Abstract
Early experience permanently alters behavior and physiology. These effects are, in part, mediated by sustained alterations in gene expression in selected brain regions. The critical question concerns the mechanism of these environmental “programming” effects. We examine this issue with an animal model that studies the consequences of variations in mother-infant interactions on the development of individual differences in behavioral and endocrine responses to stress in adulthood. Increased levels of pup licking/grooming by rat mothers in the first week of life alter DNA structure at a glucocorticoid receptor gene promoter in the hippocampus of the offspring. Differences in the DNA methylation pattern between the offspring of high- and low-lickinglgrooming mothers emerge over the first week of life; they are reversed with cross-fostering; they persist into adulthood; and they are associated with altered histone acetylation and transcription factor (nerve growth factor-induced clone A [NGFIA]) binding to the glucocorticoid receptor promoter. DNA methylation alters glucocorticoid receptor expression through modifications of chromatin structure. Pharmacological reversal of the effects on chromatin structure completely eliminates the effects of maternal care on glucocorticoid receptor expression and hypothalamic-pituitary-adrenal (HPA) responses to stress, thus suggesting a causal relation between the maternally induced, epigenetic modification of the glucocorticoid receptor gene and the effects on stress responses in the offspring. These findings demonstrate that the structural modifications of the DNA can be established through environmental programming and that, in spite of the inherent stability of this epigenomic marker, it is dynamic and potentially reversible.
Keywords: maternal behavior, glucocorticoid receptor, stress response, DNA methylation, gene expression, histone acetylation, NGFIA (nerve growth factor-induced clone A)
Abstract
Una experiencia precoz altera en forma permanente la conducta y la fisiología. Estos efectos son mediados, en parte, por alteraciones que se sustentan en la expresión génica de regiones cerebrales específicas. La pregunta central se refiere al mecanismo de estos efectos en la "programación" ambiental. En este artículo se examina este tema con un modelo animal que estudia las consecuencias de las variaciones en las interacciones entre la madre y la cría en el desarrollo de diferencias individuales en la respuesta conductual y endocrina al estrés durante la adultez. Un aumento en la conducta de aseo con lamidos a las crías por las ratas madre durante la primera semana de vida altera la estructura del ADN del gen promotor del receptor de glucocorticoides en el hipocampo de las crías. Diferencias en el patrón de metilación del ADN entre las crías de madres con altas y bajas conductas de aseo con lamidos aparecen en la primera semana de vida; éstas pueden revertir mediante la adopción cruzada, pueden persistir a lo largo de la adultez y pueden asociarse con una alteración de la acetilación de histona y de la fijación del factor de transcripción (clon A inducido por el factor de crecimiento neural [NGFIA]) al promotor del receptor de glucocorticoides. La metilación del ADN altera la expresión del receptor de glucocorticoides a través de modificaciones en la estructura de la cromatina. La reversión de los efectos en la estructura de la cromatina producida farmacológicamente elimina completamente los efectos de los cuidados maternos en la expresión del receptor de glucocorticoides y en la respuesta del eje hipotálamo-hipófisis-adrenal (HHA) al estrés, lo que sugiere una relación causal entre la modificación epigenética del gen del receptor de glucocorticoides, inducida por la madre, y los efectos en la respuesta al estrés en las crías. Estos hallazgos demuestran que las modificaciones estructurales del ADN se pueden establecer mediante programación ambiental y que, a pesar de la estabilidad intrínseca de este marcador epigenómico, éste es dinámico y potencial mente reversible.
Abstract
Les expériences précoces de la vie modifient en permanence le comportement et la physiologie. Ces effets sont en partie, dus à des transformations prolongées de l'expression génique dans des régions cérébrales sélectionnées. La question principale concerne le mécanisme de ces effets «programmants» environnementaux. Nous examinons ce problème sur un modèle animal qui étudie les conséquences des variations des interactions mère-enfant sur le développement des différences individuelles dans les réponses au stress, endocrines et environnementales, à l'âge adulte. Un léchage/toilettage intense de petits de rats par leur mère dans la première semaine de vie modifie la structure de l'ADN au niveau du gène promoteur d'un récepteur glucocorticoïde dans l'hippocampe de la descendance. Les différences de schémas de méthylation de I'ADN entre les descendants de mères fortement ou faiblement lécheuses apparaissent après la première semaine de vie; elles sont réversibles avec l'échange des mères; elles persistent à l'âge adulte et sont associées à une modification de l'acétylation de l'histone et du facteur de transcription (facteur de croissance nerveux induit par clone A [NGFIA]) en liaison avec le promoteur du récepteur glucocorticoïde. La méthylation de I'ADN modifie l'expression du récepteur glucocorticoïde par les changements de structure de la chromatine. L'inversion pharmacologique induite par les changements de structure de la chromatine élimine complètement les effets des soins maternels sur l'expression du récepteur glucocorticoïde et les réponses au stress hypothalamo-adrénalo-pituitaires (HAP), suggérant donc une relation causale entre les modifications épigénétiques du gène du récepteur glucocorticoïde induites par la mère et les effets sur les réponses au stress dans la descendance. Ces résultats démontrent que les changements structuraux de l'ADN peuvent se constituer par l'intermédiaire d'un environnement programmant et que, malgré la stabilité inhérente de ce marqueur épigénomique, elles sont dynamiquement et potentiellement réversibles.
Epidemiological studies reveal the importance of family function and early life events as predictors of health in adulthood.1 As adults, victims of childhood physical or sexual abuse, emotional neglect, family conflict, and conditions of harsh, inconsistent discipline are at considerably greater risk for mental illness, as well as for obesity, diabetes, and heart disease.2-17 These difficult conditions, in part, define the developmental origin of mental illness in adolescence and adult life.
“Stress diathesis” models suggest that adversity in early life alters the development of neural and endocrine systems in a manner that predisposes individuals to disease in adulthood. The relation between the quality of the early environment and health in adulthood appears to be mediated by parental influences on the development of neural systems that underlie the expression of behavioral and endocrine responses to stress.1,18-22 Adversity or decreased quality of parental investment increases the magnitude of emotional, autonomic, and hypothalamicpituitary-adrenal (HPA) responses to stress in adulthood. These models are constructed on two principal assumptions: (i) prolonged activation of neural and hormonal responses to stress can promote illness; (ii) early environmental events influence the development of these responses. There is strong evidence in favor of both ideas. In humans, forms of parenting that enhance the risk of chronic illness in the offspring increase endocrine and autonomic responses to stress in adulthood.22-26 There is considerable evidence for comparable effects in primates27-29 and rodents.28,30 Moreover, prolonged exposure to elevated levels of stress hormones, including corticotropin-releasing factor (CRF), catecholamines (most notably norepinephrine), and glucocorticoids promote the development of a diverse range of high-risk conditions, such as visceral obesity, hypertension, and insulin intolerance, or overt pathology, including diabetes, depression, drug addiction, and multiple forms of coronary heart disease.31-33 The clinical risks associated with prolonged activation of the HPA and autonomic systems are a logical consequence of the otherwise adaptive stress response. In response to neural signals associated with the stressor, there is an increased release of glucocorticoids from the adrenal gland and catecholamines, particularly norepinephrine from the sympathetic system. The combined actions of these hormones increase the availability of energy substrates, such as those derived from lipid and glucose metabolism, in order to maintain normal cellular output and organ efficiency. These actions protect against catastrophes such as hypotensive shock. These hormones, along with the central CRF and catecholamines, also act on multiple brain regions to increase vigilance and fear and enhance avoidance learning and fear conditioning, which reduces the chances of further encounters with the offending conditions. It is likely that such responses evolved to meet the demands of acute stressors, and that the physiological costs associated with short-term activation are minimal in otherwise healthy individuals. The high-risk conditions are associated with chronic stress and persistent activation of stress hormones.
Support for the basic elements of stress diathesis models appears compelling. Adversity during perinatal life alters development in a manner that seems likely to promote vulnerability, especially for stress-related diseases. Diathesis describes the interaction between development, including the potential influence of genetic factors, and the prevailing level of stress in predicting health outcomes. Such models have considerable appeal, and could potentially identify both the origins and the nature of vulnerability derived from either epigenetic influences, such as early family life, or genomic variations.27,34 For developmentalists the critical questions are (i) how early experience might “program” individual differences in stress responses; and (ii) whether such effects are reversible.
The development of individual differences in stress responses
In the late 1950s and early 1960s the pages of Science and Nature were frequently dedicated to articles reporting the effects of postnatal handling on the development of responses to stressors.35-37 The handling paradigm involves a brief (ie, ~15 min) separation of the pups from the dam that does not constitute any major deprivation of parental care. In infant rats and mice, handling during infancy decreases the magnitude of both behavioral and HPA responses to stress in adulthood. These findings demonstrated that the early environment influences the development of even rudimentary defensive responses to threat.
Le vine and others suggested that the effects of handling are actually mediated by changes in maternal care.35-37 Indeed, handling increases the licking/grooming (LG) of pups by the mother.38,39 Subsequent studies strongly support the maternal-mediation hypothesis. One approach was to examine the consequences of naturally occurring variations in maternal LG. These studies indicate that the adult offspring of high-LG mothers resembled postnatally handled animals on measures of behavioral and endocrine responses to stress, while those of low-LG mothers were comparable to nonhandled animals. Cross-fostering studies, where pups born to high-LG mothers are fostered at birth to low-LG mothers (and vice versa), suggest a direct relationship between maternal care and the postnatal development of individual differences in behavioral and HPA responses to stress.40,41 Finally, these studies suggest that variations within a normal range of parental care can dramatically alter development. As in humans, parental care need not include forms of overt abuse or extreme neglect in order to influence the development of the offspring. In large measure, this is most likely due to the fact that natural selection shaped offspring to respond to subtle variations in parental behaviors as a forecast of the environmental conditions they will ultimately face following independence from the parent.42 Environmental adversity promotes forms of parental care that enhance stress responses in the offspring. To the extent that the offspring are likely to inherit comparable conditions - a reasonable assumption up until recent times - the development of increased stress reactivity might be considered as adaptive.
Maternal car in the rat programs behavioral and HPA responses to stress
The effects of maternal care on the development of individual differences in behavioral and HPA responses to stress in the rat are mediated by alterations of the neural systems that regulate central CRF systems furnishing the critical signal for the activation of behavioral, emotional, autonomic, and endocrine responses to stressors. There are two major CRF pathways. First, a CRF pathway from the parvocellular regions of the paraventricular nucleus of the hypothalamus (PVNh) to the portal system of the anterior pituitary, which serves as the principal mechanism for the transduction of a neural signal into a pituitary-adrenal response.43-45 In responses to stressors, CRF is released from PVNh neurons into the portal blood supply of the anterior pituitary and stimulates the synthesis and release of adrenocorticotropin hormone (ACTH). Pituitary ACTH, in turn, causes the release of glucocorticoids from the adrenal gland. CRF synthesis and release are inhibited through a glucocorticoid negative-feedback system mediated by both mineralocorticoid and glucocorticoid receptors (GRs) in a number of brain regions including, and perhaps especially in, the hippocampus.46,47 CRF neurons in the amygdala project directly to the locus ceruleus and increase the firing rate of locus ceruleus neurons, resulting in increased noradrenaline release in the vast terminal fields of this ascending noradrenergic system. Thus, intracerebroventricular (ICV) infusion of CRF increases extracellular noradrenaline levels.48-52 The amygdaloid CRF projection to the locus ceruleus52-56 is also critical for the expression of behavioral responses to stress.57-64 Hence, the CRF neurons in the PVNh and amygdala serve as important mediators of both behavioral and endocrine responses to stress.
We examine the relation between maternal care and the development of stress responses using a rather simple model of naturally occurring variations in maternal behavior over the first 8 days after birth.65 We characterize individual differences in maternal behavior through direct observation of mother-pup interactions in normally reared animals. These observations reveal considerable variation in maternal LG of pups (Figure 1). LG includes both body as well as anogenital licking.66 We then simply define mothers according to the frequency of pup LG, ie, high- or low-LG mothers. For the sake of most of the studies described here, high- and low-LG mothers are females whose scores on pup LG are ±1 SD above (high) or below (low) the mean for their cohort. Importantly, high- and low-LG mothers do not differ in the amount of contact time with pups; differences in the frequency of LG do not occur simply as a function of time in contact with pups. High- and low-LG mothers raise a comparable number of pups to weaning, and there are no differences in the weaning weights of the pups, suggesting an adequate level of maternal care across the groups. These findings also suggest that we are examining the consequences of variations in maternal care that occur within a normal range. Indeed, the frequency of both pup LG is normally distributed across large populations of lactating female rats.65
Figure 1. Lactating female Long-Evans rat nursing litter in arched-back posture while licking/grooming an individual pup.
