peptide crosslinks with defined structures. To address this challenge, we have developed methodologies that facilitate the crosslinking of a variety of peptides to predetermined sites within DNA [5,6,8,9]. This methodology is based on a chemistry of interactions between aldehydic functions of modified oligodeoxynucleotides and nucleophilic groups of amino acids (Figure 1A). Earlier studies have demonstrated that peptides, which were composed of a single aromatic residue flanked by basic ones, such as lysine-tryptophan-lysine, bind to DNA both electrostatically and by a mechanism that involves a stacking of DNA bases with the aromatic residue [14–16]. Incubation of these tripeptides with DNAs containing abasic sites resulted in the formation of single-strand breaks [15–17], in which a transient Schiff base intermediate was formed between the peptide α-amino group and C1 of the ring-opened deoxyribose sugar, leading to β-elimination at the abasic site [5]. When such reactions were carried out in the presence of a reducing agent, such as sodium cyanoborohydride, the Schiff base intermediate could be trapped to give an irreversible crosslink [5]. Using this approach, various peptides have been site-specifically conjugated to DNA via an abasic site to modify the DNA backbone [5,8]. Similar methodology has been utilized later to attach peptides to the N2 position of deoxyguanosine (dG) via the ring-opened, aldehydic form of the γ-hydroxypropano deoxyguanosine (γ-HOPdG) adduct [6,8,9]. Among a variety of peptides tested, lysine-tryptophan-lysine-lysine (KWKK) appeared to be the most efficient in trapping reactions [5]; thus, this tetrapeptide has been chosen to generate crosslinks for the present investigation.
Figure 1.
(A) Chemistry of formation of aldehyde-mediated DNA–peptide crosslinks. The product of reaction between an aldehydic function and a nucleophilic group is a Schiff base and is formed reversibly; however, the Schiff base intermediate can be trapped with a reducing agent such as sodium cyanoborohydride to give an irreversible crosslink. (B) Deoxyguanosine adducts. (C) Deoxyadenosine adducts. The γ-HOPdG and γ-HOPdA adducts are shown as equilibria between the ring-closed and ring-opened, aldehydic forms. Reduced γ-HOPdG-mediated peptide crosslink, reduced γ-HOPdG, reduced γ-HOPdA-mediated peptide crosslink, and reduced γ-HOPdA exist exclusively in the opened structure.
The γ-HOPdG adduct (Figure 1B) is a major deoxyguanosine adduct induced following exposure to acrolein [18], a known mutagen [19–21] and a possible carcinogen [22]. As a free nucleoside and in single-stranded DNA, γ-HOPdG exists primarily in its ring-closed form [23,24]. In double-stranded DNA, the equilibrium shifts to the opened structure, as evidenced from nuclear magnetic resonance studies [9,25,26]. When γ-HOPdG is placed opposite deoxycytidine, the modified base adopts a conventional anti orientation around the glycosidic bond and participates in a standard Watson-Crick pairing. In contrast, propano deoxyguanosine (PdG), which is structurally similar to the ring-closed form of γ-HOPdG, is not capable of Watson-Crick base-pairing. Nuclear magnetic resonance analyses of PdG-containing oligodeoxynucleotides showed, that modified base adopts a syn orientation around the glycosidic bond and hybridizes with the complementary deoxycytidine via the Hoogsteen edge [27]. Mutagenic properties of γ-HOPdG have been investigated in bacterial and mammalian cell lines [28–32]. In wild-type mammalian cells, the adduct caused single base substitutions at an overall frequency of 7.4–11.0% when a single-stranded, site-specifically modified vector was replicated [31,32]; however, it was significantly less miscoding (≤ 1% base substitution) when a double-stranded vector was utilized [29,30]. Interestingly, the permanently ring-closed PdG adduct was almost equally mutagenic, regardless of whether a single- or double-stranded vector was used [29,30,33]. Collectively, these structural and mutational data suggest that the ring-opened and ring-closed forms of γ-HOPdG have different effects on fidelity of translesion synthesis, with the ring-opened form being less mutagenic.
Among aldehydes that induce DNA–protein crosslinks, acrolein is one of the most potent [34]. Currently, molecular mechanisms responsible for the generation of acrolein-induced crosslinks are not well understood. One feasible pathway may involve formation of Schiff base complexes between aldehydic DNA adducts, such as the ring-opened γ-HOPdG, and lysine ε-amino groups of DNA-associated proteins. Support for such a model was obtained by observation of efficient trapping of histones with γ-HOPdG-containing oligodeoxynucleotides under reducing conditions [35]. In addition, the bifunctional glycosylase/abasic site lyase human NEIL1 forms a transient Schiff base complex with DNA substrates, utilizing an N-terminal secondary amine as the reactive nucleophile [36,37]. Since the α-amino group of the N-terminus is the preferred reactive nucleophile in KWKK-catalyzed reactions [5,6], our model DNA–peptide crosslinks most closely resemble structures that might be formed between aldehydic moieties in DNA and a subset of glycosylase/abasic site lyases.
Here, mutagenic properties of DNA–peptide crosslinks in mammalian cells (COS-7) have been studied using a single-stranded pMS2 shuttle vector [38]. Specifically, the hypothesis has been tested that mutational outcome of replication past a crosslinked peptide depends on the site of its attachment within DNA structure. The KWKK peptide was covalently conjugated to either the N2 position of dG (DNA minor groove site) via the ring-opened γ-HOPdG (Figure 1B) or the N6 position of deoxyadenosine (dA) (DNA major groove site) via the ring-opened form of the γ-hydroxypropano deoxyadenosine (γ-HOPdA) adduct (Figure 1C). For comparative purposes, mutagenic properties of γ-HOPdG and γ-HOPdA, as well as the ring-opened, reduced derivatives of these adducts (Figure 1B, C) have been also investigated. The latter adducts were chosen as reasonable models of the open chain forms of γ-HOPdG and γ-HOPdA.
2. Materials and Methods
2.1. Materials
COS-7 cells were purchased from the American Type Culture Collection (Manassas, VA). XL1-Blue E. coli strain was obtained from Stratagene (La Jolla, CA). The pMS2 phagemid shuttle vector was a gift from Dr. M. Moriya (State University of NY, Stony Brook, NY). M13KO7 helper phage was purchased from Invitrogen (Carlsbad, CA). T4 polynucleotide kinase, T4 DNA ligase, T4 DNA polymerase, uracil DNA glycosylase, and EcoRV restriction endonuclease were obtained from New England BioLabs (Beverly, MA). S1 nuclease and proteinase K were obtained from Fermentas (Hanover, MD). [γ-32P]-ATP (6000 Ci/mmol) was purchased from Amersham Biosciences (Piscataway, NJ). The peptide KWKK was synthesized by Sigma-Genosys (Haverhill, UK). Sodium borohydride and sodium cyanoborohydride were obtained from Sigma (St. Louis, MO). Media, supplements, and reagents for the COS-7 culturing and transfection were from GIBCO, Invitrogen (Carlsbad, CA). P-6 Bio-Spin columns were purchased from Bio-Rad (Hercules, CA). Amicon Ultra centrifugal filter devices were obtained from Millipore (Billerica, MA).
