Abstract
We detected HIV-1 DNA in pure populations of perivascular macrophages, parenchymal microglia, and astrocytes, isolated using laser microdissection from brain tissue of five untreated individuals who died in the presymptomatic stage of infection from non-HIV causes. HIV-1 DNA was detected in the three cell populations, most consistently in perivascular macrophages, without evidence of productive infection. The percentage of PCR reactions detecting HIV-1 DNA in perivascular macrophages correlated inversely with peripheral blood CD4 counts. These findings demonstrate that brain cell reservoirs of latent HIV-1 exist before pathological HIV encephalitis and suggest that perivascular macrophage trafficking of latent virus into the brain increases with immunosuppression.
Establishing whether a specific cellular site of latent HIV-1 infection exists within the brain before the development of pathological HIV encephalitis (HIVE) is critical to understanding the nature of the central nervous system (CNS) HIV reservoir. The brain cell types or type within which virus is latent (DNA detection in the absence of detectable viral protein) has not been established. Studies have been hampered by the limited availability of autopsy brain tissue from individuals who have died before end-stage HIV disease and by techniques that lack both cell specificity and sensitivity for low copy viral DNA detection.
We studied a unique cohort of HIV-positive, presymptomatic individuals who died before pathological evidence of HIVE and used microdissection techniques to obtain pure cell populations from specific brain regions. Genomic DNA extracted from astrocytes, perivascular macrophages, and parenchymal microglia were used as a template in a highly sensitive, triple-nested PCR to amplify and sequence a 119-bp fragment of the HIV-1 gag gene. These techniques achieved a specificity and sensitivity not previously possible and enabled us to detect latent HIV-1 in resident brain cells, suggesting that cellular viral reservoirs exist in the brain before onset of HIVE.
Materials and Methods
Brain Pathology
Autopsy brain tissue blocks were obtained from the Medical Research Council HIV Tissue Resource (Edinburgh) and were examined according to guidelines endorsed by the Alfred Hospital Human Ethics committee (HREC #81/07). Five individuals with untreated, established HIV-1 infection who died from HIV-unrelated causes were examined. They had no evidence of HIVE, nor of any other HIV-related neuropathology at autopsy. Similarly, there was no evidence of any AIDS-defining infection, malignancy, or neurological disease.1 These individuals were therefore considered to be presymptomatic, with a spectrum of immunodeficiency. Brain tissue from two HIVE and two HIV-negative individuals were used as positive and negative controls, respectively. HIVE was diagnosed pathologically by productive infection of HIV-1 (as evidenced by p24 expression) of perivascular macrophages with or without giant cells with evident microglia activation and/or microglial nodules. Brains were routinely fixed, sectioned, and embedded. Using clean microtome blades, sections (5 μm thick) from the occipital cortex were cut, placed onto charged glass slides, and air-dried overnight before immunohistochemical staining.
Cell and HIV-1 Identification
Routine H&E staining was performed. A 1:50 dilution of a glial fibrillary acidic protein (GFAP) antibody (Zymed Laboratories, South San Francisco, CA) was used to detect astrocytes. A 1:200 dilution of a CD68 antibody (DakoCytomation, Glostrup, Denmark) was used to detect cells of macrophage lineage. Perivascular macrophages and parenchymal microglia were distinguishable by shape and location. A 1:40 dilution of p24 antibody (DakoCytomation) was used to detect HIV-1 protein. All antibodies were used with the avidin-biotin complex method and a diaminobenzidine hydrogen peroxide product as the colorimetric substrate. In situ proximity ligation assay (Duolink II; Olink Bioscience, Uppsala, Sweden) was also performed for single HIV-1 protein recognition using a 1:40 dilution of p24 antibody (DakoCytomation), according to the manufacturer's protocol. Tissue sections were assessed by a neuropathologist (C.A.M.) and with Image-Pro Plus 6.0 image analysis software (MediaCybernetics, Bethesda, MD) for immunopositive cells, as described previously.2
Laser Microdissection
Perivascular macrophages, parenchymal microglia, and astrocytes within the white matter from the occipital region of the cerebral cortex were identified as described above and were laser dissected (P.A.L.M. Microlaser Technologies, Oberkochen/Bernried, Germany) from uncovered IHC stained sections on slides and collected as pure cell populations for subsequent analysis. The occipital region provided a representative view of the immune reaction to HIV-1 in the brain.2 At least 200 cells spanning the entire tissue section were collected from each of the three cell populations, in duplicate from each individual.3,4 Neurons were microdissected from adjacent cortical regions to provide internal negative controls, as described previously.3 Genomic DNA was extracted using the Arcturus PicoPure DNA extraction kit (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol.
