Abstract
Jarid2 is required for the genomic recruitment of the polycomb repressive complex-2 (PRC2) in embryonic stem cells. However, its specific role during late development and adult tissues remains largely uncharacterized. Here, we show that deletion of Jarid2 in mouse epidermis reduces the proliferation and potentiates the differentiation of postnatal epidermal progenitors, without affecting epidermal development. In neonatal epidermis, Jarid2 deficiency reduces H3K27 trimethylation, a chromatin repressive mark, in epidermal differentiation genes previously shown to be targets of the PRC2. However, in adult epidermis Jarid2 depletion does not affect interfollicular epidermal differentiation but results in delayed hair follicle (HF) cycling as a consequence of decreased proliferation of HF stem cells and their progeny. We conclude that Jarid2 is required for the scheduled proliferation of epidermal stem and progenitor cells necessary to maintain epidermal homeostasis.
Keywords: epidermal homeostasis, epidermal stem cells, Jarid2, polycomb group proteins, polycomb repressive complex-2
Introduction
Polycomb (PcG) proteins are global transcriptional repressors that balance cell lineage choices during embryonic development (Surface et al, 2010). Misregulation of PcG proteins impinges on different aspects of tumour onset and progression of various types of human neoplasias (Sauvageau and Sauvageau, 2010). The two major polycomb complexes in mammals, polycomb repressive complex 1 (PRC1) and 2 (PRC2), interact with chromatin to sequentially establish distinct post-translational modifications of histone tails, which determine the degree of transcriptional repression of specific loci. The mammalian PRC2 complex is established by the interactions of the Ezh2 (or Ezh1), Eed, Suz12, and RBAP46/48 subunits (Cao et al, 2002; Czermin et al, 2002; Kuzmichev et al, 2002; Cao and Zhang, 2004; Kirmizis et al, 2004; Margueron et al, 2008; Shen et al, 2009). Accumulation at transcriptional start sites of the methyltransferase Ezh2 subunit, which trimethylates lysine 27 of histone H3 (to give H3K27me3), correlates with transcriptional repression. The PRC1 complex, which is comprising the CBX, RING1, BMI, and PH proteins (Levine et al, 2002), contributes to promoter silencing by catalysing the mono-ubiquitination of histone H2A on lysine 119. It has been shown that the chromodomains of the CBX subunits have an affinity for H3K27me3, which would allow for the simultaneous occupancy of PRC1 and PRC2, leading to further transcriptional repression (de Napoles et al, 2004; Wang et al, 2004; Kouzarides, 2007).
Several publications have recently shown that Jarid2, the founding member of the Jumonji/JmjC domain-containing family of proteins, is necessary for the recruitment of PRC2 to promoters in ES cells (Peng et al, 2009; Shen et al, 2009; Landeira et al, 2010; Li et al, 2010; Pasini et al, 2010). However, whereas the role of Jarid2 in the genomic recruitment of PRC2 is undisputed, its impact on PRC2 activity is less clear (Landeira and Fisher, 2011; Herz and Shilatifard, 2010). In this sense, this interaction has been reported by some to inhibit (Peng et al, 2009; Shen et al, 2009), and by others to potentiate (Li et al, 2010; Pasini et al, 2010), the H3K27me3 methyltransferase activity of PRC2. In addition, although the reduced genomic recruitment of PRC2 in Jarid2-depleted ESCs translated into more-or-less pronounced changes in the transcript levels of polycomb target genes, it was accompanied by either no, or only subtle, changes in H3K27me3 enrichment at their promoter regions. Furthermore, Jarid2 also recruits PRC1 to the promoters of lineage-commitment genes and which are poised for expression during the initial steps of ESC differentiation (Landeira et al, 2010).
Deletion of PRC2 subunits in ESCs causes profound changes in the expression of lineage-commitment genes, resulting in early lethality (Sauvageau and Sauvageau, 2010; Surface et al, 2010). However, less is known about the function of PRC2 in adult tissues. Deletion of Ezh2 causes defects in proliferation and differentiation of haematopoietic, pancreatic, and brain progenitors (Su et al, 2003; Chen et al, 2009; Majewski et al, 2010; Pereira et al, 2010). With respect to the epidermis, deletion of Ezh2 results in progenitors that have a reduced proliferative potential and undergo premature differentiation during embryonic development (Ezhkova et al, 2009). In addition, deletion of both Ezh2 and Ezh1 results in a progressive hair loss and in hyperproliferation of basal epidermal cells (Ezhkova et al, 2009, 2011). However, the mechanism by which PRC2 is recruited to specific loci in adult tissues, and which factors modulate its function, is still poorly understood. In addition, a role for Jarid2 plays in the genomic recruitment of PRC2 in adult tissues has not been described yet.
Here, we have analysed the effects of deleting Jarid2 on late murine epidermal development, as well as on adult epidermal homeostasis. We have characterized the role of Jarid2 over murine epidermal and hair follicle (HF) embryonic development, postnatal morphogenesis, and adult homeostasis. In addition, we have studied whether epidermal deletion of Jarid2 had any effect over PRC2 activity.
Results and discussion
Epidermal deletion of Jarid2 does not affect embryonic development of the epidermis
We conditionally deleted Jarid2 expression in murine epidermis by crossing Jarid2loxP/loxP mice with transgenic mice expressing Cre recombinase under the regulation of the basal layer keratinocyte-specific keratin-14 promoter (hereafter referred to as Jarid2 cKO mice; Mysliwiec et al, 2006). We verified by genomic PCR, RT–qPCR of Jarid2 transcript, and western immunoblotting that Jarid2 was efficiently deleted in newborn epidermal keratinocytes of Jarid2 cKO mice (Supplementary Figure S1A, S1C and S1D). In addition, we verified that Cre expression was homogenous throughout the entire epidermal compartment by crossing Jarid2 cKO mice with ROSA26-YFP transgenic mice (Supplementary Figure S1B).
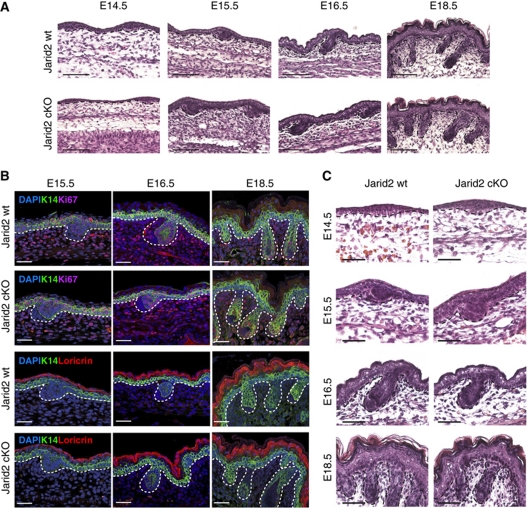
The keratin-14-Cre transgene drives Cre expression during development of embryonic ectoderm at post-coitum day 13.5 (E13.5), coinciding with the onset of embryonic epidermal stratification. Therefore, our mouse model allowed us to study whether Jarid2 deletion had any effect on epidermal development. Histological analysis of the epidermis of E14.5–E19.5 Jarid2wt/wt/K14Cre (wt mice), and Jarid2 cKO littermate embryos, did not reveal any changes in epidermal stratification, thickness, and overall morphology (Figure 1A). The expression of proliferation markers (Ki67), basal keratin-14 (K14), and terminal differentiation genes (loricrin) was indistinguishable between wt and Jarid2 cKO mice (Figure 1B).
