Abstract
Epstein-Barr virus (EBV) belongs to the gamma subfamily of herpes viruses, among the most common pathogenic viruses in humans worldwide. The viral ribonucleotide reductase small subunit (RNR R2) is involved in the biosynthesis of nucleotides, the DNA precursors necessary for viral replication, and is an important drug target for EBV. RNR R2 generates a stable tyrosyl radical required for enzymatic turnover. Here, the electronic and magnetic properties of the tyrosyl radical in EBV R2 have been determined by X-band and high-field/high-frequency electron paramagnetic resonance (EPR) spectroscopy recorded at cryogenic temperatures. The radical exhibits an unusually low g1-tensor component at 2.0080, indicative of a positive charge in the vicinity of the radical. Consistent with these EPR results a relatively high C-O stretching frequency associated with the phenoxyl radical (at 1508 cm−1) is observed with resonance Raman spectroscopy. In contrast to mouse R2, EBV R2 does not show a deuterium shift in the resonance Raman spectra. Thus, the presence of a water molecule as a hydrogen bond donor moiety could not be identified unequivocally. Theoretical simulations showed that a water molecule placed at a distance of 2.6 Å from the tyrosyl-oxygen does not result in a detectable deuterium shift in the calculated Raman spectra. UV/VIS light spectroscopic studies with metal chelators and tyrosyl radical scavengers are consistent with a more accessible dimetal binding/radical site and a lower affinity for Fe2+ in EBV R2 than in Escherichia coli R2. Comparison with previous studies of RNR R2s from mouse, bacteria, and herpes viruses, demonstrates that finely tuned electronic properties of the radical exist within the same RNR R2 Ia class.
Introduction
Ribonucleotide reductase catalyzes the conversion of ribonucleotides to the corresponding deoxyribonucleotides in all living organisms via a radical-based chemical mechanism, thereby providing and controlling the pool of precursors necessary for DNA synthesis and repair [1], [2], [3], [4]. Based on differences in the generation of the free radical, the amino acid sequence, and the overall quaternary structure [1], RNRs can be grouped into three different classes (I, II, and III). Class I RNR, the most common class, found in almost all eukaryotic organisms is oxygen dependent and consists of two subunits, designated R1 and R2 [2]. These can assemble into enzymatically active homodimeric tetramers (R12R22) and higher order oligomers [5], [6], [7]. Most R2s generate a stable tyrosyl radical in the vicinity of a dimetal-oxygen cluster [3], [6]. This radical transfers from the radical dimetal site in R2, over a distance of ca. 35 Å, to the active site of the R1 subunit, where it forms a thiyl radical. Its transfer likely is facilitated by a conserved array of hydrogen bonded amino acids [2], [8], [9]. Herpes viruses are ubiquitous eukaryotic pathogens infecting a large variety of animal species. They share similar architecture with a double-stranded DNA genome encased within a proteinaceous cage. Many herpes viruses, including those of the α-and γ-subfamilies, encode for an active RNR in their genomes. Most likely, the viral DNA replication relies on de novo synthesis of deoxyribonucleotides by the viral RNR under conditions in which the host cell RNR is inactive, such as in non-dividing cells [10]. Herpes viral R2 subunits have limited similarity in amino acid sequence compared with mammalian and bacterial homologues. Yet, previous studies of the RNR form herpes simplex virus (HSV) 1 and 2 indicated a similar catalytic mechanism as in Escherichia coli class Ia RNR [11], [12]. Epstein-Barr virus (EBV) is classified in the gamma subfamily of herpes viruses and represents one of the most common pathogenic viruses in humans worldwide. EBV R2 has low sequence similarity with mouse R2 (27.0% sequence identity and 53.7% sequence similarity (as calculated with the blosum62 matrix ), human p53R2 (26.7% and 52.7%), herpes simplex virus (HSV) 1 (36.7% and 59.6%), HSV 2 (36.4% and 60.9%) and E. coli R2 (27.0% and 53.7%). Amino acid sequence alignments indicate that the equivalent amino acid for the conserved metal coordinating aspartate in most R2s is a glutamate (Glu61 in EBV R2). The virus may induce development of several diseases such as infectious mononucleosis, and is associated with neoplasms, including lymphomas and carcinomas [13]. In this context, the viral RNR is an important drug target for EBV [14], [15]. Certain small organic molecules are potential inhibitors of herpes viral RNRs and thus can be employed for the treatment of herpes viral infections by targeting the R2 tyrosyl radical site. For example, hydroxyurea (HU), a potent reductant, diminishes the EBV genome in Burkitt's lymphoma cell lines in vitro, whereas prolonged exposure may lead to HU resistance in some cell lines [16]. Generally, an understanding of the specificity of inhibitors requires detailed knowledge of the accessibility, reactivity and the electronic and magnetic characteristics of the radical site [17]. The tyrosyl radical from several different R2s have been characterized, showing that the magnetic properties of the radical unit are strongly correlated with the dihedral angles of its tyrosyl β-methylene protons [18], [19], [20], [21] (Figure 1). Applying high-field/high-frequency electron paramagnetic resonance (HF-EPR) allows measurement of the g-tensor anisotropy of the radical center with high accuracy. Previous studies of tyrosyl radicals demonstrated that the g-values are sensitive probes for the local electrostatic environment. HF-EPR measurements showed that there are clear differences in the g-tensor anisotropy between different RNR R2s from class Ia [22], [23], [24]. Such differences are assumed to arise from variations in the hydrogen bonding interaction with a nearby hydrogen for both the mouse R2 radical and the YD radical from photo system II (PS II) [22]. In this work the physicochemical properties of the tyrosyl radical from EBV R2 were characterized experimentally and theoretically with the use of X-band, HF-EPR, resonance Raman (rRaman) spectroscopy and density functional theory (DFT). Further, we report ultraviolet/visible light spectroscopic (UV/VIS) experiments probing the dissociation of iron – in different oxidation states – and the radical with metal chelators and radical scavengers, respectively. We compare our results with previous observations for RNR R2s from mouse, bacteria, and other herpes viruses and discuss possible implications.
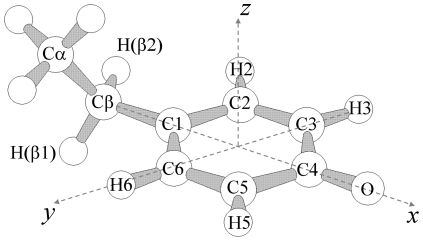
Figure 1. The tyrosyl radical.
Molecular sketch of a tyrosyl radical with the definition of the dihedral angle θ used to describe the orientation of the α-carbon (C6-C1-Cβ-Cα). An angle of θ = 0° corresponds to placing the α-carbon in the plane of the phenoxyl ring (xy), and θ = 90° corresponds to an orientation perpendicular to the plane of the ring.
Results and Discussion
Low temperature EPR spectra of the EBV R2 tyrosyl radical at two different microwave frequencies
The free tyrosyl radical from metal-free EBV R2 reconstituted with Fe2+ has been studied in the temperature range T = 3–30 K at two EPR microwave frequencies, 9.6 (X-band) and 285 GHz (high-frequency/field, HF-EPR). The formation of the tyrosyl radical in EBV R2 occurs with ∼1 radical per R2 dimer, which is similar to our earlier studies on human and mouse p53R2 and mouse R2 under similar reconstitution conditions. These values are higher than those obtained for HSV 1 and HSV 2 R2, which were calculated to be 0.3–0.6 radical per R2 dimer [21], [25], [26], [27], [28], [29].
