Abstract
Introduction
Saliva is a body fluid that holds promise for use as a diagnostic fluid for detecting diseases. Salivary proteins are known to be heavily glycosylated and are known to play functional roles in the oral cavity. We identified N-linked glycoproteins in human whole saliva, as well as the N-glycoproteins in parotid, submandibular, and sublingual glandular fluids.
Materials and Methods
We employed hydrazide chemistry to affinity enrich for N-linked glycoproteins and glycopeptides. PNGase F releases the N-peptides/proteins from the agarose-hydrazide resin, and liquid chromatography–tandem mass spectrometry was used to identify the salivary N-glycoproteins.
Results
A total of 156 formerly N-glycosylated peptides representing 77 unique N-glycoproteins were identified in salivary fluids. The total number of N-glycoproteins identified in the individual fluids was: 62, 34, 44, and 53 in whole saliva, parotid fluid, submandibular fluid, and sublingual fluid, respectively. The majority of the N-glycoproteins were annotated as extracellular proteins (40%), and several of the N-glycoproteins were annotated as membrane proteins (14%). A number of glycoproteins were differentially found in submandibular and sublingual glandular secretions.
Conclusions
Mapping the N-glycoproteome of parotid, submandibular, and sublingual saliva is important for a thorough understanding of biological processes occurring in the oral cavity and to realize the role of saliva in the overall health of human individuals. Moreover, identifying glycoproteins in saliva may also be valuable for future disease biomarker studies.
Keywords: Proteomics, Mass spectrometry, Isoelectric focusing, N-linked glycoproteins, Whole saliva, Parotid saliva, Submandibular saliva, Sublingual saliva, Disease biomarker
Introduction
Glycosylation is a posttranslational modification that plays an important role in many cellular processes such as protein conformation, folding, transport, targeting, and stability [1]. Cell growth, differentiation, cell–cell communication, cellular trafficking, immune response, microbial pathogenesis, and other biological processes are influenced by glycosylation [2, 3]. Protein glycosylation is often found to vary with disease progression [4–6]. It is not surprising to note that a significant number of known and Food and Drug Administration-approved biomarkers for diseases are glycoproteins [7], including CA 15-3 [8] and c-erb-2 [9] for breast cancer, prostrate-specific antigen [10] for prostrate cancer, CA125 [11] for ovarian cancer, and carcinoembryonic antigen for ovarian, prostrate, and colon cancers [12]. Glycoproteins represent an interesting subproteome that can be mined to improve the chances for discovering putative robust, specific, and selective biomarkers for diseases.
Biofluids such as serum/plasma, saliva, and urine are excellent starting points for such biomarker discovery studies. Human saliva is gaining popularity as a body fluid for diagnostic purposes. Its ready availability, ease of collection, and ease of storage makes it a potential ideal candidate for use as a diagnostic medium. Whole saliva has contributions from three primary salivary glands: the parotid (PA), submandibular (SM), and sublingual (SL) glands, in addition to other minor contributions (e.g., from numerous minor glands in the lip, cheek, tongue, and palate, microbes, epithelial cells, nasal, and bronchial secretions, and serum products).
Many types of posttranslational modifications have been observed on salivary proteins [13, 14]. Some of the common modifications include glycosylation, phosphorylation, sulfation, acylation, deamidation, and proteolysis. Glycosylation is a posttranslational modification that has important functions in the oral cavity such as lubrication and protection of oral cavity and teeth from oral pathogens, chemicals, and mechanical wear and tear. Some of the salivary glycoproteins involved in lubrication include proline-rich glycoprotein and mucins. Proline-rich glycoproteins are primarily secreted by the PA glands and are N-glycosylated. The oligosaccharides make up to 50% of their weight and are responsible for their lubricating properties [15]. Mucins are also heavily O-glycosylated, which explains their viscous nature and their profound role in oral lubrication [16, 17]. The sugars also help mucins bind to surfaces in the mouth to protect them from chemicals, wear and tear, and microbes [18]. Other functions of salivary glycoproteins include binding to oral pathogens and eliminating them from the oral cavity; proline-rich glycoproteins, mucins, and salivary agglutinins have an especially important part in these functions. Proline-rich glycoproteins bind Fusobacterium nucleatum that causes periodontal diseases and may be responsible for its clearance from the mouth [19]. Mucins also bind and aggregate bacteria such as Heliobacter pylori and flush them from the oral cavity [20]. Salivary agglutinins efficiently adhere to and help rid the oral cavity of Streptococcus mutans [21]. In addition, agglutinins have shown an influenza A-neutralizing effect [22]. After secretion of saliva from the salivary glands into the mouth, glycoproteins may undergo deglycosylation. This might have interesting consequences for oral pathogens and the function of glycoproteins. Many bacteria are known to deglycosylate glycoproteins such as mucins and utilize the released sugars for their growth [23]. Some Streptococcus species secrete neuraminidases that cleave the terminal sialic acid from mucins. This affects the ability of mucins to bind and aggregate bacteria such as Streptococcus sanguis [24]. The removal of sialic acid may also help bacteria bind to sugars on glycoproteins and colonize in the oral environment [14].
Mapping the salivary glycoproteome will be an important step toward using saliva for disease diagnosis. Whole saliva is far easier to collect compared to ductal saliva, and whole saliva will likely have more applications for disease biomarker studies compared to PA, SM, and SL saliva. However, mapping the N-glycoproteome of PA, SM, and SL saliva is important for a thorough understanding of biological processes occurring in the oral cavity and to realize the role of saliva in the overall health of human individuals.
Several methods for global glycoprotein profiling of biofluids have been explored to date, and many of the same techniques have also been applied to enrich for disease biomarkers. Some of the common methodologies tested include lectin affinity purification [25], hydrazide-based chemical derivatization for glycoprotein capture, and hydrophilic affinity capture methods. Lectins are a class of proteins found in plants, bacteria, fungi, and animals that bind to specific oligosaccharides on glycoproteins [26]. Whereas the use of a single lectin can help to enrich for a small group of glycoproteins, the use of multiple lectins increases the chances for isolating a broad spectrum of glycoproteins. This method, termed multilectin affinity chromatography, has been used to map glycoproteins in the plasma and serum [27–29]. To enrich for glycoproteins to which lectins do not have a strong specificity, the use of serial lectin affinity chromatography (SLAC) has been reported [30]. Because there are few reported lectins with sufficient selectivity for O-linked glycoproteins, SLAC has been used to isolate and identify O-linked glycopeptides in human serum [30]. The use of a broad-spectrum lectin such as concanavilin A was used to deplete N-linked glycoproteins and to enrich for O-linked glycoproteins. Jacalin is a lectin that has affinity for the GalNAc core of O-linked glycans but also for high mannose-type sugars found in N-linked glycoproteins. The subsequent application of the N-glycoprotein-depleted sample to a Jacalin column improved recovery and identification of O-linked glycoproteins [30]. Lectin affinity chromatography has also been employed to search for disease biomarkers in human plasma and serum [31, 32].
Phenyl boronic acid is considered a pseudolectin because it covalently attaches to cis-diol containing groups on the sugars of glycoproteins. This bond is fairly stable under alkaline conditions. Thus, boronic acids can be utilized to select for a broader group of glycoproteins including N-, O-, and C-linked glycoproteins. Lectins and boronic acid can be combined in a single experiment to enrich for glycoproteins [33]. Sparbier and coworkers used a combination of lectin and boronic acid magnetic beads to affinity select glycoproteins from human serum [34].
Hydrophilic affinity-based capture methods have been extended to isolate glycoproteins from complex mixtures. Because sugars are very hydrophilic, glycoproteins lend themselves well to being captured onto a hydrophilic stationary phase. Several hydrophilic materials have been tested in the past. Some of them include carbohydrate gel matrices such as sepharose and cellulose and hydrophilic interaction chromatography (HILIC) columns. Cellulose and sepharose matrices were successfully employed to isolated glycoproteins from human serum [35]. HILIC using a resin containing a polyhydroxyethyl aspartamide group has been used to separate sialylated glycopeptides from recombinant human interferon-γ [36]. Hagglund and coworkers used a resin with a covalently bound neutral, zwitterionic sulfobetaine functional group to perform zwitterion chromatography–HILIC (ZIC-HILIC). Glycopeptides were separated via ZIC-HILIC from nonglycopeptides in trypsin digest mixtures and were then partially deglycosylated to identify glycoproteins in human plasma [37, 38].
Larsen and coworkers published an elegant method to isolate sialic acid-containing glycoproteins in plasma and whole saliva. Their method employs titanium dioxide, which has an affinity for acidic groups. TiO2 has been used commonly to purify and measure phosphoproteins [39]. However, it also binds to sialic acid-containing glycoproteins. After removing phospho-groups on phosphopeptides by alkaline phosphatase treatment, the peptide mixture was passed through the TiO2 column to isolate sialated glycopeptides, and the glycopeptides were analyzed subsequently by mass spectrometry (MS). Using this method, sialated proteins in plasma and whole saliva were identified [40].
Zhang and coworkers published a landmark paper describing the isolation of formerly N-linked glycopeptides from complex mixtures using hydrazide chemistry [41]. This method has gained popularity over the past few years. Sugars on glycoproteins are first oxidized and immobilized onto an agarose–hydrazide resin. The resin is then washed to remove nonspecifically bound proteins. The proteins bound to the resin are digested with trypsin, and tryptic peptides not covalently bound to the column are removed by extensive washing. The bound tryptic N-glycopeptides are then released using the enzyme PNGase F, and the formerly N-glycosylated peptides are analyzed by MS. The identification of the formerly N-glycosylated peptide is confirmed by the presence of an N–X–S/T motif on the peptide sequence (where X denotes any amino acid residue except proline) and the conversion of asparagines at the site for glycosylation to aspartic acid resulting in a mass increase of +1 Da. This method has since been widely applied toward the study of the N-glycoproteome of various human body fluids including plasma [42], serum [43], whole saliva [44], and cerebrospinal fluid [45]. In 2007, two independent groups published slightly modified versions of the original method described by Zhang et al. [41]. The similarity between the two methods was that the proteins initially were digested to tryptic peptides before being coupled onto the agarose–hydrazide resin. In the method published by Sun et al., excess sodium periodate, used to oxidize sugars on glycopeptides, was quenched with sodium sulfite. The method was used to identify unique proteins from microsomal fractions of the cisplatin-resistant ovarian cancer cell lines [46]. Alternately, Tian et al. removed remaining sodium periodate by desalting the oxidized glycopeptides using a C18 column [47]. This technique was successfully used to isolate N-linked glycopeptides from mouse plasma including some proteins that are found in low abundance in human plasma [48]. There are also differences in the wash step to remove nonspecifically bound peptides from the agarose–hydrazide resin, but the basic outline of the two procedures remains similar.
Our group previously reported, using the method of Zhang et al. [41], a global N-glycoproteome analysis of whole-saliva proteins using the hydrazide capture technique and MS, identifying 84 formerly N-glycosylated peptides from 45 glycoproteins [44]. Larsen et al. used the TiO2 enrichment strategy to isolate the whole salivary sialome and identified 97 sites of N-linked glycosylation [40]. In this paper, we extend the salivary glycoprotein catalogue using the modified hydrazide capture method of Sun et al. [46], i.e., the capture of tryptic N-glycopeptides and MS measurement of formerly N-linked glycopeptides. In addition, we compare for the first time the N-linked glycoproteins identified in PA, SM, and SL saliva.
Materials and Methods
Chemicals and Reagents
The chemicals were mostly purchased from Sigma (St. Louis, MO, USA), unless stated otherwise. Affigel Hz hydrazide gel, coupling buffer, and dithiothreitol (DTT) were obtained from Bio-Rad (Hercules, CA, USA). Tris-(2-carboxyethyl)-phosphine (TCEP) and trifluoroacetic acid (TFA) were obtained from Pierce (Rockford, IL, USA). Glycerol-free PNGase F was obtained from New England Biolabs (Ipswich, MA, USA). Sequencing-grade trypsin was procured from Promega (Madison, WI, USA).
Saliva Collection
Whole saliva was collected from healthy nonsmoking adults in the morning, at least 2 h after the last intake of food. The mouth was rinsed with water immediately before the collection. Whole saliva was collected and placed on ice. Protease cocktail inhibitor (1 µL/mL of whole saliva) was added to saliva immediately after collection to minimize protein degradation. Whole saliva was then centrifuged at 12,000 rpm at 4°C for 10 min. The supernate was collected and stored at −80°C. The pellet was saved for future analysis.
For collecting PA, SM, and SL saliva, 10 adult subjects of various ethnic and racial backgrounds, ranging in age from 22 to 30 years, were recruited. Saliva collection took place on a monthly basis and was performed between the hours of 9 and 11 a.m. Stimulated PA, SM, and SL saliva were collected by the repeated application of an aqueous citric acid solution (2%). PA saliva was obtained as the ductal secretion by using a cup-like device [49]. Separate SM and SL secretions were acquired by using a saliva collector, described by Wolff and coworkers, that was fitted with a sterile 100-µL-pipette tip [50]. Collection volumes over a 10-min period ranged from 500 to 2,000 µL for PA saliva, 50 to 100 µL for SL saliva, and 100 to 500 µL for SM saliva. PA, SM, and SL saliva from five subjects were pooled before carrying out the experiments.
Solution Isoelectric Focusing Fractionation
The procedure for solution isoelectric focusing (IEF) fractionation has been described previously [44]. Briefly, proteins in whole saliva were precipitated by mixing with four times the volume of cold ethanol and incubating at −20°C overnight. The mixture was centrifuged at 13,000 rpm for 15 min at 4°C. The pellet was resuspended in Zoom 2D protein solubilizer (Invitrogen, Carlsbad, CA, USA), protease inhibitor (Roche Diagnostic, Indianapolis, IN, USA), Tris base, DTT, and water and sonicated on ice. The pH of the solution was adjusted to pH 8.5–8.7 with 1 M Tris base and then incubated for 15 min at room temperature with shaking. Proteins were alkylated with 99% dimethylacrylamide (DMA) at room temperature for 30 min, as suggested by the protocols from Invitrogen. To quench excess DMA, DTT was added and incubated at room temperature for 5 min. After centrifuging the sample for 30 min at 13,400 rpm at 4°C, the supernate was collected. The protein concentration was determined by the noninterfering protein assay (Geno Technology, St. Louis, MO, USA). The concentration was measured to be approximately 1.5 mg/mL.
The solution of proteins in the Zoom-2D solubilizer (1.5 mg/mL, 400 µL) was diluted to a final concentration of 0.6 mg/mL in diluted buffer (Zoom IEF denaturant, Zoom focusing buffer pH 3–7, Zoom focusing buffer, pH 7–12, and 5 µL 2M DTT). Zoom IEF fractionation was performed in the standard format (pH 3.0 to 10). The diluted sample was loaded into each of the five chambers of the fractionator and subjected to solution-phase IEF. Five fractions (pI 3–4.6, 4.6–5.4, 5.4–6.2, 6.2–7.0, and 7.0–10.0) were obtained after the procedure. Proteins from each fraction were precipitated by mixing with 70% acetone, incubating at −20°C for 3–4 h, and centrifuging at 13,000 rpm for 30 min.
Glycoprotein Enrichment
Proteins from saliva that were not previously pI fractionated were isolated by precipitation using ethanol. Proteins from Zoom IEF-separated fractions were obtained by acetone precipitation. The salivary proteins isolated by either method were resuspended in coupling buffer (100 mM sodium acetate, 150 mM sodium chloride, pH 5.5). Sodium periodate was added to a final concentration of 15 mM. The solution was incubated in the dark at room temperature for 1 h. Glycerol was added to a final concentration of 20 mM to quench any excess sodium periodate remaining in the solution. The mixture was incubated for 15 min with mixing at room temperature. To remove remaining sodium periodate, the solution was dialyzed using a 3.5-kDa dialysis cassette against 1× coupling buffer at 4°C overnight. The hydrazide resin was equilibrated by washing with 3 vol of water and 6 vol of coupling buffer. The proteins were added to the resin and coupled overnight by incubating overnight at room temperature with shaking. The resin was then allowed to settle, and the supernate containing uncoupled nonglycoproteins was discarded. The resin was washed six times with urea buffer A containing 8 M urea, 200 mM Tris, 0.05% sodium dodecyl sulfate, and 5 mM ethylenediamine tetraacetic acid (pH 8.3). The proteins on the resin were reduced with a solution of 10 mM TCEP in urea buffer A. The reduced proteins were alkylated with 50 mM iodoacetamide in urea buffer. The resin was then washed six times with urea buffer B (1 M urea, 25 mM Tris, pH 8.3). The resin was resuspended in urea buffer B. Trypsin was added to the solution, and the proteins attached to the resin were digested at 37°C overnight with shaking. The nonglycopeptides released by trypsin digestion were removed by washing three times with 1.5 M NaCl, 80% acetonitrile (ACN)/0.1% TFA, methanol, and water and six times with 100 mM ammonium bicarbonate. The resin was resuspended in 100 mM ammonium bicarbonate. The N-linked glycopeptides were released by adding PNGase to the resin and incubating at 37°C overnight. The resin was washed twice with 80% ACN solution. The washes were combined, and the released glycopeptides were dried by vacuum centrifugation. The peptides were then resuspended in 0.1% formic acid (FA) and analyzed by one-dimensional (1D) liquid chromatography (LC)–MS/MS.