Figure 1.
Epidermal deletion of Jarid2 does not affect embryonic development of the epidermis. (A) Haematoxylin–eosin staining revealed no abnormalities in epidermal embryonic development of Jarid2 cKO mice. At least three control (K14Cre/Jarid2wt/wt) and cKO mice were analysed for each time point shown. Scale bar: 100 μm. (B) Immunofluorescence of Ki67 and loricrin indicates that proliferation and differentiation are not modified during embryonic epidermal development upon Jarid2 deletion. Scale bar: 50 μm. (C) Higher magnification of skin sections shows a normal architecture for the embryonic hair follicles in absence of Jarid2. Scale bar: 50 μm.
Embryonic HFs start to form from basal epidermal progenitors that invaginate around E15.5, to form hair placodes (Schneider et al, 2009). From E15.5 to birth, hair placodes grow inwards to give rise to embryonic HF structures, which contain embryonic stem cells that remain present throughout the entire life of the organism. No changes in the number, spacing, or morphology of the HF placodes were observed in Jarid2 cKO embryos, or in any of the subsequent HF embryonic morphogenesis stages from E15.5 until birth (Figure 1C). Additionally, HF placodes from wt and Jarid2 cKO embryos expressed similar levels of proliferation markers and basal keratin-14 (Figure 1B).
Therefore, Jarid2 is dispensable for late embryonic epidermal development and HF morphogenesis. Interestingly, contrary to the expression pattern of Ezh2 in embryonic epidermis (Ezhkova et al, 2009), the expression of Jarid2 was the same in basal and suprabasal epidermal progenitors isolated from embryonic epidermis at E16.5 and E18.5 by FACS on the basis of their differential expression of integrin α6 (α6bright and α6dim, respectively; Supplementary Figure S2). Given that epidermal deletion of Ezh2 resulted in premature expression of terminal differentiation markers during embryonic epidermal development (Ezhkova et al, 2009), it is likely that the Ezh2-containing PRC2 complex regulates this process independently of Jarid2.
Deletion of Jarid2 results in enhanced postnatal epidermal differentiation
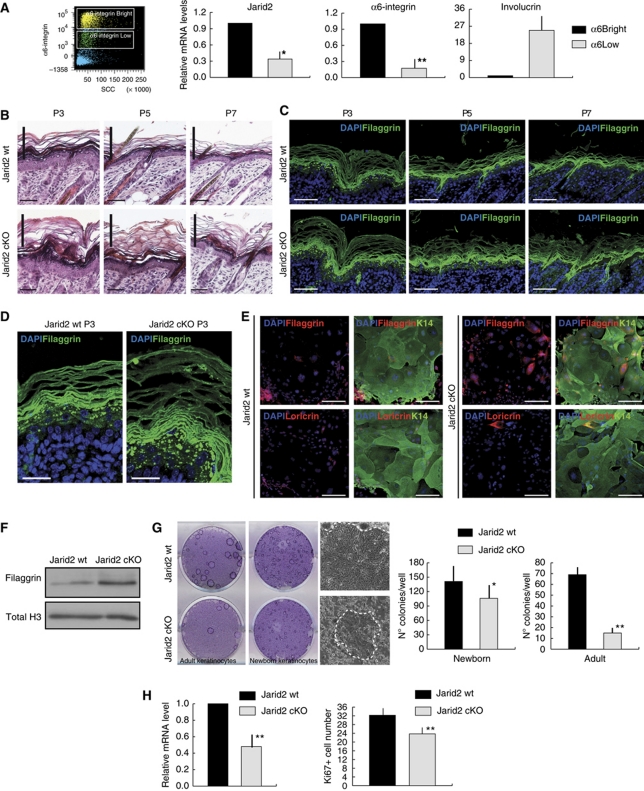
We next studied whether deletion of Jarid2 affected early postnatal epidermal morphogenesis. Expression of Jarid2 decreases as embryonic stem cells differentiate (Peng et al, 2009; Shen et al, 2009; Landeira et al, 2010; Li et al, 2010; Pasini et al, 2010). Intriguingly, although the expression of Jarid2 was similar between basal and suprabasal embryonic epidermal keratinocytes (Supplementary Figure S2), FACS sorting by α6 integrin expression revealed that basal epidermal progenitors isolated from neonatal P0 epidermis (with high α6; α6bright) had high Jarid2 transcript levels, while suprabasal differentiated cells (α6low) exhibited decreased Jarid2 transcript levels (Figure 2A).
Figure 2.
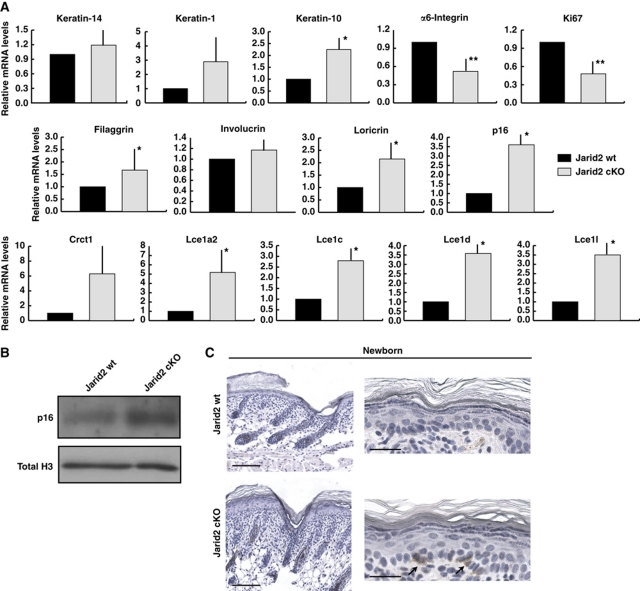
Jarid2 deletion results in enhanced postnatal epidermal differentiation. (A) Differential expression of Jarid2 in basal and suprabasal interfollicular epidermal cells, from P1 wild-type mice, that were FACS sorted on the basis of their cell surface levels of α6-integrin (bright, dim, and low). Data shown represent the mean average; vertical bars represent the s.e.m. N=12; *P< 0.05 and **P<0.01. A representative dot-plot of the sorting strategy is shown in the left panel. (B) Jarid2 cKO neonatal mice showed increased differentiation and cornification. P3, P5, and P7 are shown. Differences in the thickness of the cornified layer between control and cKO mice are highlighted with a vertical bar. Scale bar: 50 μm. N >2 wt and 2 cKO for time points. (C) The interfollicular epidermis of Jarid2 cKO mice contained a thicker filaggrin-positive layer than control mice. Scale bar: 50 μm. (D) Higher magnification of filaggrin staining of Jarid2 wt and Jarid2 cKO mice highlights the differences in the thickness of the granular layer. Scale bar: 25 μm. (E) Expression of filaggrin and loricrin is increased in newborn mouse keratinocytes isolated from Jarid2 cKO mice compared with control littermates. Keratin-14 and DAPI (green and blue fluorescence, respectively) are shown as counterstains. Pictures are representative of three independent experiments from three different litters of mice. Scale bar: 100 μm. (F) Western blot showing increased filaggrin expression in Jarid2 cKO newborn keratinocytes. Total H3 is used for normalization. (G) Deletion of Jarid2 reduced the clonogenic potential of newborn (P0) and adult (8-weeks old) primary mouse keratinocytes. Representative colonies from control and Jarid2 cKO mice are shown. Quantification of the colonies number per well is reported in the right. Values are presented as mean+s.e.m.; *P<0.05 and **P<0.01; N=3. (H) The interfollicular epidermis of Jarid2 cKO mice is less proliferative than that of control mice. Left panel: sorted α6-integrin bright cells from P1 Jarid2 cKO mice express lower transcript levels of Ki67 than their control littermates (n=14 wt, n=12 cKO; the vertical bar represents the s.e.m.; **P<0.05). Right panel: Quantification of Ki67+ cells in the interfollicular epidermis of Jarid2 wt and Jarid2 cKO newborn mice. N=4 mice for each genotype; vertical bar represents s.e.m.; **P<0.05.
