Abstract
Stromal fibroblasts perform important roles in cancer development and progression. Overexpression of Twist1, a basic helix-loop-helix transcription factor, is often associated with aggressive behavior in many tumors. In this study, we investigated Twist1 expression patterns in gastric stromal fibroblasts and cancer cells using a monoclonal Twist1 antibody after validating the effectiveness of four commercial Twist1-specific antibodies. Twist1 expression was more frequently observed in gastric cancer-associated fibroblasts (CAFs) than in other cancer cells but was otherwise rarely expressed in noncancerous tissue. In laser capture microdissection of stromal fibroblasts, Twist1 immunopositive fibroblasts exhibited significantly increased Twist1, fibroblast-specific protein 1, and CXCL14 mRNA expression. Furthermore, Twist1 mRNA expression showed a significant linear correlation with that of platelet-derived growth factor receptors β and α. We found that conditioned media from Twist1-expressing skin and lung fibroblasts significantly promote invasion of gastric cancer cells in vitro. In 195 gastric cancer samples, CAF Twist1 expression was associated with tumor size, invasion depth, and lymph node metastasis. Twist1 was also associated with poor prognosis in patients with gastric cancer, particularly in those with the diffuse type. In conclusion, CAFs in gastric cancer frequently have altered Twist1 expression, and increased Twist1 expression in fibroblasts contributes to the progression of cancer cells and poor patient survival.
Fibroblasts are one of the most common connective tissue cell types, and their main function is to maintain the extracellular matrix microenvironment and connective tissue structural integrity. Fibroblasts are also important in the regulation of epithelial differentiation through epithelial and stromal cross-talk.1 Fibroblasts have also been shown to have a profound influence on the development and progression of carcinomas.2,3
Fibroblasts exhibit a wide range of activation. On one end of the spectrum, normal fibroblasts show thin, wavy, and small spindle cell morphologic features, whereas fully activated fibroblasts, sometimes referred to as myofibroblastic cells, are large and plump spindle-shaped mesenchymal cells that often express α-smooth muscle actin (SMA).2,4 Of these myofibroblastic cells, fibroblasts that are found in association with cancer cells are known as cancer-associated fibroblasts (CAFs).2 Such stromal fibroblasts may contain an abundance of prognostic information and are a source of many potential new tumor biomarkers, which are useful in evaluating the prognosis for a given tumor.5
Twist1 is a transcription factor that belongs to the basic helix-loop-helix transcription factor family. Initially, Twist1 was shown to be essential in the development of mesodermally derived tissues, including muscle and osteogenic cell lineages.6,7 During development, Twist1 plays a key role in the cellular transdifferentiation process, known as the epithelial-mesenchymal transition (EMT). A recent report showed that Twist1 is overexpressed in a variety of human tumors.8 In addition, Twist1 was reported to promote cancer metastasis and was correlated with poor prognoses.9,10
There have been few studies on the role of Twist1 expression in gastric cancer. Rosivatz et al11 reported that Twist1 mRNA was overexpressed in diffuse-type adenocarcinoma, as assessed by quantitative real-time PCR analysis of 48 formalin-fixed paraffin-embedded (FFPE) gastric cancer tissues. Yan-Qi et al12 found that Twist1 protein expression was increased in diffuse-type gastric carcinoma and was correlated with lymph node metastasis in 76 cases of gastric cancer using immunohistochemistry. However, the cellular location of Twist1 protein expression has been described differently in various studies; it was reported to be cytoplasmic in some studies12,13 and nuclear in others.14–16 In addition, studies investigating Twist1 expression in gastric cancer using the same antibody (H81) showed mostly cytoplasmic staining. As such, the reported clinical significance and expression patterns of Twist1 vary, depending on the antibody that was used in a particular study. Furthermore, little is known about the prognostic significance of Twist1 expression in gastric cancer due to small-scale patient cohorts. Therefore, in this study we first thoroughly validated four different commercially available antibodies for Twist1. We then used the best-performing antibody to study Twist1 expression in gastric cancer tissues and found, unexpectedly, that Twist1 expression was more abundant in CAFs than in the cancer cells themselves. Finally, we evaluated the prognostic significance of Twist1 expression in gastric cancer cells, as well as in CAFs, in a larger cohort of patients with gastric cancer.
Materials and Methods
Cell Lines
Thirteen gastric cancer cell lines (SNU5, SNU16, SNU216, SNU484, SNU601, SNU620, SNU638, SNU719, MKN1, MKN28, MKN74, KATOIII, and AGS), HeLa, and two human fibroblast cell lines (skin fibroblast CCD986sk and lung fibroblast IMR90) were maintained in Dulbecco's modified Eagle's medium (DMEM) with high glucose (Life Technologies, Grand Island, NY) supplemented with 10% heat-inactivated fetal bovine serum (Life Technologies), 100 mg/mL of penicillin G, and 50 μg/mL of streptomycin (Life Technologies) at 37°C in a humidified atmosphere containing 5% CO2. All cell lines were purchased from the Korean Cell Line Bank (Seoul, Korea).
RNA Extraction and Quantitative Real-Time RT-PCR
Total RNA was extracted by standard phenol-chloroform extraction and isopropanol precipitation methods. The cDNA was synthesized from 2 μg of total RNA using Superscript II RNA Reverse Transcriptase (no. 11904-018; Invitrogen, Carlsbad, CA), according to the manufacturer's protocol.
Real-time RT-PCR was performed using the ABI 7900 HT Fast Real Time PCR system (Applied Biosystems, Foster City, CA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal loading control. PCR was conducted using the SYBR Green PCR master mix (Applied Biosystems). Thermal cycling conditions were as follows: 1 step for 10 minutes at 95°C followed by 40 cycles at 95°C for 15 seconds and at 60°C for 1 minute. Each reaction was performed in duplicate. PCR product quality was monitored using post-PCR melt-curve analysis. CT, the fractional cycle number at which the amount of amplified target reached a fixed threshold, was determined. The Twist1 concentration ratio was calculated using 2−ΔCt (ΔCt = ΔCTwist1 -ΔCGAPDH). The primers used were as follows: Twist1, forward primer (5′-GGCTCAGCTACGCCTTCTC-3′) and reverse primer (5′-TCCTTCTCTGGAAACAATGACA-3′); osteopontin, forward primer (5′-CTCCATTGACTCGAACGACTC-3′) and reverse primer (5′-CAGGTCTGCGAAACTTCTTAGAT-3′); SMA, forward primer (5′-CAGGGCTGTTTTCCCATCCAT-3′) and reverse primer (5′-ACGTAGCTGTCTTTTTGTCCC-3′); CXCL14, forward primer (5′-CGCTACAGCGACGTGAAGAA-3′) and reverse primer (5′-TTCCAGGCGTTGTACCACTTG-3′); Fibroblast Activation Protein (FAP), forward primer (5′-ACGGCTTATCACCTGATCGG-3′) and reverse primer (5′-AATTGGACGAGGAAGCTCATTT-3′); fibroblast-specific protein 1 (FSP1), forward primer (5′-GATGAGCAACTTGGACAGCAA-3′) and reverse primer (5′-CTGGGCTGCTTATCTGGGAAG-3′); platelet-derived growth factor receptor (PDGFR) α, forward primer (5′-AACCGTGTATAAGTCAGGGGA-3′) and reverse primer (5′-GCATTGTGATGCCTTTGCCTT-3′); PDGFRβ, forward primer (5′-TGATGCCGAGGAACTATTCATCT-3′) and reverse primer (5′-TTTCTTCTCGTGCAGTGTCAC-3′); and GAPDH, forward primer (5′-GCACCGTCAAGGCTGAGAA-3′) and reverse primer (5′-AGCATCGCCCCACTTGATT-3′).
