Abstract
The main aim of this article is to discuss the current state of in vivo brain imaging methods in the context of the diagnosis and treatment of mental illness. The background to current practice is discussed, and the new methods introduced which may have the capacity to increase the relevance of magnetic resonance imaqinq, particularly functional magnetic resonance imaging, for clinical application. The main focus will be on magnetic resonance imaging, but many of the comments have a general relevance across imaging modalities.
Keywords: imaging, MRI, PET, brain, structure, function, analysis, diagnosis
Abstract
El objetivo principal de este articulo es analizar la situación actual de ios métodos de neuroimágenes in vivo en el contexto del diagnóstico y del tratamiento de las enfermedades mentales. Se discute el fundamento de la práctica actual y los nuevos métodos introducidos, los que pueden tener la capacidad de aumentar la importancia de las imágeries de resonancia magnética, en especial de la resonancia funcional, para la aplicación clinica, El foco principal estará en las imágenes de resonancia magnética, pero muchos de los comentarios tienen una relevancia general para las distintas modalidades de imágenes.
Abstract
Cet article vise principalement à examiner la place actuelle des méthodes d'imagerie cérébrale in vivo dans le contexte du diagnostic et du traitement des maladies mentales: une discussion du cadre des pratiques actuelles est proposée, ainsi qu'une introduction aux nouvelles méthodes qui pourraient augmenter la pertinence de l'imagerie par résonance magnétique, plus particulièrement l'IRM fonctionnelle,en ce qui concerne ses applications cliniques. Le sujet principal en est l'IRM, mais un grand nombre des commentaires sont également valables pour d'autres méthodes d'imagerie.
Background: structural and functional brain imaging
A brain imaging method could be defined as any experimental technique that allows human (or animal) brain structure or function to be studied, preferably in vivo in the current context. Such a method should ideally produce accurate timing (in the case of functional imaging) and spatial localization (for both structural and functional imaging) of cerebral function, structure, or changes in these properties of the brain. The method should be minimally invasive and repeatable (to facilitate use in treatment monitoring and development of therapeutic strategies). Current structural magnetic resonance imaging (MRI) has good spatial resolution, is noninvasive, and meets the above criteria well for structural analysis. In contrast, no single technique currently in existence would meet all these criteria in the case of functional imaging, but the most common widely used methods are electroencephalography (EEG), positron emission tomography (PET), and functional magnetic resonance imaging (fMRI). Of these three methods, EEG has been available for the longest time (but arguably not so as a viable mapping method). PET has been available for the secondlongest period (in the order of four decades), and fMRI is the newest widely used technique. PET is arguably the most invasive (involving radioisotope administration) and EEG makes the closest approach to measuring neuronal activity directly (but has rather poor spatial mapping properties). As the location of cerebral activity and changes in activity associated with changes in brain state (either experimentally or illness-determined) seems to have been the priority in most of the research to date, fMRI has emerged as the most widely used functional brain mapping method.
Structural MRI (sMRI) has been a common tool for the investigation of trauma and disease -related brain changes for some considerable time, but fMRI is a more recent addition to the MRI armory of methods. It has been available for a little less than two decades, since Ogawa et al1 first coined the term BOLD (blood oxygen level-dependent) contrast for what has become the most widely used approach in use today. At first sight, BOLD imaging has a number of shortcomings. At what is still the most common field strength in MR scanners in clinical use (1.5 Tesla), the signal changes following neural activation are only a few percent. There are also a host of artifacts that can interfere with the signal, most notably head motion. The BOLD “signal” is also not a direct readout of neuronal electrical activity, but rather a downstream consequence of this activity, dependent on the response of the circulatory system. Finally, there is still a dispute about exactly what neural changes underlie the BOLD response (for a recent viewpoint on some of these issues, see Logothetis2). Despite all these apparent problems, BOLD fMRI has revolutionized the study of human brain activity. It is noninvasive (does not require administration of radioisotopes), can be performed repeatedly on the same individuals, and uses equipment that is increasingly widely available. There have been tens of thousands of papers published in which fMRI has been used to investigate a vast array of aspects of human brain function.
