Abstract
Nodal signaling, mediated through SMAD transcription factors, is necessary for pluripotency maintenance and endoderm commitment. We identified a new motif, termed SMAD complex-associated (SCA), that is bound by SMAD2/3/4 and FOXH1 in human embryonic stem cells (hESCs) and derived endoderm. We demonstrate that two basic helix–loop–helix (bHLH) proteins—HEB and E2A—bind the SCA motif at regions overlapping SMAD2/3 and FOXH1. Furthermore, we show that HEB and E2A associate with SMAD2/3 and FOXH1, suggesting they form a complex at critical target regions. This association is biologically important, as E2A is critical for mesendoderm specification, gastrulation, and Nodal signal transduction in Xenopus tropicalis embryos. Taken together, E proteins are novel Nodal signaling cofactors that associate with SMAD2/3 and FOXH1 and are necessary for mesendoderm differentiation.
Keywords: Nodal, SMAD2/3, FOXH1, E2A, HEB, ChIP-seq
The commitment of embryonic cells toward the endodermal lineage involves a complex interplay of signaling networks. This transition is governed in large part by the Nodal signaling pathway, which is necessary for endoderm commitment in all vertebrates (Schier 2003). In mice and zebrafish, genetic mutations in Nodal ligands lead to dramatic defects in mesendoderm formation (Conlon et al. 1994; Varlet et al. 1997; Feldman et al. 1998, 2000). In the frog Xenopus laevis, inhibition of Nodal signaling by overexpression of antagonists results in loss of mesendoderm, thus inhibiting the morphogenetic movements of gastrulation (Hemmati-Brivanlou and Melton 1992; Osada and Wright 1999; Piccolo et al. 1999; Agius et al. 2000; Eimon and Harland 2002). Similar phenotypic effects occur when members of the Nodal signal transduction pathway, including SMAD2 and FOXH1, are ablated, demonstrating key roles for this branch of the Nodal pathway in endoderm commitment (Weinstein et al. 1998; Pogoda et al. 2000; Hoodless et al. 2001).
As Nodal signaling is essential for endoderm commitment, its fundamental mechanism of signal transduction has been well characterized. Two receptor-regulated SMADs (R-SMADs), SMAD2 and SMAD3, transduce Nodal signals by phosphorylation through the Activin receptor ActRIB (Hoodless et al. 1999). This activation results in heteromeric complex formation with another SMAD protein, SMAD4, which translocates to the nucleus, forming a larger complex with the winged helix protein FOXH1 (Labbe et al. 1998; Hoodless et al. 1999). Although all SMADs can bind directly to DNA, evidence favors a model in which SMADs cooperate with other DNA-binding proteins to form high-affinity, specific interactions (Attisano and Wrana 2000).
Despite the importance of Nodal signaling for development and disease, and a tremendous body of literature characterizing the interactions and nuclear dynamics of SMAD proteins, relatively few SMAD2/3 cofactors have been identified. Currently, only a handful of potential cofactors besides FOXH1 are known: Paired-like homeodomain transcription factors of the Mix family mediate Activin-induced transcription by interacting with the effector domain of Smad2 (Germain et al. 2000); the histone demethylase Jmjd3 has been shown to be recruited by Smad2/3 to the Nodal locus (Dahle et al. 2010); the pluripotency factor NANOG has been shown to interact directly with SMAD2/3 to maintain pluripotency in human embryonic stem cells (hESCs) (Xu et al. 2008; Vallier et al. 2009); and EOMES—a transcription factor essential for mesendoderm formation—has been reported to interact directly with SMAD2/3, having overlapping DNA-binding domains genome-wide (Teo et al. 2011). As the roles played by SMAD proteins are extensive, it is likely that this diversity of function is mediated by more complex protein associations than are currently appreciated. We hypothesize that genome-wide investigation of SMAD-binding sites might reveal previously unknown patterns or motifs that would identify candidate cofactors responsible for dynamic SMAD activity.
In this study, we use chromatin immunoprecipitation (ChIP) and high-throughput sequencing (ChIP-seq) technology to identify novel sequence motifs associated with SMAD complex binding in both hESCs and derived endoderm. This analysis reveals the canonical SMAD and FOXH1 motifs and a novel motif previously unassociated with Nodal signaling. Using biochemical and functional approaches, we show that the helix–loop–helix (HLH) proteins E2A and HEB cobind this motif with the SMAD/FOXH1 complex, and that these proteins interact. Importantly, we demonstrate that E2A is necessary for mesendoderm specification and gastrulation in Xenopus tropicalis embryos. Taking these data together, we propose a role for E2A and HEB as novel Nodal signaling components.
Results
A novel motif is enriched at SMAD/FOXH1-associated regions
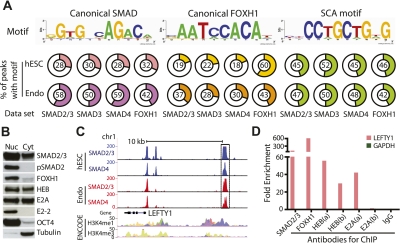
In a recent study, we used ChIP-seq to generate genome-wide occupancy maps for the Nodal signaling factors SMAD2/3, SMAD3, SMAD4, and FOXH1 in both hESCs and derived endoderm (Kim et al. 2011). Here, we sought to identify novel SMAD complex cofactors by performing de novo motif discovery on the SMAD/FOXH1 genomic targets (Supplemental Fig. 1A). We identified three nonrepetitive motifs that were consistently enriched in all data sets (SMAD2/3, SMAD3, SMAD4, and FOXH1) and in both cell types, hESCs and endoderm (Fig. 1A). The first and second motifs contain the canonical SMAD- and FOXH1-binding sites, respectively, confirming their genome-wide cooperativity in regulating Nodal signaling (Attisano et al. 2001) and further validating the antibodies used for ChIP. The third motif, CCTGCTG, has not previously been shown to associate with any of the SMAD/FOXH1 complex proteins (Fig. 1A). We hereafter refer to this element as the SCA (SMAD complex-associated) motif.
