Abstract
Little is known about how particle-specific proteins are assembled on spliceosomal small nuclear ribonucleoproteins (snRNPs). Brr2p is a U5 snRNP-specific RNA helicase required for spliceosome catalytic activation and disassembly. In yeast, the Aar2 protein is part of a cytoplasmic precursor U5 snRNP that lacks Brr2p and is replaced by Brr2p in the nucleus. Here we show that Aar2p and Brr2p bind to different domains in the C-terminal region of Prp8p; Aar2p interacts with the RNaseH domain, whereas Brr2p interacts with the Jab1/MPN domain. These domains are connected by a long, flexible linker, but the Aar2p–RNaseH complex sequesters the Jab1/MPN domain, thereby preventing binding by Brr2p. Aar2p is phosphorylated in vivo, and a phospho-mimetic S253E mutation in Aar2p leads to disruption of the Aar2p–Prp8p complex in favor of the Brr2p–Prp8p complex. We propose a model in which Aar2p acts as a phosphorylation-controlled U5 snRNP assembly factor that regulates the incorporation of the particle-specific Brr2p. The purpose of this regulation may be to safeguard against nonspecific RNA binding to Prp8p and/or premature activation of Brr2p activity.
Keywords: pre-mRNA splicing, protein interaction, protein phosphorylation, protein structure, spliceosome, yeast
Small ribonucleoproteins (RNPs) are major components of several RNA processing machineries in eukaryotic cells, including spliceosomes, which catalyze the removal of noncoding intervening sequences (introns) from precursor messenger RNAs (pre-mRNAs) and the ligation of the neighboring coding regions (exons) to generate mature mRNA. Canonical small nuclear RNPs (snRNPs), such as the U1, U2, U4, and U5 snRNPs of the major spliceosome, contain a set of seven common Sm proteins bound at a uridine-rich Sm site in the snRNAs, forming the Sm core RNPs (Pomeranz Krummel et al. 2009; Weber et al. 2010). In metazoans, Sm core RNPs are assembled via an elaborate pathway involving nucleo–cytoplasmic shuttling and two multiprotein machineries, the Prmt5 and the SMN complexes (for review, see Kolb et al. 2007; Chari et al. 2009). In addition, each snRNP contains a variable number of particle-specific proteins. The final stages of metazoan snRNP biogenesis are thought to take place in nuclear Cajal bodies, at least in the case of the U2 snRNP (Nesic et al. 2004). However, little is known about how the specific proteins are assembled.
Spliceosomes assemble de novo on the substrate pre-mRNAs by stepwise recruitment of the snRNPs and many additional splicing factors that are not stably associated with snRNPs (for review, see Wahl et al. 2009). During the cycle of assembly, activation, catalysis, and disassembly, the spliceosome is repeatedly remodeled with the help of eight conserved RNA-dependent ATPases/RNA helicases and one G-protein (for review, see Staley and Guthrie 1998). Each remodeling step is associated with changes in the macromolecular composition and in the protein–protein, protein–RNA, and RNA–RNA interaction networks of the spliceosome (Wahl et al. 2009). During the splicing cycle, several snRNPs are also profoundly reorganized. For example, the human U5 snRNP enters the spliceosome as a 20S particle that is part of the U4/U6–U5 tri-snRNP, in which the U4 and U6 snRNAs are extensively base-paired. During spliceosome activation, the U4 and U6 snRNAs are separated and all U4/U6-specific proteins are removed to make U6 snRNA available as part of the active site(s) (Staley and Guthrie 1998; Wahl et al. 2009). Upon catalytic activation, several components of the multiprotein Prp19 complex (the nineteen complex, NTC, in yeast) become stably associated with U5 snRNP. As a consequence, U5 snRNP is released as a 35S particle from the post-splicing complex, from which the 20S form has to be recycled via an unknown pathway (Makarov et al. 2002). Thus, snRNP assembly, restructuring, and reassembly are intimately tied to the splicing process itself.