Amplification and DNA Sequence Analysis of HIV-1 GAG
HIV-1 gag sequences were amplified from genomic DNA prepared from laser dissected, captured cell populations. KAPA2G robust hot start DNA polymerase (Kapa Biosystems, Boston, MA) was used to generate a 119-nucleotide (nt) gag fragment in a highly sensitive triple-nested PCR strategy using 40 cycles per round, as described previously.3,5 Primers used were designed against a highly conserved gag region from the HIV-1 clade B consensus sequence. The nucleotide (nt) numbers represent their position relative to the HIV-1 NL4.3 strain: round 1: forward 5′-AGAACCAAGGGGAAGTGACA-3′ nt 1476-1496; reverse 5′-TTGGACCAACAAGGTTTCTGT-3′ nt 1761-1741. Round 2: forward 5′-CCCTTCAGGAACAAATAGGATG-3′ nt 1514–1536; reverse 5′-GAAGCTTGCTCGGCTCTTAG-3′ nt 1718-1699. Round 3: forward 5′-TCCACCTATCCCAGTAGGAGAA-3′ nt 1548-1569; reverse 5′-AGGGTTCCTTTGGTCCTTGT-3′ nt 1666-1647.
The sensitivity of the triple-nested PCR was determined using a plasmid-containing HIV DNA of known concentration as a template in the triple-nested PCR.5 Cell populations were collected in duplicate, and the triple-nested PCR was performed on each cell population at least 10 times to determine the frequency of detection. Integrity of DNA was established by performing a similar highly sensitive PCR analysis of cellular GAPDH levels, as described previously.3 PCR products were sequenced at the Applied Genetic Diagnostics laboratory, Melbourne University (Parkville, Australia), and were aligned against the clade B consensus sequence using DNAMAN 6.0 software (Lynnon, Pointe-Claire, QC, Canada).
Statistical Analysis
Quantitative data were analyzed using nonparametric tests (Mann–Whitney U-test, Spearman rank correlation) in the Stata 10.1 software package (StataCorp, College Station, TX).
Results
Clinical details are summarized in Table 1. Neuropathological analysis detected microglial and astrocytic reactivity in the absence of HIV-1 p24 protein in brain tissue from the five cases (Figure 1). Image analysis confirmed increased microglia and astrocyte activation within cases, compared with HIV-1 negative controls (P = 0.05). Activation of all cell types within HIVE was increased, compared with the cases and HIV-1-negative groups (P = 0.05) (Table 1).
Table 1.
Clinical Backgrounds, Immunohistochemical Detection, and PCR Detection of HIV-1 gag DNA in Brain Cell Populations of Study Subjects
| Subject code no. | Sex | Age (years) | Cause of death | Hepatitis |
CD4 T-cell count⁎ | Clinical neurological status | Detection of HIV-1 gag DNA [n/N (% detection rate)†] |
Cell count‡ (no.) |
IHC staining§ (%) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | C | PVM | Micro | Astro | PVM | Micro | Astro | PVM, Micro | Astro | ||||||
| HIV-positive, encephalitic | |||||||||||||||
| 75 | M | 29 | Sepsis | + | − | 0 | HIVD | 3/11 (27) | 1/11 (9) | 2/18 (11) | 7.40 | 29.80 | 9.80 | 0.143167 | 0.814018 |
| 138 | M | 59 | Sepsis | − | − | 6 | HIVD | 4/12 (33) | 1/12 (8) | 4/24 (17) | 7.00 | 24.80 | 9.20 | 0.118800 | 0.742597 |
| Mean | 30 | 9 | 14 | 7.20¶ | 27.30¶ | 9.50⁎ | 0.130984¶ | 0.778308¶ | |||||||
