Abstract
The analysis of genetic variation in candidate genes is an issue of central importance in pharmacogenomics. The specific approaches taken will have a critical impact on the successful identification of disease genes, the molecular correlates of drug response, and the establishment of meaningful relationships between genetic variants and phenotypes of biomedical and pharmaceutical importance in general. Against a historical background, this article distinguishes different approaches to candidate gene analysis, reflecting different stages in human genome research. Only recently has it become feasible to analyze genetic variation systematically at the ultimate level of resolution, ie, the DNA sequence. In this context, the importance of haplotype-based approaches to candidate gene analysis has at last been recognized; the determination of the specific combinations of variants for each of the two sequences of a gene defined as a haplotype is essential. An up-to-date summary of such maximum resolution data on the amount, nature, and structure of genetic variation in candidate genes will be given. These data demonstrate abundant gene sequence and haplotype diversity. Numerous individually different forms of a gene may exist. This presents major challenges to the analysis of relationships between genetic variation, gene function, and phenotype. First solutions seem within reach. The implications of naturally occurring variation for pharmacogenomics and “personalized” medicine are now evident. Future approaches to the identification, evaluation, and prioritization of drug targets, the optimization of clinical trials, and the development of efficient therapies must be based on in-depth knowledge of candidate gene variation as an essential prerequisite.
Keywords: genetic variation, DNA sequence, candidate gene, single nucleotide polymorphism, haplotype, drug response
Abstract
El análisis de la variación genética en genes candidates es un tema de gran importancia en la farmacogenómica. Los abordajes específicos que se adopten tendrán un impacto crítico en la identificación exitosa de genes enfermos, en los correlatos moleculares de la respuesta a fármacos y en el establecimiento de relaciones significativas entre variantes genéticas y fenotipos de importancia biomédica y farmacéutica en general. Este artículo describe el contexto histórico y desarrolla diferentes opciones sobre el análisis de un gen candidato, lo que refleja las diferentes etapas en la investigación del genoma humano. Sólo recíentemente ha sido posible analizar sistemáticamente la variación genética con el máximo nivel de resolución, es decir, la secuencia del ADN. En este contexto, considerando que sólo recientemente se ha reconocido la importancia de aproximaciones basadas en haplotipos para el análisis de genes candidatos, resulta esencial la determinación de combinaciones específicas de variantes para cada una de las dos sequencias de un gen que determinan el haplotipo. Se entregará un resumen actualizado de la información más precisa acerca de la cantidad, naturaleza y estructura de la variación genética de los genes candidatos. Esta información es una prueba de la diversidad de secuencias de genes y de haplotipos existentes. Pueden existir numerosas formas individualmente diferentes de un gen. Este representa grandes desafíos para el análisis de las relaciones entre la variación genética, la función del gen y el fenotipo. Hay soluciones preliminares que parecen estar al alcance de la mano. Actualmente resultan evidentes las implicancias para la farmacogenómica y la medicina “personalizada” de la variación que ocurre en forma natural. Futuras aproximacíones para la identificación, evaluación y prioridad de fármacos blanco, para la optimización de ensayos clínicos y para el desarrollo de terapias eficientes deben estar basadas en un conocimiento en profundidad de la variación de genes candidatos como un prerequisito esencial.
Abstract
L'analyse de la variation génétique des gènes candidats est un problème d'une importance capitale en pharmacogénomique. Les approches spécifiques suivies auront un impact crucial pour le succès de l'identification des gènes responsables d'une maladie, les corrélats moléculaires de la réponse au médicament et l'établissement de relations significatives entre les variantes génétiques et les phénotypes ayant une importance biomédicale et pharmaceutique en général. Sur un arrière-fond historique, cet article distingue différentes approches de l'analyse du gène candidat, reflétant diverses étapes de la recherche sur le génome humain. Ce n'est que récemment qu'il est devenu possible d'analyser systématiquement la variation génétique au niveau ultime de la résolution, c'est-à-dire la séquence ADN. Dans ce contexte, l'importance des approches de l'analyse du gène candidat basées sur l'haplotype a enfin été reconnue; il est essentiel de déterminer les combinaisons spécifiques des variantes pour chacune des deux séquences géniques, que l'on définit comme un haplotype. Nous donnerons un résumé à jour de telles données de résolution maximale sur la quantité, la nature et la structure de la variation génétique des gènes candidats. Ces données démontrent une grande diversité dans les séquences géniques et les haplotypes. Il peut exister de nombreuses formes individuelles différentes d'un gène. Ceci représente un défi majeur pour l'analyse des relations entre variation génétique, fonction du gène et phénotype. Des solutions préliminaires semblent être à notre portée. Les implications des variations survenant spontanément sont maintenant évidentes pour la pharmacogénomique et la médecine « personnalisée ». De futures approches pour l'identification, l'évaluation et l'établissement de la liste des priorités des cibles médicamenteuses, l'optimisation des essais cliniques et le développement de traitements efficaces doivent être basées sur une connaissance approfondie des variations des gènes candidats comme condition préalable essentielle.
These past two decades, research on the molecular mechanisms mediating the effects of pharmacological substances has been marked by enormous progress. The first, important steps were the purification and isolation of receptor proteins, the existence of which had until then been hypothesized on the basis of their characteristic pharmacological effects. The next, major steps were the cloning of the genes encoding these proteins1 and the discovery of a much greater multiplicity at the DNA level underlying the pharmacologically defined effects; many more receptor subtypes were found to exist at the DNA level than had originally been proposed on the basis of pharmacological classification.2 The availability of the gene sequences provided the basis for protein structural models. For instance, the gene family of G protein-coupled receptors - a major model system that includes the majority of pharmacologically defined receptors2,3 and represents more than 50% of all drug targets4 - features the characteristic structural motif of seven transmembrane-spanning domains connected by intracellular and extracellular loops and terminal regions exposed at both faces of the membrane.3 The structural model allowed investigation of the molecular basis of receptor functions, such as ligand binding, signal transduction via G protein-coupling, and regulation (for example, of desensitization).5
First, analyses of sequcnce-structure-function relationships were performed. In order to correlate specific components of receptor function with specific amino acids, the effects of mutations introduced into the “wild type” sequence by in vitro site-directed mutagenesis were examined. It was demonstrated that DNA sequence differences caused differences in receptor function. Mutations were shown (i) to significantly affect, the ability of the receptor to bind ligands with a characteristic specificity and affinity; (ii) to activate characteristic and specific effectors; and (iii) to undergo functional regulation.6 At this stage, mutations were conceived primarily as the result, of experimental intervention and as an important tool for analyzing the functional content of DNA sequence information. The possibility that mutations might, occur as natural phenomena that confer a spectrum of natural functional variations was simply not part, of the picture or even an acknowledged hypothesis. For as long as one could think of, pharmacological effects were conceived as specific, uniform values, which were defined by a mean value (the average of all individual values) and a standard error (an indication of the extent of deviation of the individual values from the mean, ie, the usual scattering of these values). Such variability was supposed to reflect, deviation from the true value as a result of confounding parameters, which introduced the errors in the process of measurement. At its extreme, the mean value described an effect, that did not, apply to any of the individuals who participated in the experiment.
A gradual change in concept, to the conscious notion of individual variability at the pharmacological, clinical, and molecular level, and the acceptance of variation as the frame of reference and object of research did not emerge until the very early days of the Human Genome Project. On the basis of vision more than fact, it was hypothesized that differences in UNA sequence - the most basic level of molecular information - were related to individual genetic differences in drug response.6,7 The hypothesis of a biochemical individuality of man and its relationship to pharmacogenetic phenomena had already been raised in the early 1900s8 and first observations of individual differences in the response to the same drug had been made by Pythagoras as early as the fifth century BC.9 However, these observations were generally considered exceptions from the rule. Only in the past few years, has the picture substantiated that variations may frequently occur naturally and a broad spectrum of normal variation, reflecting naturally diverse individual gene and genome sequences, may exist, giving rise to subtle functional differences. This change in concepts marks the beginning of the end of Mendel's world,10 which was filled with rare mutations that caused discrete protein effects and gross, visible phenotypic effects.
Progress in human genome research transforms genetic variation into a central research theme
Major developments in the Human Genome Project have catalyzed a dramatic change in picture, transforming the analysis of genetic variation and its implications for disease causation and individually different, drug response into a major research theme. Pharmacogenomics, the vision of a “personalized” medicine and the development of prescriptions with a personal touch, has become the focus of attention and a widely discussed topic.11-14 Such progress was in particular spurred by the development, of cloning and high-throughput sequencing technologies,15 the availability of a draft sequence of the human genome,16,17 and consequently, access to all human genes and their regulators, transcripts, and proteins as the basis for disease gene and drug target, discovery. With defined reference sequences of genes and genomes, sequence comparisons within and between species became feasible and, consequently, the identification of differences in DNA sequence, so-called single nucleotide polymorphisms (SNPs).18 For the first, time, human genome variation data were generated on a large scale, resulting in the establishment, of SNP maps19 and public variation databases. Thus, it was for the first time possible to study the amount, nature, and structure of human genetic variation on a large scale.20-23 For this purpose, different, approaches were taken, ie, completely different, approaches to resolution, which led to completely different pictures of genetic variation.
In the first, series of studies, the structure of genetic variation (specifically the pattern and extent of linkage disequilibrium [LD] between SNPs) was assessed on a genome-wide scale. Common SNPs, with frequencies of the minor allele >5% to >30%, were randomly generated or extracted from databases at distances of 1.3 to 15 kb, and genotyped in limited numbers of individuals. As a result, SNPs were found to cooccur, ie, exist in blocks of strong LD, within genomic regions that extended up to about 60 to 100 kb in populations of European descent.20-23
These specific combinations of closely linked SNP alleles (haplotypes) were separated by regions of recombination, indicating a haplotype block structure of the human genome.20-23 Because the strong LD between SNPs appeared to result in a striking lack of genetic diversity, only a limited number of haplotypes, two to five per block, were observed, accounting for 75% to 98% of all chromosomes.
At the other end of the extreme, a number of studies were performed to systematically analyze genetic variation at the ultimate level of resolution, ie, the DNA sequence. Defined candidate genes, DNA segments of several kilobascs, were comparatively sequenced in larger numbers of individuals.24-34 These first, studies reflect, as closely as possible the molecular truth. They revealed abundant gene sequence diversity,31,35 about one SNP every 160 to 180 bp, and revised the classical measures of genetic variability.35-37 They also demonstrated unpredictable patterns of LD even within short distances of several hundred basepairs, much higher numbers of haplotypes, sometimes exceeding a hundred, and much more complex haplotype structures38 than suggested by the previous studies. To conclude, the higher the resolution, the higher the variability, and the more complex the picture.39 It is now important to develop a critical awareness for such differences in resolution. It is important, to know where one stands relative to the virtual optimum, maximum resolution, and to be able to put results into perspective. This is particularly important in order to make inferences on the validity of genotype-phenotype relationships as they have been established in the studies of interest.