The critical question concerns the potential consequences of these differences in maternal behavior for the development of behavioral and neuroendocrine responses to stress. As adults, the offspring of high-LG mothers show reduced plasma ACTH and corticosterone responses to acute stress by comparison to the adult offspring of lowLG mothers.39,67 Circulating glucocorticoids act at GR sites in corticolimbic structures, such as the hippocampus, to regulate HPA activity. Such negative-feedback effects commonly target CRF synthesis and release at the level of the PVNh. Enhanced GR activation in the hippocampus is associated with decreased hypothalamic CRF levels. The high-LG offspring showed significantly increased hippocampal GR mRNA expression, enhanced glucocorticoid negative-feedback sensitivity, and decreased hypothalamic CRF mRNA levels.39 Moreover, the magnitude of the corticosterone response to acute stress was significantly correlated with the frequency of both maternal LG (r=-0.61) during the first week of life, as was the level of hippocampal GR mRNA and hypothalamic CRF mRNA expression (all r values >0.70).39
The offspring of the high- and low-LG mothers also differ in behavioral responses to stress.40,41,68 As adults, the off spring of the high-LG mothers show decreased startle responses, increased exploration in novel, uncertain environments, and shorter latencies to eat food provided in a novel environment. The offspring of low-LG mothers also show greater burying of an electrified probe in the defensive burying paradigm,68 which involves an active response to a clearly defined threat. The offspring of the high-LG mothers exhibit decreased CRF receptor levels in the locus ceruleus and increased γ-aminobutyric acid (GABAA)/benzodiazepine (BZ) receptor levels in the basolateral and central nucleus of the amygdala, as well as in the locus ceruleus,41,69 and decreased CRF mRNA expression in the central nucleus of the amygdala (Francis, Diorio, and Meaney, unpublished data). BZ agonists suppress CRF expression in the amygdala.70 Predictably, stress-induced increases in PVNh levels of noradrenaline, which are normally stimulated by CRF, are significantly higher in the offspring of the low-LG offspring.71
Maternal care during the first week of life is associated with stable individual differences in GABAA receptor subunit expression in brain regions that regulate stress reactivity. The adult offspring of high-LG mothers show significantly higher levels of GABAA/BZ receptor binding in the basolateral and central nuclei of the amygdala, as well as the locus ceruleus. These findings provide a mechanism for increased GABAergic inhibition of amygdala-locus ceruleus activity. Importantly, maternal care also affects the behavioral sensitivity to acute BZ administration. The offspring of high-LG mothers show an increased anxiolytic response to acute BZ administration.
Recent studies41 suggest that variations in maternal care might actually permanently alter the composition of the GABAA receptor complex in the offspring. The G ABAA receptor is comprised of five individual protein subunits that collectively form a functional CI- channel mediating GABA-induced neuronal inhibition in the adult brain. There are over 20 individual subunits and variation in the function of the GABAA receptor is associated with differences in the nature of the subunits comprising the receptor. Of particular interest are the a and y subunits, the presence of which defines a BZ binding site. The offspring of the high-LG mothers show increased levels of the mRNAs for the γ1 and γ2 subunits, both of which contribute to the formation of a functional BZ binding site. Such differences are not unique to the γ subunits. Levels of mRNA for the α subunit of the GABAAA/BZ receptor complex are significantly higher in the amygdala and locus ceruleus of high-LG compared with low-LG offspring. The α1 subunit appears to confer higher affinity for GABA, providing the most efficient form of the GABAA receptor complex, through increased receptor affinity for GABA. The adult offspring of the low-LG mothers actually show increased expression of the mRNAs for the α3 and α4 subunits in the amygdala and the locus ceruleus. Interestingly, the GABAA/BZ receptor composed of the α3 and α4 subunits show reduced affinity for GABA, by comparison with those containing an α subunit. Moreover, the α4 subunit does not contribute to the formation of a BZ receptor site. These differences in subunit expression are highly specific to the amygdala and the locus ceruleus; no such differences are apparent in the hippocampus, hypothalamus, or cortex. Thus, differences in GABAA/BZ receptor binding are not simply due to a deficit in subunit expression in the offspring of the low-LG mothers, but of an apparently “active” attempt to maintain a specific GABAA/BZ receptor profile in selected brain regions.
These findings suggest that the behavior of the mother toward her offspring can “program” behavioral and neuroendocrine responses to stress in adulthood. These effects are associated with sustained changes in the expression of genes in brain regions that mediate responses to stress, and form the basis for stable individual differences in stress reactivity. These findings provide a potential mechanism for the influence of parental care on vulnerability/resistance to stress-induced illness over the lifespan.
Cross-fostering studies: evidence for direct maternal effects
Individual differences in behavioral and neuroendocrine responses to stress in the rat are associated with naturally occurring variations in maternal care. Such effects might serve as a possible mechanism by which selected traits are transmitted from one generation to another. Indeed, low-LG mothers are more fearful in response to stress than are high-LG dams.72 Individual differences in stress reactivity are apparently transmitted across generations: fearful mothers beget more stress-reactive offspring. The obvious question is whether the transmission of these traits occurs only as a function of genomic-based inheritance. If this is the case, then the differences in maternal behavior may simply be an epiphenomenon, and not causally related to the development of individual differences in stress responses. The issue is not one of inheritance, but the mode of inheritance.
The results of cross-fostering studies with the offspring of low- and high-LG mothers provide evidence for a nongenomic transmission of individual differences in stress reactivity and maternal behavior.40 The critical groups of interest are the biological offspring of low-LG mothers fostered onto high-LG dams, and vice versa. The limited cross-fostering design did not result in any effect on group differences in maternal behavior. Hence, the frequency of pup LG across all groups of high-LG mothers was significantly higher than that for any of the low-LG dams, regardless of litter composition. The biological offspring of low-LG dams reared by high-LG mothers were significantly less fearful under conditions of novelty than were the offspring reared by low-LG mothers, including the biological offspring of high-LG mothers.40 Subsequent studies reveal similar findings for hippocampal GR expression and for the differences in both the α1 and γ2-GABAA receptor subunit expression in the amygdala.41 These findings suggest that individual differences in patterns of gene expression and behavior can be directly linked to maternal care over the first week of life.
Molecular basis for the effect of maternal care on HPA responses to stress
Molecular biologists have characterized a class of intracellular proteins, termed transcription factors, which are rapidly synthesized in response to extracellular signals and subsequent changes in intracellular second-messenger systems, and which then serve to alter gene transcription. Transcription factors thus provide the molecular interface between gene and environmentally induced changes in cellular activity. The challenge for understanding the pathways by which maternal care alters gene expression is to describe the relevant extracellular and intracellular signals, including the target transcription factors.
Both postnatal handling, which increases maternal LG (see above), and rearing by a high-LG mothers enhance serotonin (5-hydroxytryptamine [5-HT]) turnover in the hippocampus in day-6 rat pups.73,74 Interestingly, postnatal handling results in specific increases in 5-HT in the hippocampus and prefrontal cortex, where GR expression is increased.74 5-HT levels in the hypothalamus, septum, and amygdala are unaffected; GR levels in these regions are not altered by handling. Thus, the sensory input associated with maternal LG selectively alters 5-HT activity in specific brain regions.
The obvious question is whether the increase in 5-HT might directly influence GR gene expression. This issue is remarkably difficult to address with in vivo studies, in which pharmacological manipulations targeting a specific neurotransmitter system inevitably alter other systems, as well as systems in other brain regions. This issue begs an in vitro approach in which the relevant system, the hippocampal neurons, can be examined in a cell culture system. In vitro, the treatment of primary hippocampal cell cultures with 5-HT increases GR expression and this effect is mediated by 5-HT7 receptor activation.75-77 The 5-HT7 receptor is positively coupled to cyclic adenosine monophosphate (cAMP) and GR expression in cultured hippocampal neurons is also significantly increased after treatment with 8-bromo-cAMP (a stable cAMP analog) or with various doses of the specific 5-HT7 receptor agonists, such as 5-carboxamidotryptamine (5-CT). For all conditions, the effect on GR expression is apparent only after 4 days of treatment, a seemingly obscure fact whose importance will later become evidence. The effect of 5-CT on GR expression is blocked by methiothepin. Likewise, 5-CT produces a significant increase in cAMP levels and the effect is blocked by methiothepin. Pindolol, which binds to the 5-HT1A, but not the 5-HT7 receptor, has little effect (see also reference 76). These results further implicate the 5-HT7 receptor. The intracellular effects of cAMP are commonly mediated by cyclic nucleotide-dependent protein kinases (PKA) and, predictably, a PKA inhibitor (H8) blocks the effects of 5HT or cAMP on hippocampal GR expression. Over the course of these studies, we found that other serotonergic agonists (quipazine, TFMPP [1-(trifluoromethylphenyl) piperazine], and DOI [(+/-)-2,5-demethoxy-4-iodoamphetamine]) could partially mimic the 5-HT effect on GR levels and, in all studies, the magnitude of the serotonergic effect on cAMP concentrations is highly correlated (r=0.97) with that on GR expression.78 This observation is consistent with the idea that the effect of 5-HT on GR expression in hippocampal neurons is mediated by a 5-HT7 receptor via activation of cAMP Importantly, both postnatal handling and increased maternal LG increase hippocampal concentrations of both cAMP and PKA in the rat pup. The conclusion of these studies provides the identification of an extracellular signal, 5-HT, and an intracellular, secondary messenger system, cAMP-PKA. Importantly, the in vivo effects of postnatal handling are blocked with compounds that serve as 5-HT7 receptor antagonists.
The in vitro hippocampal cell culture system mimics the in vivo world with surprising authenticity. The increase in GR levels in cultured hippocampal neurons following 5-HT treatment persists following 5-HT removal from the medium; for as long as the cultures can be maintained, there is a sustained increase in GR levels as long as 50 days beyond the removal of 5-HT from the medium. Thus, 5-HT can act directly on hippocampal neurons to increase GR expression, and the effect of 5-HT on GR expression is observed in hippocampal culture cells mimics the long-term effects of early environmental events. These findings provide an in vitro “programming” model. Activation of cAMP pathways can regulate gene transcription through effects on a number of transcription factors, including, of course, the cAMP-response element binding protein (CREB) via an enhanced phosphorylation of CREB. In this instance, the second-messenger system alters the activity of the transcription factor, through enzymatic modification and phosphorylation, rather than production. CREB regulates gene transcription through pathways that involve the transcriptional cofactor, CREB -binding protein (CBP). Primary hippocampal cell cultures treated with 8-bromo-cAMP, 5-CT, or 5-HT show a significant increase in CBP expression.
The 5-HT7 receptor is positively coupled to adenylyl cyclase, and thus the activation of cAMP In vivo, both handling and increased maternal LG result in an increased level of hippocampal cAMP concentrations and the activation of PKA over the first week of postnatal life.76 Activation of PKA results in the tissue-specific induction of a number of transcription factors. The day6 offspring of high-LG mothers or pups of the same age exposed to handling show increased hippocampal expression of NGFIA (nerve growth factor-induced clone A, also known as zif-268, krox-24, egr-1, and zenk) (Weaver IGC et al, unpublished results).79 In vitro, 5-HT increases NGFIA expression in cultured hippocampal neurons and the effect of 5-HT on GR expression in hippocampal cultures is completely blocked by concurrent treatment with an oligonucleotide antisense directed at the NGFIA mRNA.80 The antisense is a synthetic strand of nucleotides that hybridizes with the native mRNA and prevents transcription of the NGFIA protein. These studies serve to identify a relevant transcription factor and to link the activation of the transcription factor NGFIA to the activation of GR expression in response to 5-HT.
Maternal LG results in an increased expression of NGFIA, which in turn might then regulate GR expression. Other rodent models examining environmental regulation of hippocampal GR expression also suggest a correspondence between NGFIA levels and GR expression.81,82 In each case, increased levels of NGFIA are associated with enhanced GR expression. However, the critical site for GR regulation remains to be defined.
