Abstract
We currently rely on large randomized trials and meta-analyses to make clinical decisions; this places us at a risk of discarding subgroup or individually specific treatment options owing to their failure to prove efficacious across entire populations. There is a new era emerging in personalized medicine that will focus on individual differences that are not evident phenomenologically. Much research is directed towards identifying genes, endophenotypes, and biomarkers of disease that will facilitate diagnosis and predict treatment outcome. We are at the threshold of being able to predict treatment response, primarily through genetics and neuroimaging. In this review we discuss the most promising markers of treatment response and adverse effects emerging from the areas of pharmacogenetics and neuroimaging in depression and schizophrenia.
Keywords: personalized medicine, pharmacogenetics, neuroimaging, antipsychotic, antidepressant, treatment response, adverse effect, depression, schizophrenia
Abstract
Actualmente se cuenta con numerosos ensayos controlados, randomizados y meia-análisis para tomar decisiones clínicas; lo que nos coloca en riesgo de descartar opciones terapéuticas específicas para un subgrupo de pacientes o en forma individual debido a su fracaso en la demostración de eficacia en grandes poblaciones. En la medicina personalizada está emergiendo una nueva era que se focalizará en las diferencias individuales, las que no son evidentes fenomenológicamente. Hay mucha investigatión orientada a identificar genes, endofenotipos y biomarcadores de enfermedad lo que facilitará el diagnóstico y la prediccíon de la evolucíon terapéutica. Estamos en el umbral de la capacidad de predecir la respuesta al tratamiento, fundamentalmente a través de la genética y de las neuroimágenes. En esta revisión se analizan los marcadores más prometedores de la respuesta terapéutica y de los efectos adversos, que están surgiendo desde la farmacogenética y de las neuroimágenes en la depresión y la esquizofrenia.
Abstract
Nous nous appuyons actuellement sur de grandes études contrôlées randomisées et sur des metaanalyses pour prendre des décisions cliniques, ce qui risque d'écarter des options thérapeutiques spécifiques individuelles ou de sous-groupes à cause de leur incapacité à prouver leur efficacité dans la population toute entière, La médecine personnalisée, stratégie d'apparition récente, a pour objectif de se pencher sur des différences individuelles qui ne sont pas évidentes sur le plan phénoménologique. Afin de faciliter le diagnostic et de prévoir l'évolution des traitements, la recherche se dirige vers l'identification des gènes, des endophénotypes et des biomarqueurs de la maladie. Nous sommes sur le point de pouvoir prédire la réponse au traitement, en particulier grâce à la génétique et à la neuro-imagerie. Cet article se propose d'examiner les marqueurs les plus prometteurs de la réponse au traitement et des effets indésirables issus de la pharmacogénétique et de la neuro-imagerie dans la dépression et la schizophrénie.
Since the serendipitous discoveries of chlorpromazine and imipramine, the precursors of current antipsychotics and antidepressants, respectively we have made arguably few strides towards the improvement of clinical outcomes. Our major gains have been in the area of pharmacotherapy acceptance and tolerability. The development of serotonin reuptake inhibitors has dramatically reduced the side effects and lethality in overdose of commonly prescribed antidepressants. Similarly, second-generation antipsychotics have significantly decreased the incidence of extrapyramidal symptoms (EPS), including tardive dyskinesia and parkinsonism, but at the same time have increased the long-term likelihood of mortality and morbidity secondary to adverse metabolic effects.
We remain in an era of uncertainty with regard to the underpinnings of individual variability in order to preemptively differentiate treatment responders from nonresponders. Our current evidence-based medicine relies on large randomized control trials and meta-analyses - average medicine, which ignores individual differences. This dependence on large group analyses places us at a risk of discarding subgroup-specific treatment options owing to their failure to prove efficacious across entire populations.1
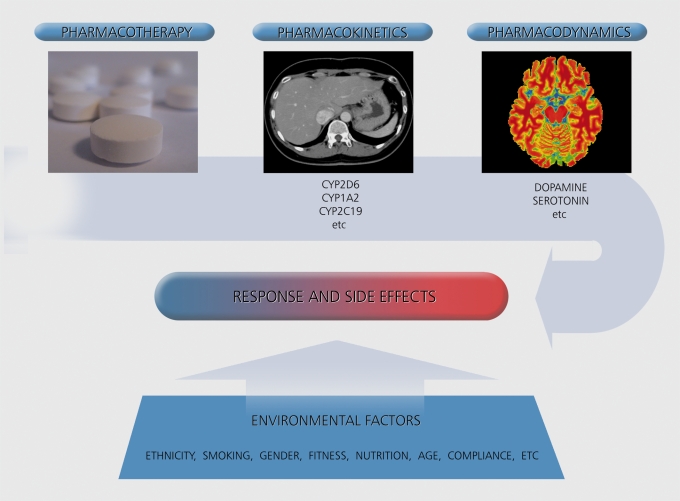
There is a new era emerging in psychiatry of personalized medicine that will focus on individual differences not evident phenomenologically. Much research is directed towards the identification of genes, endophenotypes, and biomarkers of disease that will facilitate diagnosis and predict treatment outcome. Pharmacogenetic studies that explore the role of an individual's genetic makeup in determining the effectiveness of pharmacotherapy are of increasing interest. The rationale for the hypothesized role of pharmacogenetics is based on observations made in family and twin studies, where closely related relatives tend to show similar response or side-effect patterns (reviewed in ref 2). All proteins, including those involved in the metabolism and central effects of pharmaceuticals, can differ as a result of naturally occurring variability in the DNA sequence of the associated gene. This has led investigators to study gene variants for their association with antipsychotic drug outcome. Gene variants (ie, alleles or polymorphisms) that code for the enzymes responsible for drug metabolism can affect pharmacokinetics, and therefore the amount of drug available in the bodyto elicit a response. In addition, gene variants can affect pharmacodynamics, the therapeutic effect of a drug in the target organ (Figure 1 , “PK and PD”). Representative examples in breast cancer research include the overexpression of the ITER2 gene, a positive predictor of response to the drug trastuzumab (Herceptin), and the predictive value of active cytochrome P450 (CYP) 2D6 alleles in tamoxifen discontinuation.3
Figure 1. The influence of pharmacokinetic, pharmacodynamic and environmental factors on pharmacotherapy response and side effects (source: www.silvermedia.ca).
In psychiatry, we lack basic laboratory investigations to diagnose mental illness, let alone genetic advances to guide treatment. A primary goal of current research is to characterize the etiologies and biological susceptibilities of heterogeneous, complex conditions, such as depression and psychosis. We are at the threshold of being able to predict treatment response, primarilythrough genetics and neuroimaging.
Personalized medicine in psychiatry is a broad topic. As such, we will confine our review to the most promising markers of treatment response and adverse effects emerging from the areas of pharmacogenetics and neuroimaging in depression and schizophrenia.
Genetics of antipsychotic drug metabolism, response, and side effects in schizophrenia
Antipsychotic drugs remain the cornerstone of treatment in schizophrenia. However, more than 20% of patients do not initially respond to treatment with drug therapy.4 In addition to lack of response, many patients discontinue their medication due to side effects, which can have serious and devastating consequences.5
In the following sections we discuss the genetics of antipsychotic drug metabolism, response, and side effects in schizophrenia.
Genetics of antipsychotic drag metabolism
The vast majority of antipsychotic drugs are metabolized by the liver enzymes CYP2D6 and CYP2C19, which play critical roles in determining plasma drug levels. Gene variants that confer altered enzymatic activity influence plasma drug levels, and therefore can predict effective drug doses and potential side effects.
The CYP2D6 gene codes for an enzyme that is responsible for metabolizing the majority of antipsychotic medications.6 This enzyme shows genetic variability in activity and is highly polymorphic, with over 70 single nucleotide polymorphisms (SNPs) and copy number variations (CNVs).7 These variations can influence antipsychotic drug activity and a patient's ability to metabolize them. Individuals can be classified, based on their gene polymorphisms, as poor (PM), intermediate (IM), extensive/normal (EM), or ultrarapid drug metabolizers (UM).8 The frequency of PMs, IMs, EMs, and UMs varies across ethnicities. For example, Europeans show the highest frequency of CYP2D6 PMs and African-Americans show the highest frequency of CYP2D6 UMs.9
In theory, the risk of side effects may be higher in individuals with compromised drug metabolism capabilities because of higher drug plasma levels.10 Alternatively, drug plasma levels may be lower and medications, as a result, less efficacious in individuals with high enzymatic activity. 11 The vast majority of individuals will have no or little impaired enzyme activity (ie, are IM or EM). However, it may be extremely valuable for those individuals who show impaired (PM) or markedly increased activity (UM) to have this information taken into consideration when selecting antipsychotic medication, determining appropriate dosage, or interpreting plasma levels in the context of drug monitoring.