Comprehensive knowledge on amount, nature, and structure of genetic variation: an essential prerequisite
This article first provides an overview of methods and approaches to the analysis of genetic variation as they have developed over time, reflecting a gradual transition from the indirect, random assessment of variations basically guided by chance, to the increasingly systematic and complete resolution of defined candidate gene regions. The emphasis on the historical dimension should facilitate the distinction of different, and currently coexisting, approaches. Second, the importance of a whole gene sequence-based, systematic analysis of genetic variation and its underlying haplotype structures will be outlined. Third, a state-of-the-art summary of present data describing genetic variation in candidate genes - its amount, nature, and structure at the highest, possible level of resolution to date will be given. These data reveal an abundant, sequence diversity as well as complex haplotype structures. This demonstrates at the experimental level that it is essential to resolve genetic variation and its underlying structures as systematically as possible, in order to design successful association studies and establish meaningful relationships with gene function and phenotype. The implications of given natural variability for pharmacogenomics and a personalized medicine will then be summarized in the following section. Finally, the tremendous challenges posed by both variability and the complex nature of pharmacogenetically relevant, traits will be addressed and first, solutions and future perspectives outlined. Because this article addresses basic issues regarding the nature and interpretation of genetic variability in candidate genes as the central unit, of analysis in pharmacogenomics, it complements the articles by Ackcnhcil and Weber40 and Morris-Roscndahl and Fiebich41 in this volume.
Approaches to the analysis of genetic variation and genotype-phenotype relationships
It is essential to keep the historical dimension in mind, which has shaped approaches to the analysis of genetic variation in disease and, importantly, the concepts about, how to establish links between genotype and phenotype. This will allow putting past and present, approaches and the results they generated into perspective.39 For most, of the time, a comprehensive analysis of the entire variation given in candidate genes has been neither feasible nor practicable, nor efficient.
Even though the sequences of numerous candidate genes of interest had become available in the late 1980s, the first, systematic candidate gene analyses were not performed until the late 1990s. The methods at hand were indirect, ie, the variations were detected without directly analyzing DNA sequence. The variations were selected randomly, ie, without emphasis on specific functionally relevant, gene regions. They were selected out of context, ie, given variation in the other parts of the gene were not issues of primary relevance. What was feasible and what mattered was to be able to detect any polymorphism(s) at all in and around the gene to be able to test the candidate gene hypothesis. The limited availability of technologies to access genetic variation restricted the number of detectable polymorphisms and determined the type of variants identified. What counted were the ease and robustness of typing and the numbers and frequencies (inf ormativeness) of the alleles in order to be able to perform informative association studies. For years, the variable sites utilized for such studies were largely represented by restriction fragment length polymorphisms (RFLPs), different, kinds of repeat, markers such as microsatcllites, short, tandem repeat (STR), or variable number of tandem repeats (VNTR) markers. The presence of variation within the restriction site of an enzyme or the presence of a repeat marker anywhere in the gene region were chance events that illustrate the randomness of these approaches.
Later, the analysis of SNPs, the most, frequent, type of variation in the human genome, gained center stage. These were, in the early to mid 1990s, mostly identified by application of polymerase chain reaction (PCR)based mutation scanning methods, such as singlestranded conformation polymorphism (SSCP) detection or denaturing gradient gel electrophoresis (DGGE), which were supposed to detect, 80% to 95% of all variants. In the optimal case, they were found to cause a functionally significant amino acid exchange, which would allow the direct testing of potentially causative alleles.18 In the late 1990s, when the Human Genome Project was in progress, SNPs were generated randomly at. large scale in vitro and in silico.19,42-44 They were identified by (i) sequencing sequence-tagged sites (STSs) from random genomic sequence and expressed sequence tags (ESTs) primarily representing untranslated regions of genes44; (ii) shotgun sequencing of genomic fragments42; and (iii) analysis of clone overlaps by the International Human Genome Sequencing Consortium.16,42 Any discrepancies in sequences were considered potential SNPs. Only about. 5% of the SNPs in the SNP map were discovered in studies that analyzed genes as compared to randomly generated genomic fragments.19,45 Comparison of SNPs that were detected by systematically scanning16 or resequencing a substantial number of candidate genes, eg, a total of 318 genes in the largest study performed to date,33 showed that public SNP databases contained only 2% to 25% of those genie SNPs.
Given this historical background, the approach taken in the majority of studies was to evaluate single polymorphisms or SNPs in and around the gene, one at a time, for association with the disease.20,39,47 Importantly, polymorphisms were conceived as genetic markers that would allow inference of an unobserved causative allele,18,48 which could not have been identified due to the restricted analysis range or insufficient, depth of analysis. In this approach, all polymorphisms, SNPs, or any other of the classes of polymorphisms mentioned above, were conceptually equivalent, irrespective of their specific functional significance.48
Thus, the major rationale underlying all genetic mapping by association approaches is that a marker allele exists in strong LD with the unobserved causative allele, which indicates the presence of the disease allele.48 This rationale essentially underlies all present, approaches to association analysis, given that information on genetic variation in genes and genomes remains widely incomplete and relies on subset, approaches. In order to enhance the heterozygosity - and hence informativeness of the markers defining a gene region - and have greater power to map unobserved causative variants by LD,48 several polymorphisms (of any class) were combined to construct haplotypes, which are defined in this context as the specific combinations of - desirably independent - alleles at two or more polymorphic sites on an individual chromosome.39 Again, the combination of markers that was used to construct haplotypes was primarily selected on the basis of availability, practicality, and heterozygosity, ie, the result of random screening procedures. An important preassumption implied in the use of single or several markers was that, these would represent, underlying LD structures, even at distances of several kilobascs, and hence be appropriate to capture the disease variant.
To conclude, previous approaches to the analysis of candidate genes have not been based on systematic assessment, of given candidate gene variation. Consequently, the variants chosen for analysis actually represented randomly selected variants and, obviously, only a subset, of the naturally existing variants. On the basis of such a traditional single SNP approach, numerous association studies have been performed, particularly in the field of psychiatric genetics. One major characteristic of these studies is that, they generated a vast, body of controversial results; a typical example is the story of the dopamine D2 receptor gene and alcoholism.49
In the late 1990s, it became feasible to systematically evaluate genetic variation at the ultimate level of resolution, ie, the DNA sequence, as demonstrated in a series of comparative sequencing studies. 24,25,27-33 These studies revealed presence of abundant sequence diversity and far more complex underlying LD structures than had previously been anticipated.24,25,27,29,31-33,38,50 This substantially changed the view of the amount, pattern, and structure of genetic variability in genes and genomes.35
Evidence of abundant, sequence diversity began to raise doubts about the validity of traditional single SNP approaches.30,38 Apart, from theoretical considerations,29 it was shown that multiple variants can exist, within genes and that, the combinations of variants on each of the two copies of a gene (haplotypes) should become the focus of analysis. First, systematic comparative sequencing studies demonstrated that the analysis of haplotypes defined by the grouping and interaction of several variants rather than any individual SNP were correlated with complex phenotypes, such as drug response and common disease.24,29,51 Finally, when evidence for a haplotype structure of the human genome was obtained, it was explicitly recognized that, single SNP-based candidate gene approaches may be statistically weak and have no clear end point; true associations may be missed because of the incomplete information provided by individual SNPs; negative results exclude particular SNPs as playing a role, but cannot exclude a gene.20 This was the beginning of the end of single SNP approaches; haplotype-based approaches to candidate gene analysis and disease gene discovery had at last, become the state of the art.39
The systematic analysis of candidate genes: a necessary precondition to establish links to gene function, disease, and drug response
The importance of haplotypes: context matters
Only the entire gene sequence, given in its individually variable forms, can be correlated with the function, regulation, and expression of the protein and, ultimately, phenotype. “Since it, is the entire gene and its encoded protein that act as the units of function potentially affecting a phenotype (and ultimately allow initial conclusions on disease mechanisms), we must, analyze the entire sequences of the individual genes including their regulatory and intronic regions. It is therefore essential in diploid organisms (such as humans) to determine the specific combinations of all given gene sequence variants for each of the two chromosomes defined here as haplotypes.”29 Thus, a systematic approach to candidate gene analysis necessarily implies the determination of the haplotype pairs underlying each individual genotype. In this context, it is important, to note that this definition of gene-based functional haplotypes should be distinguished from other haplotype categories39 (which, in part, have also been utilized above), which generally refer to combinations of SNPs or any markers that may be located throughout genes,48 extend over any chromosomal regions, or identify (in the most, recent, definition) sets of markers in LD within a block of chromosomal sequence (haplotype blocks).20,39 It is also important, to note that, current (mixed diploid) direct sequencing methods allow determination of genotype, but not phase, ie, the assignment of the SNPs to one of the two chromosomes.
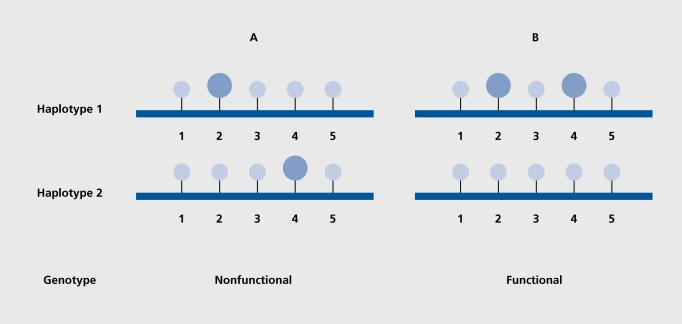
The correct determination of the molecular haplotypes underlying each genotype in a given sample is essential to make conclusions about the functionality of both forms of the gene, and establish relationships between gene variation and gene function in general.52-54 For instance, mutations that reside on the same chromosome (cis) may leave the function of the other gene copy intact. If, however, the two mutations reside on two different, chromosomes (trans), they may inactivate both gene copies (Figure 1) .53,55
Figure 1. Haplotype pairs of two individuals for a gene bearing multiple single nucleotide polymorphisms (SNPs). In this case, the phase of the coding SNPs determines the genotype. Even though the two individuals A and B are both heterozygous at the variable site positions 2 and 4, individual B expresses the gene correctly and individual A does not. Typical single SNP scoring methods would fail to distinguish between the two individuals, specifically between the two haplotype pairs. It is important to note that current (mixed diploid) direct sequencing methods allow determination of genotype, but not differentiation of phase, ie, the specific location of each SNP on one of the two chromosomes. Reproduced from reference 55: Hoehe MR, Timmermann B, Lehrach H. Haplotypes and the systematic analysis of genetic variation: disease genes, drug targets and pharmacogenomics [in German], Biospektrum. 2002;8:478-485. Copyright (c) 2002, Elsevier.
This example also demonstrates that the selection of single SNPs out, of context would not allow distinction between different underlying haplotype pairs and, ultimately, between high- and low-risk alleles. The importance of analyzing genetic variation in candidate genes systematically and comprehensively is further demonstrated by the fact, that SNPs located in one segment of the gene may, in three-dimensional (3D) space and with the DNA structural model in mind, interact with SNPs located in quite distant segments of the gene; distance in terms of linear sequence may be equivalent to proximity in space. In light of the sequence-structure-function relationship, all variants will have to be identified since any variant may have functional impact, whether it is considered essential or redundant with regard to indication of the underlying LD structure of the gene (its genetic marker function). In light, of a sequence-structure-function paradigm, the haplotype as defined above represents the immediate correlate for the individual, functional, or dysfunctional protein(s) it encodes, as well as related regulatory sequences. These gene-based functional haplotypes are of immediate relevance for pharmacogenomics: as potential disease gene correlates and/or drug targets; and as the basis for drug target characterization, evaluation, prioritization, and diagnostic test, development.