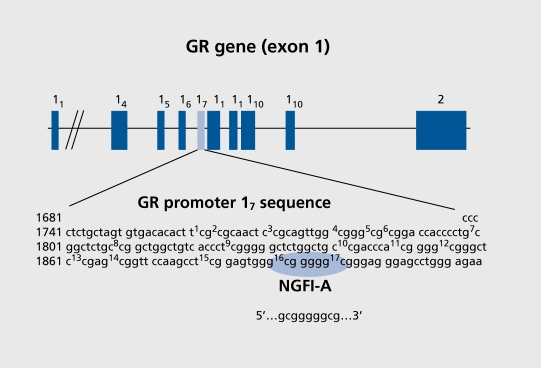
These findings provide a platform for the study of direct gene-environment interactions. However, the important missing piece is the identification of the relevant DNA target. We assumed that a potential target for regulation is the regulatory region of the GR gene. Regulatory regions contains sequences that alter the activity of the gene, such as promoters or suppressors, and are commonly found in front (or upstream) of the coding region of the gene that actually produces the protein. We identified and characterized several new GR mRNAs cloned from rat hippocampus (Figure 2).83 All mRNAs encode a common protein, but differ in their 5 '-leader sequences presumably as a consequence of alternative splicing of, potentially, several different sequences from the 5' noncoding exon 1 region of the GR gene. In this case, the variation in the mRNAs reflects the different promoters that are spliced onto the coding region during transcription to create diverse GR mRNAs. The promoter, while spliced onto the mRNA, does not alter the translational phase by which mRNA is “translated” into the amino acid sequence that defines the protein product. The alternate exon 1 sequences are unlikely to alter the amino acid sequence of the GR protein; there is an “stop” codon present immediately 5' to the translation initiation site in exon 2, common to all the mRNA variants. Hence, only the coding region is actually translated into protein. Of the alternate exon 1 sequences identified, four correspond to exon 1 sequences previously identified in mouse, exons 11,15, 19, and 110 84,85 Most alternative exons are located in a 3-kb CpG island upstream of exon 2 that exhibits substantial promoter activity in transfected cells (Figure 2). Ribonuclease protection assays demonstrate significant levels of six alternative exon 1 sequences in vivo in the rat, with differential expression in the liver, hippocampus, and thymus presumably reflecting tissue-specific differences in promoter activity. The different promoters respond to different signals, which forms the basis for tissue-specific laterations in gene expression. Simply put, it is the process by which environmental or hormonal signals can alter GR expression in one region of the body, without affecting expression in another. Hippocampal RNA contains significant levels of the exon 17-containing GR mRNA variants expressed at undetectable levels in liver and thymus. These studies thus identify a brain-specific GR promoter, the exon 17 sequence.
Figure 2. Map of the noncoding exon 1 region of the glucocorticoid receptor (GR) gene cloned from rat hippocampus.83 The sequence of the critical exon 17 region is provided below, highlighting the NGFIA (nerve growth factor-induced clone A) consensus sequence. The 5' CpG site is differentially methylated as a function of maternal care. Reproduced from reference 83: McCormick JA, Lyons V, Jacobson MD, et al. 5' Heterogeneity of glucocorticoid receptor messenger RNA is tissue specific: differential regulation of variant transcripts by early life events. Mol Endocrinol. 2000;14:506-517. Copyright © 2000. The Endocrine Society.
In transient transfection experiments, a construct encoding the entire regulatory region of the GR gene, including eight of the alternate exon 1 sequences and the splice acceptor site within the intron 5' of exon 2, was fused to a lucif erase reporter gene. The lucif erase gene is activated by the coupled promoters and its activity thus reflects the ability of the regulatory sites to activate gene transcription - hence the term reporter gene. Fusion to the socalled reporter gene permits a measure of the degree to which individual sequences can potentially influence gene expression. This alteration in activity results from various sequences originating at any point within the regulatory region and, we presume, represents the sum of the activity of individual promoters on the genomic DNA fragment. In subsequent studies examining the potency of the individual promoters, we found that the relative activity of the individual exon 1 sequences is similar, with one notable exception, the exon 17 promoter sequence. The fused exon 17 has the highest transcriptional activity of any single promoter construct. More recent studies confirm the transactivational effect of NGFIA at the exon 17sequence. We used a cotransfection model with human embryonic kidney (HEK) cells (intentionally aiming as far from the neural target as possible) with an NGFIA expression vector and an exon 17-luciferase construct. Cotransfection of the NGFIA vector and the exon 17-luciferase construct resulted in a robust increase in luciferase activity, reflecting NGFIA-induced activation of transcription through the exon 17 promoter. These later studies reveal not only the ability of the exon 17 promoter to drive gene expressions, but that it does so in response to an increased NGFIA signal. Recall that an NGFIA antisense completely blocks the effects of 5-HT on GR expression in hippocampal cell cultures.80
Interestingly, the activity of the exon 17 promoter is altered by postnatal handling, which increases GR expression in the hippocampus. Handling selectively elevated GR mRNA containing exon 17; there is, for example, no effect on exon 110.85 Predictably, maternal care also affected the expression of GR splice variants: variants containing the exon 17 sequence were also significantly increased in the adult offspring of high-LG mothers (Weaver IGC et al, unpublished results). Thus, transcriptional activation of the GR gene in the hippocampus during adulthood is altered by maternal care over the first week of life.
The exon 17 promoter sequence of the GR gene contains guanine-cytosine nucleotides, so-called GC boxes (GCGGGGGCG), which form the core consensus site (ie, a DNA binding site) for NGFIA (Figure 2).86 Thus, increases in NGFIA induced by maternal LG could increase transcription from the exon 17 promoter leading to increased GR mRNA. We previously found that handling increased the binding of NGFIA to a promoter sequence for the human GR promoter containing an NGFIA consensus sequence. Since neonatal handling increases maternal LG, these finding suggest that naturally occurring variations in maternal behavior might regulate GR expression in neonatal offspring through a 5HT-induced increase in NGFIA expression, and the subsequent binding of NGFIA to the exon 17 promoter. Recent findings support this idea, including studies using chromatin immunoprecipitation (ChIP) assay in which the in vivo formation of protein-DNA complexes are examined using cross-linking with paraformaldehyde perfusion and subsequent precipitation from soluble hippocampal samples using specific antibodies. Protein binding, defined by the specificity of the antibody, to specific DNA sequences is then quantified following polymerase chain reaction (PCR) amplification with targeted primers and Southern blotting. PCR allows for identification of precise DNA sequences and Southern blotting permits quantification of those same sequences. The experiment provides information on the amount of a specific DNA sequence bound to a specific protein. The charm of this approach is the ability to directly examine the interaction of specific proteins with specific DNA sequences at the time the biological sample is obtained. ChIP analysis of hippocampal samples from postnatal day-6 pups reveals dramatically increased NGFIA binding to the exon 17 promoter in the offspring of high-LG compared with low-LG mothers.67 These findings confirm that maternal care regulates the binding of NGFIA to the exon 17 promoter sequence in pups.
These findings suggest that maternal LG in the neonate increases NGFIA expression in the hippocampus and NGFIA binding to the exon 17 promoter. NGFIA might then increase GR expression in hippocampal neurons, and these findings might then provide a mechanism for the effect of maternal care over the first week of life. However, while there are striking differences in NGFIA expression in the offspring of high- and low-LG mothers at day 6 of postnatal life, hippocampal NGFIA expression in adulthood is unaffected by maternal care: there is no difference in hippocampal NGFIA expression in the adult offspring of high- and low-LG dams. We are thus left with the defining question of early experience studies: how are the effects of early life events sustained into adulthood?
Epigenetic programming of stress reponses
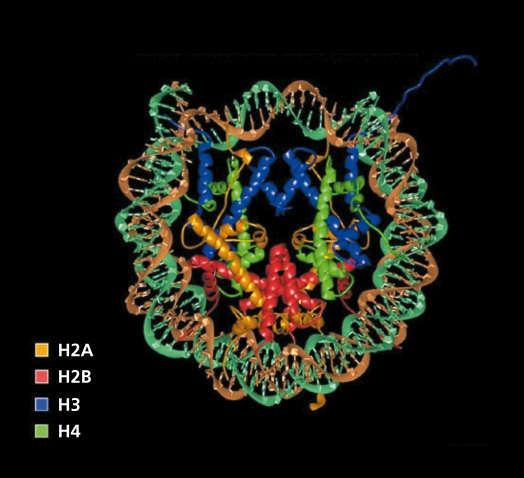
While we are all familiar with linear models of DNA and protein-DNA interactions where protein-DNA interactions occur, it would seem, in the absence of any obstruction, such models ignore the fact that most of the DNA is tightly packaged into nucleosomes that involve a close relationship formed by DNA wrapped around a core of histone proteins (Figure 3).87 The actual formation of a nucleosome is 146 bp of DNA with a histone octamer core. The conformation or structure of the histone-DNA configuration regulates gene expression.88
Figure 3. Nucleosome core particle: ribbon traces for the 146-bp DNA phosphodiester backbones (brown and turquoise) and eight histone protein chains.87 The configuration is maintained, in part, through electrostatic bonds between the positively charged histones and negatively charged DNA. The N-terminal histone 3 tail (in blue) is a major site for enzymatic modification. Acetylation of the lysine residues in proximity to the DNA neutralizes the histone charge and opens the configuration permitting transcription factor binding. Reproduced from reference 87: Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of nucelosome core particle at 2. 8 A resolution. Nature. 1997;389:251-260. Copyright© 1997. Nature Publishing Group.
The relation between DNA and histone is maintained, in part, by electrostatic bonds between positively charged histones and the negatively charged DNA. This chromatin structure commonly precludes transcription factor binding to DNA and underscores the importance of enzymes that modify histone-DNA interactions. Most modifications of the nucleosome occur on amino acid residues along the histone tail that protrudes through the DNA, and is thus vulnerable to enzymatic modification (Figure 3). The relevant histone modifications include acetylation, phosphorylation, ribosylation, and methylation. Each of these modifications can alter the interaction between the histones and the DNA, and thus alter gene expression. Our focus is on histone acetylation, which is closely associated with gene expression.
One class of such proteins, histone acetyltransferase (HAT),89 catalyze the acetylation of selected amino acids, on the protruding histone tails, most commonly histone 3 (H3). Positively charged amino acids such as lysine and arginine are the common targets for acetylation. Histone acetylation modifies the histone-DNA relation. Acetylation of the lysine (K) residue on H3 neutralizes the positively charged histone, opening the histone-DNA relationship, and facilitating transcription factor binding to DNA. Thus, H3-K9 acetylation is a marker of active gene transcription. Many known transcriptional cofactors (proteins that enhance gene expression), such as CBP, are HATs. Interestingly, CBP is activated in hippocampal cell cultures in response to 5-HT or cAMP treatment. Histone acetylation is dynamic and is regulated by histone deacetylases (HDACs). HDACs block histone acetylation and suppress gene expression. Thus, chromatin structure can be viewed as dynamic and clearly subject to modification through intracellular signals that trigger either HATs or HDACs downstream.90-92 The study of histone acetylation provides a remarkable advance in our understanding of the dynamic and complex regulation of gene expression (see reference 88 for a review). Nevertheless, histone modifications are generally transient, enduring for minutes to hours. Such events are not the basis for the persistent effects of early life events on gene expression.
The chemistry of DNA methylation
In addition to chromatin, which provides the functional environment for the DNA, the DNA molecule itself is chemically modified by the addition of methyl residues at the 5' position of the cytosine rings in the CG sequence, resulting in methylated cytosine.93,94 Cytosine methylation, while chemically a rather simple modification, has remarkable importance for gene activity, or expression. Methylation of DNA is common in early development, is associated with gene silencing, and is assumed to be the mechanism for events such as parental imprinting, where the allele derived from one parent is silent. Moreover, DNA methylation is maintained by carbon-carbon bonds and therefore highly stable. Unlike the more transient histone modifications that redefine chromatin structure, DNA methylation is a reasonable candidate mechanism for environmental programming of gene expression.
What distinguishes DNA methylation in vertebrate genomes is the fact that not all CGs within a common sequence are methylated in any given cell type.95 Different CGs are methylated in different cell types, generating cell type-specific patterns of methylation. Thus, the DNA methylation pattern confers a cell-specific identity upon the genome. Such variation is presumably related to cell-specific patterns of gene expression. Since DNA methylation is part of the chemical structure of the DNA itself, it remains long after all other proteins and epigenomic markers are degraded and thus it has extremely important diagnostic potential.96,97 It was originally believed that the DNA methylation pattern is established during development and is then maintained faithfully through life by the maintenance DNA methyltransf erase.95,98 The DNA methylation reaction was believed to be irreversible and that the only way methyl residues were lost was through replication in the absence of DNA methyltransferase, resulting in the loss of the cytosine methylation in the daughter cell,93,94 a mechanism that is not applicable to postmitotic cells such as neurons. However, recent findings together with the data reviewed here support an alternative model: one that suggests that the DNA methylation pattern is dynamic and is an equilibrium of methylation and demethylation reaction.99,100We propose that DNA methylation is a reversible, like any other biological signal, and could potentially change in response to environmental and physiological signals.99-101 The notion that DNA methylation is reversible in postmitotic cells has immense implications on our understanding the potential role of DNA methylation in marking gene expression in the brain.