Glycopeptide Enrichment
The method used followed that described by Sun et al. [46]. Proteins from whole saliva, PA, SM, and SL saliva were precipitated using cold ethanol. The pellets were resuspended in 50 mM ammonium bicarbonate buffer (pH 7.8) with sonication. TCEP was added to a final concentration of 10 mM at room temperature to reduce disulfide bonds (TCEP is preferred over DTT in this case because elevated temperatures required by DTT–disulfide reduction often causes protein precipitation from salivary fluids). The solution was incubated at room temperature for 30 min with shaking. Iodoacetamide (50 mM) was added to alkylate the reduced cysteines, and the mixture was incubated at room temperature for 30 min with shaking. Afterwards, DTT (25 mM) was added to the solution to quench any remaining unreacted iodoacetamide. The mixture was incubated at room temperature for 30 min. Trypsin was added (1:50 by weight) to digest the proteins in solution, and the solution was incubated at 37°C overnight. The tryptic peptide mixture was then acidified with FA to bring the pH below 4.0. The tryptic peptides were desalted using a C18 Sep-Pak reverse-phase (RP) column (Waters, Milford, MA, USA). Peptides were then dried and resuspended in coupling buffer (100 mM sodium acetate, 150 mM sodium chloride, pH 5.5). Sodium periodate was added to a final concentration of 10 mM to oxidize cis-diol groups on sugars. The mixture was incubated in the dark at room temperature for 1 h. Any remaining unreacted sodium periodate was quenched with sodium sulfite to a final concentration of 20 mM. The solution was incubated at room temperature for 10 min. The mixture was then added to agarose–hydrazide resin to couple the peptides to the resin, and the coupling reaction was allowed to proceed overnight at room temperature. The agarose-hydrazide resin was transferred to a Handee minispin column (Pierce, Rockford, IL, USA) and washed with 1.5 M NaCl, 80% ACN, methanol, water, and freshly prepared 100 mM ammonium bicarbonate solution. The resin was incubated overnight at 37°C with PNGase F in 100 mM ammonium bicarbonate to release formerly N-glycosylated peptides. Afterwards, the resin was washed twice with 80% ACN solution. The washes were combined, and the released glycopeptides were dried in the vacuum centrifuge. The formerly N-glycosylated peptides were then resuspended in 1% FA and analyzed by 1D- and two-dimensional (2D) LC-MS/MS.
Liquid Chromatography–Tandem Mass Spectrometry
LC-MS/MS and 2D LC-MS/MS were performed on a QSTAR XL (QqTOF) mass spectrometer (Applied Biosystems, Foster City, CA, USA) equipped with a nanoelectrospray (Protana, Odense, Denmark) interface and an LC Packings/Dionex (Sunnyvale, CA, USA) nano-LC system.
For 1D LC-MS/MS, the nano-LC was equipped with a set of homemade precolumns (75 µm× 10 mm) and a column (75 µm× 150 mm) packed with Jupiter Proteo C12 resins (particle size 4 µm, Phenomenex, Torrance, CA, USA). The peptides were dried and redissolved in 0.1% FA solution. For each LC-MS/MS run, typically, 6 µL sample solution was loaded onto the precolumn. The precolumn was washed with the loading solvent containing 0.1% FA for 4 min before the sample was injected onto the LC column. The eluents used for the LC were water containing 0.1% FA (A) and 95% ACN/water containing 0.1% FA (B). The flow was 200 nL/min. The following analytical LC gradient was used for analyzing the formerly N-glycosylated peptides obtained by the glycopeptide enrichment method: 3–21% B for 36 min, 21–35% B for 14 min, and 36–80% B for 4 min and held at 80% B for 10 min. The column was equilibrated with 3% B for 16 min before the next run. The gradient used for analyzing formerly N-glycosylated peptides obtained by the glycoprotein enrichment method was 3–35% B for 72 min and 35–80% B for 18 min and maintained at 80% B for 9 min.
For the online 2D LC-MS/MS, 20 µL of sample solution was loaded onto a strong cation exchange (SCX) precolumn (Luna SCX resin, particle size 5 µm, 150 µm× 5 mm, Phenomenex) before transfer to a RP precolumn (Jupiter Proteo C12 resin, particle size 4 µm, 150 µm× 5 mm, Phenomenex) and RP analytical column (Jupiter Proteo C12 resin, particle size 4 µm, 75 µm× 150 mm, Phenomenex). Seven concentrations of ammonium acetate solutions (50, 100, 200, 400, 600, 1,000, and 2,000 mM) were injected (4 µL) onto the SCX precolumn for the step gradient elution of peptides.
The RP precolumn was used to preconcentrate and desalt each peptide fraction eluted from the SCX column prior to nano-RP LC separation. SCX fractions were loaded onto the RP precolumn with the following gradient: 3% B for 6 s, 6–24% B for 18 min, 24–36% B for 6 min, and 36–80% B for 2 min and maintained at 80% B for 8 min. The column was equilibrated with 3% B for 15 min prior to the next run. SCX fractions were separated on the analytical RP column (200 nL/min) with the following gradient: 3% B for 5 min, 3–6% B for 6 s, 6–24% B for 18 min, 24–36% B for 6 min, and 36–80% B for 2 min and held at 80% B for 8 min.
For online MS/MS analyses, a Proxeon nanobore stainless steel online emitter (inner diameter=30 µm) was used for electrospraying with the voltage set to 1,900 V. Peptide product ion mass spectra were recorded during LC-MS/MS by information-dependent analysis on the QSTAR XL mass spectrometer. Argon was employed as the collision gas. Collision energies for maximum fragmentation were automatically calculated using empirical parameters based on the charge and mass-to-charge ratio of the precursor peptide.
Protein identification was accomplished utilizing the Mascot database search engine (Matrix Science, London, UK). For search the sequence databases, the following variable modifications were set: carbamidomethylation of cysteines, oxidization of methionines and enzyme-catalyzed conversion of asparagines to aspartic acid at the site of carbohydrate attachment on asparagines, conversion of N-terminal glutamate and aspartate to pyro-Glu, and cyclization of N-terminal cysteine. DMA modification of cysteine was set as one of the parameters in experiments where Zoom IEF fractionation had been done prior to glycoprotein pulldown. Searches for peptides obtained by the glycoprotein enrichment method were performed against the Human IPI database. For the searches, one missed tryptic cleavage was tolerated, and the peptide and MS/MS mass tolerance was set as 0.3 Da. A Mascot score of greater than 20 was considered a significant match. All matching peptide MS/MS spectra were manually examined to verify the accuracy of the identification. Positive protein identification was based on standard Mascot criteria for statistical analysis of the LC-MS/MS data. For samples obtained by the glycopeptide enrichment method, searches were done against the Human IPI database (version 3.39) and its reverse decoy database. Only peptides above Mascot’s homology or identity threshold were considered. The validity of the formerly N-glycosylated peptides were confirmed by the presence of a consensus N–X–(S/T) where X is any amino acid except proline and the conversion of asparagines to aspartic acid at the site of former N-glycosylation resulting in a mass change of +1 Da. With MS/MS tolerances set to 0.3 Da, a 1-Da difference caused by deamidation of Asn to Asp is readily detected from measurements performed on the QTOF mass spectrometer.
Results
Isolation of N-linked Glycoproteins
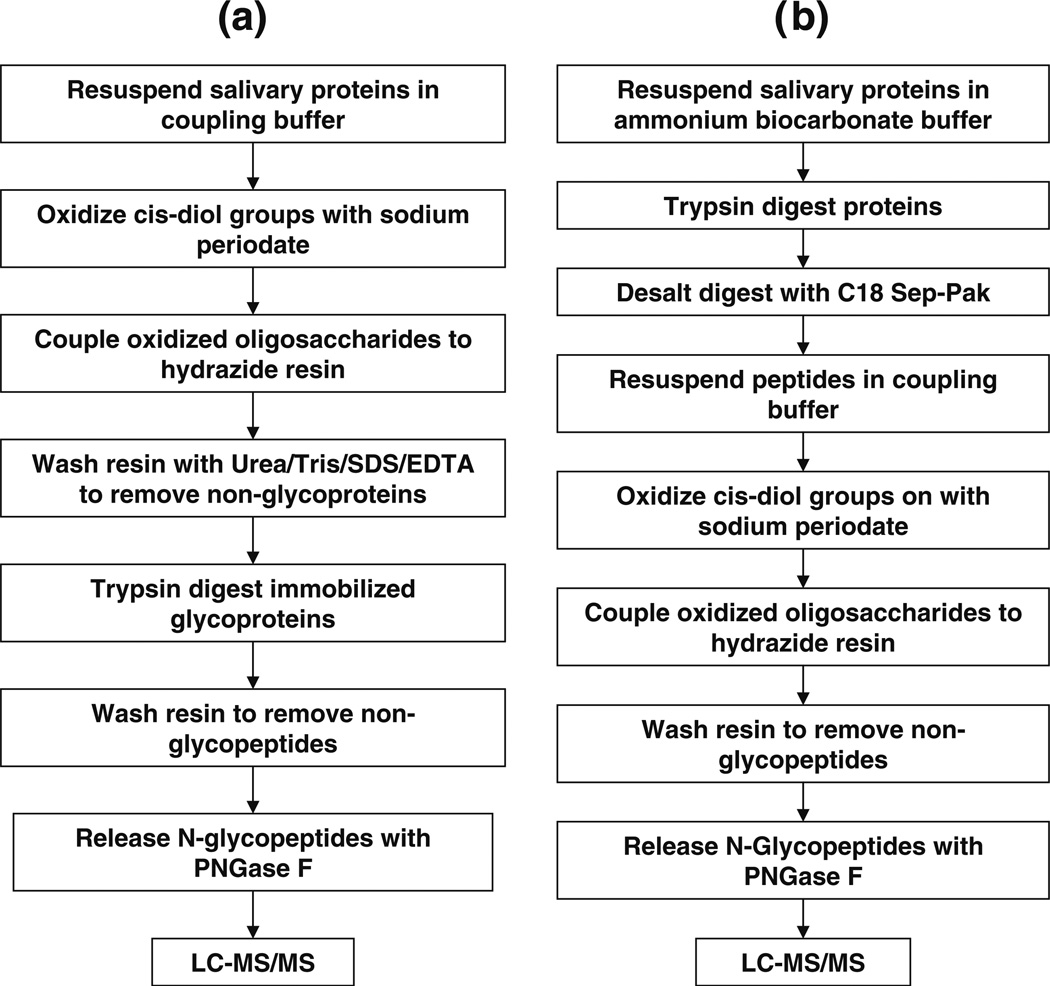
In our previous study on salivary N-linked glycoproteins [44], we used the agarose–hydrazide glycoprotein pulldown technique of Zhang et al. [41] to characterize human whole saliva (we will henceforth refer to this technique as the “Zhang method”). Proteins from whole saliva were isolated in two different ways. In the first method, proteins in whole saliva were precipitated with ethanol, and glycoprotein pulldown proceeded thereafter. The second method involved the preseparation of proteins into five different pI fractions of 3–4.6, 4.6–5.4, 5.4–6.2, 6.2–7, and 7–10, by solution IEF fractionation. The proteins in each pI fraction were then precipitated using acetone. The steps employed for glycoprotein pulldown in our previous study using the Zhang method were: (a) resuspending whole salivary proteins in the coupling buffer, (b) oxidizing saccharides on the glycoproteins with sodium periodate, (c) quenching excess sodium periodate with glycerol and removing any remaining sodium periodate by dialyzing against coupling buffer, (d) coupling glycoproteins to the agarose–hydrazide resin, (e) washing nonglycosylated proteins, (f) digesting proteins attached to the hydrazide resin with trypsin, (g) washing away the nonglycopeptides, (h) releasing the formerly N-glycosylated proteins from the hydrazide resin with PNGase F, and (i) identifying the formerly N-glycopeptides by 1D LC-MS/MS (Fig. 1a). A total of eight repetitions were performed for the glycoprotein pulldown experiments where saliva proteins were previously precipitated by ethanol precipitation. Five repetitions of the IEF fractionation followed by glycoprotein pulldown were performed.
Fig. 1.
Steps involved in the isolation of formerly N-glycosylated peptides using the a Zhang method and b the Sun method
In this current study, we applied a modified version of the glycoprotein pulldown method [46] to extend our catalogue of whole-saliva N-linked glycoproteins and developed a new catalogue of N-glycoproteins from segregated PA, SM, and SL fluids (we will refer to this modified technique as the “Sun method” in this paper). The new method entails proteolytic digestion of proteins prior to coupling to the agarose–hydrazide resin. The other difference lies in that after the oxidation of sugars, any unreacted sodium periodate is quenched with sodium sulfite. This obviates the need to remove sodium periodate by dialysis or using a desalting column as performed in the previous study. In this current study, the salivary proteins were precipitated using ethanol. Thus, the steps to isolate N-glycoproteins are: (a) resuspending salivary proteins in ammonium bicarbonate buffer, (b) digesting proteins with trypsin, (c) desalting the tryptic peptides using a C18 column, (d) oxidizing sugars on glycopeptides with sodium periodate, (e) quenching any remaining sodium periodate with sodium sulfite, (f) coupling glycopeptides onto the agarose–hydrazide column, (g) washing away nonglycopeptides, (h) releasing formerly N-linked glycopeptides with PNGase F, and (i) identifying formerly N-linked glycopeptides using 1D and 2D LC-MS/MS (Fig. 1b). A total of eight 1D LC-MS/MS runs were carried out. Some of the samples were combined, and four additional 2D LC-MS/MS experiments were performed. Data obtained from both 1D and 2D LC-MS/MS runs were combined. In both methods, the formerly N-glycosylated peptide identifications were validated by the presence of the consensus N–X–(S/T) motif and conversion of asparagines to aspartic acid at the site of N-glycosylation. We applied the Sun method to identify N-glycoproteins in whole saliva, PA, SM, and SL saliva.
Identification of N-Glycoproteins in Whole Saliva
In our previous study on N-linked glycoproteins in human whole saliva, we reported 84 formerly N-linked glycopeptides from 44 unique N-glycoproteins (Table 1) [44]. Using the newer Sun method, we identified 80 formerly N-glycosylated peptides from 46 unique N-linked glycoproteins (Table 1). However, comparing our previously identified N-glycoproteins using the Zhang method and new data set obtained by the Sun method, we found that 42 formerly N-glycosylated peptides from 28 N-glycoproteins were identified by both methods, 42 formerly N-glycosylated peptides from 16 N-glycoproteins were identified by the Zhang method uniquely, and 38 formerly N-glycopeptides from 18 N-glycoproteins were identified uniquely by the Sun method. Combining all of the N-glycoproteins identified by both methods, we have thus far identified 122 formerly N-glycosylated peptides from 62 unique N-glycoproteins (Fig. 2a,b). Figure 2c shows the LC-MS/MS spectrum of a formerly N-glycosylated peptide GDQLILNLNN*ISSDR (where the asterisk denotes the site of N-glycosylation) from Isoform 1 of long palate, lung, and nasal epithelium carcinoma-associated protein 1 identified in whole saliva using the Sun method, whereas this formerly N-glycopeptide was not detected in PA, SM, or SL saliva (vide infra).
Table 1.