Estimated dose adjustments for antipsychotics have been described based on an individual's metabolizer status.12 CYP2D6 and CYP2C19 diagnostic testing is FDA approved with the Roche AmpliChip®, but is also available at decreasing costs every year through other companies. Importantly, results from genotyping analyses are only one factor affecting drug plasma levels and should be considered in conjunction with other important criteria, such as comedication, smoking, and diet.6
Genetics of antipsychotic treatment response
Another important focus of investigation has been antipsychotic drug response in schizophrenia. The firstgeneration studies exploring the genetics of antipsychotic treatment outcome were published in the early to mid 1990s. They were performed with small sample sizes and included patients treated mainly with clozapine, but not exclusively. The most interesting findings, albeit mixed, were obtained for the serotonin 2A (5-HT2A) and the dopamine 2 (DRD2) receptor gene polymorphisms.13 These results suggested that the effect size of these polymorphisms is low and that other factors, including other genes and gene variants, are likely to be involved.
Second-generation studies have included larger samples, more sophisticated analyses, and multiple polymorphisms, which allow for the investigation of haplotypes and genome -wide associations. These continue to produce promising results for the 5-HT2A and DRD2 gene polymorphisms. A comprehensive analysis which included 12 DRD2 gene polymorphisms in a sample of 232 well-characterized subjects identified protective haplotypes in both Europeans and African- Americans.14 A review by Arranz et al concluded that the -141C/T polymorphism in the DRD2 gene is of particular significance due to its association with treatment outcome in two independent samples.2 A more recent meta-analysis of almost 700 individuals supported the association between the -141C/T polymorphism and antipsychotic drug response.15 Intriguingly, this polymorphism is located in the promoter region and could have regulatory effects in addition to functional relevance. Although the role of the DRD2 gene in antipsychotic response is not conclusive, these findings are of particular interest since D2 is the main target of antipsychotics.16
5-HT2A is proposed to be involved in the unique therapeutic action of clozapine.17 Two studies with sufficient statistical power have demonstrated a role for the structural 5-FIT2A His452Tyr polymorphism in predicting clozapine response.18,19 Significant associations have also been described in at least a dozen other genes, such as DRD3, DRD4, 5-HT1A, 5-HT2C, 5-HT6, 5-HTT, BDNF, COMT, GNB3, MDR1, MTHFR, NEF3, NRG1, RGS4 and TNF-alpha. 2,20,21
Of note, the first whole genome-wide association study of antipsychotic drug response was recently conducted by Sullivan et al.21 This approach involves no a priori hypotheses of candidate genes or gene variants, and as a result makes it difficult to interpret the significance of results in the context of adequately controlling for multiple variable testing. No significant findings have been reported thus far.
Also of note, only a few studies have tested for a direct association between CYP450 gene polymorphisms and drug response. These have yielded mostly negative results.2,22
Overall, some interesting findings exist in the area of genetics and antipsychotic response. However, many associations are not conclusive and represent a small fraction of the total variance of treatment outcome. Because the entire genome and candidate gene variability have not been fully explored, more robust observations are expected with the utilization of DNA sequencing techniques. The category “treatment response” may be too broad an outcome measure in genetic studies of heterogeneous conditions. Studies that target specific symptoms, such as neurocognitive and verbal memory scores, may yield more convincing findings.21,23
Genetics of antipsychotic-induced side effects
Antipsychotics can induce a variety of side effects, such as involuntary movements (eg, tardive dyskinesia) and weight gain, both of which appear to be genetically determined.24,25 Compared with phenotypes like treatment response, an analysis of genetic factors associated with side effects may offer several advantages. First, side effects are often more closely related to plasma levels, which can sometimes be predicted by gene variants involved in drug metabolism. Second, compared with treatment response the occurrence of side effects maybe more closely related to specific pharmacodynamic relevant receptors. Third, some side effects such as weight gain can be assessed more easily and reliably as compared with complex phenotypes, such as treatment response.
In a prototypical study of its time, Pollock et al26 prospectively distinguished poor P450 2D6 metabolizers from EM among a group of elderly patients suffering from dementia treated with perphenazine. The poor metabolizers had significantly greater side effects than the 40 extensive metabolizers.
In the case of tardive dyskinesia, previous reports have indicated that CYP1A2 may be of importance.27 Other studies which focused on the Ser9Giy variant of the DRD3 gene reported significant associations, which were supported in two meta-analyses.28,29
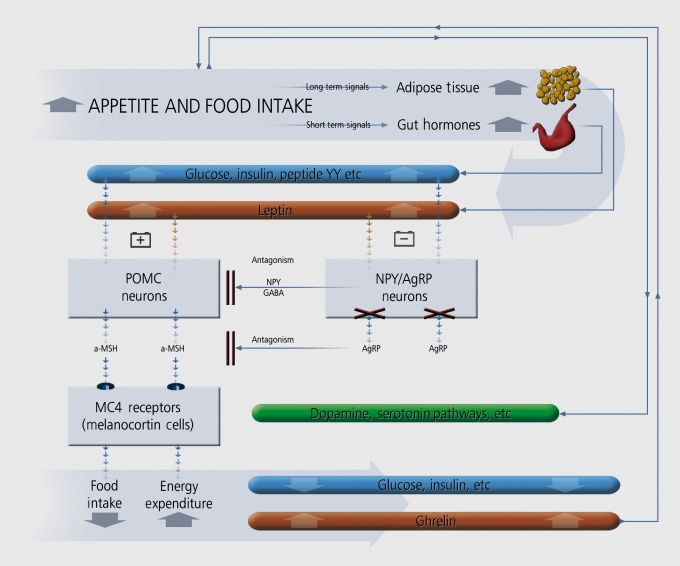
Several interesting studies have now been published regarding the genetics of antipsychotic-induced weight gain. The CYP2D6 gene has been associated with increasing weight. In pharmacodynamic analyses, the most consistent findings involve the promoter polymorphisms of the 5-HT2C gene and the leptin gene. Both genes are involved in energy and fat metabolism in studies of humans and animals (reviewed in ref 30, Figure 2). Further interesting findings are reported in the ADRA2A and SNAP-25 genes, with replications in independent samples.31-35
Figure 2. The interaction between peripheral molecules and central pathways modulating food/energy intake. AgRP, Agouti related protein, GABA, gamma aminobutyricacid, MC4, melanocortin receptor 4, NPY, neuropeptide, POMC, proopiomelanocortin, α-MSH, alpha melanocyte stimulating hormone, (source: www.silvermedia.ca) Adapted from ref 30: Muller DJ, Kennedy JL. Genetics of antipsychotic treatment emergent weight gain in schizophrenia. Pharmacogenomics. 2006;7:863-887. Copyright © Future Medicine Ltd, 2006.
In summary, studies assessing the genetic underpinnings of side effects to antipsychotic medications have yielded interesting findings, although effect sizes for single genes (or gene variants) are small.
