Abstract
To elucidate pathogenic molecules in keloids, microarray analysis was performed using RNAs extracted from keloid-derived fibroblasts and normal skin-derived fibroblasts from the same patient with a typical keloid. Among 11 up-regulated extracellular matrix genes, cartilage oligomeric matrix protein (COMP) was most prominently increased. Up-regulation of COMP mRNA and protein was confirmed in the keloid tissue by quantitative RT-PCR and Western blot. Using immunohistochemistry, we compared 15 keloids and 6 control normal tissues using a COMP-specific antibody and found that COMP stained positively in 10 keloids (66.7%), whereas no staining was observed in normal tissues, demonstrating the ectopic expression of COMP in keloids. Comparing keloids smaller or larger than 10 cm2, the larger keloids were significantly more intensely stained with the COMP-specific antibody. Because COMP reportedly accelerates collagen type I fibril assembly, we examined whether extracellular type I collagen deposition is altered by silencing COMP mRNA by small interfering RNA (siRNA). Immunocytochemistry showed at 96 hours after transfection with COMP siRNA that the extracellular deposition of type I collagen was decreased compared to that observed with control siRNA. Further, COMP knockdown decreased amount collagens type I to V in the medium and on the cell surfaces. Our data suggest that COMP facilitates keloid formation by accelerating collagen deposition, thus providing a new therapeutic target.
Keloids are raised skin lesions with redness, pain, and itching, often caused by trauma, burns, or surgical invasion. They grow larger beyond the size of the original wounds, causing not only esthetic but also mental distress.1 Keloid pathogenesis basically involves excessive wound healing with prolonged inflammation. Histopathologically, the infiltration of inflammatory cells, as well as irregular and excess accumulations of extracellular matrix components (eg, collagen, fibronectin, elastin, and proteoglycans) are observed. The molecular aberrant mechanisms in keloids can be categorized into three groups: i) extracellular matrix proteins and their deposition and degradation, ii) cytokines and growth factors, and iii) apoptotic pathways.2 To explore keloid pathogenesis, differences between keloid-derived fibroblasts (KDFs) and normal skin-derived fibroblasts (NDFs) have been investigated. KDFs, showing reduced growth-factor requirement,3 proliferate and migrate at a faster rate than NDFs.4 Moreover, KDFs are resistant to corticosteroid in terms of growth response 5 and further down-regulation of types I, III, and V collagen6,7, elastin,8 connective tissue growth factor, and insulin-like growth factor-binding protein 39 gene expression. In addition, KDFs are reportedly refractory to phorbol esters and prostaglandin E2.10 These basic findings recapitulate well clinical resistance of keloids to various treatments.11,12 Therefore, we consider that research using KDFs and NDFs is a powerful strategy to characterize the pathological mechanism of keloids as a clue to provide new targets for treatment.
To explore novel target molecules, microarray analyses of keloids have been performed and reported the up-regulated expression of many genes related to the cell cycle,13 intercellular signaling14 and the extracellular matrix,9,15,16 and the down-regulated expression apoptosis-related genes.14 In this study, using microarray analysis of KDFs and NDFs from the same patient, we identify and characterize a new potentially pathogenic gene, which encodes cartilage oligomeric matrix protein (COMP), because most previous studies used keloid and normal fibroblasts or tissues from different persons, our methodology may avoid individual difference and bias.
COMP, also referred to as thrombospondin 5, is a 524-kDa homopentameric, noncollagenous, extracellular matrix glycoprotein, which is found in cartilage, tendons, and ligaments and the growth plate.17 Its carboxyterminal globular domain binds to type I, II, and IX collagens, fibronectin,18–20 and aggrecan,21 and thereby accelerates fibrillogenesis through the promotion of matrix component assembly.22 In addition, the coiled-coil domain stores and delivers hydrophobic ligands, such as retinoid and vitamin D.23,24 These studies suggest that COMP potentially exerts a wide range of biological functions. Furthermore, recent evidences are accumulating that COMP is both a pathogenic factor and biomarker in scleroderma,25–30 indicating that this molecule can be involved in pathogenesis of other fibrosing diseases. Additionally, existence of COMP in horse scar was previously reported,31 but the role of COMP in scar or keloid is still unknown. These results prompted us to study the potential pathogenic role of COMP in keloid formation.
Materials and Methods
Keloid and Normal Control Tissues
Keloid tissues were obtained from 8 males and 7 females with a mean age of 50 years (range, 15 to 80 years) (Table 1). Normal skin as control samples was obtained from four males and two females, with a mean age of 59.2 years (range, 2 to 80 years). Among them, three were the uninvolved normal skin specimens around keloids from patients 1, 2, and 3 in Table 1. The three other normal samples were obtained from nonkeloid patients who underwent benign skin tumor excisions (two, inguinal; one abdomen).
Table 1.
Clinical Characteristics of the Patients from Whom the Samples Were Obtained
| Patient no. | Age/Sex | Site of lesion |
|---|---|---|
| >10 cm2 | ||
| 1 | 68 M | Chest |
| 2 | 60 M | Chest |
| 3 | 80 F | Neck |
| 4 | 71 M | Inguinal |
| 5 | 80 F | Retroauricular |
| <10 cm2 | ||
| 6 | 52 M | Shoulder |
| 7 | 47 M | Face |
| 8 | 55 F | Earlobe |
| 9 | 65 M | Earlobe |
| 10 | 26 F | Earlobe |
| 11 | 15 F | Earlobe |
| 12 | 32 M | Retroauricular |
| 13 | 22 M | Retroauricular |
| 14 | 33 F | Earlobe |
| 15 | 44 F | Abdomen |
F, female; M, male.
Isolation and Culture of Human Dermal Fibroblasts
Using the explant technique, we isolated KDFs from patient 1, 2, and 6 and NDFs from patients 1 and 2 in Table 1. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Nissui Pharmaceutical, Tokyo, Japan) supplemented with 10% fetal calf serum (FCS) (JRH Biosciences, Lenexa, KS), l-glutamine (4 mmol/L), penicillin (50 units/mL), and streptomycin (50 mg/mL) at 37°C under a humidified atmosphere of 95% air and 5% CO2. The cells were subcultured using 0.025% trypsin (usually 4 weeks after the beginning of primary cell cultures). KDFs in the third to sixth passage were used for these experiments. For the microarray, third passage KDFs were used. These studies were approved by the Medical Ethical Committee of Osaka University and conducted according to the Declaration of Helsinki Principles.
Microarray Analysis
For comprehensive comparison of gene expression differences between the keloid and the control tissues, microarray analysis used RNAs extracted from KDFs and from NDFs derived from a clinically typical keloid on the chest and the surrounding uninvolved skin from the same patient, respectively. Total RNA was extracted from KDFs and NDFs using a Qiagen RNeasy kit (Qiagen, San Diego, CA). Briefly, 2 μg total RNA was then reverse transcribed to cDNA with T7 oligo d(T) primer (Affymetrix, Inc., Santa Clara, CA). The cDNA synthesis products were used for in vitro transcription reactions containing T7 RNA polymerase and biotinylated nucleotide analog (pseudouridine base) cRNAs. The labeled cRNA products were then fragmented and loaded on GeneChip Human Genome U133 Plus 2.0 arrays (Affymetrix, Inc.), and hybridized according to the manufacturer's protocol. Streptavidin-Phycoerythrin (Molecular Probes, Eugene, OR) was used as the fluorescent conjugate to detect hybridized target sequences. Raw intensity data from the GeneChip arrays were analyzed using GeneChip Operating Software (Affymetrix, Inc.).