Comparison of N-linked glycoproteins identified in whole saliva using the Zhang method and the Sun method
| IPI Accession number |
Protein name | MW (Da) | Formerly N-glycosylated peptide sequencea |
Formerly N-glycosylated peptides identified |
Nonformerly N-glycosylated peptides identified |
||
|---|---|---|---|---|---|---|---|
| Zhang methodb |
Sun methodb |
Zhang method |
Sun method |
||||
| IPI00002818 | Splice isoform 1 of kallikrein 11 | 27,448 | CAN*ITIIEHQK | No | Yes (35) | – | 0 |
| IPI00003351 | Extracellular matrix protein 1 | 60,635 | HIPGLIHN*MTAR | No | Yes (43) | – | 0 |
| IPI00004573 | Polymeric-immunoglobulin receptor | 83,232 | VPGN*VTAVLGETLK | Yes (101) | Yes (113) | 18 | 3 |
| WNN*TGCQALPSQDEGPSK | Yes (72) | Yes (62) | |||||
| IIEGEPNLKVPGN*VTAVLGETLK | Yes (49) | Yes (112) | |||||
| LSLLEEPGN*GTFTVILNQLTSR | Yes (60) | Yes (99) | |||||
| ANLTNFPEN*GTFVVNIAQLSQDDSGR | Yes (70) | Yes (108) | |||||
| QIGLYPVLVIDSSGYVNPN*YTGR | Yes (95) | Yes (112) | |||||
| VPGN*VTAVLGETLKVPCHFPCK | No | Yes (50) | |||||
| IPI00007244 | Myeloperoxidase | 83,815 | SCPACPGSN*ITIR | Yes (46) | No | 2 | – |
| SYN*DSVDPR | Yes (41) | No | |||||
| IPI00011229 | Cathepsin D | 44,524 | GSLSYLN*VTR | No | Yes (49) | – | 0 |
| IPI00855918 | Mucin 5, subtype B, tracheobronchial | 590,122 | VVLLDPKPVAN*VTCVNK | Yes (60) | Yes (80) | 37 | 0 |
| AQGLVLEASN*GSVLINGQR | Yes (118) | Yes (120) | |||||
| GN*CTYVLMR | Yes (32) | Yes (39) | |||||
| AFGQFFSPGEVIYN*K | No | Yes (72) | |||||
| AFGQFFSPGEVIYN*KTDR | Yes (49) | Yes (72) | |||||
| DIECQAESFPN*WTLAQVGQK | Yes (82) | Yes (86) | |||||
| QVNETWTLEN*CTVAR | Yes (47) | Yes (67) | |||||
| LDGPTEQCPDPLPLPAGN*CTDEEGICHR | Yes (65) | Yes (30) | |||||
| FGN*LSLYLDNHYCTASATAAAAR | Yes (49) | No | |||||
| LPYSLFHN*NTEGQCGTCTNNQR | Yes (32) | No | |||||
| IPI00012887 | Cathepsin L | 37,540 | YSVAN*DTGFVDIPK | No | Yes (57) | – | 0 |
| IPI00013972 | Carcinoembryonic antigen-related cell adhesion molecule 8 | 38,130 | LFIPN*ITTK | Yes (47) | No | 0 | – |
| IPI00019943 | Afamin | 69,024 | DIENFN*STQK | Yes (41) | No | 0 | – |
| IPI00020091 | Alpha-1-acid glycoprotein 2 | 23,588 | QNQCFYN*SSYLNVQR | Yes (90) | No | 0 | – |
| IPI00020487 | Extracellular glycoprotein lacritin | 14,237 | QFIEN*GSEFAQK | Yes (32) | No | 1 | – |
| IPI00020986 | Lumican | 38,405 | LHINHNN*LTESVGPLPK | No | Yes (59) | – | 0 |
| IPI00021891 | Splice Isoform 1 of Fibrinogen gamma chain | 51,479 | DLQSLEDILHQVEN*K | No | Yes (58) | – | 0 |
| VDKDLQSLEDILHQVEN*K | No | Yes (70) | |||||
| IPI00022395 | Complement component C9 | 63,133 | AVN*ITSENLIDDVVSLIR | No | Yes (56) | – | 0 |
| IPI00022417 | Leucine-rich alpha-2-glycoprotein | 38,944 | KLPPGLLAN*FTLLR | Yes (62) | Yes (72) | 1 | 0 |
| IPI00022429 | Alpha-1-acid glycoprotein 1 | 24,777 | QDQCIYN*TTYLNVQR | Yes (105) | No | 0 | – |
| IPI00022431 | Alpha-2-HS-glycoprotein | 39,300 | AALAAFNAQNN*GSNFQLEEISR | Yes (64) | Yes (26) | 1 | – |
| VCQDCPLLAPLN*DTR | Yes (40) | No | |||||
| IPI00022463 | Serotransferrin | 77,000 | CGLVPVLAENYN*K | No | Yes (69) | 9 | 0 |
| QQQHLFGSN*VTDCSGNFCLFR | Yes (62) | No | |||||
| IPI00022488 | Hemopexin | 51,643 | ALPQPQN*VTSLLGCTH | Yes (56) | Yes (58) | 1 | 0 |
| SWPAVGN*CSSALR | Yes (47) | No | |||||
| IPI00022974 | Prolactin inducible protein | 16,562 | TFYWDFYTN*R | Yes (50) | No | 5 | – |
| IPI00023673 | Galectin-3 binding protein | 65,289 | ALGFEN*ATQALGR | Yes (83) | Yes (80) | 2 | 0 |
| AAIPSALDTN*SSK | Yes (64) | Yes (70) | |||||
| GLN*LTEDTYKPR | No | Yes (49) | |||||
| EPGSN*VTMSVDAECVPMVR | No | Yes (41) | |||||
| TVIRPFYLTN*SSGVD | Yes (57) | No | |||||
| YKGLN*LTEDTYKPR | Yes (50) | No | |||||
| IPI00025023 | Lactoperoxidase | 80,237 | KPSPCEFIN*TTAR | Yes (34) | Yes (53) | 8 | 0 |
| IVGYLNEEGVLDQN*R | Yes (77) | Yes (89) | |||||
| LRN*LSSPLGLMAVNQEVSDHGLPYLPYDSK | No | Yes (51) | |||||
| IPI00025753 | Desmoglein 1 | 113,644 | TGEIN*ITSIVDR | Yes (36) | No | 0 | – |
| IPI00025846 | Splice isoform 1 of desmocollin-2 | 99,899 | NGIYN*ITVLASDQGGR | No | Yes (88) | 1 | 0 |
| AN*YTILK | Yes (49) | Yes (48) | |||||
| AN*YTILKGNENGNFK | Yes (26) | No | |||||
| LKAIN*DTAAR | Yes (31) | No | |||||
| IPI00027412 | Carcinoembryonic antigen-related cell adhesion molecule 6 | 37,214 | LQLSNGN*MTLTLLSVK | Yes (63) | No | 0 | – |
| IPI00027486 | Carcinoembryonic antigen-related cell adhesion molecule 5 | 76,748 | TLTLFN*VTR | Yes (42) | No | 0 | – |
| IPI00031019 | Cystatin-related epididymal spermatogenic protein | 16,265 | KLKPVN*ASNANVK | Yes (43) | No | – | – |
| IPI00031121 | Carboxypeptidase E | 53,117 | DLQGNPIAN*ATISVEGIDHDVTSAK | No | Yes (69) | – | 0 |
| GN*ETIVNLIHSTR | No | Yes (43) | |||||
| IPI00031547 | Desmoglein 3 | 107,435 | NTGDIN*ITAIVDR | No | Yes (80) | 0 | 0 |
| LPAVWSITTLN*ATSALLR | Yes (39) | No | |||||
| DSTFIVN*K | Yes (45) | No | |||||
| IPI00032258 | Complement C4 | 192,650 | GLN*VTLSSTGR | Yes (59) | Yes (56) | 0 | 0 |
| IPI00032292 | Metalloproteinase inhibitor 1 | 23,156 | FVGTPEVN*QTTLYQR | Yes (88) | Yes (101) | 2 | – |
| SHN*RSEEFLIAGK | Yes (29) | No | |||||
| IPI00060143 | Protein FAM3D | 24,947 | GLNIALVN*GTTGAVLGQK | Yes (56) | Yes (83) | 0 | 0 |
| IPI00166729 | Alpha-2-glycoprotein 1, zinc | 34,223 | DIVEYYNDSN*GSHVLQGR | Yes (91) | Yes (102) | 13 | 0 |
| FGCEIENN*R | Yes (57) | No | |||||
| FGCEIENN*RSSGAFWK | Yes (58) | No | |||||
| IPI00171411 | Golgi membrane protein 1 | 49,768 | AVLVNN*ITTGER | Yes (80) | Yes (68) | 1 | 0 |
| LQQDVLQFQKN*QTNLER | No | Yes (56) | |||||
| IPI00178926 | Immunoglobulin J chain | 18,087 | EN*ISDPTSPLR | Yes (55) | Yes (75) | 6 | 0 |
| IIVPLNNREN*ISDPTSPLR | Yes (56) | Yes (68) | |||||
| IPI00218460 | Isoform 3 of attractin | 153,687 | IDSTGN*VTNELR | No | Yes (59) | – | 0 |
| IPI00242956 | Fc fragment of IgG binding protein | 571,718 | VVTVAALGTN*ISIHKDEIGK | No | Yes (53) | 11 | 1 |
| VITVQVAN*FTLR | Yes (58) | Yes (50) | |||||
| VTVRPGESVMVN*ISAK | No | Yes (54) | |||||
| KVTVRPGESVMVN*ISAK | No | Yes (34) | |||||
| YLPVN*SSLLTSDCSER | Yes (98) | Yes (46) | |||||
| IPI00291410 | Isoform 1 of long palate, lung, and nasal epithelium carcinoma-associated protein 1 | 54,878 | DHN*ATSILQQLPLLSAMR | No | Yes (87) | – | 0 |
| GDQLILNLNN*ISSDR | No | Yes (87) | |||||
| IPI00291488 | Splice isoform of WAP four-disulfile core domain protein 2 | 12,984 | TGVCPELQADQN*CTQECVSDSECADNLK | Yes (94) | No | 0 | – |
| IPI00295105 | Carbonic anhydrase VI | 35,343 | LENSLLDHRN*K | No | Yes (38) | – | 0 |
| IPI00296099 | Thrombospondin 1 | 129,330 | VVN*STTGPGEHLR | Yes (44) | No | 0 | – |
| IPI00296654 | Bactericidal permeability-increasing protein-like 1 | 49,100 | LGATPVAMLHTNN*ATLR | Yes (39) | Yes (71) | 5 | 0 |
| LLAAAN*FTFK | Yes (64) | Yes (66) | |||||
| SDDNLLN*TSALGR | Yes (81) | No | |||||
| IPI00297910 | Tumor-associated calcium signal transducer 2 | 35,687 | HRPTAGAFN*HSDLDAELR | No | Yes (34) | – | 0 |
| IPI00298082 | Calcium-activated chloride channel protein 2 | 101,283 | DSFDDALQVN*TTDLSPK | Yes (38) | No | 0 | – |
| IPI00298237 | Tripeptidyl-peptidase I | 61,708 | FLSSSPHLPPSSYFN*ASGR | No | Yes (65) | – | 0 |
| IPI00298828 | Beta-2-glycoprotein I | 38,273 | VYKPSAGN*NSLYR | Yes (40) | Yes (74) | 0 | 0 |
| IPI00299547 | Neutrophil gelatinase-associated lipocalin | 22,774 | EDKSYN*VTSVLFR | Yes (28) | No | 3 | |
| SYN*VTSVLFR | Yes (55) | No | |||||
| IPI00299729 | Transcobalamin I | 48,164 | ADEGSLKN*ISIYTK | Yes (50) | Yes (51) | 3 | 0 |
| MN*DTIFGFTMEER | Yes (32) | Yes (81) | |||||
| AQKMN*DTIFGFTMEER | Yes (49) | No | |||||
| IPI00300786 | Alpha-amylase, salivary | 57,731 | NVVDGQPFTNWYDN*GSNQVAFGR | Yes (108) | Yes (129) | 16 | 9 |
| IPI00304557 | Short palate, lung, and nasal epithelium carcinoma associated protein 2 | 26,995 | AEPIDDGKGLN*LSFPVTAN*VTVAGPIIGQIINLK | Yes (55) | No | 0 | – |
| GLNLSFPVTAN*VTVAGPIIGQIINLK | Yes (53) | No | |||||
| IPI00328960 | Similar to carcinoembryonic antigen-related cell adhesion molecule 1 | 91,568 | FVTAGSN*VTLR | No | Yes (31) | – | 0 |
| IPI00553177 | Alpha-1-antitrypsin | 48,208 | YLGN*ATAIFFLPDEGK | No | Yes (100) | – | 0 |
| IPI00328960 | Predicted: hypothetical protein XP_085831 | 91,568 | TPASN*ISTQVSHTK | Yes (44) | No | 0 | – |
| IPI00333140 | Delta notch-like EGF repeat containing transmembrane | 78,438 | LVSFEVPQN*TSVK | Yes (43) | Yes (40) | 1 | – |
| IPI00335356 | IGHM protein | 49,526 | GLTFQQN*ASSMCVPDQDTAIR | No | Yes (40) | – | 0 |
| IPI00374315 | Uncharacterized protein C6orf58 | 37,902 | IILN*QTAR | Yes (52) | Yes (5) | 1 | 0 |
| MGMYKIILN*QTAR | Yes (72) | Yes (63) | |||||
| IPI00419215 | Alpha-2-macroglobulin-like protein 1 | 78,438 | LGHIN*FTISTK | No | Yes (35) | – | 0 |
| IPI00641229 | Ig alpha-2 chain C region | 56,111 | LAGKPTHVN*VSVVMAEVDGTCY | Yes (56) | Yes (97) | 9 | 1 |
| LSLHRPALEDLLLGSEAN*LTCTLTGLR | No | Yes (109) | |||||
| TPLTAN*ITK | Yes (57) | No | |||||
| IPI00400826 | Clusterin | 57,796 | LAN*LTQGEDQYYLR | Yes (64) | Yes (88) | 0 | 0 |
| IPI00418512 | DMBT1 | 259,409 | LVNLN*SSYGLCAGR | Yes (95) | Yes (89) | 3 | 0 |
| QADN*DTIDYSNFLTAAVSGGIIK | Yes (110) | Yes (112) | |||||
| CSGN*ESYLWSCPHK | Yes (38) | No | |||||
| IPI00431645 | HP protein | 31,362 | NLFLN*HSEN*ATAK | Yes (59) | Yes (63) | 3 | 0 |
| VVLHPN*YSQVDIGLIK | No | Yes (74) | |||||
| MVSHHN*LTTGATLINEQWLLTTAK | No | Yes (68) | |||||
| IPI00168728 | FLJ00385 protein (fragment) | 56,075 | EEQFN*STFR | No | Yes (51) | – | 0 |
| IPI00550991 | Alpha-1-antichymotrypsin | 50,596 | FN*LTETSEAEIHQSFQHLLR | No | Yes (52) | – | 0 |
N* indicates the site of former N-linked glycosylation
Mascot score listed in parentheses
Fig. 2.
Comparison of a formerly N-glycosylated peptides and the b N-glycoproteins they represent identified in whole saliva using the Zhang method and the Sun method. c MS/MS mass spectrum of a doubly charged peptide, GDQLILNLNN*ISSDR (m/z 837; asterisk denotes the site of N-glycosylation), from isoform 1 of long palate, lung, and nasal epithelium carcinoma (PLUNC)-associated protein 1 measured from whole saliva
The seemingly poor overlap in protein identification between the two methods could be attributed to several factors. The method of collection of whole saliva has been the same in both studies. However, the glycoprotein pulldown experiments in our previous paper were performed with saliva collected from only one subject [44]. In this current study, saliva from five individual subjects was pooled prior to carrying out the experiments. The identification of proteins found exclusively using the Sun method and not by the Zhang method could be explained by the fact that in the latter method, the precipitated proteins were resuspended in the coupling buffer. Some proteins may not be readily soluble in this buffer and may have been lost in the subsequent steps. Even for proteins that are solubilized in the coupling buffer, carbohydrates may not be well exposed, and oxidation and the ensuing agarose–hydrazide coupling may be inefficient. The identification of peptides exclusively by the Zhang method (and not by the Sun method) could be because of the hydrophilic nature of glycopeptides and their poor retention on the C18 columns. However, we observed that the number of nonformerly N-glycosylated peptides identified were fewer for the Sun method compared to the Zhang method, i.e., the number of nonglycosylated proteins binding to the resin was reduced (Table 1). For the previous analysis of whole saliva using the Zhang method, we measured 163 nonglycosylated peptides. In the current study using the Sun method, we detected only 14 nonglycosylated peptides in whole saliva. This appears to be a significant advantage of the N-glycopeptide capture method over the glycoprotein capture.
