Abstract
Time-of-flight Secondary ion mass spectrometry (ToF-SIMS) provides a method for the detection of native and exogenous compounds in biological samples on a cellular scale. Through the development of novel ion beams the amount of molecular signal available from the sample surface has been increased. Through the introduction of polyatomic ion beams, particularly C60, ToF-SIMS can now be used to monitor molecular signals as a function of depth as the sample is eroded thus proving the ability to generate 3D molecular images. Here we describe how this new capability has led to the development of novel instrumentation for 3D molecular imaging while also highlighting the importance of sample preparation and discuss the challenges that still need to be overcome to maximise the impact of the technique.
Introduction
No analytical technique is perfect, however amongst the imaging mass spectrometries the potential of time-of-flight secondary ion mass spectrometry (ToF-SIMS) to provide label free imaging of molecular species on a sub-cellular scale is extremely exciting and provides a myriad of possibilities for studying cellular metabolomics, disease progression and prevention/treatment due to its high lateral resolution, the chemical specificity of mass spectrometry and high sensitivity to label free bio-molecules.1,2 Energetic (usually 10s of keV) primary ions can be focused to sub-micron spots and used to interrogate the surface of almost any sample ejecting species up to 1 or 2 kDa in ideal cases. Using a ToF mass spectrometer allows all the ejected secondary ions to be detected in parallel and a full mass spectrum is recorded at each pixel potentially allowing the technique to be used in a ‘discovery’ modality. Generally labelling is not employed although incorporation of mass spectrometric tags including the use of isotopically enriched compounds can provided added information and has been used to great effect for cellular imaging using dynamic SIMS particularly by Lechene and co-workers.3 The data from the ToF-SIMS image is examined retrospectively; mass spectra can be extracted from regions of interest in the image or the distribution of ion signals in the mass spectrum can be visualised. Here we describe how advances in ion beam and mass spectrometer development are helping us to realise the potential for ToF-SIMS analysis of biological cells.
SIMS has been used extensively to examine a wide range of samples and is used routinely in the micro-electronics industry for probing the distribution of dopants in silicon. Benninghoven showed in the early 1970s that the technique could also be used to provide molecular type information of the chemistry of a sample surface.4 Surface scientists utilised the technique in the study of heterogeneous catalysis on both industrial samples, such as automobile catalytic converters5, and also environmental in the study of ozone depletion reactions6. Obviously the complexity of the ToF-SIMS analyses, and the associated data, are very much greater for biological samples compared to traditional targets for SIMS analysis such as semi-conductors and polymers. To meet the demands for analysing such challenging samples considerable effort has been made to develop novel ion beams with which to probe the sample surface. The goal of such endeavours has been to improve the sensitivity of the technique thus enabling higher resolution chemical imaging while particularly increasing the yield of higher mass, more chemically characteristic secondary ions which aid the interpretation of the complex mass spectrum. The simplest way of increasing the secondary ion yield/primary ion is to increase the mass of the incoming projectile; bigger atom – bigger bang!
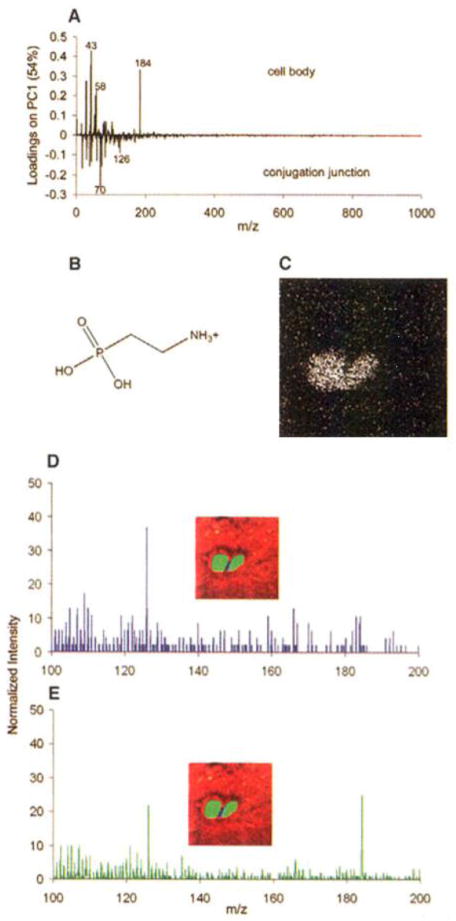
An excellent example is provided by Ostrowski et al. who have demonstrated the use of ToF-SIMS to image the changes in lipid composition during Tetrahymena mating using an In+ primary ion beam.7 During mating the Tetrahymena (common fresh water protozoa) form a series of highly curved fusion pores at the conjugation site that allow the passage of micronuclei between the cells. Such radical changes in the shape/structure of the membrane would be expected to involve a change in the lipid composition in that area. SIMS imaging of the tetrahymena was performed following freeze fracture of the cells and inspection of the conjugation site which as hypothesised, showed a change in the lipid composition. A reduction in the signal from phosphocholine (m/z 184) was observed. Principal components analysis was performed on the image and a species was identified at m/z 126 that was associated with the conjugation junction. The peak at m/z 126 was assigned to the headgroup of a group of cone-shaped non-lamellar 2-aminoethylphosphonolipids (2-AEP). The localisation of such lipids at the conjugation site is consistent with the formation of the high curvature structures (Figure 1).