Genetics of antidepressant response and drug metabolism in depression
Major depressive disorder (MDD) is one of the fourth major causes of disability worldwide, with tremendous socioeconomic consequences36 Adverse early life events are major predictors of later development of MDD, though genetic factors also appear to have a significant influence (37% heritability in twin studies). Antidepressants are the cornerstone in treating depression; however, only 50% to 70% of the patients respond to initial therapy, and less than 40% patients achieve full remission.37 Furthermore, efficacy of an antidepressant is often only apparent after treating for 4 to 8 weeks. A reliable tool to predict antidepressant response would be of great service to the clinician, leading to greater efficacy and rapidity of response. Pharmacogenetics offers an individually tailored alternative to the trial and error prescription regime. Concordance for antidepressant response has been observed in family studies implicating the role of genetic factors.38,39
Genetics of antidepressant drug metabolism
The therapeutic level achieved by antidepressants is heavily influenced by the metabolic activity of the CYP450 enzymes. CYP2D6 is involved in the metabolism of most tricyclic antidepressants (TCAs) and some SSRIs. Functional polymorphisms lead to varying degrees of metabolic activity that influence plasma drug levels, and allow for the categorization of distinct phenotypes (see Genetics of antipsychotic drug metabolism section above).6,40,41 The UM phenotype is associated with increased clearance of antidepressants and lack of response.42-44 Accordingly, the PM phenotype is reported to lead to increased adverse events with antidepressant treatment.45,46
Some evidence suggests that functional polymorphisms in the CYP2C19 gene also influence serum levels of antidepressants metabolized by this enzyme. UM (CYP2C19*17/*17) exhibited the lowest concentrations of escitalopram, whereas patients with the PM genotype (CYP2C19*2 or *3) exhibited the highest serum levels.47
Genetics of antidepressant treatment response
The selection of candidate genes for investigation is based on the hypothesized association with pharmacological targets of antidepressants. The ability of earlier antidepressants to increase the availability of monoamines within the synapse by either blocking monoamine reuptake (eg, imipramine) or inhibiting monoamine oxidase (eg, iproniazid) led to the monoamine-deficiency hypothesis of depression. As a result, several genes from the monoaminergic systems (eg, serotonin, noradrenaline, and dopamine receptors and transporters) have been investigated for their association with response to antidepressant treatment.48
Among these, the serotonergic system is the most widely investigated. Genetic variation within the serotonin transporter (5-IIydroxytryptamin transporter, 5HTT; SLC6A4) is suspected of conferring a vulnerability to anxiety and affective disorders. 5-HTT is the principal site of initial action for several antidepressants including selective serotonin reuptake inhibitors (SSRIs).49 Polymorphisms within the promoter region were described shortly after the original isolation of the SLC6A4 cDNA on chromosome 17ql2 by Lesch et al.50
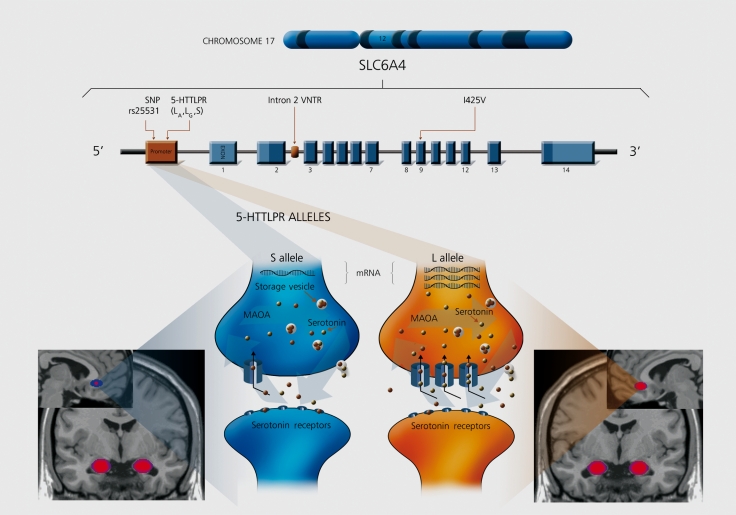
In particular, a site approximately! 200 bp 5* of the first exon of the SLC6A4 gene involves a 22 bp repetitive sequence consisting of two subtypes, a short (S) allele with 14 copies and a long (L) allele with 16 copies.51 This variation is frequently referred to as the serotonin transporter-linked polymorphic region (5-HTTLPR).The S allele is associated with a reduction of function as compared with the L allele. Cells homozygous for the L variant can have up to 66% more 5-1 ITT mRNA expression, greater serotonin transporter density in platelet and neuron cell membranes, and two times the serotonin uptake than cells with the S/S genotype (Figure 3).52-57
Figure 3. Serotonin transporter gene (SLC6A4) and function. 5-HTTLPR x PFOAmygdala endophenotype interaction. Allelic variation of the serotonin transporter (5-HTT), including the serotonin-transporter-gene-linked polymorphic region (5-HTTLPR), the variable number of tandem repeats (Intron 2 VNTR), rs25531 single nucleotide polymorphism (SNP), and the missense variant He425Val (I425V). The long (L) allele (orange) of 5-HTTLPR produces significantly less 5-HTT mRNA and protein expression, than the short (S) allele (blue), leading to higher concentrations of serotonin in the synaptic cleft. 5-HTTLPR s allele carriers show significantly less functional coupling between the amygdala and perigenual anterior cingulate cortex than LA individuals. MAOA, monoamine oxidase A. (source: www.silvermedia.ca) .
The L variant is generally associated with a better antidepressant response in Caucasian patients.58 In a metaanalysis by Serre tti et al, L carriers had better response and remission rates within 4 weeks of antidepressant treatment when compared with subjects with the SS genotype. Conversely, in an investigation of the STAR*D sample treated with citalopram (total n=1659) no association was observed between the 5-IITTLPR polymorphism and treatment tolerance or outcome.59 In another analysis of the STAR*D sample a significant association was found between the L allele and remission in white nonhispanic patients.60
Recently, the Genome Based Therapeutic Drugs for Depression (GENDEP) study61 found that the L allele was associated with better response to escitalopram. A significant interaction was identified between 5FITTLPR, drug and gender, with the effect concentrated in males. Of note, the single nucleotide polymorphism (SNP) rs2020933, found at the 5' end of the 5-IITTLPR gene, also influenced treatment outcome in this study.
A common A>G functional polymorphism within the L allele has also been identified.51 The G variant of this polymorphism (LG) shows transcription levels similar to the S allele, whereas the A genotype (LA) shows higher expression levels. In the STAR*D study they reported a significant association between the LA allele and reduced adverse events in the white nonhispanic population, but not with treatment outcome.59
The influence of 5-HTTLPR on antidepressant response is quite robust to ethnic differences although significant heterogeneity exists in Asian samples.62 In contrast to Caucasian subjects, Asians carrying the S allele have been reported to respond better to antidepressants, although findings are mixed (see refs 37, 58, 63).
Another gene of active investigation is HT2RA, which codes for the 5-HT2A receptor, a target of both antidepressant medications and second -generation antipsychotics. A polymorphism rs7997012 found in the second intron was significantly associated with citalopram response in the STAR*D study64 In addition to this variant, the A1438G polymorphism also showed evidence of association with treatment outcome. Participants who were homozygous for the A allele had an 18% absolute risk reduction of having no treatment response compared with those homozygous for the Gallele. This finding appeared specific to white subjects. Conversely, the GENDEP study61 failed to replicate this association with rs7997012, and found that the G allele of another polymorphism, rs9316233, was associated with escitalopram response. Inconsistent findings have also been reported for the Callele of the T102C polymorphism.58 Despite the lack of consistent findings for a specific polymorphism moderating response, the FIT2RA gene as a whole appears to be of importance in depression outcome.
Many other genes associated with the different monoaminergic systems that are either inconsistently associated with antidepressant response or that have produced contradictory results are reviewed in detail elsewhere (see refs 58, 63). These include HTR1A, TPH1, TPH2, MAOA, MAOB, COMT, DAT1, SLC6A3, D2, D3, D4, NET1, SLC6A2, ADRA2A, AD RBI, G protein, beta polypeptide 3.
Brain-derived neurotrophic factor (BDNF) is an important peptide abundantly expressed in limbic structures. BDNF is critical for axonal growth, neuronal survival, and synaptic plasticity. The observations of stress-induced decrease in hippocampal BDNF levels and increase in BDNF-mediated signaling following chronic treatment with antidepressants suggest a possible role in depression and its treatment:48,65,66 However, inconsistent and regionspecific effects have also been observed.48 A functional coding SNP rs6265 causes a Valine to Methionine change at codon 66, which leads to impaired intracellular trafficking and secretion of the mature BDNF protein. Carriers of the Met allele have significantly lower hippocampal volume than subjects homozygous for the Val allele.67 Although several studies have found an association between the Met allele and antidepressant response,63''68 the sample sizes were small, and the results have been inconsistent.61 In addition to the Val66Met allele, a polymorphism in the 5' untranslated region of the BDNF gene (rs61 888800) was associated with antidepressant response in Mexican-American subjects.69
This observation requires replication.
Early life stress and deregulation of the hypothalamicpituitary-adrenal (IIPA) axis are also linked with depression treatment outcome;48,70 One of the important genes that has emerged from the UFA axis is FKBP5 (FK506 binding protein 51), a cochaperone of 90 kDa heat shock protein, which regulates glucocorticoid receptor sensitivity. Carriers of the TT genotype of rsl360780 polymorphism in intron 2 of FKBP5 were demonstrated to have a better treatment outcome than other genotypes.71 This observation was replicated in a separate sample in the same study, and in two other independent studies. Smaller investigations of Spanish and Korean populations failed to reproduce this association (see ref 72).
Genetics of antidepressant-induced side effects
Side effects of antidepressant treatment have emerged as important reasons for medication discontinuation and non compliance. The first-generation TCAs and monoamine oxidase inhibitors (MAOIs) were primarily associated with sedation, weight gain, and anticholinergic side effects, including dry mouth, blurred vision, cardiac effects, and death by overdose. The newer antidepressants, including SSRIs and SNRIs, have better and safer side-effect profiles, but tend to cause nausea, diarrhea, nervousness, agitation, insomnia, and sexual side effects.
Similar to studies of antidepressant response, the candidate genes extensively investigated in relation to antidepressant induced side effects are from the serotonergic system. The presence of the 5-HTTLPR L allele is generally associated with fewer treatment related side effects. Negative studies are also reported in the literature. A recent meta-analysis found the L allele conferred protection against antidepressant side effects for all antidepressants (OR 0.64) ,63 the significance of which became more robust when analyzed with SSRI-induced side effects only. The same meta-analysis found that the presence of the -1438 G/G polymorphism of HTR2A increased the risk of antidepressant side effects (OR 1.91). Several other pharmacodynamic genes were investigated with contradictory results.63
Although strongly related to drug plasma levels, fewimportant discoveries have been made linking pharmacokinetic associated genes and antidepressant-induced side effects. CYP2D6 polymorphisms are reported to influence the emergence of TCA and MAOI side effects, but not the majority of the newer antidepressants (for details see ref 72). Thus, the clinical utility of pharmacokinetic genes predicting side effects remains limited. To date, the most promising observations of an association between genes and antidepressant side effects have come from the 5-HTTLPR polymorphism.
