Abstract
In this study, 145 peaches and nectarines displaying typical brown rot symptoms were collected from multiple provinces in China. A subsample of 26 single-spore isolates were characterized phylogenetically and morphologically to ascertain species. Phylogenetic analysis of internal transcribed spacer (ITS) regions 1 and 2, glyceraldehyde-3-phosphate dehydrogenase (G3PDH), β-tubulin (TUB2) revealed the presence of three distinct Monilinia species. These species included Monilinia fructicola, Monilia mumecola, and a previously undescribed species designated Monilia yunnanensis sp. nov. While M. fructicola is a well-documented pathogen of Prunus persica in China, M. mumecola had primarily only been isolated from mume fruit in Japan. Koch's postulates for M. mumecola and M. yunnanensis were fulfilled confirming pathogenicity of the two species on peach. Phylogenetic analysis of ITS, G3PDH, and TUB2 sequences indicated that M. yunnanensis is most closely related to M. fructigena, a species widely prevalent in Europe. Interestingly, there were considerable differences in the exon/intron structure of the cytochrome b (Cyt b) gene between the two species. Morphological characteristics, including spore size, colony morphology, lesion growth rate, and sporulation, support the phylogenetic evidence suggesting the designation of M. yunnanensis as a new species. A new multiplex PCR method was developed to facilitate the detection of M. yunnanensis and differentiation of Monilinia spp. causing brown rot of peach in China.
Introduction
China is the primarily producer of peaches (Prunus persica L. Batsch) worldwide providing approximately 43% of the world production. In addition, peaches and other agronomically important Prunus spp. are believed to have originated from Western China. Indeed, ancient records and archaeological findings, indicate that the domestication of P. persica may have occurred as early as 3000 BC [1]. Aside from P. persica, other Prunus spp. originating from Western China include Prunus davidiana (Carr.) Franch., Prunus ferganensis (Kost. and Rjab) Kov. and Kost., and Prunus kansuensis Rehd., reflecting great diversity of Prunus spp. in the region. In addition to serving as the origin of domesticated stone fruit, Western China is, likely to serve as the evolutionary origin of pathogens that cause diseases of Prunus spp.
The economically most important diseases of stone fruit are blossom blight and brown fruit rot caused by Monilinia spp. The earliest reports of brown rot on stone fruit in China were made in the 1920s [2]. At this time, fungal species were identified solely on the basis of morphology and as such were classified as Monilinia fructigena and Monilinia laxa [2], [3], [4], [5]. It was not until the 21st century that the diversity of Monilinia species in China was more extensively characterized [6], [7]. However, many PCR-based diagnostic tools used to distinguish Monilinia spp. in Europe and the Americas, failed to differentiate the morphological species believed to be present in China [8] suggesting that other undescribed species may be present. To date, however, completely thorough investigations into the species causing brown rot of peach in China have not been undertaken. Please note that although only anamorph of several species are presented and discussed in the manuscript, we will use the teleomorph designation, Monilinia, when referring to the genus in order to be consistent with the nomenclature preference of recent literature on this organism. We will however, refer to the anamorph genus name Monilia when describing the new species.
To date, only three species of Monilinia have been found to occur on Prunus species worldwide; Monilinia fructicola (G. Winter) Honey, Monilinia fructigena (Aderhold & Ruhland) Honey, and Monilinia laxa (Aderhold & Ruhland) Honey. While M. fructicola is widespread in the Americas, and parts of Europe and Asia [9], M. laxa and M. fructigena are the primary species causing brown rot of peach in Europe [10]. However, all three Monilinia spp. have also, been reported in China [2], [3], [4], [5], [6], [11]. In addition to the three ubiquitous species, two additional species were reported recently in China. These include M. polystroma, which was documented to cause brown rot of Malus spp., Pyrus spp. and Prunus spp. (but not Prunus persica) [7], [12], and M. mumecola, which was initially isolated in Japan from mume (Prunus mume) in 1982 [13], [14], and later classified as a separate species [15]. M. mumecola was described as the causal agent of the brown rot of papaya in Hubei, China in 2009 [16].
Given the evolutionary history of Prunus sp, and the extensive history of Monilinia species reporting and reclassification in Asia, there is potential for both the identification of new species, and improved taxonomic delineation of existing species. Hence, an in-depth investigation of species prevalence using modern phylogenetic tools could be warranted to better understand disease concerns as they relate to species identity in the intensive modern production systems of peach and nectarine in China. The objectives of this study were to: 1) Survey and describe the Monilinia species attacking peach in the primary production regions of China; 2) morphologically and phylogenetically characterize a subset of isolates representing each species identified from each region; and 3) develop a rapid, PCR-based method to differentiate the previously documented, and newly described Monilinia species attacking peach in China.
Materials and Methods
Ethics statement
Hundreds of samples were collected in this study and sampling was always conducted with the approval of the owners of the fields.
Collection of Monilinia isolates
Peach and nectarine (Prunus persica var. nectarine) fruit sporulating with Monilinia were collected from twenty-nine commercial orchards and one experimental orchard from nearly all of the major peach production provinces in China including Beijing (23 isolates), Shandong (12 isolates), Zhejiang (4 isolates), Fujian (11 isolates), Shanxi (4 isolates), Gansu (3 isolates), Hubei (36 isolates) and Yunnan (52 isolates). Single spore isolates were obtained as described previously with slight modifications [17]. Briefly, individual conidia dispersed on potato dextrose agar (PDA; 200 ml juice from 200 g potato, 20 g dextrose, and 18 g agar L−1) were excised with a thin glass needle under a COIC XSZ-4G compound light microscope (ChongQing Opitical & Electrical Instrument Co., Ltd, Chongqing, China) and transferred to clean PDA. After conidia germinated, individual colonies were transferred but only one single-spore isolate was maintained for each infected fruit sample. All of the 145 isolates collected were identified to species by morphological observation and PCR identification (data now shown). Twenty isolates were selected for phylogenetic and 11 for morphological analysis, respectively with an approximately equal number representing each species identified. The selected isolates represented all of the different in vitro phenotypes and geographical locations included in this study (Table S1). For long-term storage, isolates were stored at −20°C on filter paper (Haier, Shandong, China) until further use. Briefly, isolates were allowed to grow on filter paper discs (5 mm in diameter) placed on PDA and incubated at 22°C in darkness. After 4 days, discs with mycelium were removed and placed into a desiccator with silica gel for 7 days. Discs were transferred into 1.5 ml sterile centrifuge tubes with desiccated silica gel at the bottom, and stored at −20°C. For each experiment in the study, a new culture was started from a stored filter disc.
An additional 18 isolates of known species (Table S1) used as morphological and phylogenetic standards for M. laxa and Chinese M. fructicola were either on hand or provided by Dr. Imre J. Holb (Department of Plant Protection, University of Debrecen, Debrecen, Hungary) and Dr. Xili Liu (College of Agriculture and Biotechnology, China Agricultural University), respectively. In total, 37 isolates (Table S1) representing five species and three continents were used for phylogenetic analysis, and a further subset of this collection was selected for morphological and pathological characterization. An isolate of Botryotinia fuckeliana from a Hubei province peach tree, and an isolate of Sclerotinia sclerotiorum from a Hubei province canola (Brassica napus L.) plant were used to validate the genera specificity of diagnostic primer sets.
DNA extractions
All representative isolates (Table S1) selected for phylogenetic analysis were grown on PDA at 22°C for 5 days in the dark. Single agar plugs containing actively growing mycelium were taken from the periphery of the advancing colonies and transferred to 250-ml flasks containing 40 ml of PDB (200 ml juice from 200 g potato and 20 g dextrose per liter). Flasks were shaken at 120 rpm for 4 days at 22°C. The mycelium was then removed from the broth, rinsed under sterile deionized water, and genomic DNA was subsequently extracted using the Easypure Plant Genomic DNA Extraction Kit (TransGen Biotech, Beijing, China) according to the manufacturer's instruction.
Sequencing of the ITS regions and G3PDH and TUB2 gene fragments
From representative isolates (Table S1), the ITS regions and fragments of the G3PDH and TUB2 gene were sequenced for phylogenetic analysis. The ITS1-5.8S-ITS2 region was amplified from genomic DNA with primer pair ITS1/ITS4 [18]. Based on G3PDH gene sequences from M. fructicola (EF367148) and M. fructigena (AJ705043), a primer pair Mon-G3pdhF/Mon-G3pdhR was designed to amplify a 786 bp fragment (approximately 70%) of the G3PDH gene. Similarly, primer pair Mon-TubF1/Mon-TubR1 was designed based on the TUB2 sequences from M. fructicola (AY283679) and M. laxa (AY349149) to amplify a 1630 bp fragment (approximately 92%) of the TUB2 gene. All primers used in this study are listed in table 1. PCR amplification of both genes was performed for all isolates in 50 µl reaction volumes containing 1× PCR buffer (TransGen Biotech, Beijing, China), 20 ng template DNA, 0.4 µM of each primer, 200 µM of each dNTP, 2.5 unit of Easy Taq DNA polymerase (TransGen Biotech, Beijing, China). All amplifications were performed in an “iCycler” thermal cycler (Bio-Rad Laboratories Inc., Hercules, CA). The amplification parameters for the ITS, consisted of an initial denaturation at 94°C for 3 min followed by 30 cycles of 94°C for 1 min, 55°C for 1 min and 72°C for 2 min, and a final elongation step of 72°C for 5 min. The parameters for amplifying G3PDH and TUB2 gene fragments were largely identical with ITS amplification, except for the annealing temperature, which was reduced to 50°C. PCR products were extracted for sequencing from agarose gels using a DNA Gel Extraction Kit (Axygen, Hangzhou, China) according to the manufacturer's instructions. Sequencing was conducted at Beijing Genomics Institute (BGI; Shenzhen, China).