Figure 1.
The conjugation junction contains elevated amounts of 2-AEP. Mass spectra from pixels were generated by selecting the pixels of interest using software written in house. (A) Loadings plot from principal components analysis comparing the mass spectra of the cell bodies and the conjugation junction. (B) 2-AEP headgroup fragment corresponding to m/z 126. (C) SIMS image for m/z 126. (D) Mass spectrum from the pixels along the conjugation junction, the blue pixels where the cells touch in the inset. (E) Mass spectrum from the pixels in the cell bodies, as illustrated by the green pixels from the cells in the inset. Adapted with permission from reference 7.
Cluster and Polyatomic Ion Beams
Even more beneficial than increasing the mass of the primary ion is the so-called cluster effect. Cluster ion guns have been introduced that fire Aun and Bin (n is usually between 1 and 5)8,9 and take advantage of an effect first observed in the 1970’s - there is a non-linear increase in the secondary ion yield as a function of particle nuclearity.10,11,12 Of particular benefit for the imaging of biological samples the increase in secondary ion yield is especially pronounced for the molecular type ions m/z >200. Concurrently a number of groups were exploring the use of polyatomic primary ions with the same aims. Initial studies using small carbon clusters13 and SF5/614,15 were superseded by the introduction of C60 for routine SIMS analysis16. The polyatomic ion beams provided not only the non-linear increase with nuclearity, but also provided a reduction in the accumulation of ion beam induced chemical damage in the sample.
Conventional mono-atomic and even the metal ‘cluster’ ion beams deposit energy deep into the sample below the point of impact that leads to loss of molecular information. Hence the extraction of molecular information from a sample is limited to low primary ion beam dose, normally such that only 1% of the sample surface is impacted by a primary ion17. This dose commonly referred to as the ‘static limit’ hence the term static-SIMS is used to distinguish this modality from dynamic-SIMS where high ion beam flux is used to erode a sample while monitoring the secondary ion signals of atomic and/or small fragment species, predominantly in the semi-conductor industry.
The impact polyatomic ion beams have had on organic material analysis by SIMS has been so great that the properties of these ion beams have even been described as ‘magical’ in the scientific literature!18 However any mysticism has been removed, and the low damage accumulation has been explained, through analytical models19,20 and molecular dynamics simulations of the ion/surface interaction.21 As the polyatomic ion impacts the surface of the sample it disintegrates into its constituent atoms resulting in the simultaneous impact of many relatively low energy atoms. The result is that the energy is deposited in the upper layers of the sample with very little disruption of the sub-surface. If a second ion hits the same spot intact molecular species are still detected
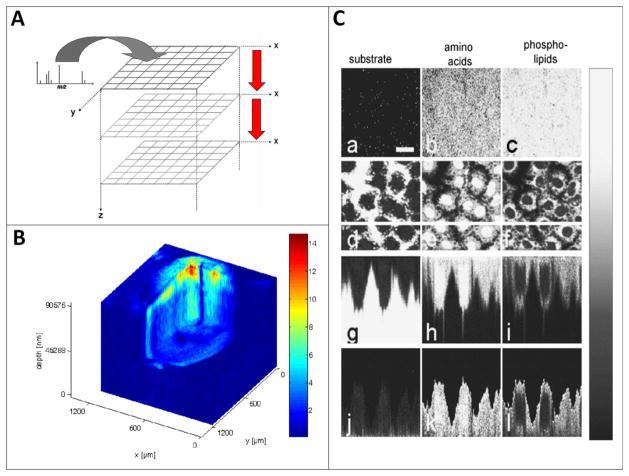
A reduction in chemical damage accumulation during the SIMS analysis has extremely significant benefits for biological analysis. Imaging can now be performed using much higher ion beam dose, thus increasing the signal in each pixel by sampling a small volume of material (a voxel) leading to improved image contrast. A further more exciting option is the generation of 3 dimensional molecular images by acquiring and reconstructing a stack of 2D images as the specimen is eroded by the primary ion beam as illustrated in Figure 2A.
Figure 2.
1st examples of 3D biological imaging using ToF-SIMS. A. A stack of images is acquired using interleaved analyse/etch cycles. B shows the distribution of a triacylglyceride species over the mass range m/z 815 960 imaged during the erosion of approximately 100 μm of material from a Xenopus laevis oocyte using C60+.
