Abstract
Background information. Intestinal absorption of alimentary lipids is a complex process ensured by enterocytes and leading to TRL [TAG (triacylglycerol)-rich lipoprotein] assembly and secretion. The accumulation of circulating intestine-derived TRL is associated with atherosclerosis, stressing the importance of the control of postprandial hypertriglyceridaemia. During the postprandial period, TAGs are also transiently stored as CLDs (cytosolic lipid droplets) in enterocytes. As a first step for determining whether CLDs could play a role in the control of enterocyte TRL secretion, we analysed the protein endowment of CLDs isolated by sucrose-gradient centrifugation from differentiated Caco-2/TC7 enterocytes, the only human model able to secrete TRL in culture and to store transiently TAGs as CLDs when supplied with lipids. Cells were analysed after a 24 h incubation with lipid micelles and thus in a state of CLD-associated TAG mobilization.
Results. Among the 105 proteins identified in the CLD fraction by LC-MS/MS (liquid chromatography coupled with tandem MS), 27 were directly involved in lipid metabolism pathways potentially relevant to enterocyte-specific functions. The transient feature of CLDs was consistent with the presence of proteins necessary for fatty acid activation (acyl-CoA synthetases) and for TAG hydrolysis. In differentiated Caco-2/TC7 enterocytes, we identified for the first time LPCAT2 (lysophosphatidylcholine acyltransferase 2), involved in PC (phosphatidylcholine) synthesis, and 3BHS1 (3-β-hydroxysteroid dehydrogenase 1), involved in steroid metabolism, and confirmed their partial CLD localization by immunofluorescence. In enterocytes, LPCAT2 may provide an economical source of PC, necessary for membrane synthesis and lipoprotein assembly, from the lysoPC present in the intestinal lumen. We also identified proteins involved in lipoprotein metabolism, such as ApoA-IV (apolipoprotein A-IV), which is specifically expressed by enterocytes and has been proposed to play many functions in vivo, including the formation of lipoproteins and the control of their size. The association of ApoA-IV with CLD was confirmed by confocal and immunoelectron microscopy and validated in vivo in the jejunum of mice fed with a high-fat diet.
Conclusions. We report for the first time the protein endowment of Caco-2/TC7 enterocyte CLDs. Our results suggest that their formation and mobilization may participate in the control of enterocyte TRL secretion in a cell-specific manner.
Keywords: 3-β-hydroxysteroid dehydrogenase, apolipoprotein A-IV, Caco-2/TC7 cell, cytosolic lipid droplet, enterocyte, lysophosphatidylcholine acyltransferase 2 (LPCAT2), proteome
Abbreviations: 3BHS1, 3-β-hydroxysteroid dehydrogenase 1; ABHD5, α/β-hydrolase-domain-containing protein 5; ACSL3, acyl-CoA synthetase long-chain 3; ApoA-IV, apolipoprotein A-IV; CCT-α, choline-phosphate cytidylyltransferase A; CE, cholesterol ester; CLD, cytosolic lipid droplet; DAPI, 4′,6-diamidino-2-phenylindole; DGAT, diacylglycerol acyltransferase; DGE, diacylglyceryl ether; emPAI, exponentially modified protein abundance index; ER, endoplasmic reticulum; GM130, Golgi matrix 130; HSD17B11, 17-β-hydroxysteroid dehydrogenase type 11; HSP60, heat-shock protein 60 kDa; LC-MS/MS, liquid chromatography coupled with tandem MS; LDH, lactate dehydrogenase; LPCAT2, lysophosphatidylcholine acyltransferase 2; MALDI, matrix-assisted laser-desorption ionization; MGAT, monoacylglycerol acyltransferase; MGLL, monoacylglycerol lipase; TAG, triacylglycerol; MTTP, microsomal TAG transfer protein; NSDHL, NAD(P)-dependent steroid dehydrogenase-like; OA, oleic acid; PC, phosphatidylcholine; PDI, protein disulfide-isomerase; PFA, paraformaldehyde; PL, phospholipid; PLIN, perilipin; TOF, time-of-flight; TRL, TAG-rich lipoprotein; VLDL, very-low-density lipoprotein
Introduction
The intestinal absorption of dietary lipids is a highly specialized and complex process. In the lumen of the upper part of the small intestine, TAGs (triacylglycerols), which are the main dietary lipids, are hydrolysed by the pancreatic carboxyl ester hydrolase into fatty acids and 2-monoglycerides that are absorbed by enterocytes, which resynthesize TAG at their ER (endoplasmic reticulum) membrane. Newly synthesized TAGs are in part used for the assembly of the intestine-specific TRLs (TAG-rich lipoproteins), i.e. chylomicrons, in the secretory compartment. This occurs through the fusion of an ApoB (apolipoprotein B) molecule stabilized by lipidation via MTTP (microsomal TAG transfer protein), with a lipid droplet formed independently in the ER lumen. Other small exchangeable apolipoproteins, such as ApoA-IV, associate with chylomicrons (Iqbal and Hussain, 2009). Part of the newly synthesized TAG is stored as CLDs (cytosolic lipid droplets; Buschmann and Manke, 1981; Robertson et al., 2003). In enterocytes, it has been shown that a dynamic accumulation and depletion of TAG in CLDs occur during the process of fat absorption (Zhu et al., 2009). In human differentiated Caco-2 enterocytes, which produce TRL when supplied with lipid micelles (Chateau et al., 2005; Luchoomun and Hussain, 1999), we previously demonstrated that TAG stored as CLDs can be mobilized to contribute to TRL production (Chateau et al., 2005) and that this TAG partition can be modulated by nutrients, e.g. glucose (Pauquai et al., 2006) or polyphenols (Vidal et al., 2005). The cellular mechanisms responsible for the formation of CLDs and their mobilization to produce TRL, and thus the connection between storage and secretion of TAG, are still poorly understood.
On their surface, CLDs have a protein endowment that has been characterized in various mammalian cell types, including CHO K2 (Chinese-hamster ovary K2) cells, human squamous epithelial carcinoma cells (A431), 3T3-L1 adipocytes, mammary epithelial cells and hepatic cells (for a review, see Hodges and Wu, 2010). It includes members of the PLIN (perilipin) family (previously known as PAT family proteins), i.e. the structural proteins of CLDs, as well as proteins involved in many cellular functions, such as lipid metabolism, intracellular traffic or signalling. The proteome of CLDs emerges as variable depending on the cell type. For example, PLIN-1 is found specifically on the adipocyte lipid droplet, PLIN-5/OXPAT is expressed in cells that have a high capacity for fatty acid oxidation, such as cardiac muscle cells and PLIN-2/ADRP and PLIN-3/TIP47 are ubiquitous (for a review, see Wolins et al., 2006). Therefore it can be hypothesized that the repertoire of proteins associated with CLDs has a functional impact on the metabolism of a given differentiated cell type.
To date, PLIN-2/ADRP and PLIN-3/TIP47 are the only known proteins associated with CLDs in enterocytes (Lee et al., 2009). The objective of the present study was to characterize the proteome of enterocyte CLDs in order to determine whether intestinal specificities exist in these organelles and to gain insights into their possible role in the balance between TAG storage and lipoprotein production.
Results
Accumulation of CLDs in mouse jejunum and in Caco-2/TC7 enterocytes after lipid supply
As a first approach, we characterized the localization of neutral lipids in enterocytes of mouse intestinal villi as a function of time after olive oil gavage.
Morphological analysis showed that, in control mice, the jejunum enterocytes were mostly devoid of CLDs (Figure 1A–1C) and no lipids could be visualized within the Golgi apparatus (Figure 1D). By contrast, 1 h after olive oil gavage, the cytoplasm of jejunum enterocytes was full of lipid droplets (Figures 1E–1H), which were localized at the apical pole of enterocytes above the nucleus and were larger at the tip (Figure 1E) than in the lower part of the villi (Figure 1F). Electron microscopic analysis showed that the large lipid droplets were not surrounded by a membrane and were thus cytosolic (Figure 1H). Despite the huge amount of cytosolic TAG observed 1 h after gavage, none could be visualized 20 h later (Figure 1I–1K), highlighting the transient nature of this accumulation. At the same time, the Golgi apparatus clearly contained lipids, indicating active lipoprotein secretion (Figure 1L).
Figure 1. Time-dependent distribution of lipid droplets in mouse jejunum after an olive oil gavage.
At 4 h after food withdrawal, mice received or not (control) an olive oil bolus (150 μl) by gavage. Control mice were killed 5 h after food withdrawal (A–D) and mice that received a lipid bolus were killed 1 h (E–H) or 20 h (I–L) after gavage. Jejunum was processed for lipid staining by the imidazole-buffered osmium tetroxide method and examined by optical microscopy after Toluidine Blue staining (A, E, F, I; scale bar, 5 μm) or by electron microscopy (B–D, G, H, J–L; scale bar, 2 μm). CLDs are indicated by arrows at the optical microscopy level and labelled as CLDs at the electron microscopy level. Red asterisks label the Golgi apparatus and N indicates the nuclei.
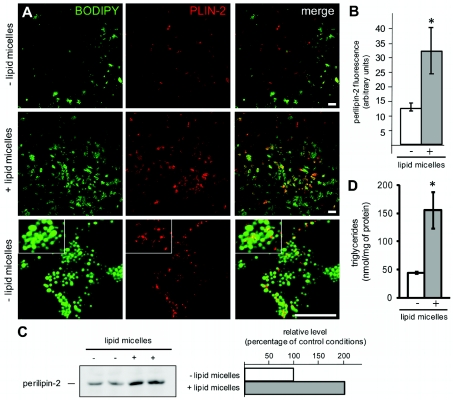
We have previously shown that human Caco-2/TC7 enterocytes, a clone that derives from the parental Caco-2 cell line, is able to produce TRL and store TAG as CLDs when supplied with lipid micelles (Chateau et al., 2005; Vidal et al., 2005). After a 24 h incubation with lipid micelles, fatty acid uptake and TAG synthesis are completed, TAG are almost entirely stored as CLDs and, although they are mobilized, the lipoprotein secretion rate is low, as shown in our previous studies (Chateau et al., 2005; Pauquai et al., 2006). Under these conditions, confocal microscopy analysis revealed that the lipid droplets, visualized with Bodipy 493/503, were mainly localized at the basal pole of the cells (see Supplementary Figures S1A and S1B at http://www.biolcell.org/boc/103/boc1030499add.htm) and were decorated with PLIN-2 in discrete patches (Figure 2A and Supplementary Figure S1A). CLDs appeared as clusters of various sizes (Figure 2A, bottom panel), which were larger than and more numerous after the lipid supply as compared with control conditions (Figure 2A, compare the middle and top panels). PLIN-2 labelling increased as well in lipid-loaded cells compared with control cells (Figure 2A). This was confirmed by quantification of the mean fluorescence of PLIN-2 labelling (Figure 2B) and by immunoblots of cell lysates probed for PLIN-2 (Figure 2C). Finally, the increased level of intracellular TAG content in Caco-2/TC7 cells after lipid supply was confirmed by biochemical quantification (Figure 2D).
