Abstract
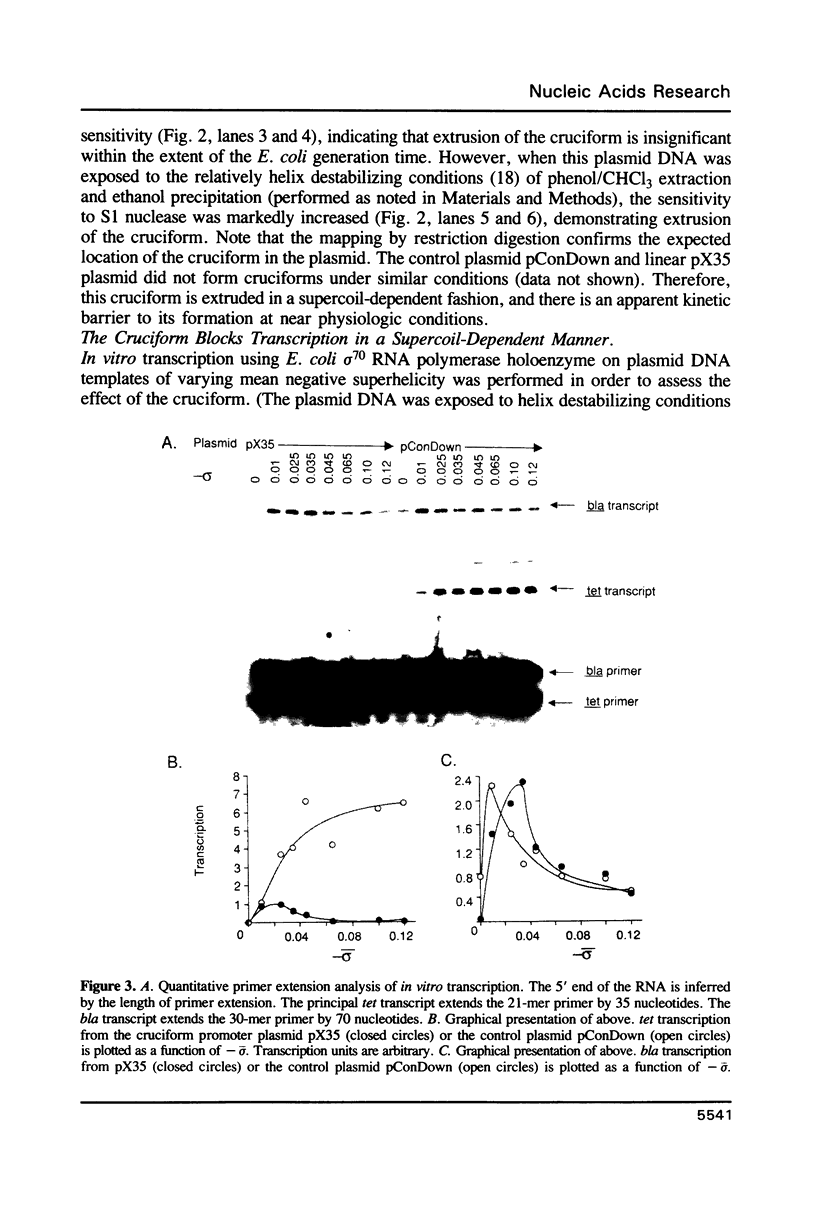
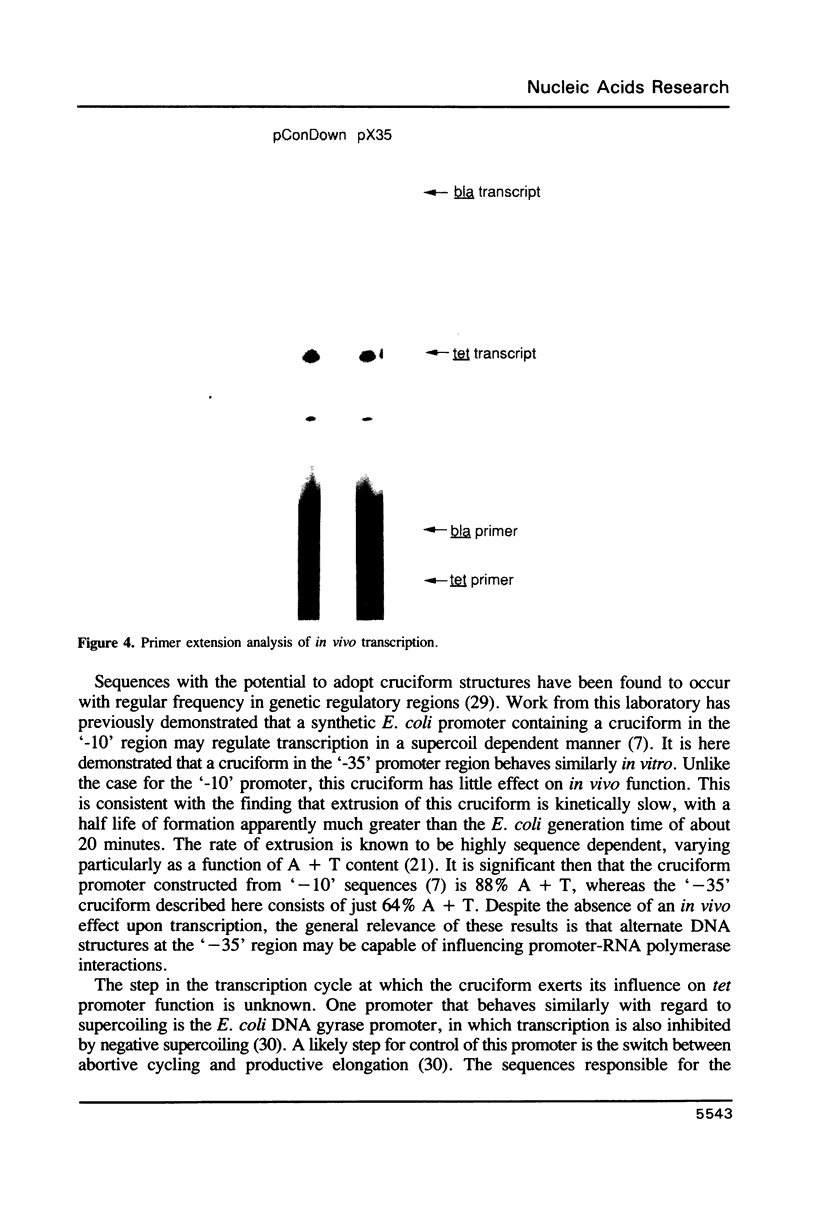
A promoter with the potential to adopt a 50 basepair (bp) cruciform spanning from -19 to -69 has been constructed in the plasmid pBR322 tetracycline resistance gene (tet) by forming an inverted repeat from '-35' sequences. Compared to a control promoter, the sequence of this cruciform promoter differs only by a 22 bp insertion between -48 and -69, upstream from the usual location of promoter sequences. The cruciform is extruded in a supercoil-dependent manner, and transcription from this promoter in vitro by RNA polymerase decreases as the negative supercoil density of the plasmid DNA increases. In contrast, transcription from the control promoter increases with negative supercoiling. Thus, DNA secondary structure in the '-35' region can affect promoter-polymerase interaction. The tet promoter cruciform also influences expression of the pBR322 beta-lactamase gene (bla). This apparently results when extrusion of the cruciform reduces the superhelicity of the plasmid molecule to a level that is below the optimum for expression from the bla promoter, illustrating one mechanism for how DNA secondary structure may effect action-at-a-distance. Transcription from both promoters in vivo does not differ from controls, suggesting that this cruciform is not generated to a significant extent intracellularly, most probably as a result of the slow kinetics of extrusion.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aleixandre V., Blanco M. Heterogeneity in the level of ampicillin resistance conferred by pBR322 derivatives with different DNA supercoiling. Mol Gen Genet. 1987 Aug;209(1):56–60. doi: 10.1007/BF00329836. [DOI] [PubMed] [Google Scholar]
- Bianchi M. E., Beltrame M., Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989 Feb 24;243(4894 Pt 1):1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- Blaho J. A., Larson J. E., McLean M. J., Wells R. D. Multiple DNA secondary structures in perfect inverted repeat inserts in plasmids. Right-handed B-DNA, cruciforms, and left-handed Z-DNA. J Biol Chem. 1988 Oct 5;263(28):14446–14455. [PubMed] [Google Scholar]
- Brahms J. G., Dargouge O., Brahms S., Ohara Y., Vagner V. Activation and inhibition of transcription by supercoiling. J Mol Biol. 1985 Feb 20;181(4):455–465. doi: 10.1016/0022-2836(85)90419-x. [DOI] [PubMed] [Google Scholar]