Table 1. Primers used for PCR amplification and sequencing.
| Primer Name | Primer Sequence (5′→3′) | Description |
| Mon-G3pdhF | ACGGTCAATTCAAGGGTGAT | To amplify the partial fragment of G3PDH in Monilinia spp |
| Mon-G3pdhR | ATCGAAGATGGAGGAGTGGT | To amplify and sequence the G3PDH fragment |
| Mon-TubF1 | ATGCGTGAGATTGTACGTAT | To amplify and sequence the β-tubulin fragment in Monilinia spp |
| Mon-TubR1 | GTACCAATGCAAGAAAGCCT | Same as Mon-TubF1 |
| ITS1a | TCCGTAGGTGAACCTGCGG | To amplify ITS region |
| ITS4a | TCCTCCGCTTATTGATATGC | To amplify and sequence ITS region |
| PRC Laxa-F1 | ATGAGAATTTTTAAAAGTCATCCC | Amplified and sequenced fragment 1 |
| PRC Laxa-R1 | CTAATGTTCTAGGTGCTCTG | Same as PRC Laxa-F1 |
| PRC Laxa-F2 | GCGTGATGTTAACAATGGATG | Amplified and sequenced fragment 2 |
| Check R3 | CAGGAACAGGCAGAATACA | Amplified fragment 2 |
| KES 1238b | AGCTTTCCTGGGTTTGTCAAA | Amplified and sequenced fragment 3 |
| KES 1261b | TCCAATTCATGGTAYAGCACTCATA | Amplified fragment 3 |
| PRC Laxa-F3 | GCAACTGTGATCACCAACCT | Amplified and sequenced fragment 4 |
| P450intron6-2-revc | AGTTCAACTCAGATCTAAAGATACCTC | Same as PRC Laxa-F3 |
| P450intron6-2-fwdc | AGGTGAGTAGGAAATACAGATAAATG | Amplified and sequenced fragment 5 |
| PRC Laxa-R4 | TTATCTACTAGGCTTTTC | Same as P450intron6-2-fwd |
| PRCmon-F | ATCTCCAACGCTTCTTGCAC | Specific primer for Monilinia spp. from G3PDH |
| PRCmon-R | CTTCTTGACGACAGCCTTGA | Same as PRCmon-F |
| Cola-F | CTGTATGATGACCGAGAAGG | Species-specific primer for M. fructicola |
| Ensis-F | GGAAACCAAGTGGTTGAGAT | Species-specific primer for M. yunnanensis |
| Mume-F | AAAGGTAGAAGACATCTTAAGG | Species-specific primer for M. mumecola |
| Mon-R | ATCTCCAAGATCCGTGAGGAG | Common reverse primer for M. fructicola, M. yunnanensis, and M. mumecola from β-tubulin gene |
White et al 1990.
Miessner and Stammler 2010.
Hily et al 2010.
Construction of phylogenetic trees
Phylogenetic analysis on the representative isolates (Table S1) was performed for the noncoding renal region (ITS1-5.8S-ITS2) and a combined data matrix of the two coding loci (G3PDH and TUB2) respectively. Multiple alignments were conducted using DNASTAR (DNASTAR Inc., Nevada City CA) and CLUSTAL X 1.81 [19], [20]. For constructing the ITS phylogenetic tree, sequences of B. fuckeliana (FJ791158) was used as out-group reference species. Strain SAS56 of B. fuckeliana (GenBank accessions: AJ705006 and Z69263) was used as the outgroup species in the construction of the G3PDH and TUB2 phylogenetic trees. Maximum parsimony (MP) method and neighbor-joining (NJ) method were used to carry out phylogenetic constructions using MEGA version 4.0 [21]. For the MP tree, the following settings were used: heuristic search using close neighbor interchange (CNI; level = 1), and branch swapping method with initial trees generated by random addition (10 reps). A maximum composite likelihood model was used to generate the NJ tree. A complete deletion option was used to treat gaps/missing data and the reliability of clusters was evaluated by bootstrapping with 1000 replicates.
Cloning and analysis of cytochrome b sequences from M. yunnanensis and M. mumecola
To provide further evidence that M. yunnanensis and M. mumecola were phylogenetically distinct from known Monilinia species of peach, the mitochondrial cytochrome b (Cyt b) genes were cloned and sequences were compared to those of known species. Genomic DNA was extracted from 1-week-old mycelial cultures of M. yunnanensis (isolate YQG10-6c) and M. mumecola (isolate ML-1a) using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations. Total RNA was isolated from 5-day-old mycelial cultures of M. yunnanensis (isolates SBG10-3a, YKG10-61c, KY-1, QJ-2a and YQG10-6c) and M. mumecola (ML-1a and HXL10-1a) using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. RNA integrity was verified by resolving on a 1% agarose gel. Quantification of all nucleic acids was carried out using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA). First-strand cDNA synthesis was conducted using 500 ng total RNA from each of the M. yunnanensis and M. mumecola isolates with an oligo-dT primer and Superscript III reverse transcriptase (Invitrogen) in a final volume of 10 µl according to the manufacturer's recommendations. Subsequent amplifications of Cyt b-specific cDNAs were performed using 1 µL cDNA and the Platinum PCR Supermix High Fidelity system (Invitrogen) in a final volume of 25 µL. Primers MoniliniaP450ATG-fwd (ATG AGA ATT TTT AAA AGT CAT CCC T) and MoniliniaP450STOP-rev (TTA TCT ACT AGG CTT TTC TTT AGT TAA TAC) were designed to amplify the Cyt b sequence from translational start codon to translational stop codon based on the previously published Cyt b sequence from the closely related B. fuckeliana (GenBank accession AB262970). In each case, reactions were incubated at 94°C for 2 min, followed by 32 cycles of 94°C for 30 s, 55°C for 30 s and 68°C for 1.5 min, with a final elongation step of 68°C for 5 min.
The full-length Cyt b gene (from translational start codon to translational stop codon) was amplified from genomic DNA of M. yunnanensis (YQG10-6c) in a single PCR reaction using primers MoniliniaP450ATG-fwd and MoniliniaP450STOP-rev. PCR amplifications were carried out with approximately 200 ng template DNA using the Platinum PCR Supermix High Fidelity system. Thermal parameters utilized were identical to those described for the cloning of Cyt b cDNA, except for the elongation time, which was increased to 13 min, and the number of cycles, which was increased to 35.
In the case of M. mumecola (isolate ML-1a), five successive PCR reactions were performed to achieve the full-length Cyt b gene sequence. The five primer pairs for the reactions are listed in Table 1. Thermal parameters for amplifying each fragment were as follows with 35 cycles each: 20 sec at 95°C, 30 sec at 52°C, and 5 min at 68°C (Fragments 1 and 4); 40 sec at 94°C, 50 sec at 50°C, and 3 min at 72°C (Fragments 2 and 5); 15 sec at 95°C, 30 sec at 55°C, and 5 min at 68°C (Fragment 3).
All PCR products were gel-purified using the Wizard SV Gel and PCR Clean-Up system (Promega, Madison, WI) and were cloned into the pGEM-T easy vector (Promega). All sequencing was carried out using Big Dye Terminator chemistry and AmpliTaq-FS DNA Polymerase (Applied Biosytems, Foster City, CA) using the Applied Biosystems Automated 3730xl DNA Analyzer at the Cornell University DNA Sequencing facility in Ithaca, NY. In the case of the Cyt b gene sequence, a primer walking strategy was utilized for sequencing due to its large size (a complete listing of the primer sequences are available upon request). All nucleotide sequences were aligned manually using Clustal W [22] and BioEdit version 7.0.8.0 [23], and were compared to previously reported sequences using BLAST [24].
