Abstract
The cost of a diploid human genome sequence has dropped from about $70M to $2000 since 2007- even as the standards for redundancy have increased from 7x to 40x in order to improve call rates. Coupled with the low return on investment for common single-nucleotide polymorphisms, this has caused a significant rise in interest in correlating genome sequences with comprehensive environmental and trait data (GET). The cost of electronic health records, imaging, and microbial, immunological, and behavioral data are also dropping quickly. Sharing such integrated GET datasets and their interpretations with a diversity of researchers and research subjects highlights the need for informed-consent models capable of addressing novel privacy and other issues, as well as for flexible data-sharing resources that make materials and data available with minimum restrictions on use. This article examines the Personal Genome Project's effort to develop a GET database as a public genomics resource broadly accessible to both researchers and research participants, while pursuing the highest standards in research ethics.
Keywords: Personal Genome Project, personal genomics, DNA sequencing technology, whole-genome sequencing, phenome, envirome, microbiome, GET data set, open consent, public genome, ELSI
Abstract
El costo de una secuencia del genoma humano diploide se ha reducido desde cerca de 70 millones de dólares a 2000 dólares desde 2007, aunque los estándares de la redundancia han aumentado de 7 a 40 veces para mejorar los índices de demanda de genotipo. Junto con el bajo retorno de inversión para los polimorfismos de nucleótidos únicos comunes, esta situatión ha causado un aumento significativo del interés en correlacionar las secuencias genómicas con una completa información ambiental y de rasgos (GAR). El costo de las fichas médicas electrónicas, de las imágenes y de la información microbiológica, inmunológica y conductual también está reduciéndose rápidamente. El compartir tal conjunto de información y sus interpretaciones con una diversidad de investigadores y sujetos de investigación pone de relieve la necesidad de contar con modelos de consentimiento informado capaces de estar orientados hacia nuevos temas de privacidad y otros, además de flexibilizar los recursos de datos compartidos que permitan disponer de materiales e información con mínimas restricciones de uso. Este artículo examina el esfuerzo del Proyecto de Genoma Personal para desarrollar una base de datos de GAR como un recurso de genómica pública ampliamente accesible tanto a investigadores como a participantes de las investigaciones, respetando los estándares más elevados de la ética de la investigación.
Abstract
Le coût de séquençage d'un génome diploïde humain a chuté de 70 millions de dollars à 2 000 dollars depuis 2007, bien que les standards de redondance aient augmenté de 7 à 40 fois afin d'améliorer le taux d'identification des bases. Associé au faible retour sur investissement des polymorphismes de simples nucléotides (SNP), cette situation explique l'intérêt accru pour la corrélation des séquences des génomes avec des données complètes environnementales et de traits (GET). Les coûts des enregistrements numériques médicaux, de l'imagerie et des données microbiennes, immunologiques et comportementales chutent aussi rapidement. Le partage de telles bases de données GET intégrées et de leurs interprétations avec un grand nombre de chercheurs et de sujets de recherche souligne la nécessité de modèles de consentement éclairé nécessaires à cette nouvelle protection des données personnelles et autres problématiques, en plus des besoins de flexibilité des ressources requises pour le partage des données, permettant en plus une utilisation peu restrictive de ces matériels et données. Cet article analyse les efforts du Projet du Génome Personnel afin de développer une base de données GET en tant que ressource génomique publique, largement accessible à la fois aux chercheurs et aux participants à la recherche, tout en respectant les standards les plus élevés de l'éthique de la recherche.
The drawing of a new decade is an appropriate time to reflect on the tremendous progress that has been made in human genomic research. In 2010, with wholegenome sequencing becoming increasingly affordable, the promise of large-scale human genomic research studies involving hundreds, thousands, and even hundreds of thousands of individuals is rapidly becoming a reality. The next generation of human genomic research will occur on a scale that would have been nearly unfathomable at the start of the last decade, when the publication of the Human Genome Project's first draft results was still pending.
When the Human Genome Project published its draft results on June 26, 2000, it published a compound human genome sequence containing genetic information from several volunteers. Seventy percent of the final sequence was obtained from one anonymous individual, while the remaining 30% came from a number of different individuals. From the first amalgamated human genome sequence - which was refined in 2003 and continues to be updated and refined to this day - private and public research efforts have gone on to sequence numerous individual human genomes with increasing speed and detail and decreasing time and cost. The acceleration of whole-genome sequencing in the research context necessitates new perspectives and models that enable scientists and society to learn as much as possible from this rapidly expanding dataset while still respecting important ethical, legal, and social norms.
The Personal Genome Project (PGP),1 an ambitious research study directed by faculty members in the Department of Genetics at Harvard Medical School, aims to recruit as many as 100 000 informed participants to contribute genomic sequence data, tissues, and extensive environmental, trait, and other information to a publicly accessible and identifiable research database.
In this review we describe the Personal Genome Project itself, focusing on its unique structural features and the rationale behind the project's design. We also elucidate the changing scientific and social landscape that makes the PGP's model of open consent and public data access increasingly important to the furtherance of human genomic research.
The PGP's mission
In contrast to research studies that focus on small subsets of traits within narrowly defined human populations exhibiting single diseases, the PGP was conceived with an expansive mission. From the outset, the mission of the project (Table I) has been to develop a broad-based, longitudinal, and participatory research study that will facilitate a comprehensive understanding of the project's participants at the genomic level and beyond.
Table I . PGP's Mission Statement, available at: htttp://www.personalgenomes.org/mission.html.1 .
| The Personal Genome Project's Mission Statement |
| The mission of the Personal Genome Project is to encourage the development of personal genomics technology and practices that: |
| • are effective, informative, and responsible |
| • yield identifiable and improvable benefits at manageable levels of risk |
| • are broadly available for the good of the general public |
| To achieve this mission we will build a framework for prototyping and evaluating personal genomics technology and practices at increasing scales. In support of this goal, we will: |
| • develop a broad vision for how personal genomes may be used to improve the understanding and management of human health and disease |
| • provide educational and informational resources for improving general understanding of personal genomics and its potential |
| • recruit individuals interested in obtaining and openly sharing their genome sequences, related health and physical information, and reporting their experiences as a participant of the project on an ongoing basis |
| • develop technologies to improve the accessibility of personal genome sequencing |
| • foster dialog with research communities, industries, and public and governmental bodies with interests in personal genomics, and related ethical, legal, and social issues (ELSI) |
| • develop tools for interpreting genomic information and correlating it with personal medical and biological information |
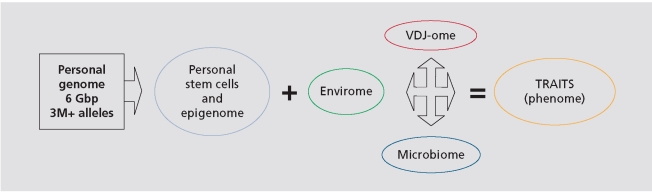
The PGP is constructed with the recognition that our desire to truly understand the genesis of most complex human traits - from dread diseases to the talents and quirks that make us each uniquely human - could only be satisfied by examining genomic information in context and by surrounding it with the richest possible data from the widest possible array of supplemental sources. By supplementing genomic sequence data with the collection and analysis of tissues and extensive environmental and trait data, and by making these data publicly accessible to researchers worldwide, the PGP aims to improve understanding of the ways in which genomes plus environments ultimately equal traits (Figure 1.)