Immunohistochemistry
Skin specimens were taken from the surgical specimen, fixed in 10% formaldehyde and paraffin embedded. Tissue sections on the slides were deparaffinized and then heated in an autoclave in 10% citrate buffer for 15 minutes at 121°C to activate epitope structures in the tissue. They were then incubated with a 1:20 dilution of monoclonal rat anti-human COMP IgG (MCA1455, AbD Serotec, Oxford, UK) overnight at 4°C. The slides were then washed 3 times in Tris-buffered saline with Tween 20 (TBST) for 5 minutes each and incubated for 10 minutes with a biotinylated rabbit anti-rat immunoglobulin (E0468, Dako, Glostrup, Denmark). The slides were then washed 3 times in Tris-buffered saline with Tween 20 (TBST) for 5 minutes each, followed by treatment with peroxidase-conjugated streptavidin. Before staining the keloid samples, we confirmed negative staining of control normal dermis to avoid nonspecific staining because of the high concentration of the antibody. Reaction products were visualized by diaminobenzidine, and tissue sections were counterstained with Mayer's hematoxylin. The score of staining intensity was estimated using the percentage of positive constituents as follows: 0%, −; 1 to 5%, ±; 6 to 50%, +; >50%, ++.
Immunocytochemistry
Cultured human fibroblasts were grown on Lab-Tek chamber slides (Electron Microscopy Sciences, Hatfield, PA) and at subconfluency the cells were transiently transfected with control and COMP siRNA as mentioned as follows. Twenty-four hours later, the cells were re-fed with fresh 10% FCS DMEM containing 50 mg/mL ascorbic acid. After incubation for 72 hours, the cells were fixed in 4% paraformaldehyde for 10 minutes at room temperature and permeabilized with 0.1% Triton X-100. COMP was detected by incubation with rat anti-human COMP IgG (1:50) (sc-59941, Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4°C, followed by Alexa fluor 488 conjugated goat anti-rat IgG. Extracellular type I collagen was detected with rabbit anti-human collagen type 1 alpha 1 (1:50) (sc-28657, Santa Cruz Biotechnology) overnight at 4°C, followed by Alexa fluor 594 conjugated donkey anti-rabbit IgG. Nuclei were stained with Hoechst33342. The cells were observed by confocal laser scanning microscope FluoView 1000-D (Olympus, Tokyo, Japan).
Transfection of KDFs with an siRNA against COMP
COMP siRNA and a negative control siRNA were prepared and provided by Ambion 32. The nucleotide sequences of sense and antisense COMP siRNAs used were: 5′-GGAGGACUCAGACCACGAUdTdT-3′ and 5′-AUCGUGGUCUGAGUCCUCCdTdG-3′. KDFs were transiently transfected with 100 nmol/L siRNA against COMP using Amaxa nucleofection kits (Lonza Cologne AG, Cologne, Germany) for adult human dermal fibroblasts according to the manufacturer's protocol. After the transfection, cells were cultured in medium. In the preliminary experiments, we confirmed that successful transfection of siRNA is constantly obtained by co-transfection of expression vector of GFP.
Real-Time Quantitative RT-PCR
Total RNAs were isolated from keloid and uninvolved normal tissue from patient 1 in Table 1 using the acid guanidium thiocyanate-phenol-chloroform method and the purity of the extracted RNA was calculated with the optical densities at 260 and 280 nm. The cDNA was reverse-transcribed from 1 mg total RNA from each sample using M-MLV Reverse Transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. The oligonucleotide primers used were as follows: COMP designed by primer 3 software-sense primer 5′-TGGGCCCGCAGATGCTTC-3′ and antisense primer 5′-CCGTCTTCCGTCTGGATG-3′, glyceraldehyde-3-phosphate dehydrogenase (G3PDH) as an internal control-sense primer 5′-CCCATCACCATCTTCCAG-3′ and antisense primer 5′- CCTGCTTCACCACCTTCT-3′. Real-time PCR was conducted using Power SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA), according to the manufacturer's instructions. The final PCR mixture contained 0.5 μL each of forward and reverse primers (final concentration of each, 500 nmol/L), 25 μL of Power SYBR Green PCR Master Mix (Applied Biosystems), and 1 μL of sample (equivalent to 50 ng RNA). Real-time PCR was performed with an ABI Prism 7900 HT machine (Applied Biosystems) and universal cycling conditions (2 minutes at 50°C, 10 minutes at 95°C, 40 cycles of 15 seconds at 95°C, and 1 minute at 60°C). Cycle threshold values were determined by automated threshold analysis with ABI Prism version 1.0 software (Applied Biosystems). The amplification efficiencies were determined by serial dilution and calculated as E = exp-−1/m, where E is the amplification efficiency and m is the slope of the dilution curve. Dissociation curves were recorded after each run.
Western Blotting
Tissues from keloid and normal skin were thoroughly homogenized in 20 mmol/L HEPES, pH 7.2, containing 1% Nonidet P-40, 0.4 M NaCl and aprotinin and were centrifuged to extract the proteins into supernatants. The protein samples were mixed with 2 × SDS-PAGE buffer, heat denatured with a reducing agent and then loaded onto 4 to 12% Tris-glycine gels. Cultured cells were lysed in 100 μL 2 × SDS-PAGE buffer in reducing conditions. All samples (5 mg/lane) were heated and loaded on 7.5% Tris-glycine gels. After electrophoresis, proteins on the gels were transferred to nitrocellulose membranes. The membranes were briefly stained with Ponceau S, photographed, and blocked in 5% milk in TBST buffer for 1 hour and then incubated for 1 hour with antibodies against COMP (1:100) (sc-59941, Santa Cruz Biotechnology) or β-actin, and then washed 3 times for 5 minutes each with TBST, followed by a 1 hour incubation with horseradish peroxidase conjugated secondary antibody. All membranes were developed using the ECL Plus Western Blotting Detection System (GE Healthcare, Buckinghamshire, England). Quantification and densitometric analysis was performed using Image J software (National Institutes of Health, Bethesda, MD).
Assay for Measuring Amount of Collagen
At 24 hours after transfection of the control and CONP siRNA into the subconfluent KDFs at 6-well plates, we put 1 mL fresh DMEM supplemented with 10% FCS containing 50 mg/mL ascorbic acid. For measuring amount of collagen, at 72 hours after the medium change, the conditioned media were harvested. We used the Sircol Collagen Assay Kit (Biocolor Ltd., Carrickfergus, UK) to measure type I to V collagens according to the manufacturer's instructions. The 1 mL of conditioned media were mixed with 200 mL isolation and concentration reagent in the kit and incubated at 4°C overnight. After centrifugation, pellets were re-dissolved in 1 mL of Sircol dye reagent (Biocolor Ltd.) which specifically binds to collagens. After centrifugation, pellets were suspended in 1 mL of the alkali compound included in the kit and collagen concentration was then determined by spectrophotometric absorbance at 540 nm as compared with a standard curve. After aspiration of the culture supernatant, extracellular matrix formed on the cells was treated with 1 mL 0.5 M cold acetic acid for 24 hours and the amount of collagens in the extract was determined with the Sircol Collagen Assay Kit (Biocolor Ltd) as previously mentioned. The amounts of collagens were determined and expressed as relatives of each control transfected with the negative control siRNA.
