Abstract
Posttransplantation lymphoproliferative disorders (PTLD) are associated with Epstein-Barr virus (EBV) and activate the NF-κB pathway. B-cell activating factor (BAFF) modulates cell growth and survival in non-Hodgkin's lymphomas. However, there are few studies of EBV, BAFF/BAFF-R signaling, and NF-κB1 and NF-κB2 pathway activation in PTLD. Diffuse large B-cell lymphomas (DLBCL) in two different clinical contexts, immunocompetent patients (DLBCL/IC; n = 30) or posttransplantation solid-organ recipients (DLBCL/PTLD; n = 21), were characterized histogenically as germinal center (GC) or non–germinal center (NGC). Expression of BAFF, BAFF-R, and NF-κB proteins p50 and p52 and the presence or absence of EBV were compared in these clinical contexts. Regardless of the GC or NGC pattern of DLBCL, BAFF-R was expressed in 37% of DLBCL/IC but in only 4.8% of DLBCL/PTLD. p52 was expressed in DLBCL/PTLD/NGC (12 of 19 cases) as compared with DLBCL/IC/NGC (0 of 18 cases). This pattern might be related to the presence of EBV and latent membrane protein 1 because p52 expression was observed primarily in EBV-positive DLBCL/PTLD cases expressing latent membrane protein 1. Thus, the activation profile or NGC pattern of DLBCL/PTLD was not associated with BAFF/BAFF-R expression, whereas nuclear p52 related to NF-κB2 pathway activation might be linked to EBV.
Posttransplantation lymphoproliferative disorders (PTLD) are among the worst prognostic complications after solid-organ transplantation.1 These lymphoid proliferations are characterized by their heterogeneity. Several categories of PTLD have been identified by the World Health Organization (World Health Organization classification). Among these, the B-cell monomorphic subcategory fulfills the criteria for diffuse large B-cell lymphomas (DLBCL) described in immunocompetent individuals.2 Studies of the histogenesis of PTLD also reflect this heterogeneity.3 Several studies of histogenesis origin using immunohistochemistry expression of CD10, BCL6, MUM1/IRF4, and CD138 demonstrated two patterns, germinal center (GC) and non–germinal center (NGC), including late GC/early post-GC and post-GC.4–7 Epstein-Barr virus (EBV)–positive cases were primarily associated with the NGC phenotype.6 EBV-negative PTLD are primarily described as late complications and are more aggressive than EBV-positive PTLD.8,9
EBV is a member of the herpesvirus family and is usually associated with mononucleosis and a wide variety of malignant lesions.10,11 Latent infection leads to expression of nine proteins including latent membrane proteins LMP1, LMP2A, and LMP2B and nuclear proteins EBNA1, EBNA2, EBN3A, EBN3B, and EBN3C, as well as the small nonencoded RNAs EBER1 and EBER2.8 Therefore, the transcriptional activator protein EBNA2 has been described as essential for B-cell transformation and LMP1 expression. The cytoplasmic C-terminal–located CTAR1 and CTAR2 regions of LMP1 activate the NF-κB2 and NF-κB1 pathways, respectively (canonical and non-canonical pathways).12
NF-κB is a dimeric transcription factor that is regulated by the IκB protein family. Two pathways have been identified: the canonical pathway (NF-κB1), which generates a p50/p65 dimer through Iκbα degradation by IKKβ, and the non-canonical pathway (NF-κB2) leading to p52/RelB dimer release via involvement of IKKα. These two pathways lead to translocation into the nucleus of these various dimers to modulate several genes.13 To date, several publications have reported constitutive expression of p65 in DLBCL subtypes.14,15
BAFF (B cell–activating factor), a member of the tumor necrosis factor family, increases the amount of peripheral B cells and is implicated in development of autoimmune diseases in a transgenic mice model overexpressing BAFF.16,17 Moreover, BAFF modulates cell growth and survival in multiple myeloma and B-cell chronic lymphocytic leukemia.18,19 BAFF is expressed in non–Hodgkin's lymphoma–like mantle and marginal zones, follicular lymphoma, and DLBCL.20
BAFF-R, also known as BR3, TNFRSF13C, or CD268, is a type III transmembrane protein that is considered the principal receptor of BAFF leading to the survival and maturation of primary B cells.21 BAFF-R is the only specific receptor of BAFF.22 Expression of BAFF-R was first detected in tonsils, and demonstrated strong staining on B cells of the mantle zone and weak staining of GC B lymphocytes.23 Similar staining has been also described in other studies. Furthermore, several non–Hodgkin's lymphomas have been investigated through BAFF-R expression, which was recently exhibited in most lymphoproliferative disorders.24–26
BAFF-R signaling activates NEMO-independent processing of the NF-κB2 pathway.27 Moreover, LMP1 activates the BAFF gene promoter via NF-κB. The ability of BAFF to increase cell survival and of BAFF-R and LMP1 to activate the NF-κB pathway, and the strong association of EBV in PTLD constitute at least three reliable reasons to study the role of EBV, NF-κB2, and BAFF-R in PTLD.
To analyze some differences in BAFF-R expression according to the activation level of B cells, we tested the expression of BAFF and BAFF-R in 51 DLBCL cases from the general population (30 patients) and immunodeficient patients (21 with PTLD). All cases were characterized according to morphologic features, immunophenotype, and GC or NGC histogenesis pattern. In EBV-positive cases, LMP1, EBNA2, and ZEBRA, an immediate replicative protein, were tested. The detection of nuclear or cytoplasmic expression of p50 and p52 proteins to analyze the activation of NF-κB pathways demonstrated a correlation between p52 expression and the presence of EBV, as well as latency II or III, ie, expression of LMP1 in PTLD.