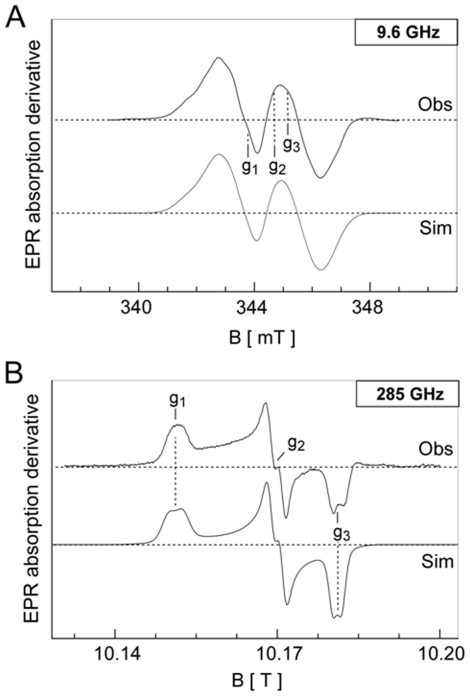
The observed first-derivative EPR spectra are shown in Figure 2A and 2B, respectively. The X-band EPR envelope of EBV R2 (Figure 2A, Obs) is similar to the spectra observed for HSV 1 and mouse R2 as well as for HSV 2 R2 [12], [23], [24], [27], [29], [30]. As expected, the anisotropic g-tensor components are poorly resolved under this microwave frequency. The HF-EPR (285 GHz) spectrum and g iso from X-band on the other hand clearly show the g-tensor anisotropy with values at g 1 = 2.0080, g 2 = 2.0043, and g 3 = 2.0021 (Figure 2B, Obs). The spin concentration of the tyrosyl radical was determined as one spin equivalent per dimer, and remained constant in the temperature range examined. The observed total resonance width (ΔB∼6.5 mT) is characterized by a composite resonance line, due to the presence of several anisotropic hyperfine splitting components (A), in analogy with previous analyses of the mouse and HSV 1 R2 spectra at 9.6 GHz. This phenomenon arises from the interaction of the unpaired spin on the phenolate with magnetically different hydrogen nuclei of the radical carrying tyrosine backbone as well as the β1,2-H protons (Figure 1 and Figure 3). The EPR signal line-shape and resonance features depend on the rotational configuration of the tyrosyl ring. Therefore, spectra simulations that fit both the X-band and high-field EPR envelopes (Figure 2A and 3B, Sim), followed by comparison with values from the literature, allowed an estimate for the rotational angle θ of ∼30° (see Scheme 1 for the definition of the θ rotational angle and the DFT generated spin density at UB3LYP/6-311++G** of the radical model in Figure 3). According to previous theoretical DFT studies on model tyrosyl radical systems such an angle falls at a local minimum in the potential energy curve as described by the rotational motion of the phenoxyl plane with respect to the α,β-carbons [18]. The simulation parameters used are collected in Table 1, together with previously determined g, A and θ values for the tyrosyl radical from E. coli R2 [23], [31], mouse R2 [23], [24], [27], [32], [33], HSV 1 R2 [23], [24], [27], [32], Salmonella typhimurium R2 [34] and PS II YD [22], [34] included for comparison. Previous work demonstrated that measurement of the g-value anisotropy can be used as a probe for the presence of a hydrogen bond to the phenol oxygen of the tyrosyl radical. The absence of a hydrogen bond, as occurring for the tyrosyl radical in E. coli R2, results in a large g 1-value of ca. 2.0090 [22], [23], [31]. In contrast, when a positive charge is located in close vicinity such that a hydrogen bond is formed with the tyrosyl phenol oxygen, the spin density on the phenolic moiety is lowered proportionally (Figure 3B), resulting in smaller g 1-values [18], [20], [22], [23], [32], [33], [35], [36]. In the crystal structure of the diferric (Fe3+-O2−-Fe3+) form of mouse R2 without a tyrosyl radical, two water molecules are present in close proximity of the tyrosyl radical site [37]. Furthermore, in the Y107•-radical site of the Y122F/W107Y double mutant from E. coli RNR R2, the radical forms a strong H-bond and displays a low g 1-value (g 1 = 2.0069 [23]). In the latter case, one of the water molecules in the crystal structures is located substantially closer to the tyrosyl radical than found in mouse R2 [23], [38]. The g 1-value of p53R2 [39] has not been reported, but is indicated in chapter 1 in reference ([2]) to have a low g 1-value (it is possible that the diiron center shows a mixed valence signal). Therefore, the perturbation of the g-tensor values of the tyrosyl radical in R2s usually originates from the local electrostatic environment [40]. The energies of the excited states associated with the radical unit, which are characterized by the nonbonding molecular orbitals of the phenoxyl oxygen, can be modified in a way that spin-orbit mixing occurs between low-lying excited states with the ground state, leading to alterations in the g-anisotropy. The sensitivity of the g 1-values for the tyrosyl radical to the local chemical environment must also be reflected in electrochromic shifts in infrared (IR), Raman (see Figure 3) and rRaman spectra. The observed g 1-value of the tyrosyl radical in EBV R2 is lower than that of E. coli R2, but slightly higher than of both mouse R2 and HSV 1 R2. Therefore, in EBV R2 spin-mixing and a hydrogen bond to the tyrosyl radical can be present. If a hydrogen bond is present, the strength of the latter must be weaker than those observed in mouse and HSV 1 R2. However, the molecular basis of such an interaction cannot be determined by simple continuous-wave EPR experiments alone (vide infra). Davies D-electron nuclear double resonance (ENDOR) studies of mouse and HSV 1 R2 demonstrated that, while both display identical g 1 values, the distance (d) between the exchangeable H/D (probably originating from a water molecule) is slightly shorter in HSV 1 R2 (d = 1.86 Å) compared with mouse R2 (d = 1.89 Å) [27]. Thus, if a similar interaction in EBV R2 were present, the expected distance to the hydrogen should be slightly larger (1.9≤d≤2.6 Å). Other moieties, than an exchangeable hydrogen from a water or hydroxide molecule, such as amino acids sidechains from the protein located in proximity of the tyrosyl radical site could contribute to the decrease of the g 1-component. As found for example in PS II YD (D2-Tyr160 from the cyanobacterium Synechocystis), the low g 1-value of the tyrosyl radical arises from positive charges located close to the phenol-oxygen (see Table 1). In this case, the hydrogen bonding interaction is not formed with a water or hydroxide molecule, but with a neighbouring histidine residue. Its imidazole moiety can serve as an acceptor for the phenolic proton upon oxidation, forming a hydrogen bond to the phenolic oxygen of the neutral tyrosyl radical as demonstrated by combined mutagenesis studies and Mims electron spin echo-ENDOR measurements [41]. Thus, any shift of the g 1-tensor component derived from HF-EPR measurements must be analyzed with caution as an indication for the presence of a water or hydroxide molecule in close proximity of the tyrosyl radical.
Figure 2. X-band and HF-EPR spectra of EBV R2.
Low-temperature EPR spectra of the tyrosyl radical of EBV R2. A protein concentration of ∼140–150 µM was used for these measurements. (A) The upper spectrum (Obs) shows the 9.6 GHz (X-band) resonance envelope recorded at T = 26 K with a microwave power of 0.08 mW and modulation amplitude 0.4 mT, and the lower spectrum shows its computer simulation (Sim) (B) The upper spectrum (obs) shows the 285 GHz resonance envelope recorded at T = 15 K with a modulation amplitude of 0.4 mT and microwave power in the µW range, and the lower spectrum its computer simulation (Sim). The derived spin-Hamiltonian parameters are listed in Table 1.
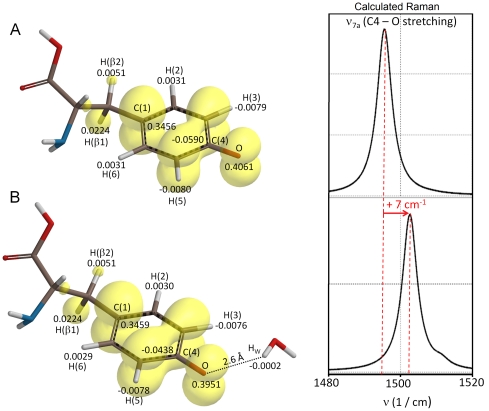
Figure 3. Spin density distribution on the tyrosyl radical in EBV.
The yellow spin spin density surfaces of the tyrosyl radical obtained by density functional theory (DFT/UB3LYP/6-311++G(d,p) in gas phase (0.0015 au isovalue), for the neutral forms after geometry optimization (the dihedral angle θ was constrained to 30°) in the absence (A) and in the presence (B) of a water molecule located close to the phenoxyl oxygen. The isosurfaces show the larger positive spin density on H(β1) with respect to the H(β2) proton. The numerical values (au) derived from Natural Atomic Populations (NAP) analyses are also given for the H(3,5) and H(2,4) protons as well as for the C4 and the tyrosyl oxygen atoms. This level of theory gives a calculated C(4)-O bond length of (A) 1.252 Å and (B) 1.256 Å. The theoretically derived Raman spectra (DFT/UB3LYP/6-311++G(d,p), 3 cm−1 Lorentzian line band-width) are shown on the right side of the panel, and highlight the frequency shift associated to the C(4)-O stretching vibration (ν7a, Wilson notation) when the phenoxyl oxygen interacts with a water molecule.
Table 1. The g–tensor, hf–tensors (A, mT), line–width tensor (LW, mT), and the rotational θ angle of the α-carbon (°, deg) for the tyrosyl radical in Class Ia, Class Ib RNRs and the YD radical in photosystem II.
| EPR-tensors | EBV R2 | HSV1 R2 | E. coli R2 | Mouse R2 | S. typhimurium R2 | PS II YD |
| g 1 | 2.0080 | 2.0076 | 2.0090 | 2.0076 | 2.0089 | 2.0076 |
| g 2 | 2.0043 | 2.0043 | 2.0046 | 2.0043 | 2.0043 | 2.0043 |
| g 3 | 2.0021 | 2.0022 | 2.0022 | 2.0022 | 2.0021 | 2.0022 |
| H3,5 Axx | −0.80/−0.70* | −0.28/−0.43* | −0.96 | −0.91/−0.73* | −1.15 | −0.91 |
| H3,5 Ayy | −0.50/−0.40* | −0.28/−0.18* | −0.28 | −0.44/−0.48* | −0.25 | −0.26 |
| H3,5 Azz | −0.30/−0.20* | −0.47−/0.64* | −0.70 | −0.66/−0.58* | −0.71 | −0.70 |
| Hβ1 Axx | 2.60 | 2.28 | 1.96 | 2.14 | 1.05 | 0.72 |
| Hβ1 Ayy | 1.80 | 2.07 | 2.12 | 1.90 | 0.74 | 1.05 |
| Hβ1 Azz | 1.40 | 1.78 | 1.96 | 2.15 | 0.95 | 0.72 |
| Hβ2 Axx | 0.40 | 0.39 | 0.18 | 0.95 | <0.3 | 0.19 |
| Hβ2 Ayy | 0.30 | 0.25 | −0.07 | 0.25 | <0.3 | 0.19 |
| Hβ2 Azz | 0.30 | 0.28 | −0.07 | 0.57 | <0.3 | 0.51 |
| LWx | 0.38 | 0.33 | - | 0.45 | 0.58 | - |
| LWy | 1.0 | 0.33 | - | 0.35 | 0.53 | - |
| LWz | 1.1 | 0.33 | - | 0.44 | 0.37 | - |
| θ | 30 | 35 | 30 | 35 | 75 | 82 |
| Ref. | This work# | [27], [33] | [31], [40], [65] | [33] | [34] | [22], [34] |
*Non-equivalent H3 and H5 protons.