Figure 1. Genome + Environment = Traits (GET) equation. Envirome: the totality of environmental influences; VDJ-ome: the DNA sequences of the entire repertoire of an individual's immunoglobulin and T-cell receptors, which reflect a lifetime of antigenic exposures; Microbiome: the billions of commensal, symbiotic, and pathogenic micro-organisms that share our body space; Epigenome: the totality of programmed biochemical and structural modifications to genomic DNA that regulate organism or phenotype development. (see overview in Table III) .
The PGP is more than just a research repository. In addition to its publicly accessible research database, the PGP, which is supported by the nonprofit PersonalGenomes.org, also works to disseminate genomic technology and knowledge at a global level, thereby producing tangible and widely available improvements in the understanding and management of human health and disease. The PGP also finds itself at the forefront of discourse surrounding the ethical, legal, and social issues (ELSI) associated with large-scale whole-genome sequencing, particularly in the areas of privacy, informed consent, and data accessibility. The PGP is, and is intended to be, a research project that is constantly in progress, exploring the boundaries of human genomic research in a way that produces maximal advances in scientific understanding and public understanding and well-being, while striving to reach beyond what is minimally required to satisfy its ethical, legal, and social obligations to its participants. In the sections that follow we report on unique aspects of the PGP relating to technology development, integrative genomics, and human subject research protocols, as well as describe the development and current state of the PGP.
Key developments in human genome sequencing
The PGP derives its impetus and importance from historic breakthroughs in understanding and analysis of DNA. DNA comprises only a very small fraction of a cell (~3% dry weight E. coli), and its role as the molecule primarily responsible for transmission of genetic traits was not recognized until a series of discoveries beginning in the 1940s. The emergence in 1953 of a clear concept of DNA as a double-helical structure comprising a pair of complementary strings of four elementary bases (the nucleotides A, C, G, and T) crystallized interest in determining the DNA sequences of genes and the sequence differences responsible for disease, and set the stage for over four decades of development of ever more efficient and comprehensive sequencing methods. Table II describes this history by a set of milestones that take one from the early beginnings of DNA sequencing up through delivery of draft human genome sequences in 2001 to 2003. In the 38 years between 1965, when Robert Holley and colleagues at Cornell and the US Department of Agriculture sequenced a 77 nt RNA gene after 4 years of effort, and 2003, when the public Human Genome Project (HGP) declared that it had met its goals regarding delivery of a ~3Gbp human genome sequence, the size of DNA sequence that could be accommodated by sequencing technology improved ~30 million-fold.
Table II. Development of DNA sequencing.
| Date | Event | Size of sequence (bp) | Reference |
| 1957 | First sequence mutation identified responsible for disease | 1 amino acid (sickle cell vs normal hemoglobin) | (Ingram 19572) |
| 1965 | First sequence of a single complete gene | 77 bases | (Holley, Apgar et al 19653) |
| 1976-1977 | Sequencing of first viral genomes | 3562 bases (MS2 RNA phage) 5375 bases (φ X174 DNA phage) | (Fiers, Contreras et al 19764; Sanger, Air et al 19775) |
| 1975-1977 | Maxam/Gilbert and Sanger DNA sequencing methods | (Sanger and Coulson 19756; Maxam and Gilbert 19777; Sanger, Nicklen et al 19778) | |
| 1994 | First commercial bacterial genome sequence | 1.7Mbp (Helicobacter pylori) | (Nature Genetics, May 19969) |
| 1995 | First published bacterial genome sequence | 1.83Mbp (Haemophilus influenzae) | (Fleischmann, Adams et al 199510) |
| 1998-2000 | Genome sequences of first animals | 100Mbp (Caenorhabditis elegans) 120Mbp (Drosophila melanogaster) | (C. elegans Sequencing Consortium 1998,11 Adams, Celniker et al 200012) |
| 2001 | Two draft sequences of human genome | ~3Gbp | (Lander, Linton et al 2001,13 Venter, Adams et al 200114) |
| 2003 | Completion of public Human Genome Project | (Collins, Morgan et al 200315) |
Post-HGP sequencing - towards whole diploid genomes
Notably, the HGP had delivered only a single human genome sequence that was a composite built from a small number of deidentified individuals, while the competing nonpublic human genome project merged in data from an identified individual (Craig Venter); both were haploid estimates. As recognized from the beginning of the HGP, many additional resources would be needed to understand the functions of the genes laid out in these “reference” human genomes, and to identify the sequence differences between individuals that contribute to individual traits, health, and disease. Indeed, as the HGP ended, projects were already under way to identify large numbers of genetic differences from the HGP-derived reference genome in different human populations that could subsequently be analyzed using low-cost array methods in large numbers of individuals, a strategy that has since given rise to more than 480 published genome-wide association studies.16,17 At the same time, however, interest was rising in the second approach: to significantly improve DNA sequencing technology to a point where an individual's entire genome could be sequenced at very low cost. A combination of two kinds of arguments were advanced supporting this approach, focusing on functional utility and economics, respectively.