The hallmark of DNA methylation patterns is the correlation between chromatin and the DNA methylation pattern: its importance for gene expression. Active chromatin is usually associated with unmethylated DNA, while inactive chromatin is associated with methylated DNA.91,102,103 The reiation between DNA methylation, and chromatin structure (referring primarily to the relation between histone proteins and the DNA) has important implications for our understanding of the function of DNA methylation, as well as the processes responsible for generating, maintaining, and altering DNA methylation patterns under physiological and pathological conditions. It was originally believed that DNA methylation precedes and is dominant over chromatin structure.104 Methylation was thought to be generated independently of chromatin structure. Over the course of development, methylation patterns were believed to be laid down shortly after cell replication and to then determine chromatin structure and gene expression. The DNA methylation pattern is proposed to guard the genome from random noise and drift.
Methylated DNA attracts methylated DNA binding proteins, which recruit a cluster of proteins referred to as repressor complexes, which include histone deacetylases that result in inactive chromatin and the silencing of gene expression.105,106 The model positioning DNA methylation as driving chromatin inactivation is pervasive. Nevertheless, new data suggest that the state of chromatin structure can also determine DNA methylation and that chromatin can affect DNA methylation in both directions triggering either de novo DNA methylation or demethylation.107-109 These data revise the classic model of a DNA methylation pattern that is determined during development and maintained through life, and adopt a more dynamic view of the DNA methylation pattern as an interface between the dynamic environment and the static genome. Thus, although DNA methylation is an extremely stable signal, it can be altered later in life when there is a sufficiently stable and consistent signal to activate the chromatin. Transient changes in cellular function and chromatin structure are not accompanied by changes in DNA methylation. The relation between chromatin state and DNA methylation forms a molecular link through which environmental signals might alter DNA methylation in specific genes in postmitotic neurons. Environmental signals trigger cellular signaling pathways, the downstream consequence of which is activation of trans-acting factors, such as transcription factors. These trans-acting factors recruit HATs to the target gene resulting in increased histone acetylation, chromatin opening, and increased accessibility of the DNA to demethylases. Since methylation of cytosine is an extremely stable chemical bond on DNA, this modification will remain stable for years. For methylation signals to serve as stable markers, they should not be responsive to transient chromatin noise or short-term signals. The mechanism proposed here also allows for a reversal of the methylation marker by a similar intense change in chromatin structure later in life.99,110 This model has important implications on our understanding of how environmental signals, such as variations in maternal care, might stably alter glucocorticoid gene expression. DNA methylation marks genes for silencing by a number of mechanisms. The first mechanism is indirect and links DNA methylation to inactive chromatin structure. A region of methylated DNA juxtaposed to regulatory regions of genes attracts different members of a family of methylated DNA binding proteins, such as methylCpG-binding protein, MeCP2, which recruits HDACs105,106 and histone methyltransferases111 to methylated genes.91,112 This results in a modification of chromatin around the gene precipitating an inactive chromatin structure. A different mechanism, which is relevant to our discussion here, involves direct interference of a specific methylated CpG residing within a response element for a transcription factor with the interaction of a transcription factor, such as the inhibition of binding of cMyc to its response element when it is methylated.113 Essentially, the methylated cytosine serves as a mutation of the recognition element, functionally reducing the binding affinity of the response element for its transcription factor. A third mechanism involves a combination of binding of a methylated DNA binding protein and inhibition of activity of a transcription factor.114 While the first mechanism is dependent on the general density of methyl cytosines within the region associated with a gene rather than methylation of a specific CpG, the second mechanism requires a discrete methylation event and is relevant to the mechanism proposed here.
The important consideration is the stability of cytosine methylation, which is preserved by covalent carbon-carbon bonds and could therefore serve as a long-term genomic “memory” of early experience influencing chromatin structure and GR expression in offspring of highand low-LG mothers. GR gene expression is increased throughout the hippocampus in the adult offspring of high-LG compared with low-LG mothers.39 The exon 17 GR promoter sequence appears to be significantly more active in the adult offspring of high-LG compared with low-LG mothers and was therefore the focus of initial studies of possible maternal effects on DNA methylation. To test the hypothesis that maternal care alters the DNA methylation mark of the GR promoter, we67 examined the level of methylation across the entire exon 17 GR promoter sequence in the hippocampus using the sodium bisulfite (NaBis) mapping technique in the adult offspring of high- and low-LG mothers. NaBis treatment of DNA samples converts nonmethylated cytosines to uracils, which are then detected as thymidine on subsequent sequencing gels.115 Methylated cytosines are unaffected by NaBis and the differences in methylation status are thus apparent and easily quantifiable on sequencing gels. We found significantly greater methylation of the exon 17 GR promoter sequence in the offspring of the low-LG mothers. These findings are consistent with the hypothesis that maternal effects alter DNA methylation patterns in the offspring.
To determine whether DNA methylation of specific target sites on the GR promoter change in response to maternal care, we mapped the differences in methylation of individual cytosines, focusing on a region around the NGFIA consensus sequence within the exon 17 promoter. The results reveal significant differences in the methylation of specific regions of the exon 17 GR promoter sequence. Notably, the cytosine within the 5' CpG dinucleotide of the NGFIA consensus sequence (Figure 2) is always methylated in the offspring low-LG mothers, and rarely methylated in the offspring of high-LG dams. This is consistent with site-specific DNA methylation silencing of the GR promoter.
To directly examine a causal relation between maternal behavior and DNA methylation changes within the exon 17 GR promoter, we67 performed an adoption study in which the biological offspring of high- or low-LG mothers were cross-fostered to either high- or low-LG dams within 12 hours of birth.40,41 These studies could rule out either a purely traditional genetic or a prenatal basis for the variation in DNA methylation in the offspring of high- versus low-LG offspring. Cross-fostering the biological offspring of high- or low-LG mothers produced a pattern of exon 17 GR promoter methylation associated with the rearing mother.67 The cytosine within the 5' CpG dinucleotide of the NGFIA consensus sequence is hypomethylated following cross-fostering of offspring of low- to high-LG dams, with no effect at the cytosine within the 3' CpG dinucleotide. Thus, the pattern of methylation of the cytosine within the 5' CpG dinucleotide of the NGFIA consensus sequence within the exon 17 GR promoter of the biological offspring of low-LG mothers cross-fostered to high-LG dams is indistinguishable from that of the biological offspring of high-LG mothers. The reverse is true for the offspring of high-LG mothers fostered to low-LG dams. These findings suggest that variations in maternal care alter the methylation status within specific sites of the exon 17 promoter of the GR gene and represent the first demonstration of a DNA methylation pattern established through a behavioral mode of programming. Essentially, these findings provide a direct example of epigenetics: a modification of the genome that does not involve an alteration in sequence, and is thus distinctive from what is thought to be Lamarckian transmission (which would involve a change in sequence transmitted through genetic inheritance). This example is also distinct from parental imprinting, a well-established paradigm of inheritance of an epigenetic marker, that requires germ-line transmission.116,117
Site-specific methylation of the 5' CpG dinucleotide of the NGFIA response element blocks transcription factor binding
The obvious question concerns the functional importance of such differences in methylation. DNA methylation affects gene expression either by attracting methylated DNA-binding proteins to a densely methylated region of a gene or by site-specific interference with the binding of a transcription factor to its recognition element.91,112 Our data showing site-specific differences in methylation of the cytosine within the 5' CpG dinucleotide of the NGFIA response element suggests alterations in the ability of the NGFIA protein to bind to its response element. We118 determined the in vitro binding of increasing concentrations of purified recombinant NGFIA protein119 to its response element under different states of methylation using the electrophilic mobility shift assay (EMSA) technique with four 32P-labelled synthetic oligonucleotide sequences bearing the NGFIA binding site that was either (i) nonmethylated; (ii) methylated in the 3' CpG site; (hi) methylated in the 5' CpG site; (iv) methylated in both sites; or (v) mutated at the two CpGs with an adenosine replacing the cytosines. NGFIA formed a protein-DNA complex with the nonmethylated oligonucleotide, while the protein is unable to form a complex with either a fully methylated sequence or a sequence methylated at the 5' CpG site. NGFIA binding to its response element was only slightly reduced with the sequence methylated at the 3' CpG site. The results indicate that while methylation of the cytosine within the 5' CpG dinucleotide reduces NGFIA protein binding to the same extent as methylation in both CpG sites, methylation of the cytosine within the 3' CpG dinucleotide only partially reduces NGFIA protein binding. These data support the hypothesis that methylation of the cytosine within the 5' CpG dinucleotide in the NGFIA response element of the exon 17 GR promoter region in the offspring of low-LG mothers inhibits NGFIA protein binding.
This is an important finding for our understanding of the processes by which maternal care programs hippocampal GR expression and thus HPA responses to stress. While there are substantial differences in differences in NGFIA expression between the offspring of high- and low-LG mothers in early postnatal life, no such differences are apparent in adulthood. Our hypothesis is that the cytosine methylation in the response element for NGFIA interferes with NGFIA binding to the GR exon 17 promoter. We therefore predicted that the reduced cytosine methylation in the adult offspring of high-LG compared with low-LG mothers would result in greater NGFIA binding to the exon 17 promoter. This prediction was confirmed using a ChIP assay (described above) examining in vivo formation of protein-DNA complexes in hippocampal tissue from adult animals.67 The results indicated a threefold greater binding of NGFIA protein to the hippocampal exon 17 GR promoter in the adult offspring of high-LG compared with low-LG mothers. Using the same tissue samples and an antibody against the acetylated form of H3, we67 found dramatically increased acetylated H3 association with the exon 17 GR promoter in the offspring of the high-LG mothers. As described above, histone acetylation is associated with active states of gene expression. These findings are therefore consistent with the idea of increased NGFIA binding to the exon 17 promoter, enhanced histone acetylation, and increased GR transcriptional activation.
We confirmed that DNA methylation inhibits the ability of NGFIA to activate the exon 17 promoter using a transient cotransfection assay in HEK293 cells. The HEK293 cells are not of neural origin and thus allow us to measure the transcriptional consequences of interaction of NGFIA with either a methylated or nonmethylated version of the GR exon 17 promoter per se, independent of the complications associated with other neuronal signals. We used transfection technology to introduce into the HEK cells (i) a viral vector containing the NGFIA gene, to produce a intracellular signal usually inactive in HEK cells; and (ii) an exon 17-luciferase reporter construct. This genomic construct that included the exon 17 promoter sequence fused with a luciferase reporter gene (the level of the easily measured luciferase activity is used as a measure of exon 17 promoter activity). Cotransfection of the NGFIA expression vector significantly increases luciferase activity; however, this effect is dramatically reduced if the CpG dinucleotides within the exon 17 sequence are methylated. Moreover, the effect of NGFIA on transcription through an exon 17-luciferase reporter construct was almost completely abolished with a point mutation at the 5' cytosine (a cytosine to adenosine mutation). Taken together, these findings suggest that an “epimutation” at a single cytosine within the NGFIA consensus sequence alters the binding of NGFIA and might therefore explain the sustained effect of maternal care on hippocampal GR expression and HPA responses to stress.
How does maternal care alter cytosine methylation?
Maternal behavior could either inhibit de novo methylation or stimulate demethylation. To address this question, we67 performed a simple developmental study of the methylation pattern of GR exon 17 promoter from embryonic day 20 to day 90 (a fully, sexually mature adult rat). High- and low-LG mothers differ in the frequency of pup LG only during the first week of life. Importantly, this period corresponds to the appearance of the difference in DNA methylation in the offspring in studies using NaBis mapping to precisely map the methylation status of the cytosines within the exon 17 GR promoter over multiple developmental time points. This analysis demonstrates that just 1 day before birth, on embryonic day 20, the entire exon 17 region is completely unmethylated in both groups. Strikingly, 1 day following birth (postnatal day 1) the exon 17 GR promoter is de novo methylated in both groups. The 5' and 3' CpG sites of the exon 17 GR NGFIA response element in the offspring of both high- and low-LG mothers, which exhibit differential methylation later in life, are de novo methylated to the same extent. These data show that both the basal state of methylation and the first wave of de novo methylation after birth occur similarly in both groups. Whereas it is generally accepted that DNA methylation patterns are formed prenatally and that de novo methylation occurs early in development, there is at least one documented example of postnatal de novo methylation of the HoxA5 and HoxB5 genes.120 Since similar analyses are not documented for other genes, it is unknown yet whether changes in methylation are common around birth or whether they are unique to this GR promoter. One aspect of these findings that is important is that of the complete absence of cytosine methylation on embryonic day 20. Since the majority of the pyramidal cells of Amnion's Horn are born between embryonic days 16 and 20, it seems unlikely that methylation patterns, at least on the exon 17 promoter of the GR, are generated at the time of DNA replication and cell division, as would normally be the case with imprinted genes.