Figure 1.
Identification of the SMAD complex-associated (SCA) motif. (A) Sequence motifs and their frequency in each cell type (hESC or endoderm) and for each transcription factor. (B) Western blot analysis showing the expression of HEB, E2A, E2-2, SMAD2/3, pSMAD2, FOXH1, and OCT4 in the nuclear (Nuc) and cytoplasmic (Cyt) fraction of hESCs. Tubulin is a loading control as well as cytoplasmic maker. (C) LEFTY1 genomic locus showing SMAD2/3 and SMAD4 peaks from ChIP-seq databases in hESCs (hESC, blue) and endoderm (Endo, red). The dotted box indicates the region containing SCA motif. The scale bar on top shows upstream 10 kb from the LEFTY1 transcription start site (TSS). H3K4me1 and H3K4me3 peaks identified from ENCODE Project (hg 18). (D) HEB and E2A bind to the LEFTY1 enhancer (dotted box in Fig. 1C). Rabbit IgG was used instead of antibody for negative control ChIP, and GAPDH intronic region was used as a negative control primer. Two different antibodies (a and b) were used for both HEB and E2A.
We next determined the frequency of motif usage and the functional significance of these patterns in both hESCs and endoderm. In hESCs, the SCA motif is bound by all SMAD proteins at twice the frequency of the SMAD or FOXH1 canonical sequences. In endoderm, it is also used extensively at 42%∼50% of all target sequences (Fig. 1A). When we examined the genomic location of these motifs, we found that all three have a surprisingly similar pattern of distribution in hESCs and endoderm (Supplemental Fig. 1B). In hESCs, motif usage is associated with gene bodies including exons and introns. In endoderm, all motifs are used primarily in regions of intergenic spaces as well as gene bodies. To investigate whether the presence or absence of the SCA motif within a SMAD complex target is functionally significant, we first rigorously defined a SMAD target region as one where all four transcription factors (SMAD2/3, SMAD3, SMAD4, and FOXH1) were bound within a 1-kb window (Supplemental Fig. 2A). This analysis resulted in 813 SMAD/FOXH1 targets in hESCs and 1270 targets in endoderm, of which 556 (68%) and 776 (61%) contained the SCA motif, respectively. We next used GREAT to assess whether the SMAD complex is enriched near different functional classes of genes depending on the presence of the SCA motif. GREAT inputs a list of ChIP-seq peaks, associates these peaks with neighboring genes, and then runs functional ontologies, much like gene ontology (GO) (McLean et al. 2010). In hESCs, SMAD targets containing the SCA motif are enriched near genes involved in pattern formation and development, whereas targets without the motif are enriched near genes involved in regulation (Supplemental Fig. 2B). In endoderm, SMAD targets are enriched near genes required for endoderm formation regardless of whether the SCA motif was present, but targets without the motif were specifically enriched near genes involved in cell migration (Supplemental Fig. 2C). These predicted functional differences, taken together with the frequent presence of the SCA motif in SMAD/FOXH1 complex ChIP-seq data sets, led us to hypothesize that it is a sequence-specific binding site for an important Nodal cofactor.
To identify candidate factors that bind the SCA motif, we scanned the TRANSFAC database (version 11.3) for similar DNA-binding motifs that have been previously characterized. Five candidate transcription factors—E2A, E2-2, HEB, MyoD, and MASH1—were identified that bind to motifs similar to SCA. Based on microarray data generated from both hESCs and endoderm (Kim et al. 2011; Pan et al. 2011), we determined that, of these candidates, only E2A, E2-2, and HEB were expressed at high levels. Furthermore, we show that HEB, E2A, and E2-2 are expressed as proteins in both hESCs and endoderm. Fractionation of hESCs into nuclear and cytoplasmic components demonstrate that HEB and E2A are expressed in both compartments (Fig. 1B), suggesting nuclear activity. As these three proteins represent a distinct classification of basic HLH (bHLH) proteins and are expressed in hESCs and endoderm, we hypothesized that they may play key roles in SMAD2/3 targeting activities (Caudy et al. 1988; Murre et al. 1989; Henthorn et al. 1990; Hu et al. 1992).
HEB and E2A bind LEFTY1 SCA region
As HEB and E2A are present within the nucleus in both hESCs and endoderm and are predicted to associate with SCA motif, we tested whether HEB and E2A associate at this predicted motif in the LEFTY1 genomic locus. To this end, we first selected a SCA motif region bound strongly by SMAD2/3, SMAD4, SMAD3, and FOXH1, which lies 10 kb upstream of LEFTY1 (Fig. 1C, indicated as dotted box). This region also contains a consensus FOXH1 motif and binds both H3K4me1 and H3K4me3 in many other cell lines (ENCODE Project [hg 18], http://genome.ucsc.edu/ENCODE), indicating potential enhancer activity. We then performed ChIP-qPCR using antibodies against SMAD2/3, FOXH1, HEB, and E2A to pull down chromatin from hESCs and assessed binding to this region by amplifying the SCA enhancer near LEFTY1. As a negative control, we used a region from an intron of GAPDH not bound by any member of the SMAD complex. We used two different commercially available antibodies against HEB and E2A (“a” and “b”) to rule out antibody cross-reactivity. While the two antibodies against HEB (a and b) and a single antibody against E2A (a) were capable of ChIP, we were not able to perform ChIP using the E2A (b) antibody (Fig. 1D). Overall, we show that SMAD2/3, FOXH1, HEB, and E2A all strongly associate with the LEFTY1 enhancer region containing the SCA motif.