Brain imaging and psychiatry
When MRI technology first became available to psychiatry and neurology, one of the primary aims was to use this new technology to establish what have often been described as the “neural correlates” of various mental disorders, ie, to determine the location and magnitude of changes in brain structure or function compared with subjects from a suitable reference population. This would facilitate the identification of “biomarkers” (objective quantifiable changes in brain function) of the mental disorder in question. The longer-term aim was then to use these biomarkers to test the effects of drug treatment or behavioral therapy, ie, to use them as quantitative measures of the effectiveness of treatment in restoring “normality”
As a research enterprise, the application of neuroimaging with the above aims has resulted in a very large number of studies and an impressive number of research publications in many of the major psychiatric and neuroscience journals, particularly in the case of fMRI. In 2003, barely a decade after the appearance of fMRI as a viable imaging tool, it was possible to list, in a book entitled Neuroimaging in Psychiatry3 produced by a number of my colleagues in London as well as eminent researchers from other centers, hundreds of research papers involving MR (as well as a large number from longer established methodologies such as PET). Since 2003 the knowledge base in this area has continued to expand at an impressive rate and, reading the literature to date, one might well conclude that fMRI has had a considerable impact on our understanding of abnormalities in brain function and structure. However, one might ask a different but no less important question. Standing as we do, almost two decades after the appearance of fMRI and having (as we do) access to widely available and reasonably reliable methods of analyzing brain imaging data, has brain imaging started to make an impact on the clinical issues of interest? Has brain imaging materially altered the pressing issues of the diagnosis and treatment of brain disorders?
This issue was the subject of a recent editorial in the British Journal of Psychiatry by Bullmore et al4: “Why psychiatry can't afford to be neurophobic.” One of the issues raised in that article is “the reality of psychiatric practice in the UK, where there is currently agreed to be no clear role for neuroimaging, biomarkers or genetic testing.” The main question in relation to imaging is why the large number of research studies have not been translated into clear beneficial effects in clinical practice. This is clearly a complex and multifactorial issue, but the aim of the present contribution is to examine the simple question of whether neuroimaging is asking the appropriate questions of the data to maximize its relevance to psychiatry and drug discovery and development. For an interesting discussion of the general issue of using neuroimaging to understand brain function, see ref 5.
Background to current approaches to MRI analysis
In order to understand how the current approaches to MRI analysis have arisen, one needs to return to the period before fMRI was widely available, and brain activation and analysis was mainly undertaken using PET. Hie technique, known as region of interest (ROI) analysis, was the earliest to be employed and consisted, as its name suggests, of picking, a priori, a region or regions of the brain which were proposed, on the basis of previous findings or hypotheses to respond to the experimental task being studied. Typically, data would be averaged over the ROI(s) and the change in blood flow related to task performance would be studied, preferably with reference to a control (nonresponding) region or regions. This method remains arguably the simplest and one of the most statistically powerful approaches to studying changes in brain function and structure when the areas involved are well known or strongly predicted a priori. However, universal application of this method would entail a complete knowledge of all the brain regions involved in normal brain functions of interest, and (in psychiatry) when brain function or structure is abnormal. Given that we are still far from such a state of knowledge, more exploratoryapproaches were, and still are, needed in many cases. Ideally, these methods needed to be able to explore activity changes at the limit of resolution of the brain images (ie, at voxel level). In the late 1980s and early 1990s, Karl Friston and his colleagues at the Hammersmith hospital in London began to develop methods for the analysis of changes in brain activation over the whole brain, an endeavor which led to the development of the package known as statistical parametric mapping (SPM - for details see http://www.fiLion.ucl.ac.Uk/spm/doc/#history). This package, freely available to researchers since 1991, has become the most widely used approach for wholebrain analysis of functional imaging data. In order to achieve a principled approach to the problem, SPM developed a sophisticated way of dealing with the obviouslysevere multiple comparison problem inherent in performing tens of thousands of statistical tests, one at each voxel.6 This approach, using the statistical theory of Gaussian random fields,7 has earned Karl Friston deserved recognition for revolutionizing the analysis of brain imaging data. With the appearance of fMRI, the SPM package was rapidly adapted to deal with the rather different characteristics of the new data sets. Somewhat later, the possibility of similarly analyzing structural changes voxel by voxel led to the development of what is now known as voxel-based morphometry or VBM. SPM was rapidly applied to large numbers of structural and functional brain imaging projects. It is the method of choice when changes need to be investigated over the whole brain, either because there is no strong prior hypothesis about the areas that need to be studied, or because the distributed nature of the expected changes makes ROI-based analysis very challenging. On a more prosaic note, it also removes much of the tedium (and potential error) of manually defining ROIs on large, high-resolution MRI data sets. Anyone who attempted to analyze structural MRI data prior to the appearance of VBM might speculate that the automated nature of this technique might have led many researchers to take this route, even when an ROI analysis might have been possible.