Western Blotting Analysis
Cell lysates were prepared using an SDS lysis solution. Protein concentration was measured using a bicinchoninic acid protein assay kit (Pierce, Rockford, IL). Equal amounts of protein were separated by electrophoresis on a 12.5% SDS-PAGE gel. The proteins were electrotransferred from the gel to a nitrocellulose membrane, which was then blocked with a 5% nonfat milk solution for 1 hour and then incubated with primary Twist1-specific antibodies (Table 1), including ab50887 (mouse monoclonal, clone: Twist2C1a; Abcam, Cambridge, UK), ab50581 (rabbit polyclonal; Abcam), 4119S (rabbit polyclonal; Cell Signaling Technology, Beverly, MA), and H81 (rabbit polyclonal; Santa Cruz Biotechnology, Santa Cruz, CA) for 2 hours at room temperature. β-Actin–specific antibody (1:1000; Santa Cruz) was used as an internal loading control. The membrane was incubated with goat anti-rabbit secondary antibodies that were detected by an enhanced chemiluminescence detection system (Amersham Biosciences Europe, Freiburg, Germany) according to the manufacturer's instructions.
Table 1.
Characteristics of Twist1-Specific Antibodies
| Catalog no. | Type | Dilution (WB) | Cellular localization | Company | Recommend for applications |
|---|---|---|---|---|---|
| H81 | Rabbit polyclonal | 1:50 (1:1000) | NA | Santa Cruz Biotechnology, Santa Cruz, CA | WB, IP, IF, ELISA |
| Ab50887 | Mouse monoclonal | 1:100 (1:1000) | Nuclear | Abcam, Cambridge, UK | ChIP, Dot, ICC/IF, IHC-P, WB |
| Ab50581 | Rabbit polyclonal | 1:100 (1:1000) | Nuclear | Abcam | ICC/IF, IHC-P, WB |
| 4119S | Rabbit polyclonal | 1:100 (1:1000) | NA | Cell Signaling Technology, Beverly, MA | WB, IP |
ChIP, chromatin immunoprecipitation; Dot, dot blot; ELISA, enzyme-linked immunosorbent assay; ICC, immunocytochemistry; IF, immunofluorescence; IHC-P, immunohistochemistry-formalin/paraformaldehyde-fixed paraffin-embedded tissue; IP, immunoprecipitation; NA, not applicable; WB, Western blot.
Transient siRNA Transfection
The ablation of Twist1 was performed by transient transfection of cells with small interfering RNA (siRNA) duplex oligos synthesized by SamCheonLee Ltd. (Seoul, Korea). Control siRNA (Scrambled RNA; Genolution, Seoul, Korea) and Twist1-specific siRNA (sense: 5′-GAUGGCAAGCUGCAGCUAUTTdTdT-3′ and antisense: 5′- AUAGCUGCAGCUUGCCAUCTTdTdT-3′) were transfected using G-Fectin reagent (Genolution). Three days after transfection, the cells were collected for Western blotting analysis.
DNA Extraction and Sodium Bisulfite Modification
Cultured cells were placed in a 1.5-mL Eppendorf tube containing 540 μL of lysis buffer with 10 mmol/L Tris (pH 8), 0.1 mmol/L EDTA, 2% SDS, and 60 μL of 20 mg/mL of proteinase K (Promega, Madison, WI). The samples were incubated for 20 hours at 60°C. DNA was then isolated by standard phenol-chloroform extraction and isopropanol precipitation methods. Bisulfite conversion was performed with an EpiTect Bisulfite Kit (Qiagen) as described by the manufacturer. For each conversion reaction, 1 μg DNA was used, and after conversion and purification, DNA was eluted in 20 μL of distilled water.
MSP Analysis
The methylation status of the Twist1 gene was determined using a methylation-specific nested PCR (MSP) method. Primer sequences used in the first amplification were as follows: for methylated DNA, forward 5′-CGTTAGGGTTCGGGGGC-3′ and reverse 5′-ACCTAACCCGAACGACCG-3′; and for unmethylated DNA, forward 5′-GTTGTGTTAGGGTTTGGGGGT-3′ and reverse 5′-ACCTAACCCGAACGACCG-3′. Primer sequences used in the second amplification were as follows: for methylated DNA, forward 5′-GGGGCGTTGTTCGTATGTTTC-3′ and reverse 5′-CACCCGCCTCCTAACCG-3′; and for unmethylated DNA, forward 5′-TTTGGGGGTGTTGTTTGTATGTTTT-3′ and reverse 5′-TCCACCCACCTCCTAACCA-3′. The first and second amplification PCR conditions consisted of one cycle for 5 minutes at 94°C followed by 35 cycles of 94°C (30 seconds), 58°C (30 seconds), and 72°C (1 minute) with a final elongation for 7 minutes at 72°C using a Hot-start PCR premix (Intron, Seoul, Korea). Amplified PCR products were resolved in a 2% agarose gel using ethidium bromide for detection.
Bisulfite-Modified DNA Sequencing Analysis
For amplification and sequencing of bisulfite-modified DNA to determine methylation status, primers were designed as follows: forward 5′-TGTGTAGAAGTTGTTGTTATTGTTG-3′ and reverse 5′-TACAAACTTAAAAACTCTTATACCTCC-3′. These primers were designed to amplify both methylated and unmethylated DNA. The PCR conditions consisted of an initial denaturation for 4 minutes at 94°C followed by 40 cycles of 94°C (30 seconds), 60°C (30 seconds), and 72°C (30 seconds), with a final elongation for 7 minutes at 72°C using the Hot-start PCR premix (Intron). Amplified PCR products were cloned into the pGEM-T Easy vector (Promega) and directly sequenced using a Taq dideoxy terminator cycle sequencing kit using an ABI 3730 DNA sequencer (Applied Biosystems).
Cell Pellet Array of Gastric Cancer Cell Lines
Cells were trypsinized, fixed for 1 hour in 4% formalin, and then centrifuged and resuspended in 0.8% agarose. Gel plugs containing fixed tumor cells were then processed through gradient alcohols before being cleared in xylem and washed multiple times in molten paraffin. Once processed, the cells were embedded in paraffin and then arrayed in 0.6-mm cores using a manual tissue arrayer (MTA-1; Beecher Instruments, Sun Prairie, WI).
Tissue Specimens and Tissue Microarray Construction
A total of 332 FFPE gastric tissue samples (from 195 gastric cancer patients, 23 healthy individuals, 17 chronic gastritis patients, 47 intestinal metaplasia patients, 25 low-grade dysplasia patients, and 25 high-grade dysplasia patients) were included in this study. Gastric cancer cases were divided into three subtypes by the Lauren classification, and 84 diffuse types, 100 intestinal types, and 11 mixed types were identified. A cylindrical core (3 mm in diameter) was removed from each FFPE tissue block to construct the tissue microarray. All samples were obtained from January 1994 to December 2006 from the Chungbuk National University Hospital and the Samsung Medical Center. All patients provided written informed consent according to institutional guidelines. None of the patients had preoperative chemotherapy or radiotherapy. Clinical and pathological reports were reviewed for age, sex, tumor size, histologic grade, Lauren classification, invasion depth (pT), and nodal status (pN). pTNM classifications were designated according to the 2002 American Joint Committee on Cancer staging manual guidelines.17
Immunohistochemical Analysis
Microslide sections were deparaffinized with xylene, hydrated using a diluted alcohol series, and immersed in 0.3% H2O2 in methanol to quench endogenous peroxidase activity. Sections were then treated with TE buffer (10 mmol/L Tris and 1 mmol/L EDTA, pH 9.0) for antigen retrieval. To reduce nonspecific staining, each section was treated with 4% bovine serum albumin in PBS with 0.1% Tween 20 for 30 minutes. Sections were then incubated with primary antibodies in PBS with 0.1% Tween 20 containing 3 mg/mL of goat globulin (Sigma) for 1 hour at room temperature, followed by three successive rinses with a wash buffer. Sections were then incubated with an anti-mouse/rabbit polymer kit (Envision Plus; Dako, Carpinteria, CA) for 30 minutes at room temperature. The chromogen used was 3,3′-diaminobenzidine (Dako). Sections were counterstained with Meyer's hematoxylin. Primary antibodies used were Twist1-specific antibodies, pancytokeratin-specific antibody (1:500, Dako), and SMA-specific antibody (1:1000; Dako). Dual immunohistochemical staining was performed with 2 primary antibodies: mouse Twist1-specific monoclonal (ab50887, stained brown) and rabbit polyclonal SMA-specific antibody (stained red). We evaluated Twist1 expression in epithelial cells and stromal fibroblasts separately in 332 FFPE gastric tissues.
