Abstract
Hundreds of genome-wide association studies have been performed in recent years in order to try to identify common variants that associate with complex disease. These have met with varying success. Some of the strongest effects of common variants have been found in lateonset diseases and in drug response. The major histocompatibility complex has also shown very strong association with a variety of disorders. Although there have been some notable success stories in neuropsychiatric genetics, on the whole, common variation has explained little of the high heritability of these traits. In contrast, early studies of rare copy number variants have led rapidly to a number of genes and loci that strongly associate with neuropsychiatric disorders. It is likely that the use of whole-genome sequencing to extend the study of rare variation in neuropsychiatry will greatly advance our understanding of neuropsychiatric genetics.
Keywords: genome-wide association study; rare variant; neuropsychiatric; schizophrenia; sequencing, rare variant, neuropsychiatric, schizophrenia , sequencing
Abstract
En los últimos años se han realizado cientos de estudios de asociación del genoma completo tratando de identificar variantes comunes que se asocien con enfermedades complejas, los que han tenido logros variables. En enfermedades de aparición tardía y en la respuesta a fármacos se han encontrado algunos de los efectos más potentes de variantes comunes. El complejo mayor de histocompatibilidad también ha mostrado una asociación muy fuerte con una variedad de trastornos. Aunque han existido algunos casos destacados de éxito en la genética neuropsiquiátrica, en conjunto, la variación común ha explicado sólo parte de la alta herencia de estos rasgos. Por otra parte, los estudios iniciales de variantes raras del número de la copia han conducido rápidamente a asociaciones potentes entre un número de genes y loci con trastornos neuropsiquiátricos. Es posible que el empleo de la secuenciación de todo el genoma se extienda al estudio de variaciones raras en neuropsiquiatría y se progrese enormemente en la comprensión de la genética neuropsiquiátrica.
Abstract
Ces dernières années, des centaines d'études d'association sur le génome entier ont tenté d'identifier des variants communs associés aux maladies complexes, ceci avec un succès mitigé. Certains des effets les plus marqués des variants communs ont été retrouvés dans les maladies à début tardif et dans la réponse au médicament. Le complexe majeur d'histocompatibilité a montré également une très forte association avec différents troubles. Malgré quelques succès notables en génétique neuropsychiatrique, dans l'ensemble, la très haute héritabilité de ces caractères a été peu expliquée par les variants communs. Au contraire, les premières études de variations rares du nombre de copies ont permis rapidement d'affirmer une forte association de nombreux gènes et loci à des maladies neuropsychiatriques. Il est probable que l'utilisation du séquençage du génome entier pour améliorer l'étude des variations rares en neuropsychiatrie va permettre de faire avancer de manière significative notre compréhension de la génétique neuropsychiatrique.
In recent years, hundreds of genetic association studies have sought to explore the relationship between common genetic variation and disease, biological characteristics, or drug response. The basic premise of these studies is that the diseases (or traits) are not caused by single gene variants of strong effect, such as, for instance, sickle-cell anemia or cystic fibrosis, but rather that some “manageable” number of common variants have an important influence on the trait under question. Part of the motivation for this perspective is the “common disease, common variant” (CDCV) theory1,2 Once a genetic variant has been found to be associated, there are a number of possible uses for the information. If the effect of the genetic variant is strong enough, perhaps in combination with lifestyle or other environmental factors, it might be used to predict risk of the disease. Alternatively, the associated variant(s) may be used to try to predict response to a particular medication. Finally, if the effect size of the genetic variant is very small and thus not useful for either of these purposes, it may still be of use in identifying a disease-associated gene or genetic pathway that could illuminate disease pathophysiology or implicate new therapeutic targets. Here we review the current status of genome-wide association studies, with a particular focus on neuropsychiatric disorders.
Genome-wide association studies
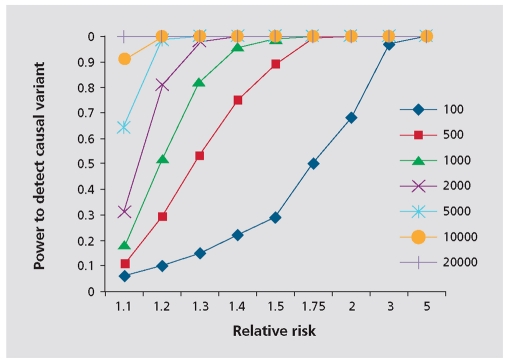
Genome-wide association studies (GWAS), are a way of performing genetic association studies without prior hypotheses about which genes are likely to be involved. To do this, arrays of single-nucleotide polymorphisms (SNPs) that cover the whole genome are used. Although there are thought to be approximately 10 million common SNPs in the genome,3 it is not necessary to genotype each one of these individually to get information about most of them. This is because, due to the way that human populations have migrated and genetic variants have arisen, many of the variants are associated with each other or “linked.” Thus, in European and Asian populations, if you genotype one variant, you are gaining information about 10 to 20 other variants simultaneously. This is called “tagging” (the genotyped variants “tag” the ungenotyped, linked variants), and was brought to the genome-wide scale by the HapMap project, which has genotyped millions of common SNPs in four populations to create a detailed map of how common genetic variants relate to one another.3-5 A significant motivation for the HapMap project was the idea that common variants make up an important part of the genetic contribution to common diseases (the CDCV hypothesis). While some theoretical arguments were marshaled in support of this hypothesis - and indeed, even before the HapMap project a handful of examples were known - there was no way to know a priori how general the CDCV hypothesis might turn out to be. For this reason, a systematic investigation of common variation was judged by much of the community (including these authors) but not all6 to be a sensible beginning to the study of human disease genetics. The result is that a true genome-wide study can be performed by actually genotyping as few as 300 000 to 1 million SNPs.7,8 However, because so many tests are being performed, it is necessary to obtain a very strongly significant P value to be sure that the result is really significant. This is known as “genome-wide significance” and the consensus is that this should be about 108 or less.9 Because the effects sizes of common variants are generally small, it is usually necessary to include a large number of subjects in the study in order to have the power to detect a genome-wide significant P value (Figure 1.)