Using a liquid metal ion beam for analysis combined with C60+ for etching sub-cellular 3D structure can be observed in panel C where mass-resolved secondary ion images of the surface of normal rat kidney cells before any sputter cycle had been applied (a–c) and within the cell after the 45th sputter cycle (d–f) images are based on the following secondary ions: (a), (d), (g) and (j): Na+; (b), (e), (h) and (k): amino acid fragment ions (pooled signal for m/z 30, 44, 70, 101, 110, 136); (c), (f), (i) and (l): phospholipid fragment ions (pooled signal for m/z 58, 166, 184, 760). (g), (h) and (i) are cross-sectional images in the plane of the white horizontal marker line in (d), (e) and (f) while (j), (k) and (l) are the same data but with the interface with the silicon wafer substrate set to be flat and truncated at that interface in order to display the cells with a more realistic shape. The scale bar in (a) corresponds to 20 μm. Adapted with permission from references 23 and 24.
3D molecular imaging, following the introduction of polyatomic ion beams began with a number of exploratory studies on drug/polymer blends22 and simple cellular systems. Fletcher et al. demonstrated the initial application of ToF SIMS using 40 keV C60+ ions to sputter etch and image Xenopus laevis oocyte, thus characterizing chemical changes in 3 dimensions of a biological cell with minimal sample preparation/intervention.23 X. laevis oocyte is a well-established cell model that has been extensively used in many branches of experimental biology and pharmacological research. It is a large single cell about 0.8–1.3 mm in diameter, the cellular compartments of which are so large that they are accessible to cell biologists for manipulation and visualization. In addition, X. laevis oocyte is remarkably resistant to osmotic changes. This facilitated the preparation of the cells for mass spectral imaging as they are reasonably resistant to washing of exogenous substances using de-ionized water. Although the handling of the sample was simplified due to the size of the specimen the variation in topography over the analysis area resulted in reduced mass resolution and meant that over the course of the analysis only the outer section of the oocyte could be consumed. Despite this the sample provided clear proof of principle for molecular depth profiling of biological cells using C60+. Persistent secondary ion signals were observed through the analysis, where over 75 μm of material was eroded, for a range of species including cholesterol and diacylglycerides associated with previously identified, by extraction and MS analysis, lipid species known to be abundant in X. laevis oocyte membranes (Fig. 2B).
The C60+ beam, arising from a gas phase electron impact source, is more difficult to finely focus than an LMIG while maintaining enough current to enable analysis on a useful time scale. Sometimes a useful compromise is to use a liquid metal ion gun (LMIG) for the imaging cycles of the experiment and a polyatomic ion beam for the interleaved etching periods. Breitenstein et al. employed this method of analysis for the 3D ToF-SIMS imaging of normal rat kidney (NRK) cells.24 Bi3+ ions were used for the imaging while C60+ was used for the etching. In this experiment the imaging capability of the Bi3+ allows the sample to be imaged with sub-cellular resolution in 3D. Figure 2C shows the cells clearly outlined against the substrate while the combination of amino acid associated peaks also highlight the cells while the nuclear region can clearly be defined by the absence of pooled phospholipid signal.
These initial results were very encouraging but also highlighted inadequacies with the conventional ToF-SIMS instrumentation. The imaging was performed with low mass resolution, using a longer than normal primary ion pulse, with a non-imaging high mass resolution mass spectrum required for accurate peak assignment. This is because in high resolution ToF-SIMS imaging too rapid pulsing and/or bunching of the primary ion beam can prevent the realisation of the full focus of the ion beam and the low ion currents from the focused and apertured ion guns means that the experiment time becomes much longer. It is therefore desirable to increase the number of primary ions in each shot thus increasing the duty cycle of the experiment in terms of ion dose over time. This mode of operation obviously has a detrimental effect on the quality of the mass spectrum. Also the useful spatial resolution in molecular imaging SIMS is often limited by the amount of signal from a single pixel. The dual beam approach to depth profiling can help speed up the acquisition, as it is usually possible to deliver more ion beam current into a smaller spot, but each image/layer of the depth profile is then restricted to analysis in the static regime. The etching cycle not only allows 3D molecular imaging but is essential for the removal of subsurface damage caused by the LMIG.