Identification of N-Glycoproteins in Parotid, Submandibular, and Sublingual Fluids
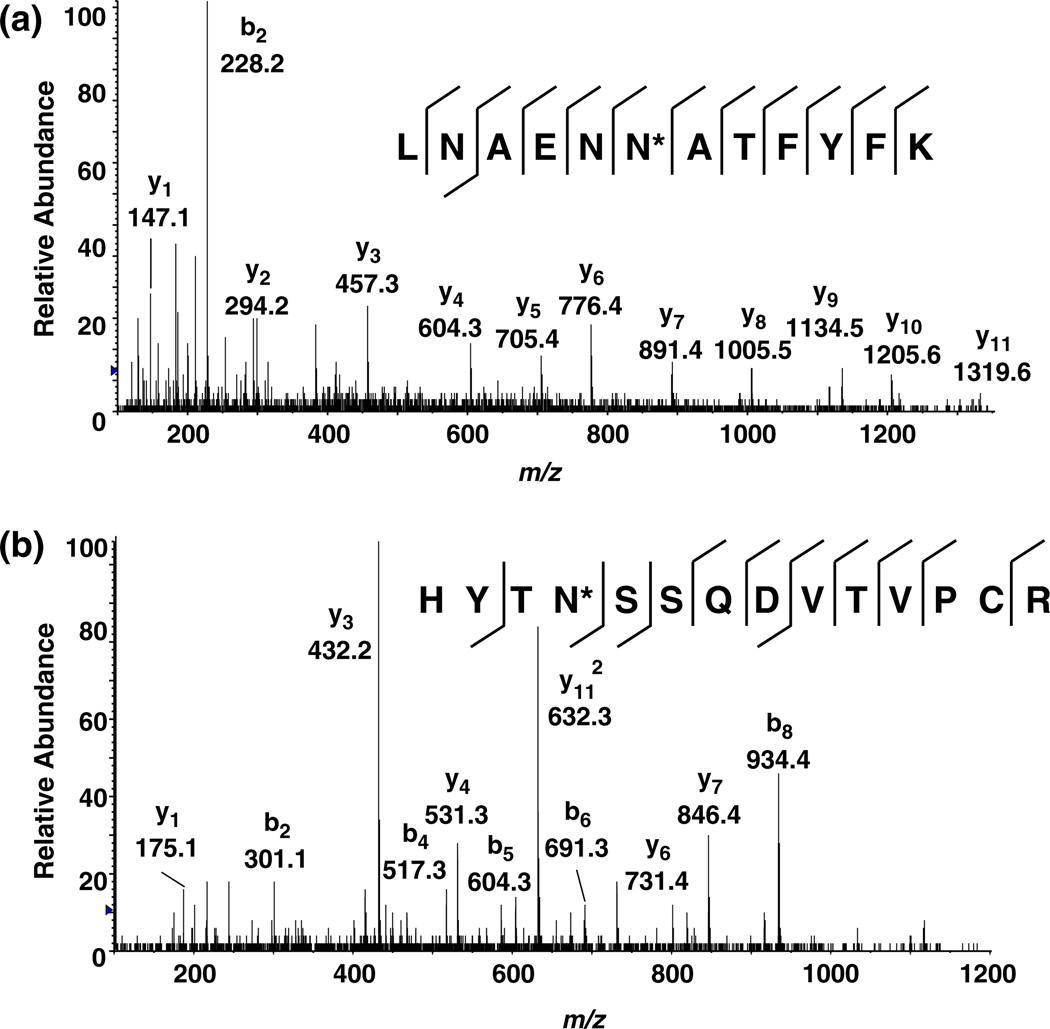
To extend our catalogue of salivary N-glycoproteins, we examined the salivary secretions of the three main salivary glands: PA, SM, and SL. Whole saliva is likely to include contributions from the salivary glands and other sites in the oral cavity. Moreover, the collection and processing of saliva from the PA, SM, and SL glands is slightly different from whole saliva, as the stimulation of secretion by the application of citric acid to the tongue was not used, and the centrifugation of saliva to remove food debris was not required. We applied the Sun method to identify N-glycoproteins from PA, SM, and SL fluids. The total number of formerly N-glycopeptides and the N-linked glycoproteins they represent identified from PA, SM, and SL fluids are 62/34 (peptides/protein), 80/44, and 98/53, respectively (Tables 2, 3, and 4). A comparison of glycoproteins isolated from the three glands is shown in Fig. 3a,b. Only 42 formerly N-glycosylated peptides from 25 N-glycoproteins were found in all three glands. There were a significant number of N-glycoproteins that were unique to the saliva secretions of the individual glands. Figure 4a shows the MS/MS spectrum of a doubly charged formerly N-glycosylated peptide (LNAENN*ATFYFK, m/z 716.8) from splice isoform 1 of kininogen. This formerly N-glycosylated peptide was found only in the PA fluid and not in whole saliva, SM, or SL gland saliva. Figure 4b shows the MS/MS spectrum of a triply charged peptide (HYTN*SSQDVTVPCR, m/z 555.6) from the hypothetical protein DKFZp686C02220. This formerly N-glycosylated peptide was observed in SM saliva but not in whole saliva, PA, or SL gland fluids.
Table 2.
N-linked glycoproteins identified in parotid salivary fluid
| IPI accession number |
Protein name | MW (Da) | Formerly N-glycosylated peptide sequencea |
Formerly N-glycosylated peptide scoreb |
Nonformerly glycosylated peptides identified |
|---|---|---|---|---|---|
| IPI00004573 | Polymeric-immunoglobulin receptor | 83,262 | AN*LTNFPEN*GTFVVNIAQLSQDDSGR | 100 | 3 |
| GLSFDVSLEVSQGLGLLN*DTK | 74 | ||||
| IIEGEPNLKVPGN*VTAVLGETLK | 118 | ||||
| LSLLEEPGN*GTFTVILNQLTSR | 118 | ||||
| QIGLYPVLVIDSSGYVNPN*YTGR | 101 | ||||
| VPGN*VTAVLGETLK | 115 | ||||
| VPGN*VTAVLGETLKVPCHFPCK | 54 | ||||
| WN*NTGCQALPSQDEGPSK | 48 | ||||
| IPI00011229 | Cathepsin D | 44,524 | GSLSYLN*VTR | 68 | 0 |
| IPI00012887 | Cathepsin L | 37,540 | YSVAN*DTGFVDIPKQEK | 42 | 0 |
| IPI00017601 | Ceruloplasmin | 122,128 | EN*LTAPGSDSAVFFEQGTTR | 69 | 0 |
| EHEGAIYPDN*TTDFQR | 49 | ||||
| IPI00021891 | Splice Isoform 1 of Fibrinogen gamma chain | 51,479 | VDKDLQSLEDILHQVEN*K | 53 | 0 |
| IPI00022417 | Leucine-rich alpha-2-glycoprotein | 38,944 | KLPPGLLAN*FTLLR | 78 | 0 |
| IPI00022431 | Alpha-2-HS-glycoprotein | 39,300 | AALAAFNAQNN*GSNFQLEEISR | 113 | 0 |
| VCQDCPLLAPLN*DTR | 50 | ||||
| IPI00022463 | Serotransferrin | 77,000 | QQQHLFGSN*VTDCSGNFCLFR | 100 | 0 |
| CGLVPVLAENYN*K | 53 | ||||
| IPI00022488 | Hemopexin | 51,643 | ALPQPQN*VTSLLGCTH | 73 | 0 |
| IPI00023673 | Galectin-3 binding protein | 65,289 | ALGFEN*ATQALGR | 108 | 0 |
| AAIPSALDTN*SSK | 58 | ||||
| GLN*LTEDTYKPR | 48 | ||||
| YKGLN*LTEDTYKPR | 35 | ||||
| IPI00025023 | Lactoperoxidase | 80,237 | ASLTN*VTDPSLDLTSLSLEVGCGAPAPVVR | 78 | 0 |
| IVGYLNEEGVLDQN*R | 97 | ||||
| KPSPCEFIN*TTAR | 53 | ||||
| LRN*LSSPLGLMAVNQEVSDHGLPYLPYDSK | 65 | ||||
| IPI00032258 | Complement C4-A | 192,650 | GLN*VTLSSTGR | 57 | 0 |
| IPI00032292 | Metalloproteinase inhibitor 1 | 23,156 | FVGTPEVN*QTTLYQR | 95 | 0 |
| IPI00032328 | Splice Isoform 1 Of Kininogen | 71,900 | LNAENN*ATFYFK | 71 | 0 |
| IPI00168728 | FLJ00385 Protein (Fragment) | 56,111 | EEQFN*STFR | 51 | 0 |
| IPI00178926 | Immunoglobulin J chain | 18,087 | EN*ISDPTSPLR | 63 | 0 |
| IIVPLNNREN*ISDPTSPLR | 57 | ||||
| IPI00291262 | Clusterin | 52,461 | LAN*LTQGEDQYYLR | 95 | 0 |
| MLN*TSSLLEQLNEQFNWVSR | 96 | ||||
| IPI00291410 | Isoform 1 of long palate, lung, and nasal epithelium carcinoma-associated protein 1 | 52,408 | DHN*ATSILQQLPLLSAMR | 33 | 0 |
| IPI00295105 | Carbonic anhydrase VI | 35,343 | GLN*MTGYETQAGEFPMVNNGHTVQIGLPSTMR | 41 | 0 |
| IPI00296099 | Thrombospondin 1 | 129,330 | VVN*STTGPGEHLR | 52 | 0 |
| IPI00296654 | Bactericidal/permeability-increasing protein-like 1 | 49,100 | LGATPVAMLHTNN*ATLR | 73 | 0 |
| IPI00298497 | Fibrinogen beta chain | 55,892 | GTAGNALMDGASQLMGEN*R | 42 | 0 |
| IPI00298828 | Beta-2-glycoprotein | 38,273 | VYKPSAGN*NSLYR | 79 | 0 |
| IPI00299729 | Transcobalamin I | 48,164 | ADEGSLKN*ISIYTK | 61 | 0 |
| MN*DTIFGTMEER | 59 | ||||
| IPI00300786 | Salivary alpha-amylase | 57,731 | NVVDGQPFTNWYDN*GSNQVAFGR | 112 | 13 |
| IPI00333140 | Delta-notch-like EGF repeat-containing transmembrane | 78,438 | LVSFEVPQN*TSVK | 52 | 0 |
| IPI00335356 | IGHM protein | 60,966 | GLTFQQN*ASSMCVPDQDTAIR | 63 | 0 |
| IPI00400826 | Clusterin | 57,796 | LAN*LTQGEDQYYLR | 96 | 0 |
| MLN*TSSLLEQLNEQFNWVSR | 96 | ||||
| IPI00418512 | DMBT1 | 78,398 | LVNLN*SSYGLCAGR | 90 | 0 |
| QADN*DTIDYSNFLTAAVSGGIIK | 123 | ||||
| IPI00431645 | HP protein | 31,362 | MVSHHN*LTTGATLINEQWLLTTAK | 73 | 0 |
| NLFLN*HSENATAK | 44 | ||||
| VVLHPN*YSQVDIGLIK | 104 | ||||
| IPI00478003 | Alpha-2-macroglobulin | 163,175 | LGHIN*FTISTK | 32 | 0 |
| IPI00550991 | Alpha-1-antichymotrypsin | 50,596 | FN*LTETSEAEIHQSFQHLLR | 59 | 0 |
| IPI00641229 | Ig alpha-2 chain C region | 53,868 | LAGKPTHVN*VSVVMAEVDGTCY | 92 | 1 |
| LSLHRPALEDLLLGSEAN*LTCTLTGLR | 96 | ||||
| IPI00553177 | Alpha-1-antitrypsin | 46,737 | YLGN*ATAIFFLPDEGK | 112 | 0 |
| ADTHDEILEGLNFN*LTEIPEAQIHEGFQELLR | 48 | ||||
| IPI00643034 | Splice Isoform 1 of Phospholipid transfer protein | 54,705 | IYSN*HSALESLALIPLQAPLK | 60 | 0 |
| VSN*VSCQASVSR | 100 | ||||
| GKEGHFYYN*ISEVK | 43 | ||||
| GAFFPLTERN*WSLPNR | 44 |
N* indicates the site of former N-linked glycosylation
Mascot score listed in parentheses
Table 3.
N-linked glycoproteins identified in submandibular salivary fluid
| IPI accession number |
Protein name | MW (Da) | Formerly N-glycosylated peptide sequencea |
Formerly N-glycosylated peptide scoreb |
Nonformerly glycosylated peptides identified |
|---|---|---|---|---|---|
| IPI00003351 | Extracellular matrix protein 1 | 60,635 | HIPGLIHN*MTAR | 62 | 0 |
| IPI00004573 | Polymeric-immunoglobulin receptor | 83,262 | AN*LTNFPEN*GTFVVNIAQLSQDDSGR | 105 | 2 |
| IIEGEPNLKVPGN*VTAVLGETLK | 108 | ||||
| LSLLEEPGN*GTFTVILNQLTSR | 97 | ||||
| QIGLYPVLVIDSSGYVNPN*YTGR | 127 | ||||
| VPGN*VTAVLGETLK | 116 | ||||
| VPGN*VTAVLGETLKVPCHFPCK | 52 | ||||
| WN*NTGCQALPSQDEGPSK | 45 | ||||
| IPI00010697 | Isoform alpha-6X1X2B of integrin alpha-6 | 126,539 | EINSLN*LTESHNSR | 31 | 0 |
| IPI00011229 | Cathepsin D | 49,768 | GSLSYLN*VTR | 68 | 0 |
| IPI00012887 | Cathepsin L | 37,540 | YSVAN*DTGFVDIPKQEK | 45 | 0 |
| IPI00017601 | Ceruloplasmin | 122,128 | EHEGAIYPDN*TTDFQR | 63 | 0 |
| IPI00022431 | Alpha-2-HS-glycoprotein | 39,300 | AALAAFNAQNN*GSNFQLEEISR | 41 | 0 |
| IPI00022488 | Hemopexin | 51,643 | ALPQPQN*VTSLLGCTH | 27 | 0 |
| IPI00023673 | Galectin-3 binding protein | 65,289 | AAIPSALDTN*SSK | 58 | 0 |
| ALGFEN*ATQALGR | 84 | ||||
| GLN*LTEDTYKPR | 60 | ||||
| EPGSN*VTMSVDAECVPMVR | 44 | ||||
| IPI00025023 | Lactoperoxidase | 80,237 | KPALGAAN*R | 46 | 0 |
| KPSPCEFIN*TTAR | 51 | ||||
| ASLTN*VTDPSLDLTSLSLEVGCGAPAPVVR | 96 | ||||
| IVGYLNEEGVLDQN*R | 100 | ||||
| LRN*LSSPLGLMAVNQEVSDHGLPYLPYDSK | 58 | ||||
| IPI00031121 | Carboxypeptidase E | 53,117 | DLQGNPIAN*ATISVEGIDHDVTSAK | 49 | 0 |
| GN*ETIVNLIHSTR | 44 | ||||
| IPI00032292 | Metalloproteinase inhibitor 1 | 23,156 | FVGTPEVN*QTTLYQR | 96 | 0 |
| IPI00060143 | Protein FAM3D | 24,947 | GLNIALVN*GTTGAVLGQK | 73 | 0 |
| IPI00166729 | Alpha-2-glycoprotein 1, zinc | 34,223 | FGCEIENN*R | 48 | 3 |
| DIVEYYN*DSN*GSHVLQGR | 66 | ||||
| IPI00168728 | FLJ00385 protein (fragment) | 56,111 | EEQFN*STFR | 51 | 0 |
| IPI00171411 | Golgi membrane protein 1 | 42,917 | AVLVNN*ITTGER | 70 | 0 |
| IPI00178926 | Immunoglobulin J chain | 18,087 | EN*ISDPTSPLR | 59 | 0 |
| IIVPLNNREN*ISDPTSPLR | 77 | ||||
| IPI00218413 | Biotinidase | 61,093 | NPVGLIGAEN*ATGETDPSHSK | 58 | 0 |
| IPI00242956 | Fc fragment of IgG binding protein | 571,718 | LLISSLSESPASVSILSQADN*TSK | 92 | 0 |
| VITVQVAN*FTLR | 62 | ||||
| VTVRPGESVMVN*ISAK | 30 | ||||
| VVTVAALGTN*ISIHKDEIGK | 67 | ||||
| IPI00291410 | Isoform 1 of long palate, lung, and nasal epithelium carcinoma-associated protein 1 | 52,408 | DHN*ATSILQQLPLLSAMR | 31 | 0 |
| IPI00296099 | Thrombospondin-1 | 129,300 | VVN*STTGPGEHLR | 70 | 0 |
| IPI00295105 | Carbonic anhydrase VI | 35,343 | GLN*MTGYETQAGEFPMVNNGHTVQIGLPSTMR | 43 | 0 |
| IPI00296654 | Bactericidal/permeability-increasing protein-like 1 | 49,100 | LLAAAN*FTFK | 74 | 0 |
| LGATPVAMLHTNN*ATLR | 68 | ||||
| SDDNLLN*TSALGR | 56 | ||||
| IPI00297910 | Tumor-associated calcium signal transducer 2 | 35,687 | HRPTAGAFN*HSDLDAELR | 43 | 0 |
| IPI00299729 | Transcobalamin I | 48,164 | ADEGSLKN*ISIYTK | 64 | 0 |
| MN*DTIFGFTMEER | 92 | ||||
| IPI00298828 | Beta-2-glycoprotein 1 | 38,273 | VYKPSAGN*NS | 0 | |
| IPI00300786 | Salivary alpha-amylase | 57,731 | NVVDGQPFTNWYDN*GSNQVAFGR | 79 | 6 |
| IPI00304557 | Short palate, lung, and nasal epithelium carcinoma associated protein 2 | 26,995 | GLNLSFPVTAN*VTVAGPIIGQIINLK | 84 | 2 |
| IPI00328460 | Transmembrane protease, serine 11B | 46,235 | MLNAFQNSSIYK | 64 | 0 |
| LLPNAN*GSNVQLQLK | 80 | ||||
| IPI00328960 | Similar to LOC147645 protein | 91,568 | FVTAGSN*VTLR | 69 | 0 |
| IPI00333140 | Delta-notch-like EGF repeat-containing transmembrane | 78,438 | LVSFEVPQN*TSVK | 48 | 0 |
| IPI00335356 | IGHM protein | 64,998 | GLTFQQN*ASSMCVPDQDTAIR | 60 | 0 |
| IPI00374315 | Hypothetical LOC389429 | 37,902 | IILN*QTAR | 63 | 0 |
| IPI00384872 | UDP-glucuronosyltransferase 1–7 | 59,779 | YTGTRPSNLAN*NTILVK | 46 | 0 |
| IPI00400826 | Clusterin | 57,796 | LAN*LTQGEDQYYLR | 81 | 0 |
| IPI00418512 | DMBT1 | 193,867 | LVNLN*SSYGLCAGR | 101 | |
| QADN*DTIDYSNFLTAAVSGGIIK | 93 | ||||
| IPI00419215 | Alpha-2-macroglobulin like protein 1 | 161,105 | LGHIN*FTISTK | 49 | 0 |
| QGN*GTFVQTDKPLYTPGQQVYFR | 72 | ||||
| IPI00423461 | Hypothetical protein DKFZp686C02220 | 54,126 | HYTN*SSQDVTVPCR | 55 | 0 |
| IPI00431645 | HP protein | 45,177 | NLFLN*HSENATAK | 39 | 0 |
| VVLHPN*YSQVDIGLIK | 57 | ||||
| IPI00550991 | Alpha-1-antichymotrypsin | 50,566 | YTGN*ASALFILPDQDK | 36 | 0 |
| IPI00553177 | Alpha-1-antitrypsin | 46,737 | YLGN*ATAIFFLPDEGK | 108 | 0 |
| ADTHDEILEGLNFN*LTEIPEAQIHEGFQELLR | 67 | ||||
| IPI00641229 | Ig alpha-2 chain C region | 53,868 | LAGKPTHVN*VSVVMAEVDGTCY | 81 | 0 |
| LSLHRPALEDLLLGSEAN*LTCTLTGLR | 110 | ||||
| TPLTAN*ITK | 57 | ||||
| IPI00643034 | Splice isoform 1 of phospholipid transfer protein | 54,705 | VSN*VSCQASVSR | 90 | 0 |
| IYSN*HSALESLALIPLQAPLK | 64 | ||||
| IPI00855918 | Mucin 5, subtype B, tracheobronchial | 590,122 | AFGQFFSPGEVIYN*K | 80 | 1 |
| AQGLVLEASN*GSVLINGQR | 141 | ||||
| DIECQAESFPN*WTLAQVGQK | 86 | ||||
| GN*CTYVLMR | 65 | ||||
| LDGPTEQCPDPLPLPAGN*CTDEEGICHR | 24 | ||||
| LPYSLFHN*NTEGQCGTCTNNQR | 20 | ||||
| QVN*ETWTLEN*CTVAR | 68 | ||||
| VVLLDPKPVAN*VTCVNK | 81 |
N* indicates the site of former N-linked glycosylation
Mascot score listed in parentheses
Table 4.