Laser Capture Microdissection of Stromal Fibroblasts
Stromal fibroblasts were selectively procured from H&E-stained slides using a laser microdissection device (ION LMD-II; JungWoo International Co., Seoul, Korea). Five cases with Twist1 immunopositive fibroblasts and 9 cases with Twist1 immunonegative fibroblasts were included. Then, total RNA was extracted and cDNA was synthesized to evaluate mRNA expression level as described previously.
Lentivirus Transduction and Preparation of Conditioned Media
IMR90 (lung fibroblast) and CCD986sk (skin fibroblast) cells were transduced with lentivirus expressing human Twist1 and a Green fluorescent protein (GFP) control vector. Vector packaging was obtained by transfection of 293T cells. CCD986sk and IMR90 cells were infected with lentiviral vectors and were selected with hygromycin.
IMR90 and CCD986sk cells were grown to 70% to 80% confluence in 6-well plates in DMEM/10% fetal calf serum. The medium was changed to serum-free DMEM and cells were cultured for 48 hours. Media were collected and centrifuged at 1000 × g for 10 minutes, and the supernatant was collected.
Invasion and Migration Assay
In vitro Matrigel invasion assays were performed using 6.5-mm Costar Transwell chambers (8-Am pore size; Costar, Corning, NY). Transwell filters were coated with the appropriate Matrigel (1 mg/mL) (Becton Dickinson, Franklin Lakes, NJ), and after solidification at 37°C, 1 × 105 MKN28 cells were seeded onto the Matrigel. After incubation with conditioned media for 2 days, the filter was gently removed from the chamber, and noninvasive cells on the upper surface were removed by wiping with a cotton swab. Cells that invaded the Matrigel and attached to the lower surface of the filter were fixed with methanol and stained with H&E solution. The number of cells attached to the lower surface of the polycarbonate filter was counted at 10 high-power fields under a light microscope. The migration assay was conducted in the same manner as the invasion assay, except for Matrigel coating. Medium containing either 10% fetal bovine serum or no serum was used for the positive and negative controls, respectively. Each cell type was assayed in triplicate.
Statistical Analysis
Correlations were examined using Pearson's χ2, Fisher's exact, Mann-Whitney U, or Spearman's tests, as appropriate. Overall survival was determined using the Kaplan-Meier method, and survival curves were compared using the log-rank test. Survival was measured from the date of surgery. The Cox proportional hazards model was used to evaluate the association between clinicopathologic factors and survival. The hazard ratio and its 95% confidence interval were assessed for each factor. All tests were two-sided, and P < 0.05 was considered statistically significant. Statistical analysis was performed using SPSS statistical software, version 17 (SPSS Inc, Chicago, IL).
Results
Twist1 Expression in Cell Lines and Validation of Twist1 Antibodies
Within the 14 cancer cell lines and 2 normal human fibroblast cell lines tested, quantitative real-time RT-PCR revealed that a high expression of Twist1 mRNA was present only in the SNU484 gastric cancer cell line (Figure 1A). Therefore, SNU484 was used as a positive control for all other assays to assess Twist1 expression. Of the four Twist1 antibodies used for immunostaining in FFPE tissues using a cell pellet array of available cell lines, only the ab50887 antibody selectively stained in the cell block section of SNU484 cells with nuclear staining (Figure 1B). The other antibodies (ab50581, 4119S, H81) stained not only the cell block section from SNU484 but also sections from all other cell lines with mainly cytoplasmic staining, with or without additional nuclear staining.
Figure 1.
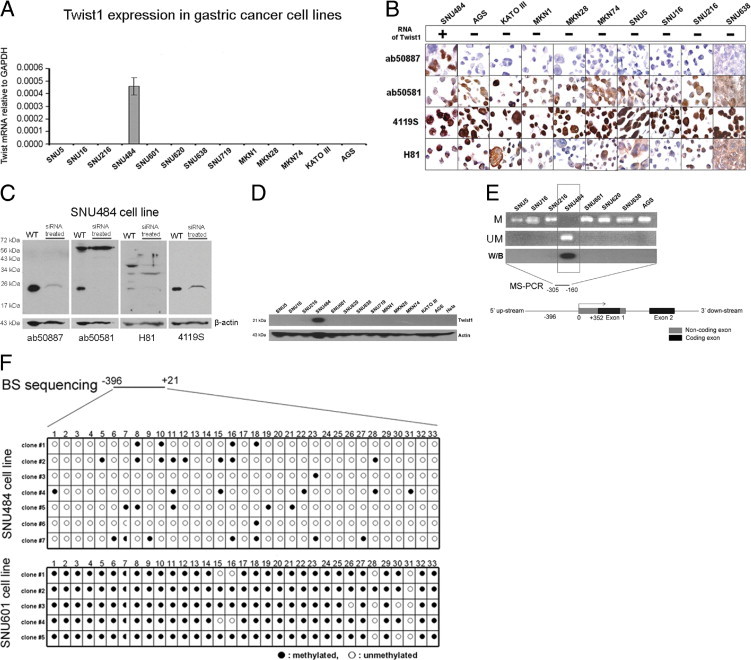
Twist1 expression in cell lines. A: Real-time RT-PCR revealed that only SNU484 cells showed a high level of Twist1 mRNA among the 13 gastric cancer cell lines. GAPDH was used as the internal control. B: Of the four Twist1 antibodies (ab50887, ab50581, 4119S, H81) used for immunostaining of FFPE tissues from cell pellets of gastric cancer cell lines, only the ab50887 antibody selectively stained SNU484 cell nuclei. C: Western blot analysis revealed that all four antibodies recognized Twist1, but nonspecific bands appeared in the SNU484 cell line when ab50581 and H81 were used. Depletion of Twist1 by siRNA led to the weakening or disappearance of Twist1 bands for all four antibodies. D: Western blot analysis revealed that only the SNU484 cell line showed a high level of Twist1 protein expression. E: Methylation-specific PCR analysis showed that Twist1 was unmethylated in the SNU484 cell line, whereas all other cell lines showed only methylated products. F: Bisulfite DNA sequencing performed on a region composed of 33 CpG dinucleotides in the SNU484 and SNU601 cell lines demonstrated that SNU484 cells had more than 70% CpG dinucleotides unmethylated in this region, whereas the same region in SNU601 cells was highly methylated.
Western blot analysis revealed that all four antibodies recognized Twist1, but nonspecific bands appeared when ab50581 and H81 antibodies were used, and depletion of Twist1 by siRNA transfection led to the weakening or disappearance of Twist1 bands for all four antibodies (Figure 1C).
Considering that all cell lines other than SNU484 were negative for Twist1 mRNA and that Western blotting showed Twist1 protein expression only in SNU484 cells (Figure 1D), the ab50581, 4119S, and H81 antibodies are considered to show nonspecific immunostaining in FFPE tissues; on the basis of these results, it was concluded that use of any of these three antibodies for immunostaining of FFPE tissue of gastric cancer was inappropriate and that the ab50887 antibody was the only suitable choice for use in further studies.
Twist1 Overexpression in Gastric Cancer Cell Lines Is Associated with Hypomethylation of Its Promoter
To identify the Twist1 expression status in gastric cancer cell lines and to reveal the regulating mechanism of Twist1 expression, MSP and bisulfite-modified sequencing were performed. MSP showed that the Twist1 promoter was unmethylated in SNU484 cells but was methylated in all other cancer cell lines (Figure 1E). Bisulfite-modified sequencing was performed to identify the methylation status of each of the 33 CpG dinucleotides covering the promoter region of Twist1 in SNU484 and SNU601 cell lines. More than 70% of CpG dinucleotides in this region were unmethylated in SNU484 cells, whereas almost all CpG dinucleotides (up to 97%) of the same region were methylated in SNU601 cells (Figure 1F). These results are highly consistent with RT-PCR and Western blot results.