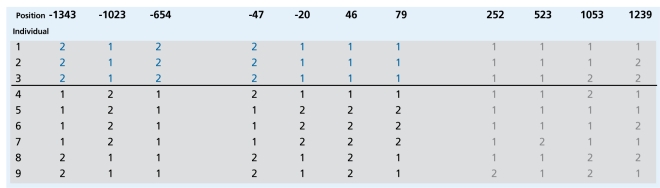
The importance of obtaining complete sequence information on each individual form of the gene is also demonstrated by the example of the human β2-adrenergic receptor gene (Figure 2).55,56 This gene, the product of which represents a key component, of the central and peripheral autonomous nervous systems, is an important candidate for a spectrum of diseases including neuropsychiatrie and cardiovascular disorders. It is also the target, for most, commonly prescribed drugs.6 Comparative sequence analysis of the gene including its regulatory and coding sequences in several hundred individuals resulted in the discovery of a total of 15 variants,55 four of which induced an amino acid mutation and were each shown to be functionally significant in vitro.57-59 In addition, a number of variants were identified in the 5' regulatory region. In a preliminary casecontrol study, individuals who carried a. specific combination of seven variants (haplotype) (blue in Figure 2) were significantly more frequently earning a predisposition to essential hypertension.55 This potential risk profile included three SNPs in the 5' regulatory region, and one SNP in the 5' untranslated region (5'UTR) at position -20, and three amino acid mutations in the leader peptide (-47) and amino terminal of the receptor protein (46 and 79).
Figure 2. Haplotypes of the human β2-adrenergic receptor gene and identification of genetic risk profiles. This figure represents, from left to right, the specific alleles at each of 11 variable positions (relative to the translation initiation site) in this gene for a subgroup of nine individuals from the Bergen Blood Pressure Study.55,56 1: Identical with reference sequence; 2: Different from reference sequence. The individual haplotypes are given by these specific combinations of 11 alleles as they occur throughout the gene. The three individuals at the top, who are genetically predisposed to essential hypertension, show potential risk haplotypes, which have in common a specific combination (“pattern”) of alleles at the first seven positions (marked in blue). This also demonstrates that the most frequently analyzed variant at position 46 (Arg>Gly) and the combination of the three in vitro functionally significant receptor mutations at positions -47, 46, and 79 are both insufficient to distinguish individuals at risk. Modified from reference 55: Hoehe MR, Timmermann B, Lehrach H. Haplotypes and the systematic analysis of genetic variation: disease genes, drug targets and pharmacogenomics [in German], Biospektrum. 2002:8:478-485. Copyright (c) 2002, Elsevier.
The variant at position 46 relative to the translation initiation site induces a functionally significant Arg>Gly exchange60 and was the most, frequently used variant in association studies; the combination of the three mutations at positions -47, 46, and 79 were used in some association studies.61 Neither of these were found to distinguish between high- and no-risk alleles. The last, four variants (marked in gray) had no impact statistically, which beautifully reflects biology: these variants affected the third base and were silent mutations.55 Moreover, this example illustrates that complete sequence analysis is necessary to focus subsequent functional experiments on all variants of potential functional significance. Of those seven variants in LD, one, several, or all variants in interaction may contribute to functional differences. Finally, this example demonstrates the complexity of functional annotation, given that, regulatory and coding variants occur in combinations.
Gene-based functional haplotypes versus gene-based complex genetic markers
The definition of a gene-based functional haplotype that requires complete DNA sequence information in all individuals is, admittedly, somewhat, futuristic at this stage of human genome research. In many cases, reality may allow different stages of approximation only. In this regard, the recently performed comparative gene sequencing studies24-26-28,31,33 mark major progress, because their approach to haplotype-based candidate gene analysis is significantly more systematic and comprehensive than the single SNP or combination-of-marker approaches used earlier. They also reflect different, stages of approximation to completeness at an early stage. Sequencing at this stage is still too laborand cost-intensive. It is generally not feasible to sequence all functionally relevant regions of the entire gene (even if they were all known) in every member of a defined population in order to identify all given variants and their frequencies.
Just to give some impression of the scale of the undertaking: in order to systematically analyze genetic variation in a typical G protein-coupled receptor gene including regulatory, exonic and intronic sequences (exon-intron boundaries) in 250 cases and controls, about 1.7 finished megabases (ie, about, twice the amount of raw sequence data to obtain maximum accuracy) need to be generated,29 comparable to the size of a bacterial genome. For completeness of genomic organization, we should refer to examples that have demonstrated, for instance, a disease-related regulatory variant, about 14 kb 5_ upstream of the translation initiation codon or regulatory elements in intronic sequences of extensive lengths.39
Thus, functionally important regions of the gene can at this stage be included in as representative a way as possible. Present approaches may still miss what we term the causative variant(s).
Thus, in practice, we are still dealing (at a comparatively advanced level) with marker or subset approaches, where identified variants represent, only a selection of all naturally existing variants. Against, this background, the genebased haplotypes will be categorized as complex genetic markers.39 A critical question is then whether the subsets of variation extracted do in fact validly represent, given LD and haplotype structures of a gene. In this case, the resulting gene-based haplotypes have been shown to be superior as markers in comparison to any single SNP, because they contain more information (heterozygosity) than any of the individual markers, or SNPs that comprise them.33,48 Multiple correlations with neighboring, or embedded, unobserved variants may be possible. Thus, a multisite gene-based haplotype (higher-order marker) will have greater power than any individual SNP to detect, an unobserved - but evolutionarily linked - variable causative site.48 Such haplotype signatures may, moreover, have significantly greater power to predict disease risk and drug response than any individual SNP within a gene.24,29,48,51,62 In the overall process of disease gene identification, it merits serious consideration to restrict, investigations in the first, pass to haplotype marker screening, the apparently less investment-intensive marker approach.
It should nevertheless be emphasized that if an association is found, the ultimate challenge to generate complete sequence information will remain. Subsequent in-depth comparative sequence analysis of entire gene sequences in all patients and controls will be indispensable to search for the presence - or exclude it - of any yet unidentified variant(s) in LD and extract the subset, of all variants in LD. These have genetically equivalent, properties63 and are supposed to contain a subset, of variants that will be biologically significant. As illustrated by the example of the human β2- adrenergic receptor gene, these comprehensive analyses may often not directly result in the identification of the causative variant(s), but may help locate the region of interest.32 The function of specific nucleotide sites will have to be assigned in subsequent functional studies in vitro and in vivo; one, several, or all of the variants in LD may be functionally significant, and interact. Any genetic analyses can at best, result, in testable biological hypotheses on the molecular causes of gene dysfunction. The true challenges remain biological after all.
Finally, there is yet another motivation for the systematic analysis of complete candidate gene sequences: we may have to allow - free of preassumptions - for any scenario of genetic variation predisposing to disease and individually different, drug response. The spectrum of polymorphic profiles may include any variant, or combinations of variants (patterns), that may interact to determine those functional variations that, are related to phenotypic variation. Common variants may play a role, rare mutations may add up, and variants may occur in similar or different, haplotype frames.29 In the light of a functional haplotype approach, which ultimately establishes the link between haplotypes, protein structure, function, and dysfunction, each haplotype matters. Rare ones will have to be included, because they may well add up to a significant fraction of the same (similar) protein isoform,25 generally confer functional similarity, or share some common pattern.29
State of the art: genetic variation in candidate genes
Overview of comparative sequencing and variation scanning studies
Complete sequence data from a number of nuclear loci first, became available in 1997,27 providing gradually more comprehensive information on given DNA sequence variation within defined segments of DNA. This allows a preliminary synthesis of the amount, nature, and organization of DNA sequence variation as given at the ultimate level of resolution.24 This also allows an insight into gene-based haplotype structures and their complexity at the DNA sequence level. Altogether, about 20 comparative sequencing studies have been performed,24-34,39 which have (i) explicitly addressed genetic variation in defined candidate genes; (ii) analyzed most, or substantial parts, of the entire gene; and (iii) determined in addition the structure of genetic variation, ie, the gene-based haplotypes, by application of molecular genetic and/or in silico methods, and/or inclusion of family information. These studies have described interindividual DNA sequence variation in a total of 331 genes; of those, 13 studies have focused on one candidate gene each, such as the β-globin gene,27 lipoprotein lipase (LPL),31 melanocortin 1 receptor (MC1R),64 pyruvate dehydrogenase A1 (PDHA1)28, angiotensin-convcrting enzyme (ACE),32 β2-adrenergic receptor (ADRB2),24 µ-opioid receptor (OPRM1),29 apolipoprotein E (APOE),25,30 caspase recruitment domain-containing protein 15 (NOD2 or CARD15, respectively),65-67 monoamine oxidase A (MAO-A) 26 genes, and the hemochromatosis locus (HFE) 34 Two studies have analyzed three genes each.68 Specifically, CAPN10, GPR35, and RNPEPL169 were resequenced as an integral part, of a general disease gene cloning procedure. In the currently most comprehensive study, a total of 313 genes including a number of G protein-coupled receptor genes, were systematically resequenced.33 In some of these studies 5' regulatory, 3', exonic, and intronic regions were examined24,25,27,29,30,32,33; while others addressed exonic and intronic regions26,28,31,66,67 and coding regions.64,65,68,69
These comparative sequencing studies usually included several different populations with total sample sizes between 10 and 494 individuals and populations of between 4 and 494 individuals. In a recent, report, analyses of genes in more than 500 individuals were described.70 Contiguous UNA segments in the range of 1.1 kb68 up to 9.7,31 24,32 and about 66 kb69 were resequenced; in a number of the described studies, the genomic regions covered were larger than the indicated segments sequenced, due to the specific genomic organization of the genes. On average, about 6.4 kb per gene (range about 1 kb)68 to about 24 kb32 were resequenced. For a more detailed description of these studies, including specific data, see reference 39.