Figure 2. TAG and PLIN-2 content in Caco-2/TC7 cells after incubation with lipid micelles for 24 h.
Caco-2/TC7 cells were cultured for 3 weeks on semi-permeable filters for differentiation and then supplied (+) or not (−; control conditions) with lipid micelles for 24 h. (A) Cells were stained with Bodipy 493/503 (green) and labelled for PLIN-2 (red; scale bars, 10 μm). Lower panels show higher magnifications and the insets display magnified areas. Acquisitions were made from the basal pole of the cells, where most of the lipid droplets are localized (see also Supplementary Figures S1A and S1B). (B) PLIN-2-associated fluorescence was quantified from (A) and expressed in arbitrary units.*P<0.05 compared with control cells. (C) Immunoblotting of PLIN-2 in cell lysates (40 μg of protein) and quantification. Results are expressed as percentage of control. (D) TAG content in cell lysates. Results shown are the means±S.E.M. for five independent experiments performed at least in duplicate.*P<0.05 when compared with control cells.
Isolation and characterization of the CLDs from differentiated Caco-2/TC7 enterocytes
Isolation of lipid droplets from the jejunum by sucrose-gradient centrifugation was unsuccessful, mainly because the small intestine is a tissue with a complex architecture, containing many different cell types, and because the enterocytes of the isolated epithelial cell layer were very resistant to nitrogen disruption, the method of choice for cell lysis while keeping intact the lipid droplets. We thus used differentiated Caco-2/TC7 cells for our future studies. Moreover, since rinsing the top fraction leads to a loss of lipid droplet-associated proteins, including PLIN-2, as described by others (Bulankina et al., 2009), we isolated lipid droplets from Caco-2/TC7 cells by a single flotation step of the 1000 g cell homogenate supernatant through a sucrose gradient.
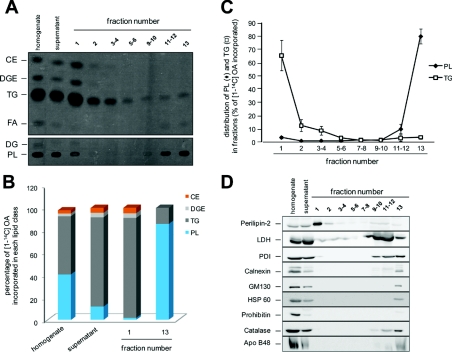
The different fractions recovered from the sucrose gradient were characterized for lipids and proteins (Figure 3). To evaluate the degree of enrichment of TAG, lipid droplet fractionation was performed from differentiated Caco-2/TC7 cells incubated for 24 h with lipid micelles supplemented with [1-14C]OA (oleic acid). Lipids from all gradient fractions as well as from the homogenate and the supernatant were extracted and lipid classes were separated by TLC. As expected, TAG were mainly recovered in fraction 1, the lowest-density fraction, and PL (phospholipids) in fraction 13, which contains cell membranes (Figure 3A). Along with TAG, fraction 1 was also enriched in DGE (diacylglyceryl ether) and CE (cholesterol ester), both neutral and highly hydrophobic lipids, which are thus probably bathed in the TAG core of the lipid droplets. DGE is a TAG analogue formed from batyl alcohol, a stable ether analogue of 2-monoacylglycerol that we use to evaluate the contribution of the MGAT (monoacylglycerol acyltransferase) pathway to TAG synthesis (Pauquai et al., 2006). The percentage of [1-14C]OA incorporated into TAG increased from 53.4±3% in the cell homogenate to 80.2±0.6% in the supernatant loaded on the gradient and to 89.6±0.6% in fraction 1 (Figure 3B; see Supplementary Table S1 at http://www.biolcell.org/boc/103/boc1030499add.htm). Interestingly, compared with the PL/TAG ratio, the DGE/TAG and the CE/TAG ratios remained remarkably constant in these samples, indicating a co-enrichment of these hydrophobic lipids with TAG, the major component of the lipid droplet core (Supplementary Table S1). Conversely, the percentage of [1-14C]OA incorporated into PL decreased from 41.1±3.2% in the cell homogenate to 11.4±0.8% in the supernatant and to 1.3±0.1% in fraction 1. Overall, in fraction 1, the [1-14C]OA incorporated into neutral lipids (TAG+DGE+CE) accounted for more than 98% of the total radioactivity incorporated and only for less than 2% in PL, as described earlier (Bartz et al., 2007). Finally, 65.5±11.5% of the TAG and 3.6±1% of the PL loaded on the sucrose gradient were recovered in the top fraction (Figure 3C). As expected, PLIN-2 was mainly recovered in fraction 1 (Figure 3D). Overall, the lowest density fraction 1 was highly enriched in TAG and PLIN-2, two markers of lipid droplets.
Figure 3. Lipid and protein analysis of sucrose gradient fractions prepared from Caco-2/TC7 cells.
Caco-2/TC7 cells were cultured on semi-permeable filters for 3 weeks for differentiation and then supplied with lipid micelles for 24 h. For lipid analysis, lipid micelles were supplemented with [1-14C]OA. Cell homogenates were centrifuged for 10 min at 1000 g and the supernatant was fractionated on a sucrose gradient. The top to bottom fractions (1–13) were analysed for lipids and proteins. (A) Autoradiography of a representative TLC of radiolabelled lipids extracted from cell homogenates, supernatants and each fraction of the sucrose gradient (FA, fatty acids; DG, diacylglycerol). (B) The radioactive bands from TLC were excised and the radioactivity was quantified by scintillation counting to evaluate the incorporation of [1-14C]OA into lipids. Results were expressed for each fraction as percentage of total [1-14C]OA incorporated into each lipid class. (C) Distribution of PL (◆) and TAG (□) in the sucrose gradient fractions. Results are expressed as percentage of total [1-14C]OA incorporated. (D) Immunoblotting of PLIN-2 (lipid droplet marker), LDH (cytosolic marker), PDI and calnexin (ER markers), GM130 (Golgi matrix protein marker), HSP60 and prohibitin (mitochondrial markers) and catalase (peroxisome marker). Immunoblotting of ApoB48 was performed to detect any contamination of the lipid droplet fraction with TRLs. Equal volumes of homogenate and supernatant were loaded on gels to evaluate the recovery/loss of material after the 1000 g centrifugation step. The same percentage of each fraction of the sucrose gradient was loaded on gels, except for fractions 1 and 2, which were 2-fold loaded in order to evaluate the presence of organelle markers with a greater sensitivity.
A set of organelle markers was analysed by Western blotting to evaluate the relative purity of the isolated CLDs (Figure 3D). The lipid-droplet-containing fraction was slightly positive for LDH (lactate dehydrogenase), an abundant cytosolic enzyme, and for PDI (protein disulfide-isomerase), a luminal ER protein that has been frequently identified in CLD fraction (Hodges and Wu, 2010). Markers of other compartments, such as calnexin (ER), GM130 (Golgi matrix 130; Golgi apparatus), HSP60 (heat-shock protein of 60 kDa) and prohibitin (mitochondria) and catalase (peroxisomes), were not detected in fraction 1, although they were easily visualized in the bottom fractions. Finally, we analysed the fractions for the presence of ApoB48, which is the non-exchangeable TRL-associated intestinal form of human ApoB. ApoB48 was not detected in the top fraction, while it was indeed detected in the membrane-containing bottom fraction (fraction 13), indicating that CLDs were not contaminated with TRL. Overall, although we cannot rule out a slight contamination with cytosolic proteins, our results indicate that PLIN-2- and TAG-enriched fraction 1 was devoid of other organelles. We thus undertook the identification of the CLD-associated proteins through a proteomic approach.
Identification of proteins associated with CLDs isolated from Caco-2/TC7 cells, by LC-MS/MS (liquid chromatography coupled with tandem MS)
In initial experiments, proteins were separated by SDS/10% PAGE and the bands were excised for analysis by LC-MS/MS after in-gel trypsin digestion. The proteins that were identified had the expected molecular mass, indicating that no proteolysis occurred during the CLD purification process. Due to the high sensitivity and resolution of LC-MS/MS, a method that can identify more than 100 proteins from a gel slice, identification of peptides by LC-MS/MS was performed from unfractionated protein samples run on a stacking gel and subjected to in-gel trypsin digestion. This proteomic approach allowed the identification of 105 proteins (Table 1). The normalized emPAI (exponentially modified protein abundance index) of the identified proteins ranged from 6.97 to 0.01. The emPAI formula is based on the number of observed peptides normalized by the theoretical number of observable peptides per protein and offers an estimation of their relative abundance within a mixture (Ishihama et al., 2005; Shinoda et al., 2010). In other words, a 10-fold difference in emPAI between two proteins indicates an approximately 10-fold difference in their amount. As expected, the most abundant protein in CLDs was PLIN-2 (emPAI of 6.97).
Table 1. List of identified (by LC-MS/MS) proteins associated with cytosolic lipid droplets isolated from Caco-2/TC7 cells.
Caco-2/TC7 cells were cultured on semi-permeable filters for 3 weeks for differentiation and then supplied with lipid micelles for 24 h. Cell homogenates were centrifuged for 10 min at 1000 g and the supernatant was fractionated on a sucrose gradient. The top fraction, containing the cytosolic lipid droplets, was analysed by LC-MS/MS for protein identification. Proteins were identified with at least two unique peptides. A Mascot search was performed allowing 1 ‘max. missed cleavage’, 20 p.p.m. ‘peptides mass tolerance’, 0.3 ‘fragment mass tolerance’ and against Homo sapiens taxonomy (20401 sequences). Mascot search results were filtered by P=0.01 and false discovery rate below 1.6%. The proteins were identified in at least two out of three independent experiments. The proteins already reported in mammalian CLD proteome are indicated (a, reported in the recent review of Hodges and Wu, 2010; b, Moessinger et al., 2011). Proteins are grouped by function or localization and, inside a group, proteins are ranked by decreasing emPAI.