N-linked glycoproteins identified in sublingual salivary fluid
| IPI accession number |
Protein name | MW (Da) | Formerly N-glycosylated peptide sequencea |
Formerly N-glycosylated peptide scoreb |
Nonformerly glycosylated peptides identified |
|---|---|---|---|---|---|
| IPI00000877 | Hypoxia-upregulated protein | 111,266 | VFGSQN*LTTVK | 79 | 0 |
| LSALDNLLN*HSSMFLK | 63 | ||||
| IPI00002818 | Isoform 1 of kallikrein 11 | 27,448 | CAN*ITIIEHQK | 36 | 0 |
| IPI00003351 | Extracellular matrix protein 1 | 60,635 | HIPGLIHN*MTAR | 65 | 0 |
| HKHIPGLIHN*MTAR | 37 | ||||
| IPI00004573 | Polymeric-immunoglobulin receptor | 83,262 | AN*LTNFPEN*GTFVVNIAQLSQDDSGR | 106 | 1 |
| GLSFDVSLEVSQGPGLLN*DTK | 49 | ||||
| IIEGEPNLKVPGN*VTAVLGETLK | 115 | ||||
| LSLLEEPGN*GTFTVILNQLTSR | 114 | ||||
| QIGLYPVLVIDSSGYVNPN*YTGR | 122 | ||||
| VPGN*VTAVLGETLK | 105 | ||||
| VPGN*VTAVLGETLKVPCHFPCK | 82 | ||||
| WN*NTGCQALPSQDEGPSK | 54 | ||||
| IPI00010697 | Isoform alpha-6X1X2B of integrin alpha-6 | 41,000 | EIN*SLN*LTESHNSR | 32 | 0 |
| IPI00011229 | Cathepsin D | 44,524 | GSLSYLN*VTR | 68 | 0 |
| IPI00021891 | Splice isoform 1 of fibrinogen gamma chain | 51,549 | DLQSLEDILHQVEN*K | 46 | 0 |
| VDKDLQSLEDILHQVEN*K | 58 | ||||
| IPI00022229 | Apolipoprotein B-100 | 515,241 | FVEGSHN*STVSLTTK | 29 | 0 |
| IPI00022431 | Alpha-2-HS-glycoprotein | 39,300 | AALAAFNAQNN*GSNFQLEEISR | 104 | 0 |
| IPI00022463 | Serotransferrin | 77,000 | QQQHLFGSN*VTDCSGNFCLFR | 82 | 0 |
| CGLVPVLAENYN*K | 43 | ||||
| IPI00022488 | Hemopexin | 51,643 | ALPQPQN*VTSLLGCTH | 63 | 0 |
| IPI00022974 | Prolactin-inducible protein | 16,562 | TFYWDFYTN*R | 43 | 0 |
| IPI00023673 | Galectin-3 binding protein | 65,289 | AAIPSALDTN*SSK | 58 | 0 |
| ALGFEN*ATQALGR | 94 | ||||
| GLN*LTEDTYKPR | 39 | ||||
| YKGLN*LTEDTYKPR | 36 | ||||
| IPI00025023 | Lactoperoxidase | 80,237 | ASLTN*VTDPSLDLTSLSLEVGCGAPAPVVR | 93 | 1 |
| IVGYLNEEGVLDQN*R | 110 | ||||
| KPSPCEFIN*TTAR | 74 | ||||
| LRN*LSSPLGLMAVNQEVSDHGLPYLPYDSK | 57 | ||||
| IPI00025846 | Splice isoform 1 of desmocollin-2 | 99,899 | NGIYN*ITVLASDQGGR | 65 | 0 |
| AN*YTILK | 32 | ||||
| IPI00027192 | Procollagen-lysine | 83,497 | EQIN*ITLDHR | 38 | 0 |
| IPI00030847 | Transmembrane 9 superfamily protein member 3 | 67,843 | IVDVN*LTSEGK | 60 | 0 |
| IPI00031121 | Carboxypeptidase E | 53,117 | GN*ETIVNLIHSTR | 44 | 0 |
| IPI00031547 | Desmoglein 3 | 107,435 | LPAVWSITTLN*ATSALLR | 80 | 0 |
| IPI00032258 | Complement C4 | 192,650 | GLN*VTLSSTGR | 55 | 0 |
| IPI00032292 | Metalloproteinase inhibitor 1 | 23,156 | FVGTPEVN*QTTLYQR | 87 | 0 |
| IPI00060143 | Protein FAM3D | 24,947 | GLNIALVN*GTTGAVLGQK | 116 | 0 |
| IPI00166729 | Alpha-2-glycoprotein 1, zinc | 34,223 | FGCEIENN*R | 50 | 0 |
| IPI00168728 | FLJ00385 protein (fragment) | 56,111 | EEQFN*STFR | 51 | 0 |
| IPI00171411 | Golgi membrane protein 1 | 49,768 | AVLVNN*ITTGER | 89 | 0 |
| LQQDVLQFQKN*QTNLER | 43 | ||||
| IPI00178926 | Immunoglobulin J chain | 18,087 | IIVPLNNREN*ISDPTSPLR | 62 | 0 |
| EN*ISDPTSPLR | 55 | ||||
| IPI00242956 | IgGFc binding protein | 571,718 | LLISSLSESPASVSILSQADN*TSKK | 40 | 0 |
| VTVRPGESVMVN*ISAK | 47 | ||||
| IPI00291410 | Isoform 1 of long palate, lung, and nasal epithelium carcinoma-associated protein 1 | 54,878 | DHN*ATSILQQLPLLSAMR | 46 | 0 |
| IPI00295105 | Carbonic anhydrase VI | 35,343 | GLN*MTGYETQAGEFPMVNN GHTVQIGLPSTMR | 39 | 0 |
| IPI00296099 | Thrombospondin 1 | 129,330 | VVNSTTGPGEHLR | 73 | 0 |
| IPI00296654 | Bactericidal/permeability-increasing protein-like 1 | 49,100 | LLAAAN*FTFK | 72 | 0 |
| LGATPVAMLHTNN*ATLR | 66 | ||||
| IPI00297910 | Tumor-associated calcium signal transducer 2 | 35,687 | HRPTAGAFN*HSDLDAELR | 65 | 0 |
| IPI00298082 | Calcium-activated chloride channel protein 2 | 101,288 | AAN*SSVPPITVNAK | 44 | 0 |
| HSN*GSYSAFGER | 51 | ||||
| DSFDDALQVN*TTDLSPK | 67 | ||||
| IPI00298828 | Beta-2-glycoprotein I | 38,273 | VYKPSAGN*NSLYR | 79 | 0 |
| IPI00299729 | Transcobalamin I | 48,164 | ADEGSLKN*ISIYTK | 73 | 0 |
| MN*DTIFGFTMEER | 96 | ||||
| IPI00300786 | Salivary alpha-amylase | 57,731 | NVVDGQPFTNWYDN*GSNQVAFGR | 93 | 1 |
| IPI00303333 | Platelet receptor GI24 | 33,901 | HGLESASDHHGN*FSITMR | 41 | 0 |
| IPI00304557 | Short palate, lung, and nasal epithelium carcinoma associated protein 2 | 26,995 | GLN*LSFPVTAN*VTVAGPIIGQIINLK | 82 | 2 |
| IPI00328460 | Transmembrane protease, serine 11B | 46,235 | LLPNAN*GSNVQLQLK | 83 | 0 |
| MLNAFQN*SSIYK | 59 | ||||
| IPI00328960 | Predicted: hypothetical protein XP_085831 | 91,568 | FVTAGSN*VTLR | 50 | 0 |
| IPI00332845 | Leucine-rich repeats and calponin homology (CH) domain containing 2 | 69,610 | KLPPGLLAN*FTLLR | 77 | 0 |
| IPI00335356 | IGHM protein | 49,526 | GLTFQQN*ASSMCVPDQDTAIR | 47 | 0 |
| IPI00374315 | Uncharacterized protein C6orf58 | 37,902 | IILN*QTAR | 62 | 0 |
| IPI00384872 | UDP-glucosyltransferase 1–7 | 59,779 | YTGTRPSNLAN*NTILVK | 33 | 0 |
| IPI00400826 | Clusterin | 57,796 | LAN*LTQGEDQYYLR | 58 | 0 |
| MLN*TSSLLEQLNEQFNWVSR | 99 | ||||
| IPI00418512 | DMBT1 | 193,867 | LVNLN*SSYGLCAGR | 99 | 0 |
| QADN*DTIDYSNFLTAAVSGGIIK | 92 | ||||
| IPI00419215 | Alpha-2-macroglobulin like protein 1 | 149,965 | QGN*GTFVQTDKPLYTPGQQVYFR | 72 | 0 |
| LGHIN*FTISTK | 39 | ||||
| HSN*GSYSAFGER | 51 | ||||
| IPI00431645 | HP protein | 31,362 | VVLHPN*YSQVDIGLIK | 53 | 0 |
| NLFLN*HSEN*ATAK | 41 | ||||
| IPI00550991 | Alpha-1-antichymotrypsin | 50,596 | FN*LTETSEAEIHQSFQHLLR | 58 | 0 |
| IPI00641229 | Ig alpha-2 chain C region | 53,868 | LAGKPTHVN*VSVVMAEVDGTCY | 97 | 3 |
| LSLHRPALEDLLLGSEAN*LTCTLTGLR | 104 | ||||
| IPI00553177 | Alpha-1-antitrypsin | 46,737 | ADTHDEILEGLNFN*LTEIPEAQIHEGFQELLR | 51 | 0 |
| YLGN*ATAIFFLPDEGK | 101 | ||||
| IPI00643034 | Splice isoform 1 of phospholipid transfer protein | 54,705 | GKEGHFYYN*ISEVK | 40 | 0 |
| GAFFPLTERN*WSLPNR | 41 | ||||
| IYSN*HSALESLALIPLQAPLK | 47 | ||||
| VSN*VSCQASVSR | 110 | ||||
| IPI00855918 | Mucin 5, subtype B, tracheobronchial | 590,122 | AFGQFFSPGEVIYN*K | 75 | 0 |
| AFGQFFSPGEVIYN*KTDR | 82 | ||||
| AQGLVLEASN*GSVLINGQR | 137 | ||||
| DIECQAESFPN*WTLAQVGQK | 84 | ||||
| FGN*LSLYLDN*HYCTASATAAAAR | 99 | ||||
| GN*CTYVLMR | 46 | ||||
| LDGPTEQCPDPLPLPAGN*CTDEEGICHR | 28 | ||||
| LPYSLFHN*NTEGQCGTCTNNQR | 31 | ||||
| QVNETWTLEN*CTVAR | 70 | ||||
| VVLLDPKPVAN*VTCVNK | 77 |
N* indicates the site of former N-linked glycosylation
Mascot score listed in parentheses
Fig. 3.
Comparison of a formerly N-glycosylated peptides and the b N-glycoproteins they represent identified in PA, SM, and SL salivary fluids
Fig. 4.
MS/MS mass spectra of a doubly charged peptide, LNAENN*ATFYFK (m/z 716.8; asterisk denotes the site of N-glycosylation) from splice isoform 1 of kininogen from parotid fluid, and b triply charged peptide, HYTN*SSQDVTVPCR (m/z 555.6) from hypothetical protein DKFZp686C02220 from submandibular fluid
Comparison of N-Glycoproteins in Whole Saliva and Parotid, Submandibular, and Sublingual Fluids
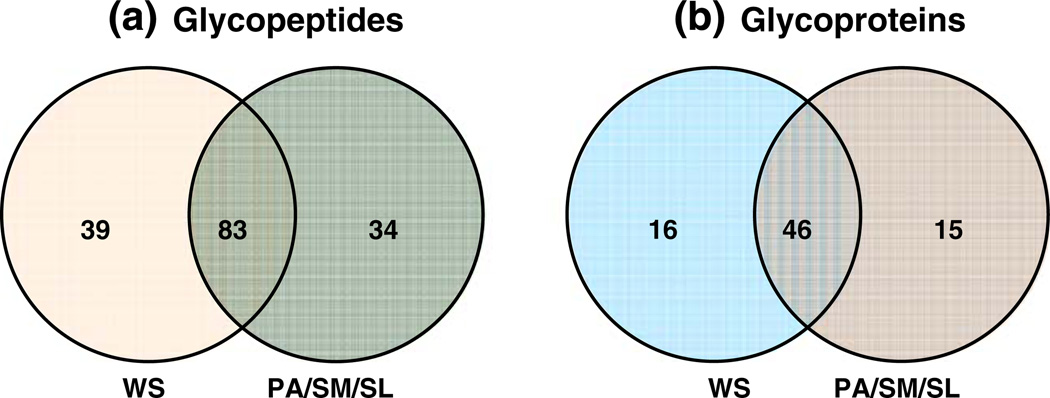
Whole saliva is a complex mixture. It has contributions not only from the three main salivary glands but also from other minor salivary glands located in the mouth. Nonsalivary secretions in whole saliva include gingival crevicular fluid, bronchial and nasal secretions, and blood derivatives that might enter the mouth by cuts or abrasions. Other components of whole saliva include microbes such as bacteria, fungi, and viruses, food, and other extrinsic substances and from the lining of the mouth [51]. It is expected that the glycoprotein profile will differ between that of whole saliva and PA, SM, and SL saliva. A total of 83 formerly N-glycosylated peptides from 46 N-glycoproteins were found to be common in whole saliva and PA, SM, and SL fluids combined. Thirty-nine formerly N-glycosylated peptides from 16 N-glycoproteins were unique to whole saliva, and 34 formerly N-glycosylated peptides from 15 N-glycoproteins were found in PA, SM, or SL and not detected in whole saliva (Fig. 5a,b). Of the glycoproteins that were unique to whole saliva and not found in PA, SM, and SL fluids, three proteins were not detected in our previous global proteome analysis of PA, SM, and SL fluids [52]. These proteins include lumican precursor, complement component C9, and cystatin-related epididymal spermatogenic protein. Of these, complement component C9 has been detected in whole saliva in our previous studies (unpublished data), but the other two proteins were not detected previously in our global whole-saliva proteome analysis. Table 5 is a combined listing of all the salivary N-glycoproteins identified. The detection of N-glycoproteins that are unique to whole saliva is not surprising because whole saliva is a mixture of various components. Some glycoproteins detected in whole saliva may not originate from the salivary glands. Additionally, some proteins originating from the salivary glands may have undergone posttranslational modifications after being secreted into the oral cavity. These formerly glycosylated peptides are observed only in whole saliva. However, the detection of N-glycoproteins unique to PA, SM, and SL saliva is somewhat surprising. It may be explained by the fact that the glycosylation might have been lost upon secretion into the mouth by the action of enzymes. Whole saliva has a plethora of oral bacteria that secrete enzymes to deglycosylate glycoproteins to alter the properties of the protein altogether or to utilize the sugars for their growth [14]. Glycoproteins from the salivary glands may be also sufficiently diluted in whole saliva, so that they fall below the detection limit of our glycoprotein pulldown method. It is also possible that these set of glycoproteins are secreted only upon stimulation of the glands and not otherwise.