The gist of the functional arguments was that sequencing of individuals is intrinsically more informative and flexible than array-based interrogation of known sites of variation and that, variation aside, any improvements in sequencing cost and capability could be quickly applied to numerous general aspects of biology that are critical to understanding gene function, traits, and health and disease.18,19 The relative advantages of sequencing have long been recognized. Unlike array analyses, sequencing: (i) does not require variations to be preidentified; (ii) can more readily accommodate more complex variations than single nucleotide changes and very short inserts or deletions; and (iii) need not focus on variations that are common in large populations vs rare or unique variations. In consequence, as sequencing technology has improved, it has increasingly been integrated into association studies of variation.20-23
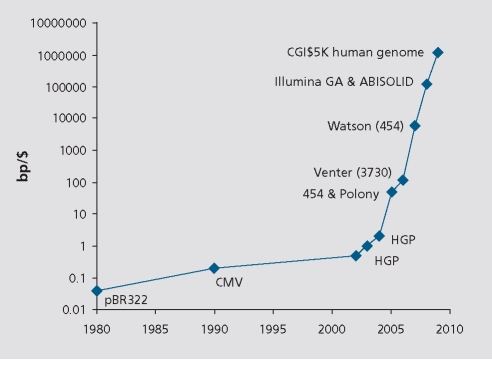
However, these advantages of sequencing were counterbalanced by their high cost, a situation well illustrated by the $3 billion US cost of the HGP itself. It is here that economic arguments were advanced suggesting that dramatic improvements in sequencing were feasible that might ultimately enable an individual's genome to be sequenced for 1000 to 10 000 USD.18 On an empirical level, sequencing technology has appeared to exhibit a historical trend of exponentially decreasing costs with time as measured by sequenced base pairs per dollar at a given error rate, a situation frequently compared with “Moore's Law” in computing,24 which noted that computing power measured by the integrated circuit transistor density doubled roughly every 2 years at constant cost (Figure 2).18,25 To get genome sequencing costs down to $1000 would require cost and throughput improvements of an additional 4 to 5 orders of magnitude, so the question of economic feasibility ultimately turned on whether new methods could enable this very large improvement.
Figure 2. Exponential trend of sequencing costs in base pairs per USD (bp/$), a trend often compared with Moore's Law (see text). See ref 25 for details.
Here, the HGP again gave grounds for optimism, for even though the HGP itself only achieved 100-fold improvements, it achieved this largely by refining, miniaturizing, and robotically scaling up, but not fundamentally changing, a Sanger sequencing method initially developed over 20 years earlier (Table II). If such methods were capable of 100-fold improvement, considerably greater improvements might be expected from more radically changing sequencing chemistry, signal generation and detection, and instrumentation in ways that could integrate some of the vast advances in chemistry and enzymology, optics and electronics, materials science, microfabrication, and process control that had accrued over the preceding 20 years and been put to good use in many other fields. The HGP also directly provided an important resource for realizing this strategy: the reference human genome sequence itself, as this could serve as a template against which reads obtained by new technologies could be located, allowing new human genomes to be assembled at least initially by “resequencing” vs de novo assembly. This reduces the burden on new sequencing methods by allowing them to generate useful data with shorter reads and higher base call error rates than would generally be needed for de novo assembly, although de novo assembly of genomes using new sequencing technology remains an important goal.
Next-generation sequencing
Researchers were quick to work out sequencing approaches along the lines indicated in these arguments, and commercial products emerged soon, giving rise to next- generation sequencing (NGS). Soon granting agencies promised funding for support, and a ~10M USD competition was announced for rapid, accurate genomic sequencing, generating increased coalescence around target goals for dramatic improvements to sequencing technology.26,27,28 Detailed reviews and comparisons of NGS approaches have been published.18, 29,30
Among the earliest NGS methods were polony sequencing (the Polonator) and 454 Life Sciences.31,32,33 Both methods amplify DNA templates onto microbeads that are packed onto two-dimensional arrays for sequencing, thereby achieving enormous economies of scale compared with Sanger sequencing, and each achieved ~25 fold better cost per bp compared with HGP (Figure 2). However, each uses different sequencing chemistry and arraying technology, giving rise to many technical tradeoffs. Together they proved the general point that great improvements in sequencing efficiency were indeed within reach, but also that the precise character and degree of improvement would depend closely on the novel technologies employed and the ingenuity with which they could be integrated. A second wave of development introduced methods by Illumina and ABI that, by very different means, have improved the utility and costs, (Figure 2)34,35 and hence use of these systems is becoming widespread for both large scale and “deep” sequencing applications, and both are under continuous development.
Two complete cancer genomes were recently sequenced, one with each platform.36,37 Further rounds of innovation have yielded a diverse set of newer NGS methods. For instance, a number of “single-molecule” sequencing methods are now available or in development. These methods avoid the need to make thousands to millions of copies of DNA template molecules on microbeads or surfaces to assure that sequencing operations generate sufficient signal to read individual bases accurately, and instead use highly sensitive optics to detect bases at the single molecule level; this allows even denser packing of DNA templates and further efficiencies in sequencing chemistry. While Helicos Biosciences has commercialized a singlemolecule system that simply arrays single template molecules on a surface and uses sequencing cycle similar to the methods above, Pacific Biosciences is developing a system in which enzymes and templates are tethered to the bottom of nanofabricated wells and which monitors the signals generated by sequencing chemistry in realtime vs artificial cycles.38,39 Here, the nanofabricated wells enable substantially increased accuracy of single molecule base incorporation events. Finally, on another track, the company Complete Genomics, Inc has developed a method whereby very compact self-assembling amplicons of template DNAs called “nanoballs” are flowed onto a nanofabricated grid of ~300nm spots at 700 to 1300 nm center-to-center distances. Three complete human genomes were sequenced with this method (as of January 2010) with an average consumable cost of $4400 and as low as $1500 for 40X coverage.40
Towards affordable personal genomes
These developments suggest that technology capable of meeting the cost target of $1000 or less for a diploid human genome sequence is within reach. Indeed, the indepth resequencing of individual human genomes has now been demonstrated several times by NGS developers to demonstrate that their methods have come of age. There are now published full genome sequences for at least seven individuals,40 with some having been sequenced by more than one method. There are also tens - and perhaps hundreds - of additional unpublished or partly published genomes (see, eg, refs 36,37), while the lower-coverage 1000 Genomes Project20,21 continues. Clearly, the age of personal genomics is now close at hand.
The PGP
As described in the first section, one of the PGP's central aims is to develop a publicly available, fully consented database containing comprehensive human genome and phenome data for its research participants. Such integrated datasets are fundamental drivers of progress in functional genomics and enable systems biology-based insights into the mechanisms of human health and disease.41 PGP studies will look beyond inherited genomes to include somatic and epigenetic variation data, as well as relevant microbiome, transcriptome, immunity-reflecting “VDJ-ome” and phenome data to develop comprehensive profiles. By developing high-resolution data profiles for each participant, and multiplying that by a large (up to 100 000) participant population, the PGP will also generate valuable data describing the kinds and distributions of variation that exist in populations. Although an improved understanding of human health and disease is a central aim of the PGP, its focus is considerably broader and will enable research into the social and behavioral sciences using personal genomic data. Finally, the PGP's flexible study protocol and public and distributed approach to research enables it to keep pace with sequencing and other technological advances while simultaneously driving these developments.
