Abstract
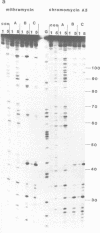
The preferred binding sites for mithramycin on three different DNA fragments have been determined by hydroxyl radical footprinting. Sequences which appear as one long protected region using DNAase I as a footprinting probe are resolved into several discrete binding domains. Each drug molecule protects three bases from radical attack, though adjacent regions show attenuated cleavage. Mithramycin and the other related compounds induce similar footprinting patterns and appear to recognise GC rich regions with a preference for those containing the dinucleotide step GpG. The ability of each such site to bind the drug depends on the sequence environment in which it is located. The data are consistent with mithramycin binding to the DNA minor groove.
Full text
PDF

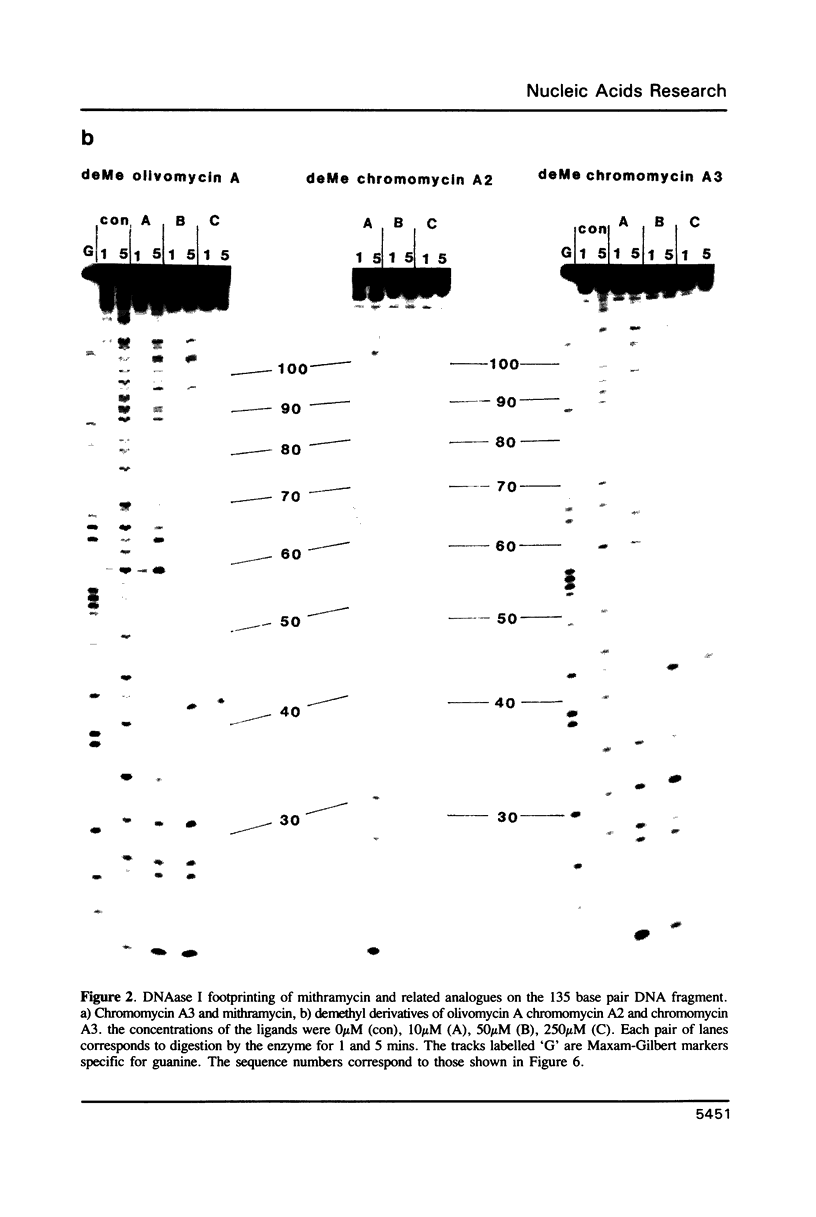
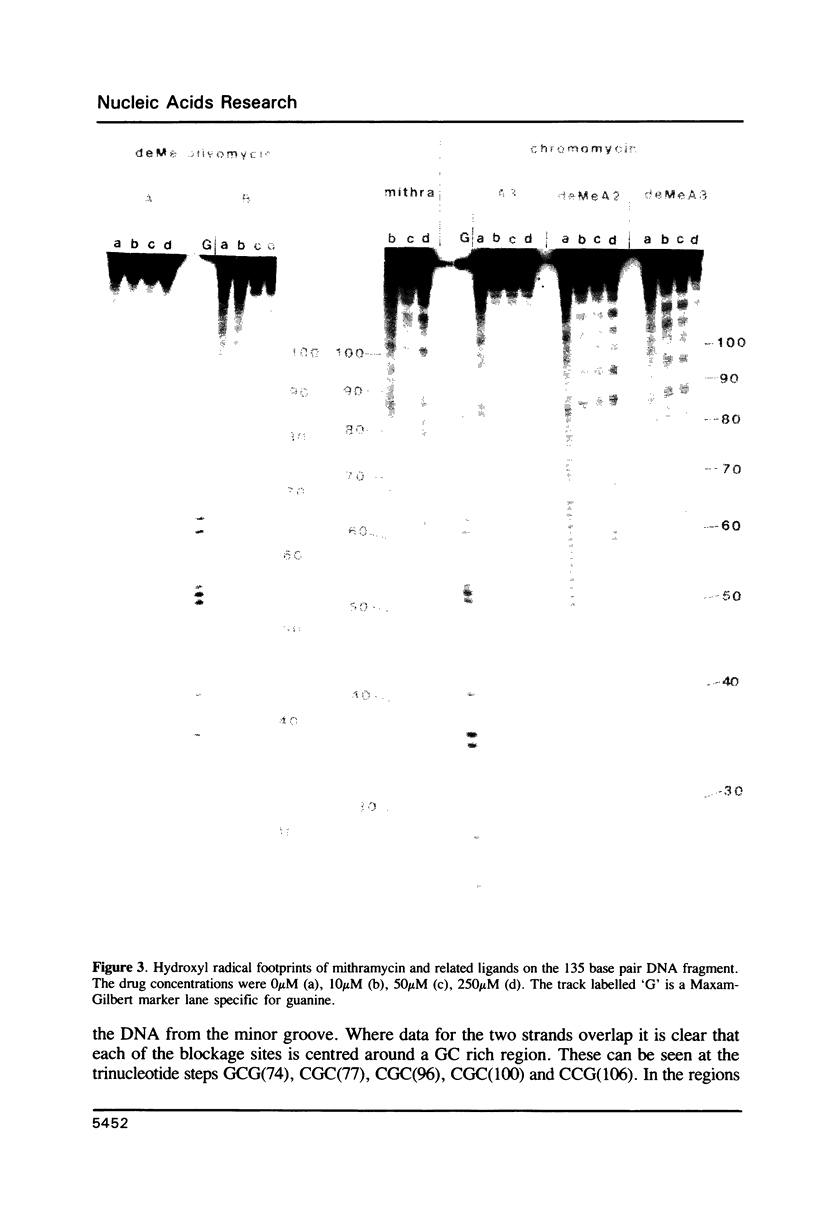

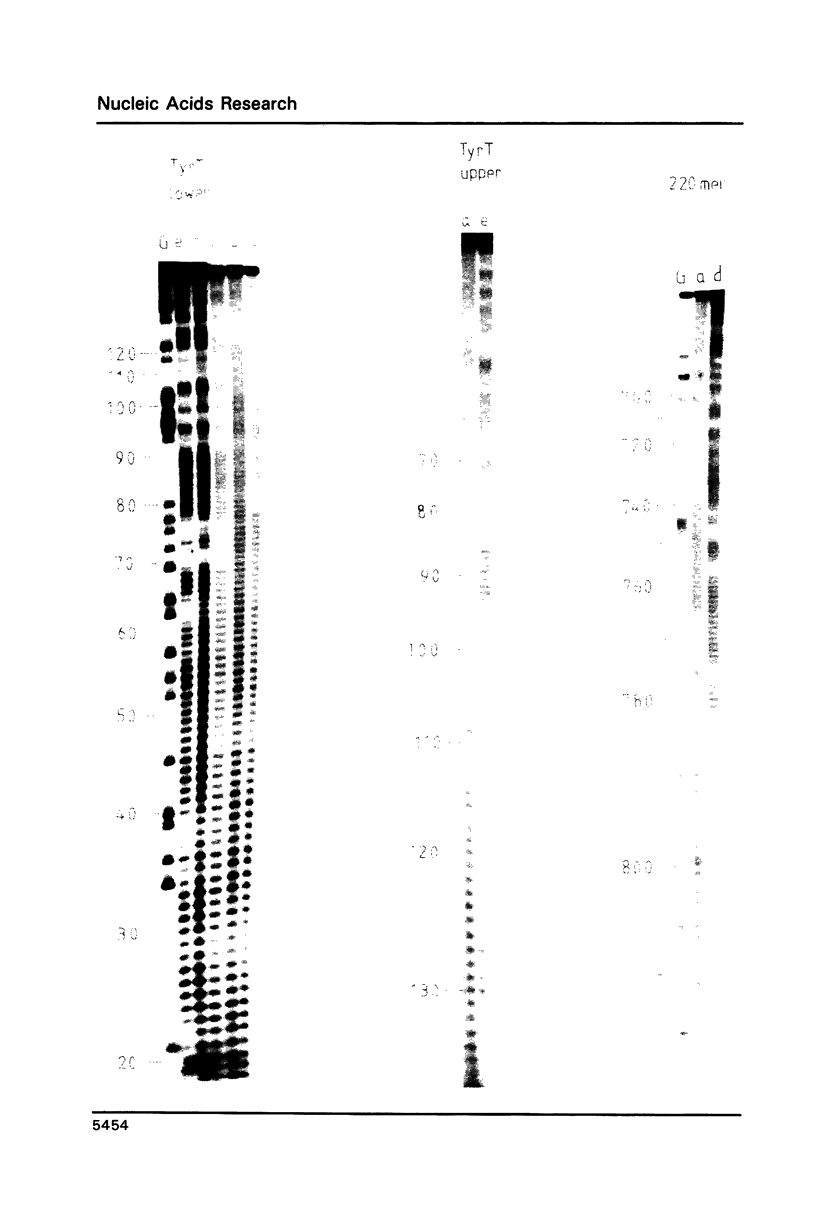

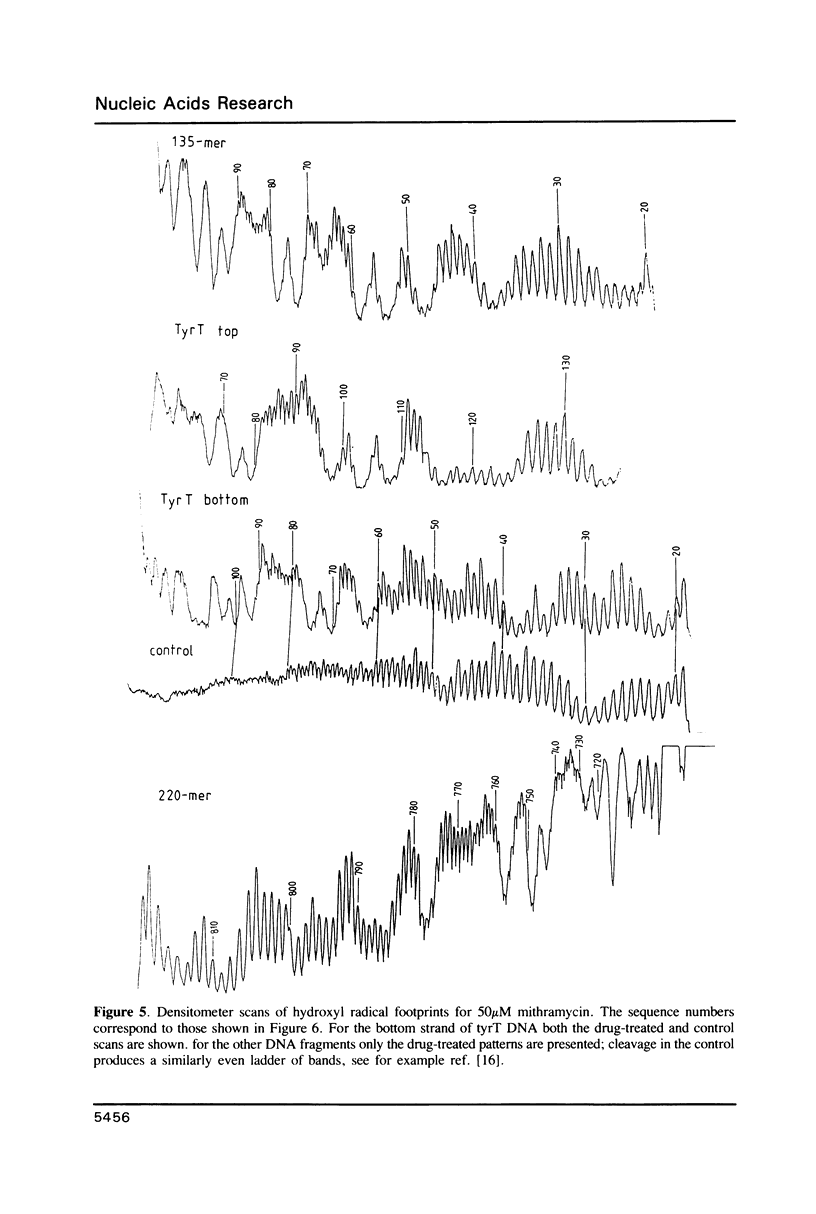



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Behr W., Hartmann G. Spektralphotometrische Untersuchungen über die Wechselwirkungen zwischen Chromomycin A3 und Nucleinsäuren. Biochem Z. 1965 Dec 31;343(4):519–527. [PubMed] [Google Scholar]