Materials and Methods
Patients
Fifty-one DLBCL cases were collected. Patients enrolled in the present study were followed up in two solid-organ transplantation centers, the Bicêtre and Paul Brousse University hospitals. All lymphomas in this series were monomorphic B-cell lymphomas according to the criteria of the World Health Organization classification. Written informed consent was obtained from all included patients. The DLBCL cases comprised two subgroups: 30 immunocompetent patients with DLBCL (DLBCL/IC) and 21 solid-organ recipients with DLBCL (DLBCL/PTLD). Patient clinical data are given in Table 1. Most DLBCL cases, either DLBCL/IC or DLBCL/PTLD, were localized in lymph nodes (34 of 51). Mean age of patients with DLBCL/PTLD at diagnosis of lymphoma was 39 years (range, 1 to 74 years), and of patients with DLBCL/IC was 61.5 years (range, 18 to 92 years).
Table 1.
Clinical Data in Patients with DLBCL
| Patients | Age at diagnosis (year) | Sex | Graft type | Delay of lymphoma occurrence (mo) | Localization | Histologic type |
|---|---|---|---|---|---|---|
| 1 | 35 | M | NR | NR | Lymph nodes | DLBCL/IC |
| 2 | 55 | F | NR | NR | Cavum | DLBCL/IC |
| 3 | 32 | M | NR | NR | Tonsils | DLBCL/IC |
| 4 | 63 | F | NR | NR | Lymph nodes | DLBCL/IC |
| 5 | 81 | F | NR | NR | Lymph nodes | DLBCL/IC |
| 6 | 81 | F | NR | NR | Lymph nodes | DLBCL/IC |
| 7 | 65 | M | NR | NR | Lymph nodes | DLBCL/IC |
| 8 | 60 | M | NR | NR | Lymph nodes | DLBCL/IC |
| 9 | 75 | M | NR | NR | Lymph nodes | DLBCL/IC |
| 10 | 92 | F | NR | NR | Lymph nodes | DLBCL/IC |
| 11 | 18 | M | NR | NR | Lymph nodes | DLBCL/IC |
| 12 | 73 | M | NR | NR | Lymph nodes | DLBCL/IC |
| 13 | 52 | M | NR | NR | Lymph nodes | DLBCL/IC |
| 14 | 51 | F | NR | NR | Lymph nodes | DLBCL/IC |
| 15 | 85 | M | NR | NR | Lymph nodes | DLBCL/IC |
| 16 | 75 | M | NR | NR | Lymph nodes | DLBCL/IC |
| 17 | 82 | F | NR | NR | ORL | DLBCL/IC |
| 18 | 77 | F | NR | NR | Lymph nodes | DLBCL/IC |
| 19 | 79 | F | NR | NR | Lymph nodes | DLBCL/IC |
| 20 | 58 | F | NR | NR | ORL | DLBCL/IC |
| 21 | 80 | F | NR | NR | Lymph nodes | DLBCL/IC |
| 22 | 34 | F | NR | NR | Kidney | DLBCL/IC |
| 23 | 46 | F | NR | NR | Cavum | DLBCL/IC |
| 24 | 58 | F | NR | NR | Lymph nodes | DLBCL/IC |
| 25 | 71 | M | NR | NR | Lymph nodes | DLBCL/IC |
| 26 | 53 | M | NR | NR | Lymph nodes | DLBCL/IC |
| 27 | 58 | M | NR | NR | Gut | DLBCL/IC |
| 28 | 56 | M | NR | NR | Gallbladder | DLBCL/IC |
| 29 | 60 | M | NR | NR | Lymph nodes | DLBCL/IC |
| 30 | 89 | M | NR | NR | Lymph nodes | DLBCL/IC |
| 31 | 51 | M | Liver | 109 | Liver | DLBCL/PTLD |
| 32 | 16 | M | Liver | 5 | Cavum | DLBCL/PTLD |
| 33 | 22 | M | Liver | 1 | Lymph nodes | DLBCL/PTLD |
| 34 | 51 | M | Lung | 6 | Lung | DLBCL/PTLD |
| 35 | 64 | F | Kidney | 114 | Lymph nodes | DLBCL/PTLD |
| 36 | 3 | F | Liver | 27 | ORL | DLBCL/PTLD |
| 37 | 2 | F | Liver | 6 | Liver | DLBCL/PTLD |
| 38 | 5 | F | Liver | 32 | Tonsil | DLBCL/PTLD |
| 39 | 56 | M | Liver | 109 | Lymph nodes | DLBCL/PTLD |
| 40 | 3 | F | Liver | 2 | Cavum | DLBCL/PTLD |
| 41 | 66 | F | Kidney | 259 | Small intestine | DLBCL/PTLD |
| 42 | 39 | M | Kidney | 84 | Lymph nodes | DLBCL/PTLD |
| 43 | 74 | M | Liver | 49 | Lymph nodes | DLBCL/PTLD |
| 44 | 2 | F | Liver | 7 | Tonsils | DLBCL/PTLD |
| 45 | 54 | M | Kidney + pancreas | 5 | Brain | DLBCL/PTLD |
| 46 | 3 | M | Liver | 5 | Lymph nodes | DLBCL/PTLD |
| 47 | 49 | M | Kidney + pancreas | 162 | Lymph nodes | DLBCL/PTLD |
| 48 | 1 | M | Liver | 10 | Tonsil | DLBCL/PTLD |
| 49 | 36 | F | Kidney | 152 | Peritoneum | DLBCL/PTLD |
| 50 | 53 | F | Kidney | 192 | Lymph nodes | DLBCL/PTLD |
| 51 | 59 | M | Kidney | 81 | Brain | DLBCL/PTLD |
DLBCL, diffuse large B-cell lymphoma; DLBCL/IC, DLBCL in immunocompetent patients; F, female; M, male; NR, not relevant; ORL, otorhinolaryngology; PTLD, post-transplantation lymphoproliferative disorder.