Integrated personal genomes: inherited, somatic, environmental genomics
If the PGP is to fulfill its mission to address the multidimensional complexity of human biology, it must encompass multiple interacting “-omes.” For example, a person's diet will have a profound influence upon her or his somatic gene expression as well as the genomic and proteomic activity of the person's microbiome. It will also affect the metabolome. Similarly, an individual's environmental exposures to pollutants will have a direct bearing on her or his immunological response and therefore, on the VDJ-ome. Germline alleles will affect how one metabolizes drugs, which will have myriad effects on an individual's physiological and behavioral phenotypes.
Genomes (vs exomes)
In its early phase, given the then-current cost of genomic sequencing, the PGP planned to focus on exomes rather than whole genomes as a way to affordably expand the project to large numbers of participants. Despite representing only 1% to 2% of the 6 billion base pairs in a human genome, the exome contains all protein-coding exons and therefore provides access to the majority of known functional variants.48,49,50 However, continued improvements in genomic sequencing have produced price declines that have rendered whole-genome sequencing significantly cheaper per base pair than exome sequencing. The PGP, as a result, has determined that whole-genome sequencing is cost-justified given the relatively high price of exomes and the additional information supplied by whole-genome sequences of PGP participants.51 See also Table III for the various “omes.”
Table III . The "omes." .
| Personal genome: Entire diploid human genome of a single individual representing 6 billion base pairs. |
| Exome: All exons, representing 1% to 2% of the entire human genome. |
| Phenome: Set of all traits in an organism, at all levels, or one of its subsystems, including morphology, physiology, and behavior.42,43 |
| Envirome: The totality of equivalent environmental influences contributing to all disorders and organisms.44 |
| Microbiome (human): The ecological community of commensal, symbiotic, and pathogenic microorganisms that share our body space.45 |
| VDJ-ome: The repertoire of rearranged V, D, and J genome segments present in an individuals's B and T immune cells at any given time (see Table IV). |
| Transcriptome: The set of all RNA molecules, including mRNA, rRNA, tRNA, and noncoding RNA produced in one or a population of cells.46 |
| Epigenome: The totality of programmed biochemical and structural modifications to genomic DNA that regulate organism or phenotype development. |
| Metabolome: Total set of metabolites generated by an organism, or subsystem. |
| Proteome: The entire set of proteins expressed by a genome, cell, tissue or organism at a given time under defined conditions. There are more proteins than genes.47 |
Phenomes
Detailed phenotype data is required to categorize and, ultimately, understand the phenotypes that the PGP seeks to explore. However, the vastness of the human phenome, defined as the physical totality of human traits at all levels, from the molecular to the behavioral, will require new strategies that permit high-throughput trait collection while yielding accurate and standardized phenotypic data. With regard to the cellular and molecular phenotypes, the PGP collects participant tissue samples and develops cell lines that are then deposited and publicly accessible through established biobanks.52,53
As the PGP expands it is exploring Web-based, highthroughput behavioral phenotype data-collection models pioneered by leading public and private researchers. While the reliability and validity of self-reported traits is a concern, particularly for phenome research conducted online,54,55 Web-based assessments provide distinct opportunities for “dynamic phenotyping” based on a particular individual's prior genotype-phenotype associations.56 The multimodal capabilities of Web-based trait collection instruments, combined with their low cost of implementation at large scales, seem likely to accelerate the ability of studies like the PGP to effectively explore new corners of the human phenome.
The PGP is also taking advantage of recent advancements in health information technologies to assist participants and researchers alike in structuring and accessing the massive amounts of personalized data generated by the project. The emergence of online Personally Controlled Health Record (PCHR) platforms and other novel tools enables individuals to collect and manage their own health data - including health history, medication, allergy, immunization, biometric and other data types57,58,59 - and can be developed for integrated data entry, access and dissemination by both the individual and third-party researchers or data providers, including health care providers.
Enviromes
The picture of genome and phenome is incomplete without the envirome. The envirome can be described as the totality of equivalent environmental influences contributing to all disorders and organisms.60 The mode of response of an organism to the environment that is reflected in its phenotype is constrained by its unique set of genetic variations and the environmental influences on gene expression. Therefore, a comprehensive approach is required to describe the envirome systematically in conjunction with genome and phenome information. The relevant envirome data is too large and complex to be reported, managed, or analyzed manually. The creation of phenome-genome and genome-envirome networks has been suggested in order to relate phenome and envirome information to potential disease-associated genes.61
Microbiomes
Even though microbial cells are estimated to outnumber human cells in a single individual by a factor of ten, we know very little about the microbes that live in and on us, including what mixture of bacteria, viruses, and other micro-organisms constitute a “normal” human microbiome and how those organisms impact different biological states.62 Major efforts such as the Human Microbiome Project are under way to characterize the microbiota at different body sites in humans and to assess how variation in microbial communities is associated with states of health and disease.63 The PGP takes advantage of the unique availability of comprehensive participant profiles and uses them to explore interactions between host genetic and phenotypic variability alongside the genomic variation in the microbes that colonize them.64
The VDJ-ome
The Church Lab at Harvard Medical School is developing techniques for characterizing the repertoires of Band T-cell receptors in individual humans from blood samples and correlated across time with personal exposure histories, with an ultimate goal of characterizing individuals repertoires of linked VD J and VJ sequences.
These techniques will be directly applicable to PGP participants and their self-reported data, and will yield a database of unprecedented depth describing the diversity and time development of human immune responses of large numbers of individuals in their life contexts.