Fig. 5.
Comparison of a formerly N-glycosylated peptides and the b N-glycoproteins they represent identified in whole saliva and PA, SM, and SL fluids combined
Table 5.
Combined list of all N-Glycoproteins identified in human salivary fluids
| IPI accession number |
Protein name | MW (Da) | Formerly N-glycosylated peptidea | Formerly N-glycosylated peptide sequence |
Formerly N-glycosylated peptides detectedb |
|||
|---|---|---|---|---|---|---|---|---|
| WS | PA | SM | SL | |||||
| IPI00000877 | Hypoxia-upregulated protein | 111,266 | VFGSQN*LTTVK | 510–520 | No | No | No | Yes |
| LSALDNLLN*HSSMFLK | 822–837 | No | No | No | Yes | |||
| IPI00002818 | Splice isoform 1 of kallikrein 11 | 27,448 | CAN*ITIIEHQK | 163–173 | Yes | No | No | Yes |
| IPI00003351 | Extracellular matrix protein 1 | 60,635 | HIPGLIHN*MTAR | 437–448 | Yes | No | Yes | Yes |
| IPI00004573 | Polymeric-immunoglobulin receptor | 83,262 | VPGN*VTAVLGETLK | 466–479 | Yes | Yes | Yes | Yes |
| WN*NTGCQALPSQDEGPSK | 498–515 | Yes | Yes | Yes | Yes | |||
| IIEGEPNLKVPGN*VTAVLGETLK | 457–479 | Yes | Yes | Yes | Yes | |||
| GLSFDVSLEVSQGPGLLN*DTK | 118–138 | No | Yes | No | Yes | |||
| LSLLEEPGN*GTFTVILNQLTSR | 413–434 | Yes | Yes | Yes | Yes | |||
| VPGN*VTAVLGETLKVPCHFPCK | 466–487 | Yes | Yes | Yes | Yes | |||
| QIGLYPVLVIDSSGYVNPN*YTGR | 168–190 | Yes | Yes | Yes | Yes | |||
| AN*LTNFPEN*GTFVVNIAQLSQDDSGR | 82–107 | Yes | Yes | Yes | Yes | |||
| IPI00007244 | Myeloperoxidase | 83,815 | SCPACPGSN*ITIR | 315–327 | Yes | No | Yes | No |
| SYN*DSVDPR | 481–489 | Yes | No | No | No | |||
| IPI00010697 | Isoform alpha 6X1X2B of integrin alpha-6 | 126,539 | EINSLN*LTESHNSR | 925–938 | No | No | Yes | No |
| IPI00011229 | Cathepsin D | 49,768 | GSLSYLN*VTR | 257–266 | Yes | Yes | Yes | Yes |
| IPI00855918 | Mucin 5, subtype B, tracheobronchial | 590,122 | VVLLDPKPVAN*VTCVNK | 4,977–4,993 | Yes | No | Yes | Yes |
| AQGLVLEASN*GSVLINGQR | 136–154 | Yes | No | Yes | Yes | |||
| GN*CTYVLMR | 5,036–5,044 | Yes | No | Yes | Yes | |||
| AFGQFFSPGEVIYN*K | 4,888–4,902 | Yes | No | Yes | Yes | |||
| AFGQFFSPGEVIYN*KTDR | 4,888–4,905 | Yes | No | Yes | Yes | |||
| DIECQAESFPN*WTLAQVGQK | 1,547–1,567 | Yes | No | Yes | Yes | |||
| QVNETWTLEN*CTVAR | 4,956–4,970 | Yes | No | Yes | Yes | |||
| LDGPTEQCPDPLPLPAGN*CTDEEGICHR | 237–264 | Yes | No | Yes | Yes | |||
| FGN*LSLYLDNHYCTASATAAAAR | 5,054–5,076 | No | No | No | Yes | |||
| LPYSLFHN*NTEGQCGTCTNNQR | 5,153–5,172 | No | No | No | Yes | |||
| IPI00012887 | Cathepsin L | 37,540 | YSVAN*DTGFVDIPK | 217–230 | Yes | Yes | Yes | No |
| IPI00013972 | Carcinoembryonic antigen-related cell adhesion molecule 8 | LFIPN*ITTK | 284–292 | Yes | No | No | No | |
| IPI00017601 | Ceruloplasmin | 122,128 | EN*LTAPGSDSAVFFEQGTTR | 396–415 | No | Yes | No | No |
| EHEGAIYPDN*TTDFQR | 129–144 | No | Yes | Yes | No | |||
| IPI00019943 | Afamin | 69,024 | DIENFN*STQK | 28–37 | Yes | No | No | No |
| IPI00020091 | Alpha-1-acid glycoprotein 2 | 23,588 | QNQCFYN*SSYLNVQR | 87–101 | Yes | No | No | No |
| IPI00020487 | Extracellular glycoprotein lacritin | 14,237 | QFIEN*GSEFAQK | 115–126 | Yes | No | No | No |
| IPI00020986 | Lumican | 38,405 | LHINHNN*LTESVGPLPK | 121–137 | Yes | No | No | No |
| IPI00021891 | Splice isoform 1 of fibrinogen gamma chain | 51,479 | VDKDLQSLEDILHQVEN*K | 62–79 | Yes | Yes | No | Yes |
| DLQSLEDILHQVEN*K | 65–79 | Yes | No | No | Yes | |||
| IPI00022229 | Apolipoprotein B-100 | 515,241 | FVEGSHN*STVSLTTK | 3,405–3,419 | No | No | No | Yes |
| IPI00022395 | Complement component C9 | 63,133 | AVN*ITSENLIDDVVSLIR | 413–430 | Yes | No | No | No |
| IPI00022417 | Leucine-rich alpha-2-glycoprotein | 38,944 | KLPPGLLAN*FTLLR | 178–191 | Yes | Yes | No | Yes |
| IPI00022429 | Alpha-1-acid glycoprotein 1 | 2,477 | QDQCIYN*TTYLNVQR | 87–101 | Yes | No | No | No |
| IPI00022431 | Alpha-2-HS-glycoprotein | 39,300 | AALAAFNAQNN*GSNFQLEEISR | 166–187 | Yes | Yes | Yes | Yes |
| VCQDCPLLAPLN*DTR | 145–159 | Yes | Yes | No | No | |||
| IPI00022463 | Serotransferrin | 77,000 | QQQHLFGSN*VTDCSGNFCLFR | 622–642 | Yes | Yes | No | Yes |
| CGLVPVLAENYN*K | 421–433 | Yes | Yes | No | Yes | |||
| IPI00022488 | Hemopexin | 51,643 | ALPQPQN*VTSLLGCTH | 747–762 | Yes | Yes | Yes | Yes |
| SWPAVGN*CSSALR | 181–193 | Yes | No | No | No | |||
| IPI00643034 | Splice isoform 1 of phospholipid transfer protein | 54,705 | VSN*VSCQASVSR | 141–152 | No | Yes | Yes | Yes |
| GAFFPLTERN*WSLPNR | 236–244 | No | Yes | No | Yes | |||
| IYSN*HSALESLALIPLQAPLK | 395–415 | No | Yes | Yes | Yes | |||
| GKEGHFYYN*ISEVK | 56–69 | No | Yes | No | Yes | |||
| IPI00022974 | Prolactin-inducible protein | 16,562 | TFYWDFYTN*R | 97–106 | Yes | No | No | Yes |
| IPI00023673 | Galectin-3 binding protein | 65,289 | ALGFEN*ATQALGR | 64–76 | Yes | Yes | Yes | Yes |
| EPGSN*VTMSVDAECVPMVR | 188–206 | Yes | No | Yes | No | |||
| YKGLN*LTEDTYKPR | 394–405 | Yes | Yes | No | Yes | |||
| GLN*LTEDTYKPR | 396–407 | Yes | Yes | Yes | Yes | |||
| AAIPSALDTN*SSK | 542–554 | Yes | Yes | Yes | Yes | |||
| TVIRPFYLTN*SSGVD | 571–585 | Yes | No | No | No | |||
| IPI00025023 | Lactoperoxidase | 80,237 | ASLTN*VTDPSLDLTSLSLEVGCGAPAPVVR | 102–131 | No | Yes | Yes | Yes |
| IVGYLNEEGVLDQN*R | 199–213 | Yes | Yes | Yes | Yes | |||
| LRN*LSSPLGLMAVNQEVSDHGLPYLPYDSK | 320–349 | Yes | Yes | Yes | Yes | |||
| KPSPCEFIN*TTAR | 350–362 | Yes | Yes | Yes | Yes | |||
| IPI00025753 | Desmoglein 1 | 113,644 | TGEIN*ITSIVDR | 106–117 | Yes | No | No | No |
| IPI00025846 | Splice isoform 1 of desmocollin-2 | 99,899 | NGIYN*ITVLASDQGGR | 542–557 | Yes | No | No | Yes |
| AN*YTILK | 391–397 | Yes | No | No | Yes | |||
| AN*YTILKGNENGNFK | 391–405 | Yes | No | No | No | |||
| LKAIN*DTAAR | 625–634 | Yes | No | No | No | |||
| IPI00027192 | Procollagen-lysine | 83,497 | EQIN*ITLDHR | 194–203 | No | No | No | Yes |
| IPI00027486 | Carcinoembryonic antigen-related cell adhesion molecule 5 | 76,748 | TLTLFN*VTR | 555–563 | Yes | No | No | No |
| IPI00030847 | Transmembrane 9 superfamily protein member 3 | 67,843 | IVDVN*LTSEGK | 170–180 | No | No | No | Yes |
| IPI00031019 | Cystatin-related epididymal spermatogenic protein | 16,265 | KLKPVN*ASNANVK | 34–46 | Yes | No | No | No |
| IPI00031121 | Carboxypeptidase E | 53,117 | GN*ETIVNLIHSTR | 138–150 | Yes | No | Yes | Yes |
| DLQGNPIAN*ATISVEGIDHDVTSAK | 382–406 | Yes | No | Yes | No | |||
| IPI00031547 | Desmoglein 3 | 107,435 | NTGDIN*ITAIVDR | 105–117 | Yes | No | No | No |
| DSTFIVN*K | 453–460 | Yes | No | No | No | |||
| LPAVWSITTLN*ATSALLR | 535–552 | Yes | No | No | Yes | |||
| IPI00032258 | Complement C4 | 192,650 | GLN*VTLSSTGR | 1,326–1,336 | Yes | Yes | No | Yes |
| IPI00032292 | Metalloproteinase inhibitor 1 | 23,156 | FVGTPEVN*QTTLYQR | 46–60 | Yes | Yes | Yes | Yes |
| IPI00032328 | Splice Isoform 1 of kininogen | 71,900 | LNAENN*ATFYFK | 289–300 | No | Yes | No | No |
| IPI00060143 | Protein FAM3D | 24,947 | GLNIALVN*GTTGAVLGQK | 100–117 | Yes | No | Yes | Yes |
| IPI00166729 | Alpha-2-glycoprotein 1, zinc | 34,223 | DIVEYYNDSN*GSHVLQGR | 100–117 | Yes | No | Yes | No |
| FGCEIENN*R | 118–126 | Yes | No | Yes | Yes | |||
| FGCEIENN*RSSGAFWK | 118–133 | Yes | No | No | No | |||
| IPI00171411 | Golgi membrane protein 1 | 49,768 | AVLVNN*ITTGER | 113–124 | Yes | No | Yes | Yes |
| LQQDVLQFQKN*QTNLER | 143–152 | Yes | No | No | Yes | |||
| IPI00178926 | Immunoglobulin J chain | 18,087 | EN*ISDPTSPLR | 70–80 | Yes | Yes | Yes | Yes |
| IIVPLNNREN*ISDPTSPLR | 62–80 | Yes | Yes | Yes | Yes | |||
| IPI00218413 | Biotinidase | 61,093 | NPVGLIGAEN*ATGETDPSHSK | 320–340 | No | No | Yes | No |
| IPI00218460 | Splice isoform 3 of attractin | 158,537 | IDSTGN*VTNELR | 411–422 | Yes | No | No | No |
| IPI00242956 | Fc fragment of IgG binding protein | 571,718 | LLISSLSESPASVSILSQADN*TSK | 55–78 | No | No | Yes | No |
| LLISSLSESPASVSILSQADN*TSKK | 55–79 | No | No | No | Yes | |||
| VTVRPGESVMVN*ISAK | 78–95 | Yes | No | Yes | Yes | |||
| KVTVRPGESVMVN*ISAK | 79–95 | Yes | No | No | No | |||
| VVTVAALGTN*ISIHKDEIGK | 1,734–1,748 | Yes | No | Yes | No | |||
| VITVQVAN*FTLR | 2,511–2,522 | Yes | No | Yes | No | |||
| YLPVN*SSLLTSDCSER | 5,182–5,197 | Yes | No | No | No | |||
| IPI00400826 | Clusterin | 57,796 | LAN*LTQGEDQYYLR | 424–437 | Yes | Yes | Yes | Yes |
| MLN*TSSLLEQLNEQFNWVSR | 352–364 | No | Yes | No | Yes | |||
| IPI00291410 | Isoform 1 of long palate, lung, and nasal epithelium carcinoma-associated protein 1 | 54,878 | DHN*ATSILQQLPLLSAMR | 46–65 | Yes | Yes | Yes | Yes |
| GDQLILNLNN*ISSDR | 392–406 | Yes | No | No | No | |||
| IPI00291488 | Splice isoform of WAP four-disulfile core domain protein 2 | 12,984 | TGVCPELQADQN*CTQECVSDSECADNLK | 33–60 | Yes | No | No | No |
| IPI00295105 | Carbonic anhydrase VI | 35,343 | LENSLLDHRN*K | 247–257 | Yes | No | No | No |
| GLN*MTGYETQAGEFPMVNNGHTVQIGLPSTMR | 65–96 | No | Yes | Yes | Yes | |||
| IPI00296099 | Thrombospondin 1 | 129,330 | VVN*STTGPGEHLR | 1,065–1,077 | Yes | Yes | Yes | Yes |
| IPI00296654 | Bactericidal/permeability-increasing protein-like 1 | 49,100 | LGATPVAMLHTNN*ATLR | 320–336 | Yes | Yes | Yes | Yes |
| LLAAAN*FTFK | 91–100 | Yes | No | Yes | Yes | |||
| SDDNLLN*TSALGR | 287–299 | Yes | No | Yes | No | |||
| IPI00297910 | Tumor-associated calcium signal transducer 2 | 35,687 | HRPTAGAFN*HSDLDAELR | 160–177 | Yes | No | Yes | Yes |
| IPI00298082 | Calcium activates chloride channel protein 2 | 101,288 | AAN*SSVPPITVNAK | 586–559 | No | No | No | Yes |
| DSFDDALQVN*TTDLSPK | 802–818 | Yes | No | No | Yes | |||
| N*VSILIPENWK | 75–85 | No | No | No | Yes | |||
| IPI00298237 | Tripeptidyl-peptidase I | 61,708 | FLSSSPHLPPSSYFN*ASGR | 429–447 | Yes | No | No | No |
| IPI00298497 | Fibrinogen beta chain | 55,892 | GTAGNALMDGASQLMGEN*R | 377–395 | No | Yes | No | No |
| IPI00298828 | Beta-2-glycoprotein I | 38,273 | VYKPSAGN*NSLYR | 155–167 | Yes | Yes | Yes | Yes |
| LGN*WSAMPSCK | 251–261 | Yes | No | No | No | |||
| IPI00299547 | Neutrophil gelatinase-associated lipocalin | 22,774 | EDKSYN*VTSVLFR | 64–76 | Yes | No | No | No |
| SYN*VTSVLFR | 83–92 | Yes | No | No | No | |||
| IPI00299729 | Transcobalamin I | 48,164 | ADEGSLKN*ISIYTK | 209–222 | Yes | Yes | Yes | Yes |
| MN*DTIFGFTMEER | 368–380 | Yes | Yes | Yes | Yes | |||
| N*ISIYTK | 216–222 | No | No | Yes | Yes | |||
| AQKMN*DTIFGFTMEER | 365–380 | Yes | No | No | No | |||
| IPI00300786 | Salivary alpha-amylase | 57,731 | NVVDGQPFTNWYDN*GSNQVAFGR | 414–436 | Yes | Yes | Yes | Yes |
| IPI00304557 | Short palate, lung, and nasal epithelium carcinoma-associated protein 2 | 26,995 | GLN*LSFPVTAN*VTVAGPIIGQIINLK | 122–147 | Yes | No | Yes | Yes |