Since the early 1990s, there have been a large number of technical developments in understanding, and dealing with, sources of error in analyzing MRI data, and many excellent packages are now available, but the main analysis approach remains a suitably corrected voxel-byvoxel exploration of whole-brain activations (or structural changes) with inferences as to which brain locations are exhibiting significant effects or changes in effect brought about by the nature of the experimental task undertaken or the membership of a particular subject group (eg, patient/control). The main approach might be termed locationist and nonconnectionist, in that it seeks to locate areas of significant response (change) but ignores, by its independent voxel-by-voxel analyses, interactions between brain regions, at least at the primary phase of analysis. Note, however, that posthoc connectivity analyses are often undertaken in the case of fMRI. Ignoring intervoxel interactions greatly simplifies the analysis, but ignores our current knowledge, suggesting that almost all significant brain activity involves network or system level behavior.
It is interesting to consider the pros and cons of this piecewise approach to the analysis of brain function on the current position of brain imaging vis à vis its uses in psychiatry and drug discovery and testing. Hie obviously positive aspects of 15 or so years of brain imaging research using (predominantly- mass- univariate) fMRI are as follows. Firstly, our knowledge of the functional neuroanatomy of the brain has been expanded considerably. Secondly, if the multiple comparison problem inherent in mass univariate analysis has been tackled in a conservative and principled fashion, the areas that we have identified should be relatively robust, as the tendency would have been to make type II rather than type I errors. On the other hand, the lack of consideration of inter-regional interactions during whole-brain activation detection will mean that we have missed some activations that might be weak but highly correlated between brain regions. In other words, we might have underreported and underdetected the distributed networks involved in many brain functions and in pathological changes in these functions. In simple terms, we have been “throwing away” useful information in the data sets during analysis. A more important consideration, central to the main theme of this article, is that, in the mass univariate analysis, there is no combination of information over the whole brain to assemble some sort of “figure of merit” indicating how well our analysis results allow us to, say, separate controls from patients, or drug responders from nonresponders. In other words, there is no indication of how well the whole-brain data would allow us to be confident that the pattern of activation was characteristic of a patient rather than a healthy subject. Instead of being able to make a prediction about an individual being healthy or not on a network-wide basis, we would instead be confined to making statements about the overall separation of groups at a particular voxel or in a particular region of interest, on the basis (typically) of a t statistic.