During the first 2 weeks after birth, strong proliferation is coupled with differentiation in the epidermis, to establish the definitive adult epidermal barrier. In addition, the first pelage emerges from the HF structures that formed during embryonic development (Schneider et al, 2009). A daily time course analysis from postnatal days 0 (P0) to 7 (P7) indicated that Jarid2 cKO mice had an expanded suprabasal granular layer and a thickened cornified envelope, compared with control littermates (Figure 2B). Expression of the interfollicular epidermis granular layer marker filaggrin was increased upon deletion of Jarid2, indicating an increased epidermal differentiation upon depletion of Jarid2 (Figure 2C and D). Enhanced expression of filaggrin was also observed by immunofluorescence and western immunoblotting in cultured primary mouse keratinocytes isolated from Jarid2 cKO P0 epidermis compared with control littermates (Figure 2E and F).
The enhanced differentiation of the interfollicular epidermis was accompanied by a reduction in the proliferative potential of basal interfollicular progenitors. In this sense, both newborn and adult keratinocytes purified from Jarid2 cKO epidermis formed fewer proliferative clones as compared with wt cells (Figure 2G). Jarid2 cKO cells not only formed fewer colonies but also displayed a morphology characteristic of terminally differentiated abortive keratinocytes, as expected from the increase expression of differentiation markers (Figure 2G). Accordingly, we scored a statistically significant reduction in the number of proliferating basal interfollicular cells, as well as the transcript levels of the proliferation marker Ki67, in Jarid2 cKO epidermis compared with wt epidermis (Figure 2H).
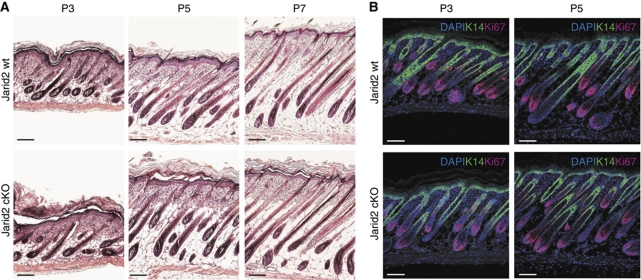
Although the interfollicular epidermis of neonatal mice was less proliferative upon depletion of Jarid2, both postnatal HF morphogenesis and pelage growth were unaffected in Jarid2 cKO mice (Figure 3A). Accordingly, proliferation along the outer root sheath and the matrix of wild-type and Jarid2 cKO HFs was undistinguishable (Figure 3B).
Figure 3.
Jarid2 does not influence postnatal hair follicle development. (A) Neonatal hair follicle morphogenesis is not altered upon the loss of Jarid2. Scale bar: 100 μm. (B) Ki67 expression (red fluorescence) was unchanged in the hair follicles of neonatal Jarid2 mice compared with control mice, but was slightly reduced in the basal IFE. Keratin-14 staining is shown as green fluorescence. Scale bar: 100 μm.
Altogether, these results indicate that loss of Jarid2 reduces the proliferative potential of basal epidermal progenitors while enhancing their differentiation, whereas it has no effect on the formation of the first wave of postnatal pelage formation.
Deletion of Jarid2 results in inefficient entry of HFs into anagen
At postnatal days P8–P10, a population of quiescent epidermal stem cells is stably established at a permanent region of the HF, located just below the sebaceous glands, termed the bulge (Nowak et al, 2008). Bulge cells remain slow cycling throughout the entire lifetime of the organism, and only undergo a reduced number of proliferative rounds during each cycle of adult hair morphogenesis (Tiede et al, 2007). After the first bout of postnatal pelage formation, all the HFs synchronously undergo a second phase of HF growth (anagen) between P20 and P31, to establish the adult pelage (Blanpain et al, 2004; Morris et al, 2004; Tumbar et al, 2004; Schneider et al, 2009). Maintenance of the interfollicular epidermis does not depend on HF stem cells (Morris et al, 2004; Tumbar et al, 2004; Levy et al, 2005), but relies on continuous proliferation of basal interfollicular epidermal cells for its maintenance during adulthood (Clayton et al, 2007). We, therefore, next studied whether deletion of Jarid2 had (i) any impact on the maintenance of the quiescent bulge stem cell population; (ii) activation of bulge stem cells during synchronous HF growth; and (iii) homeostasis of basal interfollicular epidermal progenitors.
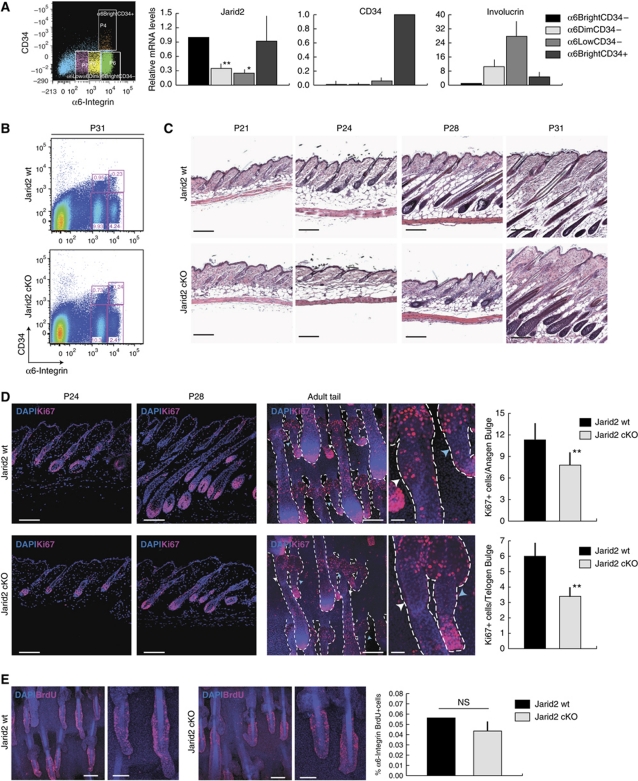
Similar to the situation in the neonatal epidermis, expression of Jarid2 in adult P19 mouse skin was highest in basal undifferentiated epidermal progenitors (α6bright/CD34neg), and lower in the progressively differentiated keratinocytes (α6dim and α6low populations; Figure 4A). Expression of Jarid2 was also gradually reduced upon differentiation (as measured by the cell surface levels of integrin α6) of adult human epidermal keratinocytes, directly isolated from foreskin samples and maintained in culture (Supplementary Figure S3).