Few studies addressed analyses of haplotype/genotypephenotype relationships against a background of high genome sequence diversity in order to test, for presence of genetic risk patterns that might predispose to drug response and complex disease.24,29,51 The others focused on evolutionary and population history issues related to the candidate genes in question .25-28-30,31,33,34 Some addressed in particular issues of DNA sequence diversity, complex LD and haplotype structures, and their potential implications for disease association studies,24-26,29,31-33,38 highlighting the tremendous challenges posed by abundant sequence diversity for disease association studies. In addition, substantial gene surveys were performed by application of variant detection arrays (VDAs). These characterized the frequency, nature, and pattern of SNPs in 75 candidate human genes for blood pressure homeostasis and hypertension,36 and 106 candidate genes relevant to cardiovascular disease, endocrinology, and neuropsychiatry.37 In a third, more recent, candidate gene survey, nine genes were scanned by application of denaturing high performance liquid chromatography (DHPLC).44
Abundant DNA sequence diversity in candidate genes
Regarding the amount of genetic variation at the DNA sequence level, these studies suggest a potentially remarkable variability in genes, ranging from about one SNP every 52 bps to about one SNP every 500 bp, at a mean spacing of one SNP every 215 bp, averaging over all gene regions included in analysis, which has sometimes been in disproportionate fractions.39 Summarizing more specifically studies that have analyzed regulatory, exonic, and intronic regions in a comparable way, an average spacing of about one SNP every 166 bp is observed; including the few studies carried out on coding regions, an average spacing of about, one SNP every 183 bp is obtained. This is in excellent agreement, with the variation data reported in the most comprehensive gene sequence survey on 313 genes; on average, about, one SNP every 185 bp was detected.33 Describing candidate gene variability in absolute numbers, a. number of variants in the range of 6 to 88 per gene was observed, an average value of about 35 variants given .25-29-31,32,34,39,64,65,68 If completely different sets of genes were considered, average values of about 12 to 15 SNPs per gene (range 0-59) were obtained.33,36,37,70
Overall, this clearly reflects a higher variability than that reported in the first gene-scanning studies, which surveyed 75 to 106 candidate genes by application of variation detection arrays; about, one SNP every 217 or 346 bp was described.36,37 These estimates of human variation among individuals also reflect a notable difference to the previously most frequently cited values of variation (between an individual's maternal and paternal genomes), ie, one sequence difference approximately every kilobase,35 and the range being one difference every 500 to 2000 bases.36,37,71 Overall, 3' UTR, exon-intron boundaries, 5' regulatory, and 5 'UTR regions appear to be more variable than coding regions, ranging from one SNP every 142 bp (3' UTR) to about one SNP every 294 bp (coding regions).33
Describing candidate gene variability by allele frequency spectra (ie, frequencies of the minor allele), about onethird of the SNPs (30%-38%) were observed only once.33,70,72 For less than one-third of the SNPs (28%32%), the frequency of the minor allele ranged between 1 % and 5%; for about 14% to 17% of the SNPs, the frequency of the minor allele ranged between 5% and 20%; and for the remaining 11% to 13%, the frequency of the minor allele ranged between 20% and 50%. 33,70 Sample sizes of analyzed studies ranged from 82 including four populations33 to an average of about 290 from one population of European descent.70 Of all SNPs, about 21 % were cosmopolitan, implying that both alleles were present, in all populations.33
Regarding the nature of genetic variability, 26% to 44% of all SNPs were found in the coding regions.33,36,37,70 Of all the coding SNPs (cSNPs) identified, 47% to 56% led to replacement, of an amino acid residue and probably impact protein function ,33,36,37,70 reflecting a high level of human protein diversity. The average gene contains about three to six cSNPs (range 0-17) and about two to three amino acid exchanges (range 0-15).33,37,70 With respect, to the functional consequences of coding region polymorphisms, the most, comprehensive survey evaluating 313 genes in 82 individuals of diverse ancestry and describing a total of 3899 SNPs,33 provided a classification of the types of changes based on Grantham values,73 which are derived from physicochemical considerations. According to these estimates, about 19% of cSNPs introduced conservative, 24% of cSNPs moderately conservative, 8% moderately radical, and about 4% radical changes; 1.5% of cSNPs introduced a premature termination codon; and about. 1% of all SNPs identified were within splice sites.33 Another large-scale gene survey showed, importantly, that, of the 75 proteins encoded by the genes that, were screened,36 83% were polymorphic at the protein level with an average heterozygosity of 17%. These values were considerably greater than classical protein studies addressing enzyme polymorphisms in humans,74 emphasizing the large degree of variation missed in those earlier studies. These protein-altering SNPs nevertheless represent, only 38% of the total number of such SNPs expected under the neutral infinite site models, demonstrating the strong role of natural (purifying) selection (eliminating 62% of replacement SNPs)75 and functional conservation on human genes.33,36,37
Variability and its variability: an intrinsic, gene-specific characteristic
An important measure to evaluate comparative surveys of sequence diversity is the nucleotide diversity of human genes, which is defined by the heterozygosity per nucleotide site.76,77 The measures used correct, for both sample size and length of region surveyed. In-depth analyses showed significant heterogeneity in nucleotide diversity and functional sequence class.33,36,37 Thus, in coding sequences, silent. SNPs showed 2.5-fold more diversity than replacement SNPs, reflecting functional constraint, and selection against changes in the protein sequence. Accordingly, heterogeneity among noncoding regions was observed: introns are about 50% more variable than 5'UTR or 3 'UTR. The greater diversity in 3 'UTR than 5'UTR and the relative patterns of noncoding sequence diversity can also be correlated with significant functional conservation of regulatory sequence. A cogent argument is that coding sequence changes are not, the only candidates for functional variation and that SNPs in proximal regulatory regions can have large phenotypic impact, too, just, as they do in evolution.36
Taken together, nucleotide diversity shows significant, variation across genes and functional class. Analyses assuming a neutral allele infinite site model showed that sequence length explained only 29% of the variation for cSNPs. Thus, gene-to-gene differences are the most, important of all factors that, contribute to such variation. An intrinsic and characteristic gene-specific diversity must, exist, as illustrated by a 15-fold variation in nucleotide diversity across genes, with coding segments being less diverse than noncoding sequences.36 Although mutation is responsible for creating SNPs, their maintenance probably depends on natural selection on coding sequences, which in turn is regulated by its precise functional role as well as meiotic recombination. This marked variability of variability in candidate genes is also illustrated by the fact that extremely invariable gene regions can also occur, with no structural mutations at all, singletons, or complete absence of any variant in coding or regulatory regions, even when genes were systematically resequenced in substantial numbers up to about 200 individuals.62,78,79 In particular, an extensive survey by Halushka et al36 showed that about, 17% of all genes were invariable at the protein level, which is in agreement with our extrapolations of a fraction of about 20% of invariable genes (Hoehe M et al, unpublished results). This may be related to certain aspects of yet, unknown or particularly high functional significance among the total gene pool, and is one of the important questions to be addressed in the future. Taken together, there is no a priori way to predict the actual natural variability of a gene; it must be empirically assessed in appropriately chosen samples in each case.
An example of the variability of variability in candidate genes, which may exist even within members of the same gene family (such as G protein-coupled receptor genes) or even within members of the same group of receptor subtypes or genes encoding endogenous receptor ligands, is given in Figure 3 80 These genes may represent prototype examples of drug targets and their potential variability, particularly with respect, to the fact that more than 50% of the total of 417 receptor targets of pharmaceutical relevance encode G protein-coupled receptors.4
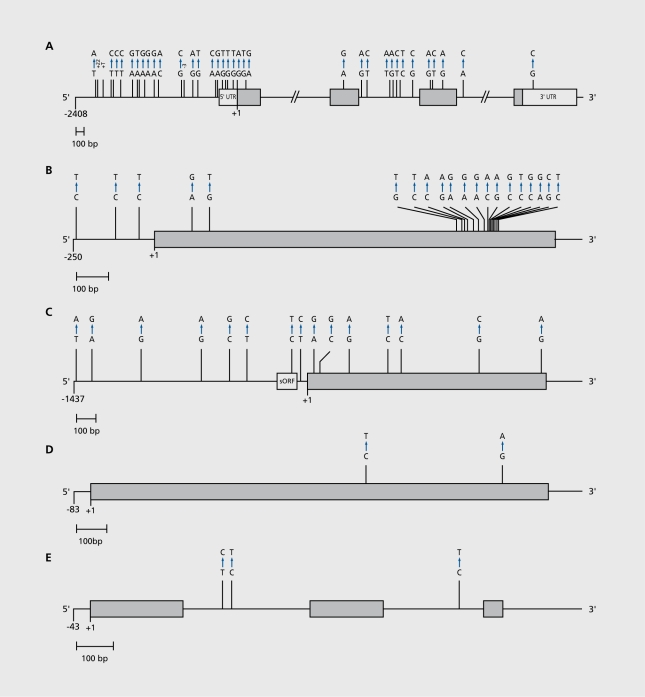
Figure 3. Polymorphic spectra of candidate genes.80 The genomic reference sequences are presented as baseline, exonic sequences as gray or white (untranslated regions [UTRs]) bars; sequences were drawn to scale, which is indicated. All gene variants are specified by nucleotide variations (substitutions, insertions, and deletions) according to the mutation nomenclature. Candidate genes had been subjected to systematic comparative sequencing. A. The human .-opioid receptor gene (OPRMT), about 6.7 kb, in 250 individuals.29 B. The human β1;-adrenergic receptor gene (ADRB1), about 2 kb, in about 80 individuals.81 C. The human β2-adrenergic receptor gene (ADRB2), about 3 kb, in 515 individuals.70 D. The human CB1 cannabinoid receptor gene (CNR1), about 1.6 kb, in about 200 individuals.62 E. The human promelanin concentrating hormone gene (PMCH), about 1.2 kb, in 141 individuals. Reproduced from reference 80: Hoehe MR, Timmermann B, Lehrach H. Human inter-individual DNA sequence variation in candidate genes, drug targets, the importance of haplotypes and implications for pharmacogenomics. Curr Pharm Biotechnol. 2003;4:351-378. Copyright (c) 2003, Bentham Science Publishers.
For instance, in the human µ-opioid receptor gene (OPRM1) (Figure 3a), a target for morphine, the classical pain killer in contemporary medicine and substance of abuse, a total of 43 variants have been identified within 6.7 kb in 250 European- and African-Americans.29 Clearly, the 5' regulatory and 5'UTR regions (one SNP every 99 bp and 73 bp, respectively) and the critical regions in intron 2 (one SNP every 110 bp) were much more variable than the coding exon (one SNP every 267 bp) and other intronic regions. Five of the six SNPs in the coding region clearly affected the encoded protein; two of those (which were relatively frequent) were located in the N-terminal, one in the third transmembrane domain, and two in the third cytoplasmic loop; all were shown to induce functional alterations.82,83
A different, picture can be observed in the human β2 adrenergic receptor gene (AD RBI), about 250 bp 5' flanking and 1434 bp coding regions of which were scanned in about 80 individuals of European descent.81 A total of 20 variants were observed, 17 of which were located in the coding region. Two variants in the N-terminal and five in the C-terminal caused an amino acid exchange (Figure 3b), which amounted to a much higher calculated density of SNPs in the coding region, about, one SNP every 84 bp. The human β2-adrenergic receptor gene (ADRB2), about 3 kb, has been resequenced in a total of several hundred individuals70; 15 variants, 8 in the 5' regulatory region including the leader peptide and 7 in the coding region, have been identified, at a roughly comparable spacing of one SNP every 175 to 200 bp.55,70 The mutation in the leader peptide and three coding SNPs, two of which were located in the N-terminal, were found to be functionally significant57-59; by far the majority of variants were highly frequent.
The human CB1 cannabinoid receptor gene (CNR1), another member of the G protein-coupled receptor gene family, was found to be remarkably invariable within and between species,62 when analyzing a total of about 200 individuals including European- and African-Americans as well as Europeans exhibiting extreme responses to cannabis use; only two silent substitutions were observed within about 1500 bp coding region. Similarly, notable invariability was observed in the coding regions of two chemokine receptor gene subtypes (Ohl et al, unpublished data). Finally, completely invariable coding exons and few SNPs in intronic regions were found in the human promelanin concentrating hormone gene (PMCH), a neuropeptide and endogenous ligand (Hoehe et al, unpublished data).
Taken together, current, approaches to describe, evaluate, and compare genetic variation in candidate genes remain in many aspects grossly insufficient and merely descriptive. They rely predominantly on the determination of frequency patterns and average values that describe and distinguish variability per se, as well as different categories of variants or functional gene sequence classes. These approaches allow, however, specific predictions of the nature and distribution of SNPs in the estimated 30 000 human genes, ie, in a study about 300-fold larger. Consequently, they may also allow extrapolations on the nature and amount of variability in potential drug targets. On the other hand, without knowledge of the specific functional variation in the genes underlying given nucleotide diversity, which will have to be based upon characterization of entire, individually different, forms of the gene and its product, the implications of the variability of candidate genes may hardly be evaluated and compared. The previous approaches to the characterization of genetic variation are in essence single SNP oriented. They do not therefore allow any conclusions on variation in its context, as represented by given haplotype structures of the gene, the very basis for functional evaluation.
A multiplicity of gene-based haplotypes
Similar observations were made for haplotypes. Potentially large numbers of haplotypes per gene as well as a notable variability of these numbers were observed. Absolute numbers of haplotypes described to date range from 2 to 8824-29,31-34,39,58,59 up to 14070; average numbers are about 14 haplotypes per gene (range 2-53) in 82 individuals from four populations in the most, comprehensive survey,33 8 haplotypes per gene (range 4-15) in about 40 to 60 individuals from one population of European descent,46 70 haplotypes per gene (range 16-140) in an average of about 309 individuals (range 141-469) from one population of European descent70; the average numbers of SNPs per gene in these studies were 12,46 12.5,33 and 31.70 Thus, the number of haplotypes appears to increase dramatically with the number of individuals and SNPs analyzed,38 and the upper end is not yet in sight; sequenced or scanned44 segments ranged in average from 2.3 kb33 to 4.9 kb70 to 15 kb per gene.46
At the other end of the spectrum is the resolution into haplotypes as described in the analysis of the LPL gene, an important, potential genetic risk factor for cardiovascular disease; about 9.7 kb of contiguous gene sequence including six of nine exons and significant, fractions of intronic sequence were systematically compared in 71 individuals from three populations.31 Eighty-eight distinct haplotypes were determined from 88 segregating sites; only three of the 88 haplotypes were detectable in all three populations and 21, 25, and 35 haplotypes were found unique to one of the three populations, respectively.38 Taken together, in analogy to genetic variation at the DNA sequence level, a notable variability of variability can be observed.
