Abstract
The hairpin ribozyme accelerates the rate of phosphodiester transfer reactions by at least 5 orders of magnitude. To achieve this rate enhancement the active site forms via a substrate helix docking event that constrains the scissile phosphate linkage and positions G8 and A38 for catalysis, both of which have been implicated as sites of proton transfer in general acid-base catalysis. To investigate the functional groups required for hairpin activity we previously reported a series of Nucleotide Analog Interference Mapping experiments [S.P. Ryder et al., RNA 7, 1454–1463]. The critical functional groups implicated in those studies were largely consistent with subsequent x-ray crystal structures, but the lack of A38 interference with 8-aza-adenosine (n8A), a pKa perturbed nucleotide analog, argued against functional base ionization at this site. This is inconsistent with a transition state crystal structure and other biochemical studies. To address this discrepancy we investigated the hairpin ribozyme with an expanded set of pKa perturbed adenosine analogs containing fluorine. A38 was the only site that showed persistent and strong interference with low pKa analogs across a variety of construct/substrate pairs. This interference pattern suggests that A38 base ionization is required for catalytic activity. The lack of n8A interference at A38, in spite of its reduced pKa, likely results from n8A stabilization of the docked state, which requires an unusual syn glycosidic base conformation at A38 for active site assembly. The fluorinated adenosine analogs are better suited to identify sites of functional ionization in systems where structural rearrangements are closely coupled to catalytic steps. All pKa reduced analogs, including those of the previous study, produce selective interference at A38 when substrates are stably bound and docked, consistent with the importance of base ionization at this site.
The hairpin ribozyme is a member of the nucleolytic class of ribozymes, a collection of small protein-independent catalytic RNAs whose members also include the hammerhead, hepatitis delta, Varkud satellite and glmS ribozymes(1). These self-cleaving RNAs catalyze a site-specific and reversible phosphodiester transfer reaction in which the 2'-hydroxyl of an upstream nucleotide attacks the adjacent phosphodiester linkage, producing cleavage products with free 5'-hydroxyl and 2',3'-cyclic phosphate termini. This concerted SN2 type reaction requires an in-line geometry of the 2'-hydroxyl, scissile phosphate, and 5'-oxygen leaving group. In the hairpin ribozyme, this scissile phosphate orientation, as well as the positioning of active site residues, is only obtained after a structural transition from an extended (undocked) to condensed (docked) state. The reaction products formed by nucleolytic ribozymes are distinct from those of self-splicing introns and RNase P, and unlike these larger ribozymes, all known members of the nucleolytic class retain catalytic activity in the absence of divalent cations(1). This observation in combination with a growing wealth of structural data for several members of the class suggests nucleolytic ribozymes may employ their nucleobases directly in catalysis. The exact nature of nucleobase contribution, either as sites of proton transfer, as the source of electrostatic stabilization, or in the positioning of substrates or bound water molecules is an unresolved question.
Several factors make the hairpin ribozyme uniquely suited for mechanistic studies. First is the observation that the hairpin readily catalyzes both cleavage and ligation, and that the reaction can be driven in either direction by the appropriate choice of construct and cleaved product sequence(2). The reversibility of the reaction is essential to the hairpin's function in the replication cycle of the tobacco ringspot virus satellite RNA(3) and allows the contribution of particular functional groups for the forward and reverse reactions to be dissected. Another attractive feature of the hairpin is that it can be reassembled from shorter oligonucleotides in a number of combinations, which allows site-specific modifications at nearly any residue. This strategy has been used extensively to prepare constructs for x-ray crystallography with a wide-variety of synthetic modifications at the scissile phosphate and at active site residues. To date nearly 30 hairpin structures have been solved in constructs prepared to mimic the pre-catalytic, post-cleavage, and transition states(1). In several of these studies the effect of base modifications on catalysis has also been measured (4–7). Finally the ability to introduce fluorophores into active constructs has allowed detailed studies of hairpin dynamics by equilibrium and single-molecule FRET(8). These studies have provided some of the most detailed descriptions to date on the connection between RNA dynamics, folding and catalysis.
Despite the success of site-specific incorporation strategies, their application is constrained by the need to synthesize and reassemble RNAs for each modification and site of interest, which limits the number of sites that can be reasonably investigated. In addition, when truncated constructs are used for site-specific incorporation they often exhibit significantly perturbed kinetic properties, even when wild-type sequences are used, an effect attributed to the decreased stability of minimal constructs. This problem is especially pronounced for the hairpin ribozyme, where the native sequence includes a four-way helical junction (4WJ), but where the majority of chemical modification experiments have been conducted in a minimal two-way junction (2WJ) construct. Although the active sites of the 2WJ and 4WJ constructs have been shown to be nearly identical by x-ray crystallography(9, 10), the 4WJ folds much faster and more stably(11, 12), favors the ligated state to a greater extent(2), exhibits a different pH dependence(13–15) and functions at physiological salt concentrations where the 2WJ construct is inactive(16).
An alternative to site-specific incorporation strategies is Nucleotide Analog Interference Mapping (NAIM). NAIM employs a series of phosphorothioate-tagged triphosphates, each carrying a chemical modification, which are introduced randomly into an RNA by transcription. Transcriptions are carried out such that any individual transcript includes at most one substitution at a random location. This pool of analog substituted RNAs is then subjected to a competitive selection and the active fraction is cleaved at the phosphorothioate tag. Gel electrophoresis of the cleaved RNAs reveals sites of analog interference (for reviews of the NAIM technique see (17–19)). There are two main advantages that NAIM has over site-specific substitutions. NAIM can rapidly assay the effect of a chemical modification at all positions simultaneously, but on a site by site basis; and the technique can be applied to any RNA sequence that can be transcribed and has a selectable function.
In two early studies of hairpin catalysis a NAIM study of the hairpin ligation reaction used 17 nucleotide analogs to study a 2WJ construct and three analogs to study a 4WJ construct(20, 21). The resulting interference pattern was highly consistent with contacts observed in the subsequent crystal structure of a pre-cleavage 4WJ construct(22). The close agreement also helped establish that two earlier NMR structures of isolated loops A and B were in inactive undocked conformations(23–25). Despite an overall high level of correspondence, the interference pattern produced by 8-aza-adenosine (n8A) at three active site residues, A9, A10, and A38 did not match the x-ray structure(25). This analog has an N1 pKa value of 2.2, more than a full pKa unit below that of A (N1 pKa = 3.5), and was used to identify sites of adenosine base ionization that are important for function (21). In the context of a 2WJ construct n8A produced strong interference at A10, which could be rescued at low pH, showed enhancement at A9, and had no effect at A38. These data suggested adenosine ionization was required at A10, with this base acting either as a site of proton transfer or as the origin of electrostatic stabilization. Although this pattern of interference did not obviously match the pre-cleavage crystal structure, where the N1 nitrogen of A10 is at least 7 Å from any atoms in the scissile linkage, it was suggested that a modest structural rearrangement in the post-cleavage or transition state would be sufficient to bring A10 into hydrogen bonding distance with the nucleophile, leaving group or scissile phosphate oxygens(25). A rearrangement of this type would also be required to explain the lack of interference at A38, which is much closer to each of these functionalities in the pre-cleavage structure(22).
Several lines of evidence now argue against this interpretation of the n8A interference data, and have motivated the current study. Since the initial report of the pre-cleavage 4WJ structure two additional structures have been reported using the same construct, one with a penta-coordinated vanadate at the location of the scissile phosphate as a transition state mimic, and one representing the post-cleavage (pre-ligation) state(9). All three structures show very similar active site geometries, suggesting a relatively rigid active site throughout the reaction and argue against a structural rearrangement sufficient to bring A10 into direct contact with either the nucleophile or leaving group(26). A large collection of crystal structures solved with 2WJ constructs also exhibit the same general active site geometry(1). In addition, two structures prepared to mimic the transition state show very short distances between the N1 of A38 and the 5'-oxygen at the cleavage site, consistent with a protonated adenosine at A38(9, 10). A number of kinetic studies have also investigated the relative importance of active site adenosines and have consistently pointed toward A38 rather than A10 as a site of functional ionization(27, 28). It has also been argued that a lack of interference at A38 from n8A substitution is not necessarily inconsistent with A38 acting as a general base for ligation reaction, given this analog's relatively modest pKa perturbation(29). Perhaps most convincing is a recent series of structural and kinetic studies with N1-deaza-adenosine incorporated site-specifically at A10 and A38 that clearly demonstrated an essential role for the N1 nitrogen of A38 but not A10(4, 5). Recent pH dependent Raman studies also indicate that the pKa of A38 is shifted toward neutrality in the folded ribozyme, further supporting its potential role as a site of proton transfer(30). Taken together, these results argue strongly for functional ionization at A38, not A10, and have inspired us to return to the hairpin ligation reaction using NAIM with additional analogs and constructs.