The proton (H) hfc signs were derived from the DFT analyses. The definition of the torsional angle θ was given in Scheme 1.
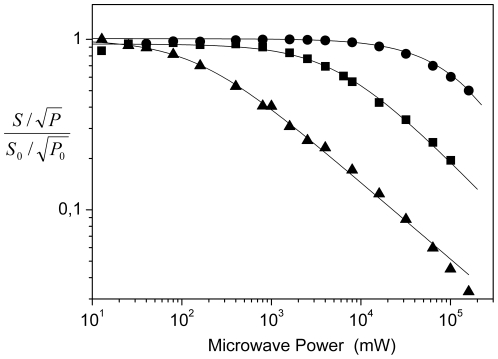
EPR relaxation measurements allow closer investigation of the chemical environment of the tyrosyl radical through observation of the progressive microwave power saturation properties of the EPR spectra in the temperature range of T = 26–100 K (Figure 4). When an effective magnetic coupling occurs between the radical and the diferric metal-oxygen cluster, the radical relaxation properties, as indexed by the half-saturation value (P 1/2), become larger than the one observed in the absence of magnetic coupling. Compared with previous studies carried out on mouse and HSV 1 R2 (with values of P 1/2 = 1.2 mW and P 1/2 = 3 mW at 30 K, respectively), the resonance saturation for the EBV R2 radical occurs at a lower applied microwave power (P 1/2 = 0.14 mW), but higher than for E. coli R2 (P 1/2 = 0.05 mW) [30], [42]. Thus, the magnetic interaction between the iron-oxygen cluster and the tyrosyl radical must be weaker than that estimated for both mouse and HSV 1 R2. The presence of a glutamate in EBV R2 (Glu61) in the amino acid sequence position of the common iron-coordinating aspartate in HSV 1 and mouse R2, may have an impact on these observed differences. Similar to E. coli R2 and S. typhimurium R2, the P 1/2 value in EBV R2 derived here is larger than that observed for a free tyrosyl radical lacking any magnetic interaction with another paramagnetic center [42]. For EBV R2 we did not succeed to trap the mixed valence (Fe2+Fe3+) complex or to detect integer spin signals, (with g values in the range 8≤g≤16) of the diferrous site with the chemical mediators employed in this study. This electronic characteristic in EBV R2 distinguishes it from those previously observed by us for mouse R2, where both signals can be detected, and from HSV 1 R2, which exhibit the mixed valence signal under mildly reducing conditions [43], [44]. The mouse R2 integer spin EPR signal can be explained by the relatively small zero field splitting parameters designated Δ or δ, making it an observable signal for X-band EPR [9], [45]. Thus, there are clear differences in the electronic characteristics of the tyrosyl radical site in EBV R2 compared with previously studied R2s [21], [27].
Figure 4. EPR microwave power saturation.
The microwave power saturation curves for EBV R2 as derived from X-Band EPR measurements performed at T = 26 K (▴), T = 50 K (▪), and T = 100 K (•). The b-values were equal to 1 for T = 26 K and 50 K and <1 for T = 100 K. P1/2 (26 K) = 0.14 mW, P1/2 (50 K) = 5 mW, P1/2 (100 K)≥150 mW. S: double integrated signal amplitude, P: microwave power, S 0 and P 0 are reference values obtained at non-saturating conditions.
Resonance Raman spectroscopic studies of the EBV R2 tyrosyl radical
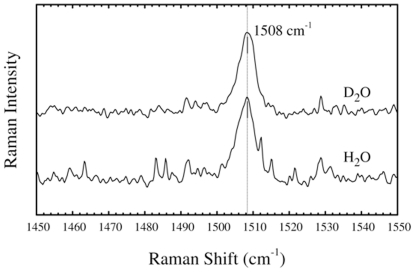
In order to investigate in more detail the basis for the low g 1-value, rRaman spectroscopy was employed. The rRaman and IR spectroscopy are excellent complementary techniques to HF-EPR spectroscopy, because even weak interactions between the tyrosyl radical and molecules in its close proximity can be detected. Tyrosyl radicals show a characteristic light absorption maximum at ca. 410 nm (see below) that is usually used for excitation in rRaman spectra of the radical. Vibrational spectroscopy can identify redox-linked structural changes associated with electron transfer reactions, as shown for the tyrosyl radical (Y122•) in E. coli RNR. For the latter such an electron transfer reaction has been proven to be coupled to a conformational change in the R2 structure [46]. One vibrational characteristic of the radical, the phenoxyl ν7a mode (Wilson notation) with components of the C4-O stretching vibrations (Figure 5), is a sensitive marker for hydrogen bonds in this context. This vibrational mode is strongly enhanced when rRaman is employed at excitation frequencies ν ex around 405–415 nm, i.e. very close to the light absorption maximum of the tyrosyl radical. Without a nearby hydrogen bonding interaction, the ν7a vibration of the tyrosyl radical has been observed between 1497 and 1501 cm−1 in E. coli R2, whereas in mouse R2 it is observed at 1515 cm−1 [47]. In mouse R2 a water molecule in close proximity (-Ophenol…H distance of 1.89 Å) with exchangeable hydrogens is present as demonstrated with pulsed ENDOR [27]. As shown in Figure 5 (lower line), the EBV R2 tyrosyl displays a C4-O stretching vibration at slightly higher energy than that observed in E. coli R2 with the ν 7a vibration maximum at 1508 cm−1, which is at lower energy than that observed for mouse R2. Therefore, our rRaman results are consistent with our HF-EPR data on EBV R2, with the tyrosyl radical displaying a g 1-value in between those observed for mouse and in E. coli R2. In EBV R2 the observation of such an intermediate frequency mode is, in principle, supportive of the existence of a hydrogen bonded tyrosyl radical with a water molecule as hydrogen bond donor. If present, such a hydrogen bond is weaker than in mouse R2.
Figure 5. Resonance Raman spectra.
The resonance Raman spectra of the tyrosyl radical in EBV R2 and the effect of D2O exchange recorded with the following settings: λ exc = 410 nm, laser power at sample ∼5 mW, 60 scan, 60 sec/scan, T = 77 K.