| AEPIDDGKGLN*LSFPVTAN*VTVAGPIIGQIINLK | 114–147 | Yes | No | No | No | |||
| IPI00553177 | Alpha-1-antitrypsin | 46,737 | YLGN*ATAIFFLPDEGK | 268–283 | Yes | Yes | Yes | Yes |
| ADTHDEILEGLNFN*LTEIPEAQIHEGFQELLR | 94–125 | No | Yes | Yes | Yes | |||
| IPI00303333 | Platelet receptor GI24 | 33,901 | HGLESASDHHGN*FSITMR | 117–134 | No | No | No | Yes |
| IPI00328460 | Transmembrane protease, serine 11B | 46,235 | MLNAFQN*SSIYK | 81–92 | No | No | Yes | Yes |
| LLPNAN*GSNVQLQLK | 102–116 | No | No | Yes | Yes | |||
| IPI00328960 | Similar to carcinoembryonic antigen-related cell adhesion molecule 1 | 91,568 | FVTAGSN*VTLR | 427–437 | Yes | No | Yes | Yes |
| TPASN*ISTQVSHTK | 140–153 | Yes | No | No | No | |||
| IPI00332845 | Leucine-rich repeats and calponin homology (CH) domain containing 2 | 69,610 | MLTYLN*ISR | 158–166 | No | No | No | Yes |
| KLPPGLLAN*FTLLR | 178–191 | No | No | No | Yes | |||
| IPI00333140 | Delta-notch-like EGF repeat-containing transmembrane | 78,438 | LVSFEVPQN*TSVK | 215–256 | Yes | Yes | Yes | No |
| IPI00335356 | IGHM protein | 49,526 | GLTFQQN*ASSMCVPDQDTAIR | 354–374 | Yes | Yes | Yes | Yes |
| IPI00374315 | Hypothetical LOC389429 | 37,902 | IILN*QTAR | 66–73 | Yes | No | Yes | Yes |
| MGMYKIILN*QTAR | 61–73 | Yes | No | No | No | |||
| IPI00384872 | UDP-glucuronosyltransferase 1–7 | 59,779 | YTGTRPSNLAN*NTILVK | 334–350 | No | No | Yes | Yes |
| IPI00641229 | Ig alpha-2 chain C region | 55,715 | LAGKPTHVN*VSVVMAEVDGTCY | 319–340 | Yes | Yes | Yes | Yes |
| LSLHRPALEDLLLGSEAN*LTCTLTGLR | 114–140 | Yes | Yes | Yes | Yes | |||
| TPLTAN*ITK | 200–208 | Yes | No | Yes | No | |||
| IPI00418512 | DMBT1 | 259,409 | LVNLN*SSYGLCAGR | 998–1,111 | Yes | Yes | Yes | Yes |
| QADN*DTIDYSNFLTAAVSGGIIK | 1,298–1,320 | Yes | Yes | Yes | Yes | |||
| CSGN*ESYLWSCPHK | 822–835 | Yes | No | No | No | |||
| IPI00419164 | Alpha-2-macroglobulin like protein 1 | 149,965 | LGHIN*FTISTK | 863–873 | Yes | No | Yes | Yes |
| QGN*GTFVQTDKPLYTPGQQVYFR | 118–140 | No | No | Yes | Yes | |||
| HSN*GSYSAFGER | 1,018–1,029 | No | No | No | Yes | |||
| IPI00423461 | Hypothetical protein DKFZp686C02220 | 54,126 | HYTN*SSQDVTVPCR | 249–262 | No | No | Yes | No |
| IPI00431645 | HP protein | 31,362 | MVSHHN*LTTGATLINEQWLLTTAK | 54–77 | Yes | Yes | No | No |
| NLFLN*HSEN*ATAK | 78–90 | Yes | Yes | Yes | Yes | |||
| VVLHPN*YSQVDIGLIK | 170–186 | Yes | Yes | Yes | Yes | |||
| IPI00168728 | FLJ00385 protein (fragment) | 56,111 | EEQFN*STFR | 285–294 | Yes | Yes | Yes | Yes |
| IPI00550991 | Alpha-1-antichymotrypsin | 50,596 | FN*LTETSEAEIHQSFQHLLR | 190–219 | No | Yes | No | Yes |
| YTGN*ASALFILPDQDK | 293–308 | No | No | Yes | No | |||
N* indicates the site of former N-linked glycosylation
WS Whole saliva, PA parotid, SM submandibular, SL sublingual
A gene ontology (GO) analysis of the N-glycoproteins identified in saliva to categorize them according to their cellular location, function, and processes is shown in Fig. 6. The majority of the N-glycoproteins are extracellular (40%; Fig. 6a). Several identified N-glycoproteins were annotated as membrane proteins (14%). Liu and coworkers employed the similar hydrazide chemistry to analyze the human plasma N-glycoproteome and observed a similar trend [42]; the majority of the plasma N-glycoproteins are extracellular or plasma membrane proteins. However, nuclear, cytoplasmic, or cytoskeletal proteins were not identified in the plasma N-glycoprotein study, whereas in our study of salivary proteins, we observed proteins from the nucleus, cytoplasm, and cytoskeleton. Figure 6b shows the GO distribution of salivary glycoproteins based on their functions. Fifty-one percent of the glycoproteins identified are involved in binding, a trend observed also in the analysis of plasma N-glycoproteins [42]. A large number of salivary N-glycoproteins also showed catalytic activity and also a role in enzyme regulation. Salivary N-glycoproteins are involved in catalytic functions such as peptidase, nuclease, hydrolase, and transferase activities. N-Glycoproteins detected in whole saliva are also involved in biological processes such as metabolism, transport, response to stimuli, response to stress, and signal transduction.
Fig. 6.
Gene ontology analysis of the a cellular distribution and b cellular functional distribution of N-glycoproteins identified in salivary fluids
Discussion
The glycoprotein pulldown method using agarose–hydrazide is an efficient technique for identifying N-linked glycoproteins in complex mixtures. We employed this method to study the whole salivary N-glycoproteome by the Zhang method [41, 44] and the Sun method [46]. This aided us to extend our list of whole-salivary N-glycoproteins that may not have been achieved by the application of one method alone. In our study of N-glycoproteins in whole saliva, several proteins were identified by the Zhang method that were not identified by the Sun method and vice versa.
In this current study, we attempted to probe deeper into the salivary proteome and extend the N-glycoprotein list by examining not only whole saliva but PA, SM, and SL fluids. Combining data sets obtained from whole saliva, PA, SM, and SL saliva, we measured 148 sites of N-linked glycosylation to date. Larsen et al. reported 97 sites of N-linked glycosylation in whole saliva [40]. An overlap of 62 sites of N-glycosylation was found in a comparison of the two studies. In a comparison of N-glycoproteins in whole saliva versus PA, SM, and SL saliva, a large number of N-glycoproteins were identified in PA, SM, and SL saliva that were not found in whole saliva and vice versa. In our study, 15 N-glycoproteins were detected in PA, SM, or SM saliva and were not found in whole saliva.
In this study, we report isolation of glycoproteins from the fluids from the SM and SL glands separately. The SM and SL glands are both located below the tongue. It was earlier believed that it is difficult to segregate the two fluids, as they often share a common salivary duct to empty the contents of the gland into the oral cavity [53]. However, other studies have shown that many individuals have separate salivary ducts for the two glands, and it is possible to collect secretions from SM and SL separately [54]. Many new devices have been fabricated that make the collection of segregated SM and SL saliva easier [50, 55]. Hu and coworkers found markers that clearly differentiate SM from SL saliva. They found cystatin C to be a specific marker for SM saliva and Muc5B and calgranulin B for SL saliva [56]. During the course of collection of SM and SL saliva for our experiments, calgranulin B was employed as a marker to ensure purity of the segregated samples. Cystatin C and calgranulin B have no sites of N-linked glycosylation and were thus not detected by our method. However, Muc5B has several known and putative sites of N-glycosylation. In our current study, we found formerly N-glycosylated peptides in SM and SL, but we found a slightly larger number of Muc5B peptides in SL compared to SM; a total of ten formerly N-glycosylated peptides were found in SL and only eight in SM. Hu et al. found no peptides from Muc5B in SM saliva in their shotgun protein identification experiments. However, we did find differences in the overall N-glycoprotein profile between SM and SL saliva (Fig. 3). As a result, we believe that collection of segregated SM and SL fluids can be accomplished and their salivary proteomes can be differentiated.
Studies on the salivary glycoproteins have potential implications on salivary biomarker discovery efforts, as saliva is gaining popularity as a diagnostic fluid for disease markers. Many attempts have been made in the past few years to use saliva to discover biomarkers for diseases localized in the oral cavity or in the head/neck region. Some diseases for which saliva has been used as a medium for biomarker discovery include oral cancer squamous cell carcinoma [57–65], head and neck squamous cell carcinoma [66–68], and Sjogren’s syndrome [69–77]. In addition, saliva has shown promise for use in the detection of many nonoral (i.e., nonproximal) systemic diseases. In patients with breast cancer, elevated levels of C-erb2 and CA15-3 have been detected in saliva of cancer patients versus control subjects [78–80]. Antibodies to the human immunodeficiency virus (HIV) have been found in saliva of HIV-positive patients; saliva-based HIV tests are now gaining popularity [81]. We believe our studies on N-glycoproteins in whole saliva will benefit future work on disease biomarker discovery.
Acknowledgments
We wish to thank Drs. Jimmy Ytterberg, Rachel Ogorzalek Loo, Shen Hu, and Melissa Sondej (UCLA) for their helpful advice. The UCLA Mass Spectrometry and Proteomics Technology Center was established with a grant from the W. M. Keck Foundation. This work was supported by a grant from the National Institutes of Health (National Institute of Dental and Craniofacial Research; U01 DE016275) to DTWand JAL, the Ruth L. Kirschstein National Service Award (GM07185), and the UCLA Fundamental Clinical Research Training Grant (T32 DE007296; to P. R).
Contributor Information
Prasanna Ramachandran, Email: pramacha@chem.ucla.edu, Department of Chemistry and Biochemistry, Molecular Biology Institute, University of California—Los Angeles, 402 Paul D. Boyer Hall, 405 Hilgard Avenue, Los Angeles, CA 90095, USA.
Pinmanee Boontheung, Email: pinmanee@chem.ucla.edu, Department of Chemistry and Biochemistry, Molecular Biology Institute, University of California—Los Angeles, 402 Paul D. Boyer Hall, 405 Hilgard Avenue, Los Angeles, CA 90095, USA.
Eric Pang, Email: ericpang@chem.ucla.edu, Department of Chemistry and Biochemistry, Molecular Biology Institute, University of California—Los Angeles, 402 Paul D. Boyer Hall, 405 Hilgard Avenue, Los Angeles, CA 90095, USA.
Weihong Yan, Email: wyan@chem.ucla.edu, Department of Chemistry and Biochemistry, Molecular Biology Institute, University of California—Los Angeles, 402 Paul D. Boyer Hall, 405 Hilgard Avenue, Los Angeles, CA 90095, USA.
David T. Wong, Email: dtww@ucla.edu, School of Dentistry, Dental Research Institute, University of California—Los Angeles, Los Angeles, CA 90095, USA.
Joseph A. Loo, Email: JLoo@chem.ucla.edu, Department of Chemistry and Biochemistry, Molecular Biology Institute, University of California—Los Angeles, 402 Paul D. Boyer Hall, 405 Hilgard Avenue, Los Angeles, CA 90095, USA; Department of Biological Chemistry, David Geffen School of Medicine, Los Angeles, CA, USA.