- Brewer B. J. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell. 1988 Jun 3;53(5):679–686. doi: 10.1016/0092-8674(88)90086-4. [DOI] [PubMed] [Google Scholar]
- Burgess R. R., Jendrisak J. J. A procedure for the rapid, large-scall purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975 Oct 21;14(21):4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- Courey A. J., Wang J. C. Cruciform formation in a negatively supercoiled DNA may be kinetically forbidden under physiological conditions. Cell. 1983 Jul;33(3):817–829. doi: 10.1016/0092-8674(83)90024-7. [DOI] [PubMed] [Google Scholar]
- Ellison M. J., Fenton M. J., Ho P. S., Rich A. Long-range interactions of multiple DNA structural transitions within a common topological domain. EMBO J. 1987 May;6(5):1513–1522. doi: 10.1002/j.1460-2075.1987.tb02394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa N., Bossi L. Transcription induces gyration of the DNA template in Escherichia coli. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9416–9420. doi: 10.1073/pnas.85.24.9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horbach E., Müller-Hill B. Insertion of d(pCpG)n.d(pCpG)n into the lacZ gene of Escherichia coli inhibits expression of beta-galactosidase in vivo. J Mol Biol. 1988 Jul 5;202(1):157–160. doi: 10.1016/0022-2836(88)90527-x. [DOI] [PubMed] [Google Scholar]
- Horwitz M. S., Loeb L. A. An E. coli promoter that regulates transcription by DNA superhelix-induced cruciform extrusion. Science. 1988 Aug 5;241(4866):703–705. doi: 10.1126/science.2456617. [DOI] [PubMed] [Google Scholar]
- Horwitz M. S., Loeb L. A. DNA sequences of random origin as probes of Escherichia coli promoter architecture. J Biol Chem. 1988 Oct 15;263(29):14724–14731. [PubMed] [Google Scholar]
- Horwitz M. S., Loeb L. A. Promoters selected from random DNA sequences. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7405–7409. doi: 10.1073/pnas.83.19.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. Bacterial chromatin. A new twist to an old story. Nature. 1986 Mar 6;320(6057):14–15. doi: 10.1038/320014a0. [DOI] [PubMed] [Google Scholar]
- Lowe P. A., Hager D. A., Burgess R. R. Purification and properties of the sigma subunit of Escherichia coli DNA-dependent RNA polymerase. Biochemistry. 1979 Apr 3;18(7):1344–1352. doi: 10.1021/bi00574a034. [DOI] [PubMed] [Google Scholar]