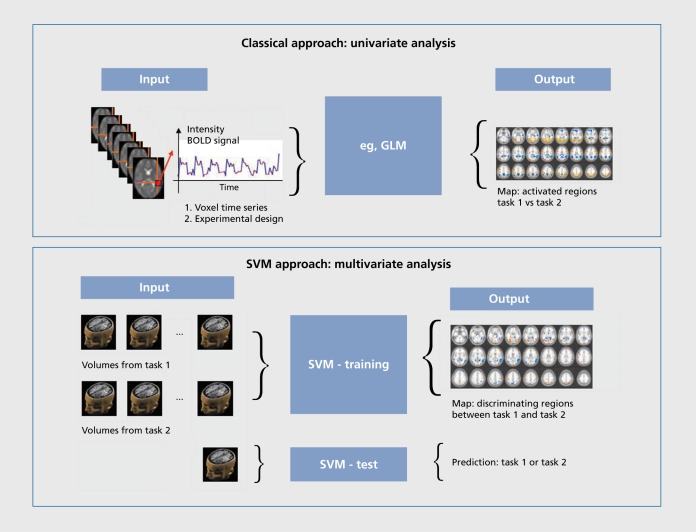
Figure 1. Data flow in a simple 2-task functional magnetic resonance imaging (fMRI) experiment (alternating blocks of each task) through traditional univariate analysis with general linear modeling (GLM) and support vector machine (SVM) analysis. The univariate approach analyses individual voxels using time points when task 1 is being performed and time points where task 2 is being performed, contrasting the two using a simple statistical test (eg, a f test) with a correction for the number of voxels analyzed. The output is a map of regions where the responses to the two tasks are significantly different. SVM-based analysis takes whole-brain volumes when task 1 is being performed and whole-brain volumes when task 2 is being performed and “trains” a computer program to associate patterns of fMRI response with each task. The outputs are a map of the regions which discriminate between the two tasks and a measure of how well the two tasks are discriminated on the basis of the whole brain data. After training, the task being performed can be predicted purely from the fMRI data. For group separation, tasks 1 and 2 can be replaced by groups 1 and 2 ( eg, patients/controls) performing a given task or structural MRI data from the two groups.
New developments in analyzing brain imaging data: machine learning methods
These observations on the mainstream fMRI analysis status quo have been made by a number of statisticians and neuroscientists in recent years.8,9 In response to the issues described above, growing interest has now come to be focused on a group of analysis techniques that have been described as “brain-reading” or “braindecoding” methods10 that belong to a broad group of techniques known collectively as machine learning.11 The basic idea of these methods is that, instead of analyzing the brain voxel by voxel, data from groups of voxels (ROI) or indeed from the whole brain, are used to train a computer program. In one set of classification methods, the most common variant of which is called the support vector machine (SVM), the program will typically find a boundary (referred to as a hyperplane in the relevant literature because it exists in high-dimensional space) between different classes of data (eg, data from patients and data from controls either from structural images of the same fMRI experiment). Once this boundary has been located, predictions can be made for data not in the training data set. For example, having trained the program to distinguish controls from depressed patients and define the optimal hyperplane to achieve this distinction, a new subject could be classified as belonging to the “patient” or the “control” class based on the relationship of their data to the hyperplane. The specificity- and sensitivity of these predictions can be examined using standardized statistical approaches. In the most common of these, the so-called “leave one out” methods, the computer program is training on all the subjects but one and tested on the remaining individual. This is repeated until all the subjects have been the “one left out.” By averaging the results across all the tests it is possible to compute the sensitivity and specificity, where sensitivity here refers to the probability of correctly classifying a patient as a patient, and specificity the probability of correctly classifying a control as a control. If the data to be analyzed are obtained with a continuous rating (depression scores) rather than a classification variable (ill/not ill), then similar programs can operate on a data regression basis, learning the association between the continuous rating and brain structure or function. As well as providing information that may clearly be of value in a clinical setting in the form of classification accuracy, which communicates the level of confidence we can have in the predictions made by this type of analysis, these “braindecoding” methods can also produce maps which indicate the levels to which different brain regions are involved in the classification accuracy that has been achieved. However, here a note of caution is in order. Unlike the maps produced by the more commonly used mass- univariate methods which can be unequivocally- interpreted in terms of the size of the effect (eg, difference in response between groups) at each voxel, the maps produced by the machine learning methods explicitly contain the effects of interactions between voxels or brain regions. In other words, a particular voxel could be important in distinguishing two groups either because there is a large difference in function or structure at that point or because there is a small difference that is highly correlated with those in many other brain regions, gaining importance from these correlations. There are two main consequences arising from this. The first is that the maps may be inherently more sensitive in depicting effects than those that we may be accustomed to seeing (though this is debated and is still undergoing detailed study). The second is that, unlike univariate maps, that can be subjected to statistical thresholding at a particular P value, thresholding these multivariate maps is more challenging, and the most effective way to accomplish this is an active area of investigation.
To summarize the above discussion, both the mass-uni variate and multivariate “brain reading” methods of analyzing MRI data can give information about the location of disease-related changes in structure or function. The univariate methods are in fact easier to interpret, but may be less sensitive in detecting small changes to distributed systems. Few would argue, however, that if properly carried out, both approaches can potentially produce useful maps.