The differences in the status of methylation of the exon 17 GR develop between the two groups emerges between postnatal day 1 and 6, which is precisely the period when differences in the maternal behavior of high- and low-LG dams are apparent. There are no differences in maternal LG between high- and low-LG mothers beyond day 8.65,69 By postnatal day 6, the 5' CpG dinucleotide of the NGFIA response element is demethylated in the highLG, but not in the low-LG group. These findings are consistent with data from the cross-fostering experiment, which illustrates that the differences between the two groups developed following birth in response to maternal behavior. The group difference in CpG dinucleotide methylation then remains consistent through to adulthood. Our findings suggest that the group difference in DNA methylation occurs as a function of a maternal behavior over the first week of life. The results of earlier studies indicated that the first week of postnatal life is indeed a critical period for the effects of early experience on hippocampal GR expression.121
The striking finding from this rather simple study was evidence of a demethylation, as opposed to the prevention of methylation. A recent paper on altered expression of interleukin-2 (IL-2) expression T lymphocytes following activation also clearly implicates an active process of demethylation in a normal differentiated somatic cell. Bruniquel and Schwartz122 found that a region in a promoter of the IL-2 gene demethylates following activation in the absence of DNA replication and results in a profound increase in the production of IL-2. These two papers provide the initial evidence for an active, environmentally driven alteration in DNA methylation in postmitotic cells. Szyf and colleagues101,123 first proposed that DNA methylation is enzymatically reversible and that DNA methylation is dynamic in fully differentiated cells. This idea remains controversial. Active demethylation was nevertheless clearly demonstrated early in embryogenesis and the parental genome undergoes replication independent, active demethylation hours after fertilization, well before the initiation of replication. Demethylation at very early stages in development has been relatively accepted, but the possibility of postnatal demethylation, especially in fully differentiated somatic cells, has been hotly disputed. However, active replication demethylation was demonstrated in Epstein-Barr virus (EB V)-infected B cells and in HEK293 cells. The HEK293 studies suggest that active replication-independent demethylation takes place in differentiated somatic cells and that it is dependent on alterations in chromatin structure.
Earlier studies from Szyf's122 laboratory extracted active DNA demethylase activity from a human lung cancer cell line and identified a protein with demethylase activity, which was cloned concurrently by Bird's group and named MBD2.123 Interestingly, the protein, MBD2, was found by Bird's group and others to also associate with a chromatin remodeling complex containing HDAC, which is involved in silencing of gene expression through the recruitment of a repressor complex. The assignment of a demethylase function to a protein that was independently discovered as a recruiter of repressor complexes triggered the expected controversy in the field and reports that MBD2 failed to produce demethylase activity. However, the observation that MBD2/demethylase expression produces the demethylation of some, but not all, promoters in a dose- and time-dependent manner has been confirmed.108,124 Clearly, the contextual factors that determine MBD2 demethylase activity remain to be fully explained. Interestingly, MBD2 increased gene expression in those instances where promoter demethylation occurred, suggesting that not all promoters respond in the same orderly manner. Indeed, the same is true for DNA methylation, which impedes the DNA binding of most, but not all transcription factors; SP1 binds to methylated DNA. Antisense knock down of MBD2 resulted in inhibition of active demethylation induced by valproate and caused hypermethylation and silencing of the prometastatic gene uPA in metastatic breast cancer cells. Another group reported that ectopic expression of MBD2/demethylase in hepatocyte cell line caused demethylation and activation of the hexokinase type 2 gene.125 Additional support for the demethylase activity of MBD2/demethylase emerges from the finding that expression of MBD2/demethylase is correlated with demethylation within the promoters of C-ERBB-2 and SURVIVIN genes in ovarian cancers126,127 and hypomethylated CMYC in gastric cancer.128 In addition, the Drosophila homolog of MBD2, dMBD2/3, formed foci that associated with DNA at the cellular blastoderm stage, concurrent with the activation of the embryonic genome, and also associated with the active Y chromosome.129
To test the hypothesis that MBD2 is associated with maternally induced demethylation, we performed an in situ hybridization assay with probes for the mRNAs of a number of methylated binding proteins at day 6 postpartum. Our analysis revealed that MBD2/demethylase expression is elevated in the hippocampus at this point in time in offspring of high-LG versus low-LG mothers. A ChIP analysis with an antiMBD2/demethylase antibody demonstrates significantly increased binding of MBD2/demethylase to the exon 17 GR promoter in day6 offspring of high-LG versus low-LG mothers. We also found increased NGFIA binding to the same sequence in day-6 offspring of high-LG offspring. We then performed a NaBis mapping of the state of methylation of the exon 17 GR promoter bound to MBD2 and precipitated in the ChIP assay with antiMBD2 antibody. If MBD2 is the demethylase involved in this process or if it is part of the demethylase complex, then MBD2-bound exon 17 sequences at day 6 should be found in the process of demethylation. Indeed, most of the MBD2bound DNA was unmethylated or partially unmethylated.
Reversal of the maternal effect on GR expression and HPA responses to stress
These findings suggest that maternal behavior produces an active demethylation process at selected and perhaps actively targeted sites. The resulting demethylation of the 5' CpG dinucleotide within the NGFIA response element of the exon 17 promoter enhances NGFIA binding to the exon 17 promoter, increasing GR gene transcription and HPA responses to stress.
These findings beg the question of how maternal high LG might activate a demethylation of the GR exon 17 promoter. A testable working hypothesis is that high LG leads to activation of NGFIA as a downstream effector of activation of a 5-HT signaling through increase cAMP and PKA. Increased NGFIA increases NGFIA binding to the GR exon 17 promoter. The interaction of NGFIA with the GR exon 17 promoter leads to increased histone acetylation and increased accessibility of the GR exon 17 promoter to demethylase resulting in DNA demethylation. In contrast, in the absence of increased NGFIA during early postnatal life, the 5' CpG site of the NGFIA response element remains methylated and significantly less sensitive to NGFIA over the life span. The methylation of the 5' CpG site is thought to preclude NGFIA binding through the participation of a repressor complex that includes methylated DNA binding proteins and HDACs. This hypothesis predicts that pharmacological activation of chromatin using HDAC inhibitors should result in activation of NGFIA binding and GR exon 17 promoter demethylation. However, the question is whether reversibility reflected in demethylation is limited to early life exclusively or whether it is possible to reverse these marks later in life as well if the appropriate signals to activate the chromatin structure are applied or by a pharmacological activation of chromatin structure. Our hypothesis is that the DNA methylation is a steady state of DNA methylation and demethylation whose direction is determined by the state of chromatin structure.99 ,110 This hypothesis predicts that both DNA methyltransferases and demethylases are present in adult neurons and that if the chromatin state is altered by either persistent physiological or pharmacological signals one should be able to change the state of methylation of a gene in postmitotic tissue, such as adult hippocampal neurons. We previously established that pharmacological activation of chromatin structure by HDAC inhibitors can trigger replication-independent active demethylation of DNA.108,130,131 We tested our hypothesis that the demethylation of the GR exon 17 promoter is driven by histone acetylation and could be activated in adult neurons as well; HDAC inhibition should reverse the effects of cytosine methylation on NGFIA binding to the exon 17 promoter, GR expression, and HPA responses to stress. We used a central infusion of adult offspring of high- or low-LG mothers with the HDAC inhibitor, trichostatin A (TSA), for 4 consecutive days. As expected, ChIP assays revealed that HDAC inhibition through TSA infusion significantly increased the level of acetylated H3 at the exon 17 site (ie, HDAC inhibition resulted in increased histone acetylation) in the offspring of lowLG mothers to levels comparable to those observed in the offspring of high-LG mothers. The increased histone acetylation is associated with enhanced NGFIA binding to the exon 17 promoter sequence and completely eliminates the effect of maternal care. As expected, enhanced NGFIA binding to the exon 17 promoter increased hippocampal GR expression. Hippocampal GR expression in the TSA-treated adult offspring of low-LG mothers was indistinguishable from that of the high-LG groups. Most important, TSA infusion eliminated the effect of maternal care on HPA responses to stress. During and following exposure to acute stress, plasma corticosterone levels in TSA-treated offspring of low-LG mothers are indistinguishable from those of TSA- or vehicle-treated high-LG mothers. There was no effect of TSA on any measure in the offspring of high-LG animals. This is understandable since under normal circumstances there is considerable H3 acetylation and NGFIA binding at the exon 17 sequence in these animals. Interestingly, TSA treatment also led to the demethylation of the 5' CpG of the NGFIA response element of the GR exon 17 promoter sequence.
These findings have important implications for our understanding of the mechanisms linking early maternal behavior and stable changes in behavior later in adulthood as well as on our understanding of the mechanisms responsible for maintaining the DNA methylation pattern in adult postmitotic tissues.
First, our data support the idea that demethylation is driven by activation of chromatin and that HDAC inhibitors produce demethylation even in nondividing cells (ie, in a replication-independent manner).
Second, our data are consistent with the hypothesis that the demethylation of GR exon 17 in offspring of highLG rats early after birth is driven by increased histone acetylation, as discussed above.
Third, these data provide evidence that molecular mechanisms that underlie the effects of early life-experience neural function are potentially reversible in adulthood. This consideration is of obvious social and therapeutic implications.
Fourth, these data provide in vivo evidence for our hypothesis that the DNA methylation pattern is dynamic even in postmitotic tissues and that its steady state is maintained by the state of chromatin acetylation.99
Finally, the data provide a framework for understanding of how environmental signals could change the DNA methylation pattern and thus the chemistry of the genome itself, even during adulthood.
Dissection of the molecular mechanisms linking maternal behavior and active demethylation of GR exon 17 promoter in the hippocampus
The data discussed above support the hypothesis that histone acetylation could produce active demethylation of the GR exon 17 promoter, yet several questions remain unanswered. How, for example, is histone acetylation targeted to the exon 17 promoter as a consequence of maternal behavior? We propose that maternal behavior stimulates 5-HT, which stimulates NGFIA, and that NGFIA then targets HATs and eventually demethylases to the GR exon 17 promoter. To dissect the different molecular components of this hypothesis, we took advantage of both hippocampal primary neuronal cell cultures as well as nonneuronal cell lines. The two systems have different strengths and could be used to test different components of the model. First, we tested the hypothesis that 5-HT acts through cAMP to produce hypomethylation. Hippocampal cell cultures treated with either 5-HT or 8-bromo-cAMP, a stable cAMP analog, show increased GR expression following 4 days of treatment. Treatment of hippocampal cells in culture with 5-HT also results in the hypomethylation of the 5' CpG dinucleotide of the NGFIA consensus sequence within the exon 17 promoter of the GR gene, with no effect at the 3' site (Weaver IGC et al, unpublished results). Treatment with 8-bromocAMP produces an even more pronounced effect on cytosine methylation at the 5' CpG site. In both studies, cultures maintained under control conditions show complete methylation of both the 5' and 3' CpG sites of the NGFIA consensus sequence. Bromodeoxyuridine labeling, which marks newly generated cells, reveals little or no cell replication in the cultures at the time of 5-HT treatment. These findings reinforce the idea that the alterations in cytosine methylation occur independently of cell replication and in response to intracellular signals associated with variations in maternal care. These cells establish that 5-HT signaling induced by maternal care triggers replication-independent changes in methylation of GR exon 17 promoter through an increase in cAMP Since increased cAMP activates NGFIA, it seems that ectopic expression of NGFIA can target the demethylation process to the GR exon 17 promoter. Indeed, there is direct evidence that NGFIA can actively target methylated DNA binding proteins to specific sites around the NGFIA response element. While this finding provides evidence for a targeting process, it does not explain the selective effect at the 5' cytosine.