Figure 1. The power to detect a causal variant that is perfectly tagged by a genotyped marker (assuming dominant model, minor allele frequency=0.2, frequency of disease is 1% and equa numbers of cases and controls). To have a good chance of detecting a variant with a relative risk of 1.2, about 2000 cases and controls are needed .
Major discoveries with GWAS
The success of GWAS has been very variable for different disease areas. Some diseases have found common variants with very strong effects, and managed to track these down to the causal variant. An inspiring example is an intronic variant in BCL11A that was found in two GWAS studies to associate with fetal hemoglobin (HbF) levels in healthy adults,10, 11 and also to modify the presentation of (3-thalassemia, and associate with HbF levels in patients with sickle-cell disease.11 This finding was soon followed up with a functional study that showed that the variant associated with high HbF12 reduced the expression of BCL11A,13 and that reduction of BCL11A expression caused increase in levels of gamma-globin in adult human red blood progenitor cells, which led to increased levels of HbF13 These findings clearly suggest that BCL11A serves as an inhibitor of HbF production and that directed repression of BCL11A could be developed as a clinical tool to ameliorate the presentation of thalassemias and sickle-cell disease. These findings in turn have led to further understanding of developmental and species-specific changes in globin regulation.14.
On the less inspirational side, however, other diseases, like hypertension, have been thoroughly and carefully investigated using huge numbers of patients and controls with very little progress.15 Here we outline some of the highest impact findings of GWAS and where (if anywhere) they have led us.
As might be expected by the laws of natural selection, there are not many common genetic variants that confer a strong predisposition to common diseases. Such variants would be expected to have been selected against, and thus maintained at low population frequencies. However, there are some phenotypes that might be expected to have dodged the purifying effects of selection. These include common diseases that do not onset until old age, and response to drugs that the body has not historically had to interact with. Accordingly, some of the strongest effects of common variants on disease have been found in association with ailments with an onset during the postreproductive years, and with drug response.
Genetic variants that affect late-onset diseases
One of the most well-known genetic risk factors is the E4 variant of the apolipoprotein E gene, ApoE, which greatly increases the risk of Alzheimer's disease (AD) and reduces the age of onset in a dose-dependent manner.16-18 The effect of this variant is so strong that it was, in fact, discovered before the GWAS era, but it has since been confirmed as the most important predictor of lateonset AD in a number of genome -wide analyses,19-22 one with fewer than 500 cases and controls reporting a P value of 1 x 10-40.21 However, despite the definitive effects of this genetic variant on AD and the length of time that we have known about it, it is still not clear how the variant mediates its effects,23 and it has not yet led to improved treatment.
One of the very earliest novel discoveries of GWAS was the association of an amino acid substitution in the complement factor H gene, CFH, with age-related macular degeneration, a very common form of blindness that affects the elderly. This genetic association was found with a tiny sample size: 96 cases and 50 controls, and carrying two copies of the risk variant increases the risk of illness up to 7 times.24 The associated variant does itself seem to be functional, changing the binding properties of the protein, although it is not yet exactly understood how the variant contributes to disease,25 nor how this can be utilized in novel treatments.
A third very strong disease-associated common genetic variant is in the LOXL1 gene in exfoliation glaucoma, another very common form of age-related blindness. The associated variant was discovered in a set of only 75 cases, and individuals homozygous for the risk haplotypes are thought to be at 700-fold increased risk of exfoliation glaucoma when compared with homozygotes of the low-risk haplotype. However, because the risk haplotype is so common, this translates to just a 2.5-fold increase risk from the population average.26 The two variants contributing to the risk haplotype are both protein-coding changes, and the same variants have now been associated with disease in multiple populations,27-40 suggesting that these are the causal variants, although the degree of penetrance, and the risk haplotype, have been reported to differ in Australia and Japan.28,29,35,37,38,41,42 Unfortunately, the very high frequency of the risk haplotype in the general population currently precludes these markers from being used to predict disease, but it is hoped that a better understanding of the role of LOXL1 in optical pathophysiology may lead to advances in treatment.40
Genetic variants that affect drug response
Genetic variants affecting drug response can have very strong effects, and often occur in the genes that would be most expected to be involved.43 Thus, pharmacogenetics was one of the more successful areas of genomics before the GWAS area, and a number of strong genetic influencers of drug response have been known for some time.44 GWAS have added at least three pharmacogenetic associations of considerable strength and importance.