Table IV . The adaptive immune system and the VDJ-ome .
| The adaptive immune system |
| The adaptive immune system enables individuals to respond to their unique exposure histories to pathogens and environmental antigens, and possibly to cancerous mutations in their own cells, by generating and modulating expression of >10 12 unique antibodies from B cells and T cell receptors.65 Antibody diversity derives from programmed stochastic rearrangements in maturing B cells of ~40 V, 23 D, and ~5 J functional genomic segments into VDJ heavy chains, and ~35 V and ~5 J segments into VJ light chains (κ or λ) in B cells, that are further randomized by somatic hypermutation; a similar process occurs in T cells.66 NGS methods are now allowing researchers to identify and analyze expressed VDJ sequences in depth.67 |
Tissue reprogramming
The PGP also applies advances in tissue reprogramming techniques to tissue samples collected from PGP participants. Cells from collected somatic tissues are reprogrammed into induced pluripotent stem (iPS) cells68 and made to differentiate into the cell types that are targeted for functional analysis. These methods enable experimental access to diverse tissue types that would otherwise be unobtainable from human subjects but are routinely analyzed in model organisms, and thus, PGP participants can effectively serve as human model organisms. By examining multiple cell types from a single individual, differences in physiological states within and between tissues can be compared within a single PGP participant and/or across the entire PGP cohort. This approach also permits researchers to elucidate connections between genetic variation and variation in other molecular traits, such as gene expression or epigenetic modifications.69 Stored fibroblast cell lines provide researchers with access to renewable supplies of different tissue types from PGP participants.
The PGP: from personal to public genomes
The potential benefits arising from large-scale and integrated human genomic datasets are immense.70 The utility of such research, however, depends upon the responsible development and widespread availability of such comprehensive datasets, which in turn depends on describing and addressing the various ethical, legal and social challenges. Those challenges include a standard set that are inherent to any research involving human subjects, as well as certain challenges that are unique to “public genomics”71 research involving publicly available, identifiable whole-genome sequence data, such as the model pioneered by the PGP. We use the term “public genomics” to denote research studies that possess the following three critical attributes.
Integrated data
The various data types, including genomic and phenomic or trait data, are accessible in a linked format, such as a PCHR or other integrated data structure. Through this explicit linkage of data it is possible to ascertain the complete list of available traits and genetic variants for any given participant. Integration also facilitates participant-researcher interactions, longitudinal study and recontact and, crucially, simultaneous investigation of the full range of complex trait associations. Although participants need not be explicitly identified, integrated data sets that include both genomic and phenomic data will be identifiable in most cases. For this reason, participants must be made explicitly aware of the probability that they will be identified with their publicly available data, rendering promises of perfect privacy, anonymity, or confidentiality impermissible within the public genomics model. However, the promise of privacy need not give way to a promise of publicity.
Open access
Data sets and tissues are made publicly available with minimal or no access restrictions (including researcher qualifications and cost), and are generally transferable outside the original research study to be utilized by and combined with data from third parties. Well-developed data structures and intellectual property licenses are important components of this characteristic. Developing datasets that are not only publicly available but also easily portable fosters the development of a genomic commons, allows data validation by third parties, and enables the use and application of data in novel contexts that may not be foreseeable at the time of collection, thereby facilitating hypothesis generation, encouraging serendipity and broadening the genomic research community.
Voluntary and informed participation
Satisfaction of the first two criteria publication of an integrated dataset in an open-access format necessitates that a premium be placed on receiving truly voluntary and informed consent from participants in public genomics research projects. Given the yet-unknown outcomes and the potential personal, familial, and social risks associated with such research, enrollment is only acceptable under an informed consent protocol that is specially designed to meet the highest standards of human research subjects protection in view of these conditions.
The study protocol
The PGP aims to produce public genomics research - and to develop and evaluate associated technologies and research - on a large and expanding scale. In October of 2008, the PGP published the first integrated set of DNA sequences, traits, and tissues collected from ten participants (the “PGP-10”) enrolled in a pilot study initiated in 2005. Today, the PGP is incrementally expanding its cohort toward 100 000 participants. More than 12 000 individuals had registered to participate in the PGP as of February 2010. In the following section we highlight significant features of the PGP study protocol as it is implemented for the enrollment of the first 100 participants (“PGP-100”) and summarized in Table V.
Table V. Overview of PGP study protocol .
Adapted from ref 52: Angrist M. Eyes wide open: the personal genome project, citizen science and veracity in informed consent. Pers Med. 2009;6:691-699 Copyright © Future Medicine 2009
| Eligibility screening | • Review and sign “mini-consent” form. |
| • Eligibility questionnaire about family circumstances and privacy preferences. | |
| • Entrance exam to ensure informed consent; includes potential risks of participating, project protocols, and basic genetics. | |
| • Review of full PGP consent form. | |
| • Submit information or delete account. | |
| Pre-enrollement | • Consent to participate. |
| • Collection of baseline trait data via questionnaire and a personal health record. Includes allergies, immunizations, medical history, medications, physical traits and measurements, diet, ethnicity/ancestry, lifestyle, and environmental exposures. | |
| • Participants asked to make a financial pledge (does not impact enrollment decisions). | |
| • Identity verification and provision of mailing address. | |
| • Submission of application for enrollment. Individuals selected to continue the enrollment process will receive an enrolment kit by mail, including saliva collection materials. | |
| Enrollment | • Participants may be interviewed by one or more PGP staff to verify identity and consent, confirm familiarity with study protocols, and/or review trait questionnaire responses. Blood samples, saliva sample, and/or skin cells may be collected. |
| • Tissue samples prepared for DNA sequencing and other biological analyses. | |
| • Participants opt-in to have their profiles made available on a publicly accessible Web site, or withdraw from the study. | |
| • Establishment, distribution and analysis of cell lines for research. | |
| Ongoing participation | • Information collected for 25 years. Participants can leave the study at any time. |
| • Data Safety Monitoring Board monitors the impacts of the PGP on enrolled participants. Quarterly emails inquire about adverse events. | |
| • Additional trait data and tissue samples may be requested periodically. |
Public genomes: adding to ELSI
The practice of public genomics poses its own challenges, especially for the organization and governance of human subjects' research, forcing us to critically reassess current frameworks and practices. In order to pursue innovative research in a responsible manner, the PGP has developed a number of project-specific tools and resources relevant to ELSI.
Open consent
The “open consent” model developed by the PGP is designed to address the set of challenges associated with the creation of datasets where it may be possible to identify individual participants with their genomic and other data. The open consent model assumes that, in such a context, conventional assurances of anonymity, privacy and confidentially are impossible and should not serve as any part of the foundation for the informed consent protocol.72,73 Due to the structure of public genomics projects such as the PGP, and their associated datasets, while privacy and confidentiality can be protected they cannot and should not be guaranteed to participants. This practice ensures veracity, which we regard as a necessary - though not sufficient - prerequisite for the exertion of substantive autonomy. It is only through veracity that the criteria underlying truly informed consent can be satisfied.