SMAD2/3, FOXH1, HEB, and E2A associate with the same SCA target regions
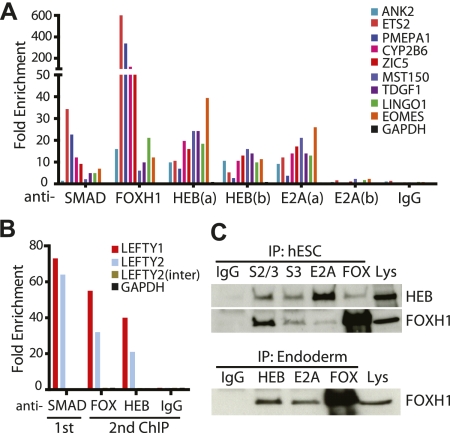
As SMAD2/3, FOXH1, HEB, and E2A associate with the identical SCA region surrounding the LEFTY1 promoter, we sought to evaluate whether this overlapping target association was more widespread. To this end, we performed ChIP-qPCR using anti-HEB and anti-E2A antibodies on nine additional SCA motif-associated target sites predicted to associate with SMAD proteins and/or FOXH1 (Fig. 2A). We observed both HEB and E2A enrichment at all interrogated SCA motif regions, except for at the negative control GAPDH intronic regions (which were not shown to bind SMADs, FOXH1, or the IgG control). Interestingly, the levels of enrichment of HEB and E2A at these regions is strikingly similar between the two HEB antibodies and the single E2A antibody.
Figure 2.
HEB and E2A interact with SMAD2/3 and FOXH1. (A) HEB and E2A bind to multiple SMAD2/3 (SMAD) and FOXH1 targets containing the SCA motif as shown by ChIP-qPCR. Rabbit IgG was used as a negative control for ChIP. GAPDH intronic region was used as a negative control for primers. (B) SMAD2/3 associates with HEB in same nucleosome using sequential ChIP. Anti-SMAD2/3 (SMAD) antibody was used to pull down hESC chromatin (1st). This was then used in a second ChIP (2nd) with anti-FOXH1 (FOX) as a positive control or anti-HEB antibody. Results were normalized against negative control GAPDH genomic region. LEFTY1 and LEFTY2 target regions were examined. LEFTY2 intergenic region (inter) was used as a negative control. (C) Co-IP of endogenous HEB or FOXH1 with SMAD2/3 (S2/3), SMAD3 (S3), E2A, and FOXH1 (FOX) in hESCs (top) or endoderm (bottom). Rabbit IgG was used for immunoprecipitation instead of antibody as a negative control.
After determining that HEB and E2A are bound to DNA regions that overlap with SMAD2/3, SMAD3, SMAD4, and FOXH1 targets, we next investigated whether SMAD2/3 and HEB associate with these regions together on the same or neighboring nucleosomes. Chromatin from hESCs was first immunoprecipitated with SMAD2/3 antibody, followed by a repeated ChIP step with either FOXH1, HEB, or nonspecific IgG antibody (Fig. 2B). We selected the strongest SMAD/FOXH1/HEB/E2A overlapping targets LEFTY1 and LEFTY2 for this sequential ChIP analysis. The intergenic region of LEFTY2 as well as GAPDH were used as negative controls. This successive ChIP analysis demonstrates that SMAD2/3, FOXH1, and HEB bind within either a single nucleosome or on adjacent nucleosomes as we sonicated DNA to generate 200- to 600-base-pair (bp) fragments (Fig. 2B).
HEB and E2A interact with SMAD2/3 and FOXH1 in hESCs and endoderm
As HEB and E2A associate with regions of DNA bound by the SMAD complex, we next asked whether these proteins interact with each other. To this end, we performed coimmunoprecipitation (co-IP) experiments. Using hESCs, we immunoprecipitated SMAD2/3, SMAD3, E2A, or FOXH1 and probed using Western blot analysis with either HEB or FOXH1 (Fig. 2C, top). In endoderm derived from hESCs, we immunoprecipitated HEB, E2A, or FOXH1 and examined whether FOXH1 was present (Fig. 2C, bottom). In both cell types, we observed an interaction between members of the SMAD/FOXH1 complex and E2A and HEB. In hESCs, HEB clearly associates with E2A, which is expected as these proteins form heterodimers. Intriguingly, HEB also pulls down SMAD2/3, SMAD3, and FOXH1. Furthermore, in both hESCs and endoderm, FOXH1 is capable of pulling down E2A and can pull down HEB as well as E2A in endoderm. To determine whether these protein associations are DNA-dependent, we performed co-IP in the presence of ethidium bromide (EtBr) to selectively inhibit any DNA-dependent protein associations in the precipitation reaction (Lai and Herr 1992). We found that the protein interactions between SMAD2/3 and FOXH1 and HEB or E2A to SMAD/FOXH1 complex were independent of DNA binding (Supplemental Fig. 3A). Overall, this strongly supports the hypothesis that E2A and HEB interact with the SMAD/FOXH1 complex and associate with regions known to be important for regulation of endoderm commitment.