Figure 4.
Deletion of Jarid2 results in inefficient entry of hair follicles into anagen in adult epidermis. (A) Jarid2 expression was reduced in suprabasal interfollicular epidermal cells, compared with basal cells, in adult wild-type mice. Adult mouse keratinocytes were sorted from the backskin of P19 mice. Data are presented as mean+s.e.m.; N=3 pools of 4 mice each; *P<0.09 and **P<0.05. (B) Jarid2 cKO mice did not show significant changes in the proportion of bulge stem cells (α6bright/CD34+) with respect to control littermates. FACS profiles shown correspond to P31 mice. N=4 Jarid2 wt and 4 Jarid2 cKO for each time point analysed. (C) Deletion of Jarid2 resulted in a delay in the onset and progression of anagen between P21 and P31. Scale bar: 200 μm. N=8 controls and 7 cKO for each time point analysed. (D) Ki67 staining in P24 and P28, both control and Jarid2 cKO mice, indicates reduced proliferation in matrix, bulge, and ORS. Scale bar: 100 μm. The right panel shows whole-mount Ki67 immunostaining of tail epidermis of control and Jarid2 cKO P24 mice. Jarid2 cKO mice showed a significant reduction in the number of Ki67+ cells in the bulge, outer root sheath and matrix of late anagen (white arrow) and early anagen (light blue arrow) hair follicles. Scale bars: 100 and 25 μm, respectively. Quantification of the number of Ki67+ cells for either anagen (upper part) or telogen (lower graph) bulge is reported. Data are presented as average+s.e.m. **P<0.05; N=20 bulges. (E) Jarid2 deletion does not affect the number of bulge label-retaining cells (LRCs) pulsed chased for 8 weeks. Scale bars: 100 and 50 μm. Right panel: measurement of the number of BrdU+ bulge cells by FACS analysis in the epidermis of 8-week-old Jarid2 wt and Jarid2 cKO mice. Data are presented as average+s.e.m. n.s. indicates non-statistically significant. N=4 Jarid2 wt and 4 Jarid2 cKO.
At adulthood, the percentage of bulge stem cells in the dorsal skin of Jarid2 cKO mice, as determined by their high expression of α6 integrin and CD34 (α6bright/CD34pos), was virtually identical to that of control mice, indicating that Jarid2 is not necessary to establish or to maintain HF bulge stem cells (Figure 4B). However, although the total number of bulge stem cells was unaffected in Jarid2 cKO mice, the mice displayed a significant delay in the onset and the progression of hair follicles into anagen from postnatal days P21 to P31, with respect to control littermates (Figure 4C). Accordingly, proliferation of hair germ cells, outer root sheath cells, and matrix cells, all of which are necessary to fuel HF anagen (Ito et al, 2004; Greco et al, 2009; Hsu et al, 2011), was significantly delayed in Jarid2 cKO mice between P21 and P31 as compared with control littermates (Figure 4D, only P24 and P28 time points are shown as representative). In addition, we observed a significant reduction in the number of Ki67+ proliferative cells in the bulge of HFs in full anagen (Figure 4D). In spite of these changes in proliferation, the HFs of Jarid2 cKO mice resumed their anagen cycle, resulting in normal, albeit delayed, hair growth. Although bulge proliferation was affected at late stages of anagen, a similar number of bulge cells were capable of stably accumulating BrdU, following an 8-week chase, in Jarid2 cKO and wt mice (Figure 4E).
Jarid2 cKO HFs were more inefficient than wt ones in becoming active not only in physiological conditions, but also when ectopically stimulated enter anagen. In this sense, Jarid2 cKO bulge and hair matrix cells were less efficient in responding to topical administration of the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA), which causes bulge hyperproliferation and ectopic anagen entry (Supplementary Figure S4A). Overall, Jarid2 cKO mice did end up responding to TPA treatment, displaying HFs in full anagen (Supplementary Figure S4B). At last, while the interfollicular epidermis of JaridKO mice was responsive to TPA, it displayed a thicker cornified layer than treated wild-type littermates and a higher expression of the differentiation marker loricrin, similar to the effect observed in the interfollicular epidermis of neonatal Jarid2 cKO mice (Supplementary Figure S4C).
These results suggest that Jarid2 activity is essential in instances where strong proliferation is required (i.e., during postnatal morphogenesis, at the peak of anagen, and during TPA-induced hyperproliferation). However, deletion of Jarid2 does not impede either normal, or ectopically induced, entry of HFs into the anagen state, indicating that Jarid2 is required for the scheduled activation of HF bulge stem cells and their progeny, rather than being essential for their proliferation per se.
Deletion of Jarid2 results in reduced PRC2 genomic occupancy, lower H3K27me3, and increased expression of PRC2-epidermal target genes in neonatal, but not adult epidermis
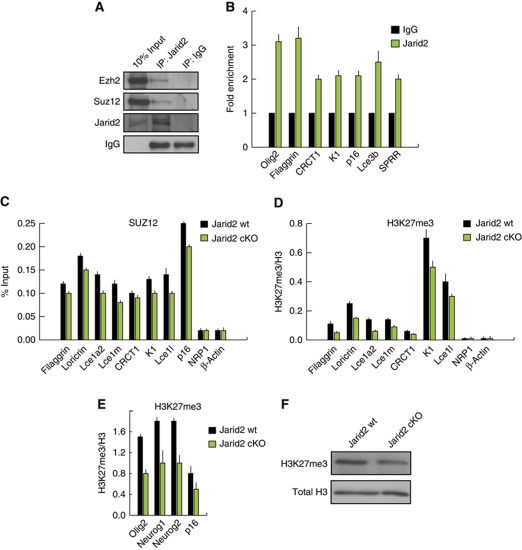
Depletion of Jarid2 in embryonic stem cells significantly impairs binding of the PRC2 core complex subunits to their genomic targets (Herz and Shilatifard, 2010; Landeira and Fisher, 2011). However, the effect of Jarid2 on the histone H3K27 methyltransferase activity of PRC2, and its consequences for the expression of polycomb targets in ES cells or adult stem cells, is still open to debate. We first confirmed that endogenous Jarid2 interacted with endogenous PRC2 (Suz12 and Ezh2) in primary mouse keratinocytes (Figure 5A). We, therefore, next tested whether epidermal deletion of Jarid2: (i) affected genomic targeting of PRC2; (ii) modified the chromatin, by affecting the H3K27 trimethylation (H3K27me3) levels; and (iii) changed the expression of epidermal differentiation genes that have previously shown to be PRC2 targets in epidermal basal progenitors (Ezhkova et al, 2009).