| HIV-positive, presymptomatic | |||||||||||||||
| 245 | M | 35 | Cirrhosis | + | + | 18 | No CI | 4/15 (27) | 1/10 (10) | 1/11 (9) | 2.70 | 9.90 | 7.20 | 0.012492 | 0.593083 |
| 205 | M | 34 | OD | + | − | 80 | No CI | 2/10 (20) | 2/18 (11) | 1/10 (10) | 1.50 | 18.10 | 4.60 | 0.012518 | 0.262532 |
| 305 | M | 34 | Cirrhosis | − | + | 229 | No CI | 2/10 (20) | 0/18 (0) | 4/29 (14) | 0.40 | 8.90 | 4.20 | 0.012617 | 0.116776 |
| 281 | F | 29 | OD | + | − | 260 | ND | 2/20 (10) | 2/20 (10) | 5/22 (23) | 2.90 | 9.80 | 1.80 | 0.026765 | 0.257186 |
| 240 | M | 40 | OD | + | + | 496 | No CI | 1/12 (8) | 6/16 (38) | 0/13 (0) | 3.10 | 10.40 | 3.20 | 0.082078 | 0.410261 |
| Mean | 17 | 14 | 11 | 2.12 | 11.42∥ | 4.2 | 0.029294∥ | 0.327968∥ | |||||||
| HIV-negative | |||||||||||||||
| 310 | M | 45 | RTA | − | − | ND | No CI | 0/10 (0) | 0/10 (0) | 0/11 (0) | 0.70 | 4.70 | 3.10 | 0.002492 | 0.056377 |
| 170 | M | 60 | Sepsis⁎⁎ | − | − | ND | No CI | 0/10 (0) | 0/10 (0) | 0/11 (0) | 0.40 | 3.20 | 2.20 | 0.004189 | 0.034862 |
| Mean | 0 | 0 | 0 | 0.55 | 3.95 | 2.65 | 0.003341 | 0.045620 | |||||||
Astro, astrocytes; CI, cognitive impairment; F, Female; HIVD, HIV-associated dementia; M, Male; Micro, parenchymal microglia; ND, no data; OD, alcohol or drug overdose; PVM, perivascular macrophage; RTA, road traffic accident.
Last recorded CD4 T-cell count.
Detection of HIV-1 gag DNA from microdissected cell populations by triple-nested PCR. The number of positive (HIV-1 gag DNA detected) PCR reactions divided by the total number of triple-nested PCRs performed on each cell population (also expressed as a percentage).
Mean of 10 fields counted under a 40χ objective.
Total area stained (expressed as a percentage).
Increased activation or cell numbers compared with corresponding brain cell populations in the HIV-positive presymptomatic group and HIV-negative group (P = 0.05).
Increased activation or cell numbers compared with corresponding brain cell populations in the HIV-negative group (P = 0.05).
Immunosuppressed (lung transplant recipient).
Figure 1.
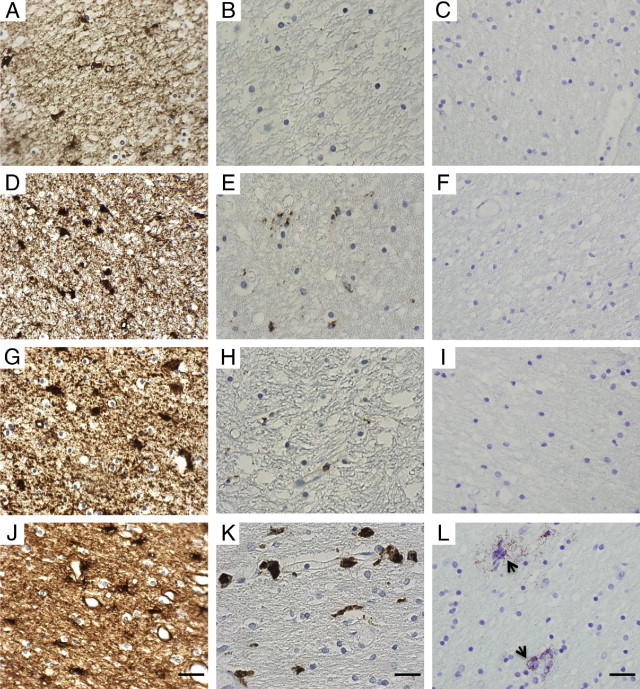
Representative photomicrographs of immunohistochemically stained sections of the white matter from the occipital region of the cerebral cortex. A–C: HIV-negative, subject code no. 310. D–F: HIV-positive presymptomatic, code no. 240. G–I: HIV-positive presymptomatic, code no. 245. J–L: HIV-positive encephalitic, code no. 138. A, D, G, and J: GFAP immunoreaction, showing baseline astrocyte numbers in controls and increased astrocyte reactivity in both HIV-positive presymptomatic and HIV-positive encephalitic cases. B, E, H, and K: CD68 immunoreaction, showing baseline microglia in controls with a minor increase in macrophage/microglial reactivity in HIV-positive presymptomatic cases, and prominent reactivity in HIV-positive encephalitic cases. C, F, I, and L:In situ proximity ligation assay for detection of individual p24 protein, showing no evidence of p24 protein (C, F, and I) or showing p24 positivity (L), indicating productive infection with HIV protein (arrows). Hematoxylin counterstain. Scale bars = 40 μm.