Treatment of KDFs with Transforming Growth Factor-β1
KDFs were grown in 10% FCS DMEM until subconfluency. Then the media were changed to DMEM without FCS for serum depletion. At 24 hours later, the cells were re-fed with fresh DMEM without FCS containing 1 ng/mL human recombinant transforming growth factor (TGF)-β1 (240B, R&D Systems, Minneapolis, MN) or a mock solution. After incubation for 24 hours, the cells were harvested and subjected to Western blot as previously mentioned.
Statistics
Statistical differences of immunohistochemical staining intensity were determined by the Mann-Whitney U-test; differences with P < 0.05 are considered significant. For other experiments, statistical differences were determined by Student's t-test; differences with P < 0.05 are considered significant.
Results
Microarray Analysis of Genes Differentially Expressed between KDFs and NDFs from the Same Patient
Initially we tried to use RNA from keloid and normal tissues for microarray analyses but we had difficulty producing high quality samples. Moreover, as previously mentioned, the prior reports demonstrated that the differences of KDFs and NDFs recapitulate well in vivo keloid pathogenesis 3 to 10. Therefore, to elucidate potentially pathogenic genes or molecules in keloids, microarray analyses were performed using RNAs extracted from KDFs and NDFs from the same patient (patient 1 in Table 1) with a clinically typical keloid on the chest (Figure 1A). The normal tissue was obtained from the surrounding normal skin. Among the 38,500 genes on the array, 374 were up-regulated (fold value, >2) and 758 were down-regulated (fold value, <0.5) in KDFs compared with NDFs. The most significantly up-regulated 31 genes are listed in Table 2. The differentially expressed genes were classified by molecular functions, possibly related to wound healing and keloid growth (Table 3). More than 10 genes were up-regulated in the molecular functions in the following gene ontology categories: extracellular matrix constituents, growth factors and their receptors, transcription factors and proteins binding with DNA, and apoptosis. In contrast, genes down-regulated in molecular functions overlapped in two categories: growth factors and their receptors and transcription factors and proteins binding with DNA. Shih et al 33 summarized previous studies and their results of microarray analyses for keloid tissues and KDFs compared with normal tissues and NDFs. As a result, they bioinformatically selected 19 valid candidate genes. Our microarray data were in agreement on the increase of aggrecan, asporin, pleiotrophin, serpin F1, and LIM and senescent cell antigen-like domains 2, and the decrease of epidermal growth factor receptor. Eleven extracellular matrix genes were up-regulated, and of those, COMP was most prominently up-regulated (Table 4).
Figure 1.
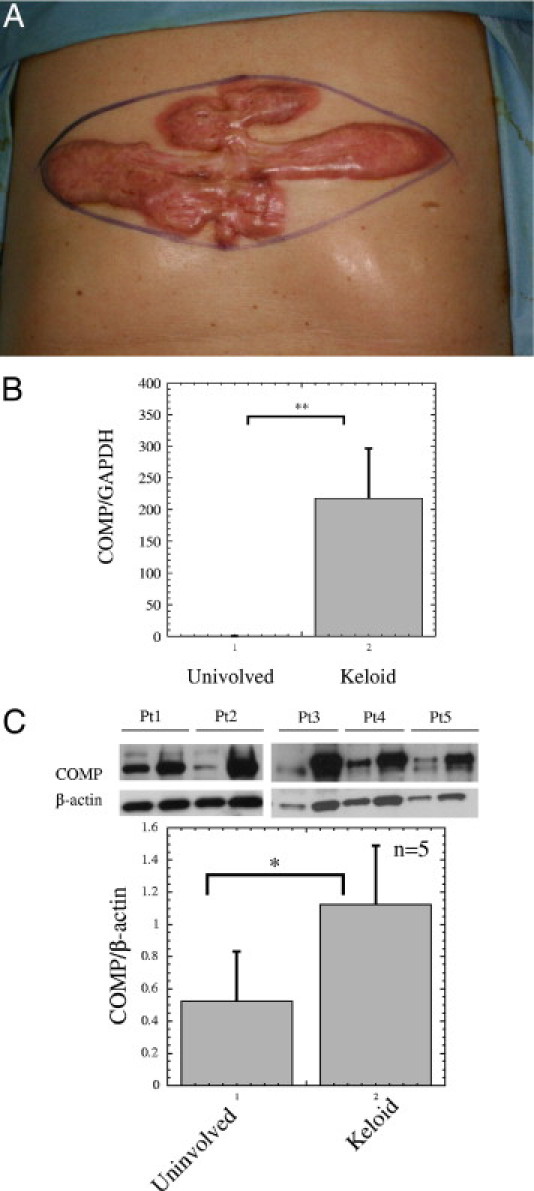
Clinical appearance of the keloid of patient 1 and cartilage oligomeric matrix protein (COMP) mRNA and protein expression in the keloid tissues. Microarray analysis was performed using RNAs extracted from keloid-derived fibroblasts (KDFs) derived from the 23 × 7.5-cm keloid and from NDFs derived from the surrounding uninvolved skin on the chest of a 68-year-old Japanese male (patient 1 in Table 1) (A). (B) COMP mRNA amount was examined by quantitative real-time RT-PCR, as mentioned in Materials and Methods. Statistical difference was determined by the Student's t-test. **P < 0.01 (C) Skin tissue from the keloid and uninvolved (normal) skin from the five patients (patients 1 to 5 in Table 1) were homogenized in 20 mmol/L HEPES, pH 7.2, containing 1% Nonidet P-40, 0.4 M NaCl, and aprotinin and were analyzed for COMP and β-actin levels by Western blot. The lower graph shows mean ± SD of COMP/β-actin protein ratios of patients 1 to 5 (n = 5). Statistical difference was determined by the Student's t-test. *P < 0.05.
Table 2.