Two distinct forms of U5 snRNP have been characterized in yeast, distinguished by the presence or absence of the Aar2 protein (Gottschalk et al. 2001; Boon et al. 2007). Aar2p was discovered as a factor involved in the splicing of some pre-mRNAs in yeast (Nakazawa et al. 1991). While Aar2p is not required for splicing per se, removal of the protein blocked repeated rounds of splicing in vitro (Gottschalk et al. 2001), suggesting that it could be involved in U5 snRNP or U4/U6–U5 tri-snRNP (re)assembly. More recently, it was shown that the Aar2p-U5 snRNP has a cytoplasmic phase (Boon et al. 2007). In addition to Aar2p and the Sm proteins, it contains the U5-specific Prp8 and Snu114 proteins, but it lacks the essential Brr2 helicase (Gottschalk et al. 2001; Boon et al. 2007) that is required for spliceosome activation (Staley and Guthrie 1998) and disassembly (Small et al. 2006). In the nucleus, Aar2p is replaced by Brr2p, which, along with the remaining U5-specific proteins, gives rise to mature U5 snRNP (Boon et al. 2007).
Here, we investigated the structural basis of Aar2p's association with Prp8p and the mechanism by which it regulates U5 snRNP biogenesis in Saccharomyces cerevisiae. We found that Aar2p forms a stable complex with the RNaseH-like domain of Prp8p, and the C terminus of Aar2p sequesters the C-terminal Jab1/MPN domain of Prp8p, which is a major interaction site of Brr2p. Thus, Aar2p directly competes with Brr2p for binding to Prp8p. Aar2p is found to be phosphorylated in vivo, including residue S253. Introduction of a phospho-mimetic mutation in Aar2p at position 253 disrupts the Aar2p–Prp8p interaction and allows Brr2p entry. Our data suggest that Aar2p acts as a phosphorylation-controlled U5 snRNP assembly factor that regulates the interaction of Brr2p, possibly to avoid nonspecific RNA binding by Prp8p and/or premature activation of Brr2p activity.
Results
Aar2p stably interacts with the RNaseH-like domain in the C-terminal region of Prp8p
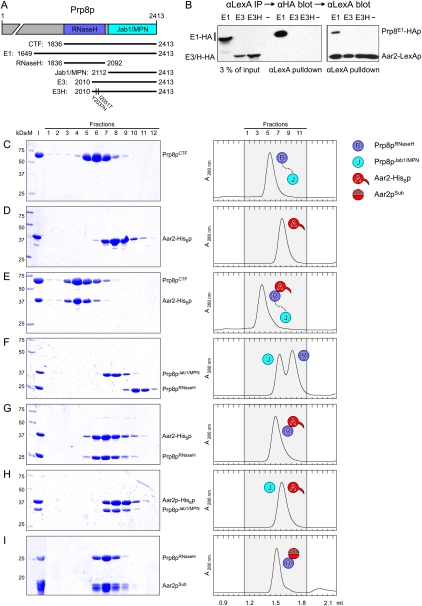
Previous work suggested that Aar2p interacts with a C-terminal region of Prp8p (Boon et al. 2006, 2007), although a direct contact between the proteins was not demonstrated. We tested interaction of full-length Aar2p fused to the LexA DNA-binding domain (in pBTM116) with HA-tagged C-terminal Prp8p fragments fused to the Gal4 activation domain (in pACTII). Prp8pE1 (residues 1649–2413) includes the RNaseH and Jab1/MPN domains, Prp8pE3 (residues 2010–2413) includes the Jab1/MPN domain but lacks most of the RNaseH domain, and Prp8pE3H (residues 2010–2413) carries two point mutations (Y2037H, I2051T) that increase its interaction with Brr2p (Fig. 1A; van Nues and Beggs 2001). As the Aar2-LexA fusion protein alone activated transcription in yeast two-hybrid assays, we tested interactions among the proteins using pull-down assays instead. Aar2-LexAp coprecipitated the Prp8E1 fusion protein but not Prp8pE3 or Prp8pE3H (Fig. 1B), indicating that Aar2p binds the C-terminal region of Prp8p and that this interaction requires sequence in the E1 fragment between residues 1649 and 2010.
Figure 1.