New Instrumentation for a New SIMS Paradigm
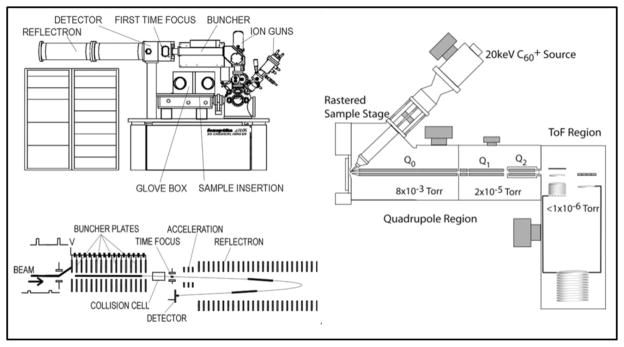
What the analyst wants is a dynamic SIMS instrument with all the advantages of the ToF analyser. Hence, we embarked on projects at Penn State and the University of Manchester to meet these demands. At Penn State a commercially available ‘Q-Star’ MALDI mass spectrometer was fitted with an adapted C60+ ion gun25. The ion beam can be used continuously while sections of the resulting secondary ion stream are pulsed orthogonally into a ToF mass analyser. Although, not without its challenges, the approach adopted at the University of Manchester was much more ambitious. A new concept of ToF-SIMS instrument, the ‘J105 - 3D Chemical Imager’ has been developed in collaboration with Ionoptika Ltd (Southampton U.K.) and uses a linear buncher placed before a quadratic field ToF mass analyser.26 The buncher squeezes a 30 cm long section of the secondary ion stream into a tight bunch of ions at the entrance to the ToF, the mass resolution of the instrument is now dependent on the quality of the bunching as opposed to the pulsing of the ion gun on a conventional instrument. Both the instruments provide a huge increase in duty cycle (x 103) facilitating 3D imaging and also large area analysis of tissue samples. 3D imaging of cells can now be performed on the J105 without the need for etching cycles where precious signal is discarded. Instead a stack of high dose images are generated, mass resolution and accuracy is also preserved in all modes of analysis as it is now de-coupled from the ion beam bombardment of the sample even on topographically challenging specimen thus improving identification of chemical species which is vital for biological analysis. The ability to perform routine 3D imaging experiments on a useful timescale has allowed systematic studies to be performed in order to investigate a number of additional challenges associated with biological analysis. Figure 3 contains schematics of the 2 DC ion beam SIMS instruments, both are also capable of performing tandem MS experiments to elucidate secondary ion structure. Tandem MS in the J105 uses a ToF-ToF configuration with the collisionally induced dissociation occurring in the collision cell between the buncher and reflectron at high energy (1–6 keV). The Q-Star performs tandem MS by low energy (10’s of eV) collisionally induced dissociation in Q2.
Figure 3.
Schematics of the J105–3D Chemical Imager (left) and the C60 equipped Q-Star (right) instruments. Both instruments can use continuous beams. The J105 bunches the secondary ion stream to a focus at the entrance to the ToF analyser while the Q-Star orthogonally pulses sections of the secondary ion stream in to a ToF analyser.
One complication for ToF-SIMS analysis of biological samples is the requirement of a high vacuum in the analysis chamber of the instrument and much work has been performed to investigate the most appropriate methods of introducing samples into the instrument while maintaining biochemical integrity for imaging. It is always desirable, and sometimes a requirement to remove excess salt from the surface of cells prior to analysis. Sjövall introduced a protocol that involved washing the samples with a volatile salt, usually ammonium formate, as this maintains osmotic pressure during the washing but evaporates away during freeze drying of the sample.27 Careful freeze drying has been shown to maintain much of the physical structure of cells analysed by ToF-SIMS, however as the majority of the compounds detected by SIMS are relatively small, and therefore highly mobile, the distribution of at least some of them would be expected to change upon drying. Flash freezing the sample in a suitable cryogenic such as liquid propane and analysing the sample in this frozen hydrated state may represent the gold standard for sample preparation for ToF-SIMS. Not only does limiting the potential for diffusion of small molecules improve image contrast, there are other potential benefits from analysing samples at low temperature that have come to light in a number of recent studies. Conlan et al.28 reported an increase in [M+H]+ signal when amino acids when analysed in a water matrix. Suppression or enhancement of a particular species in the mass spectrum based on its chemical environment (the matrix effect) is a particular challenge in all imaging mass spectrometries because it can lead to misinterpretation of the image results1 and inter alia the matrix effect has been shown to be related to the gas phase basicity of the sample components. 29 Again the excess protons may help to ameliorate this effect. Preliminary work has shown some relaxation of the matrix effect when condensing water onto a mixture of cytosine and 2,4,6-trihydroxyacetophenone (THAP) during SIMS analysis.30 Zheng et al. have also shown that when depth profiling model molecular systems, including two alternating Langmuir-Blodgett film layers and an Irganox delta-layer sample, depth resolution is improved when performing the analysis at cryogenic temperatures 31,32 An improvement in depth resolution has also been observed during the 3D imaging of frozen hydrated cells – less chemical mixing was observed at the interface between the nucleus and the cell membrane.33 To explore this effect a number of studies have examined different frozen sample preparation techniques. The low damage properties of the polyatomic beams have allowed ice condensate layers to be etched away revealing the organic sample beneath,34 while freeze fracturing a sample within the instrument can be used to remove surface ice layers and also under the right conditions, remove the upper membrane leaflet to expose internal cellular chemistry (particularly useful prior to the advent of molecular depth profiling) 35,36 Unfortunately freeze fracture and frozen hydrated analysis is often laborious with the result that most analysts still choose the easier method of freeze drying samples instead. To overcome the barrier to such analyses we have developed for the J105 – 3D Chemical Imager a simple, automated freeze fracture system that resembles as mousetrap,1 a variation of this device (the “bug trap”) has also been demonstrated for conventional ToF-SIMS instruments37. Figure 4 shows the result of the reconstruction of a ToF-SIMS analysis of freeze fractured HeLa-M cells using the mousetrap on the J105. Membrane phospholipid signal is imaged using the phosphocholine headgroup ion (m/z 184.1) in green and the nucleus is visualised by the intense adenine [M+H]+ signal (m/z 136.1) in red. The phospholipid signal has been adjusted to be slightly transparent allowing the nucleus to be seen within the cell. Several of the cells have clearly had their upper membrane layer removed during the fracture process.