Flucloxacillin-induced liver injury
Idiosyncratic drug reactions are the most common cause of liver failure in the US.45 Flucloxacillin is an antibiotic drug commonly used to treat Staphylococcus aureus infections, but it has a relatively high incidence of causing liver injury (6.1 per 100 000 users) in comparison with other antibiotics such as penicillin.46 This has previously led to restrictions on its use.46 A GWAS was performed on 51 patients with flucloxacillin-induced liver injury and 487 controls, in which a huge signal was seen for a missense polymorphism in the HCP5 gene (P= 8.710-33)47. Through linkage disequilibrium, the association was traced to the HLA-B*5701 allele, the presence of which increased the likelihood of flucloxacillin-induced liver injury by 80 times.47 Since the general frequency of the associated allele in the European population is only about 5%, and it was present in 84% of cases, this variant could potentially be used to screen out people at high risk of liver injury before flucloxacillin is prescribed. However, due to the rarity of the hepatoxicity, this would result in a high false-positive rate. A proposed alternative is to use the genotyping of this variant as a diagnostic marker in suspected cases of hepatoxicity so that the patient can be rapidly switched to alternative antibiotics.47
Statin-induced myopathy
Taking statin therapy to reduce the levels of low-density lipoprotein cholesterol has been shown to reduce the likelihood of cardiovascular events, such as heart attack and stroke.48 Occasionally, however, statins, particularly at high doses, can cause serious myopathy, which may lead to hospitalization or death.49 In August 2008 a GWAS that included only 85 cases and 90 controls revealed a SNP in the SLCOIBI gene, which accounted for more than 60% of cases of myopathy50 Carrying one C at this locus increases the risk of statin-induced myopathy by 4.5 times, and CC homozygotes have a 17fold greater risk than TT homozygotes. This has been suggested as a genetic test to identify vulnerable individuals before offering high-dose simvastatin therapy51
Hepatitis-C treatment response
One of the most recent, and perhaps the most clinically significant, of any GWAS to date is the association of a SNP close to the IL28B gene with response to treatment for hepatitis C.52 In this study, Ge et al focused on who is cured by treatment, and found that the good response genotype is associated with a greater than 80% chance of clearance in European- Americans, while the poor response genotype is associated with only about a 30% chance. A follow-up study found that the polymorphism also influences natural clearance of hepatitis C and shows very sharp geographic differentiation.53 This suggests that the variant may be common in the population because the “good response” allele conferred protection against one or more viruses and hence was positively selected. This variant is a very good candidate to use as a pharmacogenetic predictor of treatment response before beginning hepatitis C treatment, since the procedure is long and often associated with adverse effects.54
The major histocompatibility complex
Setting aside the old-age or pharmacogenetic associations, many of the strongest reported GWAS associations of common variants with common disease involve markers in the major histocompatibility complex (MHC). These associations are too extensive to discuss in detail in this review, but include autoimmune diseases, infectious diseases, neuropsychiatric disorders, and variability in normal traits such as height.55 A number of hypotheses have been put forward to explain why variants conferring disease risk at this locus have been maintained at high frequency in the population. One suggestion is that the disease-associated variants have been selected for because they confer resistance to particular infectious agents, either now or historically. An alternative hypothesis is that each locus that confers risk for one common disease is maintained at high frequency because it confers protection against one or more other common diseases. For example, the HLA gene DQB1*0602, which encodes the β chain for the HLA class II molecule DQ6, is protective against diabetes,56 but a strong risk factor for narcolepsy57 and multiple sclerosis.58
GWAS in neuropsychiatry
Neuropsychiatric traits have been among the most disappointing GWAS results. Despite many GWAS, most associated variants have either not withstood significance correction for multiple testing, or else have failed to replicate. In general, where replicable effects have been found, they have required very large sample sizes and the effects have been small.
There have been some notable success stories, however. Two GWAS have revealed strong and replicable genetic influences on restless legs syndrome (RLS), a condition characterized by an unpleasant and irresistible urge to move the legs, particularly while resting and during the evening and night. Both studies, one on Icelandic individuals and one on a more mixed European cohort, implicated BTBD9. 59,60 The European study also found an association with two other loci: MEIS1 and a locus encompassing MAP2K5/LBXCOR1 .60 The associations with MEIS1 and BTBD9 were quickly replicated in two subsequent studies,61,62 but the MAP2K5/LBXCOR1 appears to be weaker, showing a borderline significance in one study only62 Although the risk associated with MEIS1 and BTBD9 (ranging from 1.5 to 3.759,60,62,63 ,606263) is substantially lower than those described above, they do appear to be real and highly significant risk factors for RLS. Nevertheless, the biology underlying the associations remains unclear. The associated variants do not appear to have any obvious function, and a thorough search for putative functional variants in all coding exons and across intron-exon boundaries revealed no obviously causal variant.64
Another positive GWAS finding in neuropsychiatry is with narcolepsy, a disorder that causes disrupted sleep patterns, with the patient often feeling excessively tired during the day, and suffering sudden sleep attacks. PreGWAS studies had connected the disorder to an MHC class II antigen called HLA-DQB1*0602, and about 85% of narcoleptics carry this antigen.65 However, there remained unexplained heritability. Very recently, a GWAS study was done on 807 cases and 1074 controls, all positive for HLA-DQB1*0602. A significant association of three SNPs in the T cell receptor alpha locus was found, which was then replicated in the same study in 1057 further cases and 1104 controls.66 Further analysis showed a single SNP was responsible for the association, although it is not clear whether this variant is itself causal or how it may contribute to disease. This association is of particular interest because it adds considerable weight to the view that narcolepsy is an autoimmune disease, and as such, it would be the first autoimmune disease to be associated with a T-cell receptor locus. This finding also opens up the possibility of immunotherapy as a future treatment for narcolepsy.
Other neuropsychiatric diseases for which definite, replicated effects of common SNPs have been found include schizophrenia, associated with MHC markers, NRGN and TCF4 (12 945 cases and 34 591 controls, ORs=1.24, 1.15,1.23),67,68 bipolar disorder, associated with ANK3 and CACNA1C (4 387 cases and 6 209 controls, ORs=1.45 and 1.18) 69, and autism, associated with SNPs at 5p14.1 (3 101 family members, 204 cases and 6 941 controls, OR=1.19).70,71 However, all of these were discovered with very large sample sizes and account for very little of the very high heritability of these conditions.
Rare variants
Although studies of common variation in neuropsychiatric disease may be underwhelming, the opposite is true for rare variation. Although the SNP chips used for GWAS comprise only polymorphisms that are reasonably common (~≥5%), their data can be used to find other types of non-SNP variants - specifically copy number variants (CNVs) - with much lower frequency. CNVs are duplications or deletions of large stretches of DNA - ranging in size from just a few hundred base pairs to many megabases. To detect such variants, the intensity data from the SNP chips is examined to determine whether particular stretches of SNPs are less intense than expected (or absent), which would indicate a deletion, or more intense than expected, which suggests a duplication.72 Because the CNVs are identified on an individual-by-individual basis, very rare CNVs, even those present in a single individual, can be found. This has allowed us for the first time to examine the role of rare variation in common disease (albeit just a tiny fraction of the total amount of rare variant in a cohort). The majority of investigations of copy number variation to date have been in neuropsychiatric disease and, happily, they have led immediately to real, replicable and very strong associations. A summary of CNVs recently strongly associated with neuropsychiatric disease is shown in Table I.