To test the hypothesis that NGFIA targets demethylation to GR exon 17 promoter we resorted to nonneuronal cell line HEK293. Here, we can isolate the direct effect of NGFIA from other neuron-specific events that might confound the interpretation of data from the hippocampal cultures. By comparing the fate of a transiently transfected methylated GR exon 17 promoter-luciferase vector in the presence and absence of NGFIA we could better determine the specific effects of NGFIA on demethylation. Whereas in vitro methylated GR exon 17 promoter-luciferase vector remains methylated in HEK293 cells, coexpression of NGFIA results in active demethylation of a significant fraction of the transf ected plasmids. To demonstrate that this DNA demethylation requires direct contact between NGFIA and its recognition element, we performed site-directed mutagenesis of the two CGs included in the NGFIA recognition element. Our preliminary results suggest that these manipulations abolished the ability of NGFIA to activate and demethylate the GR exon 17 promoter. These experiments provide a molecular mechanism on how demethylation is triggered to specific sequences. The outstanding question is to determine how NGFIA triggers demethylation upon binding to specific sequences. One possibility is that NGFIA directly recruits a demethylase to the gene or that, as proposed before, it recruits a HAT that increases acetylation, thus increasing the accessibility to demethylase as proposed before.
Experience-dependent chromatin plasticity? Environmental variability meets epigenomic predictability
In summary, our findings suggest that shortly after birth there is a wave of de novo methylation that results in the methylation of both CpG sites within the NGFIA consensus sequence. Such events would impede the binding of NGFIA to the exon 17 promoter. However, in the offspring of the high-LG mothers, NGFIA expression is increased to the point where binding occurs despite the “low affinity” status of the binding site. The binding of NGFIA is associated with histone acetylation and the subsequent availability of the site to demethylase. In support of this idea, the treatment of the adult offspring of the low-LG mothers with TSA increases H3 acetylation and NGFIA binding (see above) and results in the demethylation of the 5' CpG site of the NGFIA consensus sequence.67 While this model remains speculative (and controversial) at this time, these findings do suggest that modifications to the DNA methylation status in fully differentiated cells are clearly possible and pharmacologically reversible, an idea that holds considerable potential therapeutic implications.
The defining question of early experience studies concerns the mechanism by which environmental effects occurring in early development are “biologically embedded” and thus sustained into adulthood (ie, so-called “environmental programming” effects). The offspring of high-LG mothers exhibit increased hippocampal GR expression from the exon 17 promoter and dampened HPA responses to stress that persist into adulthood. We propose that the differential epigenomic status of the exon 17 GR promoter in the offspring of high-LG mothers serves as a mechanism for this maternal effect. It is important to note that these findings are restricted to the study of a single promoter of but one gene in one region of the brain. The degree to which such results might generalize to other instances of environmental programming remains to be determined. Moreover, further studies are required to determine how maternal behavior alters the epigenomic status of the exon 17 GR promoter. The developmental timecourse study is critical. Recall that the 5' CpG dinucleotide of the NGFIA consensus sequence of the exon 17 promoter is methylated to the same, elevated level in the newborn offspring of high- and low-LG mothers. It is only over the first week of life that the difference emerges, with the decline in the methylation of the 5' CpG site in the offspring of high-LG, but not low-LG mothers. No such demethylation occurs at the neighboring 3' CpG site. The impressive selectivity suggests a demethyaltion process that is targeted in some manner. It is critical to define the processes by which such apparently active demethylation might occur. Regardless of these as-yet unanswered questions, these findings provide the first evidence that maternal behavior stably alters the epigenome of the offspring, providing a mechanism for the long-term effects of early experience on gene expression in the adult. These studies offer an opportunity to clearly define the nature of gene-environment interactions during development and how such effects result in the sustained “environmental programming” of gene expression and function over the life span. Finally, it is important to note that maternal effects on the expression of defensive responses, such as increased HPA activity, are a common theme in biology,132,133 such that the magnitude of the maternal influence on the development of HPA and behavioral responses to stress in the rat should not be surprising. Maternal effects on defensive responses to threat are apparent in plants, insects, and reptiles. Such effects commonly follow from the exposure of the mother to the same or similar forms of threat and may represent examples where the environmental experience of the mother is translated through an epigenetic mechanism of inheritance into phenotypic variation in the offspring. Indeed, maternal effects could result in the transmission of adaptive responses across generations.30 Epigenomic modifications of targeted regulatory sequences in response to even reasonably subtle variations in environmental conditions might serve as a major source of epigenetic variation in gene expression and function, and ultimately as a process mediating such maternal effects. We propose that epigenomic changes serve as an intermediate process that imprints dynamic environmental experiences on the fixed genome resulting in stable alterations in phenotype.
Selected abbreviations and acronyms
- ACTH
adrenocorticotropin hormone
- BZ
benzodiazepine
- CBP
CREB-binding protein
- ChIP
chromatin immunoprecipitation
- CREB
cyclic adenosine monophosphate (cAMP)-response element binding protein
- CRF
corticotropin-releasing factor
- 5-CT
5-carboxamidotryptamine
- GABA
γ-aminobutyric acid
- GR
glucocorticoid receptor
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HPA
hypothalamic-pituitary-adrenal (axis)
- 5-HT
5-hydroxytryptamine (serotonin)
- LG
licking/grooming
- PKA
protein kinase
- NaBis
sodium bisulfite
- NGFIA
nerve growth factor-induced clone A
- PVNh
paraventricular nucleus of the hypothalamus
- TSA
trichostatin A
REFERENCES
- 1.Repetti RL., Taylor SE., Seeman TE. Risky families: family social environments and the mental and physical health of offspring. Psychol Bull. 2002;128:330–366. [PubMed] [Google Scholar]
- 2.Bifulco A., Brown GW., Adler Z. Early sexual abuse and clinical depression in adult life. Br J Psychiatry. 1991;159:115–122. doi: 10.1192/bjp.159.1.115. [DOI] [PubMed] [Google Scholar]
- 3.Brown GR., Anderson B. Psychiatric morbidity in adult inpatients with childhood histories of sexual and physical abuse. Am J Psychiatry. 1991;148:55–61. doi: 10.1176/ajp.148.1.55. [DOI] [PubMed] [Google Scholar]
- 4.McCauley J., Kern DE., Kolodner K., et al. Clinical characteristics of women with a history of childhood abuse: unhealed wounds. JAMA. 1997;277:1362–1368. [PubMed] [Google Scholar]
- 5.Felitti VJ., Anda RF., Nordenberg D., et al. Relationship of childhood abuse and household dysfunction to many of the leading causes of death in adults. Am J Prevent Med. 1998;14:245–258. doi: 10.1016/s0749-3797(98)00017-8. [DOI] [PubMed] [Google Scholar]
- 6.Heim C., Newport DJ., Wagner D., Wilcox MM., Miller AH., Nemeroff CB. The role of early adverse experience and adulthood stress in the prediction of neuroendocrine stress reactivity in women: a multiple regression analysis. Depress Anxiety. 2002;15:117–125. doi: 10.1002/da.10015. [DOI] [PubMed] [Google Scholar]
- 7.Plotsky PM., Owens MJ., Nemeroff CB. Psychoneuroendocrinology of depression. Hypothalamic-pituitary-adrenal axis. Psychiatr Clin N Amer. 1998;21:293–307. doi: 10.1016/s0193-953x(05)70006-x. [DOI] [PubMed] [Google Scholar]
- 8.Montgomery SM., Bartley MJ., Wilkinson RG. Family conflict and slow growth. Arch Dis Child. 1997;77:326–330. doi: 10.1136/adc.77.4.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ammerman RT., Cassisi JE., Hersen M., van Hasselt VB. Consequences of physical abuse and neglect in children. Clin Psychol Rev. 1986;6:291–310. [Google Scholar]
- 10.Trickett PK., McBride-Chang C. The developmental impact of different forms of child abuse and neglect. Develop Rev. 1995;15:311–337. [Google Scholar]
- 11.Lissau I., Sorensen TIA. Parental neglect during childhood and increased risk of obesity in young adulthood. Lancet. 1994;343:324–327. doi: 10.1016/s0140-6736(94)91163-0. [DOI] [PubMed] [Google Scholar]
- 12.Holmes SJ., Robins LN. The influence of childhood disciplinary experience on the development of alcoholism and depression. J Child Psychol Psychiatry. 1987;28:399–415. doi: 10.1111/j.1469-7610.1987.tb01762.x. [DOI] [PubMed] [Google Scholar]
- 13.Holmes S., Robins LN. The role of parental disciplinary practices in the development of depression and alcoholism. Psychiatry. 1988;51:24–36. doi: 10.1080/00332747.1988.11024377. [DOI] [PubMed] [Google Scholar]
- 14.Gottman JM. Psychology and the study of marital processes. Ann Rev Psychol. 1998;49:169–197. doi: 10.1146/annurev.psych.49.1.169. [DOI] [PubMed] [Google Scholar]
- 15.Canetti L., Bachar E., Galili-Weisstub E., De-Nour AK., Shalev AY. Parental bonding and mental health in adolesence. Adolesence. 1997;32:381–394. [PubMed] [Google Scholar]
- 16.Parker G. Parental representations of patients with anxiety neurosis. Acta Psychiatr Scand. 1981;63:33–36. doi: 10.1111/j.1600-0447.1981.tb00647.x. [DOI] [PubMed] [Google Scholar]
- 17.Russak LG., Schwartz GE. Feelings of parental care predict health status in midlife: a 35-year follow-up of the Harvard Mastery of Stress Study. J BehavMed. 1997;20:1–11. doi: 10.1023/a:1025525428213. [DOI] [PubMed] [Google Scholar]
- 18.Seckl JR., Meaney MJ. Early life events and later development of ischaemic heart disease. Lancet. 1993;342:1236–xx. doi: 10.1016/0140-6736(93)92215-f. [DOI] [PubMed] [Google Scholar]
- 19.Newport DJ., Stowe ZN., Nemeroff CB. Parental depression: animal models of an adverse life event. Am J Psychiatry. 2002;159:1265–1283. doi: 10.1176/appi.ajp.159.8.1265. [DOI] [PubMed] [Google Scholar]
- 20.Sroufe LA. Psychopathology as an outcome of development. Develop Psychopathol. 1997;9:251–268. doi: 10.1017/s0954579497002046. [DOI] [PubMed] [Google Scholar]
- 21.Francis DD., Meaney MJ. Maternal care and the development of stress responses. CurrOpin Neurobiol. 1999;9:128–134. doi: 10.1016/s0959-4388(99)80016-6. [DOI] [PubMed] [Google Scholar]
- 22.Heim C., Newport DJ., Heit S., et al. Pituitary-adrenal and autonomic responses to stress in women after sexual and physical abuse in childhood. JAMA. 2000;284:592–597. doi: 10.1001/jama.284.5.592. [DOI] [PubMed] [Google Scholar]
- 23.Heim C., Newport DJ., Wagner D., Wilcox MM., Miller AH., Nemeroff CB. The role of early adverse experience and adulthood stress in the prediction of neuroendocrine stress reactivity in women: a multiple regression analysis. Depress Anxiety. 2002;15:117–125. doi: 10.1002/da.10015. [DOI] [PubMed] [Google Scholar]
- 24.DeBellis MD., Chrousos GP., Dom LD., et al. Hypothalamic pituitary adrenal dysregulation in sexually abused girls. J Clin Endocrinol Metab. 1994;78:249–255. doi: 10.1210/jcem.78.2.8106608. [DOI] [PubMed] [Google Scholar]