Genes Showing the Greatest Increase in Expression in KDFs Compared to NDFs in Microarray Analysis
| Gene name | Gene symbol | Fold change | UniGene ID |
|---|---|---|---|
| MRNA; cDNA DKFZp686A0837 (from clone DKFZp686A0837) | — | 78.793242 | Hs.638566 |
| Cartilage oligomeric matrix protein | COMP | 48.50293 | Hs.1584 |
| Secreted frizzled-related protein 2 | SFRP2 | 42.224253 | Hs.481022 |
| v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | KIT | 39.396621 | Hs.479754 |
| C-type lectin domain family 12, member A | CLEC12A | 36.758347 | Hs.190519 |
| Fibroblast growth factor receptor 2 | FGFR2 | 34.296751 | Hs.533683 |
| Testis expressed 9 | TEX9 | 25.992077 | Hs.511476 |
| Retinoic acid receptor responder (tazarotene induced) 2 | RARRES2 | 25.992077 | Hs.647064 |
| Transmembrane protein 16A | TMEM16A | 22.627417 | Hs.503074 |
| Transcribed locus | — | 19.698311 | Hs.662908 |
| Armadillo repeat containing, X-linked 4 | ARMCX4 | 18.379174 | Hs.399873 |
| Death-associated protein kinase 1 | DAPK1 | 17.148375 | Hs.380277 |
| cDNA FLJ38181 fis, clone FCBBF1000125 | — | 16 | Hs.143134 |
| cDNA clone IMAGE:4830514 | — | 16 | Hs.570820 |
| Colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) | CSF2RB | 16 | Hs.592192 |
| Chondrolectin | CHODL | 14.928528 | Hs.283725 |
| Dermatopontin | DPT | 14.928528 | Hs.80552 |
| Insulin-like growth factor 2 (somatomedin A) /// insulin- insulin-like growth factor 2 | IGF2 /// INS-IGF2 | 13.928809 | Hs.523414 |
| Gap junction protein, alpha 5, 40kDa | GJA5 | 12.996038 | Hs.447968 |
| Inter-alpha (globulin) inhibitor H5 | ITIH5 | 12.996038 | Hs.498586 |
| cDNA FLJ30757 fis, clone FEBRA2000468 | — | 12.996038 | Hs.662237 |
| G protein–coupled receptor 37 (endothelin receptor type B-like) | GPR37 | 12.125733 | Hs.406094 |
| Unc-5 homolog B (C. elegans) | UNC5B | 12.125733 | Hs.585457 |
| Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 | NFKBIL1 | 11.313708 | Hs.2764 |
| Hypothetical gene supported by BC008048 | LOC440934 | 10.556063 | Hs.238964 |
| Dopamine receptor D1 | DRD1 | 10.556063 | Hs.2624 |
| Signal-induced proliferation-associated 1 like 2 | SIPA1L2 | 10.556063 | Hs.268774 |
| Deleted in bladder cancer 1 | DBC1 | 10.556063 | Hs.532316 |
| Calcium/calmodulin-dependent protein kinase ID /// Hypothetical protein LOC283070 | CAMK1D /// LOC283070 | 10.556063 | Hs.600547 |
| Carboxypeptidase Z | CPZ | 10.556063 | Hs.78068 |
NDFs, normal skin-derived fibroblasts.
Table 3.
Comparison of Gene Expression Patterns between KDFs and NDFs from the Same Patient in the Microarray Analysis
| Up-regulated | Down-regulated | |
|---|---|---|
| Extracellular matrix constituents | 11 | 8 |
| Growth factors and their receptors | 18 | 17 |
| Transcription factors and proteins binding with DNA | 25 | 72 |
| Apoptosis | 15 | 0 |
| Matrix metallopeptidase | 3 | 12 |
| Cellular skeleton and movement | 3 | 9 |
| Cytokines and their binding proteins | 5 | 24 |
| Cell metabolism | 1 | 0 |
| Unknown function | 106 | 202 |
| Others | 187 | 414 |
| TOTAL | 374 | 758 |
KDFs, keloid-derived fibroblasts; NDFs, normal skin-derived fibroblasts.
Table 4.
Increased Expression of Extracellular Matrix Genes in KDFs in the Microarray Analysis
| No. | Gene name | Gene symbol | Fold change | UniGene ID |
|---|---|---|---|---|
| 1 | Cartilage oligomeric matrix protein | COMP | 48.5 | Hs.1584 |
| 2 | Elastin | ELN | 6.5 | Hs.647061 |
| 3 | Collagen, type XVIII, alpha 1 | COL18A1 | 6.5 | Hs.517356 |
| 4 | Collagen, type XV, alpha 1 | COL15A1 | 5.66 | Hs.409034 |
| 5 | Collagen, type VIII, alpha 2 | COL8A2 | 4.92 | Hs.353001 |
| 6 | Fibrillin 2 | FBN2 | 3.73 | Hs.519294 |
| 7 | Collagen, type XI, alpha 1 | COL11A1 | 3.48 | Hs.523446 |
| 8 | Fibulin 2 | FBLN2 | 3.03 | Hs.198862 |
| 9 | Collagen, type V, alpha 1 | COL5A1 | 2.46 | Hs.210283 |
| 10 | Collagen, type V, alpha 3 | COL5A3 | 2.14 | Hs.235368 |
| 11 | Aggrecan | ACAN | 2 | Hs.2159 |
KDFs, keloid-derived fibroblasts.
The increase of COMP mRNA was confirmed by real-time RT-PCR (Figure 1B). Likewise, up-regulation of COMP protein was shown in the keloid compared with normal tissue from our patients (patients 1 to 5) by Western blot (Figure 1C) (n = 5; P < 0.05). Then, we decided to further examine the potentially pathogenic role of this molecule in keloid formation.
Immunohistochemistory of COMP in Keloid Tissues
Next, we immunohistochemically compared expression level of COMP protein in 15 keloid and 6 control normal tissues, using an anti-COMP antibody. We defined staining as positive when more than 5% of the constituents were stained (0%, −; 1 to 5%, ±; 6 to 50%, +; >50%, ++, Table 5). As a result, COMP was positively stained in 10 keloids (66.7%) (Figure 2), whereas no positive staining was observed in normal tissue, demonstrating the significantly higher expression of COMP in keloid tissues (P < 0.001 by Mann-Whitney's U-test). When comparing keloids smaller or larger than 10 cm2, the larger keloids were significantly more intensely stained for COMP (P < 0.01 by the Mann-Whitney U-test) (Table 5).
Table 5.
Expression of COMP in Vivo
| Staining | − | ± | + | ++ | Total |
|---|---|---|---|---|---|
| Smaller-sized keloids | 0 | 5 | 4 | 1 | 10 |
| Larger-sized keloids | 0 | 0 | 0 | 5 | 5 |
| Normal dermis | 6 | 0 | 0 | 0 | 6 |
Fifteen keloid and 6 normal tissues were immunohistochemically stained with the cartilage oligomeric matrix protein (COMP)-specific antibody. Ten of the keloid lesions were smaller than 10 cm2 and the other 5 were larger. The score of staining intensity was estimated using the percentage of positive cells as follows: 0%, −; 1 to 5%, ±; 6–50%, +; >50%, ++. Difference of the staining score was statistically evaluated by the Mann-Whitney U-test, and as a result the score was significantly different between the keloids (n = 15) and normal dermis (n = 6; P < 0.001) and between the smaller- (n = 10) and larger-sized keloids (n = 5; P < 0.01).
Figure 2.
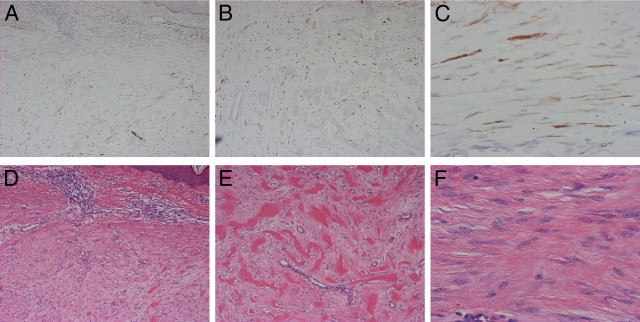
Immunohistochemical staining for cartilage oligomeric matrix protein (COMP) expression. COMP was positively stained in keloid lesions in the upper dermis (A) COMP staining; (D) H&E staining, ×200 magnification; and in the lower dermis (B) COMP staining; (E) H&E staining, ×200. COMP was detected on the wavy keloid-derived fibroblasts (KDFs); (C) COMP staining; (F) H&E staining, ×400.