2.2. Oligodeoxynucleotides Synthesis
Unmodified oligodeoxynucleotides were synthesized by the Molecular Microbiology and Immunology Research Core Facility, Oregon Health and Science University (Portland, OR). The oligodeoxynucleotides containing γ-HOPdG or γ-HOPdA adducts were prepared by the post-oligomerization methodology as described [23]. MALDI-TOF mass spectra of modified oligodeoxynucleotides were obtained at the Vanderbilt University Mass Spectrometry Resource Center on a Voyager Elite DE Instrument (PerSeptive Biosystems) in the negative ion mode using a matrix mixture of 3-hydroxypicolinic acid and ammonium hydrogen citrate. To generate the γ-HOPdG- and γ-HOPdA-derived crosslinks, a previously developed borohydride trapping methodology [6,8,9] has been employed. Briefly, 2 nmol of the oligodeoxynucleotides adducted with either γ-HOPdG or γ-HOPdA were incubated with 100 nmol of the KWKK peptide and NaCNBH3 (50 mM) in 50 mM Hepes-Na (pH 7.0) at 37 °C for 18 h and purified by PAGE. To obtain reduced forms of the γ-HOPdG and γ-HOPdA adducts, modified oligodeoxynucleotides were incubated in the presence of 25 mM NaBH4 at room temperature for 24 h. Fresh solutions of NaBH4 were added three times during incubation. Oligodeoxynucleotides were purified using P-6 Bio-Spin columns. Completeness of reduction was probed by incubation of the oligodeoxynucleotides with 100 μM KWKK in the presence of 50 mM NaCNBH3 and 50 mM Hepes-Na (pH 7.0) at 37 °C for 2 h.
2.3. Mutagenesis Assays Using Site-Specifically Modified pMS2 Vector
Mutagenesis assays were conducted using a previously developed pMS2 shuttle vector/COS-7 system [38]. Single-stranded vector DNA was prepared using XL1-Blue E. coli strain and M13KO7 helper phage according to published procedure [31,38]. Incorporation of site-specifically modified oligodeoxynucleotides into pMS2 DNA using a scaffold DNA fragment was essentially performed as described [31,38]. In its single-stranded form, the pMS2 vector forms a stem-loop hairpin structure containing an internal EcoRV restriction site. Scaffold DNAs (58-mers) were designed to have the following features: (1) the peripheral regions were complementary to the vector sequences immediately adjacent to the EcoRV site, (2) the central part was complementary to the insert sequences, and (3) all thymines were substituted by uracils. Scaffold DNA fragment was added to the vector DNA (10 pmol per preparation) at a 2-fold molar excess, and reactions were heated to 90 °C for 2 min and slowly cooled to the room temperature. In order to create a gap in the pMS2 sequence, incubation with EcoRV restriction endonuclease (5 units per preparation) was performed for 3 h at 37 °C. Single-stranded oligodeoxynucleotide inserts (50 pmol) containing either adducts or unmodified dG or dA were phosphorylated at the 5′ end by T4 polynucleotide kinase (1 unit) for 1 h at 37 °C, and added to the vector DNA. Following annealing of inserts to the scaffold, they were ligated into the vector by T4 DNA ligase (1 unit) overnight at 12 °C. Scaffold DNA was damaged by the uracil DNA glycosylase cleavage, and non-circular molecules were eliminated by the T4 DNA polymerase treatment (1 unit of each enzyme, 37 °C, 1 h incubation). To estimate yield of the resulting circular DNAs, probes were collected throughout the procedure. DNAs were separated by gel electrophoresis, stained with ethidium bromide, and quantified using Alfa-Innotech software. Emission generated by the T4 DNA polymerase-resistant ligation products was compared with that generated by a known amount of the starting single-stranded pMS2 DNA.
Transfection of pMS2 vector into COS-7 cells, isolation of progeny DNA, selection of individual clones by E. coli transformation, and differential hybridization analyses were done as described [31,38]. Briefly, cells were seeded at 5 × 105 cells per 60 mm plate and grown overnight in DMEM medium supplemented with 10% fetal bovine serum. Vector DNA (~ 100 fmol) was pre-incubated for 15 min with 30 μg of the Lipofectin reagent in 3 mL of Opti-MEM and added to the plate containing the attached COS-7 cells. Following an 18 h incubation, the culture was further grown for 48 h in a regular DMEM medium. The progeny DNAs were recovered by method of Hirt [39] and sequentially treated at 37 °C with S1 nuclease (0.1 unit) for 30 min and EcoRV (5 unit) for 1 h. To obtain individual clones, DNAs were used to transform E. coli DH5α by electroporation. Hybridization with the progeny plasmid DNA was performed utilizing the 5′-radioactivelly labeled oligodeoxynucleotide probes. Radioactive signals were visualized with a Storm 820 PhosphorImager (GE Healthcare, Piscataway, NJ). For each particular adduct, the data were collected from at least two independent experiments.