The relationship between the number of variants and number of underlying haplotypes seems to vary significantly,33,39,70 with the number of haplotypes being similar, much larger (up to eightfold)25,27,29,33,65,70 or significantly smaller32-34,64 than the number of SNPs. On average though, a linear relationship between the number of individual SNPs within a gene and the number of resulting haplotypes was observed in the most, comprehensive survey.33 In addition, a slightly higher average number of haplotypes per gene (by a factor of 1.1) than number of SNPs was observed.33 The fact that the number of haplotypes is greater than the number of SNPs indicates that, some degree of recombination and recurrent mutation may have occurred within these genes,33 which has also been emphasized in other studies.25-27,34,38
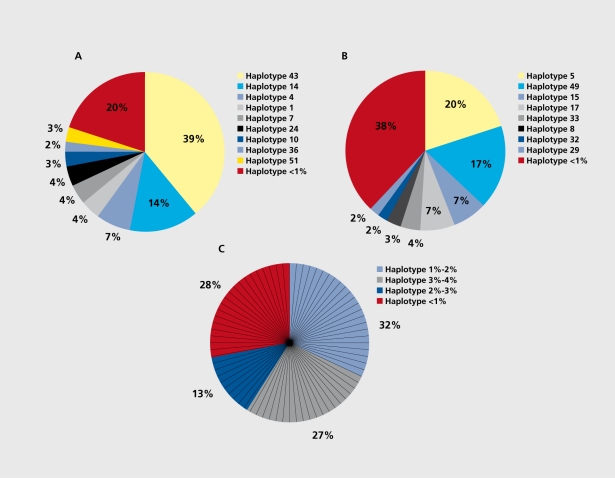
These analyses demonstrate that the decomposition of genes into different haplotypes, the so-called gene-based haplotype diversity, is remarkable. In fact, many genes do not, have one predominant haplotype at all, and the total fraction of rare haplotypes contributing to the picture may be significant. Specifically, in the largest survey performed to date,33 no single haplotype showed a. frequency ≥50% in 35% of the genes. The most, common haplotypes described ranged in frequency between 12% and 48%24-25,27,29,34-46,70; overall, the number of common haplotypes with frequencies >5% was found to be in the range of two to seven and to account for 43% to 97% of all haplotypes.24,25,27,29,34,46,70 For instance, 52 different haplotypes in a group of 172 individuals including cases and controls were predicted in the OPRM1 study29; of those, three haplotypes ranging in frequencies between 7% and 39% accounted for 60% of all haplotypes and nine haplotypes ranging in frequencies between 2% and 39% accounted for 80% of all haploypes (Figure 4). Referring to the human β2-adrenergic receptor gene, four different haplotypes at, frequencies >5% (range 7% to 20%) constitute 51% of all haplotypes (Figure 4b); considering the eight, haplotypes within a frequency range of 2% to 20%, these constitute only 62% of all haplotypes of this gene.
Figure 4. Distribution of haplotype frequencies. Each color-coded segment represents proportionately the frequency (in percent) of one specific haplotype, the corresponding haplotype numbers are given in the box; the red-colored segments contain the fraction of haplotypes with a frequency <1%. A. Haplotype frequencies of the human µ-opioid receptor gene (OPRM1), for corresponding numbers see reference 29. B. Haplotype frequencies of the human β2-adrenergic receptor gene; numbers in the box refer to a table of haplotypes predicted for 237 individuals (data not shown). C. Haplotype frequencies of the bradykinin receptor B2 gene (BDKRB2); numbers in the box refer to a table of haplotypes predicted for 234 individuals (data not shown), in this case, each color-coded major segment does not refer to a specific haplotype, but includes a number of haplotypes within a specific, defined frequency range; this is indicated by the subdivisions into multiple smaller, specific segments, each of which corresponds to one specific haplotype within the segment.
It is noteworthy that in the highest resolution comparative sequencing study performed to date on samples of 234 to 469 individuals, four to six common haplotypes at frequencies in the range of 5% to 20% were found to account for 51% to 57% of all haplotypes.70 The relative proportion of rare haplotypes (<1 %) observed amounted to 7% to 49 % ) 25,27,29,70 and ranged in absolute numbers from 14 to >100.25,27,29,70 Specifically referring to the example of OPRM1 haplotypes, 43 different rare haplotypes accounted for 20% of all haplotypes (Figure 4a). It is important, to note in this context that, potentially important, risk haplotypes were included in this class of rare haplotypes, whereas the common haplotypes occurred at similar frequencies in cases and controls.29,39 Referring to the β2-adrenergic receptor gene, for which significantly more than 100 haplotypes have been inferred to date,70 rare haplotypes accounted for 38% of all haplotypes, haplotypes <5% in fact, for 49% of all haplotypes67 (Figure 4b). A large number of rare and population-specific haplotypes have generally been observed in the majority of studies.24-29,31-34,39,64,65 At the extreme, the haplotype profile of a. gene may even be characterized by groups of relatively infrequent, haplotypes (Figure 4c), where literally no sequence haplotype at a frequency >4% existed; rather, four different groups that contain a total of 64 different, haplotypes at frequencies ranging between 3% and 4%, 2% and3%,l% and2%,and <1% may, somewhat arbitrarily, be distinguished. On the other hand, taking haplotype frequencies into consideration, cosmopolitan haplotypes accounted for nearly 82% of the total haplotypes observed, whereas population-specific haplotypes accounted for only about 8%.33
The concept of a gene revisited
There are multiple haplotypes that account for a significant, fraction of human genomic variability. The initial results clearly challenge the concept of the gene, and particularly the view that there exists one predominant form of a gene as the wild-type and various rare or mutant forms. This may well indicate the beginning of the end of Mendel's world10 and its view of the amount, nature, pattern, and structure of genetic variation. The two-allele concept, of the gene may for the time being have been nothing but the extreme and visible end of an entire spectrum, given the (until recently) limited access to genetic variation. Studies to come that will analyze continuously increasing numbers of individuals and increasingly larger, eventually complete gene regions (which may well extend up to about 100 kb and more) are likely to generate even more complex results. In brief, the concept of a gene may have to be revised completely10,29,33: the gene as a concrete molecular substrate does not exist. Genes rather appear to exist, as a spectrum of different, forms; the gene will have to be redefined as the sum of its haplotypes. The definition of a gene will have to include the positions, population specificities, and characterization of its variants, and a precise description of its haplotype structures. It is obvious that the next level of description (and the first step to reduce haplotype complexity) will be the assignment of the sequence haplotypes to the protein isoforms they constitute. Needless to say that such a revision of the concept. of the gene will have profound consequences on the analysis and classification of gene function, as well as its role as a drug target.
Genetic variability and its implications for pharmacogenomics and a personalized medicine
Knowledge on genetic variation and haplotype structures: an essential prerequisite for drug target discovery and optimization
The approaches and research data outlined above raise two major issues. First, how do the different approaches to candidate gene analysis apply to the various aspects of pharmacogenomics? Second, which conclusions and consequences should be drawn, taking into account, the recent results demonstrating potentially abundant candidate gene sequence diversity and complex haplotype structures?
With respect to the first, issue, all the entire individually variable candidate gene sequences corresponding to the gene-based functional haplotypes described earlier are the immediate correlates of pharmaceutical relevance as (i) the potential molecular correlates of the disease genes and naturally occurring different, forms of the genes, since they provide the immediate links to gene function(s) and dysfunction; and (ii) the direct objects of in vitro expression and units of functional characterization and therefore in vitro test, systems for drug action. It is these genebased functional haplotypes and their characteristics that serve as the reference substrates for target evaluation and prioritization. Because disease genes also mark functional pathways, they may serve as reference molecules for other related molecules in the affected network, which may represent, more suitable drug targets with regard to their molecular properties and genetic variability pattern.
With respect to the second issue, one major consequence to be drawn is to establish as an essential requirement of the systematic and comprehensive analysis of the entire individual gene sequences encoding the drug targets in appropriately chosen samples. It will be mandatory to determine the entire polymorphic spectra of the genes, as well as the haplotype structures underlying them. A second critical analytical task in this context, will be to evaluate to what extent potentially given complexity can be reduced to functionally distinct, haplotype classes and/or distinct protein isoforms. In-depth knowledge on the genetic variability of a drug target under consideration, especially the spectrum and frequencies of underlying haplotype structures in populations,84 will have to become an indispensable prerequisite for drug target evaluation, characterization, and prioritization. Needless to say these requirements refer to both the specific drug targets under consideration and the genes involved in drug metabolism and transport.
There is currently no a priori way of predicting the specific genetic variability in a drug target, or any other gene of pharmaceutical relevance, given its stochastic nature; each gene must, be rigorously subjected to systematic comparative sequence and haplotype analysis in populations. Extrapolating from the body of data described above, about two to seven different, haplotypes that occur most, frequently (at frequencies of the minor allele >5%) may be expected on average. This implies that the most, frequent, haplotypes amount to fractions of 16% to >50% of all haplotypes, constituting altogether about 51 % to 96% of the total of ha.pl otypes.29,33,46,70 Moreover, the numbers of rare haplotypes (frequencies of the minor allele <1%) may potentially be substantial, as outlined above. However, diversity at the sequence and haplotype level does not necessarily imply diversity at the protein level. Thus, the assignment of individual sequence haplotypes to protein isoforms will be one first, critical step towards the evaluation of the implications of given candidate gene variability. Hardly any data have been presented regarding the relationships between sequence haplotypes and protein isoforms, with the exception of the work on APOE sequence haplotypes by Fullerton et al.25 These authors demonstrated convergence of 31 different sequence haplotypes onto three different protein isoforms. Beyond that, diversity at the protein level may not necessarily imply diversity at the functional level, an issue whose clarification will be left, to the more distant future. Rare haplotypes, which per se apparently do not represent particularly favorable drug targets, may nevertheless require particular attention as potential mediators of severe side effects and may constitute significant, fractions of individual gene sequences resulting in the same protein isoform25 or share a common pattern conferring risk.29 Finally, as outlined earlier, any extreme may be possible: this may include, at the one end of the spectrum, completely invariable genes that may amount to about, 20% of all genes and, at the other end, highly decomposed genes with frequencies of numerous sequence haplotypes not exceeding 4%, for instance.
Obviously, a drug target is the more attractive if it has a low variability and decomposition into different haplotype(s) (classes) and protein isoforms. In this context, the modern version of a blockbuster drug target in the postgenome age of genetic variation would be an invariable gene. The pharmaccutically most attractive component of a proposed catalogue of all haplotypes of all genes as the ultimate biomedical resource would probably be the specific fraction containing the most, invariable genes. In reality though, we may have to concentrate on manageable variability, ie, scenarios where variability is limited or the functional implications are clearly definable. If a drug has not been tailored a priori to the target, in its variable, naturally occurring forms, incompatibilities, ineffectiveness, and adverse side effects will become apparent sooner or later. The molecular truth will eventually take its toll on both individuals and the pharmaceutical industry.