Aar2p–Prp8p interaction. (A) A schematic representation of the Prp8p constructs used in this work. For simplicity, only the RNaseH and Jab1/MPN domains are shown. (B) Aar2-LexAp interacts with Prp8-HAp fragment E1, but not with E3 or E3H. (Left panel) Yeast extracts containing Aar2-LexAp and HA-tagged Prp8pE1, Prp8pE3, or Prp8pE3H, or a control with no Prp8 fusion protein (−) were precipitated with anti-LexA antibodies followed by anti-HA Western blot. (Right panel) Subsequent probing with anti-LexA shows that Aar2-LexAp was immunoprecipitated similarly in each case. (C–I) Gel filtration analysis of the indicated proteins or mixtures. (Left) SDS PAGE analysis of eluted fractions (indicated at the top). The first two lanes in each panel show molecular mass standard (M) and the input (I). Numbers on the left indicate the molecular mass of standard proteins in kilodaltons. (Right) Elution profiles. Elution volumes are given at the bottom. Fractions analyzed by SDS PAGE (shaded area) are indicated at the top. (Aar2pSub) Subtilisin-treated Aar2p. Icons denote proteins and complexes as defined.
We next used analytical gel filtration analysis to assess the interaction between Aar2-His6p and fragments of Prp8p, both recombinantly produced in Escherichia coli. Upon mixing individually purified proteins, Aar2-His6p coeluted with Prp8p1836-2413 (covering the RNaseH and Jab1/MPN domains; herein referred to as Prp8pCTF) at the expected size of a 1:1 complex (Fig. 1C–E). The two proteins could also be coproduced and copurified via three chromatographic steps (Supplemental Fig. S1A), confirming a stable and direct interaction of Aar2-His6p and Prp8pCTF. Similarly, Aar2-His6p stably bound to Prp8p1836–2092 (Prp8pRNaseH). While Aar2-His6p also comigrated with Prp8p2112–2413 (Prp8pJab1/MPN), the elution volume was the same as for the individual proteins, showing that Aar2-His6p does not stably interact with Prp8pJab1/MPN (Fig. 1F–H). The latter finding was confirmed in a pull-down assay (Supplemental Fig. S1B). Thus, Aar2p binds directly to the C-terminal region of Prp8p, and the RNaseH domain of Prp8p is the primary interaction module for Aar2p.
Crystal structure analysis of Prp8pCTF*, Aar2p, and an Aar2p–Prp8pRNaseH complex
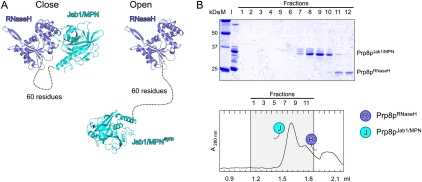
To reveal the molecular basis of the Aar2p–Prp8p interaction, we attempted to crystallize the Aar2-His6p–Prp8pCTF complex but failed. However, we could elucidate the crystal structure of Prp81836–2397 (called Prp8pCTF*) alone, using all data collected up to 3.3 Å resolution in the refinement (Fig. 2A; Supplemental Table S1; Supplemental Fig. S2A). Prp8pCTF* lacked 16 amino acids at the C terminus compared with Prp8pCTF. In that structure, a 60-residue linker between the domains lacked electron density, demonstrating its intrinsic flexibility (Fig. 2A). In the crystal structure, the unstructured linker could bridge various pairs of crystallographically equivalent domains (e.g., a closely interacting pair compared with a pair of domains in an open conformation) (Fig. 2A) and we could not unequivocally attribute a particular pair of domains to the Prp8pCTF* molecule in our crystals. However, individually produced RNaseH and Jab1/MPN domains ran as separate entities in gel filtration (Fig. 1C,F). In addition, protease cleavage in the linker of Prp8pCTF dissected RNaseH and Jab1/MPN domains that again migrated as individual proteins (Fig. 2B). These data suggest that in solution, Prp8pCTF adopts an open conformation in which the two domains are disconnected.
Figure 2.