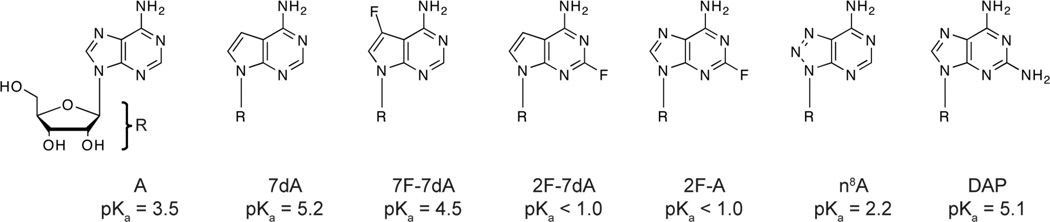
We recently developed a series of fluorine substituted adenosine analogs that produce very large changes in N1 pKa values without significant structural perturbation to the nucleobase(31). These include 7-fluoro-7-deaza-adenosine (7F-7dA), 2-fluro-7-deaza-adenosine (2F-7dA), and 2-fluoro-adenosine (2F-A). The application of these analogs in a NAIM study of the Varkud satellite ribozyme resulted in a very selective interference pattern, showing interference at only 3 of the 36 resolvable adenosine sites. Of these sites, a single adenosine, A756, exhibited an interference pattern consistent with functional ionization(31). Significantly, this site was previously shown to exhibit a pH dependent interference pattern with n8A(32), and has also been implicated as a site of proton transfer by a number of site-specific chemical modification experiments(33–35). In this study we have applied these fluorine substituted adenosine analogs to the hairpin ribozyme in an effort to clarify the dependence of interference values relative to the analog’s N1 pKa value (Figure 1).
Figure 1.
Structures and N1 pKa values of adenosine analogs used in this study(21, 31, 40).
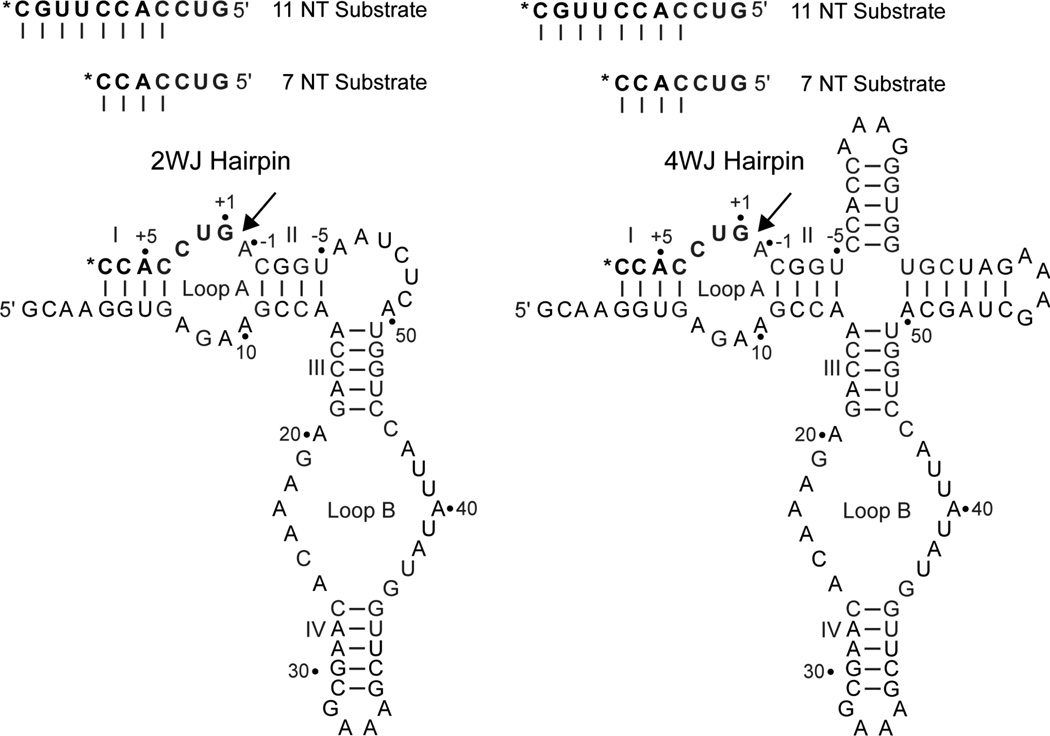
Since our initial NAIM studies a great deal has been learned regarding the influence of construct design, substrate binding affinity and reaction conditions on observed hairpin kinetics(1, 8, 36). Of greatest significance to the interpretation of NAIM data is the observation that the ligation/cleavage reaction reaches rapid equilibrium in many constructs, and that release of initially ligated substrate can also be rapid depending on the stability of the docked state. To explore the effect of construct/substrate identity on observed interference values we chose four construct/substrate pairings studied extensively by Fedor and colleagues (2, 27)(Figure 2), and conducted NAIM selections in reaction conditions that reflect those used in the majority of kinetic experiments. These NAIM selections reveal that many of the observed interferences are highly dependent on substrate binding affinity and the stability of the docked state. For constructs with stably bound and docked substrates, the observed interference pattern is highly selective and consistent with an essential role for A38 ionization in catalysis.
Figure 2.
Hairpin constructs used in this study containing minimal two-way or native four-way junctions (site of ligation indicated with an arrow). The 3'-end labeled substrates used for selections (7NT and 11NT) are also shown, and are designed to form 4 or 8 base pairs in stem I.
EXPERIMENTAL PROCEDURES
Plasmid Templates for Hairpin Ribozyme Transcriptions
2WJ and 4WJ hairpin sequences were amplified from overlapping DNA primers and cloned into pUC19 following standard procedures. An antigenomic HDV ribozyme sequence was introduced at the 3'-end of each hairpin sequence to produce the desired 2',3'-cyclic phosphate terminus. Following the HDV sequence an SspI restriction site was introduced to produce a blunt-end linearized DNA template. For the 2WJ construct the linearized template had the following sequence (promoter in bold, hairpin sequence underlined, HDV sequence uppercase, SspI site lowercase): TAATACGACTCACTATAGCAAGGTGAGAAGCCAACCAGAGAAACACAAGCGAAAGCTTGGTATATTACCTGGTACTCTAATGGCAGGGTCGGCATGGCATCTCCACCTCCTCGCGGTCCGACCTGGGCATCCGAAGGAGGACAGACGTCCACTCGGATGGCTAAGGGAGAGCCAaat.
For the 4WJ construct the hairpin sequence was replaced with the following: GCAAGGTGAGAAGCCAACCAGAGAAACACAAGCGAAAGCTTGGTATATTACCTGGTACGATCGAAAGATCGTGGTGGGAAACCACCTGGCA.
Sequenced plasmids were completely digested with SspI to form linear templates. Typical digestions contained 100ng/uL plasmid, 2U/ug SspI, and were incubated at 37°C for 2 hours. The restriction enzyme was denatured at 65°C for 20 minutes and the templates were used in transcriptions without further purification.