HEB and E2A target binding, but not localization, is Nodal-dependent
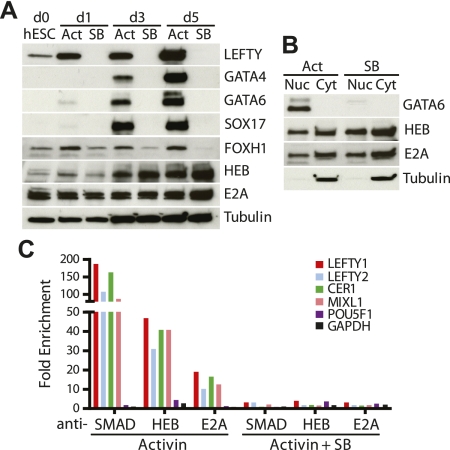
As HEB and E2A are novel members of the SMAD complex, we sought to determine whether their expression and localization is dependent on Nodal signaling. To this end, we differentiated hESCs into endoderm, but included the Nodal receptor inhibitor SB431542 throughout the differentiation time course. Activin treatment in the presence of SB431542 (SB) for 5 d completely blocked protein expression of LEFTY, GATA4, GATA6, and SOX17 (Fig. 3A). The expression of FOXH1 was gradually decreased over the course of SB431542 treatment. Conversely, the expression of HEB and E2A increased at day 5 in response to SB431542 treatment. To investigate whether this increase in protein expression was nuclear or cytoplasmic, we fractionated endoderm after 5 d in Activin or in Activin containing SB431542 and then performed Western blotting with GATA6, HEB, E2A, and Tubulin. GATA6, an endoderm marker, was detected only in the nucleus and was completely lost after SB431542 treatment (Fig. 3B). Tubulin was used as a cytoplasmic-specific marker. Interestingly, HEB and E2A were present in both the cytoplasm and the nucleus even after Nodal signaling was inhibited. The fact that HEB and E2A remain highly expressed in the nucleus after inhibition of SMAD signaling suggests that SMAD2/3 does not play an essential role in nuclear translocation of these proteins in response to Nodal signals. However, much like Jmjd3, which is present within the nucleus and recruited to Smad2/3 targets upon Nodal activation (Dahle et al. 2010), HEB and E2A may require SMAD2/3 for recruitment to specific Nodal signaling targets.
Figure 3.
HEB and E2A require Nodal for DNA association, but not localization. (A) Protein expression analysis during hESC differentiation into endoderm with Activin alone (Act) or with Activin plus the Nodal inhibitor SB431542 (SB). Western was blotted using antibodies against LEFTY, GATA4, GATA6, SOX17, FOXH1 HEB, and E2A. Tubulin was used for loading control. (B) Cellular localization of HEB and E2A with Activin alone (Act) or with Activin plus SB431542 (SB) at 5 d post-treatment. Cells were fractionated as nuclear (Nuc) and cytoplasmic (Cyt) and analyzed for the presence of GATA6, HEB, E2A, or Tubulin. (C) ChIP-qPCR using anti-SMAD2/3 (SMAD), anti-HEB, and anti-E2A antibodies on cells differentiated in Activin for 3 d and then treated cells for 48 h with Activin alone (Activin) or with Activin plus SB431542 (Activin + SB). Target regions include those of LEFTY1, LEFTY2, CER1, MIXL1, POU5F1, and GAPDH.
To determine whether HEB and E2A association with SMAD complex target regions was dependent on Nodal signaling, we exposed cells on the third day of differentiation to SB431542 and then examined HEB and E2A association with SMAD complex targets. To this end, we differentiated hESCs into endoderm by treating with high levels of Activin in low serum as previously described except that SB431542 was added at day 3 of differentiation, instead of day 0. After 48 h of treatment, we precipitated chromatin using anti-SMAD2/3, anti-HEB, and anti-E2A antibodies (Fig. 3C). We found that the association of SMAD2/3, HEB, and E2A to the Nodal DNA targets LEFTY1, LEFTY2, CER1, and MIXL1 was lost after the inhibition of Nodal signaling. Although the association of HEB and E2A to SMAD2/3 DNA targets during endoderm commitment is Nodal-dependent, we tested whether these proteins interact after the inhibition of Nodal signaling. We found that 48 h post-SB431542 treatment, FOXH1 still interacts with E2A, HEB, and SMAD2/3 (Supplemental Fig. 3B), even though the concentration of both FOXH1 and phospho-SMAD2 (pSMAD2) has greatly diminished (Supplemental Fig. 3C). This might suggest that association of the HEB/E2A and SMAD complex is independent of ongoing Nodal signaling. Overall, while HEB and E2A protein expression—nuclear or cytoplasmic—is Nodal-independent, their DNA binding—at least to SMAD/FOXH1 complex targets—is Nodal-dependent. This indicates that SMAD2/3 is not responsible for translocation of E2A or HEB into the nucleus, but may be involved in targeting these proteins to specific DNA sequences.
heb and e2a are expressed in gastrula stage X. tropicalis embryos
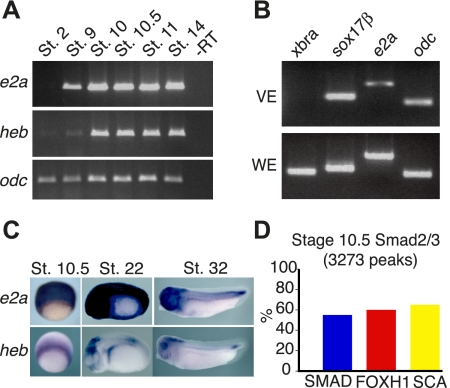
Having established that E2A and HEB can interact physically with the SMAD/FOXH1 complex and bind to the same DNA targets as SMAD2/3 and FOXH1 in hESC-derived endoderm, we next sought to determine whether E2A and HEB can function during embryogenesis in roles similar to that of other Nodal signaling components. Therefore, we turned to the frog X. tropicalis to investigate a potential role of E2A and HEB. We first asked whether heb or e2a transcripts are expressed during germ layer specification and gastrulation (Osada and Wright 1999; Agius et al. 2000; Faure et al. 2000). To this end, we investigated the expression of e2a and heb throughout gastrulation in the diploid frog X. tropicalis. In X. tropicalis embryos, e2a and heb are faintly detected in early cleavage stage embryos (maternal transcripts only, stages 2–4) but are robustly detected by RT–PCR from shortly after the onset of zygotic transcription at blastula stages through neurulation (Fig. 4A). We also investigated the expression of e2a in vegetal endoderm explants (VE), which express sox17β but not the mesodermal marker xbra (Fig. 4B). e2a is also detectable at low levels by RT–PCR in vegetal explants, suggesting it might play a role in this germ layer. To investigate the spatial expression pattern of e2a and heb, we performed in situ hybridization on gastrula stage and tailbud stage X. tropicalis embryos (Fig. 4C). e2a is strongly expressed throughout the ectoderm and mesoderm of gastrula stage (stage 10.5) embryos. Later in development, e2a is ubiquitously expressed in early tailbud stage embryos, becoming restricted to the head, somites, and lateral plate mesoderm by stage 32. Intriguingly, heb is expressed in the marginal zone (prospective mesoderm) at stage 10.5, and is found in the forebrain, lateral plate mesoderm, and posterior tailbud mesoderm at tailbud stages.