Conventional Morphologic Analysis and Immunostaining
Formalin-fixed, paraffin-embedded tissue sections from DLBCL biopsy specimens were stained using H&E for conventional morphologic analysis. The slides were reviewed by two hematopathologists (M.R. and T.L.) to reclassify cases according to the World Health Organization criteria. Immunostaining was performed on paraffin sections using an immunoperoxidase technique (ChemMate EnVision Detection Kit, DakoCytomation A/S, Glostrup, Denmark, or the Vectastain Elite ABC kit, Vector Laboratories, Inc., Burlingame, CA) according to the manufacturer's recommendations. To identify GC or NGC pattern, the following B-cell markers were tested: CD20 (clone L26; dilution 1:1950; DakoCytomation A/S), CD10 (clone 56C6; dilution 1:40; Novocastra, Newcastle-upon-Tyne, England), BCL6 (clone PG-B6p; dilution 1:40; DakoCytomation A/S), IRF4/MUM1 (clone M-17; dilution 1:400; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), and CD138 (syndecan-1; clone B-A38; dilution 1:100; Diaclone, Stamford, CT).
Using the same technique, expression of EBV latency proteins LMP1 (clone CS.1–4; dilution 1:100), EBNA2 (clone PE2; dilution 1:100), and the replicative protein ZEBRA (clone BZ.1; dilution 1:25) (all three from DakoCytomation A/S) were studied, as was expression of BAFF (clone buffy-2; dilution 1:100) and BAFF-R (clone 11C1; dilution 1:600) (both from Alexis Biochemicals Corp., Lausen, Switzerland). For BAFF and BAFF-R, cases exhibiting more than 30% of immunostained tumor cells were considered positive.5
Protein expression of p50 (dilution 1:40; Santa Cruz Biotechnology, Inc.) and p52 (dilution 1:400; Millipore Corp., Billerica, MA) was also assessed using immunoperoxidase to explore NF-κB pathways. Localization of staining was specified as nuclear (activated NF-κB pathway) or cytoplasmic.
Detection and Characterization of EBV
EBV status of all DLBCL was performed via in situ hybridization using EBER1 and EBER2 probes with the PNA In Situ Hybridization Detection Kit (DakoCytomation A/S) according to the manufacturer's recommendations. In EBV, EBER-positive cases, latency was defined as latency I (EBER-positive, LMP1-negative, or EBNA2-negative), latency II (EBER-positive, LMP1-positive, or EBNA2-negative), and latency III (EBER-positive, LMP1-positive, or EBNA2-positive).
Controls
External controls tested included reactive lymph nodes or tonsils and lymphomatous tissues known to express BAFF and BAFF-R: follicular lymphoma, 6 cases; mantle cell lymphoma, 5 cases; marginal zone B-cell lymphoma, 4 cases; MALT (mucosa-associated lymphoid tissue) lymphoma, 6 cases; and Burkitt's lymphoma, 6 cases.
Results
Characterization and Histogenesis of DLBCL/IC and DLBCL/PTLD
The definition of GC origin means a GC B-cell pattern having the expression of CD10 and/or BCL6 or activated GC B-cell pattern according to expression of at least one GC B-cell marker, either BCL6 or CD10, and at least one activation marker, either IRF4/MUM1 or CD138. The activated NGC B-cell pattern means expression of activation markers IRF4/MUM1 and/or CD138 but not GC B-cell markers.7 DLBCL/PTLD specimens generally demonstrated an activated pattern (Tables 2 and 3): of 21 cases, 19 expressed the NGC phenotype, and 2 could not be characterized because of nondetection of the four markers.28 Of 30 DLBCL/IC cases, 12 expressed the GC pattern, and 18 expressed the NGC pattern (Table 3).
Table 2.
Immunohistochemical Staining of DLBCL Samples
| DLBCL | CD10 | BCL6 | IRF4 | CD138 | BAFF | BAFF-R | EBER | LMP1 | EBNA2 | ZEBRA | P52 | P50 | Origin |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | − | + | − | − | +++ | +++ | − | NR | NR | NR | − | − | GC |
| 2 | − | + | − | − | +++ | + | − | NR | NR | NR | Nu | Nu | GC |
| 3 | + | + | − | − | +++ | + | − | NR | NR | NR | − | − | GC |
| 4 | + | − | − | − | +++ | − | − | − | NR | NR | − | − | GC |
| 5 | − | + | − | − | +++ | − | − | − | NR | NR | Nu | − | GC |
| 6 | + | + | − | - | +++ | − | − | NR | NR | NR | − | − | GC |
| 7 | + | + | − | − | ++ | − | − | NR | NR | NR | − | − | GC |
| 8 | + | + | − | − | +++ | − | − | NR | NR | NR | Nu | Nu | GC |
| 9 | + | + | − | − | + | + | − | NR | NR | NR | − | − | GC |
| 10 | + | + | − | − | +++ | − | − | NR | NR | NR | Nu | − | GC |
| 11 | + | + | - | − | +++ | +++ | − | NR | NR | NR | Nu | − | GC |
| 12 | + | + | − | − | − | +++ | − | NR | NR | NR | − | Nu | GC |
| 13 | − | + | + | − | +++ | ++ | − | NR | NR | NR | − | − | NGC |