- Behr W., Honikel K., Hartmann G. Interaction of the RNA polymerase inhibitor chromomycin with DNA. Eur J Biochem. 1969 May 1;9(1):82–92. doi: 10.1111/j.1432-1033.1969.tb00579.x. [DOI] [PubMed] [Google Scholar]
- Berman E., Brown S. C., James T. L., Shafer R. H. NMR studies of chromomycin A3 interaction with DNA. Biochemistry. 1985 Nov 19;24(24):6887–6893. doi: 10.1021/bi00345a022. [DOI] [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987 Mar 27;48(6):935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- Cons B. M., Fox K. R. Interaction of mithramycin with metal ions and DNA. Biochem Biophys Res Commun. 1989 Apr 28;160(2):517–524. doi: 10.1016/0006-291x(89)92463-7. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA structural variations in the E. coli tyrT promoter. Cell. 1984 Jun;37(2):491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- Fok J., Waring M. Breakdown of pulse-labeled ribonucleic acid in Bacillus megaterium, revealed by exposure to the antibiotics mithramycin, chromomycin, and nogalamycin. Mol Pharmacol. 1972 Jan;8(1):65–74. [PubMed] [Google Scholar]
- Fox K. R. Footprinting studies on the interactions of nogalamycin, arugomycin, decilorubicin and viriplanin with DNA. Anticancer Drug Des. 1988 Dec;3(3):157–168. [PubMed] [Google Scholar]
- Fox K. R., Howarth N. R. Investigations into the sequence-selective binding of mithramycin and related ligands to DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8695–8714. doi: 10.1093/nar/13.24.8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X. L., Patel D. J. Solution structure of the chromomycin-DNA complex. Biochemistry. 1989 Jan 24;28(2):751–762. doi: 10.1021/bi00428a051. [DOI] [PubMed] [Google Scholar]
- Keniry M. A., Brown S. C., Berman E., Shafer R. H. NMR studies of the interaction of chromomycin A3 with small DNA duplexes I. Biochemistry. 1987 Feb 24;26(4):1058–1067. doi: 10.1021/bi00378a012. [DOI] [PubMed] [Google Scholar]
- Kennedy B. J., Yarbro J. W., Kickertz V., Sandberg-Wollheim M. Effect of mithramycin on a mouse glioma. Cancer Res. 1968 Jan;28(1):91–97. [PubMed] [Google Scholar]