Personalized medicine and neuroïmagïng
A major barrier to progress in the study of complex diseases, such as schizophrenia and depression, is the heterogeneity arising from etiological and phenotypic diversity. A significant amount of neuroimaging research has been conducted to identify biomarkers or endophenotypes which may reduce the heterogeneity. Proximal markers are presumed to be less genetically complex than the clinical phenotype. The identification of intermediary phenomena and specific gene-endophenotype linkages may increase the individual variability explained by candidate genes. The validity of biomarkers and endophenotypes is contingent on their sensitivity and specificity for the disease in question.37
In the next sections we present the most promising neuroimaging markers of treatment response in depression and schizophrenia.
Neuroimaging markers of antidepressant treatment outcome
Anterior cingulate cortex
The most commonly reported finding in neuroimaging studies of depression is that increased rostral anterior cingulate cortex (rACC) activity predicts later response to depression treatment, including antidepressants,73-75 CBT,76 and sleep deprivation.77 Structural MRI measurements of the ACC have also demonstrated an association with treatment response.78 Hie ACC is implicated in numerous brain functions, likely due to its neuroanatomical position as a bridge between frontal cortical and subcortical structures.37 The rACC, primarily Brodmann area 25, is consistently reported to be hyperactive in depressed treatment responders.79 According to Mayberg et al's theory of depression, cortical-subcortical regulation shifts from the dorsolateral to the ventrolateral prefrontal cortex (PFC), which contributes to rACC hyperactivity.75 It is this region that is a target of deep brain stimulation (DBS) studies of treatment-resistant depression.80 In further support of this theory, two independent groups of researchers have identified increased pretreatment activity in rACC theta activity in responders using low-resolution electromagnetic tomography.81,87
Functional neuroimaging studies during active task conditions can facilitate distinguishing responders from nonresponders by targeting the neurocircuitry involved in depression.37 The aim is to reduce unexplained background cerebral activity (“noise”) present during the resting state, thereby increasing the signal-to-noise ratio. Common approaches involve comparing the brain regions activated during the presentation of affectively laden or sad facial expressions versus neutral or positive stimuli. In an IMRI study by Chen el al,78 participants who displayed greater pretreatment activation within ACC in response to negative versus neutral stimuli displayed the greatest response to treatment.73,74,83 Other fMRI studies have also demonstrated a relationship between pretreatment ACC activity and treatment outcome.73,74
Amygdala
Greater pretreatment amygdala activity is also associated with treatment response. Increased signal in the amygdala following the presentation of negative facial expressions is related to major depression severity84,85 and was demonstrated to predict improvement.86 Normalization of amygdala reactivity to affective stimuli is consistently reported to occur with antidepressant treatment.74,87,88 The same study that reported an association between PFC activity and response to CBT found that heightened amygdala activity to negative words also predicted response.76 Intriguingly, there is evidence to suggest that the variability in amygdala and PFC activity is moderated, in part, by the serotonin transporter gene 5-HTTLPR.
5-HTTLPR x PFC/ Amygdala endophenotype interaction
5-IITTLPR appears to have modulatory effect on emotion89 via top-down cortico-amygdala regulation.1 On the one hand, diminished cortical structure and function is associated with depression and anxiety. Results from functional brain imaging studies suggest that the S allele contributes to increased amygdala reactivity via direct anterior cingulate (ACC) -amygdala dysregulation,90,91 and indirect compensatory activation of the ventromedial prefrontal cortex (Figure J).91 Consistent with these findings, the S allele was associated with peak gray-matter volume reductions in the subgenual ACC, a structure implicated in both depression and anxiety.92 This theory is substantiated by the dense serotonergic connections between the ACC and amygdala in comparison with the relatively few amygdala connections with the ventromedial PFC.91 On the other hand, the gain of function L allele had the opposite effect of the S variant on corticoamygdala regulation.90,91 The LA/LA genotype confers a modest risk of OCD,51 which is associated with hyperfrontality, including increased ACC metabolic activity and gray matter volume.93
Hippocampus
Lower hippocampal volume is associated with depression, frequency of episodes, and chronicity of illness.94
Hippocampal volume loss, as measured with structural MRI, is also characteristic of late-life depression and may be independently influenced by the val66met BDNF (See section Genetics of antidepressant drug response) and 5-HTTLPR polymorphisms.
5-HTTLPR appears to influence the pathogenesis of depression depending on the age of onset.1 The S allele was associated with reduced hippocampal volumes in elderly subjects with early-onset depression (ie, first episode 50 years or younger), while homozygosity for the L variant predicted smaller hippocampal volumes in depressed subjects with later onset.95 By contrast, the L/L genotype appears to contribute to the relationship between late-onset depression and dementia. Reduced hippocampal volume in this context may represent the effects of subcortical ischemia in vascular cognitive impairment96 or the prodromal symptoms of depression often seen in Alzheimer's dementia.97
Although not consistently reported, some studies have shown that antidepressant treatment may prevent or even reverse hippocampal atrophy via neurogenesis.98,99 More research is required to determine the reliability of hippocampal atrophy as a predictor treatment response.37
Other regions
White matter hyperintensities on structural MRI negatively predict treatment response in late-onset depression.100,101 Fewer white matter lesions are associated with remission and maintenance of remission in late-life depression with antidepressant treatment.101 White matter disease commonly results in varied neuropsychological deficits, primarily memory, executive, and language function,102 which are associated with poor response to antidepressants.37
Although the 5-1 IT transporter is widely believed to be involved in the pathogenesis and treatment of depression, positron emission tomography (PET) studies have shown both increased and decreased binding potential of the 5-IIT transporter in the context of depression. These mixed results may reflect inter-study variation of etiology or mood state leaving it as an unreliable biomarker at present.103
Neuroimaging markers of antipsychotic treatment outcome
Neuroimaging findings predicting treatment response to antipsychotics are less robust than those for antidepressants. Both brain atrophy by various measures (eg, sulcal width, ventricle size, etc.) and rate of gray matter loss are associated with poorer treatment outcome. Ventriculomegaly104 and cortical105 and cerebellar atrophy106 were found to predict response. More recently, the extent of gray matter atrophy over time was a better predictor of outcome than baseline abnormalities.107 Neurochemical (PET) imaging offers a minimally invasive means of exploring distinct properties and cerebral distribution of neurotransmitter systems in vivo through the binding of receptor specific radiotracers. Binding potential is a principal measure in PET imaging studies that reflects both the density of available neuroreceptors and the affinity of a radiotracer to a given receptor. PET studies of dopamine 2 receptor (D9) binding potential have shown that greater than 60% occupancy is associated with increased likelihood of antipsychotic response, while greater than 80% robustly predicts EPS.108 The clinical application of neurochemical PET imaging remains limited by cost and availability, and while it has been instrumental in the reduction of antipsychotic dosing over the past decade and predicting dosing of second-generation antipsychotic drugs, it has not been applicable to the individual patients due to high intersubject variability. However, a recent line of investigation in older patients with schizophrenia has provided new evidence from neuroreceptor PET imaging that may have potential for bedside translation. These studies have suggested that measurable changes in receptor reserve with aging is associated with antipsychotic medication and that medicated older patients on a stable dose of risperidone maintain individually consistent levels of receptor occupancy, plasma concentration, and psychopathology, supporting the use of this technology in prospective studies.109 Presuming medication adherence, PET imaging data may, in the future, be used to facilitate the determination if worsening symptomatology or side effects are either due to alterations in neurochemistry or drug failure. Theoretically, this could be performed with antidepressant radiotracers specific for the serotonin transporter as well.110 In the future, PET imaging in conjunction with genetic testing for CYP 450 metabolism may help define individually tailored antipsychotic dosing schedules. UM may require higher doses of an antipsychotic to achieve the desired receptor occupancy, beyond the upper limits of what is currently defined as the normal range. Conversely, PM may require typically subtherapeutic doses to avoid developing side effects, such as EPS. Preliminary work directed towards age-specific dosing of antipsychotics has shown that EPS occurs at 50% to 70% D, receptor occupancy in the elderly, which suggests treatment efficacy occurs at even lower receptor blockade (-40% to 50%).111 In the same study, a verystrong association was observed between D, receptor occupancy and antipsychotic plasma levels. Pending replication, these results raise the possibility of predicting individualized antipsychotic dosing. Using population pharmacokinetic methodology in conjunction with neuroreceptor PET data, our group is currently investigating the predictive validity of individualized antipsychotic dosing using widely available bedside measures including plasma drug levels, drug dose, demographic factors, and concomitant medications.112
Conclusion
Personalized medicine promises the development of individually designed treatments based on the integration of all clinically relevant information, including data derived from laboratory, genetic, and imaging investigations, etc. The identification of pharmacogenetic and neuroimaging biomarkers associated with side effects and treatment response are active areas of research in psychiatry. The question is, when will genetic testing and sophisticated functional neuroimaging studies be implemented in clinical practice? With regards to genetic testing, a relative timeline can already be given. Genetic tests for critical drug-metabolizing genes, such as CYP2D6 and CYP2C19, are already available and can provide clinically useful information, potentially improving response rates and safety for those individuals who are poor or rapid metabolizers.113 It remains uncertain whether widespread genotyping prior to the onset of treatment therapy can contribute substantially to therapeutic outcome. Challenges facing the field include phenotypic and etiological heterogeneity, technological limitations, and contemporary research approaches. It seems that genetic testing for side-effect prediction has the highest likelihood of being incorporated into clinical practice. Supporting this, a test for clozapine-induced agranulocytosis is now available with satisfying sensitivity and specificity.114 Computational models that include gene variants and other factors associated with antipsychoticinduced weight gain have yielded promising results.115 Tests related to treatment response may follow through the inclusion of more sophisticated genotyping techniques (eg, sequencing) and the analysis of refined endophenotypes, such as specific symptoms or symptom clusters. Future development of algorithm-based approaches requires the integration of additional genetic and nongenetic factors.