Colony morphology, mycelial growth rate, sporulation, and conidial morphology of Monilinia species
At least four representative isolates from each species (M. yunnanensis, isolates SBG10-3a, YQG10-6c, SM09-1a, KY-1; M. fructigena, isolates SL10, Mfg2-GE-A, Mfg4-GY-A, Mfg5-SP-A; M. mumecola, isolates ML-1a, ML-1c, HWL10-13b, HXL10-1a; M. laxa, isolates EBR Ba11b, GEARI 3c2a, BEK-SZ, BSZGY-SZ-1 and M. fructicola, isolates ZM09-2a, MSA9, 0907-a, SC.Dap3, GA.Bmpc5, SC.Egpc8) were selected for in-depth morphological and physiological characterization. To obtain uniform colonies for each isolate, 6 mm-diameter plugs with actively growing mycelium were removed from the periphery of a 4-day-old colony grown on PDA and placed in the center of plastic Petri dishes (90 mm diameter) containing PDA and incubated at 22°C in the dark. Mycelial growth rates were determined on both PDA and V8 (200 ml V8 juice and 20 g agar/l) media. Growth rates of isolates were determined by incubating isolates at 22°C in darkness and measuring colony diameters (excluding the transfer plug of 6 mm) every 2 days until hyphae were within 2 mm of the edge of the 90 mm in diameter Petri dish. Growth rates were expressed as mm of growth per day and the mean of 3 replicate colonies were used to represent each isolate. Sporulation was quantified on PDA, V8, and peach fruit. Isolates were incubated on the two media at 22°C in darkness for 9 days before conidia were rinsed off in 2 ml of sterile water for spore counts. Inoculation of peach fruit for determination of sporulation was identical to the method for determining pathogencity on fruit (see below). After 9 days of incubation, 2 ml of sterile water was added to the surface of sporulating colonies and conidia were gently dislodged using sterile plastic inoculation loops. The concentration of conidia in the suspensions from media and fruit was determined using a haemocytometer, and mean counts of three replicate samples were determined for each isolate and medium (including fruit) combination. In addition of the quantification of conidia, the conidial size, and germ tube morphology were also determined for each of the representative isolates. Conidia were harvested from sporulating peach fruit and spread onto shallow PDA media (<3 mm thick for maximal optical density) using sterile cotton swabs. PDA media containing conidia was incubated at 22°C in the dark for 5 hrs, and subsequently, the length and width of conidia and germ tubes were measured with a stage micrometer using a COIC XSZ-4G compound light microscope (ChongQing Opitical & Electrical Instrument Co.) at 400× magnification. For each isolate a minimum of 100 conidia and germ tubes were measured. Least significant difference (LSD) tests were conducted using DPS Data processing system 3.01 [25].
Pathogenicity of Monilina species
The pathogenicity of representative isolates of M. yunnanensis, M. fructigena, M. mumecola, M. laxa and M. fructicola on peach was determined. Commercially mature peach fruit (cv. ‘Zhonghua 2’) of similar size were collected, surface-sterilized with 75% ethanol and rinsed with sterile water to remove any pesticide residues prior to inoculation. A 5 mm plug taken from the periphery of a 4-day-old colony grown on PDA was inserted into a 5 mm deep hole created in the periderm of the fruit using a 5 mm cork borer. Fruit were subsequently transferred into non-branded plastic plant propagation trays (l×w×h = 50×30×20 cm) and covered with a transparent plastic lid. Wet paper towels were placed on the bottom of the trays to maintain near 100% humidity. Trays were incubated at 22°C, 97% RH (relative humidity) under a 14 hr light/10 hr dark regime. Lesion diameters were measured after 2 and 4 days of incubation. Afterwards, the lid was removed and RH was adjusted to 75% to promote dehiscence of conidial chains. One day later, conidia were collected with a sterile swab and transferred into 15–20 ml distilled water. The suspension was filtered through a double layer of gauze, and the number of conidia was determined using a haemocytometer and a COIC XSZ-4G compound light microscope. Lesion development and sporulation were determined on six replicate fruit per isolate, and the entire pathogenicity assay was repeated. Statistical analyses were performed by DPS Data processing system 3.01 [25].
Evaluation of PCR-based methods to distinguish Monilinia species from China
Six PCR based methods previously developed to distinguish Monilinia spp. were evaluated for applicability to differentiate the Chinese Monilinia species pathogenic on peach. Primer sets included ITS1Mfcl/ITS4Mfcl, ITS1Mlx/ITS4Mlx, ITS1Mfgn/ITS4Mfgn [26]; IMfF/IMfR, MLF2/MLR2 [27], [28]; MO368-5, MO368-8R, MO368-10R, and Laxa-R2 [29]; IColaS/IColaAS, IGenaS/IGenaAS, and ILaxaS/ILaxaAS [30]; KES 1238/KES 1261 [31], as well as P450intron6-2-fwd/P450intron6-2-rev [32]. All PCR procedures were performed as described in the associated references.
Development of a PCR-based method to distinguish Chinese Monilinia species
Based on the aligned sequences of G3PDH from Chinese Monilinia isolates, B. fuckeliana (AM231159), and S. sclerotiorum (AJ705044), primers PRCmon-F and PRCmon-R were designed to differentiate the Monilinia species causing brown rot of peach. Primers based on TUB2 sequences were designed to distinguish Chinese Monilinia species from each other. Reverse primer Mon-R and species-specific forward primers, Cola-F, Ensis-F and Mume-F were designed to differentiate M. fructicola, M. yunnanensis and M. mumecola, respectively. Closely related B. fuckeliana and S. sclerotiorum were included in all PCR experiments as out-group negative controls [33]. PCR reactions were carried out in a volume of 25 µl containing 1× PCR buffer (TransGen Biotech, Beijing, China), 200 µM of each dNTP, 20 ng template DNA, 0.2 µM of each primer, and 1 unit of Easy Taq DNA polymerase (TransGen Biotech, Beijing, China). The PCR amplification program consisted of an initial denaturation at 94°C for 3 min followed by 35 cycles of 30 sec at 94°C, 30 sec at 58°C, and 40 sec at 72°C and a final extension step at 72°C for 5 min. Products were resolved on 1.2% agarose gel (AGAROSE G-10, GENE COMPANY, Hong Kong, China) in 0.5×TBE buffer for 1 h at 100 v. Gels were stained with ethidium bromide, and visualized using an Alphalmager® EP image acquisition system (Alpha Innotech, Santa Clara, CA, USA).
Results
Based on nucleotide sequence comparisons of the noncoding ribosomal ITS regions and the coding regions of G3PDH and TUB2 genes, and unique morphological features (see below), several Chinese isolates were found to be sufficiently distinct from other known Monilinia species as to potentially warrant classification of a new species, which we designated Monilia yunnanensis. Detailed description of the proposed species and summary to distinctive features is provided below. The remainder of the isolates was classified as either M. fructicola with highest similarity to those from the United States or M. mumecola with highest similarity to those from China.
Analysis of internal transcribed spacer (ITS) regions 1 and 2
The ITS1-5.8S-ITS2 sequences of 7 isolates (Table S1) were identical with M. mumecola (Genbank accession nos. AB125620, AB125613 and AB125614). Within each species, the ITS sequences of M. yunnanensis, M. fructigena, M. fructicola, and M. laxa isolates were 100% identical to one another. Interestingly, all of the M. mumecola isolates collected from peach had a cytosine (C) at the 442 position while all isolates from nectarine displayed a thymine (T) at this location (Fig. 1). Between the four species, there were considerable nucleotide variations. A total of eleven base pair differences were observed between M. yunnanensis and the next closest species M. fructigena. A minimum of eight base pair differences were observed between M. mumecola and the next closest species M. laxa from Europe and the United States. All M. fructicola isolates regardless of the origin had identical ITS sequences. There were 18 or 19 (depending on the origin of the isolate) variations between M. mumecola and M. polystroma, and 10 variations between M. yunnanensis and M. polystroma.
Figure 1. Sequence alignment of the ITS1 and ITS2 regions of ribosomal DNA (rDNA) of Monilinia spp.
The sequence of M. polystroma (accession no. Y17876) was obtained from Genbank. Each Symbol ‘*’ represents conserved regions of 5 bp in length. while ‘…’ represent conserved regions of 95 bp in length. “Y” in bold indicates both cytosine(C) and thymine (T) were present in this position in M. mumecola.