| Gene name | Entry name SwissProt | Protein name | Normalized emPAI | Reported in proteomic studies of lipid droplets |
|---|---|---|---|---|
| Lipid metabolism | ||||
| PLIN proteins | ||||
| PLIN2 | PLIN2_HUMAN | Perilipin-2, adipose differentiation-related protein, adipophilin | 6.97 | a |
| PLIN3 | PLIN3_HUMAN | Perilipin-3, mannose-6-phosphate receptor-binding protein, TIP47 | 3.42 | a |
| Fatty acid metabolism | ||||
| ACSL3 | ACSL3_HUMAN | Long-chain-fatty-acid-CoA ligase 3 | 2.65 | a |
| ACSL4 | ACSL4_HUMAN | Long-chain-fatty-acid-CoA ligase 4 | 0.15 | a |
| Acylglycerol metabolism | ||||
| MGLL | MGLL_HUMAN | Monoglyceride lipase | 0.67 | a |
| ABHD5 | ABHD5_HUMAN | 1-Acylglycerol-3-phosphate O-acyltransferase, abhydrolase domain-containing protein 5, CGI-58 | 0.31 | a |
| PNPLA2 | PLPL2_HUMAN | Patatin-like phospholipase domain-containing protein 2, adipose triacylglycerol lipase | 0.15 | a |
| Phospholipid metabolism | ||||
| PCYT1A | PCY1A_HUMAN | Choline-phosphate cytidylyltransferase A | 1.58 | |
| LPCAT2 | PCAT2_HUMAN | Lysophosphatidylcholine acyltransferase 2 | 0.30 | b |
| Sterol metabolism | ||||
| HSD17B11 | DHB11_HUMAN | Oestradiol 17-β-dehydrogenase 11 | 2.78 | a |
| LSS | ERG7_HUMAN | Lanosterol synthase | 2.46 | a |
| CYB5R3 | NB5R3_HUMAN | NADH–cytochrome b5 reductase 3 | 1.63 | a |
| NSDHL | NSDHL_HUMAN | Sterol-4-α-carboxylate 3-dehydrogenase, decarboxylating | 1.55 | a |
| EPHX1 | HYEP_HUMAN | Epoxide hydrolase 1 | 0.31 | |
| HSD3B1 | 3BHS1_HUMAN | 3-β-Hydroxysteroid dehydrogenase | 0.24 | |
| DHRS3 | DHRS3_HUMAN | Short-chain dehydrogenase/reductase 3 | 0.13 | a |
| DHCR7 | DHCR7_HUMAN | 7-Dehydrocholesterol reductase | 0.06 | |
| HSD17B7 | DHB7_HUMAN | 3-Oxo-steroid reductase | 0.04 | a |
| Lipoprotein metabolism | ||||
| APOA4 | APOA4_HUMAN | Apolipoprotein A-IV | 6.24 | |
| P4HB | PDIA1_HUMAN | Protein disulfide-isomerase | 4.71 | a |
| MTTP | MTP_HUMAN | Microsomal triacylglycerol transfer protein large subunit | 0.53 | |
| APOE | APOE_HUMAN | Apolipoprotein E | 0.48 | |
| Other lipidic metabolism | ||||
| FAF2 | FAF2_HUMAN | FAS-associated factor 2, UBXD8 | 1.77 | a |
| RDH10 | RDH10_HUMAN | Retinol dehydrogenase 10 | 1.24 | |
| DHRS1 | DHRS1_HUMAN | Dehydrogenase/reductase SDR family member 1 | 0.56 | a |
| ECHS1 | ECHM_HUMAN | Enoyl-CoA hydratase, mitochondrial | 0.17 | |
| SGPL1 | SGPL1_HUMAN | Sphingosine-1-phosphate lyase 1 | 0.11 | |
| TRAFFIC | ||||
| RAB7A | RAB7A_HUMAN | Ras-related protein Rab-7a | 2.78 | a |
| RAB1A | RAB1A_HUMAN | Ras-related protein Rab-1A | 1.75 | a |
| RAB5C | RAB5C_HUMAN | Ras-related protein Rab-5C | 1.66 | a |
| RAB10 | RAB10_HUMAN | Ras-related protein Rab-10 | 1.15 | a |
| RAB25 | RAB25_HUMAN | Ras-related protein Rab-25 | 1.02 | |
| RAB11A | RB11A_HUMAN | Ras-related protein Rab-11A | 0.68 | a |
| RAB15 | RAB15_HUMAN | Ras-related protein Rab-15 | 0.66 | |
| RAB6A | RAB6A_HUMAN | Ras-related protein Rab-6A | 0.20 | a |
| RAB33B | RB33B_HUMAN | Ras-related protein Rab-33B | 0.01 | a |
| ER (endoplasmic reticulum) | ||||
| ER lumen | ||||
| HSPA5 | GRP78_HUMAN | 78 kDa glucose-regulated protein | 5.76 | a |
| PDIA3 | PDIA3_HUMAN | Protein disulfide-isomerase A3 | 3.27 | a |
| CALR | CALR_HUMAN | Calreticulin | 1.97 | a |
| HSP90B | ENPL_HUMAN | Endoplasmin, heat-shock protein 90 kDa β member 1, 94 kDa glucose-regulated protein | 1.85 | a |
| PDIA6 | PDIA6_HUMAN | Protein disulfide-isomerase A6 | 1.52 | a |
| ERO1L | ERO1A_HUMAN | ERO1-like protein α | 0.82 | a |
| PDIA4 | PDIA4_HUMAN | Protein disulfide-isomerase A4 | 0.81 | a |
| SERPINH | SERPH_HUMAN | Serpin H1 | 0.38 | |
| ERP44 | ERP44_HUMAN | Endoplasmic reticulum resident protein ERp44 | 0.29 | |
| CALU | CALU_HUMAN | Calumenin | 0.28 | |
| POR | NCPR_HUMAN | NADPH–cytochrome P450 reductase | 0.27 | |
| PLOD2 | PLOD2_HUMAN | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 | 0.19 | |
| UBXN4 | UBXN4_HUMAN | UBX domain-containing protein 4, UBX domain-containing protein 2, Erasin | 0.19 | a |
| ERP29 | ERP29_HUMAN | Endoplasmic reticulum protein ERp29 | 0.19 | a |
| LRPAP1 | AMRP_HUMAN | α-2-Macroglobulin receptor-associated protein, low-density lipoprotein receptor-related protein-associated protein 1 | 0.17 | |
| ER membrane, integral protein | ||||
| CANX | CALX_HUMAN | Calnexin | 0.66 | a |
| AUP1 | AUP1_HUMAN | Ancient ubiquitous protein 1 | 0.35 | a |
| RPN1 | RPN1_HUMAN | Dolichyl-diphospho-oligosaccharide–protein glycosyltransferase subunit 1 | 0.34 | a |
| UGT1A6 | UD16_HUMAN | UDP-glucuronosyltransferase 1-6 | 0.22 | |
| DDOST | OST48_HUMAN | Dolichyl-diphospho-oligosaccharide–protein glycosyltranferase 48 kDa subunit | 0.18 | a |
| RTN4 | RTN4_HUMAN | Reticulon-4 | 0.08 | |
| LRRC59 | LRC59_HUMAN | Leucine-rich repeat-containing protein 59 | 0.17 | |
| Unknown | ||||
| PRKCSH | GLU2B_HUMAN | Glucosidase 2 subunit β | 0.13 | |
| Mitochondria | ||||
| AIFM2 | AIFM2_HUMAN | Apoptosis-inducing factor 2, apoptosis-inducing factor-like mitochondrion-associated inducer of death | 1.87 | a |
| ATP5B | ATPB_HUMAN | ATP synthase subunit β, mitochondrial | 0.51 | |
| HSPA9 | GRP75_HUMAN | Stress-70 protein, mitochondrial | 0.24 | a |
| MDH2 | MDHM_HUMAN | Malate dehydrogenase, mitochondrial | 0.20 | |
| HSPD1 | CH60_HUMAN | 60 kDa heat-shock protein, mitochondrial | 0.18 | a |
| Lysosome | ||||
| PCYOX1 | PCYOX_HUMAN | Prenylcysteine oxidase 1 | 0.30 | |
| GNS | GNS_HUMAN | N-Acetylglucosamine-6-sulfatase | 0.20 | |
| Cytosol | ||||
| cytoskeleton | ||||
| TUBB2C | TBB2C_HUMAN | Tubulin β-2C chain | 6.04 | |
| TUBB | TBB5_HUMAN | Tubulin β chain | 5.39 | |
| TUBB2A | TBB2A_HUMAN | Tubulin β-2A chain | 3.43 | |
| ACTB | ACTB_HUMAN | Actin, cytoplasmic 1 | 2.70 | a |
| TUBA1B | TBA1B_HUMAN | Tubulin α-1B chain | 2.68 | |
| ACTN4 | ACTN4_HUMAN | α-Actinin-4 | 1.02 | |
| ANXA2 | ANXA2_HUMAN | Annexin A2 | 0.86 | a |
| ANXA4 | ANXA4_HUMAN | Annexin A4 | 0.35 | |
| ELMOD2 | ELMD2_HUMAN | ELMO domain-containing protein 2 | 0.34 | a |
| SLC9A3R1 | NHRF1_HUMAN | Na+/H+ exchange regulatory cofactor NHERF1 | 0.27 | |
| SEPT2 | SEPT2_HUMAN | Septin-2 | 0.17 | |
| Carbohydrate metabolism | ||||
| TPI1 | TPIS_HUMAN | Triosephosphate isomerase | 6.85 | |
| PGK1 | PGK1_HUMAN | Phosphoglycerate kinase 1 | 1.67 | |
| ENO1 | ENOA_HUMAN | α-Enolase | 1.47 | |
| ALDOA | ALDOA_HUMAN | Fructose-bisphosphate aldolase A | 1.40 | |
| GPI | G6PI_HUMAN | Glucose-6-phosphate isomerase | 1.08 | |
| GAPDH | G3P_HUMAN | Glyceraldehyde-3-phosphate dehydrogenase | 0.76 | a |
| Cytosolic chaperones | ||||
| HSPA8 | HSP7C_HUMAN | Heat-shock cognate 71 kDa protein | 1.89 | a |
| HSPA1A | HSP71_HUMAN | Heat-shock 70 kDa protein 1 | 1.78 | a |
| HSPA6 | HSP76_HUMAN | Heat-shock 70 kDa protein 6 | 0.52 | |
| HSP90AB1 | HS90B_HUMAN | Heat-shock protein HSP 90-β | 0.20 | a |
| Other function in cytoplasm | ||||
| CKB | KCRB_HUMAN | Creatine kinase B-type | 5.18 | |
| LGALS3 | LEG3_HUMAN | Galectin-3 | 1.62 | |
| GSTP1 | GSTP1_HUMAN | Glutathione transferase P | 0.57 | |
| GSTA1 | GSTA1_HUMAN | Glutathione transferase A1 | 0.44 | |
| NDRG1 | NDRG1_HUMAN | Protein NDRG1 | 0.26 | |
| TRIO | TRIO_HUMAN | Triple functional domain protein | 0.02 | |
| Miscellaneous | ||||
| METTL7B | MET7B_HUMAN | Methyltransferase-like protein 7B | 2.97 | a |
| METTL7A | MET7A_HUMAN | Methyltransferase-like protein 7A | 1.59 | a |
| SCCPDH | SCPDH_HUMAN | Probable saccharopine dehydrogenase | 1.56 | a |
| ATP1B1 P | AT1B1_HUMAN | Sodium/potassium-transporting ATPase subunit β-1 | 0.79 | |
| CDH17 | CAD17_HUMAN | Cadherin-17 | 0.68 | |
| GPA33 | GPA33_HUMAN | Cell-surface A33 antigen | 0.55 | |
| C2orf43 | CB043_HUMAN | UPF0554 protein C2 or f43 | 0.48 | a |
| GANAB | GANAB_HUMAN | Neutral α-glucosidase AB | 0.26 | |
| ATP1A1 | AT1A1_HUMAN | Sodium/potassium-transporting ATPase subunit α1 | 0.11 | |
| EEF1A1 | EF1A1_HUMAN | Elongation factor 1-α1 | 0.09 | a |
| KPNB1 | IMB1_HUMAN | Importin subunit β-1 | 0.07 | |
| CLIC1 | CLIC1_HUMAN | Chloride intracellular channel protein 1 | 0.01 |
The identified proteins were grouped by function or localization and were ranked by decreasing emPAI within a group. Remarkably, 27 proteins (approx. 25% of the total identified proteins) were directly involved in lipid metabolism, and only 17 out of the 27 proteins were already reported as associated with CLDs (Hodges and Wu, 2010). Proteins already reported to be present in various intracellular organelles (ER, mitochondria and lysosomes) were identified. Half of them have been identified in CLD fractions in a number of previous proteomic studies (for a review, see Hodges and Wu, 2010): for example, calnexin. Cytosolic proteins were identified, and most of them could be classified as cytoskeletal proteins, chaperones and enzymes involved in carbohydrate metabolism. Many of them had already been reported in proteome studies (Hodges and Wu, 2010). Some of them, such as tubulin, could indeed be contaminants because of their high cellular abundance. However, since lipid droplets are transported through the cell using microtubules (Spandl et al., 2009), tubulin detection in the lipid droplet fraction is not surprising.