Structure of Prp8pCTF* in isolation. (A) Two possible (close and open) configurations of Prp8pCTF*. (Blue) Prp8pRNaseH; (cyan) Prp8pJab1/MPN; (dashed line) flexible linker. Two crystallographically related Jab1/MPN domains are shown with respect to the same RNaseH domain. Other, primarily open, configurations in which the distances between the domains could be bridged by the linker can be found with other symmetry-equivalent Jab1/MPN domains. (B) Gel filtration analysis of Prp8pCTF* after treatment with trypsin. Details and labeling are as in Figure 1, C–I. After treatment with trypsin, the separated domains elute as isolated molecules, indicating that they do not stably interact in solution (cf. Fig. 1F).
As the Aar2-His6p–Prp8pRNaseH complex also failed to crystallize, we suspected additional flexible elements in Aar2-His6p and subjected the protein to limited proteolysis. Subtilisin cleaved Aar2-His6p into two fragments of ∼18 kDa each, comprising residues 1–159 and 168–324 (Supplemental Fig. S1C). Thus, subtilisin removed an internal loop (residues 160–167) and the C-terminal 31 amino acids plus the tag of the protein. Gel filtration analysis indicated that the two fragments of subtilisin-treated Aar2p (Aar2pSub) remained stably associated. Like full-length Aar2-His6p, Aar2pSub bound stably to Prp8pRNaseH (Fig. 1I) but failed to interact with Prp8pJab1/MPN (data not shown).
Aar2pSub crystallized alone and in complex with Prp8pRNaseH. We first solved the structure of the complex by molecular replacement using the structure coordinates of isolated Prp8pRNaseH (Pena et al. 2008; PDB IDs: 3E9O and 3E9P). The Aar2pSub structure was subsequently solved using the coordinates of the Aar2pSub portion of the complex structure. Both structures were refined to low R/Rfree factors with good stereochemistry, using all diffraction data collected to 2.1 Å (Aar2pSub) and 1.8 Å (Aar2pSub–Prp8pRNaseH complex) resolution (Supplemental Table S1; Supplemental Fig. S2B,C).
Prp8pRNaseH grasps Aar2pSub between its thumb and fingers
Aar2pSub is organized in two globular domains, connected via a flexible loop between residues 153 and 171, part of which was cleaved during subtilisin treatment (Figs. 3A; Supplemental Figs. S2D, S3). The two domains interact closely via an extensive hydrophobic interface (53% nonpolar atoms) that buries 1504 Å2 of combined surface area (Supplemental Fig. S2E), explaining why they remained stably associated after subtilisin treatment. The N-terminal domain (NTD) contains a 10-stranded, mixed β sandwich with three helices surrounding the upper rim. The C-terminal domain (CTD) shows an all-helical fold with nine α helices, a π helix, and three 310 (η) helices. Residues 319–324 were not seen in the electron density of either Aar2pSub alone or in the complex with Prp8pRNaseH.
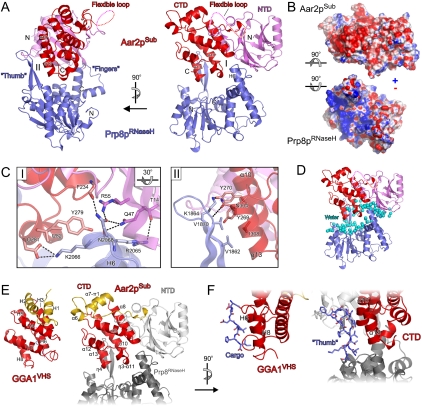
Figure 3.