These variants confer considerable risk, but they are not completely penetrant. Although the specific variants are very rare in the general population, they are occasionally seen in controls (Table I) , and where families have been examined, the variants are often inherited from unaffected or only mildly affected parents.73-77 Additionally, as can be seen in Table I , many of the variants have been associated with more than one neuropsychiatric condition. This is consistent with the characteristics of neuropsychiatrically-associated rare variants that were found before the GWAS era, such as DISC1 in schizophrenia, which associated with a range of phenotypes from psychiatrically normal to suicide, recurrent major depression, and schizophrenia.78 It seems that these variants, rather than predisposing to a specific neuropsychiatric condition, may strongly confer some sort of “neural vulnerability,” the ultimate manifestation of which depends on other interacting genetic and environmental factors. Because, to date, the only rare variants that we have been able to associate with neuropsychiatric illness are very large deletions and duplications, it is not clear whether this lack of specificity will be a general rule, or is somehow related to the size of the lesion. However, there is some evidence from the associations with common SNPs that this is a characteristic of the disease rather than the size of the associated variant. For instance, bipolar-associated common variants in CACNA1 C may also confer risk of depression and schizophrenia.79
Table I. Copy number variants (CNV) strongly associated with neuropsychiatric disorders.. Frequencies are given only when the CNV was found in a large case-control study design. *Controls may not have been carefully screened for neuropsychiatric illness. NR, not reportec; Dup, cuplication; Del, Deletion .
| CNV | Copy # linked to disease | Seen in schizophrenia patients? | Seen in autism patients? | Seen in epilepsy patients? | Sseen in patients with mental retardation? | Other disorders | Reported in controls*? | Lead candidate genes |
| 1q21.1 | Deletion and duplication | Deletion (0.23-0.29)82-84 | Yes75,94 | Yes77,83,95, unpublished data | Yes95,96 | Congenital heart disease77,97,98, micro-and macrocephaly,75,77 neuroblastoma,99, other75 | Deletion, 0.02% (8/41, 199),84 frequency for dup unclear | HYDIN paralog77; GJA84 |
| 15q11 | Deletion | Yes (0.61%)84-100 | Yes101 | No | Yes102 | Deletions of this region cause Angelman and Prader Willi syndrome | Yes, (0.19%) (79/41,194) (0.19%) | CYFIP184 |
| 15q13.1 | Duplication and deletion | Yes81,83,84,103 | NR | NR | NR | NR | no data | APBA2 |
| 15q13.3 | Deletion | Yes (0.17%-0.27%)83,84 | Yes (0.31%)84,86 | Yes (1-1.3%)73,74,110 | Yes73,106 | Various including mild developmental delay, heart defects73 | Yes, 0.02% (8/39,800)74,84 | CHRNA783,84 |
| 16p11.2 | Deletion and duplication | Yes94 | Yes (0.6%, del only,107 1% dup + del94) | Yes110,111 | Yes76,108 | Various neuropsychiatric and developmental76,108 | Yes, 0.01% del, 0,03% dup94 | SEZ6L2110,111 |
| 16p13.11 | Deletion | Yes81 | Yes112 | Yes (0.6%)unpublished data | Yes (0.5%)87,111 | NR | NR (0/3313),87,96 unpublished data but inherited from unaffected parents87 | NDEI81 |
| CNTN4 | Deletion and duplication | NR | Yes85,114,115 | NR | NR | NR | no data | |
| NRXN1 | Deletion | Yes (0.19%)81-83,120-122 | Yes85,116-118 | NR | Yes122 | NR | Yes, 0.04% (17/42054)115 |
The future for neuropsychiatric genetics
There are two, not incompatible, possible directions for neuropsychiatric genetics research. One approach is to continue searching for common variants of small effect size using much larger cohorts in the tens or hundreds of thousands. This has been suggested as a future direction for schizophrenia genetics.80 Although this will require a considerable effort, there are already established worldwide collaborations for schizophrenia,68,80 so very large collections should be achievable in the relatively near future. The disadvantages of this approach are that if such huge sample sizes are needed to discover them, the effect sizes of the associated variants must be very small (Figure 1), and they will be present at a similar frequency in unaffected controls. This makes further study of the effects of the variants very difficult or impossible. However, proponents of this approach correctly suggest that although the associated variant may have a very small effect, the gene it is in may have a big impact on disease when targeted by novel pharmaceuticals.
A second argument in favor of proceeding with GWAS in very large samples is that neuropsychiatric researchers have long expressed concern that clinical diagnostic criteria do not reflect the biological underpinnings of the disease, and that diseases such as schizophrenia may in fact represent multiple different disorders with different genetic contributors. Thus, only with very large sample sizes would one expect to obtain sufficient numbers of any one genetically homogenous subgroup to obtain a genome-wide significant association. However, as discussed above, all genetic variants that have been associated with neuropsychiatric disease so far seem to be very nonspecific. Where they are found in multiple patients with a single diagnosis (eg, schizophrenia), they do not segregate patients into any clear diagnostic categories either by disease presentation or drug response. Additionally, they tend to associate with multiple neuropsychiatric conditions.
The alternative approach is to further investigate the role of rare variants in neuropsychiatric disease. To date, the only type of rare variation that has been identifiable on a genome -wide scale has been large CNVs, and already we have found many strong associations.81-87 It is likely that when we can identify the totality of rare variation in an individual using whole-genome sequencing, many more rare variants will be found to be definitely associated with neuropsychiatric illness. Fortunately, this is rapidly becoming a reality, and the first sequencing studies in neuropsychiatric illness are already underway. For confirmation and follow-up, this approach will definitely benefit from very large cohorts collected for GWAS, but the ideal discovery samples will be rather different. With this approach, we hope to find variants with very large effect sizes and high penetrance. This means that it will be much more straightforward to understand how the variants exert their effects and what genetic and environmental factors influence them. To do this, the priority will be patients and relatives that can be reapproached for further study after potentially causal variants have been identified. Additionally, since initial sequencing attempts will be expensive, it is worth, at first at least, selecting patients who are most likely to carry highly penetrant genetic variants. These include severely ill, treatment-resistant patients88 and patients with a strong family history of mental illness. Thus, this approach benefits from close collaboration between geneticists and psychiatrists and a thorough understanding of each sequenced patient and his or her relatives.