Ligation Substrates
Ligation substrates were synthesized and deprotected according to the manufacturer's protocol (Dharmacon). Two substrates, denoted 7NT and 11NT, were used for NAIM experiments (7NT: GUCCACC, 11NT: GUCCACCUUGC). Each substrate was labeled at the 3'-end with one equivalent of [α-32P] pCp by T4 RNA ligase, purified by 20% non-denaturing PAGE and eluted into T10E0.1 overnight at 4°C. After the addition of pCp, the 7NT substrate forms four base pairs in helix I, with a 3' A•C mismatch, while the 11NT substrate forms 8 base pairs with a 3' C overhang.
Hairpin Transcription
Unligated 2WJ and 4WJ hairpin RNA was obtained by transcription of SspI linearized plasmids. Typical 100 µL transcriptions contained 40 mM Tris buffer (pH = 8.0), 15 mM MgCl2, 2 mM spermidine, 5 mM DTT, 0.005 % Triton X-100, 1 mM of each NTP, 2 µg of SspI cut plasmid, 1 unit inorganic pyrophosphatase (Sigma) and T7 RNA polymerase to a final concentration of 0.1 mg/mL. Hairpin RNAs with randomly incorporated nucleotide analogs were obtained by including the corresponding 5'-O-(1-thio)nucleoside triphosphates to final concentrations described previously(17). Transcriptions were incubated for 2.5 hours at 37°C, then heated to 70°C and slow cooled to 25°C over 45 minutes. This refolding step led to complete cleavage of the inserted HDV ribozyme, producing unligated hairpin RNAs with the desired 2',3'-cyclic phosphate at the A(−1) position. RNAs were purified by 8% denaturing PAGE and eluted at 25°C for 1 hour into T10E0.1, 250 mM NaCl, 2% w/w SDS. Elutions were PCA extracted (phenol/chloroform/isoamyl alcohol (25:24:1) saturated with 10 mM Tris–HCl (pH 8.0), 1 mM EDTA) and concentrated by ethanol precipitation, resuspended in T10E0.1 and stored at −20°C. RNA concentrations were quantified by UV absorption at 260 nM (2WJ ε260 = 8.55×105 M−1cm−1, 4WJ ε260 = 1.15×106 M−1cm−1).
Incorporation Controls
To control for the level of analog incorporation at each site unligated hairpin RNAs were radiolabeled at their 5'-end, cleaved with iodine, and loaded onto sequencing gels as follows. 20 pmole of each analog-incorporated transcript was treated with 50 units of CIP at 37 °C for 1 hour. CIP was removed with silica membrane spin columns (Qiagen) and the RNAs were eluted into T10E0.1. Each RNA was then 5'-end labeled with 20 units of T4 polynucleotide kinase (PNK) and 150 µCi of [γ-32P]ATP by incubating at 37 °C for 1 hour. Labeled RNAs were purified by 8% denaturing PAGE, visualized by autoradiography, excised from the gel, and eluted at 25°C for 1 hour into T10E0.1, 250 mM NaCl, 2% w/w SDS, then PCA extracted, ethanol precipitated, and redissolved in a minimal volume of T10E0.1. A small aliquot of each solution was scintillation counted and the remainder was diluted with T10E0.1 to provide an equal number of counts/µL. Each solution was split into two aliquots, treated with iodine (1/10 volume of 100 mM iodine in ethanol) or ethanol alone, denatured at 90 °C in formamide loading buffer (20 mM EDTA in deionized formamide) and run on a 10% denaturing PAGE sequencing gel.
NAIM Ligation Selection
Individual ligation reactions were performed with each of the analog-incorporated hairpin RNAs and 3'-end labeled substrates. Hairpin RNAs were first refolded in reaction buffer (50 mM Tris, 10 mM MgCl2, pH = 8.0) by heating to 60 °C for two minutes and cooling to 25 °C over 45 minutes. Ligation reactions were initiated by adding labeled substrate equilibrated to 25 °C in reaction buffer and mixing. Reaction volumes were typically 20 µL and contained 300 nM hairpin RNA and 20 nM 3'-end labeled substrate. Unless indicated otherwise reactions were allowed to proceed for 5 minutes, which was sufficient time to reach the ligation endpoint for all construct/substrate pairs. Reactions were quenched with formamide loading buffer and split into two aliquots. Iodine (1.0 µL of 100 mM in ethanol) was added to one aliquot to cleave phosphorothioate linkages, while the other aliquot was treated with ethanol alone to control for non-specific cleavage. Cleaved and uncleaved samples were denatured at 90 °C and run on 10% denaturing PAGE sequencing gels.
Interference Quantitation
Ligation selection gels and 5'-end labeled incorporation control gels were dried and exposed to PhosphorImager screens. Sites of interference or enhancement in activity were identified as decreases or increases in band intensity in the ligation selection gel relative to the unligated 5'-end labeled controls. Because the phosphorothioate substitution can itself lead to changes in activity a control RNA incorporating the parental 5'-O-(1-thio)nucleoside triphosphate was also included. This control was particularly important for the 2WJ hairpin construct where several sites showed significant interference or enhancement arising from the Rp phosphorothioate linkage. For positions A9 and A10 the phosphorothioate effect was so large that quantification of band intensities at these positions in the 2WJ construct was not possible, and interference values for base modified analogs were not calculated. To quantify sites of interference, line profiles were drawn through ligation selection and control lanes and peak areas calculated. From these peak areas an interference value, κ, was calculated:
where each member of the fraction is the peak area for a particular site in the ligation or 5'-end labeled gel, NαS is the parental analog, and δαS is one of the pKa perturbed analogs. A normalization factor (NF) is calculated from the ratios to account for uneven loading of the lanes. Interference values calculated in this way normalize both for the extent of analog incorporation at a particular site, and for the effect the phosphorothioate substitution at that site. Interference values between 0.5 and 2.0 are typically viewed as insignificant, while those below 0.5 are expressed as 1/κ enhancements. Because of the difficulty of accurately quantifying very weak bands interference values above 6.0 are typically treated as equivalent.
Ligation Kinetics and Chase Experiments
Ligation time courses and endpoints were measured under the same conditions as NAIM ligation reactions except hairpin RNAs transcribed without analogs were used. Chase experiments were conducted after ligation reactions with labeled substrates had reached their endpoints. For the 11NT substrate chase experiments, a solution containing unlabeled 11NT substrate in reaction buffer was added to 2WJ/7NT or 4WJ/7NT ligations in 3-fold excess relative to the hairpin RNA. For the 4WJ hairpin chase experiments 4WJ RNA was refolded and cooled to 25 °C, then added to 2WJ/7NT or 2WJ/11NT ligations in 1.05 fold excess relative to the hairpin RNA. Reaction aliquots were quenched in formamide loading buffer at the specified time points and run on 20% denaturing PAGE sequencing gels. Gels were exposed to PhosphorImager screens without drying and the percent ligation was calculated from the substrate and ligation product band intensities.