Theoretical analyses of the influence of a hydrogen-bond to a water molecule on the tyrosyl radical mode frequency and EPR spectra
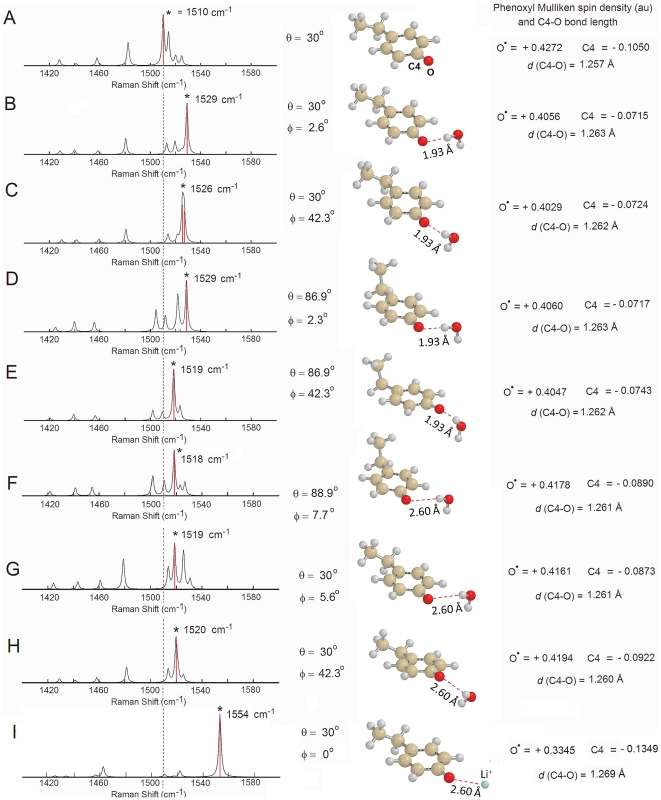
In order to test the impact of a water molecule in the proximity of the tyrosyl radical on the Raman spectra, DFT calculations were performed using a simplified model, the p-ethylphenoxyl radical. This has been demonstrated to be a good model system before [36]. The molecular backbone of the radical unit in EBV R2 is characterized by a rotational configuration of the tyrosyl ring θ constraint at 30°, as derived our EPR results. Such analyses gives an estimation of the C4-O stretching vibration trends (B3LYP/6-31G(d,p)) either in the presence or absence of a hydrogen bonded water molecule. The calculated Raman spectra are shown in Figure 6 (without enhanced resonance effects) together with the optimized structures with and without a water molecule placed close to the tyrosyl group at a distance slightly longer (1.93 Å) than that spectroscopically observed in mouse R2 (Figure 6, panel B–E). The calculated C4-O stretching vibration in the absence of a hydrogen bonded water molecule occurs at 1510 cm−1 (Figure 6A), a value identical to that obtained for the tyrosyl radical by Johnson and coworkers [48]. The calculated ν7a vibration in this simplified model closely agrees with the Fourier transform-IR analyses on the tyrosinate and tyrosyl radical by Barry and coworkers (tyrosyl radical, ν7a = 1516 cm−1), who have explicitly considered the effect of an amino acid backbone in their theoretical calculations (UB3LYP/6-31++G(d,p) as well as from our calculations using the entire tyrosyl radical backbone, computed at a higher level of theory (UB3LYP/6-311++G(d,p), ν7a = 1496 cm−1, Figure 3)) [49]. The value does not differ substantially from results obtained using the p-methoxyphenoxy radical as a model system, which employed a different level of theory (BPW91/6-31G**, ν7a = 1496 cm−1) [50]. When a water molecule is placed at a distance of 1.93 Å to the phenoxyl plane (and a φ = 2.6° after optimization), the ν7a vibration shifts significantly to longer wave number at 1529 cm−1 (Figure 6B) but slightly decreases to 1526 cm−1 (Figure 6C) if a water molecule is placed outside of the phenoxyl plane (φ∼42° after optimization). When the rotational configuration of the tyrosyl ring is lifted from the constraint of 30°, this angle after optimization, refined to slightly less than 90° (θ = 89.1°) [18]. Here a water molecule placed in the phenoxyl plane at the same distance of 1.93 Å, gives for the ν7a vibration a similar shift at 1529 cm−1 (Figure 6D) but it exhibits a larger decrease to 1519 cm−1 (Figure 6E) when is moved out of the phenoxyl plane (φ∼42° after optimization). Upon placement of the water molecule further away (2.60 Å) (Figure 6F and 6G) an effective perturbation of the ν7a vibration is reduced proportionally (ν7a = 1518 cm−1 and 1519 cm−1, respectively). This is accompanied by an increase of the phenoxyl (-O•) Mulliken spin density and Δν = 8–9 cm−1 together with a decrease in the C4-O bond length. Clearly, both properties tend to approach the structural and electronic parameters calculated without nearby water. By placing a positive charge (Li+) close to the phenoxyl oxygen (O•-Li+ = 2.60 Å, on the phenoxyl plane, φ∼0°) the perturbation of the ν7a vibration becomes nonetheless very large (Figure 6I, ν7a = 1554 cm−1) (see also Information S1). The C4-O stretching frequency (ν7a vibration) is characterized experimentally by a narrow bandwidth, of approximately 5 cm−1, as shown in Figure 5. This vibration is thus expected to be influenced by i) the relative angle of the vector between a hydrogen bonding neighboring molecule and the phenol oxygen and the phenol plane, ii) the distance of the hydrogen bonded molecule to the phenol oxygen and iii) by the conformational orientation of the phenoxyl group with respect to the protein backbone, represented here by a simple ethyl molecule. The impact of molecular conformation on the calculated g-tensor and hyperfine tensor (AH) components complemented by their further modulation due to dielectric effects associated to the medium have been discussed thoroughly in other reports, and thus are not further addressed here [35], [36], [50], [51], [52].
Figure 6. Theoretical resonance Raman modes.
Calculated Raman spectra of the tyrosyl radical model system (ethylphenoxyl radical, neutral form) in the absence (A) or presence (panel B–H) of a water molecule in close vicinity (B, C, D, and E, d = 1.93 Å) or placed further away (F–H, d = 2.60 Å) from the phenoxyl oxygen. Panel I shows the interaction with nearby positive charge (Li+). The calculations were performed by density functional theory (DFT/UB3LYP/6-31G(d,p)) in gas phase. No frequency scaling was applied and a 3 cm−1 band width (Lorentzian-line) was used. The angle φ is defined as the angle (°, deg) formed between the phenoxyl plane and the C4phen-Ophen….Hwater plane. The definition of the angle θ is given in Figure 1. The numerical values for the Mulliken spin density calculated for the phenoxyl (-O•) oxygen (positive) and the C4 atom (negative) are shown on the right-hand side of each panel. The minor bond elongation observed in the calculated C4-O bond length upon interaction of the phenoxyl oxygen with the water molecule is also shown. The asterisk (*) in each panel indicates the position of the ν7a mode. The thin dashed vertical line serves as a visual marker for the unperturbed tyrosyl radical to guide to the eyes.
Basis for the lack of deuterium shift in EBV R2 rRaman spectra
In order to probe the presence of a water molecule with exchangeable hydrogens located in proximity of the tyrosyl radical in EBV R2, we experimentally investigated the frequency shifts of the tyrosyl C4-O rRaman stretching vibration with deuterated EBV R2. In mouse R2, deuterium exchange of the protein clearly results in a 5 cm−1 shift to a lower energy of 1510 cm−1 compared with spectra of the non-deuterated protein. This effect was not observed in E. coli R2 [47]. In the former, the exchangeable hydrogens from a water molecule are located in proximity of to the phenol plane as detected by ENDOR. Interestingly, in EBV R2 a similar deuterium induced shift in the rRaman spectrum was not observed (Figure 5, upper line). Yet, this result does not preclude the presence of a water molecule as a hydrogen bond donor in close vicinity of the tyrosyl radical for the following reasons: i) Assuming a buried location of the water in the protein interior, its hydrogens may not be readily exchangeable to deuterium; ii) The distance from an immobilized water molecule to the tyrosyl residue could be too large to be detectable in our rRaman measurements. This scenario would be consistent with the theoretical Raman frequency shift calculated (Figure 6G) where a water molecule is placed close to the phenoxyl plane at a distance of 2.60 Å from the radical site. In this case, any deuterium shift is expected to be extremely small (<2 cm−1); iii) The absence of a deuterium shift could also arise from interactions with a positive charge located close (2.60 Å) to the tyrosyl radical site. However, the theoretically calculated effects on the phenoxyl oxygen spin-density and the ν7a vibration, using a Li+ placed at 2.60 Å from the phenoxyl oxygen mimicking the presence of such a charged group nearby, are too large (ν7a = 1554 cm−1, Figure 6I). Thus, the latter scenario seems unlikely; iv). The most probable explanation is a water molecule as a hydrogen bond donor in EBV R2 with similar overall structural relations to the tyrosyl radical as in HSV 1 R2. This is characterized by a distance of the water to the tyrosyl radical of >2.5 Å together with a large angle between the phenoxyl plane and the C4-Ophen… Hwater plane (φ) (Figure 6H, calculated ν7a = 1520 cm−1 and hydrogen bond C4-O…H distance of 2.60 Å). This structural organization would suitably explain the lack of an observable deuterium shift for EBV R2.
Preliminary observations of interaction of EBV R2 with radical scavenger and metal-ion chelators probed by UV/VIS spectroscopy
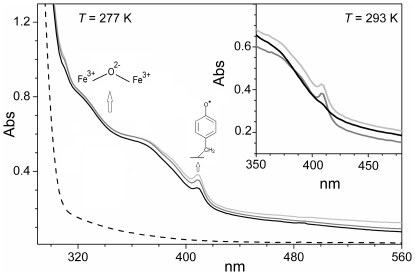
Certain radical scavengers, such as HU (used since long time in clinical treatment of some cancers), function as potent inhibitors of RNR activity and are attractive drug candidates [2] [53]. HU reduces the tyrosyl radical and large differences in reactivity of HU have been documented between E. coli R2 and both mouse R2 and HSV 2 R2s. The tyrosyl radicals of mouse R2 and HSV 2 R2 are more reactive and are scavenged substantially faster than that of E. coli R2. The radical quenching process occurs also more rapidly with some hydrophobic radical scavengers, such as p-alkoxyphenol compounds, than with HU [28], [54], [55]. Upon reconstitution of the dimetal site of metal-free EBV R2 with Fe2+ and exposure to air, relatively strong and characteristic light absorption bands are formed with distinctive absorption features for the diferric center and the tyrosyl radical are apparent at ∼320, 360 nm and ∼410 nm, respectively (Figure 7, light grey line). The strong 320–360 nm band originates from oxo-to-Fe3+ dimer charged transfer (CT) transitions. When the diferric active EBV R2 protein was incubated under aerobic conditions with HU, quenching of the tyrosyl radical signal occurred slowly at T = 277 K with the total radical quenching occurring after ∼25 min. The quenching rate however was substantially increased at higher temperature (T = 293 K), and the absorption signal associated with the tyrosyl radical disappeared after 7 min.
Figure 7. UV-VIS light absorption spectra of tyrosyl radical iron cluster formation and reduction.