It is valuable at this point, however, to consider the relationship between producing a reliable map and establishing a usable biomarker for a psychiatric illness. The concept of a biomarker contains within it the idea of classification. It associates a pattern of changes in brain structure or function with a particular mental state. This is in fact the core idea of the “brain-reading” methodologies, as stated above. However, without knowledge of the classification accuracy associated with the map of brain changes, the map itself has little value. In a distinction between two classes, a random allocation process would produce a classification accuracy of 50%. A useful biomarker would be a pattern of changes with a classification accuracy of such a level that its probability of arising by chance would be very small. We can carry out such a test very easily. Having determined the classification accuracy as described earlier, we the randomly allocate the data to the two classes of interest (thus achieving the null hypothesis of no difference between the classes) and repeat the “leave one out” testing. If we do this a very large number of times, we can establish how likely the classification process is to produce the original classification accuracy under the null hypothesis of no difference between the classes. In simple terms, we can see how far away from chance the classification lies. The further this is, the “cleaner” the separation between the groups achieved by the imaging “biomarker.”
Machine learning in current image analysis - a change of emphasis?
Although “brain reading” using machine learning methods (often also referred to as pattern classification methods) is currently arousing a good deal of interest, their use in the investigation of brain imaging is not new. In fact, they were used as long ago as the 1990s to investigate PET data.12,13 However, functional and structural brain imaging research has produced a host of new and interesting analysis methods over the last two decades. The reasons why some methods become widely used whereas others do not is a topic of considerable interest. O'Toole and colleagues8 devoted considerable space to discussing this issue and raised issues of what will move researchers out of their “comfort zone” to a new and potentially useful way of using their data. Given the availability of high-quality packages such as SPM, where mass- univariate analysis is efficiently implemented, and which are well-known and respected by neuroimagers, new methods have to be easy to use and to offer considerable added value to justify the investment in using them.
Why then does the author of the current article believe that machine learning methods may be widely taken up when many other promising methods have not? In the early 2000s considerable interest in questions of face/object recognition in the visual cortex led to some fascinating experiments. Notably, a very elegant study of face and object processing in the visual cortex by Haxby and his colleagues appeared.1“ This paper did not use machine learning methods, but introduced the idea of associating brain states (recognition of different types of object) with distributed patterns of brain activity. Shortly afterwards, in 2002, Gotland et al wrote a highly interesting account of the use of classifiers in brain imaging,15 introducing the use of the SVM, and in 2003 Cox and Savoy10 used an SVM (see above) in the same area of research as Haxby.14 It was clear from these data that information might be available in distributed patterns of brain activity that were not accessible by considering each voxel in isolation. Moreover, this information could aid classification. This was a way, not simply of locating functional or structural changes in the brain due to illness or a change in an experimental paradigm, but of using data from many voxels to explicitly classify brain data according to the group to which they belonged. A number of groups then began to realize that these ideas were much closer to the notion of a network-level biomarker than a statistically unconnected set of results from independently analyzed brain regions. Machine learning methods were soon applied to analysis of fMRI data,16,17 demonstrating the power to achieve good classification accuracies based on networks located in believable brain regions. It is but a small step from this point to the idea of automated diagnosis. In the area of structural MRI, Alzheimer's disease has been one of the major targets for this latest phase of applications of machine learning.18,19 This is perhaps understandable, given that it gives rise to both distributed and major effects on gray matter density, making it an obvious target for a multivariate classification method. The use of fMRI for diagnostic purposes has also been investigated using SVM.20,21 Machine-learning based classifiers are currently achieving accuracies of 75% to 95% using functional and structural imaging data and active research in this area is extending the armory of methods beyond categorical classification to probabilistic output using techniques such as Gaussian Process methods.22 Other techniques of interest include singleclass SVM in which the goal is outlier or novelty detection. This method has considerable promise for detection of deviations from statistical homogeneity in clinical populations.