In the lipid metabolism group, we identified PLIN family proteins, fatty acid-activating enzymes, lipolytic enzymes and two enzymes belonging to the late pathway of cholesterol biosynthesis, i.e. lanosterol synthase and NSDHL [NAD(P)-dependent steroid dehydrogenase-like; Table 1]. Among the PLIN proteins, PLIN-2/ADRP and PLIN-3/TIP47 were abundantly represented (emPAI of 6.97 and 3.42 respectively) and no other member of this family was identified. Fatty-acid-activating enzymes, which are necessary to commit fatty acids to a metabolic process, were identified and ACSL3 (acyl-CoA synthetase long-chain 3) was the main form associated with the CLD fraction. Lipolytic enzymes involved in the hydrolysis of TAG were also present. In particular, PNPLA2 (patatin-like phospholipase domain containing 2), also known as ATGL (adipose TAG lipase), which hydrolyses TAG to diacylglycerol, as well as its cofactor CGI-58, also known as ABHD5 (α/β-hydrolase domain-containing protein 5), were recovered in the CLD fraction.
We detected 11 proteins involved in lipid metabolism that were not identified previously in published CLD proteomes. These proteins are involved in the PL, sterol or lipoprotein metabolism (Table 1). For example, PCYT1A [also known as CCT-α (choline-phosphate cytidylyltransferase A)], the enzyme that catalyses the rate-limiting step in the choline pathway for PC (phosphatidylcholine) synthesis (Vance, 2008), and LPCAT2 (lysophosphatidylcholine acyltransferase 2), which re-acylates lysophospholipids by using acyl-CoA, were present in the lipid droplet fraction.
Proteins involved in the metabolism of lipoprotein were also identified in the CLD fraction of Caco-2/TC7 enterocytes. MTTP and its subunit, PDI A1, were detected as well as the exchangeable ApoA-IV. By contrast, ApoB, the constitutive non-exchangeable apolipoprotein of TRL, was not identified in the proteome. In Caco-2/TC7 cells, after a 24 h incubation with lipid micelles, lipid droplets are mainly localized at the basal pole of cells, but some of them are localized in the supranuclear region (Supplementary Figures S1A and S1B and Chateau et al., 2005). ApoB is restricted to the apical region of the cell (Supplementary Figures S1A and S1B and Morel et al., 2004) and, in this area, as we show here, lipid droplets were surrounded by PLIN-2 but devoid of ApoB (Supplementary Figure S1C). As expected, ApoB colocalized with the ER marker calnexin, and, in the perinuclear region, Bodipy-labelled lipid droplets could also be found in close vicinity of calnexin (Supplementary Figure S1D). However, ApoB was never detected in these ER-neighbouring lipid droplets.
LPCAT2 and 3BHS1 (3-β-hydroxysteroid dehydrogenase 1) expression and localization in Caco-2/TC7 cells
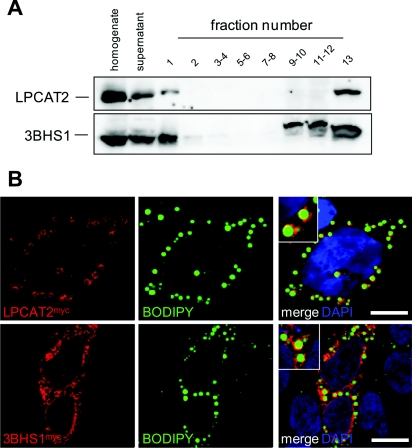
As mentioned above, some proteins involved in lipid metabolism were identified for the first time in a CLD proteome. We analysed the expression and the localization of LPCAT2, an enzyme involved in the PC metabolism, and of 3BHS1, an enzyme involved in the metabolism of steroids, because they were so far known to localize to the ER membrane (Agarwal and Garg, 2010; Morimoto et al., 2010; Wang et al., 2007) and have not been described yet in enterocytes. Western blot analysis of the sucrose gradient fractions showed that LPCAT2 and 3BHS1 were indeed mainly associated with the membrane fraction (fraction 13). However, both proteins were also detected in the lipid-droplet-containing fraction 1 (Figure 4A). Caco-2/TC7 cells were transfected with plasmids encoding myc-tagged LPCAT2 or 3BHS1, and incubated with fatty acids to induce lipid droplet formation. Confocal microscopy analysis clearly showed that LPCAT2 as well as 3BHS1 localized mainly around lipid droplets (Figure 4B).
Figure 4. Analysis of LPCAT2 and 3BHS1 distribution in Caco-2/TC7 cells.
(A) Western-blot analysis of LPCAT2 and 3BHS1 distribution along sucrose gradient fractions prepared from differentiated Caco-2/TC7 cells, as described in Figure 3. (B) Confocal microscopy analysis of Caco-2/TC7 cells transfected with LPCAT2myc (red, upper panel) or 3BHS1myc (red, lower panel) and incubated with 0.6 mM OA. Lipid droplets were stained with Bodipy (green). Insets show magnified areas of each panel. Scale bars, 10 μm.
ApoA-IV expression and localization in Caco-2/TC7 cells and in mouse jejunum
As ApoA-IV is specifically expressed by enterocytes (Green et al., 1980) and strongly up-regulated by fatty acids (Carriere et al., 2005; Lu et al., 2002), we further investigated its expression and localization.
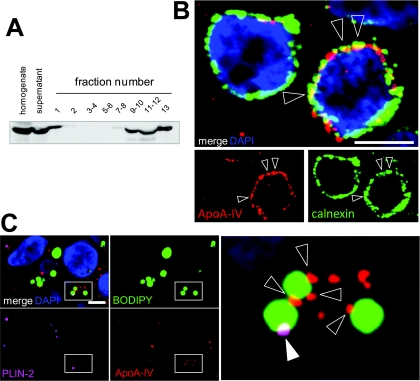
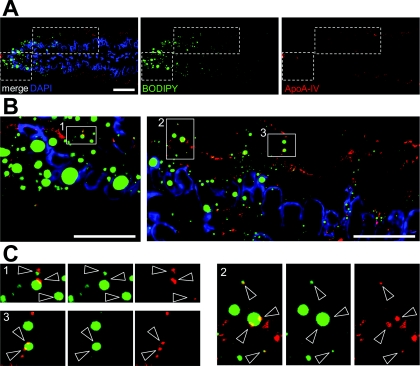
Western blot analysis from differentiated Caco-2/TC7 cells supplied with lipid micelles for 24 h confirmed the presence of ApoA-IV in the lipid-droplet-containing fraction of the gradient (fraction 1, Figure 5A). Quantification indicated that ApoA-IV in fraction 1 was below 10% of the ApoA-IV loaded on the sucrose gradient (results not shown). Since ApoA-IV is a secreted protein, it was expected to be associated with the secretory pathway organelles, i.e. ER and Golgi apparatus (Black, 2007; Gallagher et al., 2004; Green et al., 1980) and, by immunofluorescence analysis, ApoA-IV indeed co-localized partially with calnexin, an ER marker (Figure 5B). However, ApoA-IV was also detected on Bodipy-labelled CLDs (Figure 5C, empty arrowheads). Additionally, a triple labelling showed that some CLDs were positive for ApoA-IV and PLIN-2 (Figure 5C, empty and white arrowheads, respectively).
Figure 5. Intracellular distribution of ApoA-IV in Caco-2/TC7 cells.
Cells were cultured on semi-permeable filters for 3 weeks and then supplied with lipid micelles for 24 h. (A) Western-blot analysis of ApoA-IV distribution in sucrose gradient fractions prepared from differentiated Caco-2/TC7 cells as described in Figure 3. (B) Cells were labelled for ApoA-IV (red) and calnexin (green). Empty arrowheads indicate co-localizations. Nuclei were stained with DAPI (blue). Scale bar, 10 μm. (C) Cells were labelled for ApoA-IV (red), PLIN-2 (fuchsia), neutral lipids (green) and nuclei (blue). Scale bar, 5 μm. The boxed area in the merged Figure is shown magnified on the right. White and empty arrowheads indicate PLIN-2 and ApoA-IV labelling respectively at the surface of lipid droplets.
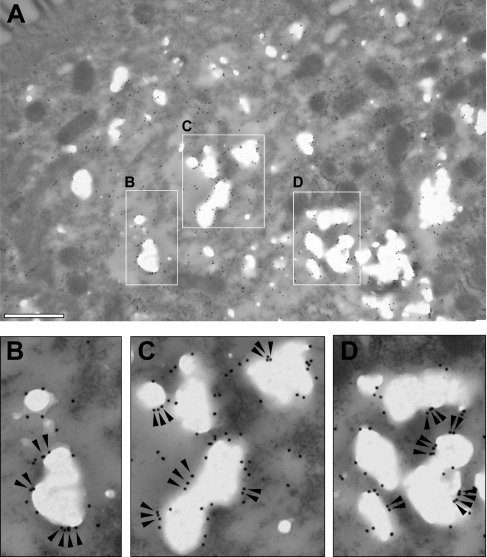
Immunoelectron microscopy analysis of ApoA-IV was therefore undertaken. As expected, immunogold labelling was widely distributed (Figure 6A), since ApoA-IV is present all along the secretory pathway, which is well developed in differentiated Caco-2/TC7 cells. Organelles of the secretory compartment (ER and Golgi apparatus) are located at the apical pole of the cells (see the ApoB distribution in the three-dimensional reconstruction on Supplementary Figure S1). Interestingly, in this area, the edges of CLDs were labelled with clusters of ApoA-IV-gold particles (Figures 6B–6D, arrowheads). Overall, our results indicate that a fraction of ApoA-IV is at the immediate vicinity of CLDs and remains associated with them after purification by density-gradient ultracentrifugation.
Figure 6. Immunoelectron microscopy analysis of ApoA-IV localization in Caco-2/TC7 cells.
Cells were cultured on semi-permeable filters for 3 weeks and then supplied with lipid micelles for 24 h. After fixation, cells were embedded in LR White resin and ultrathin sections were incubated with anti-ApoA-IV antibodies followed by 18 nm gold particle-labelled secondary antibody. (A) Low magnification of the apical part of a cell (note the cell microvilli at the top left corner of the Figure). The lipid droplets appear as electron-lucent structures. Scale bar, 500 nm. The boxed areas in (A) are shown enlarged in (B–D). Note that the gold particles gather in small clusters around the edges of the lipid droplets (arrowheads).