Triple-nested PCR was used to determine the presence of HIV-1 gag DNA within genomic DNA extracted from perivascular macrophages, parenchymal microglia, and astrocytes. The sensitivity of the triple-nested PCR was determined to be ≥1 copies of HIV-1 gag DNA per reaction. The gag region was successfully amplified from perivascular macrophages for all five cases (Table 1 and Figure 2A). The gag region was successfully amplified from astrocytes in four of the five cases (not from subject code no. 240), and from parenchymal microglial cells in four of the five cases (not from subject code no. 305). The gag region was successfully amplified in all cell populations in both HIVE individuals and was detected in a greater proportion of perivascular macrophages compared with presymptomatic individuals. HIV-1 gag DNA was not detected in cell populations from HIV-1-negative controls or cortical neurons from HIV-1-infected individuals. Sequence analysis confirmed HIV-1 gag DNA fragments were amplified from the cases specified. Mean percentage interpatient sequence variation in the gag gene is 5%, and the degree of sequence variation in gag correlates with the length of HIV-1 infection.6 Our sequence analysis in a short segment of the HIV-1 gag gene showed a mean variation from the clade B consensus sequence of 6% in the two HIV encephalitic individuals and 3% in the HIV presymptomatic cases, reflecting expected variation between patients. Resulting amino acid sequences were aligned against the HIV-1 gag consensus sequence (Figure 2C).
Figure 2.
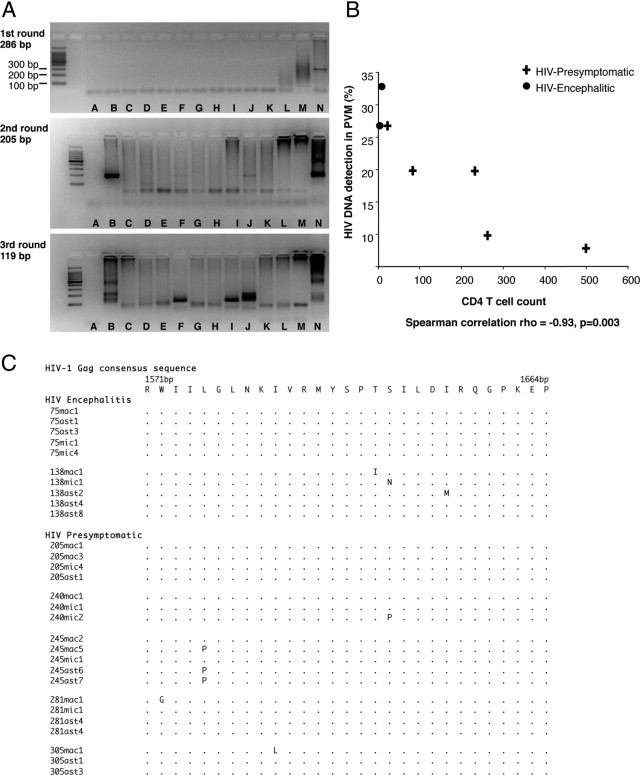
Analysis of HIV-1 gag DNA isolated from laser microdissected brain cell populations; astrocytes, perivascular macrophages (PVM), and parenchymal microglia (PM) from HIV-presymptomatic (PS) and HIV-encephalitic (HIVE) cases. A: Representative images of HIV-1 gag DNA PCR resulting in three rounds of PCR products (286, 205, and 119 bp). HIV-1 gag DNA was not consistently detected in PCR amplification from samples in which there were low levels of virus. Cell populations were therefore collected in duplicate (two collections of astrocytes per case), and the triple-nested PCR was performed on each cell population at least 10 times, to increase the frequency of detection. Not every PCR amplification from DNA of cell populations is illustrated (results of PCR amplifications of HIV-1 gag DNA from all cell populations are given in Table 1 and sequencing results are shown here in panel B). Lane A, negative PCR control (water as DNA template) carried through three rounds of PCR. Lane B, code no. 281 (PS), PVM. Lane C, code no. 305 (PS), PVM. Lane D, code no. 281, astrocytes. Lane E, code no. 305, astrocytes. Lane F, code no. 75 (HIVE), astrocytes. Lane G, code no. 75, PVM. Lane H, code no. 138 (HIVE), astrocytes. Lane I, code no. 138, PVM. Lane J, code no. 75, PM. Lane K, code no. 138, PM. Lane L, code no. 281, PM. Lane M, code no. 305, PM. Lane N, Lymph node of HIV-positive patient, positive PCR control (overamplification results in PCR product not running through the gel). B: Association between CD4 T-cell count and the frequency with which HIV-1 DNA was detected by triple-nested PCR in perivascular macrophages (PVM, expressed as a percentage) in the occipital cortex for each of the seven HIV-positive individuals. C: Amino acid sequence alignment of HIV-1 gag DNA from PCR products of laser microdissected brain cell populations. Sequences are aligned and numbered according to the HIV-1 gag consensus sequence. ast, astrocyte; mac, perivascular macrophages; mic, parenchymal microglia.