Although it is hoped that whole -genome sequencing will lead swiftly to a clearer understanding of neuropsychiatric disease, there are many challenges ahead. Not least is a very well-characterized psychiatrically normal control cohort. And, as with any new technology, there are considerable technical challenges, such as the use of whole-genome data to identify copy number variation. However, software is constantly developing and it is doubtful that these will be limiting factors for long.89 -92 There are also “genomic” challenges: there are many regions of the genome on which we tend not to focus, such as remote enhancer regions, upstream open reading frames, and chromatin binding sites, which are likely to be functional and affected by rare variation. However, using Mendelian diseases as a model, it is reasonable to expect that many of the most important variants will be in or very close to exons.93 Thus, neuropsychiatric geneticists should be able to gorge themselves on the lowhanging fruit for some time to come.
In summary, there have been many GWAS success stories in which common variants have been found to associate definitely with complex diseases. In most cases, however, the mechanism underlying the association is not well understood, and they have not yet led to strong predictive tests or to novel treatments. Neuropsychiatric disease, in particular, has so far benefited little from large-scale analysis of common variants. Use of GWAS data to examine rare copy number variants, however, rapidly led to multiple strong and highly penetrant associations with neuropsychiatric illness. However, the associated variants are not completely penetrant and tend to be associated with multiple neuropsychiatric conditions. Detailed studies of patients and their relatives will be necessary to understand what factors affect the manifestation of the phenotype. Despite this recent success, we can still only account for a very small amount of the heritability of neuropsychiatric conditions. Further investigation of rare variation using whole-genome sequencing is likely to significantly advance the field.
Contributor Information
Anna C. Need, Institute for Genome Sciences and Policy, Center for Human Genome Variation, Duke University, Durham, North Carolina, USA.
David B. Goldstein, Institute for Genome Sciences and Policy, Center for Human Genome Variation, Duke University, Durham, North Carolina, USA.
REFERENCES
- 1.Chakravarti A. Population genetics--making sense out of sequence. Nat Genet. 1999;21(1 suppl):56–60. doi: 10.1038/4482. [DOI] [PubMed] [Google Scholar]
- 2.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 3.Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 6.Terwilliger JD, Hiekkalinna T. An utter refutation of the “fundamental theorem of the HapMap”. Eur J Hum Genet. 2006;14:426–437. doi: 10.1038/sj.ejhg.5201583. [DOI] [PubMed] [Google Scholar]
- 7.Need AC, Goldstein DB. Genome-wide tagging for everyone. Nat Genet. 2006;38:1227–1228. doi: 10.1038/ng1106-1227. [DOI] [PubMed] [Google Scholar]
- 8.Johnson GC, Esposito L, Barratt BJ, et al. Haplotype tagging for the identification of common disease genes. Nat Genet. 2001;29:233. doi: 10.1038/ng1001-233. [DOI] [PubMed] [Google Scholar]
- 9.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 10.Menzel S, Garner C, Gut I, et al. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat Genet. 2007;39:1197–1199. doi: 10.1038/ng2108. [DOI] [PubMed] [Google Scholar]
- 11.Uda M, Galanello R, Sanna S, et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci U S A. 2008;105:1620–1625. doi: 10.1073/pnas.0711566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lettre G, Sankaran VG, Bezerra MA, et al. DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc Natl Acad Sci U S A. 2008;105:11 869–11 874. doi: 10.1073/pnas.0804799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sankaran VG, Menne TF, Xu J, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 14.Sankaran VG, Xu J, Ragoczy T, et al. Developmental and species-divergent globin switching are driven by BCL11A. Nature. 2009;460:1093–1097. doi: 10.1038/nature08243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charchar F, Zimmerli L, Tomaszewski M. The pressure of finding human hypertension genes: new tools, old dilemmas. J Hum Hypertens. 2008;22:821–828. doi: 10.1038/jhh.2008.67. [DOI] [PubMed] [Google Scholar]
- 16.Strittmatter WJ, Saunders AM, Schmechel D, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 18.Tsai MS, Tangalos EG, Petersen RC, et al. Apolipoprotein E: risk factor for Alzheimer disease. Am J Hum Genet. 1994;54:643–649. [PMC free article] [PubMed] [Google Scholar]
- 19.Coon KD, Myers AJ, Craig DW, et al. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer's disease. J Clin Psychiatry. 2007;68:613–618. doi: 10.4088/jcp.v68n0419. [DOI] [PubMed] [Google Scholar]
- 20.Bertram L, Lange C, Mullin K, et al. Genome-wide association analysis reveals putative Alzheimer's disease susceptibility loci in addition to APOE. Am J Hum Genet. 2008;83:623–632. doi: 10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feulner TM, Laws SM, Friedrich P, et al. Examination of the current top candidate genes for AD in a genome-wide association study. Mol Psychiatry. 2009 Jan 6;Epub ahead of print doi: 10.1038/mp.2008.141. [DOI] [PubMed] [Google Scholar]
- 22.Abraham R, Moskvina V, Sims R, et al. A genome-wide association study for late-onset Alzheimer's disease using DNA pooling. BMC Med Genomics. 2008;1:44. doi: 10.1186/1755-8794-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J, Basak JM, Holtzman DM. The role of apolipoprotein E in Alzheimer's disease. Neuron. 2009;63:287–303. doi: 10.1016/j.neuron.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boon CJ, van de Kar NC, Klevering BJ, et al. The spectrum of phenotypes caused by variants in the CFH gene. Mol Immunol. 2009;46:1573–1594. doi: 10.1016/j.molimm.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 26.Thorleifsson G, Magnusson KP, Sulem P, et al. Common sequence variants in the LOXL1 gene confer susceptibility to exfoliation glaucoma. Science. 2007;317:1397–1400. doi: 10.1126/science.1146554. [DOI] [PubMed] [Google Scholar]