- 25.Luecken LJ., Lemery KS. Early caregiving and physiological stress responses. Clin Psychol Rev. 2004;24:171–191. doi: 10.1016/j.cpr.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Pruessner JL., Champagne FA., Meaney MJ., Dagher A. Parental care and neuroendocrine and dopamine responses to stress in humans: a PET imaging study. J Neurosci. 2004;24:2825–2831. doi: 10.1523/JNEUROSCI.3422-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennett AJ., Lesch KP., Heils A., et al. Early experience and serotonin transporter gene variation interact to influence primate CNS function. Mol Psychiatry. 2002;7:118–122. doi: 10.1038/sj.mp.4000949. [DOI] [PubMed] [Google Scholar]
- 28.Higley JD., Haser MF., Suomi SJ., Linnoila M. Nonhuman primate model of alcohol abuse: effects of early experience, personality and stress on alcohol consumption. Proc Natl Acad Sci U S A. 1991;88:7261–7265. doi: 10.1073/pnas.88.16.7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suomi SJ. Early determinants of behaviour: evidence from primate studies. Br Med Bull. 1997;53:170–184. doi: 10.1093/oxfordjournals.bmb.a011598. [DOI] [PubMed] [Google Scholar]
- 30.Meaney MJ. The development of individual differences in behavioral and endocrine responses to stress. Ann Rev Neurosci. 2001;24:1161–1192. doi: 10.1146/annurev.neuro.24.1.1161. [DOI] [PubMed] [Google Scholar]
- 31.Dallman MF., Akana SF., Strack AM., Hanson ES., Sebastian RJ. The neural network that regulates energy balance is responsive to glucocorticoids and insulin and also regulates HPA axis responsivity at a site proximal to CRF neurons. Ann N Y Acad Sci. 1995;771:730–742. doi: 10.1111/j.1749-6632.1995.tb44724.x. [DOI] [PubMed] [Google Scholar]
- 32.McEwen BS. Protective and amaging effects of stress mediators. N Engl J Med. 1998;338:171–179. doi: 10.1056/NEJM199801153380307. [DOI] [PubMed] [Google Scholar]
- 33.Chrousos GP., Gold PW. The concepts of stress and stress system disorders. JAMA. 1992;267:1244–1252. [PubMed] [Google Scholar]
- 34.Caspi A., Sugden K., Moffitt TE., et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–390. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 35.Levine S. Infantile experience and resistance to physiological stress. Science. 1957;126:405–406. doi: 10.1126/science.126.3270.405. [DOI] [PubMed] [Google Scholar]
- 36.Levine S. Plasma-free corticosteroid response to electric shock in rats stimulated in infancy. Science. 1962;135:795–796. doi: 10.1126/science.135.3506.795-a. [DOI] [PubMed] [Google Scholar]
- 37.Denenberg VH. Critical periods, stimulus input, and emotional reactivity: a theory of infantile stimulation. Psychol Rev. 1964;71:335–351. doi: 10.1037/h0042567. [DOI] [PubMed] [Google Scholar]
- 38.Lee MHS., Williams Dl. Long-term changes in nest condition and pup grouping following handling of rat litters. Dev Psychobiol. 1975;8:91–95. doi: 10.1002/dev.420080113. [DOI] [PubMed] [Google Scholar]
- 39.Liu D., Tannenbaum B., Caldji C., et al. Maternal care, hippocampal glucocorticoid receptor gene expression and hypothalamic-pituitary-adrenal responses to stress. Science. 1997;77:1659–1662. doi: 10.1126/science.277.5332.1659. [DOI] [PubMed] [Google Scholar]
- 40.Francis DD., Diorio J., Liu D., Meaney MJ. Nongenomic transmission across generations in maternal behavior and stress responses in the rat. Science. 1999;286:1155–1158. doi: 10.1126/science.286.5442.1155. [DOI] [PubMed] [Google Scholar]
- 41.Caldji C., Diorio J., Meaney MJ. Variations in maternal care alter GABAA receptor subunit expression in brain regions associated with fear. Neuropsychopharmacology. 2003;28:150–159. doi: 10.1038/sj.npp.1300237. [DOI] [PubMed] [Google Scholar]
- 42.Rivier C., Plotsky PM. Mediation by corticotropin-releasing factor of adenohypophysial hormone secretion. Ann Rev Physiol. 1986;48:475–489. doi: 10.1146/annurev.ph.48.030186.002355. [DOI] [PubMed] [Google Scholar]
- 43.Plotsky PM. Pathways to the secretion of adrenocorticotropin: a view from the portal. J Neuroendocrinol. 1991;3:1–9. doi: 10.1111/j.1365-2826.1991.tb00231.x. [DOI] [PubMed] [Google Scholar]
- 44.Antoni FA. Vasopressinergic control of pituitary adrenocorticotropin secretion comes of age. Front Neuroendocrinol. 1993;14:76–122. doi: 10.1006/frne.1993.1004. [DOI] [PubMed] [Google Scholar]
- 45.Whitnall MH. Regulation of the hypothalamic corticotropin-releasing hormone neurosecretory system. Prog Neurobiol. 1993;40:573–629. doi: 10.1016/0301-0082(93)90035-q. [DOI] [PubMed] [Google Scholar]
- 46.De Kloet ER., Vregdenhil E., Oitzl MS., Joels M. Brain corticosteroid receptor balance in health and disease. Endocr Rev. 1998;19:269–301. doi: 10.1210/edrv.19.3.0331. [DOI] [PubMed] [Google Scholar]
- 47.Sapolsky RM., Romero LM., Munck AU. How do glucocorticoids influence stress responses? integrating permissive, suppressive, stimulatory, and preparative actions. Endocr Rev. 2000;21:55–89. doi: 10.1210/edrv.21.1.0389. [DOI] [PubMed] [Google Scholar]
- 48.Lavicky J., Dunn AJ. Corticotropin-releasing factor stimulates catecholamine release in hypothalamus and prefrontal cortex in freely moving rats as assessed by microdialysis. J Neurochem. 1993;60:602–612. doi: 10.1111/j.1471-4159.1993.tb03191.x. [DOI] [PubMed] [Google Scholar]
- 49.Emoto H., Yokoo H., Yoshida M., Tanaka M. Corticotropin-releasing factor enhances noradrenaline release in the rat hypothalamus assessed by intracerebral microdialysis. Brain Res. 1993;601:286–288. doi: 10.1016/0006-8993(93)91722-5. [DOI] [PubMed] [Google Scholar]
- 50.Page ME., Valentino RJ. Locus coeruleus activation by physiological challenges. Brain Res. Bull. 1994;35:557–560. doi: 10.1016/0361-9230(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 51.Valentino RJ., Curtis AL., Page ME., Pavcovich LA., Florin-Lechner SM. Activation of the locus coeruleus brain noradrenergic system during stress: circuitry, consequences, and regulation. Adv Pharmacol. 1998;42:781–784. doi: 10.1016/s1054-3589(08)60863-7. [DOI] [PubMed] [Google Scholar]
- 52.Tjoumakaris SI., Rudoy C., Peoples J., Valentino RJ., Van Bockstaele EJ. Cellular interactions between axon terminals containing endogenous opioid peptides or corticotropin-releasing factor in the rat locus coeruleus and surrounding dorsal pontine tegmentum. J Comp Neurol. 2003;466:445–456. doi: 10.1002/cne.10893. [DOI] [PubMed] [Google Scholar]
- 53.Moga MM., Gray TS. Evidence for corticotropin-releasing factor, neurotensin, and somatostatin in the neural pathway from the central nucleus of the amygdala to the parabrachial nucleus. J Comp Neurol. 1989;241:275–284. doi: 10.1002/cne.902410304. [DOI] [PubMed] [Google Scholar]
- 54.Koegler-Muly SM., Owens MJ., Ervin GN., Kilts CD., Nemeroff CB. Potential corticotropin-releasing factor pathways in the rat brain as determined by bilateral electrolytic lesions of the central amygdaloid nucleus and paraventricular nucleus of the hypothalamus. J Neuroendocrinol. 1993;5:95–98. doi: 10.1111/j.1365-2826.1993.tb00367.x. [DOI] [PubMed] [Google Scholar]
- 55.Van Bockstaele EJ., Colago EE., Valentino RJ. Corticotropin-releasing factor-containing axon terminals synapse onto catecholamine dendrites and may presynaptically modulate other afférents in the rostral pole of the nucleus locus coeruleus in the rat brain. J Comp Neurol. 1996;364:523–534. doi: 10.1002/(SICI)1096-9861(19960115)364:3<523::AID-CNE10>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 56.Gray TS., Bingaman EW. The amygdala: corticotropin-releasing factor, steroids, and stress. Crit Rev Neurobiol. 1996;10:155–168. doi: 10.1615/critrevneurobiol.v10.i2.10. [DOI] [PubMed] [Google Scholar]
- 57.Butler PD., Weiss JM., Stout JC., Nemeroff CB. Corticotropin-releasing factor produces fear-enhancing and behavioural activating effects following infusion into the locus coeruleus. J Neurosci. 1990;10:176–183. doi: 10.1523/JNEUROSCI.10-01-00176.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liang KC., Melia KR. Miserendino MJ, Falls WA, Campeau S, Davis M. Corticotropin releasing factor: long-lasting facilitation of the acoustic startle reflex. J Neurosci. 1992;12:2303–2312. doi: 10.1523/JNEUROSCI.12-06-02303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swiergiel AH. Takahashi LK. Kahn NH. Attenuation of stress-induced by antagonism of corticotropin-releasing factor receptors in the central amygdala in the rat. Brain Res. 1993;623:229–234. doi: 10.1016/0006-8993(93)91432-r. [DOI] [PubMed] [Google Scholar]
- 60.Koob GF., Heinrichs SC., Menzaghi F., Pich EM., Britton KT. Corticotropinreleasing factor, stress and behavior. Sem Neurosci. 1994;6:221–229. [Google Scholar]
- 61.Schulkin J., McEwen BS., Gold PW. Allostasis, the amygdala and anticipatory angst. Neurosci Biobehav Rev. 1994;18:385–396. doi: 10.1016/0149-7634(94)90051-5. [DOI] [PubMed] [Google Scholar]
- 62.Stenzel-Poore MP., Heinrich SC., Rivest S., Knob GF., Vale WW. Overproduction of corticotropin-releasing factor in transgenic mice: a genetic model of anxiogenic behavior. J Neurosci. 1994;14:2579–2584. doi: 10.1523/JNEUROSCI.14-05-02579.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bakshi VP., Shelton SE., Kalin NH. Neurobiological correlates of defensive behaviors. Prog. Brain Res. 2000;122:105–115. doi: 10.1016/s0079-6123(08)62133-0. [DOI] [PubMed] [Google Scholar]
- 64.Davis M., Whalen PJ. The amygdala: vigilance and emotion. Mol Psychiatry. 2001;6:13–34. doi: 10.1038/sj.mp.4000812. [DOI] [PubMed] [Google Scholar]
- 65.Champagne FA., Francis DD., Mar A., Meaney MJ. Naturally occurring variations in maternal care in the rat as a mediating influence for the effects of environment on the development of individual differences in stress reactivity. Physiol Behav. 2003;79:359–371. doi: 10.1016/s0031-9384(03)00149-5. [DOI] [PubMed] [Google Scholar]
- 66.Stern JM. Offspring-induced nurturance: animal-human parallels. Dev Psychobiol. 1997;31:19–37. doi: 10.1002/(sici)1098-2302(199707)31:1<19::aid-dev3>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 67.Weaver ICG., Cervoni N., D'Alessio AC., et al. Epigenetic programming through maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 68.Menard J., Champagne D., Meaney MJ. Maternal care alters behavioral and neural activity patterns in the defensive burying paradigm. Neuroscience. 2004;129:297–308. doi: 10.1016/j.neuroscience.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 69.Caldji C., Tannenbaum B., Sharma S., Francis D., Plotsky PM., Meaney MJ. Maternal care during infancy regulates the development of neural systems mediating the expression of behavioral fearfulness in adulthood in the rat. Proc Nat Acad Sci USA. 1998;95:5335–5340. doi: 10.1073/pnas.95.9.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Owens MJ., Vargas MA., Knight DL., Nemeroff CB. The effects of alprazoplam on corticotropin-releasing factor neurons in the rat brain: acute time course, chronic treatment and abrupt withdrawal. J Pharmacol Exp Ther. 1991;258:349–356. [PubMed] [Google Scholar]
- 71.Caldji CC., Liu D., Meaney MJ. Maternal behavior in infancy regulates the development of GABAA receptor levels and its subunit composition and GAD. Soc Neurosci Abst. 2000;179:11. [Google Scholar]
- 72.Francis DD., Champagne F., Meaney MJ. Variations in maternal behaviour are associated with differences in oxytocin receptor levels in the rat. J Neuroendocrinol. 2000;12:1145–1148. doi: 10.1046/j.1365-2826.2000.00599.x. [DOI] [PubMed] [Google Scholar]
- 73.Mitchell JB., Iny LJ., Meaney MJ. The role of serotonin in the development and environmental regulation of hippocampal type II corticosteroid receptors. Dev Brain Res. 1990;55:231–235. doi: 10.1016/0165-3806(90)90204-c. [DOI] [PubMed] [Google Scholar]
- 74.Smythe JW., Rowe W., Meaney MJ. Neonatal handling alters serotonin turnover and serotonin type 2 receptor density in selected brain regions. Dev Brain Res. 1994;80:183–189. doi: 10.1016/0165-3806(94)90103-1. [DOI] [PubMed] [Google Scholar]