Suppression of COMP by siRNA Reduces Type I Collagen Deposited on Keloid Fibroblasts
Because COMP reportedly accelerates collagen type I fibril assembly 22, we examined whether extracellular type I collagen deposition is altered by suppression of COMP function by an siRNA. At 96 hours after transfection, with the COMP siRNA or a control siRNA, the cells were immunocytochemically stained and observed by confocal microscopy. Following reduction of COMP by the specific siRNA (Figure 3B versus 3F), the extracellular deposition of type I collagen was diminished (Figure 3A versus 3E and Figure 3D versus 3H), indicating that COMP accelerates the accumulation and fibril formation of type I collagen in the extracellular matrix of keloids. Further, the colocalization of COMP and type I collagen was seen on the cell surfaces (Figure 3H). Successful knockdown of COMP protein by siRNA was confirmed by Western blot (Figure 3I). Moreover, we measured the amount of type I to V collagens in the culture medium and deposited on the KDF surfaces by the Sircol Collagen Assay Kit. As a result, knockdown of COMP by siRNA significantly decreased the collagen in the culture medium of the surface of KDF lines (patients 1, 2, and 6) by 44.0 ± 5.47% (P < 0.05) (Figure 4, lane 1 versus 2) and likewise decreased the extracellular deposition of collagens on the cell surface of the KDFs by 34.5 ± 9.17% (P < 0.01) (Figure 4, lane 3 versus 4). KDFs in the third to sixth passage were used for these experiments, but when KDFs over the 10th passage were used, the effect of COMP knockdown on the deposited type I collagen was not detected (data not shown), suggesting that characteristics of KDFs disappeared during in vitro cultivation. In these experiments, there were no significant difference between the cell number of KDFs transfected with the control and COMP siRNA.
Figure 3.
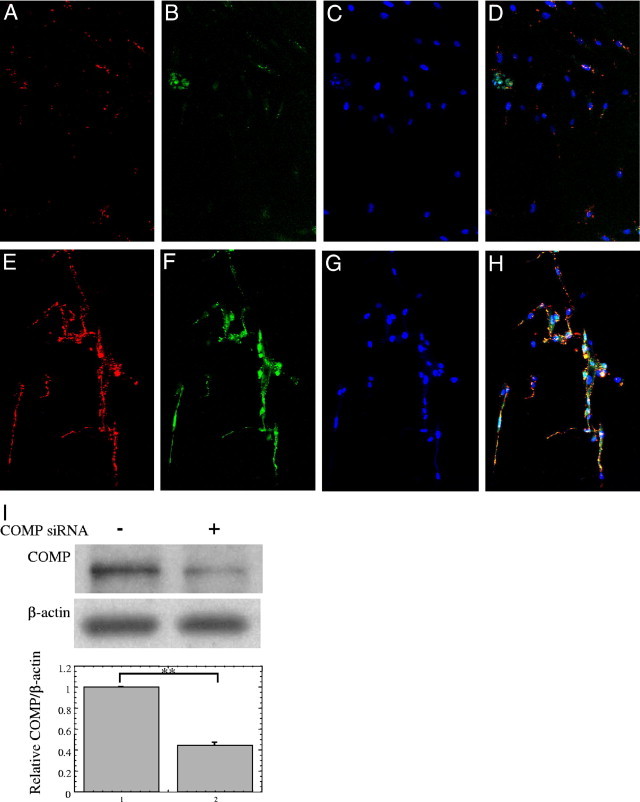
Suppression of cartilage oligomeric matrix protein (COMP) by small interfering RNA (siRNA) reduces levels of extracellular type I collagen. At 96 hours after transient transfection with the COMP siRNA (A–D) or with the control siRNA (E–H) into subconfluent keloid-derived fibroblasts (KDFs), the cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. Extracellular type I collagen was then detected with an anti-type I collagen antibody and was visualized using a red fluorophore (A and E); COMP was detected with a COMP-specific antibody and visualized using a green fluorophore (B and F), and nuclei were stained with Hoechst (C and G). A merged image (A, B, and C) is shown in (D) and an image (E, F, and G) is shown in (H). Transfection of the COMP siRNA reduces levels of extracellular type I collagen and COMP (A versus E, B versus F). Nuclei of fibroblasts were nonspecifically stained in each experiment. The distribution of type I collagen and COMP overlapped closely (H). The cells were observed by confocal laser scanning microscope and all photographs are shown at ×20 magnification. (I) KDFs transiently transfected with the control or COMP siRNA were lysed in 100 μL 2 × SDS-PAGE buffer in reducing conditions and subjected to Western blot for COMP and β-actin. The COMP/β-actin ratios were determined and expressed as the relative of COMP/β-actin of the each control sample transfected with control siRNA. The lower graph shows mean ± SD of COMP/β-actin protein ratios using the KDFs from three different patients (n = 3). Statistical difference was determined by the Student's t-test. **P < 0.01.
Figure 4.
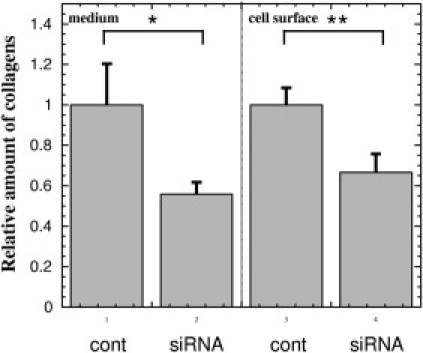
Suppression of cartilage oligomeric matrix protein (COMP) by small interfering RNA (siRNA) reduces collagens in the culture medium of keloid fibroblasts and deposited on the cell surfaces. At 24 hours after transfection of the control (lanes 1 and 3) and COMP siRNA (lanes 2 and 4) into the subconfluent keloid-derived fibroblasts (KDFs) at 6-well plates, we put 1 mL fresh Dulbecco's modified Eagle's medium (DMEM) (Nissui Pharmaceutical, Tokyo, Japan) supplemented with 10% fetal calf serum containing 50 mg/mL ascorbic acid. For assay of collagen amount, at 72 hours after the medium change, the conditioned media (lanes 1 and 2) were subjected to the Sircol Collagen Assay Kit (Biocolor Ltd., Carrickfergus, UK) to measure type I to V collagens. Briefly, 1 mL of conditioned media were mixed with 200 mL isolation and concentration reagent in the kit and incubated at 4°C overnight. After centrifugation, pellets were re-dissolved in 1 mL of Sircol dye reagent (Biocolor Ltd.). After centrifugation, pellets were suspended in 1 mL of the alkali compound and collagen concentration was determined by spectrophotometric absorbance at 540 nm. After aspiration of the culture supernatant, extracellular matrix formed on the cells was treated with 1 mL 0.5 M cold acetic acid for 24 hours and the amount of collagens in the extract was determined with the Sircol Collagen Assay Kit (lanes 3 and 4). The amounts of collagens were determined using the KDFs from three different patients (n = 3) and expressed as relatives of each control transfected with the negative control siRNA (lanes 1 and 3). Statistical difference was determined by the Student's t-test. *P < 0.05, **P < 0.01, cont, negative control siRNA.
Induction of COMP by TGF-β1 in KDFs
Since COMP is up-regulated by TGF-β1 in chondrocytes 34 and scleroderma fibroblasts 26, we examined whether COMP expression is regulated by TGF-β1 in KDFs. After serum depletion for 24 hours, COMP expression was remarkably decreased, but TGF-β1 treatment for 24 hours significantly increased COMP expression by 4.34-fold, compared with a mock treatment (Figure 5), indicating that COMP works as a pathogenic factor in keloid formation at the downstream of TGF-β.
Figure 5.