Figure 4.
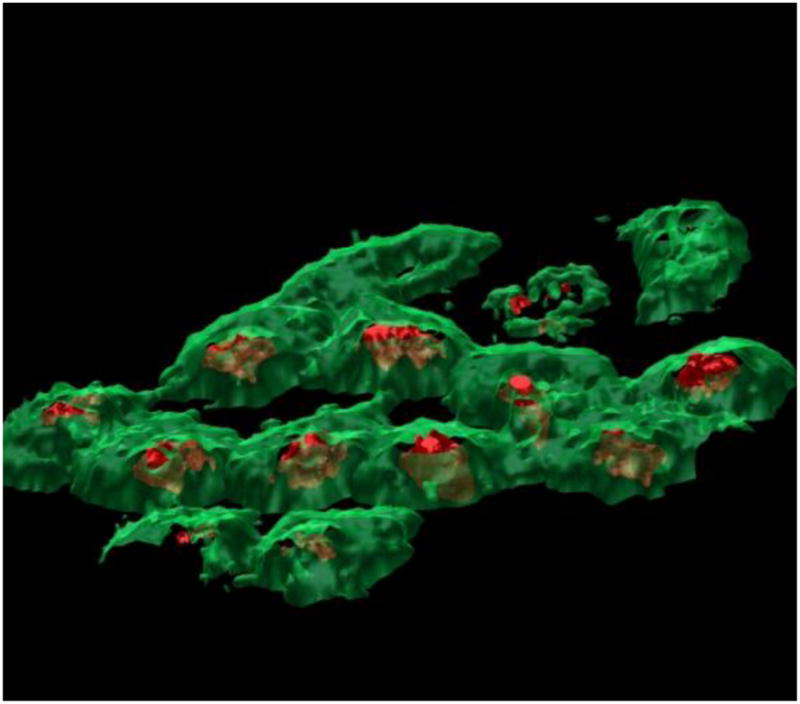
3D visualisation of the membrane (green, m/z 184.1, phosphocholine) and nucleus (red, m/z 136.1, adenine) chemistry in the frozen hydrated HeLa-M cells following the use of PCA to identify the interface between the cells and the substrate to allow more representative data reconstruction. The visualised area is 250 × 250 μm2, but inclined in order to aid 3D visualisation. Reproduced with permission from reference 33.
Conclusions and Future Direction
The imaging mass spectrometries are potentially of enormous benefit to biological research particularly cellular imaging. We have demonstrated here, and argued in a number of recent critical reviews that ToF-SIMS offers particularly powerful capabilities in this area, but there still remain a number of important challenges.38,1,2,39 Through recent forward leaps in the instrumental capabilities of ToF-SIMS 3D sub-cellular label free molecular imaging is becoming routine in a number of laboratories worldwide. Proof of concept experiments have clearly demonstrated the unique capabilities of the technique for biological analysis. The technique must now prove itself in the answering of biological questions. Cross validation with conventional, accepted approaches will no doubt be important in providing confidence in the SIMS interpretation and such experiments are under way in a number of laboratories where the imaging of fluorescent tags by ToF-SIMS has been reported.40,41 There is also a natural complementarity between SIMS and MALDI analysis, particularly for larger samples such as tissue sections and as the resolution of MALDI improves, also for (large) single cell analysis. Potentially SIMS can really provide sub-cellular 2D and 3D molecular images for which other imaging MS technique has the capability. However the accessible mass range is limited and it seems unlikely that SIMS will ever move into protein analysis. It is therefore potentially excellent for metabolites and lipids. However, the yields of ions above m/z 500 are still quite low and species of this mass are seldom detected in sufficient quantity for imaging at high resolution (as is required for cells) and this is part of the sensitivity challenge. Topography and the matrix effect are issues for all of imaging mass spectrometry and the reported results should always acknowledge the possibility that their influence may render the conclusions less certain. The matrix effect means that although MS imaging seductively seems to offer discovery mode analysis, it can never truly provide this, because we can never be sure that the absence of a signal means that the related chemistry is absent.