Twist1 Is Rarely Expressed in Gastric Cancer Cells and Is Associated with Patient Survival
We examined Twist1 expression in 332 FFPE gastric tissue samples by immunohistochemistry using the ab50887 Twist1-specific antibody. We evaluated Twist1 expression in epithelial cells and stromal fibroblasts of these tissues separately. In both epithelial cells and stromal fibroblasts, Twist1 staining was predominantly nuclear, whereas cytoplasmic staining was almost absent. Overall, Twist1 expression was generally uncommon in the epithelium, even in the carcinoma, and was detected in 57 of 195 gastric cancer tissues (29.2%) and in 2 of 47 intestinal metaplasia tissues (4.3%). Twist1 was completely absent in the normal epithelium, chronic gastritis, and low- and high-grade dysplasia (Figure 2, A and B). The diffuse-type gastric cancers showed more frequent Twist1 expression than the intestinal type; however, the difference was not significant (34.5% versus 24.0%, P = 0.116). When it was correlated with major clinical parameters, Twist1 expression in carcinoma cells was significantly associated with tumor size (P = 0.004), differentiation (P = 0.007), and lymph node metastasis (P = 0.001) (Table 2). In addition, Twist1 expression in carcinoma cells was significantly associated with poor overall survival (P = 0.0011, Figure 2C).
Figure 2.
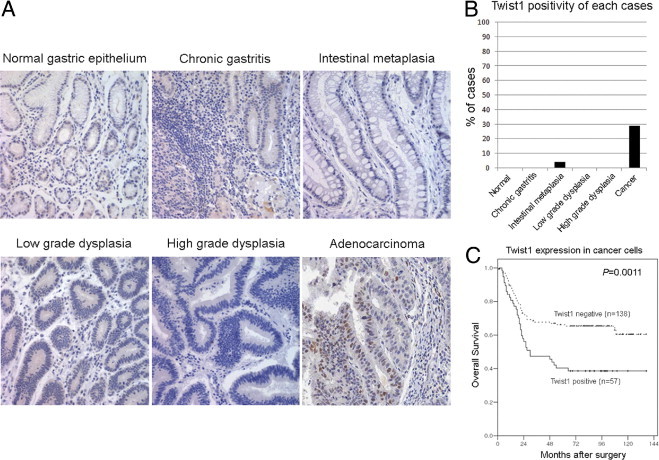
Twist1 expression in gastric tissues and in survival profiles. A: Twist1 immunostaining in human gastric tissues. The figures show nuclear expression of Twist1 in cancer cells and their stromal cells but no expression in other noncancerous tissues. B: Topography of Twist1 expression in each case showed that Twist1 expression was detected in 57 of 195 gastric cancer tissues (29.2%) and 2 of 47 intestinal metaplasia tissues (4.3%), whereas other cases, including normal epithelium, chronic gastritis, and low- and high-grade dysplasia, were negative for Twist1. C: Kaplan-Meier survival curves demonstrate the overall survival among 195 gastric cancer patients defined by Twist1 expression status in cancer cells. Patients with Twist1 expression in cancer cells show poor prognosis.
Table 2.
Relationship between Twist1 Expression of 195 Gastric Cancers and Various Clinicopathologic Factors
| Characteristic | Total | Twist1 expression in cancer cells |
Twist1 expression in cancer associated fibroblasts |
||||
|---|---|---|---|---|---|---|---|
| Negative, no. (%) | Positive, no. (%) | P value | Negative, no. (%) | Positive, no. (%) | P value | ||
| Age, years | |||||||
| <60 | 87 | 62 (71.3) | 25 (28.7) | 0.891 | 24 (27.6) | 63 (72.4) | 0.794 |
| ≥60 | 108 | 76 (70.1) | 32 (29.6) | 28 (25.9) | 80 (74.1) | ||
| Sex | |||||||
| Female | 122 | 86 (70.5) | 36 (29.5) | 0.912 | 30 (24.6) | 92 (75.4) | 0.397 |
| Male | 73 | 52 (71.2) | 21 (28.8) | 22 (30.1) | 51 (69.9) | ||
| Size, cm | |||||||
| <5 | 86 | 70 (81.4) | 16 (18.6) | 0.004 | 34 (39.5) | 52 (60.5) | <0.001 |
| ≥5 | 109 | 68 (62.4) | 41 (37.6) | 18 (16.5) | 91 (83.5) | ||
| Differentiation | |||||||
| WD | 37 | 34 (91.9) | 3 (8.1) | 0.007 | 12 (32.4) | 25 (67.6) | 0.263 |
| MD | 63 | 42 (66.7) | 21 (33.3) | 15 (23.8) | 48 (76.2) | ||
| PD | 63 | 38 (60.3) | 25 (39.7) | 13 (20.6) | 50 (79.4) | ||
| Other types⁎ | 32 | 24 (75.0) | 8 (25.0) | 12 (37.5) | 20 (62.5) | ||
| Lauren classification | |||||||
| Intestine | 100 | 76 (76.0)) | 24 (24.0) | 0.099 | 26 (26.0) | 74 (74.0) | 0.829 |
| Mixed or diffuse | 95 | 62 (65.3) | 33 (34.7) | 26 (27.4) | 69 (72.6) | ||
| pT classification | |||||||
| T1-T2 | 90 | 67 (74.4) | 23 (25.6) | 0.296 | 34 (37.8) | 56 (62.2) | 0.001 |
| T3-T4 | 105 | 71 (67.6) | 34 (33.4) | 18 (17.1) | 87 (82.9) | ||
| Lymph node metastasis | |||||||
| Negative | 71 | 60 (84.5) | 11 (15.5) | 0.001 | 32 (45.1) | 39 (54.9) | <0.001 |
| Positive | 124 | 78 (62.9) | 46 (37.1) | 20 (16.1) | 104 (83.9) | ||
MD, moderately differentiated; PD, poorly differentiated; WD, well differentiated.
Other types included signet ring cell carcinoma and mucinous carcinoma.
Twist1 Is More Frequently Expressed in Gastric Cancer Stromal Cells and Is Associated with CAF
In contrast to the epithelium, Twist1 was more abundantly expressed in stromal fibroblasts. The expression rate of Twist1 in stromal fibroblasts (cases showing ≥1/200 power field in the highest count area) was 73.3% (143/195) for gastric cancer tissue, 16% (4/25) for high-grade dysplasia, 8% (2/25) for low-grade dysplasia, 6.4% (3/47) for intestinal metaplasia, 5.9% (1/17) for chronic gastritis, and 0% (0/23) for normal gastric tissue. In all 56 tumors expressing Twist1 in cancer cells, Twist1 expression was also noted in their stromal fibroblasts. Stromal fibroblasts expressing Twist1 had mainly large, plump, and spindle-shaped cytoplasms with vesicular nuclei, which were located in the immediate vicinity of intestinal-type (Figure 3A) and diffuse-type (Figure 3B) cancer cells. These activated fibroblasts were negative for cytokeratin (Figure 3C) and showed dual expression of SMA and Twist1 (Figure 3D). To quantify Twist1 protein expression in stromal fibroblasts, we counted the number of Twist1-positive stromal fibroblasts per 200× power field in each gastric lesion. It was observed that the average number of stromal fibroblasts showing Twist1 immunopositivity significantly increased in cancerous compared with noncancerous lesions (P < 0.001, Figure 3E). However, there was no significant difference of Twist1 expression between diffuse-type and intestinal-type tissues (P = 0.121).
Figure 3.