The frequency with which HIV-1 gag DNA was detected by triple-nested PCR in the perivascular macrophages of HIV-1-positive individuals correlated inversely with the peripheral blood CD4 count (Spearman correlation, ρ = −0.93, P = 0.003) (Figure 2B).
Discussion
The optimal time to begin combination antiretroviral therapy for chronically infected individuals remains to be determined, although current guidelines recommend starting treatment when CD4 cell counts are less than 350 cells/mL.7–9 Preventing the development of HIVE requires an understanding of how, where, and when the virus accesses the brain and whether a latent viral reservoir exists within the CNS, sequestered from systemic immune surveillance. If such a reservoir is present, current therapeutics may be unable to eliminate an established CNS reservoir, allowing later reactivation. Key research priorities for HIV-1 eradication include identifying anatomical and cellular reservoirs, to allow the development of targeted strategies that eliminate virus from these sites.10
PCR studies have confirmed low levels of HIV-1 DNA in homogenized brain samples of some presymptomatic subjects without evidence of productive HIV infection.11 Nonetheless, there is still no conclusive evidence as to which brain cells are harboring virus before the onset of HIVE.12 Our ability to detect HIV-1 DNA from specific brain cell types in a cohort of individuals who died from HIV-unrelated causes during presymptomatic infection presented a unique opportunity to identify cellular viral reservoirs of the brain. Our detection of HIV-1 gag DNA in long-lived brain cells without evidence of productive infection suggests that brain cell reservoirs of latent virus exist before HIVE onset. By the time of HIVE, both perivascular macrophages and parenchymal microglia demonstrate productive HIV-1 infection in the brain, but astrocytes, which have been shown to contribute to neuropathogenesis, are not productively infected.3,13,14
Parenchymal microglia are a stable, long-lived CNS cell population that form a dense network throughout the brain. Their anatomical location and morphology is distinct from that of perivascular macrophages. Latently infected parenchymal microglia in presymptomatic HIV-1 infection may well represent a constant CNS viral reservoir. Circulating blood monocytes traffic into the perivascular space to become tissue perivascular macrophages and are constantly turned over via the systemic circulation.15–18 We previously examined brain cell-specific infection at multiple time points from acute to terminal infection in simian immunodeficiency virus (SIV) infected macaques. The study demonstrated that infected macrophages act as the Trojan horse within the brain perivascular space, with an initial increase in latent infection of perivascular macrophages and subsequent productive infection of perivascular macrophages and brain parenchymal cells concurrent with increasing plasma viral load and decreasing peripheral blood CD4 count.5 In our present study, we have similarly demonstrated increasing immunosuppression correlating with increasing latent HIV-1 infection of circulating perivascular macrophages before the onset of HIVE. Both studies suggest that perivascular macrophages may be pivotal in advancing HIV-1 brain infection at times of increasing immunosuppression and that maintaining CD4 cell levels may be crucial for controlling a critical entry point of HIV-1 from the systemic circulation into the brain. It is yet to be determined whether the onset of HIVE is due to reactivation of the microglial reservoir, influx of systemic HIV via infected macrophages, or both.