RESULTS
Construct Dependent Interference Patterns
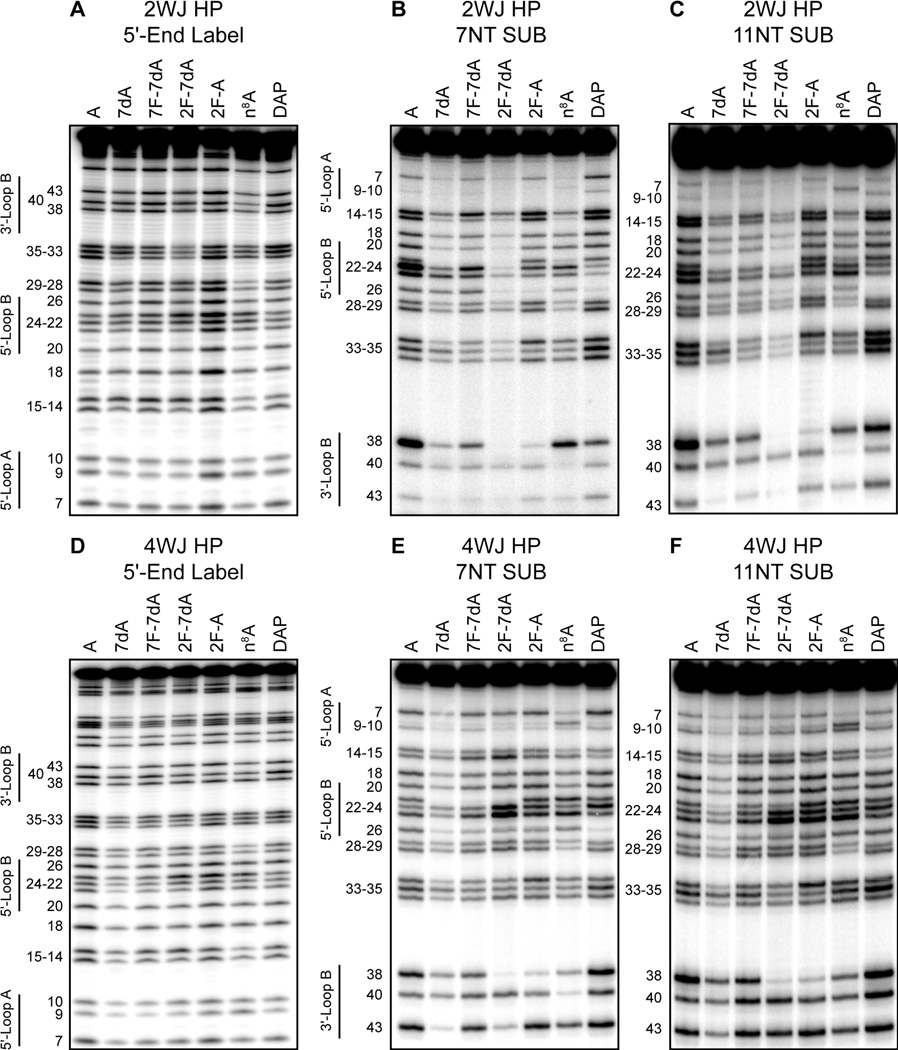
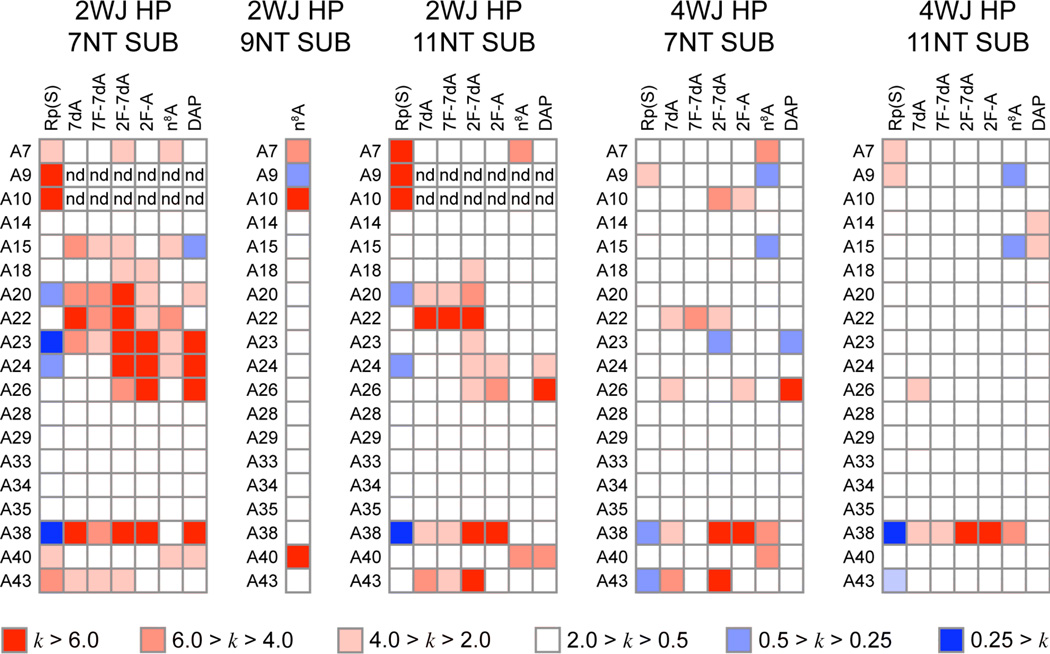
We investigated the effect of construct stability and substrate binding affinity on the observed interference patterns by conducting NAIM selections on four construct/substrate pairs under identical reaction conditions (Figure 2). Typical sequencing gels for the ligation selections and incorporation controls were of sufficient quality to calculate interference values for all analogs at adenosine sites 7–43 (Figure 3). The only sites not quantified were A9 and A10 in the 2WJ constructs, due to the large phosphorothioate effects observed at these sites in the minimal construct (Figure 3B,C). Replicate control and selection experiments for each of the analogs yielded a total of 432 average interference values at 19 sites. These data are summarized as a color coded matrix in Figure 4. As expected, none of the analogs show any interference in stem-loop IV for any of the construct/substrate pairs. In contrast the interference pattern for loop B is highly dependent on construct and substrate identity. Every analog tested showed moderate to large interferences for loop B residues in the 2WJ/7NT selections, while nearly all of these interferences were diminished in more stable construct/substrate pairs (2WJ/11NT and 4WJ/7NT), and absent in the 4WJ/11NT selections.
Figure 3.
Representative interference autoradiograms for the 2WJ construct (A–C) and 4WJ construct (D–F). (A,D) Analog incorporation controls. (B,E) Ligation selections with 7NT substrate. (C,F) Ligation selections with 11NT substrate. Each lane is labeled with the analog used and nucleotide positions are shown to the left with the regions of loops A and B indicated. The order of the bands (top to bottom) for the control and selection sequencing gels is reversed because of the labeling strategy used (see Experimental Procedures).
Figure 4.
Average interference values as a function of analog (columns) and nucleotide position (rows) for the construct/substrate pairs indicated. The first column of each matrix provides the phosphorothioate effect, which is controlled for in calculating other interference values. Interference values are color coded as indicated from strong interference (deep red) to strong enhancements (deep blue). Sites where large phosphorothioate effects prevented the calculation of interference values are marked "nd". The single column for the n8A analog in a 2WJ construct 9NT substrate selection is data taken from our previous study(21).
2-fluoro-adenosine Interference Pattern
Because the focus of this study was to re-explore the role of adenosine ionization in the hairpin reaction, the interference pattern produced by the recently developed 2F-A analog is of particular interest. This analog introduces a relatively isosteric fluorine for hydrogen substitution at C2, but possesses an extremely low N1 pKa, which we previously estimated to be below zero (the first ionization of 2F-A occurs at N7 with a pKa value of 0.84) (31). However, because all purine ring nitrogens and the exocyclic amine are retained in 2F-A, this analog produces a minimal perturbation to the various hydrogen bonding interactions of adenosine. Inspection of the 2F-A interference values for each of the hairpin construct/substrate pairs displays a clear transition in the interference pattern as more stable construct/substrate pairs are used in the selection (Figure 4). A38 is the only position that exhibits consistent interference with 2F-A regardless of construct. Even in the 4WJ/11NT selections the interference value at A38 is much larger than the typical upper bound reported (κ>6) and can be easily seen in the selection sequencing gel itself (Figure 3F). This pattern of interference suggests that the subtle structural perturbation caused by 2F-A at some sites can be overcome by increasing the stability of the construct used for ligation, while the persistent interference at A38 likely arises from the inability of 2F-A to ionize. Consistent with this interpretation is the interference produced by DAP, which introduces an amino group instead of a fluorine at C2. Inspection of Figure 4 reveals that 2F-A and DAP show similar interference patterns and that their dependence on construct/substrate identity is also similar, with the exception of interference at A38. Strong 2F-A interference persists at A38, while interference produced by DAP at A38 in the 2WJ/7NT pairing is absent in all other construct/substrate pairings. A similar trend is observed for the 2F-7dA analog, which can be viewed as a low pKa analog of 7dA. While the interference caused by 7dA is construct dependent, the 2F-7dA analog produces strong interferences for all construct/substrate pairs examined.