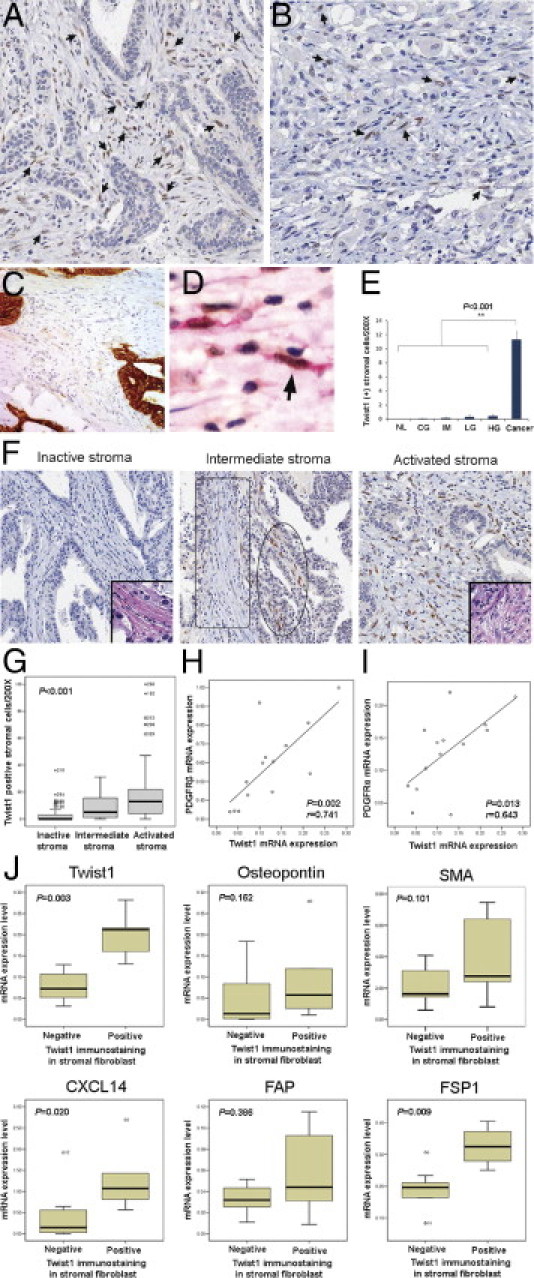
Twist1 expression versus expression of other factors in cancer tissues and stroma. Strong Twist1 nuclear expression was seen in stromal fibroblasts associated with intestinal-type (A) and diffuse-type (B) gastric cancer, which showed large and plump spindle-shaped cell morphologic features (arrows). Plump activated fibroblast cells were negative for pancytokeratin (C) and showed dual expression for SMA and Twist1 (D, arrow). E: Fibroblasts in cancer tissues showed significantly increased expression of Twist1 protein compared with fibroblasts of noncancerous lesions (P < 0.001, Mann-Whitney U-test) (NL, normal; CG, chronic gastritis; IM, intestinal metaplasia; LG, low-grade dysplasia; HG, high-grade dysplasia; CA, cancer). F: In the histologic classification of cancer stroma, an inactive stroma was predominantly composed of thin and wavy inactivated fibroblastic cells (inset, H&E stain, original magnification ×400), which showed mostly negative Twist1 immunostaining. An intermediate stroma was composed of a mix of inactive (box) and activated (circle) fibroblasts, which correlated with Twist1 expression. An activated stroma was predominantly composed of activated fibroblasts showing large and plump spindle-shaped cells (inset, H&E stain, original magnification ×400) and usually expressed Twist1 protein. G: Correlation between cancer stroma classification and the number of Twist1-positive stromal fibroblasts showed that the number of Twist1-positive fibroblasts increased according to gain of activation in the surrounding stroma (P values were calculated by Spearman's test). Twist1 mRNA expression in fibroblasts showed significant linear correlation with PDGFRβ (H) and PDGFRα (I) mRNA expression. Twist1 immunopositive fibroblasts showed significant Twist1 mRNA expression level (P = 0.003), FSP1 (P = 0.009), and CXCL14 (P = 0.020). J: Although osteopontin, SMA, and FAP were also elevated in Twist1-positive fibroblasts, these were not statistically significant.
To identify the relationships between Twist1 expression and CAF, cancer stromata were classified into three groups based on the morphologic features of the stromal fibroblasts. An inactive stroma was defined as a stroma that was composed of predominantly (>75%) thin, wavy, and inactive normal fibroblasts, whereas an activated stroma was composed of predominantly (>75%) large and plump fibroblasts. A stroma falling between these groups was classified as an intermediate stroma. We found that the category of cancer stroma and the number of Twist1-positive stromal fibroblasts were positively correlated (Figure 3F). The number of Twist1-positive fibroblasts significantly increased as more fibroblasts surrounding the cancer were activated (P < 0.001, Figure 3G). These findings indicate that Twist1 expression occurs mainly in CAFs.
To study the mRNA expression pattern of Twist1-positive fibroblasts, stromal fibroblasts around the cancer cells were collected using laser capture microdissection, and mRNA levels were quantitatively determined by real-time RT-PCR. As a result, Twist1 mRNA expression in fibroblasts showed a significant linear correlation with PDGFRβ mRNA expression (P = 0.002, correlation coefficient = 0.741, Figure 3H) and PDGFRα mRNA expression (P = 0.013, correlation coefficient = 0.643, Figure 3I). Furthermore, Twist1 immunopositive fibroblasts showed significantly enhanced levels of Twist1 mRNA (P = 0.003) and other known CAF markers, including FSP1 (P = 0.009) and the CXCL14 chemokine (P = 0.020) (Figure 3J). Osteopontin, SMA, and FAP mRNA levels were also elevated in Twist1-positive fibroblasts but without statistical significance.
Fibroblasts Expressing Twist1 Promote Invasion and Migration of Gastric Cancer Cells and Are Associated with Patient Survival
We investigated the expression of Twist1 in the CCD986sk and IMR910 human fibroblast cell lines using Western blot analyses and found that neither of the two cell lines expressed Twist1. To study the effects of Twist1-expressing fibroblasts on gastric cancer cells, we infected CCD986sk and IMR90 cell lines with lentiviral vectors containing the Twist1 gene. We confirmed that the two cell lines expressed Twist1 after infection and that Twist1 was not expressed in the control gene (GFP) infected cell lines (Figure 4A). We then obtained conditioned media from these cells and cultured the MKN28 gastric cancer cell line with this conditioned medium for 2 days and compared the invasion and migration abilities of MKN28 cells treated with these various conditioned media. In the invasion assays, we identified that conditioned media from Twist1-expressing skin fibroblasts (P = 0.018, Figure 4B) and lung fibroblasts (P = 0.019, Figure 4C) significantly promoted gastric cancer cell invasion. Migration assays also showed more frequent gastric cancer cell migration when treated with conditioned media from Twist1-expressing skin (Figure 4D) and lung (Figure 4E) fibroblasts; however, these findings were not statistically significant.
Figure 4.
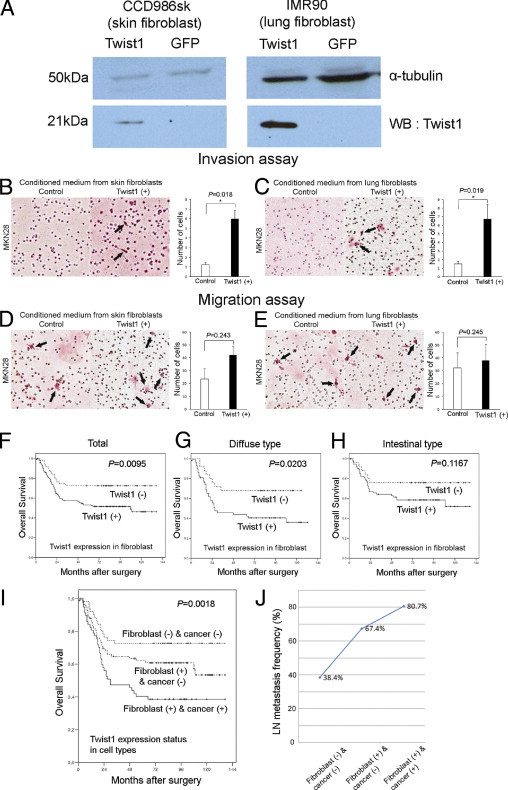
Fibroblasts expressing Twist1 promote invasion and migration of gastric cancer cells and are negatively associated with patient survival. CCD986sk and IMR90 cell lines infected with lentiviral vectors containing the Twist1 gene reveal that both cell lines expressed Twist1 after infection, and Twist1 was not expressed in control gene (GFP) infected cell lines (A). Using conditioned medium from Twist1-infected fibroblasts and control gene infected fibroblasts, Twist1-expressing skin fibroblasts (B) and lung fibroblasts (C) showed an increased number of invading gastric cancer cells (arrows) in the invasion assay. Twist1-expressing skin fibroblasts (D) and lung fibroblasts (E) showed an increased number of migrating gastric cancer cells (arrows) in the migration assay. Kaplan-Meier survival curves demonstrate that the Twist1 expression in stromal fibroblasts was significantly associated with poor overall survival (F). When all cases were stratified by Lauren classification, Twist1 expression in stromal fibroblasts was associated with poor prognosis in the diffuse-type (G), but not in the intestinal-type (H), gastric carcinoma. Kaplan-Meier estimates of overall survival for 195 patients with gastric cancer from the combined data sets according to the expression patterns of Twist1 in stromal fibroblasts and cancer cells. Overall survival rates were worst in the group showing Twist1 positivity in both fibroblasts and carcinoma cells (I). Lymph node metastasis occurred most frequently in the group showing Twist1 positivity in both fibroblasts and carcinoma cells, followed by the group that was Twist1 positive in fibroblasts but negative in carcinoma cells (P < 0.001 for trend) (J).