| 14 | − | + | + | − | +++ | + | − | NR | NR | NR | − | − | NGC |
| 15 | − | + | + | − | +++ | + | − | NR | NR | NR | − | − | NGC |
| 16 | − | − | + | − | + | + | − | NR | NR | NR | − | − | NGC |
| 17 | − | − | + | − | +++ | − | − | NR | NR | NR | − | − | NGC |
| 18 | − | − | + | − | +++ | − | − | NR | NR | NR | − | − | NGC |
| 19 | − | − | + | − | +++ | − | − | NR | NR | NR | − | − | NGC |
| 20 | − | − | + | − | +++ | − | − | NR | NR | NR | − | Nu | NGC |
| 21 | − | − | + | − | +++ | − | − | NR | NR | NR | − | − | NGC |
| 22 | − | − | + | + | +++ | − | + | + | + | − | − | Nu | NGC |
| 23 | − | + | + | − | +++ | − | − | NR | NR | NR | − | − | NGC |
| 24 | − | − | + | + | − | − | + | + | − | + | − | Nu | NGC |
| 25 | − | − | + | − | ++ | − | − | NR | NR | NR | − | − | NGC |
| 26 | − | ++ | + | − | +++ | − | − | NR | NR | NR | − | − | NGC |
| 27 | − | + | + | − | ++ | +++ | − | NR | NR | NR | − | − | NGC |
| 28 | − | − | + | − | ++ | − | − | NR | NR | NR | − | − | NGC |
| 29 | − | + | + | − | ++ | − | − | NR | NR | NR | − | − | NGC |
| 30 | − | + | + | − | + | − | − | NR | NR | NR | − | − | NGC |
| 31 | + | − | − | + | +++ | − | − | NR | NR | NR | − | − | NGC |
| 32 | − | + | + | − | +++ | − | − | NR | NR | NR | Nu | Nu | NGC |
| 33 | − | + | − | + | − | − | + | + | ND | + | Nu | ND | NGC |
| 34 | − | − | + | + | +++ | − | − | NR | NR | NR | − | Nu | NGC |
| 35 | − | − | + | − | + | − | − | NR | NR | NR | − | − | NGC |
| 36 | − | − | + | + | +++ | − | + | + | − | + | − | Nu | NGC |
| 37 | − | − | − | + | +++ | − | + | + | + | + | Nu | Nu | NGC |
| 38 | − | − | + | + | + | + | + | + | + | + | Nu | Nu | NGC |
| 39 | − | − | + | − | + | − | − | NR | NR | NR | − | ND | NGC |
| 40 | − | − | + | + | + | − | + | + | + | + | Nu | Nu | NGC |
| 41 | − | − | + | − | + | − | + | − | − | − | − | − | NGC |
| 42 | − | − | + | − | − | − | − | NR | NR | NR | − | − | NGC |
| 43 | − | − | + | − | + | − | − | NR | NR | NR | Nu | − | NGC |
| 44 | − | − | + | + | + | − | + | + | + | + | Nu | − | NGC |
| 45 | − | + | + | + | + | − | + | + | + | − | Nu | Nu | NGC |
| 46 | − | − | + | + | +++ | − | + | + | + | − | Nu | − | NGC |
| 47 | − | − | + | − | ++ | − | − | NR | NR | NR | Nu | − | NGC |
| 48 | − | − | + | + | +++ | − | + | + | − | + | Nu | − | NGC |
| 49 | − | − | + | + | +++ | − | − | NR | NR | NR | Nu | − | NGC |
| 50 | − | − | − | - | +++ | − | + | − | − | − | Nu | Nu | ND |
| 51 | − | − | − | − | +++ | − | + | + | + | + | Nu | − | ND |
Nos. 1 to 30, DLBCL/IC; 31 to 51, DLBCL/PTLD.
BAFF/BAFF-R staining, semiquantitative evaluation: +++, >60%; ++, 30% to 60%; +, <30%; −, negative. For other markers: +, >30%; −, negative.
GC, germinal center; ND, not determined; NGC, non–germinal center; NR, not relevant; Nu, nuclear staining.
Table 3.
Expression of BAFF/BAFF-R, NF-κB Proteins, and EBV in Patients with DLBCL
| Lymphoma type | Origin | No. of patients | BAFF | BAFF-R | p52 | p50 | EBV | LMP1 |
|---|---|---|---|---|---|---|---|---|
| IC | GC | 12 | 11 of 12 | 6 of 12 | 5 of 12 | 3 of 12 | 0 | ND |
| IC | NGC | 18 | 17 of 18 | 5 of 18 | 0 of 18 | 3 of 18 | 2/18 | 2/2 |
| PTLD | NGC | 19 | 17 of 19 | 1 of 19⁎ | 12 of 19⁎ | 7 of 17 | 10 of 19 | 9 of 10 |
| PTLD | ND | 2 | 2 of 2 | 0 of 2 | 2 of 2 | 1 of 2 | 2 of 2 | 1 of 2 |
GC, germinal center; IC, immunocompetent; EBV, Epstein-Barr virus; LMP1, latent membrane protein 1; ND, not determined; NGC, non–germinal center; p50 and p52 positive staining, nuclear localization (>10%); PTLD, post-transplantation lymphoproliferative disorders.
Statistically significant: P < 2.10−4 for p52, P < 0.008 for BAFF-R.
Detection and Characterization of EBV
EBV was detected in 12 of 21 DLBCL/PTLD cases (57%) versus only 2 of 30 DLBCL/IC cases that were EBV-positive. The EBV latency status was characterized in 11 of 12 DLBCL/PTLD EBV-positive cases exhibiting different types of latency: latency I, 2 cases; latency II, 3 cases; and latency III, 6 cases (Table 2).
Nine cases expressed ZEBRA: one DLBCL/IC case with latency II, and eight DLBCL/PTLD cases, of which five cases demonstrated latency III, two exhibited latency II, and latency in one case was undetermined.
Controls of BAFF and BAFF-R Immunostaining
BAFF and BAFF-R expression obtained in different lymphomatous tissues demonstrated the same positivity as has been previously described in the literature (Figure 1). In brief, all lymphomas expressed BAFF, whereas BAFF-R was expressed in follicular, mantle, and marginal zone lymphomas but was negative in Burkitt's lymphoma.24,26
Figure 1.