Structure of the Aar2pSub–Prp8pRNaseH complex. (A) Orthogonal views on the Aar2pSub–Prp8pRNaseH complex structure. (Violet) Aar2pSub NTD; (red) CTD; (dashed line) flexible linker between the domains; (blue) Prp8pRNaseH. Protein termini, selected secondary structure elements, and the Prp8pRNaseH thumb and finger elements are labeled. Roman numerals indicate the two discontinuous interfaces formed upon complex formation. (B) Book view onto the interacting surfaces of Aar2pSub and Prp8pRNaseH. Rotations of the domains relative to the right view in A are indicated by symbols. Surfaces are colored according to electrostatic potential. (Blue) Positive charge; (red) negative charge. (C) Close-up views on interfaces I (left; rotated 30° about the horizontal axis as indicated compared with the right panel of A) and II (right; same view as in the left panel of A). Selected interacting residues are shown as sticks, colored by atom type, and labeled: carbon (as for the respective protein), nitrogen (blue), and oxygen (red). Dashed lines indicate hydrogen bonds or salt bridges. (D) Water molecules (cyan spheres) blanketing the surfaces bordering the central gap in the Aar2pSub–Prp8pRNaseH complex. (E) Side-by-side view of the human GGA1 VHS domain (left) (Shiba et al. 2002; PDB ID: 1JWG) and the Aar2pSub CTD (right). The N-terminal two helices (H1 and H2 in GGA1VHS; α6 and α7-π1 in Aar2pSub) are shown in gold and are positioned differently relative to the remainder of the domains (red). The NTDs of Aar2pSub and Prp8pRNaseH are shown in light and dark gray, respectively. Proteins and selected secondary structure elements are labeled. (F) Comparison of cargo binding by the GGA1 VHS domain (left) with the interaction of the thumb region of Prp8pRNaseH with the Aar2pSub CTD (right). Cargo peptide and the thumb, respectively, are bound in an elongated conformation between two helices (H6 and H8 in GGA1VHS; α10 and α13 in Aar2pSub CTD). Ligand peptide and the Prp8pRNaseH thumb are shown as sticks and colored by atom type as in C. Proteins and selected structural elements are labeled.
As previously seen in the isolated protein (Pena et al. 2008; Ritchie et al. 2008; Yang et al. 2008), the Prp8p portion in the complex adopts a central mixed α/β fold reminiscent of RNaseH-like enzymes, with a long β-hairpin insertion (residues 1859–1875) and an additional C-terminal α-helical domain (Fig. 3A). Globally, Prp8pRNaseH resembles a mitten with the RNaseH-like core representing the palm, the β-hairpin insertion representing the thumb, and the helical appendage corresponding to the fingers. The fingers and thumb contact Aar2pSub through two discontinuous interfaces (I and II) with a large gap above the palm region in between (Fig. 3A). At interface I, the tip of the Prp8pRNaseH fingers (formed by the C terminus of helix H6 of Prp8pRNaseH) interacts with both Aar2pSub domains primarily via hydrogen bonds and ionic interactions (Fig. 3C, left). At interface II, the thumb of Prp8pRNaseH latches onto one side of the Aar2pSub CTD, running along a cleft between helices α10 and α13 with polar interactions dominating the upper part and hydrophobic interactions prevailing in the lower part of the contact (Fig. 3C, right).
Of the combined surface area, 772 Å2 is buried upon complex formation. An extended electronegative surface patch on Aar2pSub at interface II and across the gap faces a similarly extensive electropositive surface area on the thumb and palm of Prp8pRNaseH (Fig. 3B). The association of Aar2pSub with Prp8pRNaseH has hallmarks of a facultative interaction, since large parts of the interfaces are hydrophilic (30% nonpolar atoms) and thus compatible with exposure to the aqueous environment. Indeed, the surface areas of the proteins facing each other across the gap are blanketed with water molecules (Fig. 3D).
The structure of Aar2pSub in isolation is very similar to the Prp8pRNaseH-bound structure (root-mean-square deviation [RMSD] of 0.92 Å for 289 Cα atoms) (Supplemental Fig. S2F, left), with significant adjustments only in the loop between strands β5 and β6 (NTD) and in the neighborhood of helices η3 and α11 (CTD). While the β5–β6 loop undergoes an approximate rigid body movement, the region between helices η3 and α11 is structurally rearranged upon complex formation (Supplemental Fig. S2G). Similarly, there are only limited changes in the Prp8pRNaseH structure upon interaction with Aar2pSub, which entail small rigid body movements of the thumb and the tip of the fingers toward each other to grasp Aar2pSub in between (Supplemental Fig. S2F, right).