Then, to identify the prognostic significance of Twist1 expression in stromal fibroblasts in patients with gastric cancer, the cases were regarded as positive when the case showed that the number of Twist1-positive stromal fibroblasts was one or more per 200× power field in the highest stained area. Twist1 expression in stromal fibroblasts was significantly associated with tumor size (P < 0.001), invasion depth (P = 0.001), and lymph node metastasis (P < 0.001) (Table 2). Twist1 expression in stromal fibroblasts was significantly associated with poor overall survival (P = 0.0095, Figure 4F). When all cases were stratified by Lauren classification, Twist1 expression in stromal fibroblasts was associated with poor prognosis in diffuse-type (P = 0.0203, Figure 4G) gastric carcinoma but not in the intestinal type (P = 0.1167, Figure 4H). In multivariate analysis, invasion depth (P < 0.001), lymph node metastasis (P = 0.004), sex (P = 0.044), and Twist1 expression in carcinoma cells (P = 0.045) were variables of independent prognostic significance; however, Twist1 expression in stromal fibroblasts was not an independent prognostic factor (Table 3).
Table 3.
Multivariate Cox Proportional Hazards Model Predicting Survival in 195 Gastric Cancer Patients
| Characteristic | Overall survival HR (95% CI) | P value |
|---|---|---|
| Age | 1.063 (0.676–1.673) | 0.791 |
| Sex | 1.590 (1.013–2.496) | 0.044 |
| Tumor size | 1.222 (0.710–2.104) | 0.470 |
| Differentiation | 0.925 (0.713–1.199) | 0.554 |
| Lauren classification | 0.972 (0.641–1.475) | 0.893 |
| Invasion depth | 2.179 (1.453–3.269) | <0.001 |
| Lymph node metastasis | 3.288 (1.464–7.385) | 0.004 |
| Twist expression in cancer cells | 1.702 (1.012–2.862) | 0.045 |
| Twist expression in stromal cells | 0.928 (0.477–1.802) | 0.825 |
CI, confidence interval; HR, hazard ratio.
To identify the relationship between Twist1 expression in fibroblasts and carcinoma cells, patients were classified into three groups as follows: group 1, Twist1 negative in both fibroblasts and carcinoma cells; group 2, Twist1 positive in fibroblasts but negative in carcinoma cells; and group 3, Twist1 positive in both fibroblasts and carcinoma cells. Overall survival rates were worse in group 3, better in group 2, and best in group 1 (P = 0.0018, Figure 4I). Lymph node metastasis occurred most frequently in group 3, followed by group 2 and group 1 (P < 0.001 for trend, Figure 4J). These findings suggest a synergistic impact of Twist1 expression in fibroblasts and carcinoma cells on gastric cancer patient survival.
Discussion
In this study, we demonstrated that Twist1 is more frequently overexpressed in CAFs than in gastric adenocarcinoma cells and that its expression is also associated with poor prognoses. Our study revealed that the expression rate of Twist1 in gastric cancer was 29.2%, and in those cases Twist1 expression was limited to a small subset of cancer cells. This expression rate was significantly lower than that reported by Yan-Qi et al,12 who found diffuse Twist1 staining in 46.1% of 76 gastric cancer cases. In addition, the reported subcellular location of Twist1 also varied greatly. Specifically, the intracellular location of Twist1 was mostly nuclear in our study, whereas it was mainly cytoplasmic and occasionally nuclear in a previous report.12 This discrepancy can be explained by the difference in antibody used for the detection of Twist1. For example, most studies using the H81 Twist1-specific antibody showed both cytoplasmic and nuclear expression with predominantly cytoplasmic staining in various human cancer tissues,11–13,18–21 whereas a study by Valdés-Mora et al22 that used the same ab50887 Twist1-specific antibody that was used in this study showed predominantly nuclear staining in colorectal cancers.
To solve this problem, we attempted to validate the specificity of antibodies for Twist1. As a result, we found that the H81 antibody was partly nonspecific, especially in FFPE tissues. We also found that the ab50887 antibody specifically bound to Twist1 in both Western blot and immunostaining procedures. In addition, this antibody maintained its specificity even in FFPE tissue. This difference in specificity may stem from antibody recognition epitopes. Specifically, antibodies recognizing the primary sequence may only work in Western blot analyses, whereas antibodies recognizing specific three-dimensional structures may work in immunohistochemistry. On the basis of our results, the ab50887 antibody (clone: Twist2C1a) is considered to be the only one that recognizes a specific Twist1 three-dimensional structure. Furthermore, nuclear localization for Twist1 is functionally reasonable because Twist1 is a known transcription factor. According to Yuen et al,20 the nuclear expression of Twist1 may play a more positive role in promoting metastasis in prostate cancer.
There were some minor discrepancies regarding the expression of Twist1 in gastric cancer cell lines. Twist1 was reported to be up-regulated in MKN28 and AGS cells by Yan-Qi al12; however, in other studies Twist1 expression was found to be down-regulated in MKN28 cells.21,23 In this study, we found that Twist1 expression was down-regulated in MKN28 and AGS cells, of a total of 14 cell lines examined, at both the mRNA and protein levels. This discrepancy can be explained by differences in culture conditions because Twist1 expression can be induced in hypoxic conditions.24 In the current study, our results were supported by MSP and bisulfite sequencing, which confirmed our finding that Twist1 was expressed only in SNU484 cells where the CpG island of the Twist1 promoter was unmethylated. The CpG island was dominantly methylated in all other cell lines that did not express Twist1. Furthermore, the paucity of Twist1 expression in gastric cancer cell lines (1/14) was consistent with the infrequent expression of Twist1 in cancer cells of gastric cancer tissues.
Unexpectedly, we found that Twist1 expression was far more frequent in the stromal cells of gastric cancer than in the cancer cells themselves. In addition, Twist1 expression was rare in the stromal cells of noncancerous gastric tissues. A similar expression pattern of Twist1 was reported in the stromal cells of colorectal cancer in a report by Valdés-Mora et al.22 Interestingly, they used the same ab50887 Twist1-specific antibody that we used in the current study and found that staining was predominantly nuclear. The stromal cells with Twist1 expression were large and plump spindle-shaped cells and were negative for cytokeratin and, at times, positive for SMA. In addition, Twist1-positive fibroblasts showed significantly elevated FSP1 expression, and Twist1 mRNA levels showed a linear correlation with PDGFRα and PDGFRβ mRNA. Recently, FSP1 and PDGFRα have been reported as CAF markers,25,26 and stromal PDGFRβ expression in some cancers, including prostate and breast, is associated with poor patient survival.27,28 Histologic morphology of Twist1-expressing fibroblasts and the results presented here suggest that Twist1 should be considered a CAF marker in gastric cancer. This concept is also supported by the fact that Twist1 expression in stromal fibroblasts was found predominantly in cancer tissues but rarely in other noncancerous lesions.