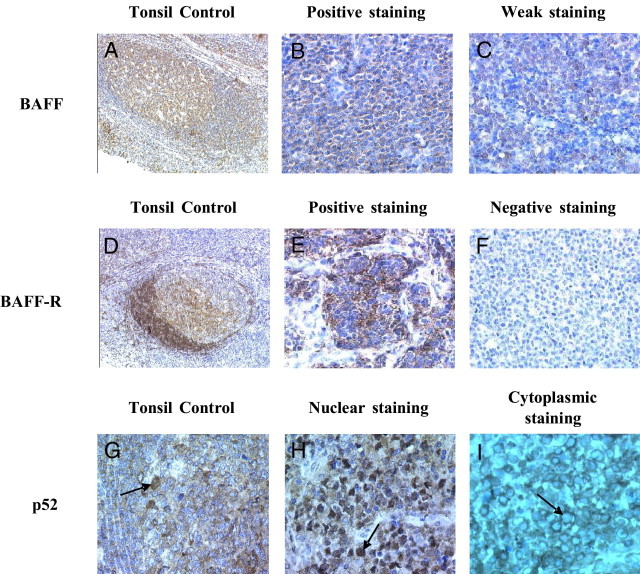
BAFF, BAFF-R, and p52 staining. A–C: BAFF staining. D–F: BAFF-R staining. G–I: p52 staining. A: Tonsil control. High expression in GC cells, weak expression in mantle cells, and heterogeneous staining in the interfollicular zone. B: Positive staining. C: Weak staining. D: Tonsil control. High expression in mantle cells and heterogeneous staining in GC cells. E: Positive staining. F: Negative staining. G: Tonsil control. Nuclear (arrow) and cytoplasmic staining in GC cells. H: Positive staining, nuclear localization (arrow). I: Negative staining, cytoplasmic localization (arrow).
Expression of BAFF and BAFF-R
BAFF expression was predominantly detected in almost all cases in the entire series of DLBCL [47 of 51 cases (92%)]. Only two DLBCL/PTLD cases exhibiting the NGC pattern and two DLBCL/IC cases exhibiting the NGC and GC patterns, respectively, did not express BAFF. Of note, regardless of the histogenesis pattern, BAFF-R was weakly expressed in only 1 of 21 (4.88%) DLBCL/PTLD cases, in contrast with DLBCL/IC cases, of which 11 of 30 (37%) were positive for BAFF-R. This distinct BAFF-R expression was statistically significant (P < 0.008).
Expression of NF-κB Pathways
In the entire series of DLBCL (DLBCL/IC or DLBCL/PTLD) demonstrating the NGC pattern (37 cases), LMP1 expression observed in 11 cases was associated with NF-κB activation, regardless of the pathway (p50 and/or p52). Among these 11 LMP1-positive cases, 8 (73%) expressed nuclear p52 protein. Nine of these 11 cases were from the subgroup DLBCL/PTLD, associated with p50 in 4 cases. In contrast, in the LMP1-negative subgroup (EBV-positive or EBV-negative), p52 nuclear expression was observed in only 4 of 26 (15%) NGC DLBCL cases (Figure 1). This difference was statistically significant (P = 0.0014). No correlation was observed between p52 and BAFF-R protein expression. p52 expression with the NGC pattern was positive in 12 of 19 cases (63%). Furthermore, none of 18 DLBCL/IC cases with the NGC pattern expressed p52 (Table 3). This distinct p52 expression was highly statistically significant (P < 2.10−4). p52 expression between DLBCL/PTLD and DLBCL/IC with the GC profile could not be compared because of the absence of GC pattern in our series of DLBCL/PTLD cases. Moreover, p52 expression seems to be associated with ZEBRA expression in DLBCL/PTLD because almost all cases expressing ZEBRA [7 of 8 cases (lsqb]87%)] demonstrated an NF-κB2–activated profile at nuclear p52 staining.
Discussion
In the present study, 51 DLBCL cases, divided into two subcategories, DLBCL/PTLD and DLBCL/IC, characterized by the level of activation of lymphomatous B cells and their histogenesis profile, ie, GC or NGC pattern, were investigated for BAFF-R and p52 expression.7 In cases demonstrating the NGC pattern, a statistically significant difference of p52 expression was observed between DLBCL/PTLD [12 of 19 cases (63%)] and DLBCL/IC (0 of 18 cases). Despite BAFF-R expression, no association with p52 expression was observed. However, regardless of the histogenesis profile (GC versus NGC), BAFF-R expression, which was rarely observed in DLBCL/PTLD [1 of 21 cases (4.8%)], was statistically different from DLBCL/IC, in which 11 of 30 cases (37%) were positive.
Among the 21 DLBCL/PTLD cases, the NGC phenotype was expressed in 19, whereas 18 of 30 DLBCL/IC cases demonstrated the NGC profile. EBV was present in 14 cases, 2 DLBCL/IC and 12 DLBCL/PTLD. All but two EBV-associated cases demonstrated the NGC histogenesis pattern. The percentage of EBV-associated DLBCL/PTLD was 57%. In the present series, this low percentage of EBV-positive PTLD can be related to the high number of PTLD cases occurring later than 24 months after transplantation, as has previously been described.29
BAFF expression was detected in 92% of the 51 DLBCL cases. This result could reflect ligand-receptor engagement as well as environmental- or cellular-secreted BAFF. Indeed, monoclonal antibodies against human BAFF recognized membrane-bound and soluble BAFF.16 The autocrine pathway could explain this expression, as described in other models such as chronic lymphocytic lymphoma and multiple myeloma.18,30 Study of BAFF-R expression was performed to respond to this hypothesis of the autocrine signaling pathway.