Open consent is therefore based on complete openness and transparency with regard to all aspects of participation, including the potential for reidentification and the reality that there may be other risks that are unidentifiable at the time of consent. Predicting all potential risks is by definition impossible and even a list of known possible risks is unlikely ever to be comprehensive.
Data sharing - and the risks of public genomes
The PGP's informed consent process begins with an extensive pre-enrollment educational examination designed to ensure a potential participant's ability to understand the specific nature of the data collected and the risks presented by public genomics research. For individuals who demonstrate the needed proficiency, the specific informed consent agreement that follows includes a lengthy but “noncomprehensive list of hypothetical scenarios that could pose risks” for participants and their families (Table VI). Participants are warned that “the complete set and magnitude of the risks that the public availability of [your genomic data] poses to you and your relatives is not known at this time.” It is crucial that participants understand that once identifying genetic and trait data and tissues are released into the public domain for the express intent of broad dissemination and use by third parties it will be, in all likelihood, impossible to effect a meaningful retraction at a later date.
Table VI . Potential risks of participation .
| Potential risks of participation in the PGP as described in the consent form (Abbreviated) |
| • The risks of public disclosure of your genetic and trait information could affect your employment, insurance and financial well-being and social interactions for you and your family. |
| • Anyone with sufficient knowledge and resources could take your DNA sequence data and/or posted trait information and use that data, with or without modification, to: (i) infer paternity or other features of your genealogy; (ii) claim statistical evidence that could affect your employment, insurance or ability to obtain financial services; (iii) claim relatedness to criminals or incriminate relatives; (iv) make synthetic DNA and plant it at a crime scene, or otherwise use it to falsely identify you; or (v) reveal the possibility of a disease or unknown propensity for a disease. |
| • Whether or not it is lawful to do so, you could be subject to actual or attempted employment, insurance, financial, or other forms of discrimination or negative treatment on the basis of the public disclosure of your genetic and trait information by the PGP or by a third party. |
| • The distribution of your cell lines could result in the creation and further distribution by a third party of additional cell lines, organs, or tissues containing your DNA for research, commercial, clinical, or other uses, including certain forms of assisted reproduction, some of which you may find objectionable or upsetting. |
| • If you have previously made available or intend to make available genetic information in a confidential setting, for example in another research study or in a clinical trial, the data that you provide as part of the PGP may be used, on its own or in combination with your previously shared data, to identify you as a participant in otherwise confidential genetic research or trials. |
The PGP's informed consent agreements and broader study protocol are developed in continuous close interaction with the Harvard Medical School Committee on Human Studies. The project is also overseen by an independent Data Safety Monitoring Board. Removing potentially disingenuous promises of anonymity, privacy, and confidentiality, while seeking to comprehensively and openly describe both known and unknown risks of participation, helps to ensure that research participants are as informed as possible about the nature of public genomics research and, simultaneously, safeguards the trustworthiness of scientists and of scientific research in general.
Return of research data to participants
Research volunteers have been traditionally treated as “objects” of study who have no intrinsic rights to the data generated by their participation.74 Today, we see that study participants are increasingly asking for access to their data75 and that available information and communication technologies have turned the return of research results into a feasible option. While some researchers adhere to the traditional viewpoint that research subjects should not or cannot receive identifiable research data, some have suggested legal and ethical grounds for finding that researchers possess the obligation to inform their participants of certain results, particularly when they are clinically actionable.76 However, defining the scenarios in which research results should be reported - and how to report such results - remains a challenging issue. The medical, financial, and psychosocial risks of disclosing variants of known and unknown clinical significance require that a careful distinction be made between those variants in which convincing clinical observational data exists and those in which disease association is less robust; a distinction that can influence both when and how to return results. Other concerns that have been voiced include the uncertainty surrounding regulations governing the return of genomics research results directly to participants, the impact of false-positive and/or false-negative results, as well as the “incidentalome,”77 and in the context of commercial direct-to-consumer testing, the concern that obtaining results could lead to a “raiding of the medical commons.”78
As new models of genomic research and commerce emerge, new mechanisms for communicating results to participants are also being explored. Many of these new models embrace a high level of involvement from their participants and, in return, may rely on some combination of education, informed consent, and intermediation to return data in a responsible fashion.79
The public genomics model adopted by the PGP utilizes the first two approaches while foregoing the third, opting to return data directly to research participants without the required intervention of an intermediary. The advantages of direct data return and participant communication are blunted by the partial shifting of the interpretative burden from the clinician to the researcher. The PGP has approached this issue by focusing on data disclosure via the Preliminary Research Report (PRR), which contains a noncomprehensive list of genetic variants present in the participant's DNA sequence data currently thought to have a likelihood of clinical relevance among individuals possessing such variants.
This preliminary identification of potentially significant variants is not intended to substitute in any way for professional medical advice, diagnosis or treatment. It leverages current knowledge by combining an evolving set of filtering algorithms and the use of existing variant databases - neither of which can be expected to have 100% accuracy in identifying truly pathogenic variants given the gaps in current scientific understanding. Participants are specifically instructed to confirm any potentially significant findings in consultation with their health care provider. It is possible that the increased rate of data return from public genomics research - as well as from commercial providers of personal genomic data - will help speed the creation of universal standards for clinical genomic interpretation that will help shift some of the interpretative burden back away from public genomics researchers.
Outlook: the PGP from 10 to 100 000
After publishing initial data from its first 10 participants in 2008, the PGP has continued to broaden the scope of the information it is collecting and publishing while simultaneously commencing the next stages of participant enrollment. From exome to whole-genome sequence data, the development and release of the GETEvidenceBase tool80 for generation of Preliminary Research Reports, and the publication of substantial scholarship based on the PGP data generated to date, the project's progress has been substantial. The PGP is now supported by PersonalGenomes.org, a 501(c)(3) non-profit charity that coordinates the international efforts of the PGP with other collaborative public genomics research projects around the world. Both the PGP and PersonalGenomes.org continue to strive to develop and disseminate genomic technologies, phenotyping strategies, and knowledge on a global scale and to produce tangible and widely available improvements in the understanding and management of human health in a responsible fashion.
Contributor Information
Jeantine E. Lunshof (Co-first author), European Centre for Public Health Genomics, FHML, Maastricht University, Maastricht, TheNetherlands; Department of Molecular Cell Physiology, VU University, Amsterdam, The Netherlands.