Figure 5.
Jarid2 modulates PRC2 activity in newborn epidermal keratinocytes. (A) Endogenous Jarid2 co-immunoprecipitates with endogenous Ezh2 and Suz12 PRC2 in newborn mouse keratinocytes. (B) Jarid2 localizes to the promoters of PRC2 targets in newborn mouse keratinocytes, as shown by ChIP-qPCR. Data are presented as fold enrichment and are representative of three independent experiments. (C) ChIP analysis in newborn mouse keratinocytes isolated from Jarid2 cKO (n=15) and control mice (n=20) indicates that the occupancy of Suz12 in the promoters of epidermal differentiation and non-epidermal-specific PRC2 targets genes, is reduced in Jarid2 cKO mice. Data are presented as percentage of input. (D) The levels of H3K27me3 are reduced in the promoters of the same cohort of genes in Jarid2 cKO cells. Data are presented as fold enrichment normalized to total histone H3. (E) The levels of H3K27me3 in non-epidermal targets of PRC2 are reduced in Jarid2cKO cells. (F) The global level of H3K27me3 is slightly reduced in Jarid2 cKO mouse keratinocytes compared with the control cells. Total H3 was used as a control of equal loading. All ChIP data shown is statistically significant with a P-value <0.05.
We first performed these analyses in keratinocytes from newborn mice, at which time Jarid2 cKO and control mice showed clear differences in interfollicular epidermal differentiation (Figure 2). Jarid2 localized to the proximal promoter region of several PRC2-target genes (both epidermal and non-epidermal specific), as determined by chromatin immunoprecipitation (ChIP) followed by quantitative PCR (Ezhkova et al, 2009; Figure 5B). We next tested whether deletion of Jarid2 would affect the occupancy of Suz12 (i.e., PRC2) and the levels of H3K27me3 in the promoter regions of epidermal differentiation genes, previously shown to be targets of PRC2 (Ezhkova et al, 2009). Jarid2 cKO neonatal keratinocytes had reduced levels of Suz12 and H3K27me3 in both epidermal differentiation genes and non-epidermal specific PRC2 targets (note that p16, Olig2, Neurog1, and Neurog2 are representative of the non-epidermal PRC2 targets; Figure 5C–E). The overall levels of H3K27me3 around the TSS of epidermal differentiation genes were considerably lower than in non-epidermal-specific PRC2 targets, in agreement with previous findings (Figure 5D; Ezhkova et al, 2009). In addition, Jarid2 cKO cells had lower levels of overall H3K27me3 than control cells (Figure 5F). This slight reduction, albeit significant, was similar to that observed upon deletion of Jarid2 both in the levels of H3K27me3 in some lineage-commitment genes, and total level of H3K27me3, in ES cells (Peng et al, 2009; Landeira et al, 2010; Li et al, 2010; Pasini et al, 2010).
As expected from our in vivo and ChIP analyses, loss of Jarid2 correlated with an increased transcription of this same group of genes involved in epidermal differentiation in purified basal (α6bright) cells isolated from newborn mice, as compared with wt littermates (Figure 6A). The same pattern was observed in keratinocytes isolated from P8 mice, a time in which increased differentiation was still evident in the epidermis of Jarid2 cKO mice (Supplementary Figure S5B). The level of Cdkn2a/p16 (Ink4a locus) transcript and protein, a bona fide target of the PRC2 complex in all the tissues studied to date, including basal interfollicular epidermal cells and HF stem cells, was also upregulated in Jarid2 cKO keratinocytes (Sauvageau and Sauvageau, 2010; Ezhkova et al, 2009, 2011; Figures 5B–D, 6A, and B). In agreement with this, some basal cells of the interfollicular epidermis, but not HFs, of Jarid2 cKO newborn mice showed expression of p16, compared with wt littermates (Figure 6C; note that we did not detect any basal cell positive for p16 in the epidermis of any of the wt mice analysed). Cdkn2a/p16 inhibits G1/S transition and is a marker of senescent cells, and its repression by PRC2 is essential to maintain the proliferative state of undifferentiated ES and tumour cells (Sauvageau and Sauvageau, 2010). Accordingly, the transcript levels of the basal marker integrin α6 and the proliferation marker Ki67 were downregulated in Jarid2 cKO cells, as a further indication of their increased differentiation and higher levels of p16, and in agreement with the epidermal phenotype we observed in Jarid2 cKO mice (Figure 6B).
Figure 6.
Deletion of Jarid2 results in enhanced expression of interfollicular epidermis differentiation genes. (A) The transcript levels (RT–qPCR) of epidermal specific and non-epidermal-specific targets of PRC2 are increased in basal interfollicular keratinocytes (α6bright/CD34− population) from Jarid2 cKO mice compared with the same wt control population. mRNA levels for HPRT were used for normalization. Data are mean+s.e.m. N=13 Jarid2 wt and 12 Jarid2 cKO; *P<0.09 and **P<0.05. (B) Western blot showing p16 protein levels in Jarid2 wt and Jarid2 cKO newborn keratinocytes. Total H3 was used for normalization. (C) Representative immunohistochemistry showing positive expression of p16 in the interfollicular epidermis of Jarid2 cKO newborn animals. Note that we did not observe any p16-positive cell in the epidermis of control mice (N=4 for wt and Jarid2 cKO mice). Scale bars: 100 μm left panel and 20 μm right panel.
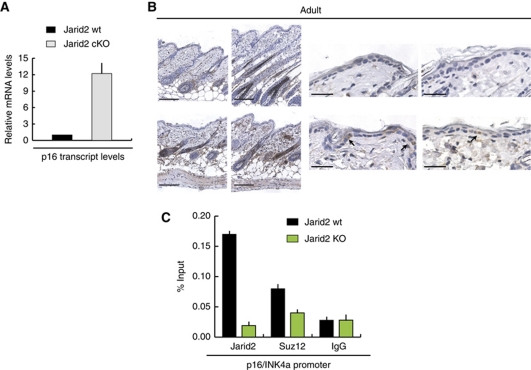
Interestingly, when the same overall molecular analysis was performed using basal (α6bright) keratinocytes isolated from the epidermis of adult (8 weeks) mice, the expression of none of the epidermal genes described above was statistically different between Jarid2 cKO and control mice (data not shown). However, we did observe a significant increase in the transcript levels of p16 in cells isolated from Jarid2 cKO compared with wt cells (Figure 7A). Enhanced transcription of the Ink4a locus resulted in positive immunostaining for p16 in the epidermis and HFs of adult Jarid2 cKO mice (Figure 7B). Accordingly, both Jarid2 and Suz12 localized to the promoter region of p16, and this occupancy was significantly reduced in Jarid2 cKO keratinocytes (Figure 7C).