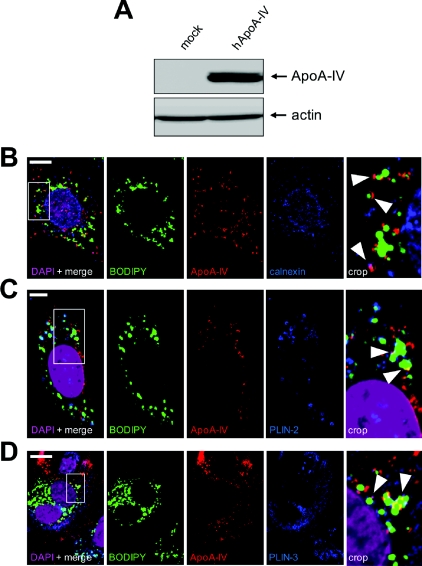
In order to estimate in vivo the relevance of these results, we analysed ApoA-IV localization in jejunum of mice fed on a high-fat diet for 2 days, stimulating lipid droplet formation (Figure 7A). As in Caco-2/TC7 cells, some ApoA-IV labelling could be detected at the close vicinity of lipid droplets in jejunum villi (Figures 7B and 7C, empty arrows). Finally, a plasmid encoding human ApoA-IV was transiently transfected in HeLa cells, which neither express ApoA-IV endogenously (Figure 8A) nor secrete TRL. Confocal microscopy showed that, interestingly, ApoA-IV associates with lipid droplets and partially co-localized with the dotty-pattern ER-marker calnexin (Figure 8B) as well as with the lipid droplet-canonical markers PLIN-2 (Figure 8C) and PLIN-3 (Figure 8D). Altogether, these data clearly indicate that ectopic expression of ApoA-IV in HeLa cells lead to its partial targeting to lipid droplets, as observed in Caco-2/TC7 enterocytes (Figures 5 and 6) and mouse intestinal cells (Figure 7).
Figure 7. Distribution of ApoA-IV in mouse jejunum.
Cryostat sections of mouse jejunum were stained for ApoA-IV (red), neutral lipids (green) and nuclei (blue). (A) Low magnification showing an entire jejunum villus. Scale bar, 20 μm. (B) Magnified view of the boxed areas in (A). Scale bar, 20 μm. (C) Magnified view of the boxed areas 1–3 in (B). Empty arrowheads show ApoA-IV labelling at the surface of lipid droplets.
Figure 8. ApoA-IV subcellular localization in HeLa cells.
HeLa cells were transfected (hApoA-IV) or not (mock) and processed for lysis and immunofluorescence analysis. (A) Immunoblotting of ApoA-IV and actin in mock or hApoA-IV transfected cells. (B–D) Cells were fixed and labelled with DAPI (violet), Bodipy (green), anti-ApoA-IV antibody (red) and indicated antibodies (blue): anti-calnexin (B), anti-PLIN-2 (C) and anti-PLIN-3 (D). Right panels show magnified areas of each acquisition and arrowheads indicate co-localizations of ApoA-IV with lipid droplets. Scale bar, 10 μm.
Discussion
We report here the first proteome analysis of CLDs isolated from differentiated Caco-2/TC7 enterocytes. Observed in enterocytes in vivo (Buschmann and Manke, 1981) and in cell culture models (Chateau et al., 2005), lipid droplets have only been recently suggested as potential players in the physiology of lipid absorption in the gastrointestinal tract (Pauquai et al., 2006; Zhu et al., 2009). The proteome of differentiated Caco-2/TC7 enterocyte CLDs, which were formed after a supply of dietary lipids, revealed several characteristics. For example, among the PLIN protein family, we identified PLIN-2/ADRP and PLIN-3/TIP47, but no PLIN-1, expressed mainly by adipocytes, or PLIN-5/OXPAT, expressed in cells that have a high capacity of fatty acid oxidation. This finding is in good agreement with recent data from mouse intestine (Lee et al., 2009). Approximately 25% of the proteins identified in the Caco-2/TC7 enterocyte CLD fraction were involved in lipid metabolism, and were mainly linked to the metabolism of TAGs, PLs, sterols and to the assembly of TRLs (Table 1). Interestingly, among these 27 proteins, 11 were newly identified proteins associated with CLDs.
The proteome included proteins necessary for TAG hydrolysis, supporting a potential dynamic role of CLDs in the control of TRL production in enterocytes. PNPLA2, which hydrolyse TAGs to diacylglycerol, and its co-activator ABHD5 were present in the proteome. ABHD5/CGI-58 was shown to facilitate the mobilization of cytoplasmic TAG and the assembly and secretion of TRL in McA RH7777 rat hepatoma cells and in HepG2 hepatic cells (Brown et al., 2007; Caviglia et al., 2009) and could play a similar role in enterocytes. Another hydrolytic enzyme, MGLL (monoacylglycerol lipase), was abundantly represented in Caco-2/TC7 enterocyte CLDs. Its function remains obscure, even though it is strongly increased in intestine of mice upon high fat feeding (Chon et al., 2007). In this study, lipid droplets were isolated from Caco-2/TC7 cells that had been incubated for 24 h with lipid micelles, thus in a state of CLD-associated TAG mobilization (Chateau et al., 2005). Our proteome results are thus in accordance with the metabolic state of the Caco-2/TC7 cells. It is of interest to note that we did not find proteins involved in the TAG biosynthetic pathway, although DGAT2 (diacylglycerol acyltransferase 2) has been previously associated with lipid droplets during TAG synthesis in fibroblasts and adipocytes (Kuerschner et al., 2008). In differentiated Caco-2 cells, the acyltransferase activities are attributed mainly to DGAT1, which was not found in our proteome analysis of lipid droplets. DGAT1 belongs to a protein family different from that of DGAT2 (Cheng et al., 2008), but DGAT2 and MGATs belong to the same protein family. In Caco-2 cells, it is known that the MGAT pathway, the main TAG synthesis pathway during lipid absorption in enterocytes, is not very active (Levy et al., 1995). It would be interesting to determine whether MGATs can localize to lipid droplets in enterocytes and whether the localization of MGLL and MGAT on the lipid droplet surface can vary depending on the metabolic state of these cells.
The protein repertoire of CLDs included ACSL3 and ACSL4, which are responsible for the activation of fatty acids prior to their entry into metabolic pathways, such as de novo lipid synthesis, fatty acid catabolism or remodelling of membranes. In human Huh7 hepatocytes, ACSL activity has been involved in local synthesis of neutral lipids in lipid droplets (Fujimoto et al., 2007) and, more recently, ACSL3, which is the most abundant ACSL in Caco-2/TC7 enterocyte lipid droplet proteome, has been shown to be specifically required for fatty acid incorporation into PC, an essential reaction for VLDL (very-low-density lipoprotein) assembly, the hepatic form of TRL (Yao and Ye, 2008). Although major differences exist, it is interesting to note that the complex and highly differentiated function of TRL assembly and secretion is shared by hepatocytes and enterocytes.
The proteome included also two enzymes involved in PC synthesis, the most abundant PL of the monolayer surrounding the TAG core of lipid droplets (Tauchi-Sato et al., 2002). PCYT1A, also known as CCT-α, catalyzes the rate-limiting step in the choline pathway for the synthesis of PC (Vance, 2008). A recent report highlighted the role of PL metabolism in the formation, utilization, size and number of lipid droplets (Guo et al., 2008). In Drosophila S2 cells, CCT1, the orthologue of mammalian CCT-α localizing in the nucleus, translocates to the lipid droplet surface upon oleate addition and its knockdown leads to fewer lipid droplets of larger size (Guo et al., 2008). Moreover, liver-specific CCT-α-knockout mice display impaired VLDL secretion in plasma (Jacobs et al., 2004). We also identified LPCAT2, which re-acylates lysophospholipids by using acyl-CoA, and confirmed its localization to lipid droplets in transfected Caco-2/TC7 cells. The lipid droplet localization of LPCAT2 was demonstrated very recently in A431 cells and Cos7 cells (Moessinger et al., 2011). In intestine, lysoPC is formed by hydrolysis of the dietary and biliary PC in the intestinal lumen to be absorbed by enterocytes. LPCAT2, which remodels PLs independently of de novo synthesis (Soupene et al., 2008), may provide an economical source of PC for membrane synthesis as well as for lipoprotein assembly in enterocytes (Vance, 2008).
Caco-2/TC7 enterocyte lipid droplets comprise enzymes involved in sterol and steroid metabolism, including NSDHL, one of the first enzymes identified on lipid droplets (Ohashi et al., 2003) and HSD17B11 (17-β-hydroxysteroid dehydrogenase type 11), which was shown to redistribute to nascent lipid droplets (Horiguchi et al., 2008). This enzyme was suggested to be involved in the metabolism of diet-derived or oxidized hydrophobic, potentially toxic molecules (Horiguchi et al., 2008). Such a detoxification function would be rather relevant in enterocytes, which are in contact with a large variety of xenobiotics. We also identified 3BHS1 and confirmed its localization to lipid droplets in Caco-2/TC7 cells. Previous studies described 3BHS1 as an ER-associated protein (Wang et al., 2007) and we show for the first time its localization to lipid droplets.
Interestingly, along with MTTP, which is required for lipoprotein production, we found that ApoA-IV is associated with the Caco-2/TC7 enterocyte CLD fraction. MTTP localizes along the secretory pathway in enterocytes (Levy et al., 2002) and was shown around lipid droplets in differentiated 3T3-L1 adipocytes (Swift et al., 2005). We demonstrated the partial localization of ApoA-IV at the surface of CLDs by confocal and immunoelectron microscopy. Recently, ApoA-V, another exchangeable apolipoprotein that is expressed exclusively in hepatocytes, has also been found associated with lipid droplets (Shu et al., 2010). ApoA-IV is strongly induced upon lipid supply (Carriere et al., 2005; Green et al., 1980; Lu et al., 2002) and has been proposed to play many functions in vivo, including regulation of food intake, gastrointestinal motility, lipoprotein formation and size control, as well as protection against lipid oxidation and atherosclerosis (Black, 2007; Lu et al., 2002; Qin and Tso, 2005; Yao et al. 2011). ApoA-IV associates with nascent chylomicrons at an early stage of their biogenesis in the ER and is then secreted on the surface of mature chylomicrons (Black, 2007; Gallagher et al., 2004; Green et al., 1980). How can this protein be associated with both CLDs and lipoproteins? The most widely accepted model for CLD biogenesis proposes that newly synthesized TAG first accumulate between the PL bilayer of the ER membrane to form a lipid droplet, which, after reaching a critical size, buds towards the cytosolic or the luminal side of the ER membrane (Murphy and Vance, 1999; van Meer, 2001). However, the proposed models evolved recently to account for the presence on CLDs of proteins that are normally localized in the ER lumen or at the ER membrane. There are increasing lines of evidence showing that lipid droplets interact with various organelles, including ER (Zehmer et al., 2009). The association of ER-associated proteins and of ApoA-IV with CLDs could occur through different mechanisms: (i) CLD production by vesicular budding or by bicelle formation from the ER membrane, leading to the presence of both membrane and luminal ER proteins (for reviews, see Ohsaki et al., 2009; Walther and Farese, 2008); (ii) CLD apposition to particular domains of the ER membrane (Robenek et al., 2006, 2009).