Our studies show microglial and astrocytic reactivity in the absence of HIV-1 p24 protein in brain tissue from presymptomatic individuals and that CD4 lymphocytic infiltration of the brain was negligible.2 Given latent virus detected within brain cells before HIVE, this low-level activation of microglia and astrocytes may indicate a response to trafficking of HIV-1 into the brain and/or a change in the replication state of the latent infection.
CNS infiltration of activated monocytes/macrophages and microglial activation have been widely reported to be essential in the development of HIV-associated neurological disorders.12,19–22 A recent study showed that risk of HIV-associated neurological disorders is associated with nadir CD4 count.23 Preventing CD4 decline by earlier initiation of combination antiretroviral therapy may reduce entry of HIV-1 infected perivascular macrophages from the systemic circulation into the brain and so reduce the likelihood of HIV-associated neurological disorders.
In conclusion, we provide evidence that brain cell reservoirs of latent HIV-1 exist before the onset of pathological HIVE and suggest that perivascular macrophages may be pivotal to brain disease progression at times of increasing immunosuppression. Therapies to prevent CNS HIV disease progression may thus need to target perivascular macrophages, before immunosuppression.
Acknowledgments
We thank Frances Carnie and the MRC HIV Brain and Tissue Bank (Edinburgh, UK) for providing samples and clinical information analyzed in this study. We also thank the staff of Anatomical Pathology, Alfred Hospital, for assistance with processing tissue samples.
Footnotes
Supported by an Australia National Health and Medical Research Council Peter Doherty fellowship (415006 to K.A.T.).
References
- 1.Centers for Disease Control and Prevention 1993 revised classification system for HIV infection and expanded surveillance case definition for AIDS among adolescents and adults. MMWR Recomm Rep. 1992;41(RR-17):1–19. [PubMed] [Google Scholar]
- 2.McCrossan M., Marsden M., Carnie F.W., Minnis S., Hansoti B., Anthony I.C., Brettle R.P., Bell J.E., Simmonds P. An immune control model for viral replication in the CNS during presymptomatic HIV infection. Brain. 2006;129:503–516. doi: 10.1093/brain/awh695. [DOI] [PubMed] [Google Scholar]
- 3.Thompson K.A., Churchill M.J., Gorry P.R., Sterjovski J., Oelrichs R.B., Wesselingh S.L., McLean C.A. Astrocyte specific viral strains in HIV dementia. Ann Neurol. 2004;56:873–877. doi: 10.1002/ana.20304. [DOI] [PubMed] [Google Scholar]
- 4.Trillo-Pazos G., Diamanturos A., Rislove L., Menza T., Chao W., Belem P., Sadiq S., Morgello S., Sharer L., Volsky D.J. Detection of HIV-1 DNA in microglia/macrophages, astrocytes and neurons isolated from brain tissue with HIV-1 encephalitis by laser capture microdissection. Brain Pathol. 2003;13:144–154. doi: 10.1111/j.1750-3639.2003.tb00014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson K.A., Varrone J.J., Jankovic-Karasoulos T., Wesselingh S.L., McLean C.A. Cell specific temporal infection of the central nervous system in a simian immunodeficiency virus model of human immunodeficiency virus encephalitis. J Neurovirol. 2009;15:300–311. doi: 10.1080/13550280903030125. [DOI] [PubMed] [Google Scholar]
- 6.Yoshimura F.K., Diem K., Learn E.H., Riddell S., Corey L. Intrapatient sequence variation of the gag gene of human immunodeficiency virus type 1 plasma virions. J Virol. 1996;70:8879–8887. doi: 10.1128/jvi.70.12.8879-8887.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panel on Antiretroviral Guidelines for Adults and Adolescents . Department of Health and Human Services; Washington, DC: January 10, 2011. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents.http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf last accessed August 6, 2011. [Google Scholar]