Little is known about BAFF-R expression relative to the level of DLBCL activation. Only one study has explored this correlation in association with Lck protein.31 Our results were similar to those of Paterson et al.31; indeed, no differences were detected between the histogenesis profile and BAFF-R expression in the DLBCL/IC subgroup. We observed that BAFF-R was expressed in 11 of 30 DLBCL/IC cases (37%), which corresponds to the mean percentage of previously published results.24,26,31 However, regardless of the histogenesis profile, BAFF-R expression was higher in DLBCL/IC (37%) than in DLBCL/PTLD (4.8%), with a statistically significant correlation. Thus, the hypothesis of the autocrine pathway through BAFF-R could be excluded. Nevertheless, receptors TACI and BCMA could also be implicated in this signaling pathway; however, their expression was not tested in the present series. The high level of BAFF expression in NHL, based on transcription enhancement through BAFF promoter, could be related to the possible effect of the polymorphism −871C→T (or other genetic variation) or to the nuclear translocation of the CD40-cRel dimer.31–34 In addition, BAFF-R weak expression could be explained by possible down-regulation of BAFF-R arising from BAFF binding.35–37
EBV activates the NF-κB pathway through LMP1 protein, and the link between this activation pathway and lymphomas has also been described.12,15 As expected, in our series, all 11 NGC DLBCL cases positive for LMP1 were associated with NF-κB activation. Among these cases, p52 was generally expressed (8 of 11 cases); however, it was positive in only 4 of 26 cases in the LMP1-negative subgroup whether EBV was or was not present. BAFF-R expression could not explain these results because there was no correlation with p52 nuclear localization, which suggests that the BAFF/BAFF-R signaling pathway could not be directly implicated in DLBCL lymphomagenesis mediated by p52 signaling. The dissociation between BAFF/BAFF-R and p52 signaling pathways could be interpreted via repression of NF-κB activity mediated by BCL6 protein expression or the posttranslational modification of RelB.38,39
The nuclear expression of p50 and/or p52 in six cases demonstrating the GC pattern in DLBCL/IC could be related to deregulation of the NF-κB pathway caused by multiple genetic events including negative and positive regulators of NF-κB, as previously reported by Compagno et al.40 The higher expression of p52 related to the NF-κB2 activation pathway in DLBCL/PTLD, which is statistically different from the NGC pattern in DLBCL/IC, could be related not only to genetic events but also to activation of NF-κB2 via LMP1.
Moreover, it must be emphasized that in seven of eight cases expressing ZEBRA in EBV-positive DLBCL/PTLD cases, p52 demonstrated nuclear staining, which suggests a relationship between the replicative cycle of EBV and p52 nuclear expression. However, this result must be confirmed in a larger series.
Thus, our results demonstrating rare expression of BAFF-R and the frequency of NF-κB2 pathway activation in DLBCL/PTLD as compared with DLBCL/IC confirm the importance of EBV, especially LMP1, and suggest other mechanisms of NF-κB2 pathway deregulation in these tumors.
Footnotes
Supported by grant R04083LS from Canceropôle-Ile de France and by Department Committee 93 of Ligue Nationale Contre le Cancer.
References
- 1.Jacobson C.A., LaCasce A.S. Lymphoma: risk and response after solid organ transplant [review] Oncology. 2010;24:936–944. [PubMed] [Google Scholar]
- 2.Jaffe E.S., Harris N.L., Stein H., Vardiman J.W. Vol 3. WHO Press; Geneva, Switzerland: 2008. WHO Classification of Tumours. (Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues). [Google Scholar]
- 3.Capello D., Rossi D., Gaidano G. Post-transplant lymphoproliferative disorders: molecular basis of disease histogenesis and pathogenesis. Hematol Oncol. 2005;23:61–67. doi: 10.1002/hon.751. [DOI] [PubMed] [Google Scholar]
- 4.Carbone A., Gloghini A., Larocca L.M., Capello D., Pierconti F., Canzonieri V., Tirelli U., Dalla-Favera R., Gaidano G. Expression profile of MUM1/IRF4: BCL-6, and CD138/syndecan-1 defines novel histogenetic subsets of human immunodeficiency virus-related lymphomas. Blood. 2001;97:744–751. doi: 10.1182/blood.v97.3.744. [DOI] [PubMed] [Google Scholar]
- 5.Hans C.P., Weisenburger D.D., Greiner T.C., Gascoyne R.D., Delabie J., Ott G., Müller-Hermelink H.K., Campo E., Braziel R.M., Jaffe E.S., Pan Z., Farinha P., Smith L.M., Falini B., Banham A.H., Rosenwald A., Staudt L.M., Connors J.M., Armitage J.O., Chan W.C. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 6.Johnson L.R., Nalesnik M.A., Swerdlow S.H. Impact of Epstein-Barr virus in monomorphic B-cell posttransplant lymphoproliferative disorders: a histogenetic study. Am J Surg Pathol. 2006;30:1604–1612. doi: 10.1097/01.pas.0000213317.59176.d2. [DOI] [PubMed] [Google Scholar]