Neuroimaging research has produced encouraging associations between imaging endophenotypes and treatment outcome, such as the 5-HTTLPR x PFC/amygdala interaction. Nonetheless, these observations lack the positive and negative predictive value required to reliably distinguish responders from nonresponders to be used clinically. Based on current research, imaging markers explain a significant, but modest, portion of the total variance. More research is required with larger, less heterogeneous samples in conjunction with other markers, eg, genotyping and electrophysiological measures. Most neuroscience research thus far involves the application of various theoretical approaches (ie, neuroimaging, genetics, neuropsychological, and physiological, etc) in isolation. The next step in the development of personalized medicine is the formation of standardized multimodal research models to better characterize markers of treatment response. Now is a time for optimism in the emerging ability of pharmacogenetics and neuroimaging to provide meaningful help to the physician in developing individually tailored treatments for complex, heterogeneous psychiatric disorders.
Selected abbreviations and acronyms
- 5-HT
serotonin
- 5-HTTLPR
serotonin transporter-linked polymorphic region
- ACC
Anterior cingulate cortex
- BDNF
brain-derived neurotrophic factor
- CYP
cytochrome P450
- PM
poor drug metabolizer
- UM
ultrarapid drug metabolizer
Contributor Information
Philip Gerretsen, Multimodal Imaging Group.
Daniel J. Müller, Centre for Addiction and Mental Health, University of Toronto, Canada; Pharmacogenetics Research Clinic, Neurogenetics Section.
Arun Tiwari, Neurogenetics Section.
David Mamo, Multimodal Imaging Group, Division of Geriatric Psychiatry.
Bruce G. Pollock, Centre for Addiction and Mental Health, University of Toronto, Canada; Division of Geriatric Psychiatry, Centre for Addiction and Mental Health, University of Toronto, Canada.
REFERENCES
- 1.Gerretsen P, Pollock BG. Pharmacogenetics and the serotonin transporter in late-life depression. . Expert Opin Drug Metab Toxicol. 2008;4:1465–1478. doi: 10.1517/17425250802560279. [DOI] [PubMed] [Google Scholar]
- 2.Arranz MJ, de Leon J. Pharmacogenetics and pharmacogenomics of schizophrenia: A review of last decade of research. . Mol Psychiatry. 2007;12:707–747. doi: 10.1038/sj.mp.4002009. [DOI] [PubMed] [Google Scholar]
- 3.Rae JM, Sikora MJ, Henry NL, et al. Cytochrome P450 2D6 activity predicts discontinuation of tamoxifen therapy in breast cancer patients. . Pharmacogenomics J. 2009;9:258–264. doi: 10.1038/tpj.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Essali A, Al-Haj HIM, Li C, et al. Clozapine versus typical neuroleptic medications for schizophrenia (review). . Cochrane Database Syst Rev. 2002;2:CD000059–xx. doi: 10.1002/14651858.CD000059.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nose M, Barbui C, Tansella M. How often do patients with psychosis fail to adhere to treatment programs? A systematic review. . Psychol Med. 2003;33:1149–1160. doi: 10.1017/s0033291703008328. [DOI] [PubMed] [Google Scholar]
- 6.de Leon J. Pharmacogenomics: the promise of personalized medicine for CNS disorders. . Neuropsychopharmacology. 2009;34:159–172. doi: 10.1038/npp.2008.147. [DOI] [PubMed] [Google Scholar]
- 7.Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): Clinical consequences, evolutionary aspects and functionary diversity. . Pharmacogenomics J. 2005;5:6–13. doi: 10.1038/sj.tpj.6500285. [DOI] [PubMed] [Google Scholar]
- 8.de Leon J, Susce MT, Murray-Carmichael E. The AmpliChip(TM) CYP450 genotyping test: Integrating a new clinical tool. . Molecular Diagnosis Ther. 2006;10:135–151. doi: 10.1007/BF03256453. [DOI] [PubMed] [Google Scholar]
- 9.Gaedigk A, Bradford D, Marcucci KA, et al. Unique CYP2D6 activity distribution and genotype-phenotype discordance in black Americans. . Clin Pharmacol Ther. 2002;72:76–83. doi: 10.1067/mcp.2002.125783. [DOI] [PubMed] [Google Scholar]
- 10.Rogers JF, Nafziger AN, Bertino JS. Pharmacogenetics affects dosing, efficacy, and toxicity of cytochrome P450-metabolized drugs. . Am J Med. 2002;113:746–750. doi: 10.1016/s0002-9343(02)01363-3. [DOI] [PubMed] [Google Scholar]
- 11.Johansson I, Lundqvist E, Bertilsson L, et al. Inherited amplification of an active gene in the cytochrome P450 CYP2D locus as a cause of ultrarapid metabolism of debrisoquin. . Proc Nat Acad Sci U S A. 1993;90:11825–11829. doi: 10.1073/pnas.90.24.11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirchheiner J, Nickchen K, Bauer M, et al. Pharmacogenetics of antidepressants and antipsychotics: The contribution of allelic variations to the phenotype of drug response. . Mol Psychiatry. 2004;9:442–473. doi: 10.1038/sj.mp.4001494. [DOI] [PubMed] [Google Scholar]
- 13.Muller DJ, De Luca V, Kennedy JL. Overview: towards individualized treatment in schizophrenia. . Drug Development Research. 2003;60:75–94. [Google Scholar]
- 14.Hwang R, Shinkai T, De Luca V, et al. Association study of 12 polymorphisms spanning the dopamine D(2) receptor gene and clozapine treatment response in two treatment refractory/intolerant populations. . Psychopharmacology. 2005;181:179–187. doi: 10.1007/s00213-005-2223-5. [DOI] [PubMed] [Google Scholar]
- 15.Malhotra AK, Zhang J-P, Lencz T. Dopamine receptor variation and clinical response to antipsychotic drug treatment. Paper presented at: 8th Pharmacogenetics in Psychiatry Meeting. xx. New York, USA; April 17-18, 2009;xx:xx–xx. [Google Scholar]
- 16.Kapur S. Psychosis as a state of aberrant salience: a framework linking biology, phenomenology, and pharmacology in schizophrenia. . Am J Psychiatry. 2003;160:13–23. doi: 10.1176/appi.ajp.160.1.13. [DOI] [PubMed] [Google Scholar]
- 17.Meltzer HY. Clinical studies on the mechanism of action of clozapine: The dopamine-serotonin hypothesis of schizophrenia. . Psychopharmacology. 1989;99:S18–S27. doi: 10.1007/BF00442554. [DOI] [PubMed] [Google Scholar]
- 18.Arranz MJ, Munro J, Owen MJ, et al. Evidence for association between polymorphisms in the promoter and coding regions of the 5-HT2A receptor gene and response to clozapine. . Mol Psychiatry. 1998;3:61–66. doi: 10.1038/sj.mp.4000348. [DOI] [PubMed] [Google Scholar]
- 19.Masellis M, Basile V, Meltzer HY, et al. Serotonin subtype 2 receptor genes and clinical response to clozapine in the schizophrenia patients. . Neuropsychopharmacology. 1998;19:123–132. doi: 10.1016/S0893-133X(98)00007-4. [DOI] [PubMed] [Google Scholar]
- 20.Campbell DB, Ebert PJ, Skelly T, et al. Ethnie stratification of the association of RGS4 variants with antipsychotic treatment response in schizophrenia. . Biol Psychiatry. 2008;63:32–41. doi: 10.1016/j.biopsych.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sullivan PF, Lin D, Tzeng JY, et al. Genomewide association for schizophrenia in the CATIE study: result of stage 1. . Mol Psychiatry. 2008;13:570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grossman I, Sullivan PF, Walley N, et al. Genetic determinants of variable metabolism have little impact on the clinical use of leading antipsychotics in the CATIE study. Genet . Med. 2008;10:720–729. doi: 10.1097/GIM.0b013e3181863239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Need AC, Dongliang G, Weale ME, et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. . PLoS Genetics. 2009;5:e1000373–xx. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller DJ, Schulze TG, Knapp M, et al. Familial occurrence of tardive dyskinesia. . Acta Psychiatrica Scandinavica. 2001;104:375–379. doi: 10.1034/j.1600-0447.2001.00401.x. [DOI] [PubMed] [Google Scholar]