Light absorption spectra of metal-free EBV R2 before (dashed-line) and after reconstitution of the diferric (Fe3+–O2−–Fe3+) metal site (light-grey line). Representative spectrum obtained under anaerobic conditions for the incubation of reconstituted EBV R2 with 4 mM HU recorded after 200 s (dark grey line) and after 20 min (black line), T = 277 K. The inset shows the resulting spectra obtained at higher temperature (T = 293 K), before (light-grey line) and after anaerobic incubation with HU (200 s, dark grey line; 400 s, black line). The protein concentration was ∼130 µM in a pH 7.5, 50 mM HEPES buffer.
In mouse R2 the reaction of the diferric center with HU leads to reduction of Fe3+ followed by rapid dissociation of Fe2+ from the protein [56]. For EBV R2, such an effect does not appear prominent (Figure 8). Thus, the reductive ability of HU towards the diferric center in EBV R2 is at an intermediate level between those observed for mouse R2 (very fast) and E. coli R2 [55] (non effective).
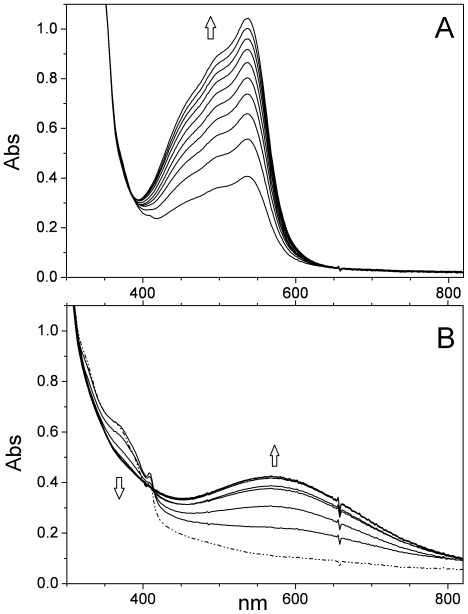
Figure 8. Reaction of reconstituted EBV R2 with scavengers.
(A) Representative light absorption spectra recorded at T = 288 K of reconstituted EBV R2 (∼130 µM, pH 7.5, 50 mM HEPES buffer) after addition of 1 mM bathophenanthrolinesulphonate and 4 mM HU, recorded in 10 min intervals under anaerobic conditions. (B) Reconstituted EBV R2 (∼130 µM) incubated with 1 mM catechol at pH 7.5 under anaerobic conditions at T = 293 K. The light absorption spectra were recorded at 30 s intervals. The dashed-dotted line represents the spectrum of the reconstituted diferric protein without any metal-ion chelator.
In order to obtain preliminary knowledge of the dynamics of this reaction in EBV R2, the reconstituted active tyrosyl-radical containing ferric protein was incubated under anaerobic conditions in the presence of HU and bathophenanthrolinesulphonate, a potent Fe2+ chelator. The Fe2+ complexation by bathophenanthrolinesulphonate was monitored following formation of an absorption band at λ max at 535 nm, as described previously [56], [57]. The complex formation process of Fe2+ occurred more slowly (Figure 8A) and in parallel with a decrease in the absorption band at 410 nm, which is associated with tyrosyl radical quenching. In order to examine this further an analogous experiment with HU and desferrioxamine, which chelates with Fe3+ with formation of a typical light absorption at ∼430 nm (53) was carried out. In contrast to mouse R2, the reaction with EBV R2 did not result in this characteristic absorption after 3 hrs (data not shown). From these preliminary results we conclude that iron dissociated from EBV R2 in the presence of HU is predominantly in the Fe2+ state. This implies that HU reacts with both the radical and Fe3+ at the diiron site in the protein, as probably also in mouse R2; and that Fe2+ is more weakly bound than Fe3+ in the dimetal site (Table 2) [54], [58].
Table 2. Summary of the UV/VIS spectroscopic experiments on EBV R2 in comparison with similar studies performed on other R2s.
| Bathophenanthroline-sulphonate | Catechol | |
| EBV R2 | + | ++ |
| E. coli R2 [56] | − | − |
| Mouse R2 [55], [56] | +++ | +++ |
| HSV2 R2 [28] | n/a | ++ |
The extent of the reactions was evaluated semi-quantitatively. The symbols indicate (+) positive reaction and (−) no reaction.
The mobilization of Fe3+ from EBV R2 by catechol, both a potent metal chelator and radical scavenger [28], was followed spectrophotometrically. The smaller chatecholate compound is a more effective Fe3+ chelator than the larger desferrioxamine, as it reacts without any addition of HU with EBV R2. The reaction resulted in a rapid decline of the absorbance at both 360 nm and 410 nm (Figure 8B) together with formation of a blue-violet catecholate to Fe3+ CT transitions around 600 nm. Thus, in these experiments with EBV R2, scavenging of the tyrosyl radical occurs concomitant with dissociation of the diferric center, similar to the analysis for HSV 2 R2 [28]. In contrast to E. coli R2, in EBV R2 catechol seems to react with the diferric center. The data imply that the reactivity of the tyrosyl radical with catechol in EBV R2 is substantially higher than for E. coli R2 and very similar to that of HSV 2 R2 (Table 2).
Conclusion
Through a combination of EPR, HF-EPR, UV/VIS and rRaman spectroscopy the electromagnetic characteristics of a spectroscopically unique tyrosyl radical from EBV R2 have been determined. The g 1-value from EPR and HF-EPR, and the rRaman shift indicate that the tyrosyl radical is hydrogen bonded, yet deuterium exchange of the protein had no effect on the rRaman spectra. Therefore, the g 1-value and rRaman results cannot be explained by a hydrogen bond with structural properties identical to that found in mouse R2. If a hydrogen bond to the tyrosyl radical is present in EBV R2, our spectroscopic data and DFT analysis indicate that it has characteristics more similar to that in HSV 1 R2, with a hydrogen outside of the phenoxyl plane. In contrast to mouse and HSV 1, where evidence for the involvement of a hydrogen bonded water molecule with exchangeable hydrogens has been observed, we could not clearly identify a water molecule as the hydrogen bond donor moiety for the tyrosyl radical in EBV R2. The observation of a spectroscopically different kind of tyrosyl radical in EBV R2 is a further indication for the variation of tyrosyl radicals of class Ia R2s in particular and in proteins in general. The preliminary UV/VIS spectroscopic studies carried out on EBV R2 showed that iron is most likely more weakly bound compared with E. coli R2, but stronger than for mouse R2. Differences can also be seen in the impact of the radical scavenger HU, which exhibits a reduced effect on the diferric center compared with mouse R2, where the reaction leads to reduction of the diferric center and dissociation of Fe2+. Our findings indicate that EBV R2 protein has evolved and includes a different kind of tyrosyl radical than in the seemingly simpler E. coli R2.
Materials and Methods
All high-grade chemicals were purchased from Sigma or Fluka unless stated otherwise.
Protein expression and purification
The BaRF1 gene (accession number YP_401656; encoding full-length EBV R2) was cloned from a B95-8 derived bacterial artificial chromosome by recombinatorial cloning (Gateway™, Invitrogen) in the laboratory of Prof. Jürgen Haas (Max von Pettenkofer Institut, Gene center, Ludwig-Maximilians University, Munich). BaRF1 was then subcloned with the LR recombination reaction into a pTH27 plasmid, with a coding sequence for an N-terminal polyhistidine tag (one letter coded amino acid sequence: MGPHHHHHHLESTSLYKKA-GSA), with the Gateway system (Invitrogen) according to the manufacturer's instructions. E. coli strain BL21 (DE3) (Stratagene) was transformed with the product from the LR reaction and small-scale soluble protein expression was verified as described [59], [60]. DNA sequencing (MWG Biotech, Germany) used the T7 forward and reverse promoter sequences in the plasmid as primers. Recombinant EBV R2 was produced in BL21 (DE3) E. coli cells in phosphate-buffered terrific broth (Formedium) supplemented with carbenicillin (Duchefa; final concentration of 50 µg/ml). Cells were grown at 37°C until an OD600 of 1–1.3. Protein expression was induced by addition of isopropyl-γ-D-thiogalactopyranoside (Anatrace; final concentration of 0.2 mM) and cells grown for ∼16 hrs at 18°C. The cells were harvested by centrifugation at 6500 rpm for 40 min. The cell pellet was resuspended in a buffer of 50 mM Na2HPO4/NaH2PO4, (Scharlau) pH 8.0, 500 mM NaCl (Scharlau), 10% (v/v) glycerol (Saveen Werner), 0.5 mM tris(2-carboxyethyl)phosphine (TCEP) (buffer A) containing 10 mM imidazole, 0.08 mg/ml lysozyme, 10 Units/ml DNase I recombinant (Roche), and ethylenediaminetetraacetic acid (EDTA)-free protease inhibitor tablets (Roche) (1 tablet/50 ml). Solubilized pellets were stored at 253 K until usage. Upon sonication on ice-bath, cell debris was removed by centrifugation at 65 000 rpm at 5°C and subsequent filtration (0.20 µm filter, Millipore) on ice. The soluble extract was passed over a 5 ml HisTrap FF Crude column (nickel sepharose 6 fast flow resin; GE Healthcare) at ∼10 ml/min flowrate, at 5°C , on an Äkta Express purification system (GE Healthcare). The column was washed with 10 column volumes of buffer A containing 10 mM imidazole followed by 10 column volumes of buffer A containing 70 mM imidazole. EBV R2 protein was eluted with buffer A containing 500 mM imidazole, with protease inhibitor tablets (1 tablet/50 ml). The combined elution fractions (volume ca. 10 ml) were gelfiltered on a Superdex S-200 HiLoad 16/60 column (GE Healthcare) on an Äkta Express system at 5°C with a buffer of 20 mM Hepes, pH 7.5, 300 mM NaCl, 10% (v/v) glycerol (buffer B), with 0.5 mM TCEP. Protein samples were concentrated with a 10 kDa molecular weight cut-off concentrator (Amicon Ultra-15, Millipore). Metalfree EBV R2 was purified analogously as described above with buffer B with 0.5 mM TCEP and 25–30 mM EDTA. The gelfiltered protein samples were left incubated 15–20 hrs in this solution, then concentrated to ∼2–3 ml, and dialysed (Slide-A-Lyzer, 7 kDa molecular weight cut-off, Pierce) twice against 5 l of buffer B at 5°C.