- 27.Fingert JH, Alward WL, Kwon YH, et al. LOXL1 mutations are associated with exfoliation syndrome in patients from the midwestern United States. Am J Ophthalmol. 2007;144:974–975. doi: 10.1016/j.ajo.2007.09.034. [DOI] [PubMed] [Google Scholar]
- 28.Hewitt AW, Sharma S, Burdon KP, et al. Ancestral LOXL1 variants are associated with pseudoexfoliation in Caucasian Australians but with markedly lower penetrance than in Nordic people. Hum Mol Genet. 2008;17:710–716. doi: 10.1093/hmg/ddm342. [DOI] [PubMed] [Google Scholar]
- 29.Hayashi H, Gotoh N, Ueda Y, Nakanishi H, Yoshimura N. Lysyl oxidaselike 1 polymorphisms and exfoliation syndrome in the Japanese population. Am J Ophthalmol. 2008;145:582–585. doi: 10.1016/j.ajo.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 30.Fan BJ, Pasquale L, Grosskreutz CL, et al. DNA sequence variants in the LOXL1 gene are associated with pseudoexfoliation glaucoma in a U.S. clinicbased population with broad ethnic diversity. BMC Med Genet. 2008;9:5. doi: 10.1186/1471-2350-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang X, Zabriskie NA, Hau VS, et al. Genetic association of LOXL1 gene variants and exfoliation glaucoma in a Utah cohort. Cell Cycle (Georgetown, Tex). 2008;7:521–524. doi: 10.4161/cc.7.4.5388. [DOI] [PubMed] [Google Scholar]
- 32.Ramprasad VL, George R, Soumittra N, Sharmila F, Vijaya L, Kumaramanickavel G. Association of non-synonymous single nucleotide polymorphisms in the LOXL1 gene with pseudoexfoliation syndrome in India. Mol Vis. 2008;14:318–322. [PMC free article] [PubMed] [Google Scholar]
- 33.Pasutto F, Krumbiegel M, Mardin CY, et al. Association of LOXL1 common sequence variants in German and Italian patients with pseudoexfoliation syndrome and pseudoexfoliation glaucoma. Invest Ophthalmol Vis Sci. 2008;49:1459–1463. doi: 10.1167/iovs.07-1449. [DOI] [PubMed] [Google Scholar]
- 34.Aragon-Martin JA, Ritch R, Liebmann J, et al. Evaluation of LOXL1 gene polymorphisms in exfoliation syndrome and exfoliation glaucoma. Mol Vis. 2008;14:533–541. [PMC free article] [PubMed] [Google Scholar]
- 35.Ozaki M, Lee KY, Vithana EN, et al. Association of LOXL1 gene polymorphisms with pseudoexfoliation in the Japanese. Invest Ophthalmol Vis Sci. 2008;49:3976–3980. doi: 10.1167/iovs.08-1805. [DOI] [PubMed] [Google Scholar]
- 36.Mossbock G, Renner W, Faschinger C, Schmut O, Wedrich A, Weger M. Lysyl oxidase-like protein 1 (LOXL1) gene polymorphisms and exfoliation glaucoma in a Central European population. Mol Vis. 2008;14:857–861. [PMC free article] [PubMed] [Google Scholar]
- 37.Mori K, Imai K, Matsuda A, et al. LOXL1 genetic polymorphisms are associated with exfoliation glaucoma in the Japanese population. Mol Vis. 2008;14:1037–1040. [PMC free article] [PubMed] [Google Scholar]
- 38.Mabuchi F, Sakurada Y, Kashiwagi K, Yamagata Z, Iijima H, Tsukahara S. Lysyl oxidase-like 1 gene polymorphisms in Japanese patients with primary open angle glaucoma and exfoliation syndrome. Mol Vis. 2008;14:1303–1308. [PMC free article] [PubMed] [Google Scholar]
- 39.Lemmela S, Forsman E, Onkamo P, et al. Association of LOXL1 gene with Finnish exfoliation syndrome patients. J Hum Genet. 2009;54:289–297. doi: 10.1038/jhg.2009.28. [DOI] [PubMed] [Google Scholar]
- 40.Wolf C, Gramer E, Muller-Myhsok B, et al. Lysyl oxidase-like 1 gene polymorphisms in german patients with normal tension glaucoma, pigmentary glaucoma and exfoliation glaucoma. J Glaucoma. 2009;In press doi: 10.1097/IJG.0b013e31819f9330. [DOI] [PubMed] [Google Scholar]
- 41.Fuse N, Miyazawa A, Nakazawa T, Mengkegale M, Otomo T, Nishida K. Evaluation of LOXL1 polymorphisms in eyes with exfoliation glaucoma in Japanese. Mol Vis. 2008;14:1338–1343. [PMC free article] [PubMed] [Google Scholar]
- 42.Tanito M, Minami M, Akahori M, et al. LOXL1 variants in elderly Japanese patients with exfoliation syndrome/glaucoma, primary open-angle glaucoma, normal tension glaucoma, and cataract. Mol Vis. 2008;14:1898–1905. [PMC free article] [PubMed] [Google Scholar]
- 43.Goldstein DB, Tate SK, Sisodiya SM. Pharmacogenetics goes genomic. Nat Rev Genet. 2003;4:937. doi: 10.1038/nrg1229. [DOI] [PubMed] [Google Scholar]
- 44.Dervieux T, Meshkin B, Neri B. Pharmacogenetic testing: proofs of principle and pharmacoeconomic implications. Mutat Res. 2005;573:180–1894. doi: 10.1016/j.mrfmmm.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 45.Ostapowicz G, Fontana RJ, Schiodt FV, et al. Results of a prospective study of acute liver failure at 17 tertiary care centers in the United States. Ann Intern Med. 2002;137:947–954. doi: 10.7326/0003-4819-137-12-200212170-00007. [DOI] [PubMed] [Google Scholar]
- 46.Li L, Jick H, Jick SS. Updated study on risk of cholestatic liver disease and flucloxacillin. Br J Clin Pharmacol. 2009;68:269–270. doi: 10.1111/j.1365-2125.2009.03454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daly AK, Donaldson PT, Bhatnagar P, et al. HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat Genet. 2009;41:816–819. doi: 10.1038/ng.379. [DOI] [PubMed] [Google Scholar]
- 48.Bowman L, Armitage J, Bulbulia R, Parish S, Collins R. Study of the effectiveness of additional reductions in cholesterol and homocysteine (SEARCH): characteristics of a randomized trial among 12064 myocardial infarction survivors. Am Heart J. 2007;1 54:815–23. doi: 10.1016/j.ahj.2007.06.034. [DOI] [PubMed] [Google Scholar]
- 49.Thompson PD, Clarkson P, Karas RH. Statin-associated myopathy. JAMA. 2003;289:1681–1690. doi: 10.1001/jama.289.13.1681. [DOI] [PubMed] [Google Scholar]
- 50.Link E, Parish S, Armitage J, et al. SLCO1B1 variants and statin-induced myopathy--a genomewide study. N Engl J Med. 2008;359:789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 51.Mitka M. Researchers worry about myopathy risk for patients taking high-dose simvastatin. JAMA. 2009;301:261–262. doi: 10.1001/jama.2008.939. [DOI] [PubMed] [Google Scholar]