- 25.Wehmeier PM, Gebhardt S, Schmidtke J, et al. Clozapine: Weight gain in a pair of monozygotic twins concordant for schizophrenia and mild mental retardation. . Psychiatry Res. 2005;133:273–276. doi: 10.1016/j.psychres.2004.02.018. [DOI] [PubMed] [Google Scholar]
- 26.Pollock BG, Mulsant BH, Sweet RA, Rosen J, Altieri LP, Perel JM. Prospective cytochrome P450 phenotyping for neuroleptic treatment in dementia. . Psychopharmacol Bull. 1995;31:327–331. [PubMed] [Google Scholar]
- 27.Tiwari AK, Deshpande SN, Rao ÂR, et al. Genetic susceptibility to tardive dyskinesia in chronic schizophrenia subjects: I. Association of CYP1A2 gene polymorphism. . Pharmacogenomics. 2005;5:60–69. doi: 10.1038/sj.tpj.6500282. [DOI] [PubMed] [Google Scholar]
- 28.Bakker PR, van Harten PN, van Os J. Antipsychotic-induced tardive dyskinesia and the ser9gly polymorphism in the DRD3 gene: a meta analysis. . Schizophrenia Res. 2006;83:185–192. doi: 10.1016/j.schres.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 29.Lerer B, Segman RH, Fangerau H, et al. Pharmacogenetics of tardive dyskinesia: combined analysis of 780 patients supports association with dopamine D3 receptor gene Ser9Gly polymorphism. . Neuropsychopharmacology. 2002;27:105–119. doi: 10.1016/S0893-133X(02)00293-2. [DOI] [PubMed] [Google Scholar]
- 30.Muller DJ, Kennedy JL. Genetics of antipsychotic treatment emergent weight gain in schizophrenia. . Pharmacogenomics. 2006;7:863–887. doi: 10.2217/14622416.7.6.863. [DOI] [PubMed] [Google Scholar]
- 31.Muller DJ, Klempan TA, De Luca V, et al The SNAP-25 gene may be associated with clinical response and weight gain in antipsychotic treatment of schizophrenia. . Neurosci Lett. 2005;379:81–89. doi: 10.1016/j.neulet.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 32.Musil R, Spellman I, Riedel M, et al. SNAP-25 gene polymorphisms and weight gain in schizophrenia patients. . J Psychiatr Res. 2008;42:963–970. doi: 10.1016/j.jpsychires.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 33.Park YM, Chung YC, Lee SH, et al. Weight gain associated with the alpha(2A)-adrenergic receptor - 1291 C/G polymorphism and olanzapine treatment. . Am J Med Genet B Neuropsychiatr Genet. 2006;141B:394–397. doi: 10.1002/ajmg.b.30311. [DOI] [PubMed] [Google Scholar]
- 34.Sickert L, Muller DJ, Tiwari AK, et al. Association of the -1291 C/Galpha2a adrenergic receptor polymorphism and antipsychotic induced weight gain in European-Americans. . Pharmacogenomics. 2009;10:1169–1176. doi: 10.2217/pgs.09.43. [DOI] [PubMed] [Google Scholar]
- 35.Wang YC, Baj YM, Chen JY, Lin CC, Lai IC, Liou YJ. Polymorphism of the adrenergic receptor alpha 2a-1291C>G genetic variation and clozapine-induced weight gain. . J Neural Transm. 2005;112:1463–1468. doi: 10.1007/s00702-005-0291-7. [DOI] [PubMed] [Google Scholar]
- 36.xx WHO. World Health Report 2001 - Mental Health: New Understanding, New Hope. Geneva, Switzerland: World Health Organization. 2001;xx:xx–xx. [Google Scholar]
- 37.Kemp AH, Gordon E, Rush AJ, Williams LM. Improving the prediction of treatment response in depression: integration of clinical, cognitive, psychophysiological, neuroimaging, and genetic measures. . CNS Spectr. 2008;13:1066–1086; quiz 87-88. doi: 10.1017/s1092852900017120. [DOI] [PubMed] [Google Scholar]
- 38.Franchini L, Serretti A, Gasperini M, Srneraldi E. Familial concordance of fluvoxamine response as a tool for differentiating mood disorder pedigrees, . j Psychiatr Res. 1998;32:255–259. doi: 10.1016/S0022-3956(98)00004-1. [DOI] [PubMed] [Google Scholar]
- 39.O'Reilly RL, Bogue L, Singh SM. Pharmacogenetic response to antidepressants in a multicase family with affective disorder. . Biol Psychiatry. 1994;36:467–471. doi: 10.1016/0006-3223(94)90642-4. [DOI] [PubMed] [Google Scholar]
- 40.Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. . Clin Pharmacol Ther. 2008;83:234–42. doi: 10.1038/sj.clpt.6100406. [DOI] [PubMed] [Google Scholar]
- 41.Kirchheiner J. CYP2D6 phenotype prediction from genotype: which system is the best? . Clin Pharmacol Ther. 2008;83:225–227. doi: 10.1038/sj.clpt.6100455. [DOI] [PubMed] [Google Scholar]
- 42.Kirchheiner J, Henckel HB, Meineke I, Roots I, Brockmoller J. Impact of the CYP2D6 ultrarapid metabolizer genotype on mirtazapine pharmacokinetics and adverse events in healthy volunteers. . J Clin Psychopharmacol. 2004;24:647–652. doi: 10.1097/01.jcp.0000145341.30547.f0. [DOI] [PubMed] [Google Scholar]
- 43.Kawanishi C, Lundgren S, Agren H, Bertilsson L. Increased incidence of CYP2D6 gene duplication in patients with persistent mood disorders: ultrarapid metabolism of antidepressants as a cause of nonresponse. A pilot study. . Eur j Clin Pharmacol. 2004;59:803–807. doi: 10.1007/s00228-003-0701-4. [DOI] [PubMed] [Google Scholar]
- 44.Schenk PW, van Fessem MA, Verploegh-Van Rij S, et al. Association of graded allele-specific changes in CYP2D6 function with imipramine dose requirement in a large group of depressed patients. . Mol Psychiatry. 2008;13:597–605. doi: 10.1038/sj.mp.4002057. [DOI] [PubMed] [Google Scholar]
- 45.Rau T, Wohlleben G, Wuttke H, et al. CYP2D6 genotype: impact on adverse effects and nonresponse during treatment with antidepressantsa pilot study. . Clin Pharmacol Ther. 2004;75:386–393. doi: 10.1016/j.clpt.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 46.Bijl MJ, Visser LE, Hofman A, et al. Influence of the CYP2D6*4 polymorphism on dose, switching and discontinuation of antidepressants. . Br J Clin Pharmacol. 2008;65:558–564. doi: 10.1111/j.1365-2125.2007.03052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rudberg I, Mohebi B, Hermann M, Refsum H, Molden E. Impact of the ultrarapid CYP2C19*17 allele on serum concentration of escitalopram in psychiatric patients. . Clin Pharmacol Ther. 2008;83:322–327. doi: 10.1038/sj.clpt.6100291. [DOI] [PubMed] [Google Scholar]
- 48.Krishnan V, Nestler EJ. The molecular neurobiology of depression. . Nature. 2008;455:894–902. doi: 10.1038/nature07455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lotrich FE, Pollock BG, Ferrell RE. Polymorphism of the serotonin transporter: implications for the use of selective serotonin reuptake inhibitors. . Am J Pharmacogenet. 2001;1:1 53–164. doi: 10.2165/00129785-200101030-00001. [DOI] [PubMed] [Google Scholar]
- 50.Lesch KP, Wolozin BL, Estler HC, Murphy DL, Riederer P. Isolation of a cDNA encoding the human brain serotonin transporter, . j Neural Transm. 1993;91:61–72. doi: 10.1007/BF01244919. [DOI] [PubMed] [Google Scholar]
- 51.Hu X-Z, Lipsky RH, Zhu G, et al. Serotonin transporter promoter gainof-function genotypes are linked to obsessive-compulsive disorder. . Am J Hum Genet. 2006;78:815–826. doi: 10.1086/503850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greenberg BD, Tolliver TJ, Huang SJ, Li Q, Bengel D, Murphy DL. Genetic variation in the serotonin transporter promoter region affects serotonin uptake in human blood platelets. . Am J Med Genet. 1999;88:83–87. [PubMed] [Google Scholar]
- 53.Heinz A, Jones DW, Mazzanti C, et al. A relationship between serotonin transporter genotype and in vivo protein expression and alcohol neurotoxicity. . Biol Psychiatry. 2000;47:643–649. doi: 10.1016/s0006-3223(99)00171-7. [DOI] [PubMed] [Google Scholar]