The push for improved signal from the SIMS experiment has not ended, the developments described in this article have improved many aspects of the technique and opened doors to new avenues of analysis but none have so far overcome the relatively low ionisation efficiencies with SIMS. Methods exist for promoting ionisation of specific chemical species by treating the sample with metals or a MALDI type matrix but these are not compatible with the new 3D imaging experiments.42,43 A method of globally increasing ionisation of secondary species is the current ‘holy grail’ in ToF-SIMS development. The presence of water (and/or ice) on28 and in the sample has shown varying enhancement for species where ion formation is through proton transfer with experiments reported where water has been squirted onto the sample during analysis44. Alternatively developments in laser post ionisation of the neutral portion of the secondary species (>99% of ejected material) have the potential to deliver huge increases in signal if the problem of photon induced fragmentation can be overcome.
Highlights.
We discuss the development of ToF-SIMS for imaging biological cells.
Novel ion beams increase the amount of information detected from a cell surface.
3 dimensional molecular images of cells can now be generated.
New instrumentation for 3D imaging is discussed.
Sample handling/preparation remains a critical step in the analysis.
Acknowledgments
The authors gratefully acknowledge the financial support of the EPSRC UK under grant EP/G045623 and the NIH USA EB-002016–17.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- *1.Fletcher JS, Lockyer NP, Vickerman JC. Developments in molecular SIMS depth profiling and 3D imaging of biological systems using polyatomic primary ions. Mass Spectrom Rev. 2011;30:142–174. doi: 10.1002/mas.20275. This comprehensive review covers fundamentals and applications of SIMS for molecular depth profiling and 3D imaging. The article includes some historical context, instrument and ion beam development with recent examples of biological cell and tissue imaging. [DOI] [PubMed] [Google Scholar]
- 2.Vickerman JC. Molecular imaging and depth profiling by mass spectrometry—SIMS, MALDI or DESI? Analyst. 2011 doi: 10.1039/c1an00008j. [DOI] [PubMed] [Google Scholar]
- *3.Lechene C, Hillion F, McMahon G, Benson D, Kleinfeld AM, Kampf JP, Distel D, Luyten Y, Bonventre J, Hentschel D, Park KM, Ito S, Schwartz M, Benichou G, Slodzian G. High-resolution quantitative imaging of mammalian and bacterial cells using stable isotope mass spectrometry. J Biol. 2006;5(20):20.1. doi: 10.1186/jbiol42. This article contains a number of stunning images generated using isotope imaging on a magnetic sector SIMS instrument. Although molecular information is not generated the potential for bioogical analysis, through careful experimental design, is still great. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benninghoven A. Die Analyse monomolekularer FestkörperoberflÄchenschichten mit Hilfe der SekundÄrionenemission. Zeitschrift für Physik. 1970;230(5):403–417. [Google Scholar]
- 5.Vickerman JC, Oaks A, Gamble H. Static SIMS studies of catalyst structure and activity. Surf Interface Anal. 2000;29:349–361. [Google Scholar]
- 6.Donsig HA, Herridge D, Vickerman JC. Static SIMS studies of reactions on mimics of Polar Stratospheric Clouds II: Low-temperature, low-pressure interactions of Cl2 and Cl2O with solid ice films. J Phys Chem A. 1998;102:2302–2308. [Google Scholar]
- *7.Ostrowski SG, Van Bell CT, Winograd N, Ewing AG. Mass Spectrometric Imaging of Highly Curved Membranes During Tetrahymena Mating. Science. 2004;305:71. doi: 10.1126/science.1099791. This excellent example of SIMS imaging of cellular systems combines high resolution imaging, frozen hydrated sample analysis and multivariate techniques to identify changes in the lipid composition at the fusion site of 2 mating tetrahymena. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies N, Weibel DE, Blenkinsopp P, Lockyer N, Hill R, Vickerman JC. Development and experimental application of a gold liquid metal ion source. App Surf Sci. 2003:203–204. 223. [Google Scholar]
- 9.Kollmer F. Cluster primary ion bombardment of organic materials. App Surf Sci. 2004;231–232:153–158. [Google Scholar]
- 10.Andersen HH, Bay HL. Nonlinear effects in heavy-ion sputtering. J Appl Phys. 1974;45(2):953. [Google Scholar]
- 11.Wittmaack K. Secondary-ion emission from silicon bombarded with atomic and molecular noble-gas ions. Surf Sci. 1979;90:557–563. [Google Scholar]