Any developments that are driven by the vision of a personalized medicine11-14 will have to be based on knowledge of the molecular diversity of potential drug targets and, generally, of any genes involved in drug action and metabolism.9,85,86 This information will be essential for decision-making processes. It will also be valuable in guiding in vitro screens and their specific experimental design. It will allow an extrapolation of drug response in population segments, as well as a correlation of in vitro and in vivo responsiveness (in conjunction with information on the genetic makeup of drug-metabolizing enzymes and competing, homologous targets). The integration of knowledge on human genetic variation into all phases of drug development, and application will be one of the pharmaceutical industry's major future tasks. Last but not least, what does the evidence for gene decomposition into multiple forms tell us about, the prospects for an individualized medicine?11-14,87 The fact, that individual sequence differences exist, does not, mean that tailoring drugs to each individual is possible or feasible. Given the remarkable genetic diversity and its challenges, this vision may seem somewhat too bold and unrealistic at this stage. However, a focus on population stratification as well as avoidance of serious harm through attention to rare profiles may merit very serious consideration.88,89
Gene-based haplotype analysis and diagnostic validity
Gene-based haplotypes seem appropriate as complex genetic markers,48 if extraction of these diagnostic markers is based on systematic analysis of underlying LD and haplotype structures in the populations. The number of SNPs necessary to validly represent, those structures may well range between one or a few48 and many (Ott, personal communication). As pointed out, randomly selected SNPs in and around the gene, even frequent ones, may not be able to distinguish between different underlying haplotypes and, importantly, not between high- and low-risk haplotypes. Thus, the selection and use of SNPs as diagnostic markers for the prediction of drug response and disease risk will have to be subjected to rigorous criteria. Finally, it is interesting to note that even for mendelian types of disease causation, numerous alleles may have to be taken into consideration.10,90
An ultimate resource for drug discovery
In view of the tens of thousands of genes existing in the human genome, many of which may not, yet be accessible to even the most, advanced approaches to predict, or annotate function, the future, bold alternative to any strategy of candidate gene selection and testing will be the simultaneous analysis of all functional haplotypes of all genes against phenotype. This may, at some point, turn out, to be even more efficient, if the appropriate technologies are at hand, than the extensive approaches to hypothesis generation and testing described above. Such an approach would represent, the haplotype-based version of the candidate gene association mapping approach proposed by Risch and Merikangas91 as the future of the genetics of complex disease, which was originally based upon a two-allele concept, of the gene,90,91 as was the catalogue of common SNPs envisaged by Lander.92 In such a catalogue of all haplotypes, each gene would be represented by the entire spectrum of individually different, forms of the gene, both the cosmopolitan33 and the population-specific ones. Such a catalogue would also include annotations of all individually variable sites including changes in regulatory, exonic, and intronic sequences (function-tagged sites)6 and, as far as possible, annotations of the entire haplotypes as functional units. In addition, a classification of these haplotypes with respect, to their functional similarity would seem a most, valuable asset to such a resource. To what, extent the necessary functional annotations will have to be achieved in silico or in vitro, remains open. Additional information of potentially great, value would be the fraction of highly invariable genes. These would represent, that, kind of drug targets that most closely would fulfil the criteria for blockbuster targets, as outlined above.
Challenges and future perspectives
The multiplicity of individual gene forms and their relation to gene function and phenotype: challenges and first solutions
These initial comparative sequencing studies have demonstrated that numerous individually different, forms of the gene may exist. Eventually, at the ultimate level of resolution including increasingly large regions of analysis and increasingly large numbers of individuals, every haplotype may become unique.38 It remains unknown whether the number of different, haplotypes may, at some point, reach a level of saturation, or whether their number will increase infinitely The molecular truth emerges: the fact that multiallelism may be the rule rather than the exception.10,90 Referring to the gene variability data presented above, the number of different, haplotypes may become unfeasibly large,29,38,90 so that, the power is not, sufficient, to detect an association of the disease phenotype or drug response with any single haplotype. Thus, this allelic complexity imposes tremendous challenges on the establishment of haplotypc/gcnotype-phenotypc relationships.29,38 The following key questions arise: how should genotype-phenotype relationships be analyzed against a background of high natural genome sequence diversity? How should the important variants be filtered from the unimportant ones? Approaches to reduce this complexity and condense information on genetic variation will be required.
Various approaches to the grouping, or classification, of haplotypes have been suggested. One major approach to reduce complexity has been the grouping of haplotypes by evolutionary relatedness as the basis for association studies; this approach has been described in detail in a previous review.93 The historical information from different, haplotypes is combined to construct a cladogram that estimates how the different, haplotypes are evolutionarily related. This allows localization of functional mutational changes in the haplotype network by identification of phenotypic contrasts between sister clades. The use of an evolutionary tree as a statistical design may become difficult, when the evolutionary history of a population may have been influenced by various forces, such as high rates of recombination, multiple mutations to high susceptibility alleles, and others.29 The reconstruction of the specific evolutionary processes in general, and the construction of evolutionary trees in the presence of recombination events in particular, may become extremely difficult. - if not. unfeasible - in most, complex genetic disease studies.
Another approach could be the extraction of the most frequent, haplotypes (>5%), which constitute - on the basis of preliminary results - about 51% to >90% of all haplotypes,“46,70 and subsequent evaluation, whether one of these haplotypes may occur significantly more frequently in cases than controls. This approach is based on the a priori assumption that common haplotypes play a major role in the genetics of common disease,23,94 which is a highly controversial topic.94-97 This approach will, however, not capture a genetic risk scenario that involves rare mutations65-68 or rare haplotypes.29 A number of examples have demonstrated that rare variants and/or haplotypes may confer genetic susceptibility to complex disease, whereas the common haplotypes did not allow distinction of cases and controls in some of these examples.39,65-68 Thus, a focus on the groups of common haplotypes from the outset, does not appear to be a sound solution.
Conceptually, another approach to cope with the multiplicity of haplotypes could be envisioned, which seems the most, promising and reasonable: the classification of haplotypes into functionally related (ideally functionally equivalent) haplotypes based on sequence-structurefunction similarity.29 Needless to say that this will by no means be less challenging than, for instance, the (reconstruction of evolutionary trees described above. However, such an approach would not rely on the reconstruction of evolutionary history with its many unknowns, but focus on the “here and now”; the given sequence would not be considered as an end point, of history, but as the information that determines structure and function of the protein. Such an approach would seem more generally applicable.
Initial approaches have been explored,29 applying a stepwise classification process, for example, a hierarchical cluster procedure. Haplotypes are grouped into ever more inclusive classes, until only one final cluster is left. This approach relics on the assumption that, the existence of functionally different, classes would be likely, if at least one class included haplotypes from cases more (or less) frequently than controls. If this is the case, the haplotypes in the different clusters can be inspected for consensus patterns. The patterns observed more frequently among individuals with the disease could be interpreted as genetic risk pattern(s).
Apart from these first attempts, the reduction of complexity through the grouping of functionally equivalent forms of the gene remains a bold vision. It seems nevertheless to represent, the ultimate approach, which would provide the basis to immediately establish the links between genetic variation, gene function, and dysfunction. Major challenges will include the development, of valid similarity measures for classification procedures that, incorporate properties that determine sequence-structure-function similarities, such as physicochemical properties. Altogether, the development, of approaches to reduce haplotype - and genotype - complexity through classification will be critical to the future of the genetics of complex disease and all aspects of pharmacogenomics as outlined above.
Genetic variation and its functional implications: the units of analysis
As indicated above, the analyses of the functional implications of candidate gene variation have been performed almost entirely with focus on single SNPs, taken out of context. Thus, the conclusions drawn may hardly be valid for the majority of naturally occurring individual gene sequences and the functions they encode. This means that the unit of functional analysis will have to change: from the previously standard single mutation analysis in vitro to the functional analysis of entire individual gene sequences or the gene-based functional haplotypes (sequence haplotypes) of a gene. The challenges are obvious, given the potentially abundant variations in all, regulatory, coding and intronic sequences. First, paradigmatic results from a functional sequence haplotype analysis in the human β2-adrenergic receptor gene show that the effects of the various SNP combinations are different, from those previously obtained with individual SNPs taken out of context, of a verified haplotype. These first results clearly support, the importance of studying SNPs in vitro within the context of a validated haplotype.24
In this example, the bronchodilator responses in vivo to β2-agonist were significantly related to haplotype pairs, but not to any individual SNP. Expression of the haplotypes associated with divergent, responsiveness clearly demonstrated that receptor m'RNA levels and receptor density in cells transfected with the haplotype associated with the greater physiological response were about 50% greater than those transfected with the lower-response haplotype.24 These results indicated that the unique interactions of multiple SNPs within a haplotype can ultimately affect, biological and therapeutic phenotype, and that individual SNPs may have poor predictive power as pharmacogenetic loci. The authors conclude from their results that, it is likely that, the biological phenotype is directed by an interaction involving transcription, translation, and protein processing, which ultimately defines the effect of these haplotypes.24 The challenges of analyzing and interpreting given genetic variation at all levels are daunting and, obviously, the true challenges will be biological. Nevertheless, the initial steps toward solutions have been taken.
Gene variability, the genetics of complex traits, and future approaches to the analysis of complex systems
The analysis of individual candidate genes constitutes an essential analytical entity, which is part of a bigger picture. The majority of diseases and individual drug response are prototypic complex traits and may involve interactions of several or multiple genes or entire gene networks with the environment.98 The complexity of the trait also arises from the fact that genetic and environmental factors may interact with each other in unpredictable ways, such that, the association between the phenotype and any single genetic factor may be imperceptible.98,99 Nonlinear interactions, including gene-environment interactions, mean that the expression of the phenotype may not be accurately predicted from knowledge of the individual effects of each of the component, factors considered alone, no matter how well understood the separate components may be.100 A full catalogue of the genetic architecture of complex phenotypes consists of a description of all the genetic and environmental factors that affect, the phenotype, along with the magnitude of their individual effects and the interactions among the factors. Clearly, this represents the particular challenge, underscores the importance of the analysis of gene-gene-environment, interactions, and implies that, potentially many different models of interactions will have to be explored.
In this context, individual variation in drug response may involve any of the gene networks that are part of the complex interplay between specific disease-associated factors, pharmacokinetics, and pharmacodynamics.9,85-87,99 These may include any of the functional pathways involved in the specific pathophysiology of the disease. Nonresponse may, for instance, be due to genetic heterogeneity in disease etiology. In this case, the drug may not, target the specific causative mechanisms active in the individual.87 Moreover, the gene encoding the specific drug target, represents the first, component, of an entire downstream pathway that controls signal transduction and elicits the cellular effects. Thus, genetic variation in any of the genes regulating this pathway may cause variation in drug response. Furthermore, numerous genes or gene families are involved in drug transport and drug metabolism, such as the genes encoding the phase I and phase II enzymes, which are expressed in the liver.9,12,85-87,99 In addition, environmental factors, such as nutrition, exercise, access to substances of abuse, etc, may influence drug response.
In the future, progress in the understanding of the molecular bases of disease and drug response is expected to come especially from advances in functional genomics as the basis for whole complex systems analysis. These advances will be based on the increasing elucidation of the function of all genes involved in all pathways constituting the relevant process. In this new approach to biological research, the same type of analyses, which are typically used to try to understand the function of single genes, are carried out on most, or all genes of the organism. Enormous amounts of information on the networks of biological processes are being generated, leading to the establishment of models of specific functional networks. Apart from deriving many novel candidate genes and drug targets of interest, this may provide yet another approach to group multiple variants in genes, in which according to functional context, the model is used as a template for classification and functional interpretation of a spectrum of gene variants. Thus, systems analysis can be extended to pose the question of whether a specific metabolic pathway involving many genes of variable variability could be involved in a specific phenotype or disease. In such an analysis, mutations in any of the genes of the pathway, each of which occurs at too low a frequency to be significant, in itself, could be pooled to increase the overall significance. The power of this approach to establish genotype-phenotype correlations will become even greater once the information on variants can be combined into functional units of increasing complexity and once these biological processes can be comprehensively modeled by systems analysis.