Figure 4.
e2a and heb are expressed during early development in X. tropicalis. (A) X. tropicalis embryos were analyzed by RT–PCR for expression of heb and e2a, from blastula stage through early neurula. Ornithine decarboxylase (odc) was used as a loading control. (B) Vegetal endoderm explants of stage 10.5 X. tropicalis express e2a at low levels, but not the mesoderm-specific gene xbra. (C) X. tropicalis embryos were analyzed by in situ hybridization for expression of e2a and heb at early gastrula stage (stage 10.5), at early tailbud stage (stage 22), and at late tailbud stage (32). (D) Bar graphs show the percentage of each motif present in Smad2/3 target regions during gastrulation in X. tropicalis as determined by ChIP-seq.
To examine whether E2A and HEB might be used biochemically to mediate Nodal signals during Xenopus gastrulation, we determined whether SMAD2/3 associates with the SCA motif in stage 10.5 X. tropicalis embryos. To this end, we performed ChIP with anti-SMAD2/3 antibody from stage 10.5 X. tropicalis embryos followed by high-throughput sequencing (ChIP-seq). This data is publically available in the Gene Expression Omnibus (GEO, accession no. GSE30146). Bioinformatic approaches revealed the same three top significant motifs as were identified in hESCs and derived endoderm: (1) the canonical SMAD motif, (2) the canonical FOXH1 motif, and (3) the SCA motif (Fig. 4D). Among the total 3273 peaks, the canonical SMAD and FOXH1 motif were associated in 1793 and 1948 peaks, respectively, whereas the SCA motif was associated with 2124 peaks. Overall, this strongly suggests that the E2A and HEB interaction with Nodal signaling is conserved during lineage commitment between humans and frogs.
Loss of E2A inhibits gastrulation in X. tropicalis
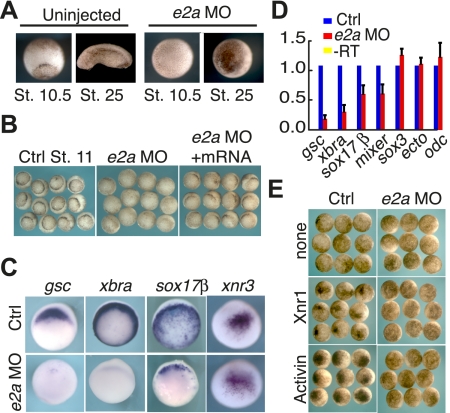
If E2A and HEB are conserved SMAD/FOXH1 cofactors, then we would expect that inhibition of E2A or HEB would affect mesendoderm commitment. To test whether E2A and/or HEB are involved in mesendoderm specification in vertebrates, we designed morpholino oligos (MOs) to inhibit translation of heb and e2a in X. tropicalis, and injected these alone or in combination into two-cell stage X. tropicalis embryos. Neither a translation blocking nor a splice-blocking heb morpholino caused noticeable morphological defects at stage 10.5 or 25 (Supplemental Fig. 4A) even when injected at high doses. However, microinjection of 20 ng of e2a MO into both blastomeres at the two-cell stage resulted in an inhibition of gastrulation (Fig. 5A). At stage 10.5, most e2a morphants showed either no evidence of bottle cells (57%, n = 215, sum of five experiments) or an isolated spot of bottle cells (38%), while a small percentage (5%) appeared morphologically normal at stage 10.5, but arrested before the blastopore became a full circle. e2a morphants fail to undergo the morphogenetic movements of gastrulation, and by stage 25, embryos appeared arrested, with no evident morphological differentiation. The e2a morpholino has a cell-autonomous effect, as embryos injected in only one blastomere at the four-cell stage have a localized inhibition of blastopore formation (Supplemental Fig. 4B).
Figure 5.
E2A is essential for gastrulation and gene expression in X. tropicalis. (A, right) X. tropicalis embryos were injected with a translation-blocking MO (e2a MO) directed against e2a, and assayed morphologically at stages 10.5 and 25. These were compared with uninjected controls (shown at left). (B) Blastopore lip formation in e2a MO-injected embryos can be restored by subsequent injection of mouse e2a mRNA. Using either in situ hybridization (C) or qRT–PCR (D), e2a MO-injected embryos were compared with controls for expression of molecular markers. qRT–PCR results represent at least four biological replicates. (E) e2a morphants and uninjected controls were injected in the animal pole at the four-cell stage with 10 pg of Activin mRNA or 40 pg of Xnr1 mRNA, and assayed for the presence of ectopic bottle cells. Representative embryos are shown at stage 11 in animal views.