- 8.Hirsch M.S. Initiating therapy: when to start, what to use. J Infect Dis. 2008;197(Suppl 3):S252–S260. doi: 10.1086/533416. [DOI] [PubMed] [Google Scholar]
- 9.Gatell J.M. When and why to start antiretroviral therapy? J Antimicrob Chemother. 2010;65:383–385. doi: 10.1093/jac/dkp487. [DOI] [PubMed] [Google Scholar]
- 10.Thomas C. Roadblocks in HIV research: five questions. Nat Med. 2009;15:855–859. doi: 10.1038/nm0809-855. [DOI] [PubMed] [Google Scholar]
- 11.Bell J.E., Busuttil A., Ironside J.W., Rebus S., Donaldson Y.K., Simmonds P., Peutherer J.F. Human immunodeficiency virus and the brain: investigation of virus load and neuropathologic changes in pre-AIDS subjects. J Infect Dis. 1993;168:818–824. doi: 10.1093/infdis/168.4.818. [DOI] [PubMed] [Google Scholar]
- 12.Anthony I.C., Bell J.E. The neuropathology of HIV/AIDS. Int Rev Psychiatry. 2008;20:15–24. doi: 10.1080/09540260701862037. [DOI] [PubMed] [Google Scholar]
- 13.Churchill M.J., Wesselingh S.L., Cowley D., Pardo C.A., McArthur J.C., Brew B.J., Gorry P.R. Extensive astrocyte infection is prominent in human immunodeficiency virus–associated dementia. Ann Neurol. 2009;66:253–258. doi: 10.1002/ana.21697. [DOI] [PubMed] [Google Scholar]
- 14.Thompson K.A., McArthur J.C., Wesselingh S.L. Correlation between neurological progression and astrocyte apoptosis in HIV-associated dementia. Ann Neurol. 2001;49:745–752. doi: 10.1002/ana.1011. [DOI] [PubMed] [Google Scholar]
- 15.Kim W.K., Alvarez X., Fisher J., Bronfin B., Westmoreland S., McLaurin J., Williams K. CD163 identifies perivascular macrophages in normal and viral encephalitic brains and potential precursors to perivascular macrophages in blood. Am J Pathol. 2006;168:822–834. doi: 10.2353/ajpath.2006.050215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim W.-K., Corey S., Alvarez X., Williams K. Monocyte/macrophage traffic in HIV and SIV encephalitis. J Leukoc Biol. 2003;74:650–656. doi: 10.1189/jlb.0503207. [DOI] [PubMed] [Google Scholar]
- 17.Bechmann I., Kwidzinski E., Kovac A.D., Simbürger E., Horvath T., Gimsa U., Dirnagl U., Priller J., Nitsch R. Turnover of rat brain perivascular cells. Exp Neurol. 2001;168:242–249. doi: 10.1006/exnr.2000.7618. [DOI] [PubMed] [Google Scholar]
- 18.Williams K.C., Corey S., Westmoreland S.V., Pauley D., Knight H., deBakker C., Alvarez X., Lackner A.A. Perivascular macrophages are the primary cell type productively infected by simian immunodeficiency virus in the brains of macaques: implications for the neuropathogenesis of AIDS. J Exp Med. 2001;193:905–916. doi: 10.1084/jem.193.8.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González-Scarano F., Martín-García J. The neuropathogenesis of AIDS. Nat Rev Immunol. 2005;5:69–81. doi: 10.1038/nri1527. [DOI] [PubMed] [Google Scholar]
- 20.Bell J.E. An update on the neuropathology of HIV in the HAART era. Histopathology. 2004;45:549–559. doi: 10.1111/j.1365-2559.2004.02004.x. [DOI] [PubMed] [Google Scholar]
- 21.Gartner S. HIV infection and dementia. Science. 2000;287:602–604. doi: 10.1126/science.287.5453.602. [DOI] [PubMed] [Google Scholar]
- 22.Glass J.D., Fedor H., Wesselingh S.L., McArthur J.C. Immunocytochemical analysis of HIV-gp41 and macrophages in HIV-associated dementia. Ann Neurol. 1995;38:755–762. doi: 10.1002/ana.410380510. [DOI] [PubMed] [Google Scholar]
- 23.Heaton R.K., Clifford D.B., Franklin D.R., Jr, Woods S.P., Ake C., Vaida F., Ellis R.J., Letendre S.L., Marcotte T.D., Atkinson J.H., Rivera-Mindt M., Vigil O.R., Taylor M.J., Collier A.C., Marra C.M., Gelman B.B., McArthur J.C., Morgello S., Simpson D.M., McCutchan J.A., Abramson I., Gamst A., Fennema-Notestine C., Jernigan T.L., Wong J., Grant I., CHARTER Group HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology. 2010;75:2087–2096. doi: 10.1212/WNL.0b013e318200d727. [DOI] [PMC free article] [PubMed] [Google Scholar]