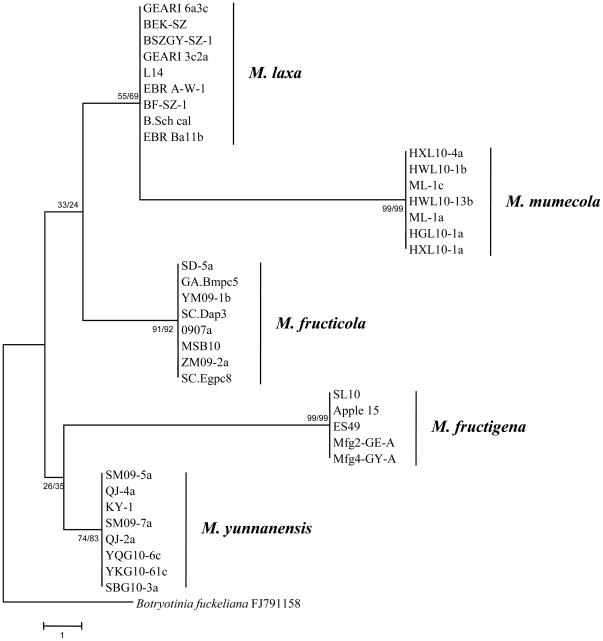
Phylogenetic analysis was conducted with Monilinia species causing brown rot of peach and nectarine in China (thus M. polystroma was excluded). The ITS data set consisted of 38 fungal isolates including the outgroup fungus B. fuckeliana. There were a total of 437 nucleotide positions included in the final dataset, 18 of which were parsimony informative. MP (Fig. 2) and NJ (data not shown) analyses resulted in similar topologies. Both phylogenetic trees revealed distinct clusters for each species. From the topology of the ITS tree, M. yunnanensis isolates were most closely related to M. fructigena, whereas M. mumecola isolates were most closely related to M. laxa. The ITS sequences for M. fructicola isolate 0907-a, M. laxa isolate BF-SZ-1 obtained from Europe , M. fructigena isolate SL10 from Europe, and M. yunnanensis isolate KY-1 were deposited in GenBank under accession numbers HQ908789, HQ908790, HQ908791, and HQ908788 respectively. The sequences of M. mumecola isolates ML-1a from peach and HXL10-1a from nectarine were submitted under accession numbers HQ908786 and HQ908787, respectively. Unfortunately, the phylogenetic tree based on ITS sequences revealed many low bootstrap values, and as such, provided a weak indication of genetic relationships between some of the clades (Fig. 2).
Figure 2. Phylogeny of the rDNA region ITS1-5.8S-ITS2.
Shown is the most parsimonious tree for 37 Monilinia isolates and one outgroup species (Botryotinia fuckeliana), with 437 characters, out of which 18 were parsimony informative. The numbers labeled at each node indicate the bootstrap (BS) percentages (N = 1000) supporting individual branches: BS value from the MP test/BS value from the neighbor-joining (NJ) test.
Construction of phylogenetic trees using G3PDH and TUB2 sequences
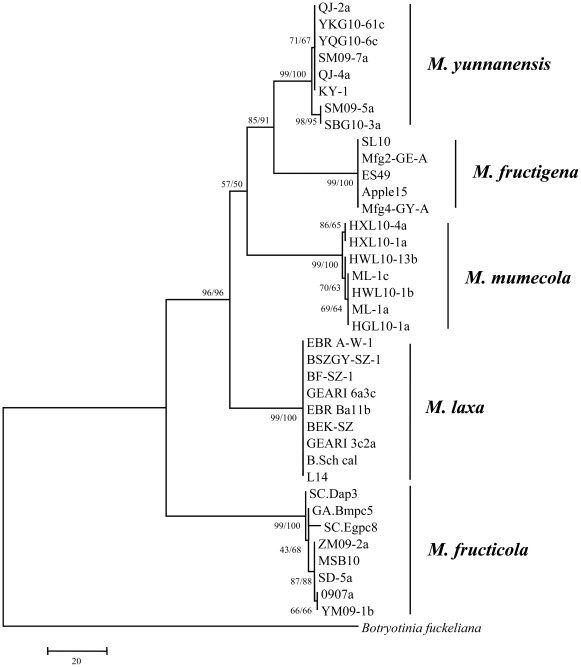
Phylogenetic trees were generated from a data set of combined partial sequences of the G3PDH and TUB2 genes from the same above-mentioned 38 isolates including the outgroup species B. fuckeliana. There were a total of 2215 nucleotide positions included in the final dataset, 172 of which were parsimony informative. MP and NJ analysis of the combined DNA sequences generated two distinct phylogenetic trees with a similar topological structure. Hence, the MP phylogeny for the combined partial G3PDH and TUB2 sequences (Fig. 3) was used for drawing conclusions regarding species. In both MP and NJ (data not shown) phylogenetic trees, isolates of the three European and North American Monilinia species (M. fructicola, M. fructigena and M. laxa), and the two Chinese species M. yunnanensis and M. mumecola formed monophyletic clades within a larger derived clade within the Monilinia genus. M. yunnanensis isolates were more closely related to European M. fructigena isolates than to M. mumecola isolates. M. fructicola was the most basal clade, while the clades of the other species were more derived but shared a single common ancestor with M. fructicola. G3PDH sequences were deposited in GenBank under accession nos. HQ908777, HQ908778 & HQ908779 for M. fructicola, HQ908781 for M. laxa from Europe, HQ908784 & HQ908785 for M. mumecola, HQ908780 for European M. fructigena, and HQ908782 & HQ908783 for M. yunnanensis. TUB2 sequences were deposited under accession nos. HQ908768, HQ908769 & HQ908770 for M. fructicola, HQ908772 for M. laxa from Europe, HQ908774, HQ908775 & HQ908776 for M. mumecola, HQ908771 for European M. fructigena, and HQ908773 for M. yunnanensis.
Figure 3. Phylogeny of 38 isolates of Monilinia spp. and Botryotinia fuckeliana.
Maximum parsimony (MP) tree inferred from the data set containing the combined DNA sequences of G3PDH and TUB2, with 2215 characters, 172 of which were informative. The numbers labeled at each node indicate the bootstrap (BS) percentage (N = 1000): BS value from the MP test/BS value from the neighbor-joining (NJ) test.
Comparison of the Cyt b sequences from Monilinia species
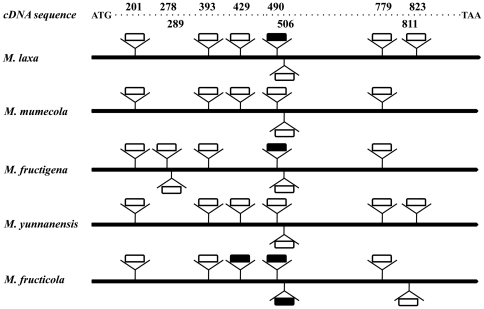
Cyt b sequences were shown phylogenetically informative among Monilinia spp. in our previous study [32]. Therefore, Cyt b genes were isolated from M. yunnanensis and M. mumecola and compared to M. fructicola, M. laxa and M. fructigena. The Cyt b gene from M. yunnanensis was 12632 bp in length with 7 introns separating the exons; the same gene from M. mumecola was 14203 bp in length with 6 introns separating exons (Genbank accession nos. HQ908793 and JN204425, respectively; Fig. 4). The Cyt b coding sequences were both 1176 bp in length and shared 99% identity.
Figure 4. Exon/intron organization of the Cyt b gene from five Monilinia species.
Schematic of Cyt b gene exon and intron organizations from Monilinia species showing intron positions and intron sequence similarities. Identical intron symbols at the same nucleotide position indicate high sequence similarity. For example, at position 490, M. laxa and M. fructigena had homologous introns, and M. mumecola, M. yunnanensis and M. fructicola had homologous introns but the two groups of introns did not share high sequence similarity.
The Cyt b gene coding sequences were identical for all five M. yunnanensis isolates (GenBank accession no. of isolate YQG10-6c is HQ908792) and shared 99% identity with M. fructigena from Europe (Genbank accession no. GU952818), 99% identity with M. laxa from the United States (GU952816), and 98% identity with M. fructicola from the United States (GU952814). The Cyt b gene coding regions were identical for the two M. mumecola isolates sequenced (GenBank accession no. for isolate ML-1a is JN572107) and shared 99% identity with M. laxa from the United States (GU952816), 99% identity with M. fructigena from Europe (GU952818), and 98% identity with M. fructicola from the United States (GU952814). The Cyt b gene structure of M. laxa, M. mumecola, M. fructigena, M. yunnanensis and M. fructicola isolates differed in exon/intron organization (Fig. 4) in that the introns showed a patchy distribution. For example, in M. yunnanensis, the positions of five of the seven introns were identical to those of other Monilinia species [32]. Intron 1 of M. yunnanensis and M. fructigena was located at position 201, and introns 2, 5, and 6 of M. yunnanensis corresponded to introns 4, 6, and 7 of M. fructigena located at positions 393, 506 and 779, respectively (Fig. 4). Despite the fact that these homologous introns have high levels of identity (>98%) with one another, the sizes of these introns clearly differed between the two species. Interestingly, intron 4 of M. yunnanensis and intron 5 of M. fructigena were located at position 490 but the sequences did not share noticeable sequence similarity (Fig. 4). Moreover, introns 3 and 7 of M. yunnanensis and introns 2 and 3 of M. fructigena were located at different positions in the coding sequence and shared no noticeable sequence similarity (Fig. 4).