Figure 7.
Deletion of Jarid2 does not affect the expression of epidermal differentiation genes, but reduces the genomic occupancy of PRC2 in the p16/INK4a locus. (A) The transcript level of Cdkn2a/p16 mRNA is elevated in basal epidermal keratinocytes isolated from 8-week-old mice compared with their wt counterparts. (B) Immunohistochemical analysis showing increased expression of p16 in the interfollicular epidermis and hair follicles in the backskin of Jarid2 cKO adult mice compared with their control littermates. Scale bars: 100 and 20 μm, respectively. (C) Jarid2 and Suz12 colocalize on the promoter of the p16/INK4a locus. ChIP was performed on primary mouse keratinocytes isolated from 8-week-old Jarid2 cKO and control mice (n=5 for each). Data are presented as percentage of the input.
Discussion
Our results indicate that Jarid2 is required for instances when robust proliferation of interfollicular and HF progenitor and stem cells is required. However, the predominant role of Jarid2 is to ensure that the proliferative and undifferentiated state occurs in schedule, but overall its activity is dispensable for proliferation in neonatal and young adult mice. At the molecular level, while loss of Jarid2 does not completely abolish PRC2 genomic occupancy and trimethylation of H3K27 at epidermal PRC2 targets, it does lead to a reduction in the level of both in neonatal epidermis. These mild effects on PRC2 function are nevertheless causal, since deletion of Jarid2 results in enhanced differentiation and reduced proliferation of epidermal progenitors. Intriguingly, Ezh1/Ezh2 and Jarid2 are not required to repress epidermal differentiation once adulthood is reached, but still exert a pro-proliferative function by repressing the expression of p16 (Ezhkova et al, 2009, 2011). Although in young adults the epidermis and HFs of Jarid2 cKO mice do not suffer from any overt phenotype (besides the delay in anagen entry), it will be interesting in the future to determine whether the upregulation of p16 translates into a premature aging of the tissue, given that p16 expression is elevated in the epidermis of old mice (Barradas et al, 2009).
The epidermal consequences of deletion of Ezh2 (Ezhkova et al, 2009) and Jarid2 (our results) share similar overall characteristics (i.e., enhanced differentiation and reduced proliferation). However, the mouse models differ in the timing and the extent of their phenotypes, suggesting that the functional overlap between PRC2 and Jarid2 is limited, and that additional factors must be required for their epidermal activities. Several lines of evidence support this. Deletion of Ezh2 results in enhanced expression of terminal differentiation markers during epidermal embryonic development (Ezhkova et al, 2009), whereas epidermal knockout of Jarid2 does not cause defects in embryonic epidermis. Conversely, the premature differentiation defects of the Ezh2KO epidermis lessen as the mice are born, which is precisely the point when these defects become visible in Jarid2 cKO mice. Additionally, although Ezh1/Ezh2 double knockout and Jarid2 cKO mice show a reduction in the proliferation of transit amplifying cells during HF growth, the former undergoes a progressive loss of HFs (Ezhkova et al, 2011), whereas the latter shows no gradual HF deterioration in young adults. Furthermore, the fact that Jarid2 and Ezh1 have been reported to not interact (Landeira et al, 2010), rules out any functional interaction among both.
That Jarid2 and PRC2 may have non-dependent functions is underscored by the fact that several independently generated, complete-knockout models of Jarid2 display various degrees of lethality as well as defects in heart, liver, neural tube closure, and haematopoietic development; this differs from the phenotypes of PRC2-subunit knockout mice (Takeuchi et al, 1995; Lee et al, 2000; Toyoda et al, 2003; Jung et al, 2005a, 2005b; Sasai et al, 2007; Takeuchi et al, 2006).
Altogether, these results indicate that Jarid2 is necessary to maintain the correct level of epidermal differentiation and the efficient activation of HF stem cells and their progeny after birth. Although Jarid2 is required for the robustness of PRC2 function in postnatal epidermal progenitors, other PRC2-independent interacting factors are likely required for Jarid2 to exert its epidermal function. It will be of interest to determine in future studies the partners of Jarid2 in epidermal progenitors, and given its role in sustaining efficient progenitor and epidermal stem cell proliferation, whether Jarid2 has a role in epidermal aging or neoplastic transformation.
Materials and methods
Generation and handling of mice
To generate Jarid2flx/flx/K14CreYFP-Rosa26 mice and their Jarid2wt/wt counterpart (referred to as Jarid2 cKO and wt, respectively), hemizygous K14CreYFP-Rosa26 mice were mated with Jarid2flx/flx mice. F1 Jarid2flx/wt/K14CreYFP-Rosa26 mice were bred with other mice of the same genotype. All the mice were kept in the C57Bl6/FvBN mixed background. After standard isolation of the genomic DNA, mice were genotyped as described (Mysliwiec et al, 2006), using primer reported in Supplementary Table I. Mice were housed under 12 h light/12 h dark cycles and SPF conditions, and all procedures were evaluated and approved by the CEEA (Ethical Committee for Animal Experimentation) of the Government of Catalonia. For 5-Bromo-2-deoxyuridine (BrdU)-labelling experiments, 100 mg/g BrdU (Invitrogen) was injected intraperitoneally and chased for 2 months. To activate epidermal proliferation, backskin and tailskin of Jarid wt and Jarid2 cKO mice were treated three times with 20 nM TPA (Sigma-Aldrich) during 1 week.
Primary keratinocyte cultures
Primary mouse keratinocytes from either newborn mice or the tailskin of adult mice were isolated as described previously (Litchi et al, 2008; Jensen et al, 2010). Cells were plated in EMEM (Lonza) containing 4% chelated fetal bovine serum (FBS), 1% penicillin/streptomycin, and 20 nM calcium, for 24 h; medium was then changed to growth medium (EMEM with 8% chelated FBS, 1% penicillin/streptomycin, EGF (10 ng/ml), and 50 nM calcium). For clonogenic assays, keratinocytes were plated in E-medium on mitomycin (Sigma-Aldrich)-treated J2P 3T3 feeder cells (Nowak and Fuchs, 2009). Primary human keratinocytes were isolated from neonatal or adult foreskin and cultured together with a feeder culture of fibroblasts (J2P-3T3) in FAD medium (one part Ham's F12 medium with three parts Dulbecco Modified Eagle Medium (DMEM), containing penicillin, streptomycin, and 18 mM adenine) supplemented with 10% FBS and a cocktail of 0.5 μg/ml hydrocortisone, 5 μg/ml insulin, 100 pM cholera enterotoxin, and 10 ng/ml EGF (all final concentrations), as described previously (Gandarillas et al, 1997). J2P feeder cells were cultured in DMEM with 10% FBS; they were treated with 4 μg/ml mitomycin before co-culturing them with primary keratinocytes.