In order to demonstrate that the e2a MO phenotype was specific, we rescued blastopore formation by overexpression of mouse e2a mRNA, which cannot be bound by the e2a MO (Fig. 5B). Embryos were first injected in both blastomeres at the two-cell stage with e2a MOs, then at the four-cell stage a subset of injected embryos were reinjected vegetally in the dorsal blastomeres with 500 pg of mouse e2a mRNA. Whereas most e2a morphants have either no blastopore (49% in this experiment, n = 71) or an isolated dorsal pinch of bottle cells (42%), rescue with mouse e2a mRNA led to a recovery of a semicircular blastopore in most embryos (60%, n = 47), while only 4% had no blastopore, and 32% had a localized pinch of bottle cells. We therefore consider that the e2a morphant phenotype is specific and can be rescued by overexpression of e2a mRNA.
Loss of E2A leads to reduced mesendoderm gene expression by inhibiting Nodal signal transduction
To better characterize the molecular basis of the e2a MO phenotype, we examined the expression of molecular markers in e2a morphants. We assessed expression of the organizer marker and Nodal target gsc, the pan-mesodermal gene xbra (T), and the endoderm markers sox17β and mixer using in situ hybridization and quantitative RT–PCR (qRT–PCR). Expression of gsc, xbra, sox17β, and mixer was significantly reduced in e2a morphants (Fig. 5C,D). Inhibition of gene expression was cell-autonomous in embryos injected in one blastomere at the four-cell stage, and was rescued by coinjection with mouse e2a mRNA (Supplemental Fig. 4B,C). Conversely, the expression of the β-catenin target gene xnr3 is unaffected at stage 9, as is expression of the ectoderm genes sox3 and ectodermin at stage 10.5, suggesting that the effects of the e2a MO are confined to Nodal-dependent cell types. The selective reduction of expression of xbra, gsc, and early endoderm genes suggests that mesoderm induction, organizer induction, and endoderm specification is impaired in e2a morphants, leading to failure of the gastrulation movements that depend on these processes.
To further investigate the relationship between the e2a MO and Nodal signaling, we performed an epistasis experiment to determine whether the presence of the e2a morpholino could inhibit ectopic Nodal signaling. To this end, we overexpressed the Nodal ligands Activin or Xnr1 in the prospective ectoderm with or without the e2a MO present. In the absence of the e2a MO, ectopic bottle cells were widely observed—95.8% of embryos expressing Activin and 93.1% of embryos expressing Xnr1 (Fig. 5E). Conversely, in the presence of the e2a MO, fewer embryos presented with ectopic bottle cells: 25% (n = 60) for Activin and 25.5% (n = 55) for Xnr1. We hypothesize that e2a normally forms an essential part of the Nodal signaling transcription factor complex, and that in the absence of e2a, the complex induces transcription from Nodal target genes with far less efficiency.
Discussion
This study presents strong genomic, biochemical, and functional evidence that E2A and HEB interact with SMAD2/3/4 and FOXH1 to regulate transcription of Nodal target genes. E2A and HEB associate with SMAD2/3 and FOXH1 at the SCA consensus site, which is functionally conserved between frogs and humans. The genomic identification of this site using the power of large sequence reads in multiple data sets provided inroads into testing the interaction of E2A, HEB, and the SMAD/FOXH1 complex. Using biochemical approaches, we show that these proteins interact in a DNA-independent manner, but then associate with similar target regions. Based on evidence presented in this study, we hypothesize that a complex consisting of E2A, HEB, SMAD2/3, and FOXH1 forms within the nucleus in response to Nodal, but that maintenance of this complex is independent of continual Nodal signaling. Overall, we suggest that E2A and HEB are key regulators of SMAD2/3-mediated transcriptional responses, and thus are fundamental Nodal cofactors that have not previously been implicated in this important developmental pathway.
While genomic and biochemical association is suggestive of a key signaling role, the phenotypic effect of knocking down e2a in X. tropicalis embryos is highly reminiscent of phenotypes resulting from perturbation of other key Nodal signaling factors, such as overexpression of a dominant-negative Nodal receptor or of the Nodal antagonists Cerberus-short and Lefty (Piccolo et al. 1999; Luxardi et al. 2010). Furthermore, we show epistatically that e2a knockdown inhibits the ability of both Activin and Xnr1 to induce bottle cell formation, strongly suggesting a key downstream role in the pathway. In the mouse, the roles of HEB and E2A and their family member, E2-2, have been extensively characterized as essential factors in hematopoiesis. The phenotypes of single-gene knockout models for E2A and HEB demonstrated that E2A was the primary E-protein member driving B-cell development, but that both E2A and HEB were required for proper T-cell development (Bain et al. 1994, 1997; Zhuang et al. 1994, 1996; Barndt et al. 1999). Interestingly, however, there is very strong evidence that these proteins are highly redundant due to their heterodimerizing abilities. Dominant-negative HEB, which can also disrupt E2A function through nonproductive heterodimer formation, causes a stronger phenotype than the heb-null mutation (Barndt et al. 2000). In B-cell development, HEB, driven by the E2A promoter, can rescue E2A loss of function (Zhuang et al. 1998). These complex genetics and the associated lethality of some compound mutants have made investigation of the roles of these proteins in early embryonic development difficult, and a role for E2A or HEB in early embryogenesis or SMAD/FOXH1 signaling has never been identified. Conditional genetic approaches to ablate several family members during gastrulation will more accurately address the role of E2A and HEB during mammalian germ layer formation. We note with interest that loss of e2a function in X. tropcialis achieves an effect on gastrulation not seen in the mouse. We hypothesize that the expansion of the Nodal pathway in frogs during evolution may have generated less redundancy between the E proteins, and we are currently testing this by evaluating compound MOs. Overall, further investigation of the mechanisms used by E2A and HEB to modulate Nodal signal transduction will elucidate new insights into how this important pathway is diversified to induce cell lineages within distinct species.