Colony morphology, mycelial growth rate, sporulation, and conidial morphology of Monilinia species
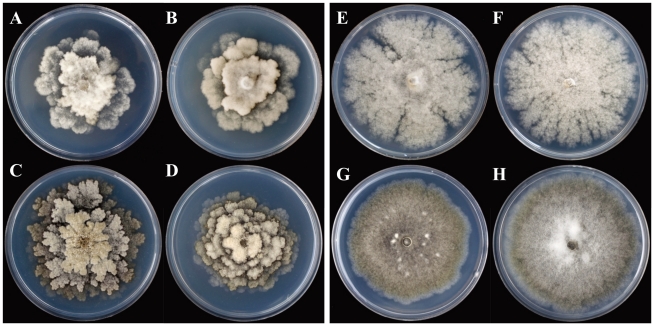
Colony morphology of M. mumecola was similar to M. laxa (Fig. 5A–D). These two species have a growth pattern distinct from any other species characterized by concentric rings of mycelium with lobbed margins. Despite a similar appearance, the colony color of M. mumecola tended to be gray-green compared to the gray M. laxa colony. Similarly, the colony color of M. yunnanensis isolates was gray-green compared to the gray M. fructigena colony (Fig. 5E–H). Fragmented radial colonies, which were frequently observed in M. fructigena isolates, were rarely observed in M. yunnanensis (Fig. 5E–H). After more than 10 days of incubation, most M. yunnanensis isolates started to develop stromata. However, stromatization was never observed in any of the M. fructigena isolates.
Figure 5. Characteristics of single spore isolates of Monilinia spp. grown on potato dextrose agar (PDA) at 22°C in darkness for 9 days.
A–B colonies of M. laxa (isolates GEARI 3c2a and BSZGY-SZ-1). C–D colonies of M. mumecola (isolates HGL10-1a and ML-1a). E–F colonies of M. fructigena (isolates Mfg2-GE-A and Mfg4-GY-A). G–H colonies of M. yunnanensis (isolates QJ-4a and SM09-5a).
The mean and range of colony growth rates of Monilinia species on PDA medium were significantly different (P<0.05) between M. fructicola and M. laxa, M. fructicola and M. mumecola and between M. fructigena and M. laxa. M. fructicola isolates displayed the highest growth rates while M. laxa exhibited the lowest. On V8 medium, M. mumecola grew significantly (P<0.05) faster than M. laxa isolates. However, no significant differences (P>0.05) in growth rate were observed between M. yunnanensis and M. fructigena. In general, colony growth rates of all test isolates were found to be higher on V8 medium compared to PDA. On PDA medium, there were no significant differences in the number of conidia produced among Monilinia species (Table 2). On V8 medium only M. fructicola consistently sporulated, and the other species groups rarely produced conidia (Table 2).
Table 2. Colony growth rate and sporulation of Monilinia isolates on PDA and V8 mediaz.
| Species | Colony growth rate (mm d−1) | Sporulationx | ||||||
| On PDA | On V8 | On PDA | On V8 | |||||
| Meany | Range | Mean | Range | Mean | Range | Mean | Range | |
| M. fructicola | 10.8±0.79a | 8.3–12.7 | 12.0±0.24a | 10.9–12.5 | 3.9±0.45a | 2.8–5.3 | 5.2±0.14 | 4.6–5.4 |
| M. yunnanensis | 8.5±0.59abc | 6.5–9.9 | 10.9±0.06ab | 10.8–11 | nd | - | nd | - |
| M. fructigena | 8.9±1.02ab | 6.8–10.8 | 10.2±0.82ab | 7.8–11.5 | nd | - | nd | - |
| M. mumecola | 6.9±1.48bc | 4.0–8.7 | 11.6±1.31a | 9.7–14.1 | 2.5±0.85a | 0–2.9 | nd | - |
| M. laxa | 6.0±0.58c | 4.5–7.3 | 8.2±0.94b | 5.4–9.5 | 1.8±0.91a | 0–3.7 | nd | - |
Average of at least 4 isolates from each species.
log-transformed number of conidia per cm2; nd = not detected.
Mean ± S.E.M (standard error of mean); values within the same column followed by the same letters are not significantly different based on the analysis of least significant difference (LSD) test at P = 0.05.
Differences in conidia size were apparent among species (Table 3). The conidia of M. mumecola were the largest on average, and those of the M. laxa were the smallest on average. The conidia of M. yunnanensis isolates were smaller on average compared with those of M. fructigena (Table 3). Consistent with that, the range of conidia sizes was unique for each of the new species and strikingly distinct from the genetically closest Monilinia relative. For example, the range of conidia size for M. yunnanensis was 10–21×7–12, whereas the range for M. fructigena was 12–31×7–17 (Table 3).
Table 3. Lesion growth rate, sporulation, and conidia size of selected Monilinia isolates on peach fruit.
| Species | Lesion growth rate (mm d−1) | Num of conidia, cm−2 | Conidia size, µm | |||
| Exp 1 | Exp 2 | Exp 1 | Exp 2 | Mean (L×W) | Range | |
| M. fructicola | 21.2±1.16cz | 20.7±0.77b | 6.3±0.11a | 6.5±0.08a | 16×10 | 10–19×7–14 |
| M. yunnanensis | 25.5±0.52ab | 27.7±0.79a | 2.6±0.34d | 2.9±0.32d | 15×9 | 10–21×7–12 |
| M. fructigena | 21.3±0.89c | 23.6±1.04b | 4.3±0.02b | 4.2±0.07b | 22×12 | 12–31×7–17 |
| M. mumecola | 22.4±0.79bc | 23.5±1.30b | 3.3±0.29cd | 3.4±0.04cd | 21×15 | 14–31×11–17 |
| M. laxa | 25.8±1.07a | 27.3±0.21a | 4.0±0.16bc | 3.9±0.09bc | 13×9 | 10–17×7–11 |
Mean ± S.E.M (standard error of mean); values within the same column followed by the same letters are not significantly different based on the analysis of least significant difference (LSD) test at P = 0.05. Values were log transformed prior to statistical analysis.
M. mumecola isolates often produced more than two germ tubes per conidium, which also appeared somewhat misshapen (Fig. 6A, B). By comparison M. yunnanensis and M. fructigena produced one or two germ tubes per conidium, and all M. laxa and M. fructicola isolates consistently produced one germ tube per conidium (Fig. 6C).
Figure 6. Germ tube morphology of M. mumecola (A–B) and M. laxa (C).
Conidia were incubated on PDA at 22°C in the dark for 6 hours. Bar = 10 µm.
Pathogenicity of Monilina species
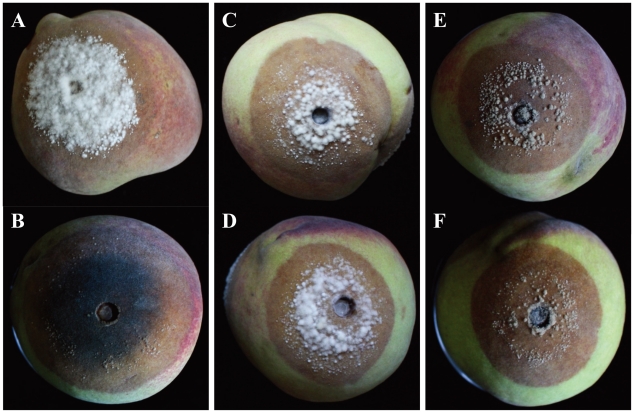
All species tested (M. yunnanensis. M. fructigena, M. mumecola, M. laxa and M. fructicola) were pathogenic and sporulated on peach fruit. Koch's postulates were fulfilled by reisolating the fungus from symptomatic fruit and re-identifying the pathogen to the species level (data not shown). In each experimental run, significant differences (P<0.05) in average lesion growth rates were observed between M. yunnanensis and M. fructigena and between M. mumecola and M. laxa (Table 3). Similarly, the average number of conidia produced on lesions by M. yunnanensis and M. fructigena were significantly different (P<0.05), but not for M. mumecola and M. laxa (Table 3). Also, lesion morphology on peach fruit differed between M. mumecola and M. laxa (Fig. 7A, B). The lesions on fruit inoculated with M. mumecola developed a whiter, denser mycelium than those of produced by isolates of M. laxa, which produced sparse aerial mycelium with a black distinct lesion margin. M. yunnanensis and M. fructigena produced indistinguishable symptoms (Fig. 7C, D), which was also the case for M. fructicola isolates from China and the US (Fig. 7E, F).
Figure 7. Symptoms of infection by Monilinia spp. on peach fruit (cv. ‘Zhonghua 2’) after 3 days of incubation at 22°C in 14 h light/10 dark regime.
A–F, lesion resulting from infection by M. mumecola, M. laxa, M. yunnanensis, M. fructigena, Chinese M. fructicola and American M. fructicola, respectively.
Evaluation of PCR-based methods to distinguish Monilinia species from China
PCR results of six methods designed to differentiate Monilinia spp. are shown in Table 4. None of the six molecular tools alone was able to distinguish all five species (M. fructicola, M. fructigena, M. laxa, M. yunnanensis and M. mumecola) from one another. M. fructicola, M. fructigena, and M. laxa isolates were reliably differentiated by the methods of Ioos et al. [26], Miessner and Stammler, [31], and Hily et al. [32]. However, neither of these methods was able to distinguish M. yunnanensis from M. fructigena. Likewise, the methods developed by Ioos et al. [26] and Ma et al. [27], [28] did not distinguish between M. mumecola and M. laxa and the method developed by Hily [32] did not distinguish M. mumecola from M. fructicola. Additionally, the methods developed by Miessner and Stammler [31], and Hily et al. [32] did not distinguish between M. yunnanensis and M. laxa (Table 4).