3. Results
3.1. Design and Preparation of Site-Specific DNA Peptide Crosslinks
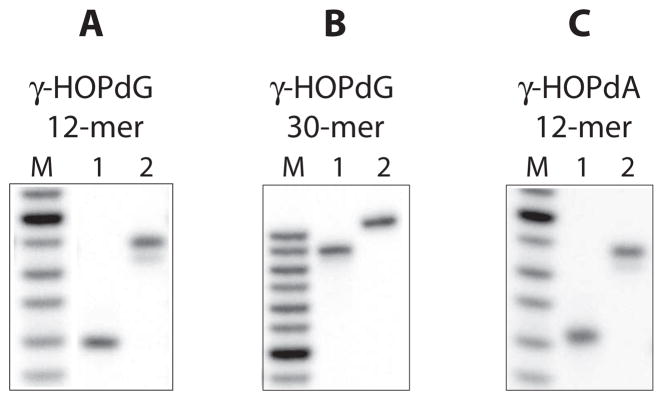
In order to covalently conjugate KWKK to the N2 position of dG (DNA minor groove site), this peptide was reacted with γ-HOPdG-containing oligodeoxynucleotides (Figure 1B) as described in the Materials and Methods according to a previously developed protocol [8]. In initial experiments, the adducted oligodeoxynucleotides were 12-mers with a sequence 5′-GCT AGC GAG TCC-3′, where underlined G is modified. Under the conditions of these experiments, approximately 80% of the DNA molecules were modified with the crosslink. However, due to inefficient recovery of the DNA–peptide crosslinked material following ethanol precipitation, only about 10% was obtained after the PAGE purification procedure. In order to improve the overall yield of the crosslinked species, 30-mer oligodeoxynucleotides were synthesized and used in subsequent experiments. Since the mutagenic properties of DNA adducts are known to be affected by local sequence context [40], the 30-mer oligodeoxynucleotides (5′-CGT AGT ACT GCT AGC GAA TTC CGT ATC CAT-3′) were designed in such a way that the deoxynucleotides immediately adjacent to the modified dG from both 5′ and 3′ sides were identical to those present in these positions in 12-mer DNAs. Using 30-mer oligodeoxynucleotides, no reduction in the efficiency of the trapping reaction was measured relative to reactions carried out using 12-mers, but recovery of the crosslinked species after the purification procedure was significantly improved (approximately 90%). Isolated γ-HOPdG-mediated DNA–peptide crosslinks were tested for purity by PAGE (Figure 2A and B). To attach KWKK to the N6 position of dA (DNA major groove site), 12-mer γ-HOPdA-containing oligodeoxynucleotides were synthesized and utilized to generate DNA–peptide crosslinks (Figure 1C). The sequence of these oligodeoxynucleotides (5′-GCT AGC AAG TCC-3′), with an exception for the modification site, was identical to that of γ-HOPdG-containing 12-mers. Previous studies have indicated that the ring-opened derivative of γ-HOPdA can be easily detected by nuclear magnetic resonance spectroscopy, even when the adduct is in a free nucleoside form [23]. It has been anticipated from these observations, that γ-HOPdA would effectively interact with various nucleophiles and, in particular, form complexes with peptides. Experiments were conducted to test for reactivity of this adduct towards KWKK using site-specifically modified oligodeoxynucleotides, and as expected, efficient formation of stable DNA–peptide conjugates in the presence of reducing agent was observed. Consistent with this prior analysis, preparation of the γ-HOPdA-derived crosslinks was easily accomplished following the procedure that had been developed for the γ-HOPdG-derived crosslinks. Isolated γ-HOPdA-mediated DNA–peptide crosslinks were tested for purity by PAGE (Figure 2C).
Figure 2.
Isolated DNA–peptide crosslinks. DNA– peptide crosslinks were obtained using either γ-HOPdG-adducted 12-mer (A), or γ-HOPdG-adducted 30-mer (B), or γ-HOPdA-adducted 12-mer (C). Following to purification, DNA–peptide crosslinks were tested for purity by PAGE under denaturing conditions (in the presence of 8 M urea). DNAs shown are: (M) oligodeoxynucleotide sizing markers, (1) oligodeoxynucleotides adducted with either γ-HOPdG or γ-HOPdA, and (2) corresponding oligodeoxynucleotides modified with the reduced KWKK crosslink.
3.2. Analysis of Mutations
To evaluate potential of DNA–peptide crosslinks and related adducts to promote mutagenesis, we inserted the modified or non-modified, control oligodeoxynucleotides into a single-stranded pMS2 vector, and replicated these vectors in COS-7 cells. Following the isolation of the progeny DNAs, individual clones were obtained by transforming the E. coli DH5α cells. Although the absolute yield of bacterial transformants varied from experiment to experiment by as much as 2-fold, we did not observe any correlation between the yield of bacterial transformants and the type of DNA replicated.
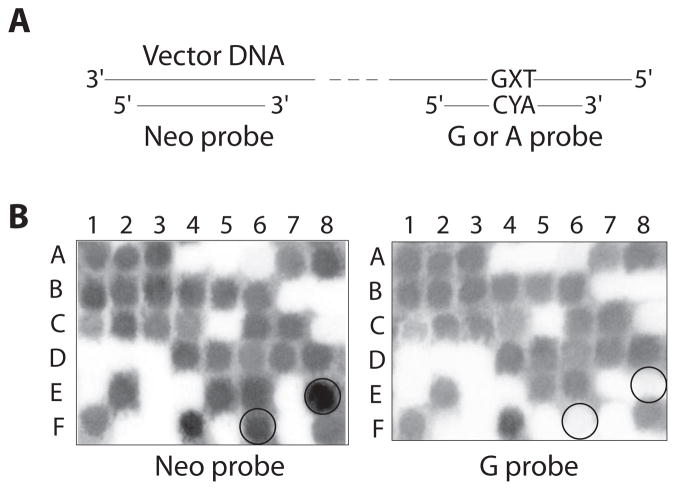
The progeny DNAs were tested for mutations by differential hybridization utilizing the 5′-radioactively labeled 19-mer oligodeoxynucleotide probes G (for γ-HOPdG-derived adducts) or A (for γ-HOPdA-derived adducts) (Figure 3A). These probes were designed to be complementary to the DNA sequences expected from non-mutagenic replication. Since replication of a single-stranded pMS2 vector is known to result in large deletions [31], an additional probe (Neo) was utilized. This 20-mer was complementary to the vector sequence located 52 nucleotides away from the site of insertion and allowed us to exclude from the analyses the progeny DNAs with deletions involving the region of interest. An example of hybridization of probes G and Neo with DNAs obtained from replication of the γ-HOPdG-mediated crosslink is shown on Figure 3B. Individual DNAs (such as E8 and F6 on the given example) that hybridized with Neo probe, but did not hybridize with the correct probe, were isolated and subjected to DNA sequencing using Neo as a primer. Majority of such DNAs contained the insert sequences bearing mutations. In addition, clones were identified that did not contain the insert sequences. Since the latter were equally represented in all DNA samples, including the progeny DNAs from non-damaged vector, and therefore, the origin of these deletions is unlikely due to the presence of the adduct, these clones were excluded from further analyses. In order to verify the accuracy of hybridization procedure, approximately 10% of clones that hybridized with the correct probe were analyzed by DNA sequencing. Among these DNAs, no single clone had any sequence alteration. Relative to previously utilized procedures [31] that rely on differential hybridization using pre-determined probes to detect mutations, our approach is more informative and has no bias; it allows for a full description of the spectrum of sequence alterations both at the adducted site and any other position in its vicinity.