- 7.Chang C.C., McClintock S., Cleveland R.P., Trzpuc T., Vesole D.H., Logan B., Kajdacsy-Balla A., Perkins S.L. Immunohistochemical expression patterns of germinal center and activation B-cell markers correlate with prognosis in diffuse large B-cell lymphoma. Am J Surg Pathol. 2004;28:464–470. doi: 10.1097/00000478-200404000-00005. [DOI] [PubMed] [Google Scholar]
- 8.Nelson B.P., Nalesnik M.A., Bahler D.W., Locker J., Fung J.J., Swerdlow S.H. Epstein-Barr virus–negative post-transplant lymphoproliferative disorders: a distinct entity? Am J Surg Pathol. 2000;24:375–385. doi: 10.1097/00000478-200003000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Paessler M., Kossev P., Tsai D., Raghunath P., Majewski M., Zhang Q., Ramalingam P., Schuster S., Tomaszewski J., Arber D.A., Hsi E., Wasik M.A. Expression of SHP-1 phosphatase indicates post–germinal center cell derivation of B-cell posttransplant lymphoproliferative disorders. Lab Invest. 2002;82:1599–1606. doi: 10.1097/01.lab.0000036873.16297.a5. [DOI] [PubMed] [Google Scholar]
- 10.McClain K.L. Epstein-Barr virus and HIV-associated diseases. Biomed Pharmacother. 2001;55:348–352. doi: 10.1016/s0753-3322(01)00092-0. [DOI] [PubMed] [Google Scholar]
- 11.Young L.S., Rickinson A.B. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 12.Wu L., Nakano H., Wu Z. The C-terminal activating region 2 of the Epstein-Barr virus–encoded latent membrane protein 1 activates NF-kappaB through TRAF6 and TAK1. J Biol Chem. 2006;281:2162–2169. doi: 10.1074/jbc.M505903200. [DOI] [PubMed] [Google Scholar]
- 13.Vallabhapurapu S., Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 14.Ramalingam P., Chu W.S., Tubbs R., Rybicki L., Pettay J., Hsi E.D. Latent membrane protein 1, tumor necrosis factor receptor-associated factor (TRAF) 1, TRAF-2, TRAF-3, and nuclear factor kappa B expression in posttransplantation lymphoproliferative disorders. Arch Pathol Lab Med. 2003;127:1335–1339. doi: 10.5858/2003-127-1335-LMPTNF. [DOI] [PubMed] [Google Scholar]
- 15.Jost P.J., Ruland J. Aberrant NF-kappaB signaling in lymphoma: mechanisms, consequences, and therapeutic implications. Blood. 2007;109:2700–2707. doi: 10.1182/blood-2006-07-025809. [DOI] [PubMed] [Google Scholar]
- 16.Schneider P., MacKay F., Steiner V., Hofmann K., Bodmer J.L., Holler N., Ambrose C., Lawton P., Bixler S., Acha-Orbea H., Valmori D., Romero P., Werner-Favre C., Zubler R.H., Browning J.L., Tschopp J. BAFF, a novel ligand of the tumor necrosis factor family, stimulates B cell growth. J Exp Med. 1999;189:1747–1756. doi: 10.1084/jem.189.11.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mackay F., Woodcock S.A., Lawton P., Ambrose C., Baetscher M., Schneider P., Tschopp J., Browning J.L. Mice transgenic for BAFF develop lymphocytic disorders along with autoimmune manifestations. J Exp Med. 1999;190:1697–1710. doi: 10.1084/jem.190.11.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novak A.J., Darce J.R., Arendt B.K., Harder B., Henderson K., Kindsvogel W., Gross J.A., Greipp P.R., Jelinek D.F. Expression of BCMA: TACI, and BAFF-R in multiple myeloma: a mechanism for growth and survival. Blood. 2004;103:689–694. doi: 10.1182/blood-2003-06-2043. [DOI] [PubMed] [Google Scholar]
- 19.Novak A.J., Bram R.J., Kay N.E., Jelinek D.F. Aberrant expression of B-lymphocyte stimulator by B chronic lymphocytic leukemia cells: a mechanism for survival. Blood. 2002;100:2973–2979. doi: 10.1182/blood-2002-02-0558. [DOI] [PubMed] [Google Scholar]
- 20.Novak A.J., Grote D.M., Stenson M., Ziesmer S.C., Witzig T.E., Habermann T.M., Harder B., Ristow K.M., Bram R.J., Jelinek D.F., Gross J.A., Ansell S.M. Expression of BLyS and its receptors in B-cell non-Hodgkin lymphoma: correlation with disease activity and patient outcome. Blood. 2004;104:2247–2253. doi: 10.1182/blood-2004-02-0762. [DOI] [PubMed] [Google Scholar]
- 21.Treml J.F., Hao Y., Stadanlick J.E., Cancro M.P. The BLyS family: toward a molecular understanding of B cell homeostasis. Cell Biochem Biophys. 2009;53:1–16. doi: 10.1007/s12013-008-9036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson J.S., Bixler S.A., Qian F., Vora K., Scott M.L., Cachero T.G., Hession C., Schneider P., Sizing I.D., Mullen C., Strauch K., Zafari M., Benjamin C.D., Tschopp J., Browning J.L., Ambrose C. BAFF-R, a newly identified TNF receptor that specifically interacts with BAFF. Science. 2001;293:2108–2111. doi: 10.1126/science.1061965. [DOI] [PubMed] [Google Scholar]
- 23.Ng L.G., Sutherland A.P., Newton R., Qian F., Cachero T.G., Scott M.L., Thompson J.S., Wheway J., Chtanova T., Groom J., Sutton I.J., Xin C., Tangye S.G., Kalled S.L., Mackay F., Mackay C.R. B cell–activating factor belonging to the TNF family (BAFF)-R is the principal BAFF receptor facilitating BAFF costimulation of circulating T and B cells. J Immunol. 2004;173:807–817. doi: 10.4049/jimmunol.173.2.807. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura N., Hase H., Sakurai D., Yoshida S., Abe M., Tsukada N., Takizawa J., Aoki S., Kojima M., Nakamura S., Kobata T. Expression of BAFF-R (BR 3) in normal and neoplastic lymphoid tissues characterized with a newly developed monoclonal antibody. Virchows Arch. 2005;447:53–60. doi: 10.1007/s00428-005-1275-6. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X., Park C.S., Yoon S.O., Li L., Hsu Y.M., Ambrose C., Choi Y.S. BAFF supports human B cell differentiation in the lymphoid follicles through distinct receptors. Int Immunol. 2005;17:779–788. doi: 10.1093/intimm/dxh259. [DOI] [PubMed] [Google Scholar]