- 75.Mitchell JB., Rowe W., Boksa P., Meaney MJ. Serotonin regulates type II, corticosteroid receptor binding in cultured hippocampal cells. J Neurosci. 1990;10:1745–1752. doi: 10.1523/JNEUROSCI.10-06-01745.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mitchell JB., Betito K., Rowe W., Boksa P., Meaney MJ. Serotonergic regulation of type II corticosteroid receptor binding in cultured hippocampal cells: the role of serotonin-induced increases in cAMP levels. Neuroscience. 1992;48:631–639. doi: 10.1016/0306-4522(92)90407-s. [DOI] [PubMed] [Google Scholar]
- 77.Laplante P., Diorio J., Meaney MJ. Serotonin regulates hippocampal glucocorticoid receptor expression via a 5-HT7 receptor. Brain Res Dev Brain Res. 2002;139:199–203. doi: 10.1016/s0165-3806(02)00550-3. [DOI] [PubMed] [Google Scholar]
- 78.Francis D., Diorio J., Laplante P., Weaver S., Seckl RJ., Meaney MJ. The role of early environmental events in regulating neuroendocrine development: moms, pups, stress and glucocorticoid receptors. Ann N YAcadSci. 1996;794:136–152. doi: 10.1111/j.1749-6632.1996.tb32517.x. [DOI] [PubMed] [Google Scholar]
- 79.Meaney MJ., Diorio J., Francis D., et al. Postnatal handling increases the expression of cAMP-inducible transcription factors in the rat hippocampus: the effects of thyroid hormones and serotonin. J Neurosci. 2000;20:3926–3935. doi: 10.1523/JNEUROSCI.20-10-03926.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hellstrom I., Diorio J., Weaver ICG., Szyf M., Meaney MJ. Ex vivo characterization of maternal programming of stress responses through modulation of the epigenome. Soc Neurosci Abstr. 2004;662:11–xx. [Google Scholar]
- 81.Mohammed AH., Henricksson BG., Söderström S., Ebendal T., Olsson T., Seckl JR. Environmental influences on the central nervous system and their implications for the aging rat. Behav Brain Res. 1993;57:183–191. doi: 10.1016/0166-4328(93)90134-c. [DOI] [PubMed] [Google Scholar]
- 82.Andrews MH., Kostaki A., Setiawan E., et al. Developmental regulation of the 5-HT7 serotonin receptor and transcription factor NGFI-A in the fetal guinea-pig limbic system: influence of G Cs. J Physiol. 2004;555:659–670. doi: 10.1113/jphysiol.2003.056705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McCormick JA., Lyons V., Jacobson MD., et al. 5' Heterogeneity of glucocorticoid receptor messenger RNA is tissue specific: differential regulation of variant transcripts by early life events. Mol Endocrinol. 2000;14:506–517. doi: 10.1210/mend.14.4.0438. [DOI] [PubMed] [Google Scholar]
- 84.Chen FH., Watson CS., Gametchu B. Multiple glucocorticoid receptor transcripts in membrane glucocorticoid receptor-enriched S-49 mouse lymphoma cells. J Cell Biochem. 1999;74:418–429. [PubMed] [Google Scholar]
- 85.Strahle U., Schmidt A., Kelsey G., et al. At least three promoters direct expression of the mouse glucocorticoid receptor gene. Proc Nat Acad Sci U S A. 1992;89:6731–6735. doi: 10.1073/pnas.89.15.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Crosby SD., Puetz JJ., Simburger KS., Fahrner TJ., Milbrandt J. The early response gene NGFI-C encodes a zinc finger transcriptional activator and is a member of the GCGGGGGCG (GSG) element-binding protein family. Mol Cell Biol. 1991;11:3835–3841. doi: 10.1128/mcb.11.8.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Luger K., Mader AW., Richmond RK., Sargent DF., Richmond TJ. Crystal structure of nucelosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 88.Turner B. Chromatin and Gene Regulation. Edinburgh, UK: Blackwell Science; 2001 [Google Scholar]
- 89.Roth SY., Denu JM., Allis CD. Histone acetyltransferases. Ann Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 90.Bird AP. Methylation talk between histones and DNA. Science. 2001;294:2113–2115. doi: 10.1126/science.1066726. [DOI] [PubMed] [Google Scholar]
- 91.Bird A., Wolffe AP. Methylation-induced repression - belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 92.Strobl JS. A role for DNA methylation in vertebrate gene expression. Mol Endocrinol. 1990;4:181–183. doi: 10.1210/mend-4-2-181. [DOI] [PubMed] [Google Scholar]
- 93.Razin A., Riggs AD. DNA methylation and gene function. Science. 1980;210:604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- 94.Wolffe AP., Jones PL., Wade PA. DNA demethylation. Proc Natl Acad Sci USA. 1999;96:5894–5896. doi: 10.1073/pnas.96.11.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Razin A., Szyf M. DNA methylation patterns. Formation and function. Biochim BiophysActa. 1984;782:331–342. doi: 10.1016/0167-4781(84)90043-5. [DOI] [PubMed] [Google Scholar]
- 96.Shi H., Yan PS., Chen CM., et al. Expressed CpG island sequence tag microarray for dual screening of DNA hypermethylation and gene silencing in cancer cells. Cancer Res. 2002;62:3214–3220. [PubMed] [Google Scholar]
- 97.Beck S., Olek A., Walter J. From genomics to epigenomics: a loftier view of life [see comment]. Nat Biotechnol. 1999;17:1144–xx. doi: 10.1038/70651. [DOI] [PubMed] [Google Scholar]
- 98.Reik W., Dean W., Walter J. Epigenetic reprogramming in mammalian development. Science. 2000;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 99.Szyf M. Towards a pharmacology of DNA methylation. Trends Pharmacol Sci. 2001;22:350–354. doi: 10.1016/s0165-6147(00)01713-2. [DOI] [PubMed] [Google Scholar]
- 100.Meaney MJ., Szyf M. Environmentally driven chromatin plasticity? Trends Neurosci. In press. 2005 doi: 10.1016/j.tins.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 101.Ramchandani S. Bhattacharya SK. Cervoni N, Szyf M. DNA methylation is a reversible biological signal. Proc Nat Acad Sci U S A. 1999;96:6107–6112. doi: 10.1073/pnas.96.11.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Razin A., Cedar H. Distribution of 5-methylcytosine in chromatin. Proc Natl Acad Sci U S A. 1977;74:2725–2728. doi: 10.1073/pnas.74.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kadonaga JT. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 104.Hashimshony T., Zhang J., Keshet I., Bustin M., Cedar H. The role of DNA methylation in setting up chromatin structure during development. Nat Genet. 2003;34:187–192. doi: 10.1038/ng1158. [DOI] [PubMed] [Google Scholar]
- 105.Nan X., Ng HH., Johnson CA., et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 106.Jones PL., Veenstra GJ., Wade PA., et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 107.Cervoni N., Detich N., Seo SB., Chakravarti D., Szyf M. The oncoprotein Set/TAF-1beta, an inhibitor of histone acetyltransferase, inhibits active demethylation of DNA, integrating DNA methylation and transcriptional silencing. J Biol Chem. 2002;277:25026–25031. doi: 10.1074/jbc.M202256200. [DOI] [PubMed] [Google Scholar]
- 108.Cervoni N., Szyf M. Demethylase is directed by histone acetylation. J Biol Chem. 2001;276:40778–40787. doi: 10.1074/jbc.M103921200. [DOI] [PubMed] [Google Scholar]
- 109.Detich N., Hamm S., Just G., Knox JD., Szyf M. The methyl donor S-adenosylmethionine inhibits active demethylation of DNA: a candidate novel mechanism for the pharmacological effects of S-adenosylmethionine. J Biol Chem. 2003;278:20812–20820. doi: 10.1074/jbc.M211813200. [DOI] [PubMed] [Google Scholar]
- 110.Szyf M. DNA methylation and cancer therapy. Drug Resistance Updates. 2003;6:341–353. doi: 10.1016/j.drup.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 111.Fuks F., Hurd PJ., Wolf D., Nan X., Bird AP., Kouzarides T. The methyl-CpGbinding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- 112.Razin A. CpG methylation, chromatin structure and gene silencing - a three-way connection. EmboJ. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Prendergast GC., Ziff EB. Methylation-sensitive sequence-specific DNA binding by the c-myc basic region. Science. 1991;251:186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- 114.Kudo S. Methyl-CpG-binding protein MeCP2 represses Sp1 -activated transcription of the human leukosialin gene when the promoter is methylated. Mol Cell Biol. 1998;18:5492–5499. doi: 10.1128/mcb.18.9.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Frommer M., McDonald LE., Miller DS., et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Reik W., Dean W., Walter J. Epigenetic reprogramming in mammalian development. Science. 2000;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 117.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2003;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 118.Weaver ICG., Diorio J., Seckl JR., Szyf M., Meaney MJ. Environmental regulation of hippocampal glucocorticoid receptor gene expression: characterization of intracellular mediators and potential genomic targets. Ann N YAcadSci. 2004;1024:182–212. doi: 10.1196/annals.1321.099. [DOI] [PubMed] [Google Scholar]
- 119.Milbrandt J. A nerve growth factor-induced gene encodes a possible transcriptional regulatory factor. Science. 1987;238:797–799. doi: 10.1126/science.3672127. [DOI] [PubMed] [Google Scholar]
- 120.Hershko AY., Kafri T., Fainsod A., Razin A. Methylation of HoxA5 and HoxB5 and its relevance to expression during mouse development. Gene. 2003;302:65–72. doi: 10.1016/s0378111902010910. [DOI] [PubMed] [Google Scholar]
- 121.Meaney MJ., Aitken DH. The effects of early postnatal handling on hippocampal glucocorticoid receptor concentrations: temporal parameters. Brain Res. 1985;354:301–304. doi: 10.1016/0165-3806(85)90183-x. [DOI] [PubMed] [Google Scholar]
- 122.Bruniquel D., Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 123.Bhattacharya SK., Ramchandani S., Cervoni N., Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 124.Detich N., Theberge J., Szyf M. Promoter-specific activation and demethylation by MBD2/demethylase. J Biol Chem. 2002;277:35791–35794. doi: 10.1074/jbc.C200408200. [DOI] [PubMed] [Google Scholar]
- 125.Goel A., Mathupala SP., Pedersen PL. Glucose metabolism in cancer: evidence that demethylation events play a role in activating type ii hexokinase gene expression. J Biol Chem. 2003;278:15333–15340. doi: 10.1074/jbc.M300608200. [DOI] [PubMed] [Google Scholar]
- 126.Hattori M., Sakamoto H., Yamamoto T. DNA demethylase expression correlates with lung resistance protein expression in common epithelial ovarian cancers. J Int Med Res. 2001;29:204–213. doi: 10.1177/147323000102900308. [DOI] [PubMed] [Google Scholar]
- 127.Hattori M., Sakamoto H., Satoh K., Yamamoto T. DNA demethylase is expressed in ovarian cancers and the expression correlates with demethylation of CpG sites in the promoter region of c-erbB-2 and surviving genes. Cancer Lett. 2001;169:155–164. doi: 10.1016/s0304-3835(01)00499-2. [DOI] [PubMed] [Google Scholar]
- 128.Fang JY., Cheng ZH., Chen YX., et al. Expression of Dnmtl, demethylase, MeCP2 and methylation of tumor-related genes in human gastric cancer. World J Gastroenterol. 2004;10:3394–3398. doi: 10.3748/wjg.v10.i23.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Marhold J., Zbylut M., Lankenau DH., et al. Stage-specific chromosomal association of Drosophila dMBD2/3 during genome activation. Chromosoma. 2002;111:13–21. doi: 10.1007/s00412-002-0188-2. [DOI] [PubMed] [Google Scholar]
- 130.Cervoni N., Detich N., Seo SB., Chakravarti D., Szyf M. The oncoprotein Set/TAF-1beta, an inhibitor of histone acetyltransferase, inhibits active demethylation of DNA, integrating DNA methylation and transcriptional silencing. J Biol Chem. 2002;277:25026–25031. doi: 10.1074/jbc.M202256200. [DOI] [PubMed] [Google Scholar]
- 131.Detich N., Bovenzi V., Szyf M. Valproate induces replication-independent active DNA demethylation. J Biol Chem. 2003;278:27586–27592. doi: 10.1074/jbc.M303740200. [DOI] [PubMed] [Google Scholar]
- 132.Mousseau TA., Fox CW. The adaptive significance of maternal effects. Trends Evol Ecol. 1998;13:403–407. doi: 10.1016/s0169-5347(98)01472-4. [DOI] [PubMed] [Google Scholar]
- 133.Cameron N., Parent C., Champagne FA., Fish EW., Ozaki-Kuroda K., Meaney MJ. The programming of individual differences in defensive responses and reproductive strategies in the rat through variations in maternal care. Neurosci Biobehav Rev. In press. 2005 doi: 10.1016/j.neubiorev.2005.03.022. [DOI] [PubMed] [Google Scholar]