FACS
The backskin of newborn or adult wt and Jarid2 cKO mice was separated from the body with a scalpel. After enzymatic separation of the dermis from the epidermis, total keratinocytes were isolated, filtered through a 70-μm cell strainer (BD Bioscience), and stained for 45 min in ice with primary antibodies. Primary antibodies used for FACS analysis and for cell sorting were α6-integrin (CD49f) coupled with PE (Serotech), APC-coupled CD34 (BD Bioscience). Non-keratinocyte lineages were excluded from our FACS sorts using the Biotin-CD31 (BD Bioscience), Biotin-CD140a (eBioscience), Biotin-CD45 (eBioscience), and Streptavidin-APC-Cy7 (eBioscience) antibodies. For cell sorting, total keratinocytes from newborn and adult mice, or primary human keratinocytes, were gated for single events and viability (DAPI incorporation), and sorted on the basis of α6-integrin expression, or both α6-integrin and CD34 expression. FACS acquisitions were done with the FACS LSR-II system (BD), and cell sorting was performed with the FACS Aria system. Analysis was performed using FACS DiVa software (BD Bioscience).
Whole-mount immunofluorescence
Preparation of tailskin and whole-mount stainings were performed as previously described (Braun et al, 2003). Primary and secondary antibodies were incubated overnight and used at the following concentrations: 1:1000 for anti-GFP (Invitrogen); 1:250 for anti-BrdU (Serotec) and anti-Ki67 (Abcam); and 1:500 for anti-rabbit and anti-rat conjugated to AlexaFluor488 or AlexaFluor594 (Molecular Probes). Nuclei were stained with DAPI (1:5000; Roche) and epidermal sheets were mounted in Mowiol. Pictures were acquired with a Leica TCS SP5 confocal microscope.
Immunohistochemistry
Samples were fixed in 4% NBF (Sigma-Aldrich) at 4°C overnight and then embedded in paraffin. Deparaffinized sections were boiled for 10 min in 0.01 M citric acid for antigen retrieval. In all, 8 μm slices of sections were permeabilized for 25 min in 0.25% Triton X-100/PBS, and then blocked for 90 min in 0.25% gelatin/PBS. Primary antibodies were incubated overnight at 4°C, and secondary antibodies were incubated for 2 h at room temperature in 0.25% gelatin/PBS. Nuclei were stained with DAPI (1:5000; Roche), and the slides were mounted in Mowiol. The antibodies were against keratin-14 (mouse, 1:200; Abcam), Ki67 (rabbit, 1:200; Abcam), filaggrin (rabbit, 1:200; Abcam), loricrin (rabbit, 1:200; Abcam), BrdU (rat, 1:100; Gibco), and GFP (rabbit, 1:100; Molecular Probes). Secondary antibodies were conjugated with AlexaFluor 488 or 594 (Molecular Probes). Images were collected with a Leica SP5 confocal microscope. For immunohistochemistry for p16 (Santa Cruz M156 sc1207, 1:150 dilution), deparaffinized sections were incubated with H202 to block endogenous peroxidase and the staining was performed as described for immunofluorescence. The immunocomplexes were stained using the ABC peroxidase method (Vector Laboratories).
RNA isolation and PCR
Cells collected by FACS were directly lysated in RLT buffer (Qiagen), and total RNA was extracted using the RNeasy Mini Kit (Qiagen). mRNA was quantified with NanoDrop, and equal amounts of mRNA were retro-transcribed using oligo(dT) and SuperscriptIII (Invitrogen). cDNAs were normalized for the expression of two housekeeping genes, HPRT-1 and Pum-1. Samples were PCR amplified using specifically designed primers (Supplementary Table II). For real-time PCR, the same primers were employed using the LightCycler System (Roche), LightCycler 3.5 software and the LightCycler DNA Master SYBR Green I reagents. Differences between samples and controls were calculated based on the 2−ΔΔCP method.
Western blot and co-immunoprecipitation
For co-immunoprecipitation and western blot, cells were lysed with 50 mM Tris–HCl pH 7.6, 300 mM NaCl, 10% glycerol, 0.2% Igepal. For co-IP, total cell lysates were incubated with 5 μg of Jarid2, Suz12, or Ezh2 antibodies. The antibody-bound fraction was purified with A-agarose beads and then probed for immunoblotting. The antibodies used for western blot are β-Tubulin I (Sigma T 7816), Suz12 (Abcam ab12073), filaggrin (Santa Cruz M-290 sc-30230), p16 (Santa Cruz M-156 sc1207), total H3 (Abcam ab1791), H3K27me3 (Millipore 07-449), Ezh2 (Cell Signaling, AC22), IgG (Abcam ab48137), Jarid2 (rabbit polyclonal, generated in house).
Chromatin immunoprecipitation
For ChIP assays from intact epidermis, 2 month-old mice were killed, and tails were incubated in 0.25% trypsin for 4 h at 37°C, to separate the dermis from the epidermis. Tail keratinocytes were extracted as described (Litchi et al, 2008; Jensen et al, 2010). Cells in suspension were crosslinked for 10 min at room temperature in 1% formaldehyde. Crosslinking reactions were stopped by adding 1.25 M glycine to a final concentration of 125 mM. Cells were centrifuged for 10 min at 4°C and then washed in cold PBS. Cells were lysed with 1.3 ml of ChIP buffer (100 mM NaCl, 50 mM Tris-Cl, pH 8.1, 5 mM EDTA, pH 8.0, 0.2% NaN3, 0.5% SDS, and 5% Triton X-100, in water) and sonicated for 10 min in a Bioruptor (Diagenode). The soluble fraction was quantified by Bradford, and 500 μg was used to immunoprecipitate transcription factors, while 100 μg was used to immunoprecipitate histones and specifically modified histone. The chromatin and antibody mixtures were incubated overnight at 4°C in total volume of 500 μl. To recover the immunocomplexes, mixtures were incubated with 30 μl of protein A or G slurry for 1 h. The immunoprecipitated material was washed three times with a low salt buffer (50 mM HEPES, pH 7.5, 140 mM NaCl, 1% Triton X-100, and protease inhibitors), and once with a high salt buffer (50 mM HEPES, pH 7.5, 500 mM NaCl, 1% Triton X-100, and protease inhibitors). Samples were decrosslinked by incubation in 100 μl of 1% SDS and 100 mM NaHCO3 at 65°C for 3 h. DNA was eluted in 200 μl of water using a PCR purification kit (Qiagen) and quantitated by real-time PCR. The specific primers used for ChIP are listed in Supplementary Table III.
Statistics
Results are presented as mean±s.e.m. Statistical significance was determined by the Student's t-test.
Supplementary Material
Acknowledgments
This work was supported by the Fondo de Investigación Sanitario (FIS, Spanish Ministerio de Sanidad) and the AGAUR (Government of Cataluña) to SAB, and by the Spanish ‘Ministerio de Educación y Ciencia’ (BFU2010-18692) and Consolider to LDC. YL was funded by the National Institute of Health (HL067050), and MRM by an AHA postdoctoral fellowship (10POST2600279). SM is funded by La Caixa International PhD Fellowship; LM is funded by CRG-Novartis Fellowship; and GP is the recipient of a FIS fellowship (Spanish Ministerio de Sanidad). We would like to thank Veronica Raker for helpful discussions preparing the manuscript, and to all the CRG core facilities and Animal Unit for technical support.