- Kersten W., Kersten H., Szybalski W. Physicochemical properties of complexes between deoxyribonucleic acid and antibiotics which affect ribonucleic acid synthesis (actinomycin, daunomycin, cinerubin, nogalamycin, chormomycin, mithramycin, and olivomycin). Biochemistry. 1966 Jan;5(1):236–244. doi: 10.1021/bi00865a031. [DOI] [PubMed] [Google Scholar]
- Koenuma M., Uchida N., Yamaguchi K., Kawamura Y., Matsumoto K. New aureolic acid antibiotics. I. Screening, isolation, characterization and biological properties. J Antibiot (Tokyo) 1988 Jan;41(1):45–52. doi: 10.7164/antibiotics.41.45. [DOI] [PubMed] [Google Scholar]
- Koenuma M., Yoshimura Y., Matsumoto K., Terui Y. New aureolic acid antibiotics. II. Structure determination. J Antibiot (Tokyo) 1988 Jan;41(1):68–72. doi: 10.7164/antibiotics.41.68. [DOI] [PubMed] [Google Scholar]
- Meyer K. Problem Abrechnung. Quintessenz J. 1981 Jun;11(6):551–558. [PubMed] [Google Scholar]
- Peterson R. C., Doering J. L., Brown D. D. Characterization of two xenopus somatic 5S DNAs and one minor oocyte-specific 5S DNA. Cell. 1980 May;20(1):131–141. doi: 10.1016/0092-8674(80)90241-x. [DOI] [PubMed] [Google Scholar]
- Portugal J., Waring M. J. Hydroxyl radical footprinting of the sequence-selective binding of netropsin and distamycin to DNA. FEBS Lett. 1987 Dec 10;225(1-2):195–200. doi: 10.1016/0014-5793(87)81156-0. [DOI] [PubMed] [Google Scholar]
- Tullius T. D. DNA footprinting with hydroxyl radical. Nature. 1988 Apr 14;332(6165):663–664. doi: 10.1038/332663a0. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Iron(II) EDTA used to measure the helical twist along any DNA molecule. Science. 1985 Nov 8;230(4726):679–681. doi: 10.1126/science.2996145. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Chromomycin, mithramycin, and olivomycin binding sites on heterogeneous deoxyribonucleic acid. Footprinting with (methidiumpropyl-EDTA)iron(II). Biochemistry. 1983 May 10;22(10):2373–2377. doi: 10.1021/bi00279a011. [DOI] [PubMed] [Google Scholar]
- Ward D. C., Reich E., Goldberg I. H. Base specificity in the interaction of polynucleotides with antibiotic drugs. Science. 1965 Sep 10;149(3689):1259–1263. doi: 10.1126/science.149.3689.1259. [DOI] [PubMed] [Google Scholar]