Materials and methods
Cell culture
H9 hESCs (WiCell) were cultured on mouse embryo fibroblast (MEF) feeder layers and transferred to Matrigel (BD Biosciences) for endoderm differentiation as described in the Supplemental Material. hESCs were induced to differentiate into endoderm using 100 ng/mL recombinant human Activin A (R&D Systems) and defined FBS (HyClone). Ten micromolar Alk4/5/7 inhibitor SB431542 (Tocris Bioscience) was added together with Activin during the differentiation to block the function of TGFβ pathway (Inman et al. 2002; Besser 2004; Xu et al. 2008).
ChIP
ChIP was carried out as described in the Supplemental Material. Immunoprecipitated DNA and whole-cell extract input DNA were purified and used as a template for qPCR using SYBR green (Bio-Rad). Fold enrichment was normalized against the GAPDH control region located in the intronic region of the GAPDH gene. Primary antibodies and primers are provided in the Supplemental Material.
Motif analysis and association of genes
Further details can be found in the Supplemental Material. Briefly, we used MEME for de novo motif discovery, STAMP to search for candidate genes that bind to the SCA motif, FIMO to scan genomic regions for the SCA motif, and GREAT for functional enrichment tests.
X. tropicalis culture, microinjections, and in situ hybridization
Embryos were generated by natural mating (for details, see the Supplemental Material). Translation-blocking (E2A, HEB) and splice-blocking (HEB) MOs (Gene Tools, LLC) were designed against X. tropicalis gene products, using the JGI genome browser and ESTs to identify transcription start sites, intron/exon boundaries, and potential polymorphisms. Unless otherwise noted, MOs were coinjected with 10 ng of fluoresceinated standard control MOs as a tracer for injection (for morpholino sequences, see the Supplemental Material). For in situ hybridization, embryos were developed to the desired stage and then fixed in MEMFA for 2–6 h at room temperature or overnight at 4°C. X. tropicalis multibasket in situ hybridization protocols were followed as described in Khokha et al. (2002). For details on plasmids used and RT–PCR primer sequences, see the Supplemental Material.
Acknowledgments
We thank Eduardo Gonzalez-Maldonado, Damek Spacek. and Nora Yucel for excellent technical assistance, and Yuqiong Pan for experimental advice. We thank Richard Harland for X. tropicalis EST clones used for in situ hybridization probe preparation. This work was funded by a grant from the California Institute of Regenerative Medicine to J.C.B and NIH NRSA fellowship F32DK089643-01 to A.E.W.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.16800511.
References
- Agius E, Oelgeschlager M, Wessely O, Kemp C, De Robertis EM 2000. Endodermal Nodal-related signals and mesoderm induction in Xenopus. Development 127: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attisano L, Wrana JL 2000. Smads as transcriptional co-modulators. Curr Opin Cell Biol 12: 235–243 [DOI] [PubMed] [Google Scholar]
- Attisano L, Silvestri C, Izzi L, Labbe E 2001. The transcriptional role of Smads and FAST (FoxH1) in TGFβ and activin signalling. Mol Cell Endocrinol 180: 3–11 [DOI] [PubMed] [Google Scholar]
- Bain G, Maandag EC, Izon DJ, Amsen D, Kruisbeek AM, Weintraub BC, Krop I, Schlissel MS, Feeney AJ, van Roon M, et al. 1994. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell 79: 885–892 [DOI] [PubMed] [Google Scholar]
- Bain G, Engel I, Robanus Maandag EC, te Riele HP, Voland JR, Sharp LL, Chun J, Huey B, Pinkel D, Murre C 1997. E2A deficiency leads to abnormalities in αβ T-cell development and to rapid development of T-cell lymphomas. Mol Cell Biol 17: 4782–4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barndt R, Dai MF, Zhuang Y 1999. A novel role for HEB downstream or parallel to the pre-TCR signaling pathway during αβ thymopoiesis. J Immunol 163: 3331–3343 [PubMed] [Google Scholar]
- Barndt RJ, Dai M, Zhuang Y 2000. Functions of E2A–HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol Cell Biol 20: 6677–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser D 2004. Expression of nodal, lefty-a, and lefty-B in undifferentiated human embryonic stem cells requires activation of Smad2/3. J Biol Chem 279: 45076–45084 [DOI] [PubMed] [Google Scholar]
- Caudy M, Vassin H, Brand M, Tuma R, Jan LY, Jan YN 1988. daughterless, a Drosophila gene essential for both neurogenesis and sex determination, has sequence similarities to myc and the achaete–scute complex. Cell 55: 1061–1067 [DOI] [PubMed] [Google Scholar]
- Conlon FL, Lyons KM, Takaesu N, Barth KS, Kispert A, Herrmann B, Robertson EJ 1994. A primary requirement for nodal in the formation and maintenance of the primitive streak in the mouse. Development 120: 1919–1928 [DOI] [PubMed] [Google Scholar]
- Dahle O, Kumar A, Kuehn MR 2010. Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Sci Signal 3: ra48 doi: 10.1126/scisignal.2000841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eimon PM, Harland RM 2002. Effects of heterodimerization and proteolytic processing on Derriere and Nodal activity: implications for mesoderm induction in Xenopus. Development 129: 3089–3103 [DOI] [PubMed] [Google Scholar]
- Faure S, Lee MA, Keller T, ten Dijke P, Whitman M 2000. Endogenous patterns of TGFβ superfamily signaling during early Xenopus development. Development 127: 2917–2931 [DOI] [PubMed] [Google Scholar]
- Feldman B, Gates MA, Egan ES, Dougan ST, Rennebeck G, Sirotkin HI, Schier AF, Talbot WS 1998. Zebrafish organizer development and germ-layer formation require nodal-related signals. Nature 395: 181–185 [DOI] [PubMed] [Google Scholar]