- 12.Johar SS, Thompson DA. Spike effects in heavy-ion sputtering of Ag, Au and Pt thin films. Surf Sci. 1979;90:319–330. [Google Scholar]
- 13.Gillen G. Microbeam Analysis. Institute of Physics Conference Series 165; Proceedings of the International Conference on Microbeam Analysis; 8–15 July 2000; London: Taylor & Francis Ltd; 2000. p. 339. [Google Scholar]
- 14.Gillen G, Roberson S. Preliminary evaluation of an SF5+ polyatomic primary ion beam for analysis of organic thin films by secondary ion mass spectrometry. Rapid Commun Mass Spectrom. 1998;12:1303–1312. doi: 10.1002/(SICI)1097-0231(19981015)12:19<1303::AID-RCM330>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 15.Kötter F, Benninghoven A. Secondary ion emission from polymer surfaces under Ar+, Xe+ and SF5+ ion bombardment. Appl Surf Sci. 1998;133:47–457. [Google Scholar]
- 16.Weibel D, Wong S, Lockyer NP, Blenkinsopp P, Hill R, Vickerman JC. A C60 primary ion beam system for ToF-SIMS: Its development and secondary ion yield characteristics. Analytical Chemistry. 2003;75:1754–1764. doi: 10.1021/ac026338o. [DOI] [PubMed] [Google Scholar]
- 17.Gilmore IS, Seah MP. Static SIMS:A study of damage using polymers. Surf Interface Anal. 1996;24(11):746–762. [Google Scholar]
- *18.Winograd N. The Magic of Cluster SIMS. Anal Chem. 2005;77:142A–149A. This highly cited article describes early work characterising the properties of polyatomic ion beams for SIMS analysis, particularly molecular depth profiling, and ties in the experimental observations with results from molecular dynamics simulations of the ion surface interaction. [Google Scholar]
- 19.Gillen G, Simons DS, Williams P. Molecular ion imaging and dynamic secondary ion mass spectrometry of organic compounds. Anal Chem. 1990;62(19):2122–2130. doi: 10.1021/ac00218a014. [DOI] [PubMed] [Google Scholar]
- 20.Cheng J, Wucher A, Winograd N. Molecular Depth Profiling with Cluster Beams. J Phys Chem B. 2006;110:8329–8336. doi: 10.1021/jp0573341. [DOI] [PubMed] [Google Scholar]
- 21.Garrison BJ, Postawa Z. Computational view of surface based organic mass spectrometry. Mass Spectrom Rev. 2008;27:289. doi: 10.1002/mas.20165. [DOI] [PubMed] [Google Scholar]
- 22.Gillen G, Fahey A, Wagner M, Mahoney C. 3D molecular imaging SIMS. Appl Surf Sci. 2006;252:6537–6541. [Google Scholar]
- *23.Fletcher JS, Lockyer NP, Vaidyanathan S, Vickerman JC. TOF-SIMS 3D biomolecular imaging of Xenopus laevis oocytes using buckminsterfullerene (C60+) primary ions. Anal Chem. 2006;79:2199–2206. doi: 10.1021/ac061370u. One of the 1st 3D bio-molecular SIMS imaging papers. A frog egg was analysed using C60 and molecular signals were monitored as a function of ion beam dose to depths approaching 100 μm. The paper highlights some of the challenges associated with topographically challenging samples and 3D image reconstruction. [DOI] [PubMed] [Google Scholar]
- *24.Breitenstein D, Rommel CE, Mollers R, Wegener J, Hagenhoff B. The chemical composition of animal cells and their intracellular compartments reconstructed from 3D mass spectrometry. Angew Chem Int Ed. 46:5332–5335. doi: 10.1002/anie.200604468. Another early example of 3D cellular imaging, this time on smaller mamallian cells, pooled signals from amino acids and phospholipids allow subcellular imaging in 3D. The authors highlight the fact that simple reconstruction of SIMS images does not give a realistic 3D image so the data must be reshaped by considering the interface between the cellular material and the sample substrate. [DOI] [PubMed] [Google Scholar]
- *25.Carado A, Passarelli MK, Kozole J, Wingate JE, Winograd N, Loboda AV. C60 SIMS with a Hybrid-Quadrupole Orthogonal time-of-flight Mass Spectrometer. Anal Chem. 2008;80:7921–7929. doi: 10.1021/ac801712s. The authors describe the coupling of a C60 ion beam system to a commercial quadrupole-ToF mass spectrometer. Increasing the duty cycle of the SIMS experiment, improving mass resolution and enabling tandem MS analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *26.Fletcher JS, Rabbani S, Henderson A, Blenkinsopp P, Thompson SP, Lockyer NP, Vickerman JC. A new dynamic in mass spectral imaging of single biological cells. Anal Chem. 2008;80:9058. doi: 10.1021/ac8015278. This paper describes the function, with initial biological imaging results from a new design of SIMS instrument using a buncher-ToF mass spectrometer. The instrument is optimised for 3D imaging of biological samples in that it includes cryogenic sample handling and is optimised for high speed, high resolution imaging. [DOI] [PubMed] [Google Scholar]