- Menzel R., Gellert M. Modulation of transcription by DNA supercoiling: a deletion analysis of the Escherichia coli gyrA and gyrB promoters. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4185–4189. doi: 10.1073/pnas.84.12.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U. R., Fitch W. M. Evolutionary selection for perfect hairpin structures in viral DNAs. Nature. 1982 Aug 5;298(5874):582–585. doi: 10.1038/298582a0. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Fontaine A. A native cruciform DNA structure probed in bacteria by recombinant T7 endonuclease. J Biol Chem. 1987 Aug 15;262(23):11364–11368. [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Transcriptional block caused by a negative supercoiling induced structural change in an alternating CG sequence. Cell. 1985 Jan;40(1):129–137. doi: 10.1016/0092-8674(85)90316-2. [DOI] [PubMed] [Google Scholar]
- Plon S. E., Wang J. C. Transcription of the human beta-globin gene is stimulated by an SV40 enhancer to which it is physically linked but topologically uncoupled. Cell. 1986 May 23;45(4):575–580. doi: 10.1016/0092-8674(86)90289-8. [DOI] [PubMed] [Google Scholar]
- Sinden R. R., Pettijohn D. E. Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc Natl Acad Sci U S A. 1981 Jan;78(1):224–228. doi: 10.1073/pnas.78.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C. K. Effects of salts, temperature, and stem length on supercoil-induced formation of cruciforms. J Biol Chem. 1983 Jun 25;258(12):7661–7668. [PubMed] [Google Scholar]
- Singleton C. K., Wells R. D. The facile generation of covalently closed, circular DNAs with defined negative superhelical densities. Anal Biochem. 1982 May 15;122(2):253–257. doi: 10.1016/0003-2697(82)90277-9. [DOI] [PubMed] [Google Scholar]
- Stefano J. E., Gralla J. D. Mutation-induced changes in RNA polymerase-lac ps promoter interactions. J Biol Chem. 1982 Dec 10;257(23):13924–13929. [PubMed] [Google Scholar]
- Tsao Y. P., Wu H. Y., Liu L. F. Transcription-driven supercoiling of DNA: direct biochemical evidence from in vitro studies. Cell. 1989 Jan 13;56(1):111–118. doi: 10.1016/0092-8674(89)90989-6. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Peck L. J., Becherer K. DNA supercoiling and its effects on DNA structure and function. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):85–91. doi: 10.1101/sqb.1983.047.01.011. [DOI] [PubMed] [Google Scholar]
- Wells R. D. Unusual DNA structures. J Biol Chem. 1988 Jan 25;263(3):1095–1098. [PubMed] [Google Scholar]
- Wu H. Y., Shyy S. H., Wang J. C., Liu L. F. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988 May 6;53(3):433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]