Jason Bobe (Co-first author), Department of Genetics, Harvard Medical School, Boston, Massachusetts, USA; PersonalGenomes.org, Boston, Massachusetts, USA.
John Aach, Department of Genetics, Harvard Medical School, Boston, Massachusetts, USA.
Misha Angrist, Institute for Genome Sciences & Policy, Duke University, Durham, North Carolina, USA.
Joseph V. Thakuria, PersonalGenomes.org, Boston, Massachusetts, USA; Harvard Medical School, Massachusetts General Hospital, Boston, Massachusetts, USA.
Daniel B. Vorhaus, PersonalGenomes.org, Boston, Massachusetts, USA; Robinson, Bradshaw & Hinson, P.A., Charlotte, North Carolina, USA.
Margret R. Hoehe (Co-last author), Department of Vertebrate Genomics, Max Planck Institute for Molecular Genetics, Berlin, Germany.
George M. Church (Co-last author), Department of Genetics, Harvard Medical School, Boston, Massachusetts, USA; PersonalGenomes.org, Boston, Massachusetts, USA.
REFERENCES
- 1.Personal Genome Project. http://www.personalgenomes.org. Accessed January 30, 2010 [Google Scholar]
- 2.Ingram VM. Gene mutations in human haemoglobin: the chemical difference between normal and sickle cell haemoglobin. Nature. 1957;180:326–328. doi: 10.1038/180326a0. [DOI] [PubMed] [Google Scholar]
- 3.Holley RW, Apgar J, Everett GA, et al. Structure of a ribonucleic acid. Science. 1965;147:1462–1465. doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- 4.Fiers W, Contreras R, Duerinck F, et al. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature. 1976;260:500–507. doi: 10.1038/260500a0. [DOI] [PubMed] [Google Scholar]
- 5.Sanger F, Air GM, Barrell BG, et al. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977;265:687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- 6.Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975;94:441–8. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- 7.Maxam AM, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanger F, Nicklen S, Cowlson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anonymous Editorial. Capitalizing on the genome. Nat Genet. 1996;13:1. doi: 10.1038/ng0596-1. [DOI] [PubMed] [Google Scholar]
- 10.Fleischmann RD, Adams MD, White O, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 11.C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 12.Adams MD, Celniker SE, Holt RA, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 13.Lander ES, Linton LM, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 14.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 15.Collins FS, Morgan M, Patrinos A. The Human Genome Project: lessons from large-scale biology. Science. 2003;300:286–290. doi: 10.1126/science.1084564. [DOI] [PubMed] [Google Scholar]
- 16.International HapmMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hindorff LA, Junkins HA, Mehta JP, Manolio TA, et al. A Catalog of Published Genome-Wide Association Studies. Accessed 30 January, 2010 Available at: http://www.genome.gov/gwastudies. [Google Scholar]
- 18.Shendure J, Mitra RD, Varma C, Church GM, et al. Advanced sequencing technologies: methods and goals. Nat Rev Genet. 2004;5:335–344. doi: 10.1038/nrg1325. [DOI] [PubMed] [Google Scholar]
- 19.Kahvejian A, Quackenbush J, Thompson JF. What would you do if you could sequence everything? Nat Biotechnol. 2008;26:1125–1133. doi: 10.1038/nbt1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.1000 Genomes Project. 2007 Meeting Report: A Workshop to Plan a Deep Catalog of Human Genetic Variation. Accessed January 30, 2010 Available at: http://www.1000genomes.org/docs/1000Genomes-MeetingReport.pdf. [Google Scholar]
- 21.Altshuler D, Daly MJ, Lander ES, et al. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siva N. 1000 Genomes project. Nat Biotechnol. 2008;26:256. doi: 10.1038/nbt0308-256b. [DOI] [PubMed] [Google Scholar]
- 23.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore GE. Cramming more components onto integrated circuits. Electronics. 1965;38:114–117. [Google Scholar]
- 25.Carr PA, Church GM. Genome engineering. Nat Biotechnol. 2009;27:1151–1162. doi: 10.1038/nbt.1590. [DOI] [PubMed] [Google Scholar]
- 26.National Human Genome Research Institute. 2004 Near-Term Technology Development for Genome Sequencing (RFA-HG-04-002. Available at: http://grants.nih.gov/grants/guide/rfa-files/RFA-HG-04002.html. Accessed 30 January, 2010. [Google Scholar]
- 27.National Human Genome Research Institute. 2004 Revolutionary Genome Sequencing Technologies -- the 1000 Genome (RFA-HG-04-003. Available at: http://grants.nih.gov/grants/guide/rfa-files/RFA-HG-04003.html. Accessed January 30, 2010. [Google Scholar]
- 28.X PRIZE Foundation. 2006 Archon X PRIZE for Genomics: X PRIZE Foundation Announces Largest Medical Prize in History. Available at: http://genomics.xprize.org/press-release/x-prize-foundation-announceslargest-medical-prize-in-history. Accessed January 30, 2010. [Google Scholar]
- 29.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 30.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 31.Ultra-Low Cost Sequencing Technology. Accessed February 5, 2010 Available at: http://arep.med.harvard.edu/Polonator/ [Google Scholar]
- 32.Shendure J, Porreca GJ, Reppas NB, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 33.Margulies M, Egholm M. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Applied Biosystems Inc 2010. Accessed January 22, 2010 ABI product literature. Available at https://docs.appliedbiosystems.com/pebiodocs/00113233.pdf. [Google Scholar]
- 35.Applied Biosystems Inc 2010. The ABI SoLID 3 System: Enabling the Next Generation of Science. Accessed January 30, 2010 Available at: http://www3.appliedbiosystems.com/cms/groups/mcb_marketing/documents/generaldocuments/cms_ 061241.pdf. [Google Scholar]
- 36.Bentley DR, Balasubramanian S, Swerdlow HP, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pleasance ED, Stephens PJ, O'Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris TD, Buzby PR, Babcock H. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 40.Eid J, Fehr A, Gray J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 41.Drmanac R, Sparks AB, Callow MJ, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 42.Church GM. The Personal Genome Project. Mol Systems Biol. 2005;1:2005.0030. doi: 10.1038/msb4100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freimer N, Sabatti C. The human phenome project. Nat Genet. 2003;34:15–21. doi: 10.1038/ng0503-15. [DOI] [PubMed] [Google Scholar]