Figure 3.
Screening for mutations. (A) Probes used for hybridization. X indicates position of deoxynucleotide that, during replication in COS-7 cells, was inserted opposite the adducted site. Y is a G or an A. Probe G was used to analyze progeny DNAs that were obtained from replication of vectors containing dG adducts. Probe A was used to analyze progeny DNAs that were obtained from replication of vectors containing dA adducts. (B) An example of hybridization: probes Neo and G were used to test individual DNA clones that were isolated following replication of vectors containing γ-HOPdG-mediated crosslink.
3.3. Mutagenic Potential of the KWKK Crosslink Mediated by the γ-HOPdG adduct
Mutagenic properties of the reduced KWKK crosslink mediated by the γ-HOPdG adduct were investigated in COS-7 cells as described above. In parallel, assays were conducted using DNAs containing non-damaged dG, γ-HOPdG, or reduced γ-HOPdG. To construct site-specifically modified vectors, either 12-mer or 30-mer inserts were utilized. Since no differences in mutational outcome of replication were observed that were dependent on the insert size, results were combined and these data are presented collectively in Table 1.
Table 1.
Comparative Mutagenesis of γ-HOPdG, Reduced γ-HOPdG, and Reduced γ-HOPdG-Mediated KWKK Crosslink in COS-7 Cells*.
| DNA modification | Correct sequence | Mutations at the modified site, number | Mutations at positions adjacent to the modified site, number | Mutations at the modified site, % | Overall sequence alterations**, % | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| G to A | G to C | G to T | Deletion | 5′C to A | 5′C to G | 5′C to T | 3′A deletion | ||||
| non- damaged dG | 229 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 | 0.0 |
| γ-HOPdG | 299 | 1 | 2 | 8 | 0 | 1*** | 0 | 0 | 2 | 3.5 | 4.2 |
| reduced γ-HOPdG | 245 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 2 | 0.8 | 2.8 |
| reduced γ-HOPdG-mediated KWKK crosslink | 311 | 2 | 9 | 17 | 1 | 2**** | 1*** | 2 | 1 | 8.4 | 9.6 |
Site-specifically modified vectors were constructed using either 12-mer or 30-mer inserts.
To calculate percentage, mutations in tandem were counted as one sequence alteration.
tandem with adducted G to T.
1 out of two mutations was in tandem with adducted G to C.
As expected, no alterations in inserted sequences were detected in the progeny DNAs when non-damaged vectors were replicated. In contrast, a variety of mutations were observed using γ-HOPdG-modified DNAs. Single base substitutions at the adducted site were most frequent (3.5%); predominantly, these were G to T transversions. In addition, other mutations were detected at low frequencies: two deletions of an A adjacent to γ-HOPdG from the 3′ side and one tandem mutation involving the adducted G and the 5′C.
As previously discussed, prior data suggested that translesion synthesis past γ-HOPdG is more accurate when the adduct enters the replication machinery in the ring-opened form [29–31]. Since our model DNA–peptide crosslink exists exclusively in the opened structure, we reasoned that reduced γ-HOPdG, which we regard as a ring-opened analogue of γ-HOPdG, would be another essential control for the comparative mutagenesis assays. When vector DNAs containing the reduced γ-HOPdG were replicated, only 0.8% of the total insertions opposite the lesion site were mutagenic (Table 1). The number of deletions of the 3′A was similar to that measured for γ-HOPdG, and transversions of the 5′C to G were observed at a low frequency.
Relative to the γ-HOPdG adduct and its ring-opened, reduced derivative, the γ-HOPdG-mediated KWKK crosslink caused mutations more frequently, with 8.4% of the translesion bypass events resulting in sequence alterations opposite the lesion site (Table 1). G to T transversions were most common, but some G to C transversions, G to A transitions, and one nucleotide deletion were also detected. In addition, single base substitutions at the 5′C, deletion of the 3′A, and tandem mutations involving the adducted G and the 5′C were observed at low frequencies.
Thus, these data revealed that during extrachromosomal DNA synthesis in COS-7 cells, the reduced γ-HOPdG-mediated KWKK crosslink was moderately mutagenic. Replication bypass of the crosslink appeared to be less accurate than past the parent unreduced γ-HOPdG adduct. The spectra of mutations caused by these two DNA lesions however were quite similar. Significantly, an ~ 10-fold higher frequency of misincorporations opposite the modified site was observed for the crosslink relative to the ring-opened, reduced γ-HOPdG adduct.
3.4. Mutagenic Potential of the KWKK Crosslink Mediated by the γ-HOPdA adduct
Mutagenic properties of the KWKK crosslink mediated by γ-HOPdA were also studied in COS-7 cells and they were compared with mutagenic properties of the parent γ-HOPdA adduct and the ring-opened, reduced γ-HOPdA.
γ-HOPdA is a minor dA adduct that is formed in DNA following acrolein exposure [41]. The data showed that replication bypass of this adduct in COS-7 cells was moderately mutagenic (Table 2), with 5.0% of the translesion synthesis events resulting in single base substitutions, predominantly A to T transversions. However, the ring-opened, reduced derivative of γ-HOPdA caused mutations at a much lower frequency (≤ 1.0%). When vectors containing the γ-HOPdA-mediated KWKK crosslink were replicated, surprisingly accurate translesion synthesis was observed. Specifically, out of 260 individual DNAs tested, only one point mutation at the site of modification (A to T transversion) and two substitutions at the 3′A position were detected.
Table 2.
Comparative Mutagenesis of γ-HOPdA, Reduced γ-HOPdA, and Reduced γ-HOPdA-Mediated KWKK Crosslink in COS-7 Cells*.
| DNA modification | Correct sequence | Mutations at the modified site, number | Mutations at positions adjacent to the modified site, number | Mutations at the modified site, % | Overall sequence alterations**, % | ||||
|---|---|---|---|---|---|---|---|---|---|
| A to C | A to G | A to T | Deletion | 5′C to G | 3′A to T | ||||
| non-damaged dA | 236 | 0 | 0 | 0 | 1 | 0 | 0 | 0.4 | 0.4 |
| γ-HOPdA | 265 | 1 | 1 | 12 | 0 | 0 | 0 | 5.0 | 5.0 |
| reduced γ-HOPdA | 288 | 0 | 1 | 1 | 0 | 1*** | 1 | 0.7 | 1.0 |
| reduced γ-HOPdA-mediated KWKK crosslink | 257 | 0 | 0 | 1 | 0 | 0 | 2 | 0.4 | 1.2 |
Site-specifically modified vectors were constructed using 12-mer inserts.