References
- 1.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 2.Collins BE, Paulson JC. Cell surface biology mediated by low affinity multivalent protein-glycan interactions. Curr Opin Chem Biol. 2004;8:617–625. doi: 10.1016/j.cbpa.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Lin X. Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 2004;131:6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- 4.Dennis JW, Granovsky M, Warren CE. Glycoprotein glycosylation and cancer progression. Biochim Biophys Acta. 1999;1:21–34. doi: 10.1016/s0304-4165(99)00167-1. [DOI] [PubMed] [Google Scholar]
- 5.Kannagi R, Izawa M, Koike T, Miyazaki K, Kimura N. Carbohydrate-mediated cell adhesion in cancer metastasis and angiogenesis. Cancer Sci. 2004;5:377–384. doi: 10.1111/j.1349-7006.2004.tb03219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobata A, Amano J. Altered glycosylation of proteins produced by malignant cells, and application for the diagnosis and immunotherapy of tumors. Immunol and Cell Biol. 2005;4:429–439. doi: 10.1111/j.1440-1711.2005.01351.x. [DOI] [PubMed] [Google Scholar]
- 7.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 8.Duffy MJ, Shering S, Sherry F, McDermott E, O'Higgins N. CA 15-3: a prognostic marker in breast cancer. Int J Biol Markers. 2000;15:330–333. doi: 10.1177/172460080001500410. [DOI] [PubMed] [Google Scholar]
- 9.Akiyama T, Sudo C, Ogawara H, Toyoshima K, Yamamoto T. The product of the human c-erb 2 gene: a 185-kilodalton glycoprotein with tyrosine kinase activity. Science. 1986;232:1644–1646. doi: 10.1126/science.3012781. [DOI] [PubMed] [Google Scholar]
- 10.Jankovic MM, Kosanovic MM. Glycosylation of urinary prostrate specific antigen in benign hyperplasia and cancer: assessment by lectin-binding partners. Clin Biochem. 2005;38:58–65. doi: 10.1016/j.clinbiochem.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 11.Lloyd KO, Yin BW, Kudryashov V. Isolation and characterization of ovarian cancer antigen CA125 using a new monoclonal antibody (VK-8): identification as a mucin-type molecule. Int J Cancer. 1997;71:842–850. doi: 10.1002/(sici)1097-0215(19970529)71:5<842::aid-ijc24>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 12.Blumenthal RD, Leon E, Hansen HJ, Goldenberg DM. Expression patterns of CEACAM5 and CEACAM6 in primary and metastatic cancers. BMC Cancer. 2007;7:2. doi: 10.1186/1471-2407-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helmerhorst EJ. Whole saliva proteolysis: wealth of information for diagnostic exploitation. Ann NY Acad Sci. 2007;1098:454–460. doi: 10.1196/annals.1384.013. [DOI] [PubMed] [Google Scholar]
- 14.Helmerhorst EJ, Oppenheim FG. Saliva: a dynamic proteome. J Dent Res. 2007;86:680–693. doi: 10.1177/154405910708600802. [DOI] [PubMed] [Google Scholar]
- 15.Hatton MN, Loomis RE, Levine MJ, Tabak LA. Masticatory lubrication. The role of carbohydrate in the lubricating property of a salivary glycoprotein-albumin complex. Biochem J. 1985;230:817–820. doi: 10.1042/bj2300817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loomis RE, Prakobphol A, Levine MJ, Reddy MS, Jones PC. Biochemical and biophysical comparison of two mucins from human submandibular–sublingual saliva. Arch Biochem Biophys. 1987;258:452–464. doi: 10.1016/0003-9861(87)90366-3. [DOI] [PubMed] [Google Scholar]
- 17.Thomsson KA, Prakobphol A, Leffler H, et al. The salivary mucin MG1 (MUC5B) carries a repertoire of unique oligosaccharides that is large and diverse. Glycobiology. 2002;12:1–14. doi: 10.1093/glycob/12.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Gabriel MO, Grunheid T, Zentner A. Glycosylation pattern and cell attachment-inhibiting property of human salivary mucins. J Periodontol. 2005;76:1175–1181. doi: 10.1902/jop.2005.76.7.1175. [DOI] [PubMed] [Google Scholar]
- 19.Prakobphol A, Murray PA, Fisher SJ. Bacterial adherence on replicas of sodium dodecyl sulfate-polyacrylamide gels. Anal Biochem. 1987;164:5–11. doi: 10.1016/0003-2697(87)90359-9. [DOI] [PubMed] [Google Scholar]
- 20.Prakobphol A, Tangemann K, Rosen SD, Hoover CI, Leffler H, Fisher SJ. Separate oligosaccharide determinants mediate interactions of the low molecular weight salivary mucin with neutrophils and bacteria. Biochemistry. 1999;38:6817–6825. doi: 10.1021/bi990145m. [DOI] [PubMed] [Google Scholar]
- 21.Ericson T, Rundergren J. Characterization of a salivary agglutinin reacting with a serotype c strain of Streptococcus mutans. Eur J Biochem. 1983;133:2023–2031. doi: 10.1111/j.1432-1033.1983.tb07456.x. [DOI] [PubMed] [Google Scholar]
- 22.Hartshorn KL, Ligtenberg A, White MR, et al. Salivary agglutinin and lung scavenger receptor cysteine-rich glycoprotein 340 have broad anti-influenza activities and interactions with surfactant protein D that vary according to donor source and sialation. Biochem J. 2006;393:545–553. doi: 10.1042/BJ20050695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Hoeven JS, cvan der Kieboom CW, Camp PJ. Utilization of mucin by oral Streptococcus species. Antonie Van Leeuwenhoek. 1990;57:165–172. doi: 10.1007/BF00403951. [DOI] [PubMed] [Google Scholar]
- 24.McBride BC, Gisslow MT. Role of sialic acid in saliva-induced aggregation of Streptococcus sanguis. Infect Immun. 1977;18:171–177. doi: 10.1128/iai.18.1.35-40.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaji H, Saito H, Yamauchi Y, et al. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat Biotechnol. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- 26.Sharon N, Lis H. History of lectins: from hemagglutinins to biological recognition molecules. Glycobiology. 2004;14:53R–62R. doi: 10.1093/glycob/cwh122. [DOI] [PubMed] [Google Scholar]
- 27.Yang Z, Hancock WS. Approach to the comprehensive analysis of glycoproteins isolated from human serum using a multi-lectin affinity column. J Chromatogr A. 2004;1053:79–88. [PubMed] [Google Scholar]
- 28.Yang Z, Hancock WS. Monitoring glycosylation pattern changes of glycoproteins using multi-lectin affinity chromatography. J Chromatogr A. 2005;1020:57–64. doi: 10.1016/j.chroma.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 29.Yang Z, Hancock WS, Chew TR, Bonilla L. A study of glycoproteins in human serum and plasma reference standards (HUPO) using multilectin affinity chromatography coupled with RPLC-MS/MS. Proteomics. 2005;5:3353–3366. doi: 10.1002/pmic.200401190. [DOI] [PubMed] [Google Scholar]
- 30.Durham M, Regnier FE. Targeted glycoproteomics: serial lectin affinity chromatography in the selection of O-glycosylation sites on proteins from the blood proteome. J Chromatogr A. 2006;1132:165–173. doi: 10.1016/j.chroma.2006.07.070. [DOI] [PubMed] [Google Scholar]
- 31.Drake RR, Schwegler EE, Malik G, et al. Lectin capture strategies combined with mass spectrometry for the discovery of serum glycoprotein biomarkers. Mol Cell Proteomics. 2006;5:1957–1967. doi: 10.1074/mcp.M600176-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Palvina T, Wakshull E, Hancock WS, Hincapie M. Combination of abundant protein depletion and multi-lectin affinity chromatography (M-LAC) for plasma protein biomarker discovery. J Proteome Res. 2007;6:662–671. doi: 10.1021/pr060413k. [DOI] [PubMed] [Google Scholar]
- 33.Monzo A, Bonn GK, Guttman A. Boronic acid-lectin affinity chromatography. 1. Simulteneous glycoprotein binding with selective or combined elution. Anal Bioanal Chem. 2007;389:2097–2102. doi: 10.1007/s00216-007-1627-y. [DOI] [PubMed] [Google Scholar]
- 34.Sparbier K, Asperger A, Resemann A, et al. Analysis of glycoproteins in human serum by means of glycospecific magnetic bead separation and LC-MALDI-TOF/TOF analysis with automated glycopeptide detection. J Biomol Tech. 2007;18:252–258. [PMC free article] [PubMed] [Google Scholar]
- 35.Wada Y, Tajiri M, Yoshida S. Hydrophilic affinity isolation and MALDI multiple-stage tandem mass spectrometry of glycopeptides for glycoproteomics. Anal Chem. 2004;76:6560–6565. doi: 10.1021/ac049062o. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Wang DIC. Quantitative analysis and process monitoring of site-specific glycosylation microheterogeneity in recombinant human interferon from Chinese hamster ovary cell culture by hydrophilic interaction chromatography. J Chromatogr B. 1998;712:73. doi: 10.1016/s0378-4347(98)00131-5. [DOI] [PubMed] [Google Scholar]
- 37.Hagglund P, Bunkenborg J, Elortza F, Jensen ON, Roepstorff P. A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J Proteome Res. 2004;3:556–566. doi: 10.1021/pr034112b. [DOI] [PubMed] [Google Scholar]
- 38.Hagglund P, Matthiesen R, Elortza F, et al. An enzymatoc deglycosylation scheme enabling identification of core fucosylated N-glycans and O-glycosylation site mapping of human plasma proteins. J Proteome Res. 2007;6:3021–3031. doi: 10.1021/pr0700605. [DOI] [PubMed] [Google Scholar]
- 39.Pinske MW, Uitto PM, Hilhorst MJ, Ooms B, Heck AJ. Selective isolation at femtomole level of phosphopeptides from proteolytic digests using 2D-Nano-LC-ESI-MS/MS and titanium oxide precolumns. Anal Chem. 2004;76:3935–3943. doi: 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- 40.Larsen MR, Jensen SS, Jakobsen LA, Heegaard NHH. Exploring the sialome using titanium dioxide chromatography and mass spectrometry. Mol Cell Proteomics. 2007;6:1778–1787. doi: 10.1074/mcp.M700086-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Zhang H, Li X, Martin DB, Aerbersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotech. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 42.Liu T, Qian WJ, Gritsenko MA, et al. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J Proteome Res. 2005;4:2070–2080. doi: 10.1021/pr0502065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H, Yi EC, Li X, et al. High throughput quantitative analysis of serum proteins using glycopeptide capture and liquid chromatography mass spectrometry. Mol Cell Proteomics. 2005;4:144–155. doi: 10.1074/mcp.M400090-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Ramachandran P, Boontheung P, Xie Y, Sondej M, Wong DT, Loo JA. Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J Proteome Res. 2006;5:1493–1503. doi: 10.1021/pr050492k. [DOI] [PubMed] [Google Scholar]
- 45.Pan S, Wang Y, Quinn JF, et al. Identification of glycoproteins in human cerebrospinal fluid with a complementary proteomic approach. J Proteome Res. 2006;5:2769–2779. doi: 10.1021/pr060251s. [DOI] [PubMed] [Google Scholar]
- 46.Sun B, Ranish JA, Utleg AG, et al. Shotgun glycopeptide capture approach coupled with mass spectrometry for comprehensive glycoproteomics. Mol Cell Proteomics. 2007;6:141–149. doi: 10.1074/mcp.T600046-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Tian Y, Zhou Y, Elliott S, Aebersold R, Zhang H. Solid-phase extraction of N-linked glycopeptides. Nat Protocols. 2007;2:334–339. doi: 10.1038/nprot.2007.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Y, Aebersold R, Zhang H. Isolation of N-linked glycopeptides from plasma. Anal Chem. 2007;79:5826–5837. doi: 10.1021/ac0623181. [DOI] [PubMed] [Google Scholar]
- 49.Lashley LS. Reflex secretion of the human parotid gland. J Exp Psychol. 1916;1:461–493. [Google Scholar]
- 50.Wolff A, Begleiter A, Moskana D. A novel system of human submandibular/sublingual saliva collection. J Dent Res. 1997;76:1782–1786. doi: 10.1177/00220345970760111001. [DOI] [PubMed] [Google Scholar]
- 51.Kaufman E, Lamster IB. The diagnostic application of saliva—a review. Crit Rev Oral Biol Med. 2002;13:197–212. doi: 10.1177/154411130201300209. [DOI] [PubMed] [Google Scholar]
- 52.Denny P, Hagen FK, Hardt M, et al. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J Proteome Res. 2008;7:1994–2006. doi: 10.1021/pr700764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sicher H, Du Brul EL. Oral anatomy. 6th ed. St. Louis, MO: C. V. Mosby; 1975. [Google Scholar]
- 54.Truelove EL, Bixler D, Merrit AD. Simplified method for collection of pure submandibular saliva in large volumes. J Dent Res. 1967;46:1400–1403. doi: 10.1177/00220345670460064301. [DOI] [PubMed] [Google Scholar]
- 55.Veerman EC, van der Keybus PA, Vissink A, Nieuw Amerongen AV. Human glandular salivas: their separate collection and analysis. Euro J Oral Sci. 1996;104:346–352. doi: 10.1111/j.1600-0722.1996.tb00090.x. [DOI] [PubMed] [Google Scholar]
- 56.Hu S, Denny P, Denny P, et al. Defferentially expressed protein markers in human submandibular and sublingual secretions. Int J Oncol. 2004;25:1423–1430. [PubMed] [Google Scholar]
- 57.Hu S, Yu T, Xie Y, et al. Discovery of oral fluid biomarkers for human oral cancer by mass spectrometry. Cancer Genomics Proteomics. 2007;4:55–64. [PubMed] [Google Scholar]
- 58.Meyer P, Zechel T. Quantitative studies of lysozymes and phosphohexose isomerase in mixed saliva in oral squamous epithelial carcinoma. HNO. 2001;49:626–629. doi: 10.1007/s001060170058. [DOI] [PubMed] [Google Scholar]
- 59.Nagler R, Bahar G, Shpitzer T, Feinmesser R. Concomitant analysis of salivary tumor markers-a new diagnostic tool for oral cancer. Clin Cancer Res. 2006;12:3979–3984. doi: 10.1158/1078-0432.CCR-05-2412. [DOI] [PubMed] [Google Scholar]
- 60.Negri L, Pacchioni D, Calabrese F, Giacomasso S, Mastromatteo V, Fazio M. Serum and salivary CEA and GICA levels in oral cavity tumors. Int J Bio Markers. 1988;3:107–112. [PubMed] [Google Scholar]
- 61.Pickering V, Jordan RC, Schmidt BL. Elevated salivary endothelin levels in oral cancer patient-a pilot study. Oral Oncol. 2007;43:37–41. doi: 10.1016/j.oraloncology.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 62.Rhodus NL, Ho V, Miller CS, Myers S, Ondrey F. NF-kappaB dependant cytokine levels in saliva of patients with oral preneoplastic lesions and oral squamous cell carcinoma. Cancer Detec Prev. 2005;29:42–45. doi: 10.1016/j.cdp.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 63.St John MA, Li Y, Zhao X, et al. Interleukin 6 and interleukin 8 as potential biomarkers for oral cavity and oropharyngeal squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2004;130:929–935. doi: 10.1001/archotol.130.8.929. [DOI] [PubMed] [Google Scholar]
- 64.Xie H, Onsongo G, Popko J, et al. Proteomics Analysis of cells in whole saliva from oral cancer patients via value-added three-dimensional peptide fractionation and tandem mass spectrometry. Mol Cell Proteomics. 2008;7:486–498. doi: 10.1074/mcp.M700146-MCP200. [DOI] [PubMed] [Google Scholar]
- 65.Zhong LP, Chen GF, Xu ZF, Zhang X, Ping FY, Zhao SF. Detection of telomerase activity in saliva from oral squamous cell carcinoma patients. Int J Oral Maxillofac Surg. 2005;34:566–570. doi: 10.1016/j.ijom.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 66.Drake RR, Cazare LH, Semmes OJ, Wadsworth JT. Serum, salivary and tissue proteomics for discovery of biomarkers for head and neck cancer. Expert Rev Mol Diagn. 2005;5:93–100. doi: 10.1586/14737159.5.1.93. [DOI] [PubMed] [Google Scholar]
- 67.Franzmann EJ, Reategi EP, Pedroso F, et al. Soluble CD44 is a potential marker for the early detection of head and neck cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:1348–1355. doi: 10.1158/1055-9965.EPI-06-0011. [DOI] [PubMed] [Google Scholar]
- 68.Franzmann EJ, Schroeder GL, Goodwin WJ, Weed DT, Fisher P, Lokeshwar VB. Expression of tumor markers hyaluronic acid and hyaluronidase (HYAL1) in head and neck tumors. Int J Cancer. 2003;106:438–445. doi: 10.1002/ijc.11252. [DOI] [PubMed] [Google Scholar]
- 69.Castro J, Jimenez-Alonso J, Sabio JM, et al. Salivary and serum deta2-microglobulin and gamma-glutamyl-transferase in patients with primary Sjogren syndrome and Sjogren syndrome secondary to systemic lupus erythematosus. Clin Chim Acta. 2003;334:225–231. doi: 10.1016/s0009-8981(03)00162-1. [DOI] [PubMed] [Google Scholar]
- 70.Grisius MM, Bermudez DK, Fox PC. Salivary and serum interleukin 6 in primary Sjogren's syndrome. J Rheumatol. 1997;24:1089–1091. [PubMed] [Google Scholar]
- 71.Haghighat N, al-Hashimi I. The status of lactoferrin and total iron binding capacity of human parotid saliva in Sjogren’s syndrome. Clin Exp Rheumatol. 2003;21:485–488. [PubMed] [Google Scholar]
- 72.Hu S, Wang J, Meijer J, et al. Salivary proteomic and genomic biomarkers for primary Sjogren’s syndrome. Arthritis Rheum. 2007;56:3588–3600. doi: 10.1002/art.22954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ryu OH, Atkinson JC, Hoehn GT, Illei GG, Hart TC. Identification of parotid salivary biomarkers in Sjogren’s syndrome by surface enhanced laser desorption/ionization time-of-flight mass spectrometry and two dimensional gel electrophoresis. Rheumatol. 2006;45:1077–1086. doi: 10.1093/rheumatology/kei212. [DOI] [PubMed] [Google Scholar]
- 74.Sfrisco P, Ostuni P, Botsios C, et al. Serum and salivary neopterin and interferon-gamma in primary Sjogren’s syndrome. Correlation with clinical, laboratory and histopathological features. Scand J Rheumatol. 2003;32:74–78. doi: 10.1080/03009740310000067. [DOI] [PubMed] [Google Scholar]
- 75.Takei M, Azuhata T, Yoshimatu T, et al. Increased soluble CD23 moleculaes in serum/saliva and correlation with the stage of sialoectasis in patients with primary Sjogren’s syndrome. Clin Exp Rheumatol. 1995;13:711–715. [PubMed] [Google Scholar]
- 76.Tishler M, Yaron I, Shirazi I, Yossipov Y, Yaron M. Increased salivary interleukin-6 levels in patients with primary Sjogren’s syndrome. Rheumatol Int. 1999;18:125–127. doi: 10.1007/s002960050070. [DOI] [PubMed] [Google Scholar]
- 77.Yuvaz S, Toker E, Bicakcigil M, Mumcu G, Cakir S. Comparative analysis of autoantibodies against a-fodrin in serum, tear fluid and saliva from patients with Sjogren’s syndrome. J Rheumatol. 2006;33:1289–1292. [PubMed] [Google Scholar]
- 78.Streckfus C, Bigler L, Dellinger T, Dai X, Kingman A, Thigpen JT. The presence of soluble c-erb-2 in saliva and serum among women with breast carcinoma: a preliminary study. Clin Cancer Res. 2000;6:2363–2370. [PubMed] [Google Scholar]
- 79.Streckfus C, Bigler L, Dellinger T, Pfeifer M, Rose A, Thigpen JT. CA15-3 and c-erb-2 presence in saliva of women. Clin Oral Invest. 1999;3:138–143. doi: 10.1007/s007840050092. [DOI] [PubMed] [Google Scholar]
- 80.Streckfus C, Bigler L, Tucci M, Thigpen JT. A preliminary study of CA15-3, c-erb2, epidermal growth factor receptor, cathepsin-D and p53 in saliva among women with breast carcinoma. Cancer Invest. 2000;18:101–109. doi: 10.3109/07357900009038240. [DOI] [PubMed] [Google Scholar]
- 81.Malamud D. Oral diagnostic testing for detecting human immunodeficiency virus-1 antibodies: a technology whose time has come. Am J Med. 1997;102:9–14. doi: 10.1016/s0002-9343(97)00032-6. [DOI] [PubMed] [Google Scholar]