In conclusion, from our proteome analysis, Caco-2/TC7 enterocyte CLDs appear as complex organelles displaying tissue-specific characteristics. The presence of proteins necessary for TAG hydrolysis is consistent with the transient feature of lipid droplets in intestine and Caco-2/TC7 enterocytes. In enterocytes, the transient accumulation of CLDs could represent a way to control TRL secretion after a lipid-rich meal. Located to lipid droplets, we also identified LPCAT2, which may provide an economical source of PC for membrane synthesis as well as for lipoprotein assembly in enterocytes. Moreover, the presence of ApoA-IV, an enterocyte-specific exchangeable apolipoprotein in the CLD fraction, may suggest a TAG connection between storage as CLDs and secretion as TRL. Overall, the CLD protein population may contribute to the control of the postprandial delivery of TRL by enterocytes. The analysis of the underlying mechanisms may now be envisaged.
Materials and methods
Antibodies and plasmids
Antibodies
Mouse monoclonal anti-PLIN-2 (AP125, for immunofluorescence) and guinea-pig anti-PLIN-3 antibodies were from Progen. Mouse monoclonal antibodies to calnexin, PDI and GM130 were from BD Biosciences. Rabbit polyclonal antibodies to LDH, prohibitin and HSP60 were obtained from Abcam. Mouse monoclonal antibodies to c-Myc (9E10 clone), LPCAT2 and 3BHS1 were from Sigma. Goat anti-ApoB antibody (for immunoflurescence) was from Chemicon International and mouse monoclonal anti-ApoB antibody (1D1, for Western blots) was obtained from the Heart Institute of the University of Ottawa (Canada). Rabbit anti-ApoA-IV antibody was provided by M. Zakin (Institut Pasteur, Paris, France) and sheep polyclonal anti-PLIN-2 (for Western blots) antibody was provided by J. McLauchlan (Targett-Adams et al., 2003).
Plasmids
For Myc-tagged human LPCAT2 plasmid, residues 141–1775 of the LPCAT2 gene were amplified by PCR from the IMAGE clone 3347690 and inserted between the EcoRV and XbaI sites of the mammalian expression vector pCDNA3.1-myc (Pasdeloup et al., 2009). GGCGATATCATGAGCCGGTGCGCCC/ATATCTAGATCAGTCATCTTTTTTGTCTGAGGACTCTCTTCATG were used as primers. For myc-HSD3B1 plasmid, residues 88–1209 of the HSD3B1 gene were amplified from IMAGE clone 4755300 and inserted between the EcoRI and NotI restriction sites of pCDNA3.1-myc. GCTGAATTCGATGGCCATGACGGGCTGG/TCAGCGGCCGCTCACTGAGTCTTGGACTTCAGGTTCTC were used as primers. The human ApoA-IV cDNA (hApoA-IV) was kindly provided by G.S. Shelness (Wake Forest University).
Animals and treatments
Male C57BL/6 mice (6–8 weeks old) were purchased from Charles River. Mice were maintained in a 12 h light/12 h dark cycle and fed with a chow diet (AO3, SAFE). All experimental procedures were in accordance with institutional regulations for the care and use of laboratory animals. To analyse the distribution of lipid droplets in jejunum as a function of time after gavage, groups of mice (three mice per group) were fasted for 4 h; then two groups were given a bolus of olive oil (150 μl) while the third one was kept as a control. At 1 h after the lipid bolus, which is the time when TAG reaches a peak in plasma (Hernandez Vallejo et al., 2009), control mice and one group that received a lipid bolus were killed for jejunum analysis. The last group was maintained without food and killed 20 h after the lipid bolus. In other experiments, mice were fed with a high-fat diet (24% lard and 3% sunflower oil) ad libitum for 2 days. Mice had free access to water during all procedures.
Cell culture, lipid supply and transfections
Caco-2/TC7 cells were plated at a density of 0.25×106 cells per small insert (23.1 mm diameter; Becton Dickinson) or 2.6×106 cells per large insert (75 mm diameter; Corning) and were grown as described previously (Chateau et al., 2005) for differentiation. After 18 days of culture, lipid micelles (2 mM sodium taurocholate, 0.6 mM OA, 0.2 mM lysoPC, 0.05 mM cholesterol and 0.2 mM 1-O-octadecyl-rac-glycerol, a stable analogue of monoacylglycerol) were prepared in a serum-free medium, as previously described (Pauquai et al., 2006), and added to the upper compartment for 24 h, a time that allows the determination of both TRL secretion and CLD accumulation (Chateau et al., 2005). When appropriate, lipid micelles were supplemented with 0.1 μCi of [1-14C]OA per ml of final medium as described previously (Chateau et al., 2005).
HeLa cells were grown at 37°C and 5% CO2 in MEM (minimal essential medium) supplemented with antibiotics, glutamine (Invitrogen) and 10% foetal calf serum (AbCys). For plasmid transfection, Caco-2/TC7 or HeLa cells were seeded on coverslips and transfected with 0.5 μg of the indicated plasmid by lipofection (Lipofectamine™ 2000; Invitrogen) according to the manufacturer's instructions. To induce lipid droplet production, 0.6 mM OA/BSA was added to the culture medium.
Cells were then processed for microscopy or cell fractionation, as indicated in the next sections, or rinsed twice with ice-cold PBS, scraped into lysis buffer (1% Triton X-100 and 5 mM EDTA in PBS) supplemented with a 2% protease inhibitor cocktail (P8340; Sigma) and kept frozen until analysis.
Jejunum processing for optical and electron microscopy
Jejunum was excised, rinsed in 0.1 M phosphate buffer (pH 7.4), cut longitudinally and laid flat in 2.5% glutaraldehyde/0.1 M cacodylate buffer (pH 7.4). Jejunum were cut into small blocks and kept for 2 h in this solution for fixation. Blocks were incubated for 1 h in 0.1 M imidazole buffer (pH 7.4) containing 2% osmium tetroxide for neutral lipid staining (Angermuller and Fahimi, 1982). All material was dehydrated with ethanol and embedded in Epon 812. Semi-thin sections were stained for 1 min with Toluidine Blue (1% in 1% sodium tetraborate) and examined by optical microscopy. Ultrathin sections were contrast-stained with 3% lead citrate for 1 min and examined with a Jeol 100 CX-II electron microscope.
Confocal fluorescence microscopy
Jejunum pieces were rinsed, cut longitudinally and laid flat in 4% (w/v) PFA (paraformaldehyde) for 30 min before embedding in Tissue-Tek. Jejunum cryosections or cells were fixed with 4% PFA, permeabilized by 0.05% saponin in PBS and incubated with primary antibodies and then rinsed in PBS. After incubation with appropriate cyanin dyes or Alexa Fluor®-conjuguated fluorescent secondary antibodies (Jackson Immunoresearch), tissue sections or cells were stained for neutral lipids by incubation with Bodipy 493/503 (Invitrogen). After nuclear staining by DAPI (4′,6-diamidino-2-phenylindole), samples were examined by laser scanning confocal microscopy (LSM 510 or LSM 710 microscope; Carl Zeiss). Fluorescence intensity was quantified using ImageJ software.
Immunoelectron microscopy
Cells were fixed with 4% PFA in cacodylate buffer for 2 h. After alcohol-graded dehydration, cells were embedded in LR White (Electron Microscopy Sciences), and ultrathin sections were incubated with rabbit anti-human ApoA-IV antibody and then with 18 nm gold particle-labelled donkey anti-rabbit IgGs (Jackson Immunoresearch). Sections were analysed in a Jeol 100CX II electron microscope. Images were captured with an Erlangshen 1000 camera and software (Gatan; Roper Scientific).
Subcellular fractionation
Lipid droplets were isolated by density-gradient centrifugation from Caco-2/TC7 cells incubated for 24 h with lipid micelles, using a protocol adapted from Yu et al. (2000) and Brasaemle and Wolins (2006). Cell layers (two 75 mm diameter inserts per centrifuge tube) were rinsed briefly (three times with cold PBS) and scraped and the volume of the cell homogenate was adjusted to 2 ml with buffer A (25 mM Tris/HCl, pH 7.4, 100 mM KCl, 1 mM EDTA and 5 mM EGTA) containing a protease inhibitor cocktail (Complete® protease inhibitor cocktail from Roche). Cells were lysed twice using a cell disruption bomb (Parr Instrument Company; 1200 lbf/in2, 10 min; 1 lbf/in2=6.9 kPa). Cell homogenates were centrifuged at 1000 g for 10 min at 15°C. The lipid droplet-containing supernatant was adjusted to 0.33 M sucrose in a final volume of 3 ml, using a 1 M sucrose solution in buffer A, and transferred into a 12 ml ultracentrifugation tube. The supernatant was then overlaid sequentially with 3 ml of 0.25 M sucrose-containing buffer A, 3 ml of 0.125 M sucrose-containing buffer A and 2 ml of 25 mM Tris/HCl (pH 7.4) containing 1 mM EDTA, 1 mM EGTA and the protease inhibitor cocktail to form a discontinuous sucrose gradient ranging from 0.33 to 0 M. Tubes were centrifuged for 2 h (150000 g, 15°C) in a Beckman SW41 rotor and 1 ml fractions were recovered from the top to the bottom (12 fractions). The pellet (fraction 13) was rinsed in PBS and suspended in 2 ml of 10 mM Tris/HCl (pH 7.4) buffer containing 0.25 M sucrose and protease inhibitors. Fractions were stored at −80°C until use.
Protein concentration and lipid analysis
Protein concentration was determined by the Bio-Rad DC protein assay with BSA as the standard. Quantification of the TAG content in cell lysates was performed using the PAP150TG kit (Biomerieux) as described previously (Pauquai et al., 2006). For the analysis of the lipid classes present in homogenates, supernatants and sucrose gradient fractions, lipids were extracted with chloroform/methanol (2:1, v/v) and separated by TLC as described previously (Chateau et al., 2005). The radioactive bands were excised and the radioactivity was quantified by scintillation counting to evaluate the incorporation of [1-14C]OA into lipids.
Western blotting
Total cell lysates and fractions isolated from sucrose gradient were fractionated by SDS/10% PAGE (5% for ApoB48) and proteins were transferred on to a nitrocellulose membrane. After incubation in TBS-T (20 mM Tris/HCl, pH 7.6, 137 mM NaCl and 0.1% Tween 20) supplemented with 10% non-fat dried skimmed milk powder, blots were probed with primary antibodies in TBS-T containing 5% non-fat dried skimmed milk powder and then with the appropriate peroxidase-conjugated secondary antibodies (Vector Laboratories). Blots were developed with ECL® reagent (Amersham Pharmacia Biotech). Bands were visualized with the Image Reader LAS-4000 (Fujifilm) and quantified using ImageJ software.