Interference at A38
The construct dependent interferences noted above highlight the need to control for subtle structural perturbations when attempting to identify sites of functional ionization. This is particularly true for A38, which exhibits a complex pattern of structural interactions in the reported crystal structures. The residue adopts an unusual syn-glycosidic conformation in the docked state, makes hydrogen bonds via its N1, N7 and N6 exocyclic amine, and stacks with the critical G(+1) residue. The new interference patterns reported in this study arise from analogs that make fluorine for hydrogen substitutions at the 2 or 7 position of adenosine or 7-deaza-adenosine (Figure 1). To control for structural effects produced by these substitutions we performed identical NAIM selections with 7dA and DAP. As with other residues in loop B the interference pattern at A38 is dependent on construct/substrate stability for a number of analogs (Figure 4). The exception is interference produced by 2F-A and 2F-7dA, both of which have N1 pKa values below 1 and exhibit interference values above 6 at A38 for all construct/substrate pairs. Of the remaining analogs 7dA, 7F-7dA, and DAP follow the general trend of decreasing interference with increasing construct/substrate stability. The observation that interference caused by 7-deaza substitutions is mitigated by stabilized constructs is readily explained by a hydrogen bond that involves this nitrogen(9, 22). Consequently the large construct independent interference observed for 2F-7dA is likely to originate from the 2-fluoro substitution rather than the 7-deaza substitution. The fact that DAP does not interfere in stabilized constructs indicates the origin of 2F-A and 2F-7dA interference is not a steric effect, suggesting the interference observed with theses analogs can be attributed to their low N1 pKa values.
The construct dependent interference pattern observed with n8A at A38 stands in stark contrast to that of the other analogs. The magnitude of n8A interference at A38 increases in the 4WJ constructs relative to the 2WJ constructs (Figure 4). This pattern of increased interference with increased construct/substrate stability is unique to n8A interference at A38, and is not observed with n8A at any other site or with any other analog at any site. The absence of n8A interference in the previous study led to the conclusion that A38 was not a site of functional ionization for hairpin catalysis(21). The detection of strong interference with n8A at A38 in stabilized 4WJ constructs is significant because it is the expected interference pattern for functional ionization at this site. Thus, in the most stable construct/substrate pair all analogs with reduced N1 pKa values show strong interference. An interesting comparison is the construct dependent interference pattern of 7dA (Figure 4). This pKa elevated analog might be expected to produce an enhancement at A38, but instead produces a strong structural interference in the 2WJ/7NT selections that is mitigated in more stable construct/substrate pairs. The analogous interpretation of the construct dependent n8A interferences at A38 is that this analog provides a structural advantage when substituted at A38 in destabilized construct/substrate pairs, while revealing an underlying catalytic deficiency in selections using the most stable construct/substrate pairs.
Equilibration of Reaction Substrates
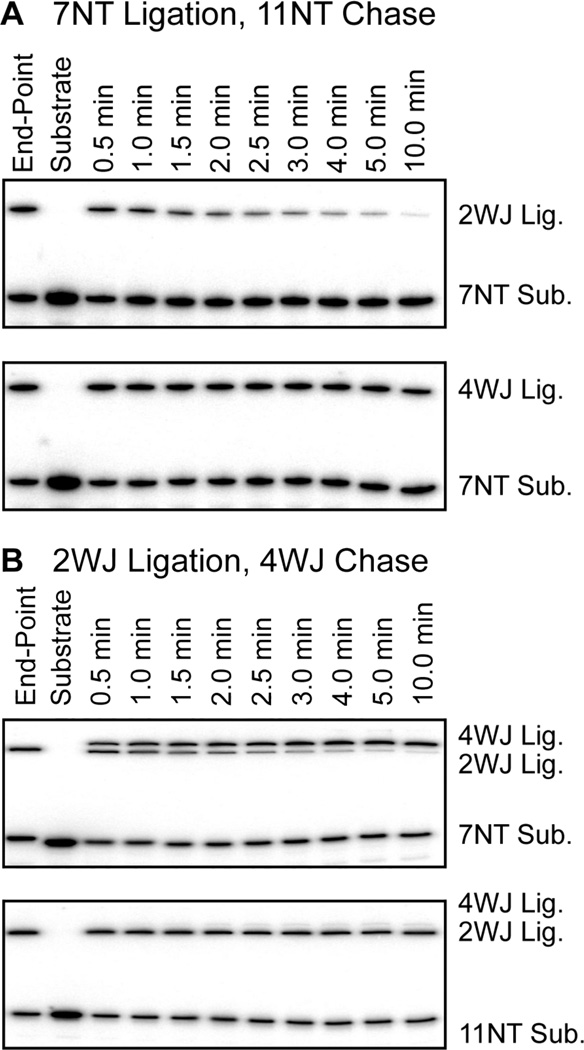
The interpretation of interference data is highly dependent on whether the steps of substrate binding, docking and ligation are reversible (see below). Based on the careful analysis of Fedor and colleagues(2, 27) we anticipated our construct/substrate pairs would differ in their ability to reverse these steps and exchange labeled substrates among members of the analog incorporated hairpin pool. To test this possibility we conducted chase experiments under the exact conditions used for NAIM selections (Figure 5). We were particularly interested in whether the reactions were reversible on the timescale of the ligation selections (5 min.). When the 2WJ construct was incubated with hot 7NT substrate, an endpoint of ~30% was reached by 5 minutes. Addition of an excess of cold 11NT substrate led to a rapid decrease of 2WJ/7NT product on a timescale similar to that of the initial ligation (Figure 5A). The equivalent experiment with the 4WJ construct showed much more rapid formation of the 4WJ/7NT ligation product, with an endpoint near 55%, but a much slower reversal of this product upon addition of cold 11NT substrate (45% 4WJ/7NT after 5 minute quench). These results roughly reproduce what has been observed previously(2), namely that the cleaved 7NT substrate readily binds and dissociates from the undocked state of both hairpin constructs(2), and that the 4WJ construct forms the docked state faster and more stably than the 2WJ construct(37). A parallel experiment in which 2WJ/7NT ligation was chased with a slight excess of 4WJ hairpin RNA yielded a comparable result, with the near compete conversion of 2WJ/7NT ligated product to 4WJ/7NT ligated product in 5 minutes (Figure 5B). However, when a 2WJ/11NT ligation was chased with 4WJ hairpin RNA no conversion to the 4WJ/11NT product was observed. This result confirms the much slower release of the 11NT substrate. Thus in NAIM ligation selections with the 11NT substrate, once substrate binds a member of the analog incorporated pool it does not release before the reaction is quenched.
Figure 5.
Hairpin ligation chase experiments. In each panel the indicated construct/substrate ligation was allowed to reach its endpoint, then chased with 11NT cold substrate (A) or 4WJ hairpin RNA (B).