In a recent demonstration of the possibilities for machine learning, Sato and his colleagues carried out an interesting experiment.23 They first trained a computer program (using a technique called maximum entropylinear discriminant analysis) to recognize the association between age and brain activation changes during performance of a motor (finger-tapping task). They were then able to predict the ages of subjects not included in the training purely from their brain activation data. If one imagines the association computed in this experiment as a biomarker for age, and then extends the logic to other areas (eg, changes in depression) one can appreciate the possibilities of the method.
Some of the most exciting possibilities of machine learning methods in clinical practice stem from the ideas raised in the two previous paragraphs. One is that we may be able to locate individual patients on a continuum of brain structural or functional abnormalities that are correlated with illness severity. This would be a great advance on simply categorizing an individual as belonging to the group of “controls” or the group of “patients.”
We would also be able to identify patients who, on the basis of their brain structure or function, appeared to be atypical of their diagnostic group. The second is that this “continuum” or probabilistic rather than categorical approach, could be extended from diagnosis to response prediction - personalization of treatment. The probability that a given patient might be a “responder” rather than a “nonresponder” based on objective measurement of brain structure or function would be a valuable adjunct to the choice and direction of treatment.
In order to make these new methods available on a wide basis, a number of groups are also actively- developing toolboxes with user-friendly interfaces. Also, in order to avoid repetition of already time-consuming image processing, these toolboxes are often being designed to accept data from widely used preprocessing streams in packages such as SPM.
Conclusion
Seventeen years ago, it was felt that fMRI might revolutionize the study of human brain activity.1,24 Arguably, this has proved to be the case. It was also felt by many that fMRI might prove to be an invaluable clinical for the investigation and treatment of mental illness. There are many who would argue that has not proved to be the case. Kosslyn in 19995 asked “If fMRI is the answer - what is the question?” With machine learning, perhaps fMRI may be able to answer more of the questions that we wish to ask.?
Selected abbreviations and acronyms
- fMRI
functional magnetic resonance imaging
- ROI
region of interest
- sMRI
structural magnetic resonance imaging
- SPM
statistical par am etric mapping
- SVM
support vector machine
REFERENCES
- 1.Ogawa S., Tank DW., Menon R., et al. Intrinsic signal changes accompanying sensory stimulation: functional brain mapping with magnetic resonance imaging. Proc Natl Acad Sci U S A. 1992;89:5951–5955. doi: 10.1073/pnas.89.13.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Logothetis N. What we can and what we cannot do with fMRI. Nature. 2008;453:869–878. doi: 10.1038/nature06976. [DOI] [PubMed] [Google Scholar]
- 3.Fu CHY., Senior C., Russell TA., Weinberger D., Murray R. . Neuroimaging in Psychiatry. London; NY: Martin Dunitz. 2003 [Google Scholar]
- 4.Bullmore E., Fletcher P., Jones PB. Why psychiatry can't afford to be neurophobic.. Br J Psychiatry. 2009;194:293–295. doi: 10.1192/bjp.bp.108.058479. [DOI] [PubMed] [Google Scholar]
- 5.Kosslyn SM. If neuroimaging is the answer -what is the question?. Phil Trans Proc Roy SocB. 1999;354:1283–1294. doi: 10.1098/rstb.1999.0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friston KJ., Frith C., Liddle PF., Frackowiak RSJ. Comparing functional (PET) images: the assessment of significant change.. J Cerebr Blood Flow. 1991;11:690–699. doi: 10.1038/jcbfm.1991.122. [DOI] [PubMed] [Google Scholar]
- 7.Adler RJ. The Geometry of Random Fields. New York, NY: Wiley. 1981 [Google Scholar]