- 52.Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:798–801. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 53.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801 . doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McHutchison JG, Lawitz EJ, Shiffman ML, et al. Peginterferon alfa-2b or alfa-2a with ribavirin for treatment of hepatitis C infection. N Engl J Med. 2009;361:580–593. doi: 10.1056/NEJMoa0808010. [DOI] [PubMed] [Google Scholar]
- 55.Hindorff LA, Junkins HA, Mehta JP, Manolio TA. A Catalog of Published Genome-Wide Association Studies. Cited 2009. Available at: www.genome.gov/gwastudies. Accessed 8/10/2009 [Google Scholar]
- 56.Erlich HA, Griffith RL, Bugawan TL, Ziegler R, Alper C, Eisenbarth G. Implication of specific DQB1 alleles in genetic susceptibility and resistance by identification of IDDM siblings with novel HLA-DQB1 allele and unusual DR2 and DR1 haplotypes. Diabetes. 1991;40:478–481. doi: 10.2337/diab.40.4.478. [DOI] [PubMed] [Google Scholar]
- 57.Matsuki K, Grumet FC, Lin X, et al. DQ (rather than DR) gene marks susceptibility to narcolepsy. Lancet. 1992;339:1052. doi: 10.1016/0140-6736(92)90571-j. [DOI] [PubMed] [Google Scholar]
- 58.Etzensperger R, McMahon RM, Jones EY, Fugger L. Dissection of the multiple sclerosis associated DR2 haplotype. J Autoimmun. 2008;31:201–207. doi: 10.1016/j.jaut.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 59.Stefansson H, Rye DB, Hicks A, et al. A genetic risk factor for periodic limb movements in sleep. N Engl J Med. 2007;357:639–647. doi: 10.1056/NEJMoa072743. [DOI] [PubMed] [Google Scholar]
- 60.Winkelmann J, Schormair B, Lichtner P, et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007;39:1000–1006. doi: 10.1038/ng2099. [DOI] [PubMed] [Google Scholar]
- 61.Vilarino-Guell C, Farrer MJ, Lin SC. A genetic risk factor for periodic limb movements in sleep. N Engl J Med. 2008;358:425–427. doi: 10.1056/NEJMc072518. [DOI] [PubMed] [Google Scholar]
- 62.Kemlink D, Polo O, Frauscher B, et al. Replication of restless legs syndrome loci in three European populations. J Med Genet. 2009;46:315–318. doi: 10.1136/jmg.2008.062992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Desautels A, Turecki G, Montplaisir J, et al. Evidence for a genetic association between monoamine oxidase A and restless legs syndrome. Neurology. 2002;59:215. doi: 10.1212/wnl.59.2.215. [DOI] [PubMed] [Google Scholar]
- 64.Vilarino-Guell C, Chai H, Keeling BH, et al. MEIS1 p.R272H in familial restless legs syndrome. Neurology. 2009;73:243–245. doi: 10.1212/WNL.0b013e3181ae7c79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mignot E. Genetic and familial aspects of narcolepsy. Neurology. 1998;50(2 suppl 1):S16–S22. doi: 10.1212/wnl.50.2_suppl_1.s16. [DOI] [PubMed] [Google Scholar]
- 66.Hallmayer J, Faraco J, Lin L, et al. Narcolepsy is strongly associated with the T-cell receptor alpha locus. Nat Genet. 2009;41:708–711. doi: 10.1038/ng.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi J, Levinson DF, Duan J, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stefansson H, Ophoff RA, Steinberg S, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ferreira MA, O'Donovan MC, Meng YA, et al. Collaborative genomewide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma D, Salyakina D, Jaworski JM, et al. A genome-wide association study of autism reveals a common novel risk locus at 5p14.1. Ann Hum Genet. 2009;73(Pt 3):263–273. doi: 10.1111/j.1469-1809.2009.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang K, Zhang H, Ma D, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang K, Li M, Hadley D, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Bon BW, Mefford HC, Menten B, et al. Further delineation of the 15q13 microdeletion and duplication syndromes: a clinical spectrum varying from non-pathogenic to a severe outcome. J Med Genet. 2009;46:511–523. doi: 10.1136/jmg.2008.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dibbens LM, Mullen S, Helbig I, et al. Familial and sporadic 15q13.3 microdeletions in idiopathic generalized epilepsy: precedent for disorders with complex inheritance. Hum Mol Genet. 2009 ;18:3626–3631. doi: 10.1093/hmg/ddp311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brunetti-Pierri N, Berg JS, Scaglia F, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bijlsma EK, Gijsbers AC, Schuurs-Hoeijmakers JH, et al. Extending the phenotype of recurrent rearrangements of 16p11.2: deletions in mentally retarded patients without autism and in normal individuals. Eur J Med Genet. 2009;52:77–87. doi: 10.1016/j.ejmg.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 77.Mefford HC, Sharp AJ, Baker C, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.St Clair D, Blackwood D, Muir W, et al. Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990;336:13–16. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- 79.Green EK, Grozeva D, Jones I, et al. The bipolar disorder risk allele at CACNA1C also confers risk of recurrent major depression and of schizophrenia. Mol Psychiatry. 2009. In press doi: 10.1038/mp.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Need AC, Ge D, Weale ME, et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS genetics. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Walsh T, McClellan JM, McCarthy SE, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 83.Stone JL, O'Donovan MC, Gurling H, et al. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stefansson H, Rujescu D, Cichon S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Glessner JT, Wang K, Cai G, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pagnamenta AT, Wing K, Akha ES, et al. A 15q13.3 microdeletion segregating with autism. Eur J Hum Genet. 2009;17:687–692. doi: 10.1038/ejhg.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hannes FD, Sharp AJ, Mefford HC, et al. Recurrent reciprocal deletions and duplications of 16p13.11: The deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J Med Genet. 2008;46:223–242. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gottesman I. Schizophrenia Genesis, the Origins of Madness. New York, NY: W.H. Freeman and Company 1991 [Google Scholar]