- Feldman B, Dougan ST, Schier AF, Talbot WS 2000. Nodal-related signals establish mesendodermal fate and trunk neural identity in zebrafish. Curr Biol 10: 531–534 [DOI] [PubMed] [Google Scholar]
- Germain S, Howell M, Esslemont GM, Hill CS 2000. Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev 14: 435–451 [PMC free article] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Melton DA 1992. A truncated activin receptor inhibits mesoderm induction and formation of axial structures in Xenopus embryos. Nature 359: 609–614 [DOI] [PubMed] [Google Scholar]
- Henthorn P, Kiledjian M, Kadesch T 1990. Two distinct transcription factors that bind the immunoglobulin enhancer microE5/κ 2 motif. Science 247: 467–470 [DOI] [PubMed] [Google Scholar]
- Hoodless PA, Tsukazaki T, Nishimatsu S, Attisano L, Wrana JL, Thomsen GH 1999. Dominant-negative Smad2 mutants inhibit activin/Vg1 signaling and disrupt axis formation in Xenopus. Dev Biol 207: 364–379 [DOI] [PubMed] [Google Scholar]
- Hoodless PA, Pye M, Chazaud C, Labbe E, Attisano L, Rossant J, Wrana JL 2001. FoxH1 (Fast) functions to specify the anterior primitive streak in the mouse. Genes Dev 15: 1257–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JS, Olson EN, Kingston RE 1992. HEB, a helix–loop–helix protein related to E2A and ITF2 that can modulate the DNA-binding ability of myogenic regulatory factors. Mol Cell Biol 12: 1031–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS 2002. SB-431542 is a potent and specific inhibitor of transforming growth factor-β superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol 62: 65–74 [DOI] [PubMed] [Google Scholar]
- Khokha MK, Chung C, Bustamante EL, Gaw LW, Trott KA, Yeh J, Lim N, Lin JC, Taverner N, Amaya E, et al. 2002. Techniques and probes for the study of Xenopus tropicalis development. Dev Dyn 225: 499–510 [DOI] [PubMed] [Google Scholar]
- Kim SW, Yoon SJ, Chuong E, Oyolu C, Wills A, Gupta R, Baker JC 2011. Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev Biol doi: 10.1016/j.ydbio.2011.06.009 [DOI] [PubMed] [Google Scholar]
- Labbe E, Silvestri C, Hoodless PA, Wrana JL, Attisano L 1998. Smad2 and Smad3 positively and negatively regulate TGFβ-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell 2: 109–120 [DOI] [PubMed] [Google Scholar]
- Lai JS, Herr W 1992. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci 89: 6958–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxardi G, Marchal L, Thome V, Kodjabachian L 2010. Distinct Xenopus Nodal ligands sequentially induce mesendoderm and control gastrulation movements in parallel to the Wnt/PCP pathway. Development 137: 417–426 [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56: 777–783 [DOI] [PubMed] [Google Scholar]
- Osada SI, Wright CV 1999. Xenopus nodal-related signaling is essential for mesendodermal patterning during early embryogenesis. Development 126: 3229–3240 [DOI] [PubMed] [Google Scholar]
- Pan Y, Ouyang Z, Wong WH, Baker JC 2011. A new FACS approach isolates hESC derived endoderm using transcription factors. PLoS ONE 6: e17536 doi: 10.1371/journal.pone.0017536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo S, Agius E, Leyns L, Bhattacharyya S, Grunz H, Bouwmeester T, De Robertis EM 1999. The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature 397: 707–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogoda HM, Solnica-Krezel L, Driever W, Meyer D 2000. The zebrafish forkhead transcription factor FoxH1/Fast1 is a modulator of nodal signaling required for organizer formation. Curr Biol 10: 1041–1049 [DOI] [PubMed] [Google Scholar]
- Schier AF 2003. Nodal signaling in vertebrate development. Annu Rev Cell Dev Biol 19: 589–621 [DOI] [PubMed] [Google Scholar]
- Teo AK, Arnold SJ, Trotter MW, Brown S, Ang LT, Chng Z, Robertson EJ, Dunn NR, Vallier L 2011. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev 25: 238–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L, Mendjan S, Brown S, Chng Z, Teo A, Smithers LE, Trotter MW, Cho CH, Martinez A, Rugg-Gunn P, et al. 2009. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development 136: 1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlet I, Collignon J, Robertson EJ 1997. nodal expression in the primitive endoderm is required for specification of the anterior axis during mouse gastrulation. Development 124: 1033–1044 [DOI] [PubMed] [Google Scholar]
- Weinstein M, Yang X, Li C, Xu X, Gotay J, Deng CX 1998. Failure of egg cylinder elongation and mesoderm induction in mouse embryos lacking the tumor suppressor smad2. Proc Natl Acad Sci 95: 9378–9383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RH, Sampsell-Barron TL, Gu F, Root S, Peck RM, Pan G, Yu J, Antosiewicz-Bourget J, Tian S, Stewart R, et al. 2008. NANOG is a direct target of TGFβ/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell 3: 196–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Soriano P, Weintraub H 1994. The helix–loop–helix gene E2A is required for B cell formation. Cell 79: 875–884 [DOI] [PubMed] [Google Scholar]
- Zhuang Y, Cheng P, Weintraub H 1996. B-lymphocyte development is regulated by the combined dosage of three basic helix–loop–helix genes, E2A, E2-2, and HEB. Mol Cell Biol 16: 2898–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Barndt RJ, Pan L, Kelley R, Dai M 1998. Functional replacement of the mouse E2A gene with a human HEB cDNA. Mol Cell Biol 18: 3340–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]