Time-Dependent Interference Values
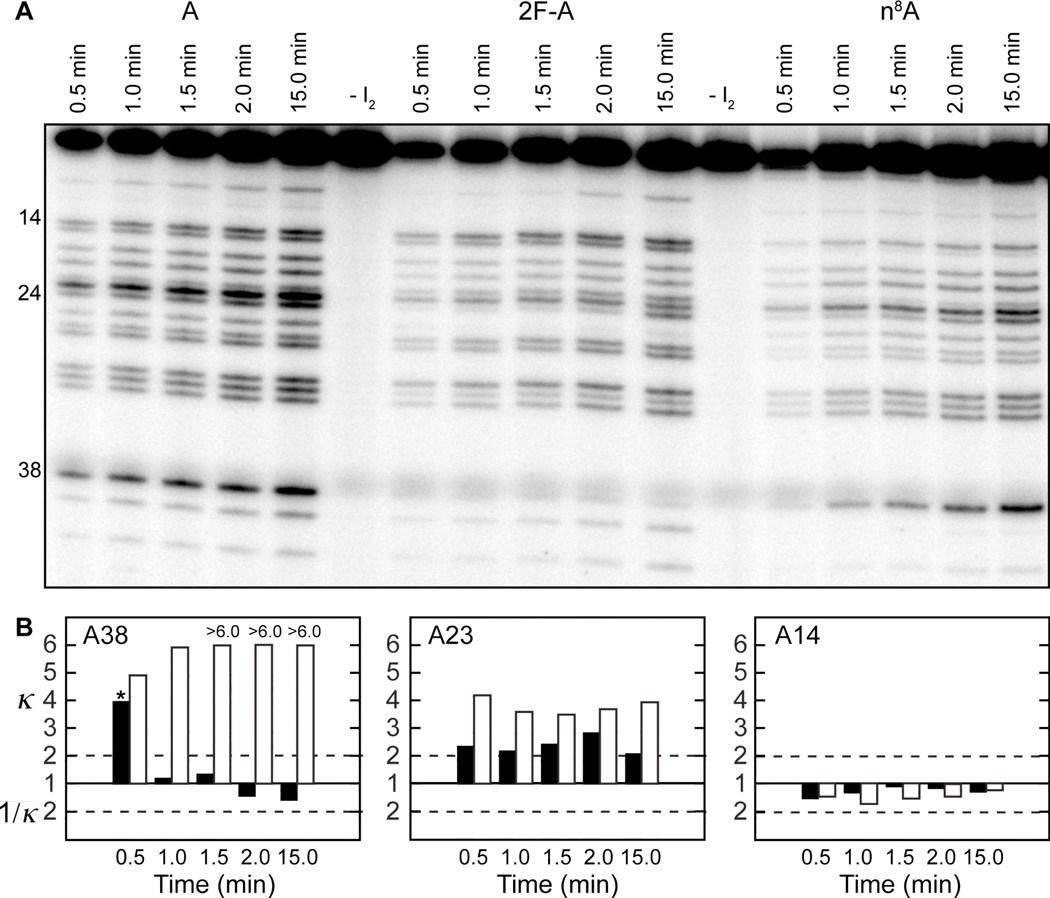
The observation that 7NT substrates rapidly exchange between members of the 2WJ hairpin RNA pool suggested interference values for the 2WJ/7NT ligation might be time dependent. To test this dependence we repeated the 2WJ/7NT NAIM selections as a function of time for the n8A and 2F-A analogs (Figure 6A). Significantly, n8A did show moderate interference at A38 in the 2WJ/7NT selection at our earliest time point (30 sec), but had no effect or even slight enhancement at longer times (Figure 6B). This is in contrast to 2F-A, which showed moderate to strong interference at A38 at all ligation time points. The time dependence of n8A interference is specific to the A38 position. For example n8A interference at A23 is consistent at all ligation time points, while the absence of n8A interference at A14 continues at all ligation time points (Figure 6B). The same consistency is seen for the 2F-A interferences at these sites (Figure 6B). Therefore, in addition to providing the expected interference at A38 in stabilized constructs, n8A also produces A38 interference at early times of the 2WJ/7NT selections. The equilibration results described above suggest that n8A interference at A38 is observed before 7NT substrates equilibrate throughout the selection pool via multiple rounds of substrate binding, ligation, cleavage and release.
Figure 6.
Time dependent NAIM study of the 2WJ/7NT construct/substrate pair. (A) Sequencing gel of ligation selections performed with the indicated analogs as a function of ligation reaction time. (B) Interference values for n8A (solid bars) and 2F-A (open bars) at the indicated residues as a function of ligation reaction time.
DISCUSSION
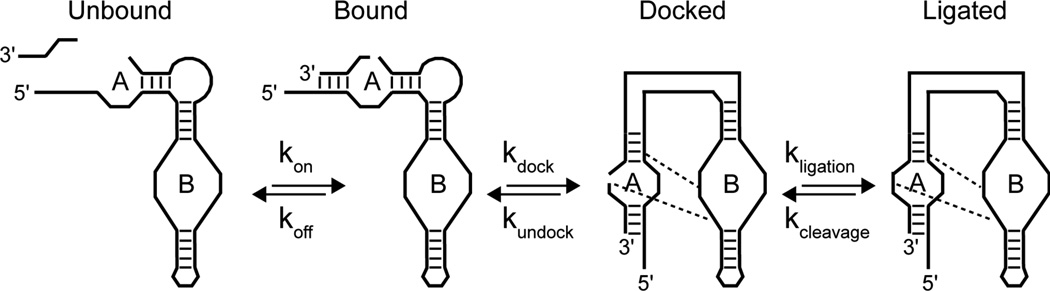
In the simplest model of hairpin ligation the reaction consists of three steps, i) binding of substrate via base pairing in helix I, ii) formation of the condensed docked state mediated by a number of specific interactions between loops A and B, and iii) the catalyzed attack of the free 5'-OH at G(+1) on the cyclic 2',3'-phosphate at the N(−1) position(8) (Figure 7). Hairpin catalyzed cleavage is simply the reverse of these steps, resulting in dissociated substrate and a cleaved hairpin RNA capable of another ligation reaction. Single molecule FRET studies have shown that the sequence of at least these steps can be obligate, indicating that bound substrates must dock before ligation and cleaved substrates must undock before release in certain constructs(38). Furthermore, the energetics of these steps are not easily separated(36), as indicated by the fact that more stably docked constructs also exhibit an internal equilibrium significantly shifted toward ligation(2). A challenge in interpreting nucleotide modification experiments is dissecting the contribution made by chemical and structural steps along the reaction pathway. This is particularly true for the hairpin ribozyme, where the rate limiting step has been shown to be construct dependent(38) and where some of the most detrimental base modifications are now known to be structural in origin(26).
Figure 7.
Minimum reaction sequence for hairpin ligation, consisting of substrate binding, the formation of a docked state requiring specific interactions between residues in loop A and B (dashed lines), and ligation between G(+1) and N(−1). All steps are reversible, with the equilibrium dependent on the energetics of substrate binding, the stability of the docked state, and the ribozyme catalyzed equilibrium between ligation and cleavage.
The introduction of nucleobase modifications further complicates this picture since substitution at one site with a given analog may introduce a defect in docking, while substitution at another site with the same analog may introduce a defect in ligation chemistry. In the context of NAIM experiments, this situation might result in strong interference with a given analog at two different sites arising from completely different origins. In an attempt to deconvolute these effects in the hairpin ligation reaction, we conducted a NAIM analysis with a structurally similar series of analogs using construct/substrate pairs expected to exhibit distinctly different kinetics in the steps prior to chemistry. The ligation chase experiments indicate that this is indeed the case under our NAIM selection conditions, and that the interference patterns observed represent a progression between two extremes. In the context of the 2WJ/7NT reaction, ligation reactions reach full equilibrium amongst the analog incorporated pool. This means any given substrate will complete a full reaction cycle (bind, dock, ligate, cleave, undock, and release) from several members of the analog incorporated pool before the reaction is quenched. In this extreme, members that stabilize the ligated docked state accumulate in the active fraction, and members deficient in either docking or chemistry are underrepresented. The ability of labeled substrates to exchange between members of the hairpin ribozyme pool through cleavage and release accentuates deficiencies in docking, since the ability of cleaved substrates to release is dependent on the formation of the undocked state for some constructs. Ligation selections carried out with the 4WJ/11NT substrate represent the opposite extreme. In these reactions labeled substrates bind members of the analog incorporated pool and are not released during the time course of the selection, whether ligated or not. Here the active fraction is determined primarily by the internal ligation/cleavage equilibrium. The 2WJ/11NT and 4WJ/7NT reaction pairs lie between these extremes. In the 2WJ/11NT pair, the docked state is destabilized relative to the 4WJ/11NT pair, but cleaved substrate is not released to equilibrate with other members of the selection pool. In the 4WJ/7NT pair, the docked state is stabilized relative to the 2WJ/7NT pair, but the undocked fraction will release cleaved substrate to equilibrate with the selection pool.