- 26.Rodig S.J., Shahsafaei A., Li B., Mackay C.R., Dorfman D.M. BAFF-R, the major B cell–activating factor receptor, is expressed on most mature B cells and B-cell lymphoproliferative disorders. Hum Pathol. 2005;36:1113–1119. doi: 10.1016/j.humpath.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Claudio E., Brown K., Park S., Wang H., Siebenlist U. BAFF-induced NEMO-independent processing of NF-kappa B2 in maturing B cells. Nat Immunol. 2002;3:958–965. doi: 10.1038/ni842. [DOI] [PubMed] [Google Scholar]
- 28.Vakiani E., Basso K., Klein U., Mansukhani M.M., Narayan G., Smith P.M., Murty V.V., Dalla-Favera R., Pasqualucci L., Bhagat G. Genetic and phenotypic analysis of B-cell post-transplant lymphoproliferative disorders provides insights into disease biology. Hematol Oncol. 2008;26:199–211. doi: 10.1002/hon.859. [DOI] [PubMed] [Google Scholar]
- 29.Leblond V., Dhedin N., Mamzer Bruneel M.F., Choquet S., Hermine O., Porcher R., Nguyen Quoc S., Davi F., Charlotte F., Dorent R., Barrou B., Vernant J.P., Raphael M., Levy V. Identification of prognostic factors in 61 patients with posttransplantation lymphoproliferative disorders. J Clin Oncol. 2001;19:772–778. doi: 10.1200/JCO.2001.19.3.772. [DOI] [PubMed] [Google Scholar]
- 30.Kern C., Cornuel J.F., Billard C., Tang R., Rouillard D., Stenou V., Defrance T., Ajchenbaum-Cymbalista F., Simonin P.Y., Feldblum S., Kolb J.P. Involvement of BAFF and APRIL in the resistance to apoptosis of B-CLL through an autocrine pathway. Blood. 2004;103:679–688. doi: 10.1182/blood-2003-02-0540. [DOI] [PubMed] [Google Scholar]
- 31.Paterson J.C., Tedoldi S., Craxton A., Jones M., Hansmann M.L., Collins G., Roberton H., Natkunam Y., Pileri S., Campo E., Clark E.A., Mason D.Y., Marafioti T. The differential expression of LCK and BAFF-receptor and their role in apoptosis in human lymphomas. Haematologica. 2006;91:772–780. [PubMed] [Google Scholar]
- 32.Novak A.J., Grote D.M., Ziesmer S.C., Kline M.P., Manske M.K., Slager S., Witzig T.E., Shanafelt T., Call T.G., Kay N.E., Jelinek D.F., Cerhan J.R., Gross J.A., Harder B., Dillon S.R., Ansell S.M. Elevated serum B-lymphocyte stimulator levels in patients with familial lymphoproliferative disorders. J Clin Oncol. 2006;24:983–987. doi: 10.1200/JCO.2005.02.7938. [DOI] [PubMed] [Google Scholar]
- 33.Novak A.J., Slager S.L., Fredericksen Z.S., Wang A.H., Manske M.M., Ziesmer S., Liebow M., Macon W.R., Dillon S.R., Witzig T.E., Cerhan J.R., Ansell S.M. Genetic variation in B-cell–activating factor is associated with an increased risk of developing B-cell non-Hodgkin lymphoma. Cancer Res. 2009;69:4217–4224. doi: 10.1158/0008-5472.CAN-08-4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou H.J., Pham L.V., Tamayo A.T., Lin-Lee Y.C., Fu L., Yoshimura L.C., Ford R.J. Nuclear CD40 interacts with c-Rel and enhances proliferation in aggressive B-cell lymphoma. Blood. 2007;110:2121–2127. doi: 10.1182/blood-2007-02-073080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sellam J., Miceli-Richard C., Gottenberg J.E., Ittah M., Lavie F., Lacabaratz C., Gestermann N., Proust A., Lambotte O., Mariette X. Decreased B cell activating factor receptor expression on peripheral lymphocytes associated with increased disease activity in primary Sjogren's syndrome and systemic lupus erythematosus. Ann Rheum Dis. 2007;66:790–797. doi: 10.1136/ard.2006.065656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nimmanapalli R., Lyu M.A., Du M., Keating M.J., Rosenblum M.G., Gandhi V. The growth factor fusion construct containing B-lymphocyte stimulator (BLyS) and the toxin rGel induces apoptosis specifically in BAFF-R–positive CLL cells. Blood. 2007;109:2557–2564. doi: 10.1182/blood-2006-08-042424. [DOI] [PubMed] [Google Scholar]
- 37.Bloom D., Chang Z., Pauly K., Kwun J., Fechner J., Hayes C., Samaniego M., Knechtle S. BAFF is increased in renal transplant patients following treatment with alemtuzumab. Am J Transplant. 2009;9:1835–1845. doi: 10.1111/j.1600-6143.2009.02710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez-Rosado A., Artiga M., Vargiu P., Sanchez-Aguilera A., Alvarez-Barrientos A., Piris M. BCL6 represses NFkappaB activity in diffuse large B-cell lymphomas. J Pathol. 2008;214:498–507. doi: 10.1002/path.2279. [DOI] [PubMed] [Google Scholar]
- 39.Neumann M., Naumann M. Beyond IkappaBs: alternative regulation of NF-kappaB activity. FASEB J. 2007;11:2642–2654. doi: 10.1096/fj.06-7615rev. [DOI] [PubMed] [Google Scholar]
- 40.Compagno M., Lim W.K., Grunn A., Nandula S.V., Brahmachary M., Shen Q., Bertoni F., Ponzoni M., Scandurra M., Califano A., Bhagat G., Chadburn A., Dalla-Favera R., Pasqualucci L. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–720. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]