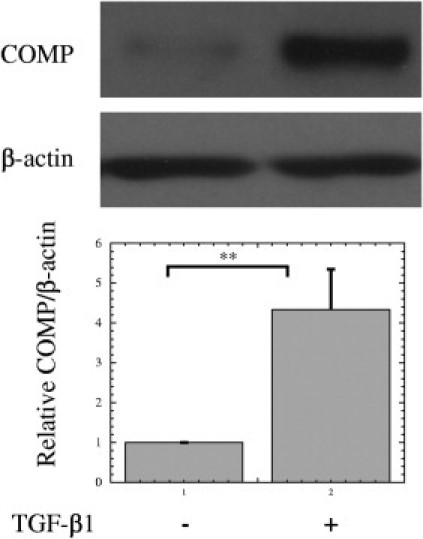
Induction of cartilage oligomeric matrix protein (COMP) by transforming growth factor (TGF)-β1 in keloid fibroblasts. Keloid-derived fibroblasts (KDFs) were grown in 10% fetal calf serum (FCS) Dulbecco's modified Eagle's medium (DMEM) (Nissui Pharmaceutical, Tokyo, Japan) until confluency. Then, the media were changed to DMEM without FCS for serum depletion and the cells were incubated for 24 hours. The cells were re-fed with fresh DMEM (Nissui Pharmacetical) without FCS containing 1 ng/mL human recombinant TGF-β1 or a mock solution. After incubation for 24 hours, the cells were harvested and subjected to Western blot. The COMP/β-actin ratios were determined and expressed as the relative of COMP/β-actin of each control sample (a mock treatment). The lower graph shows mean ± SD of COMP/β-actin protein ratios using the KDFs from three different patients (n = 3). Statistical difference was determined by the Student's t-test. **P < 0.01
Discussion
Clinically, mutations of the COMP gene cause pseudoachondroplasia and multiple epiphyseal dysplasia35,36 and this protein can be a biomarker of rheumatoid arthritis37 and osteoarthritis 38. In scleroderma, serum COMP has been reported to be significantly higher than normal controls 30 and correlate to skin involvement as measured by modified Rodnan total skin thickness score.25,28 Moreover, a pathogenic role of COMP in scleroderma has been reported.25–28,30 By the gene expression profiling of cultured skin fibroblasts, COMP mRNA level is elevated in scleroderma fibroblasts and COMP protein accumulates in scleroderma, but not normal skin by immunohistochemistry.26 Cultured scleroderma fibroblasts express higher amount of COMP than normal controls and further TGF-β1 induces a large increase of COMP mRNA and protein in vitro.26 These findings indicate that COMP plays a role in matrix deposition in scleroderma. In this study, we show the pathogenic role of COMP in another fibrotic skin disease, keloids, where up-regulated COMP facilitates the deposition of collagen and other matrix proteins, resulting in further growth of the keloid. Taken together, COMP exerts similar pathogenic roles in scleroderma and keloids as a matrix deposition promoter. Stronger staining of COMP is observed in long-standing scleroderma26 and COMP mRNA expression in lesional skin from scleroderma patients correlates with the modified Rodnan total skin thickness score 27 Similarly, our data in this study indicate that larger keloids (>10 cm2) express higher levels of COMP than smaller keloids (Table 5), suggesting that expression level of COMP can change during disease progression and be used as a biomarker of activity and severity.
Very recently, it has been reported that there are significant associations of keloid with four SNP loci in three chromosomal regions, such as 1q41, 3q22.3-−23, and 15q21.3 through a multistage genome-wide association study using 824 individuals with keloid and 3205 unaffected controls in the Japanese population.39 Human COMP gene is located on chromosome 19p13.1,40 indicating lack of association between keloid and COMP polymorphisms. Additionally, no candidate genes in Table 2 are not located on the chromosomal regions 1q41, 3q22.3-−23, and 15q21.3. Our data showed here that the expression of COMP is increased in KDFs by TGF-β1, as previously reported in chondrocytes34 and in scleroderma fibroblasts,26 suggesting that COMP plays a role in the downstream of TGF-β receptor I, a major pathogenic player in keloids. Moreover, the recent study pointed out the pathogenic involvement of epigenetically altered program, such as DNA methylation and histone acetylation in KDFs.41 Therefore, epigenetic modification of COMP gene may associate with keloids. In this regard, our data showed the persistent differences between the KDFs and NDFs from the same patient, implying an autonomous lesion-specific change of gene expression, including epigenetic or mutational alteration. These aspects may be worthy of further examination.
Previously, Naitoh et al15 detected a 5.4-fold up-regulation of COMP transcripts in keloids compared to normal skin from the same patient by microarray analysis. However, because there was no signal for COMP by Northern blotting, no further investigation was performed. Since our immunohistochemical study shows that the in vivo expression of COMP is heterogeneous and is sometimes only subtly positive, especially in small keloids, we speculate that their negative Northern blot was due to a low amount of COMP mRNA in their tissue. Likewise, in scleroderma, COMP is reportedly expressed in distinct cells.27 In this study, we used KDFs, not keloid tissues, to detect COMP mRNA and protein, and therefore succeeded in its identification. Additionally, Naitoh et al15 found the up-regulation of chondrogenesis-associated genes, such as collagen type X and VI, SOX9 and CBFA1, in keloids. In this context, we identified in this study 2.0-, 1.5-, and 1.4-fold increases of aggrecan (a major cartilage matrix component), CBFA1 and SOX9, respectively, suggesting the reprogramming of the gene expression profile toward the chondrocytic lineage in keloids.
For this microarray study, we isolated KDFs and NDFs derived from a keloid and uninvolved tissue from the same patient. Although we confirmed histopathologically that there was no fibrosis in the normal tissue, they may inherently contain abnormal characteristics. Indeed, another group14 found differential expression of caspases 6 and 14 through an alternative approach to perform microarray analyses using normal skin from keloid-prone and from keloid-resistant individuals, although we did not detect any alteration in expression of those genes. Further, when comparing our results with their microarray analysis, also we found the up-regulation of mitogen-activated protein kinase 1.
Our immunohistochemistry showed that the most COMP staining in tissues is cell associated, not consistent with a role as a matricellular protein. However, recently it has been reported that COMP promotes cell attachment via CD47 and integrins at the cell surface. Moreover, cell attachment to COMP stimulates the formation of lamellipodia and the reorganization of actin into fascin-stabilized microspikes as its downstream effect, indicating the importance of COMP as a cell surface signaling protein.42 Because our data showed that COMP expression is dominantly cell associated in the keloid, COMP suggestively accelerates matrix deposition in the pericellular space. In addition, it will be of interest to study downstream signal from COMP in the future.
Because our microarray analysis did not detect differences in collagen type I expression in KDFs versus NDFs, some concerns may be raised about the phenotypic stability of the cells. Indeed, the previous microarrays, which compared keloid tissues or fibroblasts with normal tissues or NDFs from nonkeloid different persons,13,15,16 could show the difference of collagen type I expression. However, when using KDFs and NDFs from the same keloid patient43 or KDFs and normal scar derived fibroblasts,9 the microarrays could not detect any significant difference of collagen type I expression. These contradictory results suggest that the NDFs from keloid patients have intrinsic potential to overproduce collagen type I and, after subcultivation, express it similarly to KDFs as our microarray showed.