- 54.Little KY, McLaughlin DP, Zhang L, et al. Cocaine, ethanol, and genotype effects on human midbrain serotonin transporter binding sites and mRNA levels. . Am J Geriatr Psychiatry. 1998;155:207–213. doi: 10.1176/ajp.155.2.207. [DOI] [PubMed] [Google Scholar]
- 55.Heils A, Teufel A, Petri S, et al. Allelic variation of human serotonin transporter gene expression. . J Neurochem. 1996;66:2621–2624. doi: 10.1046/j.1471-4159.1996.66062621.x. [DOI] [PubMed] [Google Scholar]
- 56.Lesch KP, Bengel D, Heils A, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. . Science. 1996;274:1527–1531. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- 57.Rausch JL. Initial conditions of psychotropic drug response: studies of serotonin transporter long promoter region (5-HTTLPR), serotonin transporter efficiency, cytokine and kinase gene expression relevant to depression and antidepressant outcome. . Prog Neuropsychopharrnacol Biol Psychiatry. 2005;29:1046–1061. doi: 10.1016/j.pnpbp.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 58.Drago A, De Ronchi D, Serretti A. Pharmacogenetics of antidepressant response: an update. . Hum Genomics. 2009;3:257–274. doi: 10.1186/1479-7364-3-3-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu XZ, Rush AJ, Charney D, et al. Association between a functional serotonin transporter promoter polymorphism and citalopram treatment in adult outpatients with major depression. . Arch Gen Psychiatry. 2007;64:783–792. doi: 10.1001/archpsyc.64.7.783. [DOI] [PubMed] [Google Scholar]
- 60.Mrazek DA, Rush AJ, Biernacka JM, et al. SLC6A4 variation and citalopram response. . Am J Med Genet B Neuropsychiatr Genet. 2009 ;150B:341–351. doi: 10.1002/ajmg.b.30816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huezo-Diaz P, Uher R, Smith R, et al. Moderation of antidepressant response by the serotonin transporter gene. . Br J Psychiatry. 2009;195:30–38. doi: 10.1192/bjp.bp.108.062521. [DOI] [PubMed] [Google Scholar]
- 62.Serretti A, Kato M, de Ronchi D, Kinoshita T. Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with selective serotonin reuptake inhibitor efficacy in depressed patients. . Mol Psychiatry. 2007;12:247–257. doi: 10.1038/sj.mp.4001926. [DOI] [PubMed] [Google Scholar]
- 63.Kato M, Serretti A. Review and meta-analysis of antidepressant pharmacogenetic findings in major depressive disorder. . Mol Psychiatry. In press. xx;xx:xx–xx. doi: 10.1038/mp.2008.116. [DOI] [PubMed] [Google Scholar]
- 64.McMahon FJ, Buerventich S, Charney D, et al. Variation in the gene encoding the serotonin 2A receptor is associated with outcome of antidepressant treatment. . Am J Hum Genet. 2006;78:804–814. doi: 10.1086/503820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Racagni G, Popoli M. Cellular and molecular mechanisms in the longterm action of antidepressants. . Dialogues Clin Neurosci. 2008;10:385–400. doi: 10.31887/DCNS.2008.10.4/gracagni. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Duman RS, Monteggia LM. A neurotrophic model for stress-related mood disorders. . Biol Psychiatry. 2006;59:1116–1127. doi: 10.1016/j.biopsych.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 67.Pezawas L, Verchinski BA, Mattay VS, et al. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. . J Neurosci. 2004;24:10099–10102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Choi MJ, Kang RH, Lim SW, Oh KS, Lee MS. Brain-derived neurotrophic factor gene polymorphism (Val66Met) and citalopram response in major depressive disorder. . Brain Res. 2006;1118:176–182. doi: 10.1016/j.brainres.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 69.Licinio J, Dong C, Wong ML. Novel sequence variations in the brainderived neurotrophic factor gene and association with major depression and antidepressant treatment response. . Arch Gen Psychiatry. 2009;66:488–497. doi: 10.1001/archgenpsychiatry.2009.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pariante CM, Lightman SL. The HPA axis in major depression: classical theories and new developments. . Trends Neurosci. 2008;31:464–468. doi: 10.1016/j.tins.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 71.Binder EB, Salyakina D, Lichtner P, et al. Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. . Nat Genet. 2004;36:1319–1325. doi: 10.1038/ng1479. [DOI] [PubMed] [Google Scholar]
- 72.Horstmann S, Binder EB. Pharmacogenomics of antidepressant drugs. . Pharmacol Ther. 2009;124:57–73. doi: 10.1016/j.pharmthera.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 73.Davidson RJ, Irwin W, Anderle MJ, Kalin NH. The neural substrates of affective processing in depressed patients treated with venlafaxine. . Am J Psychiatry. 2003;160:64–75. doi: 10.1176/appi.ajp.160.1.64. [DOI] [PubMed] [Google Scholar]
- 74.Fu CH, Williams SC, Cleare AJ, et al. Attenuation of the neural response to sad faces in major depression by antidepressant treatment: a prospective, event-related functional magnetic resonance imaging study. . Arch Gen Psychiatry. 2004;61:877–889. doi: 10.1001/archpsyc.61.9.877. [DOI] [PubMed] [Google Scholar]
- 75.Mayberg HS, Liotti M, Brannan SK, et al. Reciprocal llmblc-cortlcal function and negative mood: converging PET findings in depression and normal sadness. . Am J Psychiatry. 1 999;1 56:675–682. doi: 10.1176/ajp.156.5.675. [DOI] [PubMed] [Google Scholar]
- 76.Siegle GJ, Carter CS, Thase ME. Use of FMRI to predict recovery from unipolar depression with cognitive behavior therapy. . Am J Psychiatry. 2006;163:735–738. doi: 10.1176/ajp.2006.163.4.735. [DOI] [PubMed] [Google Scholar]
- 77.Wu J, Buchsbaum MS, Gillin JC, et al. Prediction of antidepressant effects of sleep deprivation by metabolic rates in the ventral anterior cingulate and medial prefrontal cortex. . Am J Psychiatry. 1999;1 56:1149–1158. doi: 10.1176/ajp.156.8.1149. [DOI] [PubMed] [Google Scholar]
- 78.Chen CH, Ridler K, Suckling J, et al. Brain imaging correlates of depressive symptom severity and predictors of symptom improvement after antidepressant treatment. . Biol Psychiatry. 2007;62:407–414. doi: 10.1016/j.biopsych.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 79.Bush G, Luu P, Posner Ml. Cognitive and emotional influences in anterior cingulate cortex. . Trends Cogn Sci. 2000;4:215–222. doi: 10.1016/s1364-6613(00)01483-2. [DOI] [PubMed] [Google Scholar]
- 80.Giacobbe P, Mayberg HS, Lozano AM. Treatment resistant depression as a failure of brain homeostatic mechanisms: Implications for deep brain stimulation. . Exp Neurol. 2009;219:44–52. doi: 10.1016/j.expneurol.2009.04.028. [DOI] [PubMed] [Google Scholar]
- 81.Mulert C, Juckel G, Brunnmeier M, et al. Prediction of treatment response in major depression: integration of concepts. . J Affect Disord. 2007;98:215–225. doi: 10.1016/j.jad.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 82.Pizzagalli D, Pascual-Marqui RD, Nitschke JB, et al. Anterior cingulate activity as a predictor of degree of treatment response in major depression: evidence from brain electrical tomography analysis. . Am J Psychiatry. 2001;158:405–415. doi: 10.1176/appi.ajp.158.3.405. [DOI] [PubMed] [Google Scholar]
- 83.Fu CH, Williams SC, Brammer MJ, et al. Neural responses to happy facial expressions in major depression following antidepressant treatment. . Am J Psychiatry. 2007;164:599–607. doi: 10.1176/ajp.2007.164.4.599. [DOI] [PubMed] [Google Scholar]
- 84.Abercrombie HC, Schaefer SM, Larson CL, et al. Metabolic rate in the right amygdala predicts negative affect in depressed patients. . Neuroreport. 1998;9:3301–3307. doi: 10.1097/00001756-199810050-00028. [DOI] [PubMed] [Google Scholar]