Table 4. PCR results of different diagnostic methods to distinguish Monilinia speciesz.
| Isolates | Ioos et al., 2000 | Ma et al., 2003, 2007 | Cote et al., 2004 | Gell et al., 2007 | Miessner and Stammler, 2010 | Hily et al., 2010 | ||||||||||||
| Name | Taxon | A | B | C | A | C | A | B | C | A | B | C | A | B | C | A | B | C |
| 0907a | M. fructicola | + | − | − | + | − | + | − | − | + | − | − | + | − | − | + | − | − |
| SD−5a | M. fructicola | + | − | − | + | − | − | − | − | + | − | − | + | − | − | + | − | − |
| ZM09−2a | M. fructicola | + | − | − | + | − | + | − | − | + | − | − | + | − | − | + | − | − |
| SC.Dap3 | M. fructicola | + | − | − | + | − | − | − | − | + | − | − | + | − | − | + | − | − |
| SC.Egpc8 | M. fructicola | + | − | − | + | − | − | − | − | + | − | − | + | − | − | + | − | − |
| QJ−2a | M. yunnanensis | − | + | + | − | + | − | − | − | − | − | − | − | − | + | − | − | + |
| SM09−7a | M. yunnanensis | − | + | + | − | + | − | − | − | − | − | − | − | − | + | − | − | + |
| SBG10−3a | M. yunnanensis | − | + | + | − | + | − | − | − | − | − | − | − | − | − | − | − | + |
| SL10 | M. fructigena | − | + | − | − | + | − | + | − | − | − | − | − | + | − | − | + | − |
| Mfg2−GE−A | M. fructigena | − | + | − | − | + | − | + | − | − | − | − | − | + | − | − | + | − |
| ML−1c | M. mumecola | − | − | + | + | + | − | − | − | − | − | − | − | − | − | + | − | − |
| HGL10−1a | M. mumecola | − | − | + | − | + | − | − | − | − | − | − | − | − | − | + | − | − |
| HXL10−4a | M. mumecola | − | − | + | − | + | − | − | − | − | − | − | − | − | − | + | − | − |
| BSZGY−SZ−1 | M. laxa | − | − | + | − | + | − | − | + | − | − | + | − | − | + | − | − | + |
| EBR Ba−1−1b | M. laxa | − | − | + | − | + | − | − | − | − | − | + | − | − | + | − | − | + |
A: M. fructicola–specific product; B: M. fructigena–specific product; C: M. laxa–specific product; ‘+’ and ‘−’ indicate the presence and absence of specific band patterns, respectively.
Development of a molecular tool to distinguish Chinese Monilinia species affecting peach
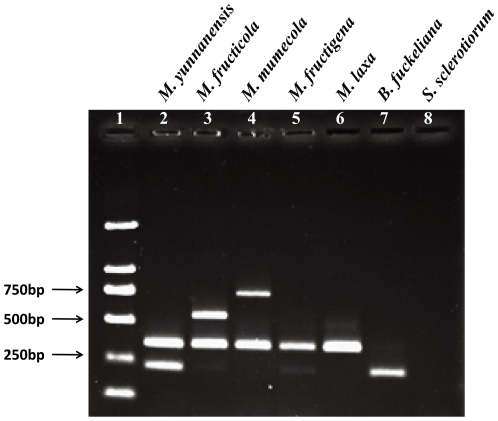
A multiplex PCR method was developed to differentiate Chinese Monilinia species on peach. Based on the G3PDH gene sequences, primer pair PRCmon-F/PRCmon-R amplified a 354 bp fragment from all Monilinia species, but not from closely related genera such as Botrytinia and Sclerotinia (Fig. 8). The cocktail also contained common reverse primer Mon-R and forward species-specific primers Cola-F, Ensis-F, and Mume-F, which produced amplicons 237 bp, 534 bp, or 712 bp in length from M. yunnanensis, M. fructicola, or M. mumecola, respectively (Fig. 8). All previously confirmed Monilinia isolates used in this study (Table S1) produced the expected amplicon sizes with this new multiplex PCR tool. All 145 isolates collected for this study from China were identified as M. fructicola, M. yunnanensis, or M. mumecola. Isolates that were collected from Beijing, Shandong, Zhejiang, Fujian and Gansu provinces were M. fructicola, isolates from Hubei province in central China were M. mumecola, and isolates from Yunnan and Shanxi provinces in Western China were M. yunnanensis. The closely related B. fuckeliana and S. sclerotiorum were also tested using these two primer sets. While there was no amplicon obtained from S. sclerotiorum, a 237 bp fragment was produced from B. fuckeliana. Although, this 237 bp amplicon was also produced in M. fructicola, B. fuckeliana produces no amplicon with the PRCmon-F1/PRCmon-R1 primer set (Fig. 8).
Figure 8. Multiplex polymerase chain reaction (PCR) results for five Monilinia spp.
Trans 2K (TransGen Biotech, Beijing, China) molecular marker is shown in the first lane. Primer pair PRCmon-F/PRCmon-R were used to produce a 354 bp fragment (lanes 2–6) from all five Monilinia species, but not from other closely related genera (Botrytinia,lane 7; Sclerotinia, lane 8). Reverse primer Mon-R and species-specific forward primers Ensis-F, Cola-F, and Mume-F, produced amplicons 237 bp (lane 2), 534 bp (lane 3), or 712 bp (lane 4) in length from M. yunnanensis, M. fructicola, or M. mumecola, respectively.
Discussion
Our phylogenetic, morphological, and cultural characterization of Monilinia isolates suggested that in our sample size there were three species causing brown rot of peach and nectarine in China, one of which we believe to be a new species M. yunnanensis. The ITS sequence is widely used in taxonomy and molecular phylogeny [34], [35], [36], and several phylogenetic analyses had been performed among and within Monilinia species on the basis of sequence differences in this region [33], [37], [38]. However, we found this locus to provide a fairly weak phylogeny of Monilinia species evidenced by the low bootstrap values resulting from the relative low number of parsimony informative positions. Hence, we constructed addiltional phylogenies from TUB2, and G3PDH, which have also revealed significant taxonomic relations in fungi [39], [40], [41], [42], [43], [44]. For examples, Couch and Kohn [39] segregated Magnaporthe oryzae from M. grisea using a multilocus gene genealogy, including the TUB2 sequences; Fournier et al. [45] distinguished two Botrytis sibling species (B. cinerea Group I and II) from grape by using DNA sequence data including the TUB2 and G3PDH genes. The ITS, TUB2, and G3PDH data in our study clearly support the designation of a new Monilinia species, M. yunnanensis. Morphological data as well as in vitro growth characteristics and pathogenicity data are concordant with the molecular evidence for the suggested species delineation.
ITS sequence data showed that M. yunnanensis was distantly related to M. polystroma, a species shown to affect Malus, Pyrus, and Prunus spp. (but not P. persica) and which was found in China only recently [7]. M. polystroma, which is most closely related to M. fructigena, was designated as a new species based on its ability to produce more stroma and based on five basepair differences in the ITS regions compared to M. fructigena [12]. In comparison, M. yunnanensis revealed eleven nucleotide variations in the ITS compared to its respective closest Monilinia relative M. fructigena.
In addition to the phylogenetic support, morphological data also distinguished M. yunnanensis from the closely related M. fructigena. For example, the colony morphology was unlike that of M. fructigena, and M. yunnanensis conidia were smaller than those of M. fructigena. Previous studies [37], [46], [47], [48] have acknowledged that what was believed to be populations of M. fructigena from Asia were different from European M. fructigena populations. Indeed, Khokhriakova [46] found differences in conidia size between M. fructigena populations from Europe and different regional populations in the former Soviet Union. The author found that the mean length and width of conidia were 19.4×11.5 µm for isolates from a European population, 17.2×11.9 µm for isolates from a central Asian population, and 18×11.4 µm for isolates from a Far Eastern population. Leeuwen [12] also made similar observations and found that conidia from Asian isolates tended to be smaller. Wormald [47] noted that a Japanese M. fructigena culture produced zones of black stromatal plates in culture, but the author never observed this phenomenon in European M. fructigena isolates. Willetts [48] found that European M. fructigena isolates produced few poorly-developed stromatic tissue, compared with Asian M. fructigena isolates. Consistent with this study, considerable numbers of stromata were produced in most M. yunnanensis isolates. Given the considerable evidence, it is likely that what was believed to be M. fructigena from Asia was likely a new species, but not realized.
The already strong phylogenetic and morphological evidence outlined in this study for delineating M. yunnanensis from M. fructigena was further supported by analysis of the cytochrome b gene sequence. The Cyt b gene exon/intron organization of M. yunnanensis was quite different from M. fructigena. Only four of the seven introns in M. yunnanensis revealed sequence identities greater than 98% compared to the corresponding introns in M. fructigena. The coding region of the Cyt b gene from M. yunnanensis showed 99.1% identity with that of M. fructigena. This high level of homology is to be expected given that the Cyt b exon sequences in general were highly conserved among the five Monilinia species for which the gene has been cloned [32]. For example, the authors found that M. fructicola and M. fructigena exhibited 97.5% sequence identity the least, while M. laxa and M. fructigena displayed more than 99.1% sequence identity.