In-gel trypsin digestion, nanochromatography and MS analysis
The 1 ml top fractions recovered from the sucrose-density-gradient ultracentrifugation were freeze-dried, dissolved in Laemmli buffer and subjected to SDS/PAGE under reducing conditions. Migration was stopped before the migration front entered the resolving gel, so that all the proteins of one sample were still contained in a single band. Gels were silver stained using a MS-compatible silver stain and protein bands were digested with trypsin as described earlier (Blouin et al., 2010).
Peptides were separated with an Ultimate3000 (Dionex) series HPLC, using a C18 trap column Acclaim pepmap1000 C18 [5 mm particles, 100 Å (1 Å=0.1 nm) pore, 300 mm i.d. (inner diameter) and 5 mm length] and an analytical column C18pepmap100 (3 mm, 15 cm length, 75 mm i.d., 100 Å). Peptides were separated on a gradient of 40 min ranging from 93% solution A (0.1% trifluoroacetic acid)/7% solution B (80% acetonitrile and 20% solution A) to 50% solution A/50% solution B. Eluted fractions were spotted on-line on a MALDI (matrix-assisted laser-desorption ionization) target using a Probot (Dionex) fraction collector. Spotted fractions were mixed 1:4 with 2 mg/ml α-CHC (α-cyano-4-hydroxycinnamate; Laser Biolabs) in 70% acetonitrile containing 0.1% trifluoroacetic acid and Glu-fibrinopeptide at 3 fmol per spot. Fractions were collected and analysed using a 4800 MALDI-TOF (time-of-flight) analyser (ABI).
Spectra acquisition and processing were performed using the 4000 series Explorer software (ABI) version 3.5.28193 build 1011 in positive reflection mode. External calibration was performed using four calibration points spotted throughout the plate, additional internal calibration being performed using the Glu-fibrinopeptide (m/z=1570.677). For each fraction, 500 MS spectra were acquired in the range of 700–4000 Da. For each MS spectrum, the eight most abundant peaks were selected for fragmentation (1000 MS/MS spectra per precursor). Global MS/MS peak lists were subjected to an in-house mascot (Matrix Science) version 2.2 search engine for protein identification (Perkins et al., 1999). The Swiss Prot 56.8 (410518 sequences; 148080998 residues) release database was used with human as species selection. Parent and fragment mass tolerances were, respectively, set to 20 p.p.m. and 0.3 Da, partial modification (oxidation) of methionine residues being allowed. A filter was applied to the search in order to reduce false positives and matching redundancies of the same peptide in several hits. Peptide score above 20 and FDR (false discovery rate) below 1.6% (P<0.01) were required. Only proteins identified with at least two unique peptides were retained. Whenever the result was ambiguous, spectra were manually checked.
The emPAI offers a label-free, approximate estimation of the relative abundance of proteins within a mixture (Ishihama et al., 2005; Shinoda et al., 2010). To compare the emPAI factor across independent biological experiments, we needed to make it independent from the amount of sample injected. The normalized emPAI factor was obtained by calculating first the average ratio of the Mascot emPAI factor of each protein divided by the total number of proteins identified in the experiment. This was then multiplied by the average number of proteins identified in the three experiments.
Statistical analysis
Results are presented as means±S.E.M. Statistical significance was evaluated using the Student's t test for unpaired data.
Author contribution
Nathalie Ly and Malik Alqub contributed to Western-blot and immunofluorescence analyses and Christophe Klein assisted with confocal microscopy analysis. Chiara Guerrera performed the MS analyses and Danielle Chateau performed the electron microscopy analyses. All other authors were involved in the conception, design and realization of the experiments, the collection, analysis and interpretation of data and the writing of the manuscript or revising it critically for important intellectual content. Sylvie Demignot was also the co-ordinator of the project.
Acknowledgements
We acknowledge François Guillonneau (Plateforme Proteomique Université Paris Descartes, Paris, France) for MS/MS analysis, Julien Bricambert for technical assistance during his training, Philippe Cardot for helpful suggestions and Zeina Chamoun for a critical reading of the paper prior to submission. We thank J. McLauchlan, M. Zakin, D. Pasdeloup and G.S. Shelness for kindly providing antibodies and plasmids. Confocal microscopy was performed using the facilities of the Centre d'Imagerie Cellulaire et de Cytométrie of UMR S 872.
Funding
Juien Bouchoux and Frauke Beilstein were the recipients of an ANRS (Agence Nationale de Recherches sur le Sida et les hépatites; Paris, France) fellowship and Thomas Pauquai was the recipient of a MENRT (Ministère de l'Education Nationale, de la Recherche, et de la Technologie) fellowship. This work was partially supported by the ANRS.
References
- Agarwal A.K., Garg A. Enzymatic activity of the human 1-acylglycerol-3-phosphate-O-acyltransferase isoform 11: upregulated in breast and cervical cancers. J. Lipid Res. 2010;51:2143–2152. doi: 10.1194/jlr.M004762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermuller S., Fahimi H.D. Imidazole-buffered osmium tetroxide: an excellent stain for visualization of lipids in transmission electron microscopy. Histochem. J. 1982;14:823–835. doi: 10.1007/BF01033631. [DOI] [PubMed] [Google Scholar]
- Bartz R., Li W.H., Venables B., Zehmer J.K., Roth M.R., Welti R., Anderson R.G., Liu P., Chapman K.D. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J. Lipid Res. 2007;48:837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- Black D.D. Development and physiological regulation of intestinal lipid absorption. I. Development of intestinal lipid absorption: cellular events in chylomicron assembly and secretion. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;293:G519–G524. doi: 10.1152/ajpgi.00189.2007. [DOI] [PubMed] [Google Scholar]
- Blouin C.M., Le Lay S., Eberl A., Kofeler H.C., Guerrera I.C., Klein C., Le Liepvre X., Lasnier F., Bourron O., Gautier J.F., et al. Lipid droplet analysis in caveolin-deficient adipocytes: alterations in surface phospholipid composition and maturation defects. J. Lipid Res. 2010;51:945–956. doi: 10.1194/jlr.M001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasaemle D.L., Wolins N.E. Isolation of lipid droplets from cells by density gradient centrifugation. Curr. Protoc. Cell Biol. 2006 doi: 10.1002/0471143030.cb0315s29. Chapter 3, Unit 3.15. [DOI] [PubMed] [Google Scholar]
- Brown J.M., Chung S., Das A., Shelness G.S., Rudel L.L., Yu L. CGI-58 facilitates the mobilization of cytoplasmic triglyceride for lipoprotein secretion in hepatoma cells. J. Lipid Res. 2007;48:2295–2305. doi: 10.1194/jlr.M700279-JLR200. [DOI] [PubMed] [Google Scholar]
- Bulankina A.V., Deggerich A., Wenzel D., Mutenda K., Wittmann J.G., Rudolph M.G., Burger K.N., Honing S. TIP47 functions in the biogenesis of lipid droplets. J. Cell Biol. 2009;185:641–655. doi: 10.1083/jcb.200812042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann R.J., Manke D.J. Morphometric analysis of the membranes and organelles of small intestinal enterocytes. II. lipid-fed hamster. J. Ultrastruct. Res. 1981;76:15–26. doi: 10.1016/s0022-5320(81)80047-0. [DOI] [PubMed] [Google Scholar]
- Carriere V., Vidal R., Lazou K., Lacasa M., Delers F., Ribeiro A., Rousset M., Chambaz J., Lacorte J.M. HNF-4dependent induction of apolipoprotein A-IV gene transcription by an apical supply of lipid micelles in intestinal cells. J. Biol. Chem. 2005;280:5406–5413. doi: 10.1074/jbc.M408002200. [DOI] [PubMed] [Google Scholar]
- Caviglia J.M., Sparks J.D., Toraskar N., Brinker A.M., Yin T.C., Dixon J.L., Brasaemle D.L. ABHD5/CGI-58 facilitates the assembly and secretion of apolipoprotein B lipoproteins by McA RH7777 rat hepatoma cells. Biochim. Biophys. Acta. 2009;1791:198–205. doi: 10.1016/j.bbalip.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateau D., Pauquai T., Delers F., Rousset M., Chambaz J., Demignot S. Lipid micelles stimulate the secretion of triglyceride-enriched apolipoprotein B48-containing lipoproteins by Caco-2 cells. J. Cell Physiol. 2005;202:767–76. doi: 10.1002/jcp.20173. [DOI] [PubMed] [Google Scholar]
- Cheng D., Iqbal J., Devenny J., Chu C.H., Chen L., Dong J., Seethala R., Keim W.J., Azzara A.V., Lawrence R.M., et al. Acylation of acylglycerols by acyl coenzyme A:diacylglycerol acyltransferase 1 (DGAT1). Functional importance of DGAT1 in the intestinal fat absorption. J. Biol. Chem. 2008;283:29802–29811. doi: 10.1074/jbc.M800494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon S.H., Zhou Y.X., Dixon J.L., Storch J. Intestinal monoacylglycerol metabolism: developmental and nutritional regulation of monoacylglycerol lipase and monoacylglycerol acyltransferase. J. Biol. Chem. 2007;282:33346–33357. doi: 10.1074/jbc.M706994200. [DOI] [PubMed] [Google Scholar]
- Fujimoto Y., Itabe H., Kinoshita T., Homma K.J., Onoduka J., Mori M., Yamaguchi S., Makita M., Higashi Y., Yamashita A., Takano T. Involvement of ACSL in local synthesis of neutral lipids in cytoplasmic lipid droplets in human hepatocyte HuH7. J. Lipid Res. 2007;48:1280–1292. doi: 10.1194/jlr.M700050-JLR200. [DOI] [PubMed] [Google Scholar]
- Gallagher J.W., Weinberg R.B., Shelness G.S. apoA-IV tagged with the ER retention signal KDEL perturbs the intracellular trafficking and secretion of apoB. J. Lipid Res. 2004;45:1826–1834. doi: 10.1194/jlr.M400188-JLR200. [DOI] [PubMed] [Google Scholar]
- Green P.H., Glickman R.M., Riley J.W., Quinet E. Human apolipoprotein A-IV. Intestinal origin and distribution in plasma. J. Clin. Invest. 1980;65:911–919. doi: 10.1172/JCI109745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Walther T.C., Rao M., Stuurman N., Goshima G., Terayama K., Wong J.S., Vale R.D., Walter P., Farese R.V. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453:657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez Vallejo S.J., Alqub M., Luquet S., Cruciani-Guglielmacci C., Delerive P., Lobaccaro J.M., Kalopissis A.D., Chambaz J., Rousset M., Lacorte J.M. Short-term adaptation of postprandial lipoprotein secretion and intestinal gene expression to a high-fat diet. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;296:G782–G792. doi: 10.1152/ajpgi.90324.2008. [DOI] [PubMed] [Google Scholar]