To calculate percentage, mutations in tandem were counted as one sequence alteration.
tandem with adducted A to T.
Thus, translesion synthesis past the γ-HOPdA-mediated KWKK crosslink in COS-7 cells was more accurate than past the parent unreduced γ-HOPdA adduct. Interestingly, it was also significantly less mutagenic than translesion synthesis past the KWKK crosslink mediated by the γ-HOPdG adduct.
4. Discussion
The aim of the current investigation was to determine the mutagenic properties of DNA–peptide crosslinks. Using site-specifically modified vectors, we not only confirmed the previous findings [2] that certain DNA–peptide crosslinks can cause mutations, but also showed that mutagenic potential of these lesions can be affected by the site of peptide attachment within DNA structure. The mutational outcome of replication past the DNA–peptide crosslinks in COS-7 cells varied significantly depending on whether the KWKK peptide was conjugated to the N2 position of dG or the N6 position of dA. Specifically, about 10-fold higher mutation frequencies were observed when the peptide was attached at the N2-dG minor groove site versus the N6-dA major groove site.
Our data on mutagenicity of DNA–peptide crosslinks are especially interesting in the context of the current model for repair of DNA–protein crosslinks. It has been hypothesized that proteolytic degradation of proteins that have become covalently attached to DNA takes place to facilitate excision of the damaged DNA fragment, presumably by nucleotide excision repair (NER) proteins [11]. A possible role for NER in processing of DNA–protein crosslinks has been examined using NER-deficient mammalian cells [11,42,43]. To induce formation of DNA–protein crosslinks in cellular DNA, either formaldehyde or transplatin was utilized. No differences between NER-deficient and NER-proficient cells were observed when removal of the formaldehyde-induced crosslinks was measured [11,43]. In contrast, removal of the transplatin-induced crosslinks appeared to be NER-dependent [42]. Collectively, these results suggest that the NER pathway can be involved in the repair of certain but not all types of DNA–protein crosslinks.
The ability of NER proteins to initiate the removal of DNA–protein and DNA–peptide crosslinks has also been tested in reconstituted in vitro systems [8,44–46]. The KWKK and a dodecapeptide attached to the DNA backbone via linkage at an abasic site and stabilized by borohydride reduction appeared to be excellent substrates for bacterial NER nuclease [8]. However, only moderate rates of excision were observed in reactions utilizing a 16-kDa protein crosslink [8,44]. When the very same crosslinks were examined with human proteins, substrate-dependant variations in efficiencies were even more significant; such that excision of DNA–peptide crosslinks was quite robust, while removal of a 16-kDa protein crosslink was not detected [45]. Similarly, no excision of the histone–trans-[PtCl2(E-iminoether)2] –DNA complexes was observed in reactions using rodent NER excinuclease [46]. These biochemical data imply that proteolytic degradation of proteins involved in DNA–protein crosslinks would significantly facilitate removal of these lesions and consequently diminish their genotoxicity. On the other hand, anticipated intermediates in this proposed mechanism, DNA–peptide crosslinks, can cause mutations and, as evident from the current study, at a considerably high frequency. Thus, we hypothesize that in order to avoid error-prone replication, proteolytic degradation of proteins that have become covalently attached to DNA and subsequent steps of DNA repair should be tightly coordinated.
Results of our work also contribute to the understanding of the molecular mechanisms of acrolein-associated mutagenicity. Due to its high reactivity, this bifunctional aldehyde can interact with DNA bases to form a multitude of DNA adducts [18,41,47–49]. Mutagenic properties of two of them, γ-HOPdG and γ-HOPdA, will be discussed below.
Among acrolein-induced adducts, the major dG adduct, γ-HOPdG [18], is perhaps the most extensively studied. Its structural characteristics and biological properties have been addressed in several investigations revealing unique features of this adduct [6,9,23–26,29–32,35,50–54]. One remarkable characteristic is that γ-HOPdG exists in two interconvertable forms, with the equilibrium shifted to the opened structure in double-stranded DNA and to the closed structure in single-stranded DNA and nucleosides [9,23–26]. Prior investigations collectively suggested that the ring-opened form of the adduct is less mutagenic [29–31,33]. Here, we used site-specifically modified single-stranded vector to directly compare mutagenic properties of γ-HOPdG and its irreversibly ring-opened derivative, the reduced γ-HOPdG. As expected, the reduced derivative appeared to be less miscoding than the parent adduct. This observation is in a good agreement with our earlier findings that reduced γ-HOPdG is an instructive lesion for several individual DNA polymerases, whereas accurate bypass of non-reduced γ-HOPdG was achieved only by sequential action of two DNA polymerases, pol τ and pol κ [32,52,53]. Taken together, these data strongly support the hypothesis that during translesion synthesis, mutations are caused primarily by the ring-closed, not the ring-opened form of the γ-HOPdG adduct. Thus, to further evaluate the mutagenic potential of γ-HOPdG, rate of the ring closure should be measured and compared with rates of replication fork progression.
Another interesting feature of γ-HOPdG is its ability to form various secondary adducts, including DNA–DNA, DNA–peptide, and DNA–protein crosslinks [6,9,26,35,50,51,54]. This raises the possibility that intracellular processing of γ-HOPdG and, in particular, DNA synthesis past the lesion site will be affected when such modifications occur. As shown here, the γ-HOPdG-mediated KWKK crosslink caused mutations more frequently than the parent adduct and much more frequently than the ring-opened, reduced derivative. Thus, secondary modifications of the hydroxypropano moiety can modulate the mutagenic properties of the γ-HOPdG adduct.
Although it is premature to speculate on identities of DNA polymerases involved in replication past the γ-HOPdG-mediated crosslink, pol κ is a likely candidate to perform this function in an error-free manner, since previous reports implicate pol κ in the non-mutagenic bypass of N2-dG adducts [55–57]. Regarding the mutagenic bypass of N2-dG lesions, data presented herein reveal that at least two structural characteristics of these adducts play a role: first, the size, and second, the ability to form the ring-closed structure involving the N1 position. Our findings suggest that mammalian replication machinery can tolerate and accurately bypass relatively small minor groove N2-dG adducts; however, chances to generate mutations increase when replication machinery encounters more bulky N2-dG adducts or cyclic 1,N2-dG adducts.