Collectively, our microarray and biochemical analyses using KDFs and NDFs from the same patient suggest that COMP, which is ectopically expressed in keloids, facilitates the formation of keloids by accelerating collagen deposition. COMP represents a novel pathogenic factor in keloid formation or growth, and provides a hopeful new therapeutic target.
Acknowledgments
We thank Dr. Motoko Naitoh (Kyoto University) for the helpful discussions and Saiko Ishii, Ayako Sato, and Naoko Yamada for their excellent technical assistance.
Footnotes
Supported in part by a research grant from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (22791720 to F.S.).
S.I. and F.S. contributed equally to this work.
References
- 1.Brown B.C., McKenna S.P., Siddhi K., McGrouther D.A., Bayat A. The hidden cost of skin scars: quality of life after skin scarring. J Plast Reconstr Aesthet Surg. 2008;61:1049–1058. doi: 10.1016/j.bjps.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 2.Shih B., Garside E., McGrouther D.A., Bayat A. Molecular dissection of abnormal wound healing processes resulting in keloid disease. Wound Repair Regen. 2010;18:139–153. doi: 10.1111/j.1524-475X.2009.00553.x. [DOI] [PubMed] [Google Scholar]
- 3.Russell S.B., Trupin K.M., Rodriguez-Eaton S., Russell J.D., Trupin J.S. Reduced growth-factor requirement of keloid-derived fibroblasts may account for tumor growth. Proc Natl Acad Sci USA. 1988;85:587–591. doi: 10.1073/pnas.85.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witt E., Maliri A., McGrouther D.A., Bayat A. RAC activity in keloid disease: comparative analysis of fibroblasts from margin of keloid to its surrounding normal skin. Eplasty. 2008;8:e19. [PMC free article] [PubMed] [Google Scholar]
- 5.Russell J.D., Witt W.S. Cell size and growth characteristics of cultured fibroblasts isolated from normal and keloid tissue. Plast Reconstr Surg. 1976;57:207–212. doi: 10.1097/00006534-197602000-00014. [DOI] [PubMed] [Google Scholar]
- 6.Russell J.D., Russell S.B., Trupin K.M. Differential effects of hydrocortisone on both growth and collagen metabolism of human fibroblasts from normal and keloid tissue. J Cell Physiol. 1978;97:221–229. doi: 10.1002/jcp.1040970211. [DOI] [PubMed] [Google Scholar]
- 7.Russell S.B., Trupin J.S., Myers J.C., Broquist A.H., Smith J.C., Myles M.E., Russell J.D. Differential glucocorticoid regulation of collagen mRNAs in human dermal fibroblasts: Keloid-derived and fetal fibroblasts are refractory to down-regulation. J Biol Chem. 1989;264:13730–13735. [PubMed] [Google Scholar]
- 8.Russell S.B., Trupin J.S., Kennedy R.Z., Russell J.D., Davidson J.M. Glucocorticoid regulation of elastin synthesis in human fibroblasts: down-regulation in fibroblasts from normal dermis but not from keloids. J Invest Dermatol. 1995;104:241–245. doi: 10.1111/1523-1747.ep12612788. [DOI] [PubMed] [Google Scholar]
- 9.Smith J.C., Boone B.E., Opalenik S.R., Williams S.M., Russell S.B. Gene profiling of keloid fibroblasts shows altered expression in multiple fibrosis-associated pathways. J Invest Dermatol. 2008;128:1298–1310. doi: 10.1038/sj.jid.5701149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myles M.E., Russell J.D., Trupin J.S., Smith J.C., Russell S.B. Keloid fibroblasts are refractory to inhibition of DNA synthesis by phorbol esters: Altered response is accompanied by reduced sensitivity to prostaglandin E2 and altered down-regulation of phorbol ester binding sites. J Biol Chem. 1992;267:9014–9020. [PubMed] [Google Scholar]
- 11.Mustoe T.A., Cooter R.D., Gold M.H., Hobbs F.D., Ramelet A.A., Shakespeare P.G., Stella M., Teot L., Wood F.M., Ziegler U.E. International clinical recommendations on scar management. Plast Reconstr Surg. 2002;110:560–571. doi: 10.1097/00006534-200208000-00031. [DOI] [PubMed] [Google Scholar]
- 12.Ogawa R. The most current algorithms for the treatment and prevention of hypertrophic scars and keloids. Plast Reconstr Surg. 2010;125:557–568. doi: 10.1097/PRS.0b013e3181c82dd5. [DOI] [PubMed] [Google Scholar]
- 13.Satish L., Lyons-Weiler J., Hebda P.A., Wells A. Gene expression patterns in isolated keloid fibroblasts. Wound Repair Regen. 2006;14:463–470. doi: 10.1111/j.1743-6109.2006.00135.x. [DOI] [PubMed] [Google Scholar]
- 14.Nassiri M., Woolery-Lloyd H., Ramos S., Jacob S.E., Gugic D., Viciana A., Romanelli P., Elgart G., Berman B., Vincek V. Gene expression profiling reveals alteration of caspase 6 and 14 transcripts in normal skin of keloid-prone patients. Arch Dermatol Res. 2009;301:183–188. doi: 10.1007/s00403-008-0880-z. [DOI] [PubMed] [Google Scholar]
- 15.Naitoh M., Kubota H., Ikeda M., Tanaka T., Shirane H., Suzuki S., Nagata K. Gene expression in human keloids is altered from dermal to chondrocytic and osteogenic lineage. Genes Cells. 2005;10:1081–1091. doi: 10.1111/j.1365-2443.2005.00902.x. [DOI] [PubMed] [Google Scholar]
- 16.Seifert O., Bayat A., Geffers R., Dienus K., Buer J., Lofgren S., Matussek A. Identification of unique gene expression patterns within different lesional sites of keloids. Wound Repair Regen. 2008;16:254–265. doi: 10.1111/j.1524-475X.2007.00343.x. [DOI] [PubMed] [Google Scholar]
- 17.Hedbom E., Antonsson P., Hjerpe A., Aeschlimann D., Paulsson M., Rosa-Pimentel E., Sommarin Y., Wendel M., Oldberg A., Heinegard D. Cartilage matrix proteins: An acidic oligomeric protein (COMP) detected only in cartilage. J Biol Chem. 1992;267:6132–6136. [PubMed] [Google Scholar]
- 18.Rosenberg K., Olsson H., Morgelin M., Heinegard D. Cartilage oligomeric matrix protein shows high affinity zinc-dependent interaction with triple helical collagen. J Biol Chem. 1998;273:20397–20403. doi: 10.1074/jbc.273.32.20397. [DOI] [PubMed] [Google Scholar]
- 19.Holden P., Meadows R.S., Chapman K.L., Grant M.E., Kadler K.E., Briggs M.D. Cartilage oligomeric matrix protein interacts with type IX collagen, and disruptions to these interactions identify a pathogenetic mechanism in a bone dysplasia family. J Biol Chem. 2001;276:6046–6055. doi: 10.1074/jbc.M009507200. [DOI] [PubMed] [Google Scholar]
- 20.Thur J., Rosenberg K., Nitsche D.P., Pihlajamaa T., Ala-Kokko L., Heinegard D., Paulsson M., Maurer P. Mutations in cartilage oligomeric matrix protein causing pseudoachondroplasia and multiple epiphyseal dysplasia affect binding of calcium and collagen I, II, and IX. J Biol Chem. 2001;276:6083–6092. doi: 10.1074/jbc.M009512200. [DOI] [PubMed] [Google Scholar]