In the future, we can also hope to gain considerable power in the establishment of the more complex genotype-phenotype relationships by the modeling of the predicted effects of any sequence variant, or combination of sequence variants, taking into account cis effects (all variants affecting the function of a specific gene on a specific chromosome in a haplotype), trans effects (complementation between the two copies of each gene on autosomes), as well as genc-gene and genc-environment interactions. It is highly likely that, the establishment of quantitative models of all of these effects and interactions will be essential to derive many of the more complex genotype-phenotype relationships, and to ultimately understand many of the complex biological and disease processes. F/ven if biology may be too complex to be understood in the classical sense, the best, we can possibly hope for is to establish models of these processes that correctly predict, all the parameters we can assess. Such systems will be a key step in being able to use the enormous amount of knowledge being generated to improve diagnosis and therapy, and ultimately guide therapy in an individual patient. Thus, hopes are high that these developments will have a major impact, on medicine and prepare the ground for the future of an optimized, patientoriented therapy.
Selected abbreviations and acronyms
- DGGE
denaturing gradient gel electrophoresis
- DHPLC
denaturing high-performance liquid chromatography
- EST
expressed sequence tag
- LD
linkage disequilibrium
- RFLP
restriction fragment length polymorphism
- SNP
single nucleotide polymorphism
- SSCP
single-stranded conformation polymorphism
- STR
short tandem repeat
- STS
sequence-tagged site
- UTR
untranslated region
- VDA
variant detection array
- VNTR
variable number of tandem repeats
MRH is grateful to H. Lehrach, Max Planck Institute for Molecular Genetics (MPI-MG), Berlin, for most valuable discussions and comments. She acknowledges B. Timmermann (MPI-MG) for technical assistance and data analysis and K. Köpke (Humbold University, Berlin) for statistical analysis. MRH was supported by a grant (01GR0155) from the BMBF (Federal Ministry for Education and Research) as part of the German National Genome Research Network (NGFN) Core.
Contributor Information
Margret R. Hoehe, Genetic Variation Program, Department of Vertebrate Genomics, Max Planck Institute for Molecular Genetics, Berlin, Germany.
Thomas Kroslak, Genetic Variation Program, Department of Vertebrate Genomics, Max Planck Institute for Molecular Genetics, Berlin, Germany.
REFERENCES
- 1.Kobilka BK., Frielle T., Dohlman HG., et al. Delineation of the intronless nature of the genes for the human and hamster pYadrenergic receptor and their putative promoter regions. J Biol Chem. 1987;262:7321–7327. [PubMed] [Google Scholar]
- 2.Bradshaw RA., Dennis E. Handbook of Cell Signaling. New York, NY: Academic Press; 2003 [Google Scholar]
- 3.Pierce KL., Premont RT., Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 4.Drews J. Genomic sciences and the medicine of tomorrow. Nat. Biotechnol. 1996;14:1516–1518. doi: 10.1038/nbt1196-1516. [DOI] [PubMed] [Google Scholar]
- 5.Liggett SB. Molecular and genetic basis of P2-adrenergic receptor function. J Allergy Clin Immunol. 1999;104:S42–S46. doi: 10.1016/s0091-6749(99)70272-1. [DOI] [PubMed] [Google Scholar]
- 6.Hoehe MR. Project Proposal. A Genotype Approach: The Investigation of Inter-individual DNA Sequence Differences in Adrenergic Receptor Genes and Their Possible Functional Implications. http://www.molgen.mpg.de/ -genetic variation program/ Accessed 21 January 2004. [Google Scholar]
- 7.Mansfield BK. The Genome Project and the pharmaceutical industry. Hum Genome News. 1990:10–11. [Google Scholar]
- 8.Garrod AE. inborn Errors of Metabolism. New York, NY: Oxford University Press; 1909 [Google Scholar]
- 9.Nebert DW. Drug-metabolizing enzymes, polymorphisms and interindividual response to environmental toxicants. Clin Chem Lab Med. 2000;38:857–861. doi: 10.1515/CCLM.2000.124. [DOI] [PubMed] [Google Scholar]
- 10.Weiss KM. Is there a paradigm shift in genetics? Lessons from the study of human diseases. Mol Phylogenet Evol. 1996;5:259–265. doi: 10.1006/mpev.1996.0019. [DOI] [PubMed] [Google Scholar]
- 11.Cohen J. Developing prescriptions with a personal touch. Science. 1997;275:776. doi: 10.1126/science.275.5301.776. [DOI] [PubMed] [Google Scholar]
- 12.Marshall A. Laying the foundations for personalized medicines. Nat Biotechnoi. 1997;15:954–957. doi: 10.1038/nbt1097-954. [DOI] [PubMed] [Google Scholar]
- 13.Marshall A. Getting the right drug into the right patient. Nat Biotechnoi. 1997;15:1249–1252. doi: 10.1038/nbt1197-1249. [DOI] [PubMed] [Google Scholar]
- 14.Roses AD. Pharmacogenetics and the practice of medicine. Nature. 2000;405:857–865. doi: 10.1038/35015728. [DOI] [PubMed] [Google Scholar]
- 15.Collins FS. Positional cloning moves from perditional to traditional. Nat Genet. 1995;9:347–350. doi: 10.1038/ng0495-347. [DOI] [PubMed] [Google Scholar]
- 16.Venter JC., Adams MD., Myers EW., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 17.Lander ES., Linton LM., Birren B., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 18.Collins FS., Guyer MS., Charkravarti A. Variations on a theme: cataloging human DNA sequence variation. Science. 1997;278:1580–1581. doi: 10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- 19.Sachidanandam R., Weissman D., Schmidt SC., et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 20.Daly MJ., Rioux JD., Schaffner SF., Hudson TJ., Lander ES. High-resolution haplotype structure in the human genome. Nat Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 21.Gabriel SB., Schaffner SF., Nguyen H., et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 22.Patil N., Berno AJ., Hinds DA., et al. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science. 2001;294:1719–1723. doi: 10.1126/science.1065573. [DOI] [PubMed] [Google Scholar]
- 23.Reich DE., Cargill M., Bolk S., et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- 24.Drysdale CM., McGraw DW., Stack CB., et al. Complex promoter and coding region β2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc Natl Acad Sci USA. 2000;97:10483–10488. doi: 10.1073/pnas.97.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fullerton SM., Clark AG., Weiss KM., et al. Apolipoprotein E variation at the sequence haplotype level: implications for the origin and maintenance of a major human polymorphism. Am J Hum Genet. 2000;67:881–900. doi: 10.1086/303070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilad Y., Rosenberg S., Przeworski M., Lancet D., Skorecki K. Evidence for positive selection and population structure at the human MAO-A gene. Proc Natl Acad Sci USA. 2002;99:862–867. doi: 10.1073/pnas.022614799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harding RM., Fullerton SM., Griffiths RC., et al. Archaic African and Asian lineages in the genetic ancestry of modern humans. Am J Hum Genet. 1997;60:772–789. [PMC free article] [PubMed] [Google Scholar]
- 28.Harris EE., Hey J. X chromosome evidence for ancient human histories. Proc. Natl Acad Sci USA. 1999;96:3320–3324. doi: 10.1073/pnas.96.6.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoehe MR., Kopke K., Wendel B., et al. Sequence variability and candidate gene analysis in complex disease: association of i opioid receptor gene variation with substance dependence. Hum Moi Genet. 2000;9:2895–2908. doi: 10.1093/hmg/9.19.2895. [DOI] [PubMed] [Google Scholar]
- 30.Nickerson DA., Taylor SL., Fullerton SM., et al. Sequence diversity and large-scale typing of SNPs in the human apolipoprotein E gene. Genome Res. 2000;10:1532–1545. doi: 10.1101/gr.146900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nickerson DA., Taylor SL., Weiss KM., et al. DNA sequence diversity in a 9.7-kb region of the human lipoprotein lipase gene. Nat Genet. 1998;19:233–240. doi: 10.1038/907. [DOI] [PubMed] [Google Scholar]
- 32.Rieder MJ., Taylor SL., Clark AG., Nickerson DA. Sequence variation in the human angiotensin-converting enzyme. Nat Genet. 1999;22:59–62. doi: 10.1038/8760. [DOI] [PubMed] [Google Scholar]
- 33.Stephens JC., Schneider JA., Tanguay DA., et al. Haplotype variation and linkage disequilibrium in 313 human genes. Science. 2001;293:489–493. doi: 10.1126/science.1059431. [DOI] [PubMed] [Google Scholar]
- 34.Toomajian C., Kreitman M. Sequence variation and haplotype structure at the human HFE locus. Genetics. 2002;161:1609–1623. doi: 10.1093/genetics/161.4.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chakravarti A. It's raining SNPs, hallelujah?. Nat Genet. 1998;19:216–217. doi: 10.1038/885. [DOI] [PubMed] [Google Scholar]
- 36.Halushka MK., Fan JB., Bentley K., et al. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet. 1999;22:239–247. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]
- 37.Cargill M., Altshuler D., Ireland J., et al. Characterization of singlenucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 38.Clark AG., Weiss KM., Nickerson DA., et al. Haplotype structure and population genetic inferences from nucleotide-sequence variation in human lipoprotein lipase. Am J Hum Genet. 1998;63:595–612. doi: 10.1086/301977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoehe MR. Haplotypes and the systematic analysis of genetic variation in genes and genomes. Pharmacogenomics. 2003;4:547–570. doi: 10.2217/14622416.4.5.547. [DOI] [PubMed] [Google Scholar]
- 40.Ackenheil M., Weber K. Differing response to antipsychotic therapy in schizophrenia: pharmacogenomic aspects. Dialogues Clin Neurosci. 2004;6:71–77. doi: 10.31887/DCNS.2004.6.1/mackenheil. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morris-Rosendahl DJ., Fiebich BL. The future of genetic testing for drug response. Dialogues Clin Neurosci. 2004;6:27–37. [PMC free article] [PubMed] [Google Scholar]
- 42.Altshuler D., Pollara VJ., Cowles CR., et al. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature. 2000;407:513–516. doi: 10.1038/35035083. [DOI] [PubMed] [Google Scholar]
- 43.Mullikin JC., Hunt SE., Cole CG., et al. An SNP map of human chromosome 22. Nature. 2000;407:516–520. doi: 10.1038/35035089. [DOI] [PubMed] [Google Scholar]
- 44.Wang DG., Fan JB., Siao CJ., et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 45.Marth GT., Korf I., Yandell MD., et al. A general approach to singlenucleotide polymorphism discovery. Nat Genet. 1999;23:452–456. doi: 10.1038/70570. [DOI] [PubMed] [Google Scholar]