In comparison to the relatively well-studied acrolein-induced dG adducts, the structural and biological properties of acrolein-dA adducts have received less attention, even though the major dA monoadduct was detected under oxidative stress conditions in the nuclei of experimental animals [58]. Here, we showed that in COS-7 cells, replication past the minor adduct, γ-HOPdA, resulted in mutations at an overall frequency of 5.0%. In contrast, the reduced γ-HOPdA adduct, that is incapable of secondary reactions, caused mutations less frequently (≤ 1%). Thus, it is possible that higher mutagenicity of unreduced γ-HOPdA is due to its ability to form secondary adducts with cellular proteins or other nucleophilic compounds. However, we favor an alternative interpretation of this observation. Considering low mutagenic potential of the γ-HOPdA-mediated KWKK crosslink and extrapolating from the γ-HOPdG adduct, we suggest that mutations are generated during replication past the ring-closed form of γ-HOPdA. Even though this adduct undergoes ring-opening more easily relative to γ-HOPdG, ring-closed structures were detected in nuclear magnetic resonance spectra [23]. Importantly, when γ-HOPdA is in its closed form, the Watson-Crick edge is masked by a dual modification in a manner similar to the ring-closed form of γ-HOPdG. Since in the major acrolein-dA adduct, the Watson-Crick edge is also dually modified, this lesion is hypothesized to be mutagenic. Thus, the mutagenic effect of acrolein may be at least partially due to formation of miscoding dA adducts.
Interestingly, the γ-HOPdA-mediated KWKK crosslink was only marginally miscoding in our system; it caused mutations at approximately 10-fold lower frequency relative to the corresponding γ-HOPdG-mediated crosslink. This observation suggests that in the case of bulky lesions, replication bypass of the major groove adducts in mammalian cells is more accurate than bypass of the minor groove adducts. Structural and biochemical analyses showed that many DNA polymerases interact with substrate DNA at the minor groove [59]. For that reason, modifications of the N2-dG are expected to inhibit the replication by a majority of individual polymerases more strongly than identical modifications of the N6-dA. However, mutational outcome of translesion synthesis in vivo is cumulative and dependent on a combination of factors including, but not limited to the extent of blockage to replicative polymerases, the fidelity of replicative polymerases, and the identities of bypass polymerases involved. Thus, further studies are needed to elucidate differential processing of the minor groove and the major groove adducts by mammalian replication machinery.
Acknowledgments
This work was supported by NIH grants CA 106858 (R.S.L.) and ES 05355 (R.S.L and C.J.R.), and Center Grant ES 000267 (C.J.R.).
We are grateful to M. Moriya for providing the pMS2 vector. We thank V. Vartanian for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhitkovich A, Voitkun V, Costa M. Glutathione and free amino acids form stable complexes with DNA following exposure of intact mammalian cells to chromate. Carcinogenesis. 1995;16:907–913. doi: 10.1093/carcin/16.4.907. [DOI] [PubMed] [Google Scholar]
- 2.Voitkun V, Zhitkovich A, Costa M. Cr(III)-mediated crosslinks of glutathione or amino acids to the DNA phosphate backbone are mutagenic in human cells. Nucleic Acids Res. 1998;26:2024–2030. doi: 10.1093/nar/26.8.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuykendall JR, Bogdanffy MS. Formation and stability of acetaldehyde-induced crosslinks between poly-lysine and poly-deoxyguanosine. Mutat Res. 1994;311:49–56. doi: 10.1016/0027-5107(94)90072-8. [DOI] [PubMed] [Google Scholar]
- 4.Hawkins CL, Pattison DI, Davies MJ. Reaction of protein chloramines with DNA and nucleosides: evidence for the formation of radicals, protein-DNA cross-links and DNA fragmentation. Biochem J. 2002;365:605–615. doi: 10.1042/BJ20020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurtz AJ, Dodson ML, Lloyd RS. Evidence for multiple imino intermediates and identification of reactive nucleophiles in peptide-catalyzed beta-elimination at abasic sites. Biochemistry. 2002;41:7054–7064. doi: 10.1021/bi020026y. [DOI] [PubMed] [Google Scholar]
- 6.Kurtz AJ, Lloyd RS. 1,N2-deoxyguanosine adducts of acrolein, crotonaldehyde, and trans-4-hydroxynonenal cross-link to peptides via Schiff base linkage. J Biol Chem. 2003;278:5970–5976. doi: 10.1074/jbc.M212012200. [DOI] [PubMed] [Google Scholar]
- 7.Copeland KD, Lueras AM, Stemp ED, Barton JK. DNA cross-linking with metallointercalator-peptide conjugates. Biochemistry. 2002;41:12785–12797. doi: 10.1021/bi020407b. [DOI] [PubMed] [Google Scholar]
- 8.Minko IG, Kurtz AJ, Croteau DL, Van Houten B, Harris TM, Lloyd RS. Initiation of repair of DNA-polypeptide cross-links by the UvrABC nuclease. Biochemistry. 2005;44:3000–3009. doi: 10.1021/bi0478805. [DOI] [PubMed] [Google Scholar]
- 9.Cho YJ, Kim HY, Huang H, Slutsky A, Minko IG, Wang H, Nechev LV, Kozekov ID, Kozekova A, Tamura P, Jacob J, Voehler M, Harris TM, Lloyd RS, Rizzo CJ, Stone MP. Spectroscopic characterization of interstrand carbinolamine cross-links formed in the 5′-CpG-3′ sequence by the acrolein-derived gamma-OH-1,N2-propano-2′-deoxyguanosine DNA adduct. J Am Chem Soc. 2005;127:17686–17696. doi: 10.1021/ja053897e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marchand C, Krajewski K, Lee HF, Antony S, Johnson AA, Amin R, Roller P, Kvaratskhelia M, Pommier Y. Covalent binding of the natural antimicrobial peptide indolicidin to DNA abasic sites. Nucleic Acids Res. 2006;34:5157–5165. doi: 10.1093/nar/gkl667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quievryn G, Zhitkovich A. Loss of DNA-protein crosslinks from formaldehyde-exposed cells occurs through spontaneous hydrolysis and an active repair process linked to proteosome function. Carcinogenesis. 2000;21:1573–1580. [PubMed] [Google Scholar]