- 8.O'Toole AJ., Jiang F., Abdi H., Penard N., Dunlop JP., Parent MA. Theoretical, statistical and practical perspectives on pattern-based classification approaches to the analysis of functional neuroimaging data. J Cogn Neurosci. 2007;19:1735–1752. doi: 10.1162/jocn.2007.19.11.1735. [DOI] [PubMed] [Google Scholar]
- 9.Haynes J-D., Reese G. Decoding mental states from brain activity in humans. Nat Rev Neurosci. 2006;7:523–534. doi: 10.1038/nrn1931. [DOI] [PubMed] [Google Scholar]
- 10.Cox D., Savoy R. Functional magnetic resonance imaging (fMRI) “brainreading”: Detecting and classifying distributed patterns of fMRI activity in human visual cortex. Neuroimage. 2003;19:61–270. doi: 10.1016/s1053-8119(03)00049-1. [DOI] [PubMed] [Google Scholar]
- 11.Hastie T., Tibshirani R., Friedman J. . The Elements of Statistical Learning: Data Mining, Inference and Prediction. New York, NY Springer-Verlag. . 2001 [Google Scholar]
- 12.Clark CM., Amann W., Martin WRW., Ty P., Hayden MR. The FDG/PET methodology for early detection of disease onset: a statistical model. J Cerebr Blood Flow Metab. 1991;11:A96–A102. doi: 10.1038/jcbfm.1991.44. [DOI] [PubMed] [Google Scholar]
- 13.Moeller JR., Strother SC. A regional covariance approach to the analysis of functional patterns in positron emission tomographic data. J Cereb Blood Flow Metab. 1991;11:A121–A135. doi: 10.1038/jcbfm.1991.47. [DOI] [PubMed] [Google Scholar]
- 14.Haxby JV., Gobbini Ml., Furey ML., Ishai PJL., Pietrini P. Distributed and overlapping representations of faces and objects in the ventral temporal cortex. Science. 2001;293:2425–2430. doi: 10.1126/science.1063736. [DOI] [PubMed] [Google Scholar]
- 15.Golland P., Fischl B., Spiridon M., et al. Discriminative analysis for imagebased studies. Lecture Notes in Computer Science. 2002:508–515. [Google Scholar]
- 16.Davatzikos C., Ruparel K., Fan Y., Shen DG., Acharya M., Longhead JW., et al. Classifying spatial patterns of brain activity with machine learning methods: application to lie detection. Neuroimage. 2005;28:663–668. doi: 10.1016/j.neuroimage.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 17.Mourao-Miranda J., Bokcle ALW., Born C., Hampel H., Stetter M. Classifying brain states and determining the discriminating activations patterns: support vector machine on functional MRI data. Neuroimage. 2005;28:980–995. doi: 10.1016/j.neuroimage.2005.06.070. [DOI] [PubMed] [Google Scholar]
- 18.Davatzikos C., Fan Y., Wu X., Shen D., Resnick SM. Detection of prodromal Alzheimer's disease via pattern classification of magnetic resonance imaging. Neurobiol Aging. 2008;29:514–523. doi: 10.1016/j.neurobiolaging.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kloppel S., Stonnington CM., Barnes J., et al. Accuracy of dementia diagnosis - a direct comparison between radiologists and a computerised method. Brain. 2008;131:2969–2974. doi: 10.1093/brain/awn239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu CHY., Mourao-Miranda J., Costafreda SG., et al. Pattern recognition of sad facial processing: towards the development of neurobiological markers in depression. Biol Psychiatry. 2007;63:656–662. doi: 10.1016/j.biopsych.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 21.Marquand AF., Mourao-Miranda J., Brammer MJ., Cleare AJ., Fu CHY. Neuroanatomy of verbal working memory as a diagnostic biomarker for depression. Neuroreport. 2008;19:1507–1511. doi: 10.1097/WNR.0b013e328310425e. [DOI] [PubMed] [Google Scholar]
- 22.Rasmussen CE., Williams CKI. . Gaussian Processes for Machine Learning. Boston, MA: MIT Press USA. 2006 [Google Scholar]
- 23.Sato JR., Thomaz CE., Cardoso TF., Fujita A., Martin Mda G., Amaro E Jr. Hyperplane navigation: a method to set individual scores in fMRI datasets. Neuroimage. 2008;42:1473–1480. doi: 10.1016/j.neuroimage.2008.06.024. [DOI] [PubMed] [Google Scholar]
- 24.Kwong KK., Belliveau JW., Chester DA., et al. Dynamic magnetic resonance imaging of human brain activity during primary sensory stimulation. Proc Natl AcadSciUSA. 1992;89:5675–5679. doi: 10.1073/pnas.89.12.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]