- 27.Sjövall P, Lausmaa J, Nygren H, Carlsson L, Malmberg P. Imaging of membrane lipids in single cells by imprintimaging time-of-flight secondary ion mass spectrometry. AnalChem. 2003;75:3429. doi: 10.1021/ac0207675. [DOI] [PubMed] [Google Scholar]
- 28.Conlan XA, Lockyer NP, Vickerman JC. Is proton cationization promoted by polyatomic primary ion bombardment during time-of-flight secondary ion mass spectrometry analysis of frozen aqueous solutions? Rapid Commun Mass Spectrom. 2006;20:1327–1334. doi: 10.1002/rcm.2446. [DOI] [PubMed] [Google Scholar]
- 29.Jones EA, Lockyer NP, Kordys J, Vickerman JC. Suppression and enhancement of secondary ion formation due to the chemical environment in static-secondary ion mass spectrometry. J Am Soc Mass Spectrom. 2007;18:1559–1567. doi: 10.1016/j.jasms.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 30.Fletcher JS, Henderson A, Biddulph GX, Vaidyanathan S, Lockyer NP, Vickerman JC. Uncovering new challenges in bio-analysis with ToF-SIMS. Appl Surf Sci. 2008;255:1264–1270. [Google Scholar]
- 31.Zheng LL, Wucher A, Winograd N. Appl Surf Sci. 2007;255:816. doi: 10.1016/j.apsusc.2008.05.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sjövall P, Rading D, Ray S, Yang L, Shard AG. Sample Cooling or Rotation Improves C60 Organic Depth Profiles of Multilayered Reference Samples: Results from a VAMAS Interlaboratory Study. J Phys Chem B. 2010;114:769. doi: 10.1021/jp9095216. [DOI] [PubMed] [Google Scholar]
- *33.Fletcher JS, Rabbani S, Henderson A, Lockyer NP, Vickerman JC. Three-dimensional mass spectral imaging of HeLa-M cells - sample preparation, data interpretation and visualisation. Rapid Commun Mass Spectrom. 2011;25:925–932. doi: 10.1002/rcm.4944. [DOI] [PubMed] [Google Scholar]
- 34.Piehowski PD, Kurczy ME, Willingham D, Parry† S, Heien ML, Winograd N, Ewing AG. Freeze-Etching and Vapor Matrix Deposition for ToF-SIMS Imaging of Single Cells. Langmuir. 2008;24(15):7906–7911. doi: 10.1021/la800292e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roddy TP, Cannon DM, Jr, Ostrowski SG, Winograd N, Ewing AG. Identification of cellular sections with imaging mass spectrometry following freeze fracture. Anal Chem. 2002;74:4020. doi: 10.1021/ac025574w. [DOI] [PubMed] [Google Scholar]
- 36.Cliff B, Lockyer N, Jungnickel H, Stephens G, Vickerman JC. Probing cell chemistry with time-of-flight secondary ion mass spectrometry: development and exploitation of instrumentation for studies of frozen-hydrated biological material. Rapid Commun Mass Spectrom. 2003;17:2163. doi: 10.1002/rcm.1169. [DOI] [PubMed] [Google Scholar]
- 37.Lanekoff I, Kurczy ME, Hill R, Fletcher JS, Vickerman JC, Winograd N, Sjövall P, Ewing AG. Time of Flight Mass Spectrometry Imaging of Samples Fractured In Situ with a Spring-Loaded Trap System. Anal Chem. 2010;82(15):6652–6659. doi: 10.1021/ac101243b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *38.Fletcher JS. Cellular imaging with secondary ion mass spectrometry. Analyst. 2009;134:2204–2215. doi: 10.1039/b913575h. [DOI] [PubMed] [Google Scholar]
- *39.Winograd N, Garrison BJ. Biological Cluster Mass Spectrometry. Annu Rev Phys Chem. 2010;61:305–322. doi: 10.1146/annurev.physchem.040808.090249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Breitenstein D, Rommel CE, Stolwijk J, Wegener J, Hagenhoff B. The chemical composition of animal cells reconstructed from 2D and 3D ToF-SIMS analysis. App Surf Sci. 2008;255:1249. [Google Scholar]
- 41.Brison J, Benoit DSW, Muramoto S, Robinson M, Stayton PS, Castner DG. ToF-SIMS imaging and depth profiling of HeLa cells treated with bromodeoxyuridine. Surf Interface Anal. 2011;43(1–2):354–357. doi: 10.1002/sia.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sjovall P, Lausmaa J, Nygren H, Carlsson L, Malmberg P. Imaging of Membrane Lipids in Single Cells by Imprint-Imaging Time-of-Flight Secondary Ion Mass Spectrometry. Anal Chem. 2003;75:3429. doi: 10.1021/ac0207675. [DOI] [PubMed] [Google Scholar]
- 43.Altelaar AFM, Van Minnen J, Jimenez CR, Heeren RMA, Piersma SR. Direct molecular imaging of Lymnaea stagnalis nervous tissue at subcellular spatial resolution by mass spectrometry. Anal Chem. 2005;77:735. doi: 10.1021/ac048329g. [DOI] [PubMed] [Google Scholar]
- 44.Mouhib T, Delcorte A, Poleunis C, Bertrand P. Organic Secondary Ion Mass Spectrometry: Signal Enhancement by Water Vapor Injection. J Am Soc Mass Spectrom. 2010;21:2005–2010. doi: 10.1016/j.jasms.2010.08.013. [DOI] [PubMed] [Google Scholar]