- 12.Mao Y, Desai SD, Ting CY, Hwang J, Liu LF. 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes. J Biol Chem. 2001;276:40652–40658. doi: 10.1074/jbc.M104009200. [DOI] [PubMed] [Google Scholar]
- 13.Desai SD, Zhang H, Rodriguez-Bauman A, Yang JM, Wu X, Gounder MK, Rubin EH, Liu LF. Transcription-dependent degradation of topoisomerase I-DNA covalent complexes. Mol Cell Biol. 2003;23:2341–2350. doi: 10.1128/MCB.23.7.2341-2350.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brun F, Toulme JJ, Helene C. Interactions of aromatic residues of proteins with nucleic acids. Fluorescence studies of the binding of oligopeptides containing tryptophan and tyrosine residues to polynucleotides. Biochemistry. 1975;14:558–563. doi: 10.1021/bi00674a015. [DOI] [PubMed] [Google Scholar]
- 15.Behmoaras T, Toulme JJ, Helene C. A tryptophan-containing peptide recognizes and cleaves DNA at apurinic sites. Nature. 1981;292:858–859. doi: 10.1038/292858a0. [DOI] [PubMed] [Google Scholar]
- 16.Behmoaras T, Toulme JJ, Helene C. Specific recognition of apurinic sites in DNA by a tryptophan-containing peptide. Proc Natl Acad Sci USA. 1981;78:926–930. doi: 10.1073/pnas.78.2.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierre J, Laval J. Specific nicking of DNA at apurinic sites by peptides containing aromatic residues. J Biol Chem. 1981;256:10217–10220. [PubMed] [Google Scholar]
- 18.Chung FL, Young R, Hecht SS. Formation of cyclic 1,N2-propanodeoxyguanosine adducts in DNA upon reaction with acrolein or crotonaldehyde. Cancer Res. 1984;44:990–995. [PubMed] [Google Scholar]
- 19.Marnett LJ, Hurd HK, Hollstein MC, Levin DE, Esterbauer H, Ames BN. Naturally occurring carbonyl compounds are mutagens in Salmonella tester strain TA104. Mutat Res. 1985;148:25–34. doi: 10.1016/0027-5107(85)90204-0. [DOI] [PubMed] [Google Scholar]
- 20.Curren RD, Yang LL, Conklin PM, Grafstrom RC, Harris CC. Mutagenesis of xeroderma pigmentosum fibroblasts by acrolein. Mutat Res. 1988;209:17–22. doi: 10.1016/0165-7992(88)90104-2. [DOI] [PubMed] [Google Scholar]
- 21.Smith RA, Cohen SM, Lawson TA. Acrolein mutagenicity in the V79 assay. Carcinogenesis. 1990;11:497–498. doi: 10.1093/carcin/11.3.497. [DOI] [PubMed] [Google Scholar]
- 22.Cohen SM, Garland EM, St John M, Okamura T, Smith RA. Acrolein initiates rat urinary bladder carcinogenesis. Cancer Res. 1992;52:3577–3581. [PubMed] [Google Scholar]
- 23.Nechev LV, Harris CM, Harris TM. Synthesis of nucleosides and oligonucleotides containing adducts of acrolein and vinyl chloride. Chem Res Toxicol. 2000;13:421–429. doi: 10.1021/tx990167+. [DOI] [PubMed] [Google Scholar]
- 24.Nechev LV, Kozekov ID, Brock AK, Rizzo CJ, Harris TM. DNA adducts of acrolein: site-specific synthesis of an oligodeoxynucleotide containing 6-hydroxy-5,6,7,8-tetrahydropyrimido[1,2-a]purin-10(3H)-one, an acrolein adduct of guanine. Chem Res Toxicol. 2002;15:607–613. doi: 10.1021/tx010181y. [DOI] [PubMed] [Google Scholar]
- 25.de los Santos C, Zaliznyak T, Johnson F. NMR characterization of a DNA duplex containing the major acrolein-derived deoxyguanosine adduct gamma-OH-1,-N2-propano-2′-deoxyguanosine. J Biol Chem. 2001;276:9077–9082. doi: 10.1074/jbc.M009028200. [DOI] [PubMed] [Google Scholar]
- 26.Kim HY, Voehler M, Harris TM, Stone MP. Detection of an interchain carbinolamine cross-link formed in a CpG sequence by the acrolein DNA adduct gamma-OH-1,N2-propano-2′-deoxyguanosine. J Am Chem Soc. 2002;124:9324–9325. doi: 10.1021/ja020333r. [DOI] [PubMed] [Google Scholar]
- 27.Singh US, Moe JG, Reddy GR, Weisenseel JP, Marnett LJ, Stone MP. 1H NMR of an oligodeoxynucleotide containing a propanodeoxyguanosine adduct positioned in a (CG)3 frameshift hotspot of Salmonella typhimurium hisD3052: Hoogsteen base-pairing at pH 5.8. Chem Res Toxicol. 1993;6:825–836. doi: 10.1021/tx00036a012. [DOI] [PubMed] [Google Scholar]
- 28.Yang IY, Hossain M, Miller H, Khullar S, Johnson F, Grollman A, Moriya M. Responses to the major acrolein-derived deoxyguanosine adduct in Escherichia coli. J Biol Chem. 2001;276:9071–9076. doi: 10.1074/jbc.M008918200. [DOI] [PubMed] [Google Scholar]
- 29.Yang IY, Johnson F, Grollman AP, Moriya M. Genotoxic mechanism for the major acrolein-derived deoxyguanosine adduct in human cells. Chem Res Toxicol. 2002;15:160–164. doi: 10.1021/tx010123c. [DOI] [PubMed] [Google Scholar]
- 30.Yang IY, Chan G, Miller H, Huang Y, Torres MC, Johnson F, Moriya M. Mutagenesis by acrolein-derived propanodeoxyguanosine adducts in human cells. Biochemistry. 2002;41:13826–13832. doi: 10.1021/bi0264723. [DOI] [PubMed] [Google Scholar]
- 31.Kanuri M, Minko IG, Nechev LV, Harris TM, Harris CM, Lloyd RS. Error prone translesion synthesis past gamma-hydroxypropano deoxyguanosine, the primary acrolein-derived adduct in mammalian cells. J Biol Chem. 2002;277:18257–18265. doi: 10.1074/jbc.M112419200. [DOI] [PubMed] [Google Scholar]
- 32.Minko IG, Washington MT, Kanuri M, Prakash L, Prakash S, Lloyd RS. Translesion synthesis past acrolein-derived DNA adduct, gamma -hydroxypropanodeoxyguanosine, by yeast and human DNA polymerase eta. J Biol Chem. 2003;278:784–790. doi: 10.1074/jbc.M207774200. [DOI] [PubMed] [Google Scholar]