- 85.Drevets WC, Videen TO, Price JL, Preskorn SH, Carmichael ST, Raichle ME. A functional anatomical study of unipolar depression. . J Neurosci. 1992;12:3628–3641. doi: 10.1523/JNEUROSCI.12-09-03628.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Canli T, Cooney RE, Goldin P, et al. Amygdala reactivity to emotional faces predicts improvement in major depression. . Neuroreport. 2005;16:1267–1270. doi: 10.1097/01.wnr.0000174407.09515.cc. [DOI] [PubMed] [Google Scholar]
- 87.Sheline Yl, Barch DM, Donnelly JM, Ollinger JM, Snyder AZ, Mintun MA. Increased amygdala response to masked emotional faces in depressed subjects resolves with antidepressant treatment: an fMRI study. . Biol Psychiatry. 2001;50:651–658. doi: 10.1016/s0006-3223(01)01263-x. [DOI] [PubMed] [Google Scholar]
- 88.Robertson B, Wang L, Diaz MT, et al. Effect of bupropion extended release on negative emotion processing in major depressive disorder: a pilot functional magnetic resonance imaging study. . J Clin Psychiatry. 2007;68:261–267. doi: 10.4088/jcp.v68n0212. [DOI] [PubMed] [Google Scholar]
- 89.Hariri AR, Drabant EM, Munoz KE, et al. A susceptibility gene for affective disorders and the response of the human amygdala. . Arch Gen Psychiatry. 2005;62:146–152. doi: 10.1001/archpsyc.62.2.146. [DOI] [PubMed] [Google Scholar]
- 90.Heinz A, Braus DF, Smolka MN, et al Amygdala-prefrontal coupling depends on a genetic variation of the serotonin transporter. Nat Neurosci. 2005;8:20–21. doi: 10.1038/nn1366. [DOI] [PubMed] [Google Scholar]
- 91.Pezawas L, Meyer-Lindenberg A, Drabant EM, et al. 5-HTTLPR polymorphism impacts human cingulate-amygdala interactions: a genetic susceptibility mechanism for depression. . Nat Neurosci. 2005;8:828–834. doi: 10.1038/nn1463. [DOI] [PubMed] [Google Scholar]
- 92.Mayberg HS. Modulating dysfunctional limbic-cortical circuits in depression: towards development of brain-based algorithms for diagnosis and optimised treatment. . Br Med Bull. 2003;65:193–207. doi: 10.1093/bmb/65.1.193. [DOI] [PubMed] [Google Scholar]
- 93.Ursu S, Stenger VA, Shear MK, Jones MR, Carter CS. Overactive action monitoring in obsessive-compulsive disorder: evidence from functional magnetic resonance imaging. . Psychol Sci. 2003;14:347–353. doi: 10.1111/1467-9280.24411. [DOI] [PubMed] [Google Scholar]
- 94.Campbell S, Marriott M, Nahmias C, MacQueen GM. Lower hippocampal volume in patients suffering from depression: a meta-analysis. . Am j Psychiatry. 2004;161:598–607. doi: 10.1176/appi.ajp.161.4.598. [DOI] [PubMed] [Google Scholar]
- 95.Taylor WD, Steffens DC, Payne ME, et al. Influence of serotonin transporter promoter region polymorphisms on hippocampal volumes in latelife depression. . Arch Gen Psychiatry. 2005;62:537–544. doi: 10.1001/archpsyc.62.5.537. [DOI] [PubMed] [Google Scholar]
- 96.Krishnan KR, Taylor WD, McQuoid DR, et al. Clinical characteristics of magnetic resonance imaging-defined subcortical ischemic depression. . Biol Psychiatry. 2004;55:390–397. doi: 10.1016/j.biopsych.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 97.Steffens DC, Payne ME, Greenberg DL, et al. Hippocampal volume and incident dementia in geriatric depression. . Am J Geriatr Psychiatry. 2002;10:62–71. [PubMed] [Google Scholar]
- 98.Sheline Yl, Gado MH, Kraemer HC. Untreated depression and hippocampal volume loss. . Am J Psychiatry. 2003;160:1516–1518. doi: 10.1176/appi.ajp.160.8.1516. [DOI] [PubMed] [Google Scholar]
- 99.Dranovsky A, Hen R. Hippocampal neurogenesis: regulation by stress and antidepressants. . Biol Psychiatry. 2006;59:1136–1143. doi: 10.1016/j.biopsych.2006.03.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Teodorczuk A, O'Brien JT, Firbank MJ, et al. White matter changes and late-life depressive symptoms: longitudinal study. . Br J Psychiatry. 2007;191:212–217. doi: 10.1192/bjp.bp.107.036756. [DOI] [PubMed] [Google Scholar]
- 101.Taylor WD, Steffens DC, MacFall JR, et al. White matter hyperintensity progression and late-life depression outcomes. . Arch Gen Psychiatry. 2003;60:1090–1096. doi: 10.1001/archpsyc.60.11.1090. [DOI] [PubMed] [Google Scholar]
- 102.Kramer-Ginsberg E, Greenwald BS, et al. Neuropsychological functioning and MRI signal hyperintensities in geriatric depression. . Am J Psychiatry. 1999;156:438–444. doi: 10.1176/ajp.156.3.438. [DOI] [PubMed] [Google Scholar]
- 103.avitz JB, Drevets WC. Imaging phenotypes of major depressive disorder: genetic correlates. . Neuroscience. 2009;164:300–330. doi: 10.1016/j.neuroscience.2009.03.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.van Os J, Fahy TA, Jones P, et al. Increased intracerebral cerebrospinal fluid spaces predict unemployment and negative symptoms in psychotic illness. A prospective study. . Br J Psychiatry. 1995;166:750–758. doi: 10.1192/bjp.166.6.750. [DOI] [PubMed] [Google Scholar]
- 105.Zipursky RB, Lambe EK, Kapur S, Mikulis DJ. Cerebral gray matter volume deficits in first episode psychosis. . Arch Gen Psychiatry. 1998;55:540–546. doi: 10.1001/archpsyc.55.6.540. [DOI] [PubMed] [Google Scholar]
- 106.assink TH, Andreasen NC, Nopoulos P, Flaum M. Cerebellar morphology as a predictor of symptom and psychosocial outcome in schizophrenia. . Biol Psychiatry. 1999;45:41–48. doi: 10.1016/s0006-3223(98)00175-9. [DOI] [PubMed] [Google Scholar]
- 107.Cahn W, Rais M, Stigter FP, et al. Psychosis and brain volume changes during the first five years of schizophrenia. . Eur Neuropsychopharrnacol. 2009;19:147–151. doi: 10.1016/j.euroneuro.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 108.auscher J, Kapur S. Choosing the right dose of antipsychotics in schizophrenia: lessons from neuroimaging studies. . CNS Drugs. 2001;15:671–678. doi: 10.2165/00023210-200115090-00001. [DOI] [PubMed] [Google Scholar]
- 109.Uchida H, Graff-Guerrero A, Mulsant BH, Pollock BG, Mamo DC. Longterm stability of measuring D(2) receptors in schizophrenia patients treated with antipsychotics. . Schizophr Res. 2009;109:130–133. doi: 10.1016/j.schres.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 110.Meyer JH, Wilson AA, Sagrati S, et al. Serotonin transporter occupancy of five selective serotonin reuptake inhibitors at different doses: an [11QDASB positron emission tomography study. . Am J Psychiatry. 2004;161:826–35. doi: 10.1176/appi.ajp.161.5.826. [DOI] [PubMed] [Google Scholar]
- 111.Uchida H, Kapur S, Mulsant BH, Graff-Guerrero A, Pollock BG, Mamo DC. Sensitivity of older patients to antipsychotic motor side effects: a PET study examining potential mechanisms. . Am j Geriatr Psychiatry. 2009;17:255–263. doi: 10.1097/JGP.0b013e318198776d. [DOI] [PubMed] [Google Scholar]
- 112.Ng W, Uchida H, Ismail Z, et al. Clozapine exposure and the impact of smoking and gender: a population pharmacokinetic study. . Ther Drug Monit. 2009;31:360–366. doi: 10.1097/FTD.0b013e31819c7037. [DOI] [PubMed] [Google Scholar]
- 113.de Leon J. Pharmacogenomics: the promise of personalized medicine for CNS disorders. . Neuropsychopharmacology. 2009;34:159–172. doi: 10.1038/npp.2008.147. [DOI] [PubMed] [Google Scholar]
- 114.alhotra AK, Athanasiou M, Reed CR, et al. Discovery of genetic markers associated with clozapine induced agranulocytosis. . Am J Med Genet B Neuropsychiatr Genet. 2005;138b:22–xx. [Google Scholar]
- 115.Lan TH, Loh EW, Wu WS, et al Performance of a neuro-fuzzy model in predicting weight changes of chronic schizophrenia patients exposed to antipsychotics. . Mol Psychiatry. 2008;13:1129–1137. doi: 10.1038/sj.mp.4002128. [DOI] [PubMed] [Google Scholar]