Based on both molecular and morphological evidence we propose to name M. yunnanensis its own species. Given that only the anamorph has been observed, we describe the anamorph of Monilia yunnanensis.
Monilia yunnanensis M.J. Hu & C.X. Luo, sp. nov
MycoBank no.: MB563122.
Etym. “yunnanensis” indicates the province (Yunnan), where the fungus was originally isolated.
Colonies on potato dextrose agar (PDA) and on V8 reaching 50–70 mm, 75–80 mm in diameter respectively after 7 d at 22°C. When grown on PDA, colonies begin pale green and become tan after 15–20 days of incubation at 22°C, conidia sparse, stromata abundant, black in color, spherical to elliptical in shape, discrete or aggregated. Conidia ovoid or limoniform, measuring 10–21×7–12 µm, av. 15×9 µm when grown on peach fruit at 22°C. Mycelial pustules are common on symptomatic fruits.
Living culture YKG10-61c (AF2011002, CCTCC) was deposited in the China Center for Type Culture Collection (CCTCC) at Wuhan University, Wuhan City, Hubei Province, China. This single-spore isolate was collected from a peach fruit in Anning City, Yunnan Province, China, on August 5, 2010.
Despite attempts (not shown), we were not able to induce apothecium production in M. yunnanensis following the method described previously [49]. Mummified peach fruit infected by M. yunnanensis were collected to induce the production of apothecia using the method for Monilinia vaccinii-corymbosi [50]. However, no apothecia were produced from fruit infected with M. yunnanensis using this method. Given that no teleomorph examples are currently available, we only describe a new anamorphic species.
In regard to M. mumecola, one of the other species in our survey, the phylogenetic and morphological data (ITS1-5.8S-ITS2 sequences, conidium size, germ tube development, and colony appearance on PDA) were consistent with the previous description of the species [15]. However, to our knowledge this is the first report of M. mumecola causing brown rot of peach and nectarine in China.
M. fructicola was the third species we routinely identified in our survey. This fungus was only recently reported a new pathogen in China [6], [11], however, a population analysis indicated that this species had been in the country long before the first report in 2005 [51]. In our study, M. fructicola was collected from most provinces investigated, which supports the conclusions made by Fan et al. (2010) that this species has been in China for a considerable period of time. In both of our phylogenetic analyses, the M. fructicola isolates from China and America clustered together and this close relationship was further supported by morphological and cultural characteristics. This species was most distantly related to M. laxa and M. mumecola.
In contrast to other studies [8], [52], we did not find evidence for the presence of M. fructigena and M. laxa on peach or nectarine in China. Although isolates were collected from nearly all major peach growing provinces including Northern China (Beijing), Eastern China (Shandong), Southeastern China (Zhejing, Fujian), Central China (Hubei), Northwestern China (Shanxi, Gansu), and Southwestern China (Yunnan), our sample size of 145 isolates may not be sufficient to rule out the existence of M. fructigena and M. laxa in China.
We developed a new PCR-based method for differentiating Monilinia spp. infecting peach in China. Aforementioned studies [8] used three molecular techniques to identify Chinese isolates of Monilinia to the species level. We've demonstrated that techniques, developed primarily to distinguish European and North American Monilinia species, do not accurately discriminate Monilinia species from China, especially M. yunnanensis. Moreover, using previously developed techniques, we've found that M. yunnanensis and M. mumecola can be misidentified as M. fructigena and M. laxa, respectively. For example, our phylogenetic analysis of the ITS, G3PDH and TUB2 sequences confirmed that M. mumecola is closely related to M. laxa, which supports of the possibility of prior misidentification. Additionally, the cultural characteristics on PDA medium, including growth rate, colony morphology, and sporulation, largely matched those described for M. laxa by Leeuwen and Leeuwen [53] and Lane [54].
The phylogenetic analysis of ITS, G3PDH and TUB2 gene sequences allows some speculation on the evolution of the Monilinia species. M. mumecola appears to be a direct descendant of M. laxa. Both phylogenetic trees also indicate a close relationship between M. yunnanensis and M. fructigena suggesting that M. fructigena may have evolved from M. yunnanensis. In contrast to a previous rDNA analysis [33], our phylogenetic analyses of G3PDH plus TUB2 gene sequences illustrated that M. fructicola may have evolved earlier than other Monilinia spp. The latter hypothesis is strengthened by the fact that the rDNA sequences contained less phylogenetically informative characters compared with the G3PDH and TUB2 sequences. A more detailed DNA sequence analysis, including additional populations, populations from other parts of world, and additional genes, would provide a more complete picture of the evolution of Monilinia species. Leeuwen [12] suggested that the ancestor of M. fructigena and M. polystroma might have evolved in the Far East as a specialized fruit pathogen, before they evolved into two distinct groups in Europe and Japan due to geographical separation. The fact that M. yunnanensis found in China appears to be more basal in the phylogeny than M. fructigena, lends some support to Leeuwen's suggestion.
In conclusion, phylogenetic, morphological, and cultural analysis of Monilinia isolates from China revealed a previously undescribed species we've designated Monilia yunnanensis, and the first report of M. mumecola on peach and nectarine. A molecular assay was developed to detect these two species and differentiate the Chinese Monilinia species pathogenic on peach including the newly described M. yunnanensis.
Supporting Information
Isolates utilized in this study.
(DOC)
Acknowledgments
The authors thank Dr Liu X. L. from Beijing, China and Holb, I. J. from Debrecen, Hungary for generously providing some isolates.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This material is based upon work supported by Program for New Century Excellent Talents in University (NCET-08-0783) and the Fundamental Research Funds for the Central Universities (Program No.2009PY010), also partially sponsored by Project supported by the National Natural Science Foundation of China (Grant No. 31071703). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Li Z. Peach germplasm and breeding in China. HortScience. 1984;19:348–351. [Google Scholar]
- 2.Xiang WN. Reference of Mycology and Plant Pathology in China. Beijing: Science Press; 1957. 322 [Google Scholar]
- 3.Tai FL. Sylloge Fungorum Sinicorum. Beijing, China: Science Press, Academia Sinica; 1979. [Google Scholar]
- 4.Wang YX, Wang G, Zeng QQ, Zhang ZY. Studies on the investigation and integrated control of brown rot disease of peach. JYunnan Agric Univ. 1998;13:29–32. [Google Scholar]
- 5.Zhuang WY. Flora Fungorum Sinicorum: Science Press, Beijing. 1998;Vol. 8:57–60. [Google Scholar]
- 6.Zhu XQ, Chen XY, Luo Y, Guo LY. First report of Monilinia fructicola on peach and nectarine in China. Plant Pathology. 2005;54:575–575. [Google Scholar]
- 7.Zhu XQ, Guo LY. First Report of Brown Rot on Plum Caused by Monilia polystroma in China. Plant Disease. 2010;94:478–478. doi: 10.1094/PDIS-94-4-0478A. [DOI] [PubMed] [Google Scholar]
- 8.Fan JY, Zhu XQ, Guo LY, Luo Y. Comparison of three molecular identification methods for Monilinia species on stone and pome fruits. Acta Phytophyl Sin. 2007;34:289–295. [Google Scholar]
- 9.CABI. Crop Protection Compendium. 2010. Commonwealth Agricultural Bureau International (CABI). Wallingford, UK.