- 33.Moriya M, Zhang W, Johnson F, Grollman AP. Mutagenic potency of exocyclic DNA adducts: marked differences between Escherichia coli and simian kidney cells. Proc Natl Acad Sci USA. 1994;91:11899–11903. doi: 10.1073/pnas.91.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuykendall JR, Bogdanffy MS. Efficiency of DNA-histone crosslinking induced by saturated and unsaturated aldehydes in vitro. Mutat Res. 1992;283:131–136. doi: 10.1016/0165-7992(92)90145-8. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez AM, Minko IG, Kurtz AJ, Kanuri M, Moriya M, Lloyd RS. Comparative evaluation of the bioreactivity and mutagenic spectra of acrolein-derived alpha-HOPdG and gamma-HOPdG regioisomeric deoxyguanosine adducts. Chem Res Toxicol. 2003;16:1019–1028. doi: 10.1021/tx034066u. [DOI] [PubMed] [Google Scholar]
- 36.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 37.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci USA. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moriya M. Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-oxoguanine in DNA induces targeted G.C-->T.A transversions in simian kidney cells. Proc Natl Acad Sci USA. 1993;90:1122–1126. doi: 10.1073/pnas.90.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 40.Kalam MA, Haraguchi K, Chandani S, Loechler EL, Moriya M, Greenberg MM, Basu AK. Genetic effects of oxidative DNA damages: comparative mutagenesis of the imidazole ring-opened formamidopyrimidines (Fapy lesions) and 8-oxo-purines in simian kidney cells. Nucleic Acids Res. 2006;34:2305–2315. doi: 10.1093/nar/gkl099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pawlowicz AJ, Munter T, Zhao Y, Kronberg L. Formation of acrolein adducts with 2′-deoxyadenosine in calf thymus DNA. Chem Res Toxicol. 2006;19:571–576. doi: 10.1021/tx0503496. [DOI] [PubMed] [Google Scholar]
- 42.Fornace AJ, Jr, Seres DS. Repair of trans-Pt(II) diamminedichloride DNA-protein crosslinks in normal and excision-deficient human cells. Mutat Res. 1982;94:277–284. doi: 10.1016/0027-5107(82)90291-3. [DOI] [PubMed] [Google Scholar]
- 43.Speit G, Schutz P, Merk O. Induction and repair of formaldehyde-induced DNA-protein crosslinks in repair-deficient human cell lines. Mutagenesis. 2000;15:85–90. doi: 10.1093/mutage/15.1.85. [DOI] [PubMed] [Google Scholar]
- 44.Minko IG, Zou Y, Lloyd RS. Incision of DNA-protein crosslinks by UvrABC nuclease suggests a potential repair pathway involving nucleotide excision repair. Proc Natl Acad Sci USA. 2002;99:1905–1909. doi: 10.1073/pnas.042700399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reardon JT, Sancar A. Repair of DNA-polypeptide crosslinks by human excision nuclease. Proc Natl Acad Sci USA. 2006;103:4056–4061. doi: 10.1073/pnas.0600538103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Novakova O, Kasparkova J, Malina J, Natile G, Brabec V. DNA-protein cross-linking by trans-[PtCl2(E-iminoether)2]. A concept for activation of the trans geometry in platinum antitumor complexes. Nucleic Acids Res. 2003;31:6450–6460. doi: 10.1093/nar/gkg863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shapiro R, Sodum RS, Everett DW, Kundu SK. Reactions of nucleosides with glyoxal and acrolein. IARC Sci Publ; 1986. pp. 165–173. [PubMed] [Google Scholar]
- 48.Smith RA, Williamson DS, Cohen SM. Identification of 3,N4-propanodeoxycytidine 5′-monophosphate formed by the reaction of acrolein with deoxycytidine 5′-monophosphate. Chem Res Toxicol. 1989;2:267–271. doi: 10.1021/tx00010a009. [DOI] [PubMed] [Google Scholar]
- 49.Smith RA, Williamson DS, Cerny RL, Cohen SM. Detection of 1,N6-propanodeoxyadenosine in acrolein-modified polydeoxyadenylic acid and DNA by 32P postlabeling. Cancer Res. 1990;50:3005–3012. [PubMed] [Google Scholar]
- 50.Kozekov ID, Nechev LV, Sanchez A, Harris CM, Lloyd RS, Harris TM. Interchain cross-linking of DNA mediated by the principal adduct of acrolein. Chem Res Toxicol. 2001;14:1482–1485. doi: 10.1021/tx010127h. [DOI] [PubMed] [Google Scholar]
- 51.Kozekov ID, Nechev LV, Moseley MS, Harris CM, Rizzo CJ, Stone MP, Harris TM. DNA interchain cross-links formed by acrolein and crotonaldehyde. J Am Chem Soc. 2003;125:50–61. doi: 10.1021/ja020778f. [DOI] [PubMed] [Google Scholar]
- 52.Washington MT, Minko IG, Johnson RE, Wolfle WT, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases iota and kappa. Mol Cell Biol. 2004;24:5687–5693. doi: 10.1128/MCB.24.13.5687-5693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolfle WT, Johnson RE, Minko IG, Lloyd RS, Prakash S, Prakash L. Human DNA polymerase iota promotes replication through a ring-closed minor-groove adduct that adopts a syn conformation in DNA. Mol Cell Biol. 2005;25:8748–8754. doi: 10.1128/MCB.25.19.8748-8754.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanchez AM, Kozekov ID, Harris TM, Lloyd RS. Formation of inter- and intrastrand imine type DNA-DNA cross-links through secondary reactions of aldehydic adducts. Chem Res Toxicol. 2005;18:1683–1690. doi: 10.1021/tx0500528. [DOI] [PubMed] [Google Scholar]
- 55.Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: the role of DNA polymerase kappa. J Biol Chem. 2004;279:53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- 56.Bi X, Slater DM, Ohmori H, Vaziri C. DNA polymerase kappa is specifically required for recovery from the benzo[a]pyrene-dihydrodiol epoxide (BPDE)-induced S-phase checkpoint. J Biol Chem. 2005;280:22343–22355. doi: 10.1074/jbc.M501562200. [DOI] [PubMed] [Google Scholar]
- 57.Choi JY, Angel KC, Guengerich FP. Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase kappa. J Biol Chem. 2006;281:21062–21072. doi: 10.1074/jbc.M602246200. [DOI] [PubMed] [Google Scholar]
- 58.Kawai Y, Furuhata A, Toyokuni S, Aratani Y, Uchida K. Formation of acrolein-derived 2′-deoxyadenosine adduct in an iron-induced carcinogenesis model. J Biol Chem. 2003;278:50346–50354. doi: 10.1074/jbc.M309057200. [DOI] [PubMed] [Google Scholar]
- 59.Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Adv Protein Chem. 2005;71:401–440. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]