Sample preparation
Protein samples of metal-free EBV R2 were concentrated up to ca. 1 mM, using the theoretical calculated absorption coefficient [61] of EBV R2 at 280 nm. Reconstitution of the dimetal-oxygen cluster and generation of the tyrosyl radical site in EBV R2 was performed by addition of Fe2+ ((NH4)2Fe(SO4)2·6H2O) in a ca. 7∶1 molar ratio to the R2 homodimer) and naturally present O2. All samples were incubated at 273 K for 10 minutes. The final volume of the EPR samples was ∼200 µL in 50 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) pH 7.5, 100 mM KCl, 20% (v/v) glycerol. Samples of active EBV R2 for resonance Raman spectroscopy were prepared in the same way as for EPR measurements, but the protein was first transferred to a buffer of 50 mM Tris (tris(hydroxymethyl)aminomethane)-HCl, pH 7.5, 100 mM KCl. Samples of active EBV R2 protein in deuterated buffer (50 mM Tris-HCl, 100 mM KCl, pD = 7.9) for resonance Raman spectroscopy were prepared by deuterating the metal-free protein with D2O (99.9%D, Cambridge Isotope Laboratories) by repeatedly diluting and re-concentrating in this buffer using Amicon Ultra-15 concentrators (Millipore) before reconstituting the diiron-oxygen cluster as explained above. The diferrous R2 samples were prepared under anaerobic conditions in air-tight vessels by several rounds of vacuum and argon exchange using the Schlenk technique. 200 µl of apo EBV R2 (150 µM) was reduced with 2 µL of 10 mM dithionite 2 µL and 5 mM methyl viologen (reductant mediator). 5 µl of the ferrous solution (42 mM) was added to fully reconstitute the dimetal site followed by incubation for 10 min at 273 K. The samples were finally transferred to anaerobic EPR tubes. Samples of EBV R2 for measurement of the mixed valence signal were prepared as described [43] both anaerobically and aerobically. 1–2 mM of dithionite was used as a reductant and 2 mM of phenazine methosulfate was used as a reductant mediator. Light absorption spectra of purified histidine-tagged EBV R2 were measured on a Hewlett-Packard 8452 diode array spectrophotometer in the wavelength range of 250–700 nm using a thermostated water bath. Experiments under anaerobic conditions were carried out in cuvettes capped with a rubber septum, which had been deaerated with argon for ≥2 hrs. Addition of deaerated solutions of EBV R2 and radical scavengers were made through a septum with gas tight syringes purged with deoxygenated water.
EPR experiments
EPR spectra were recorded at X-band on a Bruker Elexsys 560 EPR spectrometer fitted with a Bruker ER41116DM dual-mode cavity. All EPR samples contained 20% (v/v) glycerol for vitrification during the low-temperature recordings. EPR signals were measured at different microwave powers to prevent microwave power saturation and quantified by comparing double integrals of spectra with a standard of 1 mM Cu2+ EDTA in a solution of 50 mM HEPES, pH 7.5, 20% (v/v) glycerol. All spectra were measured under identical non-saturating microwave power. First-derivative EPR spectra were recorded at different microwave power (P) and at various temperatures (see graphs) to determine the microwave power at half saturation (P1/2) for each of the temperatures. The data were fit with the function S/√P = 1/1+(P/P1/2))b /2, where S denotes the double integrated signal intensity of the EPR signal, and b is a component relating to the type of relaxation. The b factor is equal to 1 for a completely inhomogenous relaxation and 3 for an entirely homogenous relaxation. The simulated EPR spectra (at X-band and at 285 GHz) were computed with the program SIM written by Weihe in order to extract numerical values of spin Hamiltonian parameters from experimental EPR spectra [62], [63].
High-field EPR measurements
The low temperature 285 GHz spectra were obtained with a 95 GHz Gunn oscillator (Radiometer Physics, Germany) coupled to a frequency tripler as the frequency source and a superconducting magnet with a maximum field of 12 T at 4.2 K (Cryogenics Consultant, UK) for the main magnetic field. The detection of light transmitted through the sample was performed with a ‘hot electron’ InSb bolometer as described [32], [34].
Resonance Raman spectroscopy
A three stage Laser system was employed as a light source for the Raman spectroscopy: a Spectra-Physics Millennia Pro 12sJS Nd∶YV04 solid state laser (6.5 W at 532 nm) pumped a Sirah Matisse TR Ti∶Sa ring Laser that produced a power of 1 W at 820 nm. This was finally doubled to 410 nm using a Spectra-Physics Wavetrain frequency doubler (550–990 nm) yielding 20 mW of laser light having a line width <4 MHz. The applied power of the 410 nm laser at the sample was ∼5 mW. For the Raman measurements, a Jobyn Yvon Horiba T64000 instrument equipped with a 410 nm Kaiser Optical holographic Super-Notch filter served as a single spectrograph in order to minimize the reduction of Raman light. The narrow bandwidth and the small shifts of the Raman peaks investigated required an entrance slit width of 100 µm and resolution grating of 3600 grooves/mm. 60 scans of 60 seconds were averaged for each Raman spectrum. Data reproducibility was evaluated by triple measurements on protein samples from different preparations. The samples with a volume of ca. 50 µl were kept in EPR tubes cooled with liquid N2 in a quartz cold finger EPR cryostat. The fluorescence that inevitably occurred was subtracted by fitting with a polynomial function, and the frequency scale was calibrated using 4-acetamidophenol. The accuracy for the frequency of the Raman peaks was then determined within an error range of ±1.1 cm−1 under these settings. The reference peak values were obtained from published tables [64].
Computational procedures
The theoretical modeling of the tyrosyl radical was carried out by density functional theory (DFT) in gas phase using a simplified system with the p-ethylphenoxyl radical (neutral form) using unrestricted B3LYP function (Exchange: 0.2000 Hartree-Fock, 0.0800 Slater and 0.7200 Becke, and correlation 0.8100 LYP and 0.1900 VWN1RPA) with the Euler-Maclaurin-Lebedev (70,302) grid and basis set 6-31G(d,p) as implemented in the computational package Spartan 08/10. The molecular structures in the presence and absence of an H-bonded water molecule were fully optimized (root mean square gradient below 10−7) with and without C6-C1-Cα-Cβ torsional angle (constraint θ) followed by frequency calculation in order to derive vibrational frequencies and Raman scattering activities (unscaled). More accurate spin-densities used for the estimation of the hyperfine AH tensor components in EBV R2 have been derived by using the entire tyrosyl radical without and with a water molecule located at 2.60 Å from the tyrosyl oxygen by UB3LYP/6-311++G(d,p) complemented by Raman frequency calculations at the same theory level. Details are provided in the Information S1 and methods section.
UV/VIS light spectrophotometric assays
Light absorption spectra of purified histidine-tagged EBV R2 were measured on a Hewlett-Packard 8452 diode array spectrophotometer in the wavelength range of 250–700 nm using a thermostated water bath. Experiments under anaerobic conditions were carried out in cuvettes capped with a rubber septum, which had been deaerated with argon for ≥2 hrs. Addition of deaerated solutions of EBV R2 and radical scavengers were made through a septum with gas tight syringes purged with deoxygenated water.
Supporting Information
Theoretical DFT Analyses.