- 10.Bryde RJ, Willetts HJ. The brown rot fungi of fruit: Their biology and control. Pergamon Press (Oxford and New York); 1977. [Google Scholar]
- 11.Hu MJ, Chen Y, Chen SN, Liu XL, Yin LF, et al. First Report of Brown Rot of Peach Caused by Monilinia fructicola in Southeastern China. Plant Disease. 2011;95:225–225. doi: 10.1094/PDIS-11-10-0779. [DOI] [PubMed] [Google Scholar]
- 12.van Leeuwen GCM, Baayen RP, Holb IJ, Jeger MJ. Distinction of the Asiatic brown rot fungus Monilia polystroma sp nov from M. fructigena. Mycological Research. 2002;106:444–451. [Google Scholar]
- 13.Nakao S. Brown rot disease of Prunus mume caused by an undescribed Monilia fungus. Kongetsu-no-noyaku. 1992;(1):92–95. [Google Scholar]
- 14.Harada Y, Sasaki M, Nakao S. On occurrence in recent years of brown rot disease on Prunus mume in Oita Pref. and its causal fungus. Ann Phytopathol Soc Jpn. 1990;59:387. [Google Scholar]
- 15.Harada Y, Nakao S, Sasaki M, Sasaki Y, Ichihashi Y, et al. Monilia mumecola, a new brown rot fungus on Prunus mume in Japan. Journal of General Plant Pathology. 2004;70:297–307. [Google Scholar]
- 16.Shao W. Etiology, occurrence and control of Papaya (Chaenomeles lagenaria) brown rot. Wuhan: Huazhong Agricultural University; 2009. 54 [Google Scholar]
- 17.Luo C-X, Hanamura H, Sezaki H, Kusaba M, Yaegashi H. Relationship between Avirulence Genes of the Same Family in Rice Blast Fungus Magnaporthe grisea. J Gen Plant Pathol. 2002;68:300–306. [Google Scholar]
- 18.White TJ, Bruns T, Lee SaTJ. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols. A Guide to Methods and Applications ed. London: Academic Press; 1990. [Google Scholar]
- 19.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends in Biochemical Sciences. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 20.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X Windows Interface: Flexible Strategies for Multiple Sequence Alignment Aided by Quality Analysis Tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 22.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 24.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Tang QY, Feng MG. DPS Data processing system: Experimental design, statistical analysis, and data mining. Beijing: Science Press; 2007. [Google Scholar]
- 26.Ioos R, Frey P. Genomic Variation within Monilinia laxa, M. fructigena, and M. fructicola, and Application to Species Identification by PCR. European Journal of Plant Pathology. 2000;106:373–378. [Google Scholar]
- 27.Ma Z, Luo Y, Michailides TJ. Nested PCR assays for detection of Monilinia fructicola in stone fruit orchards and Botryosphaeria dothidea from pistachios in California. Journal of Phytopathology-Phytopathologische Zeitschrift. 2003;151:312–322. [Google Scholar]
- 28.Ma ZH, Yoshimura MA, Holtz BA, Michailides TJ. Characterization and PCR-based detection of benzimidazole-resistant isolates of Monilinia laxa in California. Pest Management Science. 2005;61:449–457. doi: 10.1002/ps.982. [DOI] [PubMed] [Google Scholar]
- 29.Cote MJ, Tardif MC, Meldrum AJ. Identification of Monilinia fructigena, M. fructicola, M. laxa, and Monilia polystroma on inoculated and naturally infected fruit using multiplex PCR. Plant Disease. 2004;88:1219–1225. doi: 10.1094/PDIS.2004.88.11.1219. [DOI] [PubMed] [Google Scholar]
- 30.Gell I, Cubero J, Melgarejo P. Two different PCR approaches for universal diagnosis of brown rot and identification of Monilinia spp. in stone fruit trees. Journal of Applied Microbiology. 2007;103:2629–2637. doi: 10.1111/j.1365-2672.2007.03495.x. [DOI] [PubMed] [Google Scholar]
- 31.Miessner S, Stammler G. Monilinia laxa, M. fructigena and M. fructicola: Risk estimation of resistance to QoI fungicides and identification of species with cytochrome b gene sequences. Journal of Plant Diseases and Protection. 2010;117:162–167. [Google Scholar]
- 32.Hily J-M, Singer SD, Villani SM, Cox KD. Characterization of the cytochrome b (cyt b) gene from Monilinia species causing brown rot of stone and pome fruit and its significance in the development of QoI resistance. Pest Management Science. 2010;67:385–396. doi: 10.1002/ps.2074. [DOI] [PubMed] [Google Scholar]
- 33.Holst-Jensen A, Kohn L, Jakobsen K, Schumacher T. Molecular phylogeny and evolution of Monilinia (Sclerotiniaceae) based on coding and noncoding rDNA sequences. Am J Bot. 1997;84:686–701. [PubMed] [Google Scholar]
- 34.Chen Y-C, Eisner JD, Kattar MM, Rassoulian-Barrett SL, Lafe K, et al. Polymorphic Internal Transcribed Spacer Region 1 DNA Sequences Identify Medically Important Yeasts. J Clin Microbiol. 2001;39:4042–4051. doi: 10.1128/JCM.39.11.4042-4051.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baldwin BG. Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: An example from the compositae. Molecular Phylogenetics and Evolution. 1992;1:3–16. doi: 10.1016/1055-7903(92)90030-k. [DOI] [PubMed] [Google Scholar]
- 36.Iwen PC, Hinrichs SH, Rupp ME. Utilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. Medical Mycology. 2002;40:87–109. doi: 10.1080/mmy.40.1.87.109. [DOI] [PubMed] [Google Scholar]
- 37.Fulton CE, van Leeuwen GCM, Brown A. Genetic Variation among and within Monilinia species causing brown rot of stone and pome fruits. European Journal of Plant Pathology. 1999;105:495–500. [Google Scholar]
- 38.Takahashi Y, Ichihashi Y, Sano T, Harada Y. Monilinia jezoensis sp nov in the Sclerotiniaceae, causing leaf blight and mummy fruit disease of Rhododendron kaempferi in Hokkaido, northern Japan. Mycoscience. 2005;46:106–109. [Google Scholar]
- 39.Couch BC, Kohn LM. A multilocus gene genealogy concordant with host preference indicates segregation of a new species, Magnaporthe oryzae, from M. grisea. Mycologia. 2002;94:683–693. doi: 10.1080/15572536.2003.11833196. [DOI] [PubMed] [Google Scholar]
- 40.de Jong SN, Lévesque CA, Verkley GJM, Abeln ECA, Rahe JE, et al. Phylogenetic relationships among Neofabraea species causing tree cankers and bull's-eye rot of apple based on DNA sequencing of ITS nuclear rDNA, mitochondrial rDNA, and the [beta]-tubulin gene. Mycological Research. 2001;105:658–669. [Google Scholar]
- 41.Sholberg PL, Harlton C, Haag P, Levesque CA, O'Gorman D, et al. Benzimidazole and diphenylamine sensitivity and identity of Penicillium spp. that cause postharvest blue mold of apples using beta-tubulin gene sequences. Postharvest Biology and Technology. 2005;36:41–49. [Google Scholar]
- 42.Fournier E, Giraud T, Albertini C, Brygoo Y. Partition of the Botrytis cinerea complex in France using multiple gene genealogies. Mycologia. 2005;97:1251–1267. doi: 10.3852/mycologia.97.6.1251. [DOI] [PubMed] [Google Scholar]
- 43.Staats M, van Baarlen P, van Kan JAL. Molecular phylogeny of the plant pathogenic genus Botrytis and the evolution of host specificity. Molecular Biology and Evolution. 2005;22:333–346. doi: 10.1093/molbev/msi020. [DOI] [PubMed] [Google Scholar]
- 44.O'Gorman DT, Sholberg PL, Stokes SC, Ginns J. DNA sequence analysis of herbarium specimens facilitates the revival of Botrytis mali, a postharvest pathogen of apple. Mycologia. 2008;100:227–235. doi: 10.3852/mycologia.100.2.227. [DOI] [PubMed] [Google Scholar]
- 45.Fournier E, A G, AS W, S K, T G. 2006. 195 Genetic structure of the species complex Botrytis cinerea: 2006 APS-CPS-MSA joint annual meeting.
- 46.Khokhriakova TM. On the geographical formation of species in phytopathogenic ascomycetes on fruit crops in the USSR. Mikologiyai Fitopatologiya. 1978;12:154–163. [In Russian.] [Google Scholar]
- 47.Wormald H. Further studies of the brown-rot fungi. II. A contribution to our knowledge of the distribution of the species of Sclerotinia causing brown-rot. Annals of Botany. 1927;41:287–299. [Google Scholar]
- 48.Willetts HJ. The development of stromata of Sclerotinia fructicola and related species: I. In culture. Transactions of the British Mycological Society. 1968;51:625–632, IN621–IN622. [Google Scholar]
- 49.Willetts HJ, Harada Y. A review of apothecial production by Monilinia fungi in Japan. Mycologia. 1984;76:314–325. [Google Scholar]
- 50.Lehman JS, Oudemans PV. Variation and heritability of phenology in the fungus Monilinia vaccinii-corymbosi on blueberry. Phytopathology. 2000;90:390–395. doi: 10.1094/PHYTO.2000.90.4.390. [DOI] [PubMed] [Google Scholar]
- 51.Fan JY, Guo LY, Xu JP, Luo Y, Michailides TJ. Genetic Diversity of populations of Monilinia fructicola (Fungi, Ascomycota, Helotiales) from China. Journal of Eukaryotic Microbiology. 2010;57:206–212. doi: 10.1111/j.1550-7408.2009.00467.x. [DOI] [PubMed] [Google Scholar]
- 52.Xue W, Zhao X, Liu S, Zhao X, Liu Z. Protoplast preparation and regeneration of Monilia fructigena. Microbiology China. 2010;37(1):71–77. [Google Scholar]
- 53.van Leeuwen GCM, van Kesteren HA. Delineation of the three brown rot fungi of fruit crops (Monilinia spp.) on the basis of quantitative characteristics. Canadian Journal of Botany. 1998;76(12):2042–2050. [Google Scholar]
- 54.Lane CR. A synoptic key for differentiation of Monilinia fructicola, M. fructigena and M. laxa, based on examination of cultural characters. EPPO Bulletin. 2002;32:489–493. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Isolates utilized in this study.
(DOC)