The source of CAFs is unclear; however, it is likely to include recruitment of circulating fibroblast precursors, the transdifferentiation of epithelial cells into fibroblasts (EMT), and proliferation and activation of resident gastric stromal fibroblasts. Twist1-positive cancer cells are almost always accompanied by Twist1-positive CAFs. The overexpression of Twist1 in CAFs was significantly associated with poor survival in the diffuse-type gastric cancer. Because diffuse-type gastric cancer displays many features of EMT, such as cancer cell dyscohesion, and Twist1 is a major transcription factor regulating EMT, Twist1-positive CAFs can be related to EMT in diffuse-type gastric cancer. Considering all these data, at least a subset of Twist1-positive CAFs was likely to derive from carcinoma cells through EMT. However, the exact origin of Twist1-positive CAFs remains to be clarified and may be of multiple origins instead of a single one.
Regardless of their origins, our study showed that activated stromal fibroblasts frequently expressed Twist1 and that these Twist1-positive CAFs were significantly associated with poor prognosis, advanced invasion, and frequent lymph node metastasis. We also confirmed that Twist1-expressing fibroblasts promoted gastric cancer cell invasion and migration using an in vitro assay. Furthermore, we demonstrated that the patient group with Twist1 expression in both cancer cells and fibroblasts had the most unfavorable prognosis, followed by the patient group with Twist expression in only fibroblasts but not cancer cells. The patient group without Twist1 expression in both fibroblasts and cancer cells had the most favorable prognosis. However, in the multivariate Cox analysis, Twist1 expression in fibroblasts was not an independent prognostic factor, but Twist1 expression in cancer cells was an independent prognostic factor. These findings suggest that Twist1 expression in stromal fibroblasts plays an important role in the progression and aggressive behavior of gastric cancer cells with cross-talk between cancer cells and fibroblasts. There may be synergistic effects between cancer cells and CAFs, and CAFs expressing Twist1 may promote the progression and metastasis of Twist1-positive gastric cancer cells.
According to our study, conditioned media from Twist1-expressing fibroblasts significantly increased the invasion, but not migration, of gastric cancer cells. These findings suggest that invasion factors may be included in the conditioned media from Twist1-expressing fibroblasts. As an example, matrix-degrading enzymes, such as matrix metalloproteinase, may be considered. In fact, extracellular matrix degradation has been reported to be involved in Twist1 signaling,29 and activated fibroblasts are known to be a source of matrix metalloproteinases.2
Recently, many studies have shown that altered fibroblasts can contribute to cancer initiation and progression. Various mechanisms that may explain how stromal cells affect cancer cell progression include angiogenesis and stimulation of tumor growth or metastasis by CAF chemokines or growth factors.2,25–28,30,31 Indeed, Šošić et al32 reported that Twist genes are induced by tumor necrosis factor α in an NF-κB–dependent manner, and Twist regulates cytokine gene expression in fibroblasts. In this study, Twist1 expression was significantly associated with CXCL14 and PDGFR expression in fibroblasts. Recently, Eckert et al29 revealed that Twist1 induced PDGFR expression. These findings suggest that regulation of Twist1 expression is related to chemokines and PDGFR signaling, and Twist1-positive fibroblasts affect progression of cancer cells through chemokines, such as CXCL14, or PDGFR signaling in gastric cancer. It has been demonstrated that paracrine activation of PDGF receptors on fibroblasts acts as a potent signal for tumor stroma recruitment,27,33–35 and chemokines, such as CXCL14 and CXCL12, were shown to bind to receptors on epithelial cells, causing enhanced proliferation and invasion.31
Regarding Twist1 and fibroblasts, only one study has investigated the up-regulation of Twist1 in fibroblasts.36 In this study, Bridges et al36 demonstrated that Twist1 expression in fibroblasts promoted the survival and accumulation of fibroblasts in fibrotic lung disease. Twist1 was also reported to be highly expressed in mesenchymal stem cells.37 In their study, Twist1 enhanced the proliferation of mesenchymal stem cells by the induction of Id-1 and Id-2. Most studies of Twist1 in cancer have focused on its role in EMT of cancer cells; however, there are no studies regarding Twist1 in CAFs. Therefore, to our best knowledge, this is the first study investigating Twist1 in CAFs in gastric cancer. Twist1 expression in fibroblasts requires further study to reveal the more detailed mechanism.
In conclusion, our study demonstrates that CAFs in gastric cancer frequently have altered Twist1 expression and that increased Twist1 expression in fibroblasts contributes to the progression of cancer cells and poor patient survival. Twist1 should also be considered a molecular marker of altered fibroblasts in gastric cancer, and focusing on CAFs with altered Twist1 expression may improve our understanding of cancer development and the effectiveness of gastric cancer treatment.
Footnotes
Supported by a grant from the Samsung Biomedical Research Institute (SBRI-C-B0-212-1).
C.O.S. and K.-W.L. contributed equally to this study.
References
- 1.Wiseman B.S., Werb Z. Stromal effects on mammary gland development and breast cancer. Science. 2002;296:1046–1049. doi: 10.1126/science.1067431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalluri R., Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 3.Trimboli A.J., Cantemir-Stone C.Z., Li F., Wallace J.A., Merchant A., Creasap N., Thompson J.C., Caserta E., Wang H., Chong J.L., Naidu S., Wei G., Sharma S.M., Stephens J.A., Fernandez S.A., Gurcan M.N., Weinstein M.B., Barsky S.H., Yee L., Rosol T.J., Stromberg P.C., Robinson M.L., Pepin F., Hallett M., Park M., Ostrowski M.C., Leone G. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature. 2009;461:1084–1091. doi: 10.1038/nature08486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mueller M.M., Fusenig N.E. Friends or foes - bipolar effects of the tumour stroma in cancer. Nat Rev Cancer. 2004;4:839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 5.Sund M., Kalluri R. Tumor stroma derived biomarkers in cancer. Cancer Metastasis Rev. 2009;28:177–183. doi: 10.1007/s10555-008-9175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hebrok M., Wertz K., Fuchtbauer E.M. M-twist is an inhibitor of muscle differentiation. Dev Biol. 1994;165:537–544. doi: 10.1006/dbio.1994.1273. [DOI] [PubMed] [Google Scholar]
- 7.Lee M.S., Lowe G.N., Strong D.D., Wergedal J.E., Glackin C.A. TWIST, a basic helix-loop-helix transcription factor, can regulate the human osteogenic lineage. J Cell Biochem. 1999;75:566–577. doi: 10.1002/(sici)1097-4644(19991215)75:4<566::aid-jcb3>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 8.Ansieau S., Morel A.P., Hinkal G., Bastid J., Puisieux A. TWISTing an embryonic transcription factor into an oncoprotein. Oncogene. 2010;3(29):3173–3184. doi: 10.1038/onc.2010.92. [DOI] [PubMed] [Google Scholar]