(DOC)
Acknowledgments
Professor Jürgen Haas, Max-von-Pettenkofer Institute, Ludwig-Maximilians University, Munich is thanked for the BaRF1 clone encoding EBV R2. Daniel Gurmu, Sue-Li Dahlroth and Edwin Tjandra, and Renaud Vincentelli for help with protein expression and protein purification protocols respectively.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: GZ thanks the Marie Curie Intra–European Fellowship within the 7th European Community Framework Programme (PIEF–GA–2009–235237) for financial support, KKA the Research Council of Norway (Grant 177661/V30) and The Cancer Foundation of Norway, PN the Swedish Cancer Foundation and the Swedish Research Council, FS a Karolinska Institute Foundation grant and a Howard Hughes Medical Institute fellowship. The Raman instrument and laser equipment was funded by a grant from the Research Council of Norway. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Reichard P. From RNA to DNA, Why So Many Ribonucleotide Reductases. Science. 1993;260:1773–1777. doi: 10.1126/science.8511586. [DOI] [PubMed] [Google Scholar]
- 2.Andersson KK, editor. (2008) Ribonucleotide reductase. Hauppauge NY, USA: Nova Science publishers, Inc; 2008. [Google Scholar]
- 3.Andersson KK, Graslund A. Diiron-oxygen proteins. Adv Inorg Chem. 1995;43:359–408. [Google Scholar]
- 4.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 5.Fairman JW, Wijerathna SR, Ahmad MF, Xu H, Nakano R, et al. Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization. Nat Struct Mol Biol. 2011;18:316–322. doi: 10.1038/nsmb.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kashlan OB, Scott CP, Lear JD, Cooperman BS. A comprehensive model for the allosteric regulation of mammalian ribonucleotide reductase. Functional consequences of ATP- and dATP-induced oligomerization of the large subunit. Biochemistry. 2002;41:462–474. doi: 10.1021/bi011653a. [DOI] [PubMed] [Google Scholar]
- 7.Rofougaran R, Vodnala M, Hofer A. Enzymatically active mammalian ribonucleotide reductase exists primarily as an alpha(6)beta(2) octamer. J Biol Chem. 2006;281:27705–27711. doi: 10.1074/jbc.M605573200. [DOI] [PubMed] [Google Scholar]
- 8.Uhlin U, Eklund H. Structure of Ribonucleotide Reductase Protein R1. Nature. 1994;370:533–539. doi: 10.1038/370533a0. [DOI] [PubMed] [Google Scholar]
- 9.Kolberg M, Strand KR, Graff P, Andersson KK. Structure, function, and mechanism of ribonucleotide reductases. Biochim Biophys Acta. 2004;1699:1–34. doi: 10.1016/j.bbapap.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 10.Conner J, Marsden H, Clements JB. Ribonucleotide Reductase of Herpesviruses. Reviews in Medical Virology. 1994;4:25–34. [Google Scholar]
- 11.Ator MA, Stubbe J, Spector T. Mechanism of Ribonucleotide Reductase from Herpes-Simplex Virus Type-1 - Evidence for 3′ Carbon-Hydrogen Bond-Cleavage and Inactivation by Nucleotide Analogs. J Biol Chem. 1986;261:3595–3599. [PubMed] [Google Scholar]
- 12.Potsch S, Sahlin M, Langelier Y, Graslund A, Lassmann G. Reduction of the Tyrosyl Radical and the Iron Center in Protein R2 of Ribonucleotide Reductase from Mouse, Herpes-Simplex Virus and Escherichia-Coli by P-Alkoxyphenols. FEBS Letters. 1995;374:95–99. doi: 10.1016/0014-5793(95)01082-p. [DOI] [PubMed] [Google Scholar]
- 13.Thorley-Lawson DA. Epstein-Barr virus: exploiting the immune system. Nature Reviews Immunology. 2001;1:75–82. doi: 10.1038/35095584. [DOI] [PubMed] [Google Scholar]
- 14.Chodosh J, Bolder VP, Gan YJ, Belgaumi A, Sample J, et al. Eradication of latent Epstein-Barr virus by hydroxyurea alters the growth-transformed cell phenotype. J Infect Dis. 1998;177:1194–1201. doi: 10.1086/515290. [DOI] [PubMed] [Google Scholar]
- 15.Wakisaka N, Yoshizaki T, Raab-Traub N, Pagano JS. Ribonucleotide reductase inhibitors enhance cidofovir-induced apoptosis in EBV-positive nasopharyngeal carcinoma xenografts. Int J Cancer. 2005;116:640–645. doi: 10.1002/ijc.21096. [DOI] [PubMed] [Google Scholar]
- 16.Jiang R, Zhang JL, Satoh Y, Sairenji T. Mechanism for induction of hydroxyurea resistance and loss of latent EBV genome in hydroxyurea-treated Burkitt's lymphoma cell line Raji. J Med Virol. 2004;73:589–595. doi: 10.1002/jmv.20130. [DOI] [PubMed] [Google Scholar]
- 17.Wnuk SF, Robins MJ. Ribonucleotide reductase inhibitors as anti-herpes agents. Antiviral Research. 2006;71:122–126. doi: 10.1016/j.antiviral.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Himo F, Graslund A, Eriksson LA. Density functional calculations on model tyrosyl radicals. Biophys J. 1997;72:1556–1567. doi: 10.1016/S0006-3495(97)78803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jordan A, Pontis E, Atta M, Krook M, Gibert I, et al. A 2nd Class-I Ribonucleotide Reductase in Enterobacteriaceae - Characterization of the Salmonella-Typhimurium Enzyme. Proc Natl Acad Sci USA. 1994;91:12892–12896. doi: 10.1073/pnas.91.26.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svistunenko DA, Cooper CE. A new method of identifying the site of tyrosyl radicals in proteins. Biophys J. 2004;87:582–595. doi: 10.1529/biophysj.104.041046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galli C, Atta M, Andersson KK, Graslund A, Brudvig GW. Variations of the Diferric Exchange Coupling in the R2-Subunit of Ribonucleotide Reductase from 4 Species as Determined by Saturation - Recovery Epr Spectroscopy. J Am Chem Soc. 1995;117:740–746. [Google Scholar]
- 22.Un S, Atta M, Fontecave M, Rutherford AW. G-Values as a Probe of the Local Protein Environment - High-Field Epr of Tyrosyl Radicals in Ribonucleotide Reductase and Photosystem-Ii. J Am Chem Soc. 1995;117:10713–10719. [Google Scholar]
- 23.Andersson KK, Schmidt PP, Katterle B, Strand KR, Palmer AE, et al. Examples of high-frequency EPR studies in bioinorganic chemistry. J Biol Inorg Chem. 2003;8:235–247. doi: 10.1007/s00775-002-0429-0. [DOI] [PubMed] [Google Scholar]
- 24.Andersson KK, Barra AL. The use of high field/frequency EPR in studies of radical and metal sites in proteins and small inorganic models. Spectrochimica Acta Part a-Molecular and Biomolecular Spectroscopy. 2002;58:1101–1112. doi: 10.1016/s1386-1425(01)00700-4. [DOI] [PubMed] [Google Scholar]
- 25.Wei PP, Tomter AB, Rohr AK, Andersson KK, Solomon EI. Circular dichroism and magnetic circular dichroism studies of the active site of p53R2 from human and mouse: Iron binding and nature of the biferrous site relative to other ribonucleotide reductases. Biochemistry. 2006;45:14043–14051. doi: 10.1021/bi061127p. [DOI] [PubMed] [Google Scholar]
- 26.Smith P, Zhou BS, Ho N, Yuan YC, Su L, et al. 2.6 angstrom X-ray Crystal Structure of Human p53R2, a p53-Inducible Ribonucleotide Reductase. Biochemistry. 2009;48:11134–11141. doi: 10.1021/bi9001425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Dam PJ, Willems JP, Schmidt PP, Potsch S, Barra AL, et al. High-frequency EPR and pulsed Q-Band ENDOR studies on the origin of the hydrogen bond in tyrosyl radicals of ribonucleotide reductase R2 proteins from mouse and herpes simplex virus type 1. J Am Chem Soc. 1998;120:5080–5085. [Google Scholar]
- 28.Atta M, Lamarche N, Battioni JP, Massie B, Langelier Y, et al. Escherichia-Coli and Herpes-Simplex-Virus Ribonucleotide Reductase R2 Subunit - Compared Reactivities of the Redox Centers. Biochem J. 1993;290:807–810. doi: 10.1042/bj2900807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamarche N, Matton G, Massie B, Fontecave M, Atta M, et al. Production of the R2 subunit of ribonucleotide reductase from herpes simplex virus with prokaryotic and eukaryotic expression systems: Higher activity of R2 produced by eukaryotic cells related to higher iron-binding capacity. Biochem J. 1996;320:129–135. doi: 10.1042/bj3200129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mann GJ, Graslund A, Ochiai EI, Ingemarson R, Thelander L. Purification and Characterization of Recombinant Mouse and Herpes-Simplex Virus Ribonucleotide Reductase R2 Subunit. Biochemistry. 1991;30:1939–1947. doi: 10.1021/bi00221a030. [DOI] [PubMed] [Google Scholar]
- 31.Gerfen GJ, Bellew BF, Un S, Bollinger JM, Stubbe J, et al. High-Frequency (139.5 GHz) EPR Spectroscopy of the Tyrosyl Radical in Escherichia-Coli Ribonucleotide Reductase. J Am Chem Soc. 1993;115:6420–6421. [Google Scholar]