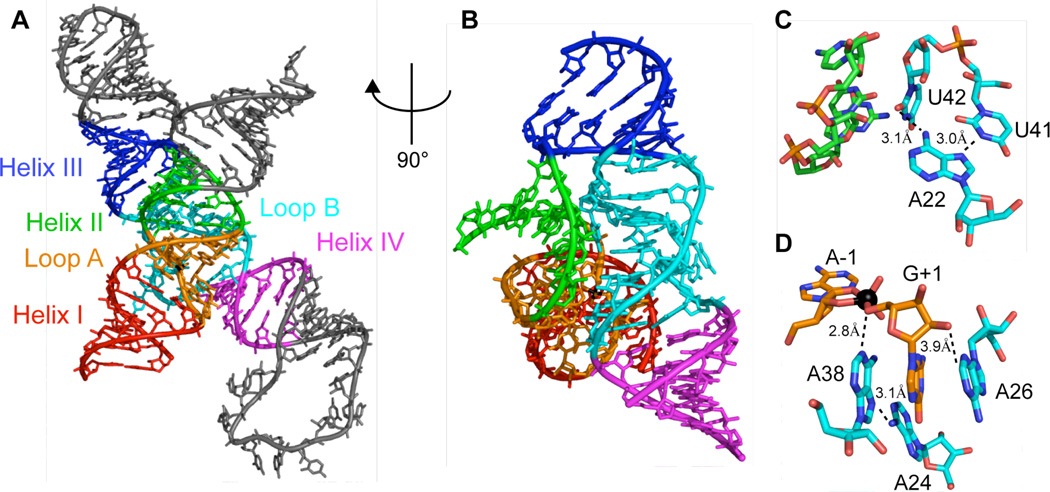
The construct dependence of the observed interferences can be used to differentiate those modifications that primarily affect docking stability from those that affect chemistry. For example, all of the adenosine residues in the A rich loop from A20 to A26 show strong interference with one or more analogs in the context of the 2WJ/7NT reaction pair. These interferences decrease systematically as more stable construct/substrate pairs are used in the selection. Tertiary interactions within the hairpin crystal structures readily explain many of these interferences. For example, the strong interference of 7-deaza substitutions at A22 likely arises from this base's involvement in the positioning of U42 at the interface of loop B and helix II in the docked state (Figure 8C). The unusual turn between U41 and U42 that allows U42 to bridge loop B and helix II relies on hydrogen bonds to the exocyclic amine and N7 nitrogen of A22. Removal of the N7 nitrogen at A22 disrupts this interaction, leading to a destabilized docked state and interference in less stable constructs. Many of the construct-dependent interferences can be explained in a similar manner by observed or anticipated contacts in the available crystal structures.
Figure 8.
(A) Crystal structure of a 4WJ construct with a vanadate transition state mimic(9), color coded by helices I-IV and loop A and B. (B) Rotated view emphasizing docking interface between loop B and helix II/loop A. (C) Involvement of A22 in orienting U42 at the loop B/helix II interface. (D) Adenosine residues in vicinity of G(+1) and the vanadate transition state mimic (black ball).
The primary goal of this study was to reinvestigate functional adenosine ionization with a new series of pKa perturbed analogs. The extreme sensitivity of interference values to construct/substrate stability suggests that interferences arising from catalytic steps are best identified in stabilized construct/substrate pairs. As noted above A38 is the only site that shows strong interferences that persist in the most stable constructs (Figure 4). The pattern of interference at A38 is consistent with N1 ionization, with all low pKa analogs showing interference at this site in the 4WJ/11NT reaction pair. However, as in our earlier study we failed to observe n8A interference in reactions with 2WJ constructs. One possible explanation for this result is that n8A substitution at A38 simultaneously slows the rate of ligation but increases the stability of the docked state. In this scenario a defect in catalysis would be compensated for by an increased lifetime in the active conformation. This advantage would be eliminated in constructs where the stability of the docked state equalizes the smaller stability differences introduced by analog substitution. The concept that ligation with n8A at A38 is kinetically slower but more favored at equilibrium in the 2WJ construct is supported by the time-dependent interference pattern seen with this analog (Figure 6B).
Taken together the construct and time dependence of n8A interference at A38 suggests functional ionization at this site was missed in our previous study because this analog stabilizes the docked state, masking its effect on catalysis in the least stable hairpin constructs. Although our NAIM data do not directly reveal the origin of this docking advantage they do suggest the effect is unique to A38, since it is the only site where n8A substitution produces a distinctive pattern of larger interferences (or smaller enhancements) in more stabilized constructs. Interestingly, A38 is also the only adenosine nucleotide that adopts a syn-glycoside conformation in any of the reported crystal structures(9, 10, 22). This rare nucleoside conformation is not observed at A38 in the isolated loop B NMR structure(24), suggesting an anti to syn transition is part of the large structural reorganization that occurs between the extended and docked states. The fact that n8A exhibits a decreased barrier of rotation to the syn conformation(39) provides a reasonable explanation for its selective advantage in destabilized constructs. Indeed, substrate helix docking can be rate limiting in minimal hairpin constructs under some conditions, with the undocked fraction existing in multiple states, one or more of which may involve an anti to syn transition at A38 along the folding pathway. The requirement of a syn conformation at A38 to form the fully docked state could also explain the interference produced by 7dA in destabilized constructs, since the observed hydrogen bond between the N7 of A38 and the N6 amine of A24 is also absent from the undocked NMR structure and acts to stabilize the syn conformation (Figure 8D).
Overall the interference data obtained in this study argue for functional ionization at A38, consistent with a wealth of structural data and a number of site-specific modification experiments. A38 was the only site that showed persistent and strong interference with low pKa analogs across a variety of construct/substrate pairs. As in our previous study we failed to observe interference with n8A at A38 in destabilized constructs. However, by employing stably docked constructs or by quenching destabilized constructs at early times, n8A interference at A38 was observed. These results suggest that n8A is deficient in the chemical step of ligation but compensates for slow chemistry by stabilizing the docked state. The origin of this docking advantage may be the increased propensity of n8A to form the required syn-glycoside conformation. In contrast, the low pKa fluoro-adenosine analogs produced strong and construct independent interference at A38, suggesting these analogs are better suited to identify sites of functional ionization in systems where structural rearrangements are closely coupled to catalytic steps.
ABBREVIATIONS AND TEXTUAL FOOTNOTES
- NAIM
Nucleotide Analog Interference Mapping
- 2WJ
two-way junction
- 4WJ
four-way junction
- NT
nucleotide
- FRET
Förster resonance energy transfer
- HDV
hepatitis delta virus
Footnotes
This work was supported in part by the National Science Foundation (MCB-0544255). I.T.S. acknowledges the NIH for a NRSA postdoctoral fellowship (GM-078764).