- 46.Johnson GC., Esposito L., Barratt BJ., et al. Haplotype tagging for the identification of common disease genes. Nat Genet. 2001;29:233–237. doi: 10.1038/ng1001-233. [DOI] [PubMed] [Google Scholar]
- 47.NIH Meeting on Developing a Haplotype Map of the Human Genome for Finding Genes Related to Health and Disease, Bethesda, Md, July 1819, 2001. [Google Scholar]
- 48.Judson R., Stephens JC., Windemuth A. The predictive power of haplotypes in clinical response. Pharmacogenomics. 2000;1:15–26. doi: 10.1517/14622416.1.1.15. [DOI] [PubMed] [Google Scholar]
- 49.Kidd KK., Pakstis AJ., Castiglione CM., et al. DRD2 haplotypes containing the Taqi A1 allele: implications for alcoholism research. Alcohol Clin Exp Res. 1996;20:697–705. doi: 10.1111/j.1530-0277.1996.tb01674.x. [DOI] [PubMed] [Google Scholar]
- 50.Templeton AR., Clark AG., Weiss KM., Nickerson DA., Boerwinkle E., Sing CF. Recomblnatlonal and mutational hotspots within the human lipoprotein lipase gene. Am J Hum Genet. 2000;66:69–83. doi: 10.1086/302699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davidson S. Research suggests importance of haplotypes over SNPs. Nat Biotechnoi. 2000;18:1134–1135. doi: 10.1038/81100. [DOI] [PubMed] [Google Scholar]
- 52.Joosten PH., Toepoel M., Mariman EC., VanZoelen EJ. Promoter haplotype combinations of the platelet-derived growth factor alpha-receptor gene predispose to human neural tube defects. Nat Genet. 2001;27:215–217. doi: 10.1038/84867. [DOI] [PubMed] [Google Scholar]
- 53.Taton TA., Mirkin CA. Haplotyping by force. Nat Biotechnoi. 2000;18:713. doi: 10.1038/77267. [DOI] [PubMed] [Google Scholar]
- 54.Wood NA., Keen LJ., TiIley LA., Bidwell JL. Determination of cytokine regulatory haplotypes by induced heteroduplex analysis of DNA. J Immunol Methods. 2001;249:191–198. doi: 10.1016/s0022-1759(00)00360-4. [DOI] [PubMed] [Google Scholar]
- 55.Hoehe MR., Timmermann B., Lehrach H. Haplotypes and the systematic analysis of genetic variation: disease genes, drug targets and pharmacogenomics [in German]. Biospektrum. 2002;8:478–485. [Google Scholar]
- 56.Timmermann B., Mo R., Luft FC., et al. Beta adrenoceptor genetic variation is associated with genetic predisposition to essential hypertension: The Bergen Blood Pressure Study. Kidney Int. 1998;53:1455–1460. doi: 10.1046/j.1523-1755.1998.00926.x. [DOI] [PubMed] [Google Scholar]
- 57.Green SA., Turki J., Innis M., Liggett SB. Amino-terminal polymorphisms of the human β2-adrenergic receptor impart distinct agonist-promoted regulatory properties. Biochemistry. 1994;33:9414–9419. doi: 10.1021/bi00198a006. [DOI] [PubMed] [Google Scholar]
- 58.Green SA., Cole G., Jacinto M., Innis M., Liggett SB. A polymorphism of the human P2-adrenergic receptor within the fourth transmembrane domain alters ligand binding and functional properties of the receptor. J Biol Chem. 1993;268:23116–23121. [PubMed] [Google Scholar]
- 59.Parola, Kobilka BK. The peptide product of a 5' leader cistron in the β2 adrenergic receptor mRNA inhibits receptor synthesis. J Biol Chem. 1994;269:4497–4505. [PubMed] [Google Scholar]
- 60.Kotanko P., Binder A., Tasker J., et al. Essential hypertension in African Caribbeans associates with a variant of the β2-adrenoceptor. Hypertension. 1997;30:773–776. doi: 10.1161/01.hyp.30.4.773. [DOI] [PubMed] [Google Scholar]
- 61.McGraw DW., Forbes SL., Kramer LA., Liggett SB. Polymorphisms of the 5' leader cistron of the human p2-adrenergic receptor regulate receptor expression. J Clin Invest. 1998;102:1927–1932. doi: 10.1172/JCI4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoehe MR., Rinn T., Flachmeier C., et al. Comparative sequencing of the human CB1 cannabinoid receptor gene coding exon: no structural mutations in individuals exhibiting extreme responses to cannabis. Psychiatr Genet. 2000;10:173–177. doi: 10.1097/00041444-200010040-00004. [DOI] [PubMed] [Google Scholar]
- 63.Rioux JD., Daly MJ., Silverberg MS., et al. Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Wat. Genet. 2001;29:223–228. doi: 10.1038/ng1001-223. [DOI] [PubMed] [Google Scholar]
- 64.Rana BK., Hewett-Emmett D., Jin L., et al. High polymorphism at the human melanocortin 1 receptor locus. Genetics. 1999;151:1547–1557. doi: 10.1093/genetics/151.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hugot JP., Chamaillard M., Zouali H., et al. Association of NOD2 leucinerich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 66.Lesage S., Zouali H., Cezard JP., et al. CARD1SINOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am J Hum Genet. 2002;70:845–857. doi: 10.1086/339432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ogura Y., Bonen DK., Inohara N., et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 68.Branson R., Potoczna N., Krai JG., Lentes KU., Hoehe MR., Horber FF. Binge eating as a major phenotype of melanocortin 4 receptor gene mutations. N Engl J Med. 2003;348:1096–1103. doi: 10.1056/NEJMoa021971. [DOI] [PubMed] [Google Scholar]
- 69.Horikawa Y., Oda N., Cox NJ., et al. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet. 2000;26:163–175. doi: 10.1038/79876. [DOI] [PubMed] [Google Scholar]
- 70.Hoehe MR., Timmermann B., Reinhardt R., Ott J., Lehrach H., Church GM. Haplotypes, in depth variation analysis, and the future of the analysis of genotype-phenotype relationships. Cold Spring Harbor Meeting. The Biology of DNA. Cold Spring Harbor, NY, February 26-March 2;. 2003:45. [Google Scholar]
- 71.Li WH., Sadler LA. Low nucleotide diversity in man. Genetics. 1991;129:513–523. doi: 10.1093/genetics/129.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sunyaev SR., Lathe WC III., Ramensky VE., Bork P. SNP frequencies in human genes an excess of rare alleles and differing modes of selection. Trends Genet. 2000;16:335–337. doi: 10.1016/s0168-9525(00)02058-8. [DOI] [PubMed] [Google Scholar]
- 73.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 74.Harris H., Hopkinson DA. Average heterozygosity per locus in man: an estimate based on the incidence of enzyme polymorphisms. Ann Hum Genet. 1972;36:9–20. doi: 10.1111/j.1469-1809.1972.tb00578.x. [DOI] [PubMed] [Google Scholar]
- 75.Eyre-Walker A., Keightley PD. High genomic deleterious mutation rates in hominids. Nature. 1999;397:344–347. doi: 10.1038/16915. [DOI] [PubMed] [Google Scholar]
- 76.Li WH. Molecular Evolution. Sunderland, Mass: Sinauer Associates; 1997 [Google Scholar]
- 77.Nei M. Molecular Evolutionary Genetics. New York, NY: Columbia University Press; 1987 [Google Scholar]
- 78.Fullerton SM., Clark AG., Weiss KM., et al. Sequence polymorphism at the human apolipoprotein All gene (APOA2): unexpected deficit of variation in an African-American sample. Hum Genet. 2002;111:75–87. doi: 10.1007/s00439-002-0763-x. [DOI] [PubMed] [Google Scholar]
- 79.Harris EE., Hey J. Human populations show reduced DNA sequence variation at the factor IX locus. Curr Biol. 2001;11:774–778. doi: 10.1016/s0960-9822(01)00223-8. [DOI] [PubMed] [Google Scholar]
- 80.Hoehe MR., Timmermann B., Lehrach H. Human inter-individual DNA sequence variation in candidate genes, drug targets, the importance of haplotypes and implications for pharmacogenomics. Curr Pharm Biotechnol. 2003;4:351–378. doi: 10.2174/1389201033377300. [DOI] [PubMed] [Google Scholar]
- 81.Podlowski S., Wenzel K., Luther HP., et al. β1-adrenoceptor gene variations: a role in idiopathic dilated cardiomyopathy?. J Mol Med. 2000;78:87–93. doi: 10.1007/s001090000080. [DOI] [PubMed] [Google Scholar]
- 82.Befort K., Filliol D., Decaillot FM., Gaveriaux-Ruff C., Hoehe MR., Kieffer BL. A single nucleotide polymorphic mutation in the human (i-opioid receptor severely impairs receptor signaling. J Biol Chem. 2001;276:3130–3137. doi: 10.1074/jbc.M006352200. [DOI] [PubMed] [Google Scholar]
- 83.Bond C., LaForge KS., Tian M., et al. Single-nucleotide polymorphism in the human i-opioid receptor gene alters p-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci USA. 1998;95:9608–9613. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wilson JF., Weale ME., Smith AC., et al. Population genetic structure of variable drug response. Nat Genet. 2001;29:265–269. doi: 10.1038/ng761. [DOI] [PubMed] [Google Scholar]
- 85.Lichter J., McNamara D. What's in a gene: using genetic information for the design of clinical trials. Curr Opin Biotechnoi. 1995;6:715–717. doi: 10.1016/0958-1669(95)80117-0. [DOI] [PubMed] [Google Scholar]
- 86.Meyer UA. Pharmacogenetics and adverse drug reactions. Lancet. 2000;356:1667–1671. doi: 10.1016/S0140-6736(00)03167-6. [DOI] [PubMed] [Google Scholar]
- 87.Ferrari P. Pharmacogenomics: a new approach to individual therapy of hypertension?. Curr Opin Nephrol Hypertens. 1998;7:217–222. doi: 10.1097/00041552-199803000-00012. [DOI] [PubMed] [Google Scholar]
- 88.Lindpaintner K. Pharmacogenetics and the future of medical practice: conceptual considerations. Pharmacogenomics J. 2001;1:23–26. doi: 10.1038/sj.tpj.6500009. [DOI] [PubMed] [Google Scholar]
- 89.Lindpaintner K. The importance of being modest - reflections on the pharmacogenetics of abacavir. Pharmacogenomics. 2002;3:835–838. doi: 10.1517/14622416.3.6.835. [DOI] [PubMed] [Google Scholar]
- 90.Terwilliger JD., Weiss KM. Linkage disequilibrium mapping of complex disease: fantasy or reality?. Curr Opin Biotechnol. 1998;9:578–594. doi: 10.1016/s0958-1669(98)80135-3. [DOI] [PubMed] [Google Scholar]
- 91.Risch N., Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 92.Lander ES. The new genomics: global views of biology. Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 93.Zhao H., Pfeiffer R., Gail MH. Haplotype analysis in population genetics and association studies. Pharmacogenomics. 2003;4:171–178. doi: 10.1517/phgs.4.2.171.22636. [DOI] [PubMed] [Google Scholar]
- 94.Reich DE., Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 95.Pritchard JK. Are rare variants responsible for susceptibility to complex diseases?. Am J Hum Genet. 2001;69:124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pritchard JK., Cox NJ. The allelic architecture of human disease genes: common disease-common variant or not. Hum Mol Genet. 2002;11:2417–2423. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- 97.Weiss KM., Terwilliger JD. How many diseases does it take to map a gene with SNPs. Nat Genet. 2000;26:151–157. doi: 10.1038/79866. [DOI] [PubMed] [Google Scholar]
- 98.Lander ES., Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 99.Persidis A. The business of pharmacogenomics. Nat Biotechnol. 1998;16:209–210. doi: 10.1038/nbt0298-209. [DOI] [PubMed] [Google Scholar]
- 100.Schork NJ., Cardon LR., Xu X. The future of genetic epidemiology. Trends Genet. 1998;14:266–272. doi: 10.1016/s0168-9525(98)01497-8. [DOI] [PubMed] [Google Scholar]