- 44.Mahner M, Kary M. What exactly are genomes, genotypes and phenotypes? And what about phenomes? J Theor Biol. 1997;186:55–63. doi: 10.1006/jtbi.1996.0335. [DOI] [PubMed] [Google Scholar]
- 45.Anthony JC, Eaton WW, Henderson AS. Looking to the future in psychiatric epidemiology. Epidemiol Rev. 1995;17:240–242. doi: 10.1093/oxfordjournals.epirev.a036182. [DOI] [PubMed] [Google Scholar]
- 46.Lederberg J, McCray AT. 'Ome Sweet 'Omics - a genealogical treasury of words. Scientist. 2001;15:8. [Google Scholar]
- 47.Transcriptome. Accessed January 31, 2010 Available at: http://en.wikipedia.org/wiki/ Transcriptome. [Google Scholar]
- 48.Proteome. Accessed January 31, 2010 Available at: http://en.wikipedia.org/wiki/Proteome. [Google Scholar]
- 49.Ng PC, Levy S, Huang J, et al. Genetic variation in an individual human exome. PLoS Genet. 2008;4:e1000160. doi: 10.1371/journal.pgen.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kryukov GV, Shpunt A, Stamatoyannopoulos JA, Sunyaev SR. Power of deep, all-exon resequencing for discovery of human trait genes. Proc Natl Acad Sci U S A. 2009;106:3871–3876. doi: 10.1073/pnas.0812824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Angrist M. Eyes wide open: the personal genome project, citizen science and veracity in informed consent. Pers Med. 2009;6:691–699. doi: 10.2217/pme.09.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coriell Cell Repositories at the Coriell Institute for Medical Research. Accessed January 31, 2010 Available at: http://www.coriell.org/ [Google Scholar]
- 54.dbGaP: the database of Genotypes and Phenotypes. Accessed 31 January, 2010 Available at: http://www.ncbi.nlm.nih.gov/gap. [Google Scholar]
- 55.Merrill RM, Richardson JS. Validity of self-reported height, weight, and body mass index: findings from the National Health and Nutrition Examination Survey, 2001-2006. Prev Chronic Dis. 2009;6:A121. [PMC free article] [PubMed] [Google Scholar]
- 56.Porter SC, Manzi SF, Volpe D, Stack AM. Getting the data right: information accuracy in pediatric emergency medicine. Qual Saf Health Care. 2006;15:296–301. doi: 10.1136/qshc.2005.017442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bilder RM, Sabb FW, Cannon TD, et al. Phenomics: the systematic study of phenotypes on a genome-wide scale. Neuroscience. 2009;164:30–42. doi: 10.1016/j.neuroscience.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.GoogleHealth. Accessed January 31, 2010 Available at: https://www.google.com/health/ [Google Scholar]
- 59.MS Healthvault. Accessed January 31, 2010 Available at: http://www.healthvault.com/. [Google Scholar]
- 60.Indivo™ The Personally Controlled Health Record. Accessed January 31, 2010 Available at: http://indivohealth.org/ [Google Scholar]
- 61.Anthony JC, Eaton WW, Henderson AS. Looking to the future in psychiatric epidemiology. Epidemiol Rev. 1995;17:240–242. doi: 10.1093/oxfordjournals.epirev.a036182. [DOI] [PubMed] [Google Scholar]
- 62.Butte AJ, Kohane IS. Creation and implications of a phenome-genome network. Nat Biotechnol. 2006;24:55–62. doi: 10.1038/nbt1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turnbaugh PJ, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peterson J. The NIH Human Microbiome Project. Genome Res. December 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sommer MO, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lefranc MP, Giudicelli V, Ginestoux C, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Number of functional IG and TR genes per haploid genome. Accessed 5 February, 2010 Available at: http://www.imgt.org/textes/IMGTrepertoire/LocusGenes/tabgenes/ human/geneNumber.html#functional. [Google Scholar]
- 68.Weinstein JA, Jiang N, White RA 3rd, Fisher DS, Quake SR. Highthroughput sequencing of the zebrafish antibody repertoire. Science. 2009;324:807–810. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park IH, Arora N, Huo H, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee JH, Park IH, Gao Y, et al. A robust approach to identifying tissuespecific gene expression regulatory variants using personalized human induced pluripotent stem cells. PLoS Genet. 2009;5:e1000718. doi: 10.1371/journal.pgen.1000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Collins F S. The case for a US prospective cohort study of genes and environment. Nature. 2004;429:475–477. doi: 10.1038/nature02628. [DOI] [PubMed] [Google Scholar]
- 72.Conley JM, Doerr AK, Vorhaus DB. Enabling responsible public genomics. Health-Matrix: Journal of Law-Medicine. In press . [PubMed] [Google Scholar]
- 73.Lunshof JE, Chadwick R, Vorhaus DB, Church GM. From genetic privacy to open consent. Nat Rev Genet. 2008; 9:406–411. doi: 10.1038/nrg2360. [DOI] [PubMed] [Google Scholar]
- 74.Overview of PGP consent forms. Accessed January 31, 2010 Available at: http://www.personalgenomes.org/consent/ [Google Scholar]
- 75.Renegar G, Webster CJ, Stuerzebecher S, et al. Returning genetic research results to individuals: points-to-consider. Bioethics. 2006;20:24–36. doi: 10.1111/j.1467-8519.2006.00473.x. [DOI] [PubMed] [Google Scholar]
- 76.Murphy J, Scott J, Kaufman D, Geller G, LeRoy L, Hudson K. Public expectationsfor return of results from large-cohort genetic research. Am J Bioeth. 2008;8:36–43. doi: 10.1080/15265160802513093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wolf SM, Lawrenz FP, Kahn JP, et al. Managing. 0J Law Med Ethics. 2008;Summer 2008:2–31. [Google Scholar]
- 78.Kohane IS, Masys DR, Altman RB. The incidentalome: a threat to genomic medicine. JAMA. 2006 12;296:212–215. doi: 10.1001/jama.296.2.212. [DOI] [PubMed] [Google Scholar]
- 79.McGuire AL, Burke W. An unwelcome side effect of direct-toconsumer personal genome testing: raiding the medical commons. JAMA. 2008;300:2669–2671. doi: 10.1001/jama.2008.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kohane IS, Mandl KD, Taylor PL, Holm IA, Nigrin DJ, Kunkel LM. Reestablishing the researcher-patient compact. Science. 2007;316:836–837. doi: 10.1126/science.1135489. [DOI] [PubMed] [Google Scholar]
- 81.Trait-o-matic. Accessed February 4, 2010 Available at: http://snp.med.harvard.edu. [Google Scholar]