References
- 1.Fedor MJ. Comparative Enzymology and Structural Biology of RNA Self-Cleavage. Ann Rev Biophys. 2009;38:271–299. doi: 10.1146/annurev.biophys.050708.133710. [DOI] [PubMed] [Google Scholar]
- 2.Fedor MJ. Tertiary structure stabilization promotes hairpin ribozyme ligation. Biochemistry. 1999;38:11040–11050. doi: 10.1021/bi991069q. [DOI] [PubMed] [Google Scholar]
- 3.Buzayan JM, Gerlach WL, Bruening G, Keese P, Gould AR. Nucleotide-Sequence of Satellite Tobacco Ringspot Virus-RNA and its Relationship to Multimeric Forms. Virology. 1986;151:186–199. doi: 10.1016/0042-6822(86)90041-3. [DOI] [PubMed] [Google Scholar]
- 4.Spitale RC, Volpini R, Heller MG, Krucinska J, Cristalli G, Wedekind JE. Identification of an Imino Group Indispensable for Cleavage by a Small Ribozyme. J. Am. Chem. Soc. 2009;131:6093–6095. doi: 10.1021/ja900450h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spitale RC, Volpini R, Mungillo MV, Krucinska J, Cristalli G, Wedekind JE. Single-Atom Imino Substitutions at A9 and A10 Reveal Distinct Effects on the Fold and Function of the Hairpin Ribozyme Catalytic Core. Biochemistry. 2009;48:7777–7779. doi: 10.1021/bi9011622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macelrevey C, Salter JD, Krucinska J, Wedekind JE. Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme. RNA. 2008;14:1600–1616. doi: 10.1261/rna.1055308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salter J, Krucinska J, Alam S, Grum-Tokars V, Wedekind JE. Water in the active site of an all-RNA hairpin ribozyme and effects of Gua8 base variants on the geometry of phosphoryl transfer. Biochemistry. 2006;45:686–700. doi: 10.1021/bi051887k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson TJ, Nahas M, Araki L, Harusawa S, Ha T, Lilley DMJ. RNA folding and the origins of catalytic activity in the hairpin ribozyme. Blood Cell Mol Dis. 2007;38:8–14. doi: 10.1016/j.bcmd.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Rupert PB, Massey AP, Sigurdsson ST, Ferre-D'Amare AR. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- 10.Torelli AT, Krucinska J, Wedekind JE. A comparison of vanadate to a 2 '-5 ' linkage at the active site of a small ribozyme suggests a role for water in transition-state stabilization. RNA. 2007;13:1052–1070. doi: 10.1261/rna.510807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan E, Wilson TJ, Nahas MK, Clegg RM, Lilley DMJ, Ha T. A four-way junction accelerates hairpin ribozyme folding via a discrete intermediate. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9308–9313. doi: 10.1073/pnas.1233536100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walter NG, Burke JM, Millar DP. Stability of hairpin ribozyme tertiary structure is governed by the interdomain junction. Nat Struct Biol. 1999;6:544–549. doi: 10.1038/9316. [DOI] [PubMed] [Google Scholar]
- 13.Nesbitt S, Hegg LA, Fedor MJ. An unusual pH-independent and metal-ion-independent mechanism for hairpin ribozyme catalysis. Chem. Biol. 1997;4:619–630. doi: 10.1016/s1074-5521(97)90247-7. [DOI] [PubMed] [Google Scholar]
- 14.Kuzmin YI, Da Costa CP, Fedor MJ. Role of an active site guanine in hairpin ribozyme catalysis probed by exogenous nucleobase rescue. J. Mol. Biol. 2004;340:223–251. doi: 10.1016/j.jmb.2004.04.067. [DOI] [PubMed] [Google Scholar]
- 15.Nesbitt SM, Erlacher HA, Fedor MJ. The internal equilibrium of the hairpin ribozyme: Temperature, ion and pH effects. J. Mol. Biol. 1999;286:1009–1024. doi: 10.1006/jmbi.1999.2543. [DOI] [PubMed] [Google Scholar]
- 16.Fedor MJ. Structure and function of the hairpin ribozyme. J. Mol. Biol. 2000;297:269–291. doi: 10.1006/jmbi.2000.3560. [DOI] [PubMed] [Google Scholar]
- 17.Suydam IT, Strobel SA. Nucleotide Analog Interference Mapping. Methods Enzymol. 2009;468:3–30. doi: 10.1016/S0076-6879(09)68001-0. [DOI] [PubMed] [Google Scholar]
- 18.Waldsich C. Dissecting RNA folding by nucleotide analog interference mapping (NAIM) Nat. Protoc. 2008;3:811–823. doi: 10.1038/nprot.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cochrane JC, Strobel SA. Current Protocols in Nucleic Acid Chemistry. Vol. 2. New York: Wiley; 2004. [DOI] [PubMed] [Google Scholar]
- 20.Ryder SP, Strobel SA. Nucleotide analog interference mapping of the hairpin ribozyme: Implications for secondary and tertiary structure formation. J. Mol. Biol. 1999;291:295–311. doi: 10.1006/jmbi.1999.2959. [DOI] [PubMed] [Google Scholar]
- 21.Ryder SP, Oyelere AK, Padilla JL, Klostermeier D, Millar DP, Strobel SA. Investigation of adenosine base ionization in the hairpin ribozyme by nucleotide analog interference mapping. RNA. 2001;7:1454–1463. [PMC free article] [PubMed] [Google Scholar]
- 22.Rupert PB, Ferre-D'Amare AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 23.Cai ZP, Tinoco I. Solution structure of loop A from the hairpin ribozyme from tobacco ringspot virus satellite. Biochemistry. 1996;35:6026–6036. doi: 10.1021/bi952985g. [DOI] [PubMed] [Google Scholar]
- 24.Butcher SE, Allain FHT, Feigon J. Solution structure of the loop B domain from the hairpin ribozyme. Nat Struct Biol. 1999;6:212–216. doi: 10.1038/6651. [DOI] [PubMed] [Google Scholar]
- 25.Ryder SP, Strobel SA. Comparative analysis of hairpin ribozyme structures and interference data. Nucleic Acids Res. 2002;30:1287–1291. doi: 10.1093/nar/30.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferre-D'Amare AR. The hairpin ribozyme. Biopolymers. 2004;73:71–78. doi: 10.1002/bip.10516. [DOI] [PubMed] [Google Scholar]
- 27.Kuzmin YI, Da Costa CP, Cottrell JW, Fedor MJ. Role of an active site adenine in hairpin ribozyme catalysis. J. Mol. Biol. 2005;349:989–1010. doi: 10.1016/j.jmb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 28.Gaur S, Heckman JE, Burke JM. Mutational inhibition of ligation in the hairpin ribozyme: Substitutions of conserved nucleobases A9 and A10 destabilize tertiary structure and selectively promote cleavage. RNA. 2008;14:55–65. doi: 10.1261/rna.716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bevilacqua PC. Mechanistic considerations for general acid-base catalysis by RNA: Revisiting the mechanism of the hairpin ribozyme. Biochemistry. 2003;42:2259–2265. doi: 10.1021/bi027273m. [DOI] [PubMed] [Google Scholar]
- 30.Guo M, Spitale RC, Volpini R, Krucinska J, Cristalli G, Carey PR, Wedekind JE. Direct Raman Measurement of an Elevated Base pK(a) in the Active Site of a Small Ribozyme in a Precatalytic Conformation. J. Am. Chem. Soc. 2009;131:12908–12909. doi: 10.1021/ja9060883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suydam IT, Strobel SA. Fluorine substituted adenosines as probes of nucleobase protonation in functional RNAs. J. Am. Chem. Soc. 2008;130:13639–13648. doi: 10.1021/ja803336y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones FD, Strobel SA. Ionization of a critical adenosine residue in the Neurospora Varkud Satellite ribozyme active site. Biochemistry. 2003;42:4265–4276. doi: 10.1021/bi020707t. [DOI] [PubMed] [Google Scholar]
- 33.Smith MD, Collins RA. Evidence for proton transfer in the rate-limiting step of a fast-cleaving Varkud satellite ribozyme. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5818–5823. doi: 10.1073/pnas.0608864104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao ZY, McLeod A, Harusawa S, Araki L, Yamaguchi M, Kurihara T, Lilley DMJ. Nucleobase participation in ribozyme catalysis. J. Am. Chem. Soc. 2005;127:5026–2027. doi: 10.1021/ja0502775. [DOI] [PubMed] [Google Scholar]
- 35.Lafontaine DA, Wilson TJ, Zhao ZY, Lilley DMJ. Functional group requirements in the probable active site of the VS ribozyme. J. Mol. Biol. 2002;323:23–34. doi: 10.1016/s0022-2836(02)00910-5. [DOI] [PubMed] [Google Scholar]
- 36.Cottrell JW, Kuzmin YI, Fedor MJ. Functional analysis of hairpin ribozyme active site architecture. J. Biol. Chem. 2007;282:13498–13507. doi: 10.1074/jbc.M700451200. [DOI] [PubMed] [Google Scholar]
- 37.Nahas MK, Wilson TJ, Hohng SC, Jarvie K, Lilley DMJ, Ha T. Observation of internal cleavage and ligation reactions of a ribozyme. Nat. Struct. Mol. Biol. 2004;11:1107–1113. doi: 10.1038/nsmb842. [DOI] [PubMed] [Google Scholar]
- 38.Zhuang XW, Kim H, Pereira MJB, Babcock HP, Walter NG, Chu S. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 39.Singh P, Hodgson DJ. 8-Azaadenosine - Crystal-Structure of Its Monohydrate and Conformational-Analysis for Rotation around Glycosyl Bond. J. Am. Chem. Soc. 1977;99:4807–4815. doi: 10.1021/ja00456a045. [DOI] [PubMed] [Google Scholar]
- 40.Saenger W. Principles of nucleic acid structure. New York: Springer-Verlag; 1984. [Google Scholar]