Author contributions: SM performed all the analysis of the mice (embryonic development and postnatal morphogenesis). GP and BK assisted SM with the handling of the mice and performing some of the experiments. LM performed all the Jarid2, Suz12, H3K27me3, and H3 ChIP experiments. YL provided the Jarid2flox/flox mice and assisted in the preparation of the manuscript. LDC, RS, and SAB wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenführ M, Maertens G, Banck M, Zhou MM, Walsh MJ, Peters G, Gil J (2009) Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev 23: 1177–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E (2004) Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118: 635–648 [DOI] [PubMed] [Google Scholar]
- Braun KM, Niemann C, Jensen UB, Sundberg JP, Silva-Vargas V, Watt FM (2003) Manipulation of stem cell proliferation and lineage commitment: visualisation of label-retaining cells in wholemounts of mouse epidermis. Development 130: 5241–5255 [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y (2004) SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell 15: 57–67 [DOI] [PubMed] [Google Scholar]
- Chen H, Gu X, Su IH, Bottino R, Contreras JL, Tarakhovsky A, Kim SK (2009) Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev 23: 975–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton E, Doupé DP, Klein AM, Winton DJ, Simons BD, Jones PH (2007) A single type of progenitor cell maintains normal epidermis. Nature 446: 185–189 [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196 [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N (2004) Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7: 663–676 [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E (2011) EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev 25: 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E (2009) Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136: 1122–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandarillas A, Watt FM (1997) c-Myc promotes differentiation of human epidermal stem cells. Genes Dev 11: 2869–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco V, Chen T, Rendl M, Schober M, Pasolli HA, Stokes N, Dela Cruz-Racelis J, Fuchs E (2009) A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell 4: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz HM, Shilatifard A (2010) The JARID2-PRC2 duality. Genes Dev 24: 857–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YC, Pasolli HA, Fuchs E (2011) Dynamics between stem cells, niche and progeny in the hair follicle. Cell 144: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Kizawa K, Hamada K, Cotsarelis G (2004) Hair follicle stem cells in the lower bulge form the secondary germ, a biochemically distinct but functionally equivalent progenitor cell population, at the termination of catagen. Differentiation 72: 548–557 [DOI] [PubMed] [Google Scholar]
- Jensen KB, Driskell RR, Watt FM (2010) Assaying proliferation and differentiation capacity of stem cells using disaggregated adult mouse epidermis. Nat Protocol 5: 898–911 [DOI] [PubMed] [Google Scholar]
- Jung J, Kim TG, Lyons GE, Kim HR, Lee Y (2005a) Jumonji regulates cardiomyocyte proliferation via interaction with retinoblastoma protein. J Biol Chem 280: 30916–30923 [DOI] [PubMed] [Google Scholar]
- Jung J, Mysliwiec MR, Lee Y (2005b) Roles of JUMONJI in mouse embryonic development. Dev Dyn 232: 21–32 [DOI] [PubMed] [Google Scholar]
- Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ (2004) Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev 18: 1592–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of Zeste protein. Genes Dev 16: 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D, Fisher AG (2011) Inactive yet indispensable: the tale of Jarid2. Trends Cell Biol 21: 74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, Brookes E, Pombo A, Fisher C, Skarnes WC, Snoek T, Bezstarosti K, Demmers J, Klose RJ, Casanova M, Tavares L et al. (2010) Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol 12: 618–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Song AJ, Baker R, Micales B, Conway SJ, Lyons GE (2000) Jumonji, a nuclear protein that is necessary for normal heart development. Circ Res 86: 932–938 [DOI] [PubMed] [Google Scholar]
- Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingstone RE (2002) The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol 22: 6070–6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy V, Lindon C, Harfe BD, Morgan BA (2005) Distinct stem cell populations regenerate the follicle and interfollicular epidermis. Dev Cell 9: 855–861 [DOI] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D (2010) Jarid2 and PRC2, partners in regulating gene expression. Genes Dev 24: 368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litchi U, Anders J, Yuspa SH (2008) Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat Protoc 3: 799–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski IJ, Ritchie ME, Phipson B, Corbin J, Pakusch M, Ebert A, Busslinger M, Koseki H, Hu Y, Smyth GK, Alexander WS, Hilton DJ, Blewitt ME (2010) Opposing roles of polycomb repressive complexes in hematopoietic stem and progenitor cells. Blood 116: 731–739 [DOI] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadi J, Woodcock CL, Dynlacht BD, Reinberg D (2008) Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell 32: 503–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, Lin JS, Sawicki JA, Cotsarelis G (2004) Capturing and profiling adult hair follicle stem cells. Nat Biotechnol 22: 411–417 [DOI] [PubMed] [Google Scholar]
- Mysliwiec MR, Chen J, Powers PA, Bartley CR, Schneider MD, Lee Y (2006) Generation of a conditional null allele of jumonji. Genesis 44: 407–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak JA, Fuchs E (2009) Isolation and culture of epithelial stem cells. Methods Mol Biol 482: 215–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak JA, Polak L, Pasolli HA, Fuchs E (2008) Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell 3: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K (2010) JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464: 306–310 [DOI] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J (2009) Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139: 1290–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JD, Sansom SN, Smith J, Dobenecker MW, Tarakhovsky A, Livesey FJ (2010) Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci USA 107: 15957–15962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai N, Kato Y, Kimura G, Takeuchi T, Yamaguchi M (2007) The Drosophila jumonji gene encodes a JmjC-containing nuclear protein that is required for metamorphosis. FEBS J 274: 6139–6151 [DOI] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G (2010) Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 7: 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider MR, Schmidt-Ullrich R, Paus R (2009) The hair follicle as a dynamic miniorgan. Curr Biol 19: R132–R142 [DOI] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH (2009) Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139: 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A (2003) Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol 4: 124–131 [DOI] [PubMed] [Google Scholar]
- Surface LE, Thornton SR, Boyer LA (2010) Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell 7: 288–298 [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Watanabe Y, Takano-Shimizu T, Kondo S (2006) Roles of jumonji and jumonji family genes in chromatin regulation and development. Dev Dyn 235: 2449–2459 [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Yamazaki Y, Katoh-Fukui Y, Tsuchiya R, Kondo S, Motoyama J, Higashinakagawa T (1995) Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev 9: 1211–1222 [DOI] [PubMed] [Google Scholar]
- Tiede S, Kloepper JE, Bodò E, Tiwari S, Kruse C, Paus R (2007) Hair follicle stem cells: walking the maze. Eur J Cell Biol 86: 355–376 [DOI] [PubMed] [Google Scholar]
- Toyoda M, Shirato H, Nakajima K, Kojima M, Takahashi M, Kubota M, Suzuki-Migishima R, Motegi Y, Yokoyama M, Takeuchi T (2003) Jumonji downregulates cardiac cell proliferation by repressing cyclin D1 expression. Dev Cell 5: 85–97 [DOI] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E (2004) Defining the epithelial stem cell niche in skin. Science 303: 359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431: 873–878 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.