- Hodges B.D., Wu C.C. Proteomic insights into an expanded cellular role for cytoplasmic lipid droplets. J. Lipid Res. 2010;51:262–273. doi: 10.1194/jlr.R003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi Y., Araki M., Motojima K. Identification and characterization of the ER/lipid droplet-targeting sequence in 17beta-hydroxysteroid dehydrogenase type 11. Arch. Biochem. Biophys. 2008;479:121–130. doi: 10.1016/j.abb.2008.08.020. [DOI] [PubMed] [Google Scholar]
- Iqbal J., Hussain M. Intestinal lipid absorption. Am. J. Physiol. Endocrinol. Metab. 2009;296:E1183–E1194. doi: 10.1152/ajpendo.90899.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- Jacobs R.L., Devlin C., Tabas I., Vance D.E. Targeted deletion of hepatic CTP:phosphocholine cytidylyltransferase alpha in mice decreases plasma high density and very low density lipoproteins. J. Biol. Chem. 2004;279:47402–47410. doi: 10.1074/jbc.M404027200. [DOI] [PubMed] [Google Scholar]
- Kuerschner L., Moessinger C., Thiele C. Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic. 2008;9:338–352. doi: 10.1111/j.1600-0854.2007.00689.x. [DOI] [PubMed] [Google Scholar]
- Lee B., Zhu J., Wolins N.E., Cheng J.X., Buhman K.K. Differential association of adipophilin and TIP47 proteins with cytoplasmic lipid droplets in mouse enterocytes during dietary fat absorption. Biochim. Biophys. Acta. 2009;1791:1173–1180. doi: 10.1016/j.bbalip.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Levy E., Mehran M., Seidman E. Caco-2 cells as a model for intestinal lipoprotein synthesis and secretion. FASEB J. 1995;9:626–635. doi: 10.1096/fasebj.9.8.7768354. [DOI] [PubMed] [Google Scholar]
- Levy E., Stan S., Delvin E., Menard D., Shoulders C., Garofalo C., Slight I., Seidman E., Mayer G., Bendayan M. Localization of microsomal triglyceride transfer protein in the Golgi: possible role in the assembly of chylomicrons. J. Biol. Chem. 2002;277:16470–16477. doi: 10.1074/jbc.M102385200. [DOI] [PubMed] [Google Scholar]
- Lu S., Yao Y., Meng S., Cheng X., Black D.D. Overexpression of apolipoprotein A-IV enhances lipid transport in newborn swine intestinal epithelial cells. J. Biol. Chem. 2002;277:31929–31937. doi: 10.1074/jbc.M201418200. [DOI] [PubMed] [Google Scholar]
- Luchoomun J., Hussain M.M. Assembly and secretion of chylomicrons by differentiated Caco-2 cells. Nascent triglycerides and preformed phospholipids are preferentially used for lipoprotein assembly. J. Biol. Chem. 1999;274:19565–19572. doi: 10.1074/jbc.274.28.19565. [DOI] [PubMed] [Google Scholar]
- Moessinger C., Kuerschner L., Spandl J., Shevchenko A., Thiele C. Human lysophosphatidylcholine acyltransferase 1 and 2 are located to lipid droplets, where they catalyze the formation of phosphatidylcholine. J. Biol. Chem. 2011;286:5599–5606. doi: 10.1074/jbc.M110.202424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel E., Demignot S., Chateau D., Chambaz J., Rousset M., Delers F. Lipid-dependent bidirectional traffic of apolipoprotein B in polarized enterocytes. Mol. Biol. Cell. 2004;15:132–141. doi: 10.1091/mbc.E03-04-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto R., Shindou H., Oda Y., Shimizu T. Phosphorylation of lysophosphatidylcholine acyltransferase 2 at Ser34 enhances platelet-activating factor production in endotoxinstimulated macrophages. J. Biol. Chem. 2010;285:29857–29862. doi: 10.1074/jbc.M110.147025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D.J., Vance J. Mechanisms of lipid-body formation. Trends Biochem. Sci. 1999;24:109–115. doi: 10.1016/s0968-0004(98)01349-8. [DOI] [PubMed] [Google Scholar]
- Ohashi M., Mizushima N., Kabeya Y., Yoshimori T. Localization of mammalian NAD(P)H steroid dehydrogenase-like protein on lipid droplets. J. Biol. Chem. 2003;278:36819–36829. doi: 10.1074/jbc.M301408200. [DOI] [PubMed] [Google Scholar]
- Ohsaki Y., Cheng J., Suzuki M., Shinohara Y., Fujita A., Fujimoto T. Biogenesis of cytoplasmic lipid droplets: from the lipid ester globule in the membrane to the visible structure. Biochim. Biophys. Acta. 2009;1791:399–407. doi: 10.1016/j.bbalip.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Pasdeloup D., Blondel D., Isidro A.L., Rixon F.J. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J. Virol. 2009;83:6610–6623. doi: 10.1128/JVI.02655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauquai T., Bouchoux J., Chateau D., Vidal R., Rousset M., Chambaz J., Demignot S. Adaptation of enterocytic Caco-2 cells to glucose modulates triacylglycerol-rich lipoprotein secretion through triacylglycerol targeting into the endoplasmic reticulum lumen. Biochem. J. 2006;395:393–403. doi: 10.1042/BJ20051359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Qin X., Tso P. The role of apolipoprotein AIV on the control of food intake. Curr. Drug Targets. 2005;6:145–151. doi: 10.2174/1389450053174541. [DOI] [PubMed] [Google Scholar]
- Robenek H., Buers I., Hofnagel O., Robenek M.J., Troyer D., Severs N.J. Compartmentalization of proteins in lipid droplet biogenesis. Biochim. Biophys. Acta. 2009;1791:408–418. doi: 10.1016/j.bbalip.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Robenek H., Hofnagel O., Buers I., Robenek M.J., Troyer D., Severs N.J. Adipophilin-enriched domains in the ER membrane are sites of lipid droplet biogenesis. J. Cell Sci. 2006;119:4215–4224. doi: 10.1242/jcs.03191. [DOI] [PubMed] [Google Scholar]
- Robertson M.D., Parkes M., Warren B.F., Ferguson D.J., Jackson K.G., Jewell D.P., Frayn K.N. Mobilisation of enterocyte fat stores by oral glucose in humans. Gut. 2003;52:834–839. doi: 10.1136/gut.52.6.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda K., Tomita M., Ishihama Y. emPAI Calc – for the estimation of protein abundance from large-scale identification data by liquid chromatography-tandem mass spectrometry. Bioinformatics. 2010;26:576–577. doi: 10.1093/bioinformatics/btp700. [DOI] [PubMed] [Google Scholar]
- Shu X., Nelbach L., Ryan R.O., Forte T.M. Apolipoprotein A-V associates with intrahepatic lipid droplets and influences triglyceride accumulation. Biochim. Biophys. Acta. 2010;1801:605–608. doi: 10.1016/j.bbalip.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soupene E., Fyrst H., Kuypers F.A. Mammalian acyl-CoA:lysophosphatidylcholine acyltransferase enzymes. Proc. Natl. Acad. Sci. U.S.A. 2008;105:88–93. doi: 10.1073/pnas.0709737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandl J., White D.J., Peychl J., Thiele C. Live cell multicolor imaging of lipid droplets with a new dye, LD540. Traffic. 2009;10:1579–1584. doi: 10.1111/j.1600-0854.2009.00980.x. [DOI] [PubMed] [Google Scholar]
- Swift L.L., Kakkad B., Boone C., Jovanovska A., Jerome W.G., Mohler P.J., Ong D.E. Microsomal triglyceride transfer protein expression in adipocytes: a new component in fat metabolism. FEBS Lett. 2005;579:3183–3189. doi: 10.1016/j.febslet.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Targett-Adams P., Chambers D., Gledhill S., Hope R.G., Coy J.F., Girod A., McLauchlan J. Live cell analysis and targeting of the lipid droplet-binding adipocyte differentiation-related protein. J. Biol. Chem. 2003;278:15998–16007. doi: 10.1074/jbc.M211289200. [DOI] [PubMed] [Google Scholar]
- Tauchi-Sato K., Ozeki S., Houjou T., Taguchi R., Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J. Biol. Chem. 2002;277:44507–44512. doi: 10.1074/jbc.M207712200. [DOI] [PubMed] [Google Scholar]
- van Meer G. Caveolin, cholesterol, and lipid droplets? J. Cell Biol. 2001;152:F29–F34. doi: 10.1083/jcb.152.5.f29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance D.E. Role of phosphatidylcholine biosynthesis in the regulation of lipoprotein homeostasis. Curr. Opin. Lipidol. 2008;19:229–234. doi: 10.1097/MOL.0b013e3282fee935. [DOI] [PubMed] [Google Scholar]
- Vidal R., Hernandez-Vallejo S., Pauquai T., Texier O., Rousset M., Chambaz J., Demignot S., Lacorte J.M. Apple procyanidins decrease cholesterol esterification and lipoprotein secretion in Caco-2/TC7 enterocytes. J. Lipid Res. 2005;46:258–268. doi: 10.1194/jlr.M400209-JLR200. [DOI] [PubMed] [Google Scholar]
- Walther T.C., Farese R.V., Jr The life of lipid droplets. Biochim. Biophys. Acta. 2008;1791:459–466. doi: 10.1016/j.bbalip.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Salavaggione E., Pelleymounter L., Eckloff B., Wieben E., Weinshilboum R. Human 3beta-hydroxysteroid dehydrogenase types 1 and 2: Gene sequence variation and functional genomics. J. Steroid Biochem. Mol. Biol. 2007;107:88–99. doi: 10.1016/j.jsbmb.2007.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolins N.E., Brasaemle D.L., Bickel P.E. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 2006;580:5484–5491. doi: 10.1016/j.febslet.2006.08.040. [DOI] [PubMed] [Google Scholar]
- Yao H., Ye J. Long chain acyl-CoA synthetase 3-mediated phosphatidylcholine synthesis is required for assembly of very low density lipoproteins in human hepatoma Huh7 cells. J. Biol. Chem. 2008;283:849–854. doi: 10.1074/jbc.M706160200. [DOI] [PubMed] [Google Scholar]
- Yao Y., lLu S., Huang Y., Beeman-Black C.C., Lu R., Pan X., Hussain M.M., Black D.D. Regulation of microsomal triglyceride transfer protein (MTTP) by apolipoprotein A-IV in newborm swine intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;300:G357–G363. doi: 10.1152/ajpgi.00353.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., Cassara J., Weller P.F. Phosphatidylinositide 3-kinase localizes to cytoplasmic lipid bodies in human polymorphonuclear leukocytes and other myeloid-derived cells. Blood. 2000;95:1078–1085. [PubMed] [Google Scholar]
- Zehmer J.K., Huang Y., Peng G., Pu J., Anderson R.G., Liu P. A role for lipid droplets in inter-membrane lipid traffic. Proteomics. 2009;9:914–921. doi: 10.1002/pmic.200800584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Lee B., Buhman K.K., Cheng J.X. A dynamic, cytoplasmic triacylglycerol pool in enterocytes revealed by ex vivo and in vivo coherent anti-Stokes Raman scattering imaging. J. Lipid Res. 2009;50:1080–1089. doi: 10.1194/jlr.M800555-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]