- 21.Chen F.H., Herndon M.E., Patel N., Hecht J.T., Tuan R.S., Lawler J. Interaction of cartilage oligomeric matrix protein/thrombospondin 5 with aggrecan. J Biol Chem. 2007;282:24591–24598. doi: 10.1074/jbc.M611390200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halasz K., Kassner A., Morgelin M., Heinegard D. COMP acts as a catalyst in collagen fibrillogenesis. J Biol Chem. 2007;282:31166–31173. doi: 10.1074/jbc.M705735200. [DOI] [PubMed] [Google Scholar]
- 23.Guo Y., Bozic D., Malashkevich V.N., Kammerer R.A., Schulthess T., Engel J. All-trans retinol, vitamin D and other hydrophobic compounds bind in the axial pore of the five-stranded coiled-coil domain of cartilage oligomeric matrix protein. EMBO J. 1998;17:5265–5272. doi: 10.1093/emboj/17.18.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozbek S., Engel J., Stetefeld J. Storage function of cartilage oligomeric matrix protein: the crystal structure of the coiled-coil domain in complex with vitamin D(3) EMBO J. 2002;21:5960–5968. doi: 10.1093/emboj/cdf628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farina G., Lafyatis D., Lemaire R., Lafyatis R. A four-gene biomarker predicts skin disease in patients with diffuse cutaneous systemic sclerosis. Arthritis Rheum. 2010;62:580–588. doi: 10.1002/art.27220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farina G., Lemaire R., Korn J.H., Widom R.L. Cartilage oligomeric matrix protein is overexpressed by scleroderma dermal fibroblasts. Matrix Biol. 2006;25:213–222. doi: 10.1016/j.matbio.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 27.Farina G., Lemaire R., Pancari P., Bayle J., Widom R.L., Lafyatis R. Cartilage oligomeric matrix protein expression in systemic sclerosis reveals heterogeneity of dermal fibroblast responses to transforming growth factor beta. Ann Rheum Dis. 2009;68:435–441. doi: 10.1136/ard.2007.086850. [DOI] [PubMed] [Google Scholar]
- 28.Hesselstrand R., Kassner A., Heinegard D., Saxne T. COMP: a candidate molecule in the pathogenesis of systemic sclerosis with a potential as a disease marker. Ann Rheum Dis. 2008;67:1242–1248. doi: 10.1136/ard.2007.082099. [DOI] [PubMed] [Google Scholar]
- 29.Skoumal M., Haberhauer G., Feyertag J., Kittl E.M., Bauer K., Dunky A. Serum levels of cartilage oligomeric matrix protein are elevated in rheumatoid arthritis, but not in inflammatory rheumatic diseases such as psoriatic arthritis, reactive arthritis: Raynaud's syndrome, scleroderma, systemic lupus erythematosus, vasculitis and Sjogren's syndrome. Arthritis Res Ther. 2004;6:73–74. doi: 10.1186/ar1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto M., Takahashi H., Suzuki C., Naishiro Y., Yamamoto H., Imai K., Shinomura Y. Cartilage oligomeric matrix protein in systemic sclerosis. Rheumatology (Oxford) 2007;46:1858–1859. doi: 10.1093/rheumatology/kem254. [DOI] [PubMed] [Google Scholar]
- 31.Smith R.K., Zunino L., Webbon P.M., Heinegard D. The distribution of cartilage oligomeric matrix protein (COMP) in tendon and its variation with tendon site, age and load. Matrix Biol. 1997;16:255–271. doi: 10.1016/s0945-053x(97)90014-7. [DOI] [PubMed] [Google Scholar]
- 32.Gagarina V., Carlberg A.L., Pereira-Mouries L., Hall D.J. Cartilage oligomeric matrix protein protects cells against death by elevating members of the IAP family of survival proteins. J Biol Chem. 2008;283:648–659. doi: 10.1074/jbc.M704035200. [DOI] [PubMed] [Google Scholar]
- 33.Shih B., McGrouther D.A., Bayat A. Identification of novel keloid biomarkers through profiling of tissue biopsies versus cell cultures in keloid margin specimens compared to adjacent normal skin. Eplasty. 2010;10:e24. [PMC free article] [PubMed] [Google Scholar]
- 34.Recklies A.D., Baillargeon L., White C. Regulation of cartilage oligomeric matrix protein synthesis in human synovial cells and articular chondrocytes. Arthritis Rheum. 1998;41:997–1006. doi: 10.1002/1529-0131(199806)41:6<997::AID-ART6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 35.Briggs M.D., Hoffman S.M., King L.M., Olsen A.S., Mohrenweiser H., Leroy J.G., Mortier G.R., Rimoin D.L., Lachman R.S., Gaines E.S. Pseudoachondroplasia and multiple epiphyseal dysplasia due to mutations in the cartilage oligomeric matrix protein gene. Nat Genet. 1995;10:330–336. doi: 10.1038/ng0795-330. [DOI] [PubMed] [Google Scholar]
- 36.Hecht J.T., Nelson L.D., Crowder E., Wang Y., Elder F.F., Harrison W.R., Francomano C.A., Prange C.K., Lennon G.G., Deere M. Mutations in exon 17B of cartilage oligomeric matrix protein (COMP) cause pseudoachondroplasia. Nat Genet. 1995;10:325–329. doi: 10.1038/ng0795-325. [DOI] [PubMed] [Google Scholar]
- 37.Tseng S., Reddi A.H., Di Cesare P.E. Cartilage Oligomeric Matrix Protein (COMP): A Biomarker of Arthritis. Biomark Insights. 2009;4:33–44. doi: 10.4137/bmi.s645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jordan J.M. Update on cartilage oligomeric matrix protein as a marker of osteoarthritis. J Rheumatol. 2005;32:1145–1147. [PubMed] [Google Scholar]
- 39.Nakashima M., Chung S., Takahashi A., Kamatani N., Kawaguchi T., Tsunoda T., Hosono N., Kubo M., Nakamura Y., Zembutsu H. A genome-wide association study identifies four susceptibility loci for keloid in the Japanese population. Nat Genet. 2010;42:768–771. doi: 10.1038/ng.645. [DOI] [PubMed] [Google Scholar]
- 40.Newton G., Weremowicz S., Morton C.C., Jenkins N.A., Gilbert D.J., Copeland N.G., Lawler J. The thrombospondin-4 gene. Mamm Genome. 1999;10:1010–1016. doi: 10.1007/s003359901149. [DOI] [PubMed] [Google Scholar]
- 41.Russell S.B., Russell J.D., Trupin K.M., Gayden A.E., Opalenik S.R., Nanney L.B., Broquist A.H., Raju L., Williams S.M. Epigenetically altered wound healing in keloid fibroblasts. J Invest Dermatol. 2010;130:2489–2496. doi: 10.1038/jid.2010.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rock M.J., Holden P., Horton W.A., Cohn D.H. Cartilage oligomeric matrix protein promotes cell attachment via two independent mechanisms involving CD47 and alphaVbeta3 integrin. Mol Cell Biochem. 2010;338:215–224. doi: 10.1007/s11010-009-0355-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen W., Fu X., Sun X., Sun T., Zhao Z., Sheng Z. Analysis of differentially expressed genes in keloids and normal skin with cDNA microarray. J Surg Res. 2003;113:208–216. doi: 10.1016/s0022-4804(03)00188-4. [DOI] [PubMed] [Google Scholar]