- 32.Barra AL, Graslund A, Andersson KK. The use of very high frequency EPR (VHF-EPR) in studies in proteins and small inorganic models. Biol Magn Res. 2004;45:145–164. [Google Scholar]
- 33.Schmidt PP, Andersson KK, Barra AL, Thelander L, Graslund A. High field EPR studies of mouse ribonucleotide reductase indicate hydrogen bonding of the tyrosyl radical. J Biol Chem. 1996;271:23615–23618. doi: 10.1074/jbc.271.39.23615. [DOI] [PubMed] [Google Scholar]
- 34.Allard P, Barra AL, Andersson KK, Schmidt PP, Atta M, et al. Characterization of a new tyrosyl free radical in Salmonella typhimurium ribonucleotide reductase with EPR at 9.45 and 245 GHz. J Am Chem Soc. 1996;118:895–896. [Google Scholar]
- 35.Svistunenko DA, Jones GA. Tyrosyl radicals in proteins: a comparison of empirical and density functional calculated EPR parameters. Phys Chem Chem Phys. 2009;11:6600–6613. doi: 10.1039/b905522c. [DOI] [PubMed] [Google Scholar]
- 36.Un S. The g-values and hyperfine coupling of amino acid radicals in proteins: comparison of experimental measurements with ab initio calculations. Magn Reson Chem. 2005;43:S229–S236. doi: 10.1002/mrc.1660. [DOI] [PubMed] [Google Scholar]
- 37.Strand KR, Karlsen S, Kolberg M, Rohr AK, Gorbitz CH, et al. Crystal structural studies of changes in the native dinuclear iron center of ribonucleotide reductase protein R2 from mouse. J Biol Chem. 2004;279:46794–46801. doi: 10.1074/jbc.M407346200. [DOI] [PubMed] [Google Scholar]
- 38.Katterle B, Sahlin M, Schmidt PP, Potsch S, Logan DT, et al. Kinetics of transient radicals in Escherichia coli ribonucleotide reductase - Formation of a new tyrosyl radical in mutant protein R2. J Biol Chem. 1997;272:10414–10421. doi: 10.1074/jbc.272.16.10414. [DOI] [PubMed] [Google Scholar]
- 39.Liu X, Xue L, Yen Y. Redox property of ribonucleotide reductase small subunit M2 and p53R2. Methods in Molecular Biology. 2008;477:195–206. doi: 10.1007/978-1-60327-517-0_15. [DOI] [PubMed] [Google Scholar]
- 40.Un S, Gerez C, Elleingand E, Fontecave M. Sensitivity of tyrosyl radical g-values to changes in protein structure: A high-field EPR study of mutants of ribonucleotide reductase. J Am Chem Soc. 2001;123:3048–3054. doi: 10.1021/ja003650b. [DOI] [PubMed] [Google Scholar]
- 41.Campbell KA, Peloquin JM, Diner BA, Tang XS, Chisholm DA, et al. The tau-nitrogen of D2 histidine 189 is the hydrogen bond donor to the tyrosine radical Y-D(center dot) of photosystem II. J Am Chem Soc. 1997;119:4787–4788. [Google Scholar]
- 42.Sahlin M, Petersson L, Graslund A, Ehrenberg A, Sjoberg BM, et al. Magnetic Interaction between the Tyrosyl Free-Radical and the Antiferromagnetically Coupled Iron Center in Ribonucleotide Reductase. Biochemistry. 1987;26:5541–5548. doi: 10.1021/bi00391a049. [DOI] [PubMed] [Google Scholar]
- 43.Atta M, Andersson KK, Ingemarson R, Thelander L, Graslund A. Epr Studies of Mixed-Valent [(Fefeiii)-Fe-Ii] Clusters Formed in the R2 Subunit of Ribonucleotide Reductase from Mouse or Herpes-Simplex Virus - Mild Chemical-Reduction of the Diferric Centers. J Am Chem Soc. 1994;116:6429–6430. [Google Scholar]
- 44.Atta M, Debaecker N, Andersson KK, Latour JM, Thelander L, et al. EPR and multi-field magnetisation of reduced forms of the binuclear iron centre in ribonucleotide reductase from mouse. J Biol Inorg Chem. 1996;1:210–220. [Google Scholar]
- 45.Strand KR, Yang YS, Andersson KK, Solomon EI. Circular dichroism and magnetic circular dichroism studies of the biferrous form of the R2 subunit of ribonucleotide reductase from mouse: Comparison to the R2 from Escherichia coli and other binuclear ferrous enzymes. Biochemistry. 2003;42:12223–12234. doi: 10.1021/bi035248q. [DOI] [PubMed] [Google Scholar]
- 46.Offenbacher AR, Vassiliev IR, Seyedsayamdost MR, Stubbe J, Barry BA. Redox-Linked Structural Changes in Ribonucleotide Reductase. J Am Chem Soc. 2009;131:7496–7497s. doi: 10.1021/ja901908j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanson MA, Schmidt PP, Strand KR, Graslund A, Solomon EI, et al. Resonance Raman evidence for a hydrogen-bonded oxo bridge in the R2 protein of ribonucleotide reductase from mouse. J Am Chem Soc. 1999;121:6755–6756. [Google Scholar]
- 48.Johnson CR, Ludwig M, Asher SA. Ultraviolet Resonance Raman Characterization of Photochemical Transients of Phenol, Tyrosine, and Tryptophan. J Am Chem Soc. 1986;108:905–912. [Google Scholar]
- 49.Range K, Ayala I, York D, Barry BA. Normal modes of redox-active tyrosine: Conformation dependence and comparison to experiment. J Phys Chem B. 2006;110:10970–10981. doi: 10.1021/jp061503f. [DOI] [PubMed] [Google Scholar]
- 50.Ivancich A, Mattioli TA, Un S. Effect of protein microenvironment on tyrosyl radicals. A high-field (285 GHz) EPR, resonance raman, and hybrid density functional study. J Am Chem Soc. 1999;121:5743–5753. [Google Scholar]
- 51.Brynda M, Britt RD. Density Functional Theory calculations on the magnetic properties of the model tyrosine radical-histidine complex mimicking tyrosyl radical YD· in photosystem II Research on Chemical. Intermediates. 2007;33:863–883. [Google Scholar]
- 52.Engstrom M, Himo F, Graslund A, Minaev B, Vahtras O, et al. Hydrogen bonding to tyrosyl radical analyzed by ab initio g-tensor calculations. Journal of Physical Chemistry A. 2000;104:5149–5153. [Google Scholar]
- 53.Shao J, Zhou B, Chu B, Yen Y. Ribonucleotide reductase inhibitors and future drug design. Curr Cancer Drug Targets. 2006;6:409–431. doi: 10.2174/156800906777723949. [DOI] [PubMed] [Google Scholar]
- 54.Mann GJ, Graslund A, Ochiai E, Ingemarson R, Thelander L. Purification and characterization of recombinant mouse and herpes simplex virus ribonucleotide reductase R2 subunit. Biochemistry. 1991;30:1939–1947. doi: 10.1021/bi00221a030. [DOI] [PubMed] [Google Scholar]
- 55.Potsch S, Sahlin M, Langelier Y, Graslund A, Lassmann G. Reduction of the tyrosyl radical and the iron center in protein R2 of ribonucleotide reductase from mouse, herpes simplex virus and E. coli by p-alkoxyphenols. FEBS Lett. 1995;374:95–99. doi: 10.1016/0014-5793(95)01082-p. [DOI] [PubMed] [Google Scholar]
- 56.Nyholm S, Thelander L, Graslund A. Reduction and Loss of the Iron Center in the Reaction of the Small-Subunit of Mouse Ribonucleotide Reductase with Hydroxyurea. Biochemistry. 1993;32:11569–11574. doi: 10.1021/bi00094a013. [DOI] [PubMed] [Google Scholar]
- 57.Atkin CL, Thelander L, Reichard P, Lang G. Iron and free radical in ribonucleotide reductase. Exchange of iron and Mossbauer spectroscopy of the protein B2 subunit of the Escherichia coli enzyme. J Biol Chem. 1973;248:7464–7472. [PubMed] [Google Scholar]
- 58.Nyholm S, Mann GJ, Johansson AG, Bergeron RJ, Graslund A, et al. Role of ribonucleotide reductase in inhibition of mammalian cell growth by potent iron chelators. J Biol Chem. 1993;268:26200–26205. [PubMed] [Google Scholar]
- 59.Cornvik T, Dahlroth SL, Magnusdottir A, Herman MD, Knaust R, et al. Colony filtration blot: a new screening method for soluble protein expression in Escherichia coli. Nature Methods. 2005;2:507–509. doi: 10.1038/nmeth767. [DOI] [PubMed] [Google Scholar]
- 60.Dahlroth SL, Lieu V, Haas J, Nordlund P. Screening Colonies of Pooled ORFeomes (SCOOP): A rapid and efficient strategy for expression screening ORFeomes in Escherichia coli. Protein Expres Purif. 2009;68:121–127. doi: 10.1016/j.pep.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 61.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to Measure and Predict the Molar Absorption-Coefficient of a Protein. Protein Science. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glerup J, Weihe H. Magnetic-Susceptibility and Epr-Spectra of Mu-Cyano-Bis[Pentaamminechromium(Iii)] Perchlorate. Acta Chem Scand. 1991;45:444–448. doi: 10.1021/ic970029c. [DOI] [PubMed] [Google Scholar]
- 63.Weihe H. SIM software. Institute of Chemistry, University of Copenhagen, Denmark.
- 64.McCreery RL, editor. (2000) Raman Spectroscopy for Chemical Analysis. New York: Wiley; 2000. [Google Scholar]
- 65.Hoganson CW, Sahlin M, Sjoberg BM, Babcock GT. Electron magnetic resonance of the tyrosyl radical in ribonucleotide reductase from Escherichia coli. J Am Chem Soc. 1996;118:4672–4679. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Theoretical DFT Analyses.
(DOC)