- 89.Alkan C, Kidd JM, Marques-Bonet T, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yoon S, Xuan Z, Makarov V, Ye K, Sebat J. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 2009;19:1586–1592. doi: 10.1101/gr.092981.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xie C, Tammi MT. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics. 2009;10:80. doi: 10.1186/1471-2105-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Korn JM, Kuruvilla FG, McCarroll SA, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet. 2003;33(suppl):228. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 94.Weiss LA, Shen Y, Korn JM, et al. Association between microdeletion and microduplication at 16p11 .2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 95.Sharp AJ, Hansen S, Selzer RR, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 96.de Vries BB, Pfundt R, Leisink M, et al. Diagnostic genome profiling in mental retardation. Am J Hum Genet. 2005;77:606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Christiansen J, Dyck JD, Elyas BG, et al. Chromosome 1q21.1 contiguous gene deletion is associated with congenital heart disease. Circ Res. 2004;94:1429–1435. doi: 10.1161/01.RES.0000130528.72330.5c. [DOI] [PubMed] [Google Scholar]
- 98.Greenway SC, Pereira AC, Lin JC, et al. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet. 2009;41:931–935. doi: 10.1038/ng.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Diskin SJ, Hou C, Glessner JT, et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987–991. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kirov G, Grozeva D, Norton N, et al. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Doornbos M, Sikkema-Raddatz B, Ruijvenkamp CA, et al. Nine patients with a microdeletion 15q11.2 between breakpoints 1 and 2 of the PraderWilli critical region, possibly associated with behavioural disturbances. Eur J Med Genet. 2009;52:108–115. doi: 10.1016/j.ejmg.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 102.Murthy SK, Nygren AO, El Shakankiry HM, et al. Detection of a novel familial deletion of four genes between BP1 and BP2 of the PraderWilli/Angelman syndrome critical region by oligo-array CGH in a child with neurological disorder and speech impairment. Cytogenetic Genome Res. 2007;116:135–140. doi: 10.1159/000097433. [DOI] [PubMed] [Google Scholar]
- 103.Kirov G, Gumus D, Chen W, et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia Hum M et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J Med Genet. 2009;46:242–248. [Google Scholar]
- 105.Helbig I, Mefford HC, Sharp AJ, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41:160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sharp AJ, Mefford HC, Li K, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kumar RA, KaraMohamed S, Sudi J, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 108.Ghebranious N, Giampietro PF, Wesbrook FP, Rezkalla SH. A novel microdeletion at 1 6p1 1 .2 harbors candidate genes for aortic valve development, seizure disorder, and mild mental retardation. Am J Med Genet A. 2007;143A:1462–1471. doi: 10.1002/ajmg.a.31837. [DOI] [PubMed] [Google Scholar]
- 109.Weber YG, Jacob M, Weber G, Lerche H. A BFIS-like syndrome with late onset and febrile seizures: suggestive linkage to chromosome 16p11.21 6q12.1. Epilepsia. 2008;49:1959–1964. doi: 10.1111/j.1528-1167.2008.01646.x. [DOI] [PubMed] [Google Scholar]
- 110.Kumar RA, Marshall CR, Badner JA, et al. Association and mutation analyses of 1 6p11.2 autism candidate genes. PloS one. 2009;4:e4582. doi: 10.1371/journal.pone.0004582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McCarthy SE, Makarov V, Kirov G, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ullmann R, Turner G, Kirchhoff M, et al. Array CGH identifies reciprocal 16p1 3.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat. 2007;28:674–682. doi: 10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- 113.Fernandez T, Morgan T, Davis N, et al. Disruption of Contactin 4 (CNTN4) results in developmental delay and other features of 3p deletion syndrome. Am J Hum Genet. 2008;82:1385. doi: 10.1016/j.ajhg.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Roohi J, Montagna C, Tegay DH, et al. Disruption of contactin 4 in three subjects with autism spectrum disorder. J Med Genet. 2009;46:176–182. doi: 10.1136/jmg.2008.057505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kirov G, Rujescu D, Ingason A, Collier DA, O'Donovan MC, Owen MJ. Neurexin 1 (NRXN1) deletions in Schizophrenia. Schizophr Bull. 2009;35:851–854. doi: 10.1093/schbul/sbp079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rujescu D, Ingason A, Cichon S, et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18:988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, et al. Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Hum Genet. 2008;83:504–510. doi: 10.1016/j.ajhg.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.ucan M, Abrahams BS, Wang K, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS genetics. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim HG, Kishikawa S, Higgins AW, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zahir FR, Baross A, Delaney AD, et al. A patient with vertebral, cognitive and behavioural abnormalities and a de novo deletion of NRXN1alpha. J Med Genet. 2008;45:239–243. doi: 10.1136/jmg.2007.054437. [DOI] [PubMed] [Google Scholar]
- 122.Friedman JM, Baross A, Delaney AD, et al. Oligonucleotide microarray analysis of genomic imbalance in children with mental retardation. Am J Hum Genet. 2006;79:500–513. doi: 10.1086/507471. [DOI] [PMC free article] [PubMed] [Google Scholar]