- 9.Puisieux A., Valsesia-Wittmann S., Ansieau S. A twist for survival and cancer progression. Br J Cancer. 2006;94:13–17. doi: 10.1038/sj.bjc.6602876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J., Mani S.A., Donaher J.L., Ramaswamy S., Itzykson R.A., Come C., Savagner P., Gitelman I., Richardson A., Weinberg R.A. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Rosivatz E., Becker I., Specht K., Fricke E., Luber B., Busch R., Höfler H., Becker K.F. Differential expression of the epithelial-mesenchymal transition regulators snail: SIP1, and twist in gastric cancer. Am J Pathol. 2002;161:1881–1891. doi: 10.1016/S0002-9440(10)64464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan-Qi Z., Xue-Yan G., Shuang H., Yu C., Fu-Lin G., Fei-Hu B., Shi-Ren S., Xu-Feng W., Jie D., Dai-Ming F. Expression and significance of TWIST basic helix-loop-helix protein over-expression in gastric cancer. Pathology. 2007;39:470–475. doi: 10.1080/00313020701570053. [DOI] [PubMed] [Google Scholar]
- 13.Song L.B., Liao W.T., Mai H.Q., Zhang H.Z., Zhang L., Li M.Z., Hou J.H., Fu L.W., Huang W.L., Zeng Y.X., Zeng M.S. The clinical significance of twist expression in nasopharyngeal carcinoma. Cancer Lett. 2006;242:258–265. doi: 10.1016/j.canlet.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 14.Gort E.H., Suijkerbuijk K.P., Roothaan S.M., Raman V., Vooijs M., van der Wall E., van Diest P.J. Methylation of the TWIST1 promoter: TWIST1 mRNA levels, and immunohistochemical expression of TWIST1 in breast cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:3325–3330. doi: 10.1158/1055-9965.EPI-08-0472. [DOI] [PubMed] [Google Scholar]
- 15.Kwok W.K., Ling M.T., Lee T.W., Lau T.C., Zhou C., Zhang X., Chua C.W., Chan K.W., Chan F.L., Glackin C., Wong Y.C., Wang X. Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res. 2005;65:5153–5162. doi: 10.1158/0008-5472.CAN-04-3785. [DOI] [PubMed] [Google Scholar]
- 16.Maestro R., Dei Tos A.P., Hamamori Y., Krasnokutsky S., Sartorelli V., Kedes L., Doglioni C., Beach D.H., Hannon G.J. Twist is a potential oncogene that inhibits apoptosis. Genes Dev. 1999;13:2207–2217. doi: 10.1101/gad.13.17.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greene F.L., Page D.L., Fleming I.D., Fritz A., Balch C.M., Haller D.G., Morrow M., editors. AJCC Cancer Staging Manual. 6th ed. Springer; New York: 2002. [Google Scholar]
- 18.Kyo S., Sakaguchi J., Ohno S., Mizumoto Y., Maida Y., Hashimoto M., Nakamura M., Takakura M., Nakajima M., Masutomi K., Inoue M. High Twist expression is involved in infiltrative endometrial cancer and affects patient survival. Hum Pathol. 2006;37:431–438. doi: 10.1016/j.humpath.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 19.Matsuo N., Shiraha H., Fujikawa T., Takaoka N., Ueda N., Tanaka S., Nishina S., Nakanishi Y., Uemura M., Takaki A., Nakamura S., Kobayashi Y., Nouso K., Yagi T., Yamamoto K. Twist expression promotes migration and invasion in hepatocellular carcinoma. BMC Cancer. 2009;9:240. doi: 10.1186/1471-2407-9-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuen H.F., Chua C.W., Chan Y.P., Wong Y.C., Wang X., Chan K.W. Significance of TWIST and E-cadherin expression in the metastatic progression of prostatic cancer. Histopathology. 2007;50:648–658. doi: 10.1111/j.1365-2559.2007.02665.x. [DOI] [PubMed] [Google Scholar]
- 21.Yang Z., Zhang X., Gang H., Li X., Li Z., Wang T., Han J., Luo T., Wen F., Wu X. Up-regulation of gastric cancer cell invasion by Twist is accompanied by N-cadherin and fibronectin expression. Biochem Biophys Res Commun. 2007;358:925–930. doi: 10.1016/j.bbrc.2007.05.023. [DOI] [PubMed] [Google Scholar]
- 22.Valdés-Mora F., Gómez del Pulgar T., Bandrés E., Cejas P., Ramírez de Molina A., Pérez-Palacios R., Gallego-Ortega D., García-Cabezas M.A., Casado E., Larrauri J., Nistal M., González-Barón M., García-Foncillas J., Lacal J.C. TWIST1 overexpression is associated with nodal invasion and male sex in primary colorectal cancer. Ann Surg Oncol. 2009;16:78–87. doi: 10.1245/s10434-008-0166-x. [DOI] [PubMed] [Google Scholar]
- 23.Luo G.Q., Li J.H., Wen J.F., Zhou Y.H., Hu Y.B., Zhou J.H. Effect and mechanism of the Twist gene on invasion and metastasis of gastric carcinoma cells. World J Gastroenterol. 2008;14:2487–2493. doi: 10.3748/wjg.14.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gort E.H., van Haaften G., Verlaan I., Groot A.J., Plasterk R.H., Shvarts A., Suijkerbuijk K.P., van Laar T., van der Wall E., Raman V., van Diest P.J., Tijsterman M., Vooijs M. The TWIST1 oncogene is a direct target of hypoxia-inducible factor-2alpha. Oncogene. 2008;27:1501–1510. doi: 10.1038/sj.onc.1210795. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J., Chen L., Xiao M., Wang C., Qin Z. FSP1+ fibroblasts promote skin carcinogenesis by maintaining MCP-1-mediated macrophage infiltration and chronic inflammation. Am J Pathol. 2011;178:382–390. doi: 10.1016/j.ajpath.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erez N., Truitt M., Olson P., Arron S.T., Hanahan D. Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an NF-kappaB-dependent manner. Cancer Cell. 2010;17:135–147. doi: 10.1016/j.ccr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- 27.Hägglöf C., Hammarsten P., Josefsson A., Stattin P., Paulsson J., Bergh A., Ostman A. Stromal PDGFRbeta expression in prostate tumors and non-malignant prostate tissue predicts prostate cancer survival. PLoS One. 2010;5(5):e10747. doi: 10.1371/journal.pone.0010747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulsson J., Sjöblom T., Micke P., Pontén F., Landberg G., Heldin C.H., Bergh J., Brennan D.J., Jirström K., Ostman A. Prognostic significance of stromal platelet-derived growth factor beta-receptor expression in human breast cancer. Am J Pathol. 2009;175:334–341. doi: 10.2353/ajpath.2009.081030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eckert M.A., Lwin T.M., Chang A.T., Kim J., Danis E., Ohno-Machado L., Yang J. Twist1-induced invadopodia formation promotes tumor metastasis. Cancer Cell. 2011;19:372–386. doi: 10.1016/j.ccr.2011.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitadai Y. Cancer-stromal cell interaction and tumor angiogenesis in gastric cancer. Cancer Microenviron. 2010;3:109–116. doi: 10.1007/s12307-009-0032-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhowmick N.A., Neilson E.G., Moses H.L. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Šošić D., Richardson J.A., Yu K., Ornitz D.M., Olson E.N. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell. 2003;112:169–180. doi: 10.1016/s0092-8674(03)00002-3. [DOI] [PubMed] [Google Scholar]
- 33.Ostman A., Heldin C.H. PDGF receptors as targets in tumor treatment. Adv Cancer Res. 2007;97:247–274. doi: 10.1016/S0065-230X(06)97011-0. [DOI] [PubMed] [Google Scholar]
- 34.Skobe M., Fusenig N.E. Tumorigenic conversion of immortal human keratinocytes through stromal cell activation. Proc Natl Acad Sci U S A. 1998;95:1050–1055. doi: 10.1073/pnas.95.3.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forsberg K., Valyi-Nagy I., Heldin C.H., Herlyn M., Westermark B. Platelet-derived growth factor (PDGF) in oncogenesis: development of a vascular connective tissue stroma in xenotransplanted human melanoma producing PDGF-BB. Proc Natl Acad Sci U S A. 1993;90:393–397. doi: 10.1073/pnas.90.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bridges R.S., Kass D., Loh K., Glackin C., Borczuk A.C., Greenberg S. Gene expression profiling of pulmonary fibrosis identifies Twist1 as an antiapoptotic molecular “rectifier” of growth factor signaling. Am J Pathol. 2009;175:235–261. doi: 10.2353/ajpath.2009.080954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isenmann S., Arthur A., Zannettino A.C., Turner J.L., Shi S., Glackin C.A., Gronthos S. TWIST family of basic helix-loop-helix transcription factors mediate human mesenchymal stem cell growth and commitment. Stem Cells. 2009;27:2457–2468. doi: 10.1002/stem.181. [DOI] [PubMed] [Google Scholar]