Part of the Aar2pSub–Prp8pRNaseH interaction resembles cargo binding by vesicular transport adaptors
Comparison of the Aar2pSub structure to known structures in the Protein Data Bank (http://www.pdb.org) revealed the NTD as a novel fold. Although it resembles diverse proteins with similar β sandwiches, no other protein has analogous interspersed helices. The CTD of Aar2pSub is most similar to the domain of Pcf11p that binds Ser2-phosphorylated heptad repeats of the C-terminal tail of RNA polymerase II (Z-score = 7.4, RMSD 3.6 Å for 112 Cα atoms) and to the related families of VHS and ENTH domains known from Golgi-ER transport adaptors and endocytic adaptor proteins, respectively (Z-score = 7.0 to the VHS domains of human GGA1 and GGA3; RMSD 3.7 Å for 111 Cα atoms; Fig. 3E). The helical stack of the CTD also resembles armadillo repeat proteins such as β-catenin or importin-β, posing the question of whether Aar2p may act as a transport adaptor for pre-U5 snRNP during nucleo–cytoplasmic shuttling. However, Prp8p has its own nuclear localization signal (Boon et al. 2007).
VHS domain proteins bind acidic cluster–dileucine motifs of their cargo proteins between two helices in an extended conformation (Fig. 3F; Misra et al. 2002; Shiba et al. 2002). This mode of cargo binding resembles the binding of the thumb region of Prp8pRNaseH to the CTD of Aar2pSub, although the atomic contacts differ in detail; the Aar2pSub–Prp8pRNaseH interaction displays a different arrangement of polar and hydrophobic interactions, and amino acids from both strands of the Prp8pRNaseH β hairpin bind to Aar2pSub. In contrast, Pcf11p binds Ser2-phosphorylated RNA polymerase II heptad repeats on the opposite side of its VHS-like domain (Meinhart and Cramer 2004).
Aar2p directly competes with Brr2p binding at the Prp8p C-terminal region by sequestering the Jab1/MPN domain
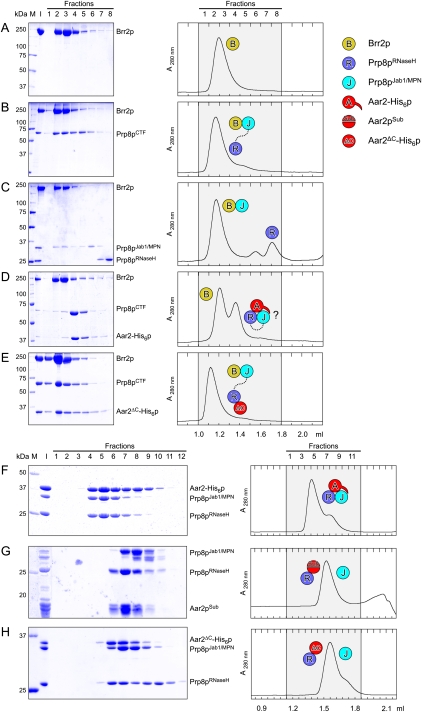
A number of studies have shown that Brr2p also interacts with the C-terminal region of Prp8p (van Nues and Beggs 2001; Liu et al. 2006; Pena et al. 2007), and it was reported that more Aar2p associates with Prp8p in cells producing less Brr2p (Boon et al. 2007). We therefore investigated whether Aar2p and Brr2p bind competitively to Prp8p. Gel filtration analysis revealed that Brr2p formed a stable complex with Prp8pCTF (Fig. 4A,B). However, unlike Aar2p, Brr2p interacted stably with Prp8pJab1/MPN but not with Prp8pRNaseH (Fig. 4C).
Figure 4.
Brr2p–Prp8p interaction and modulation by Aar2p. (A–E) Gel filtration analysis of Brr2p–Prp8p interactions. Details and labeling are as in Figure 1, C–I. Brr2p (A) comigrates with Prp8pCTF (B) and Prp8pJab1/MPN (C) but not Prp8pRNaseH (C). (C) No ternary complex Brr2p–Prp8pRNaseH–Prp8pJab1/MPN is formed. (D) Primarily free Brr2p and the binary Aar2-His6p–Prp8pCTF complex form upon mixing Brr2p, Aar2-His6p, and Prp8pCTF; therefore, the Prp8pJab1/MPN domain may be unavailable to Brr2p (indicated by question mark [?]). (E) Since the Aar2ΔC-His6p still binds the Prp8p RNaseH domain but does not sequester the Jab1/MPN domain (see H), the latter remains available for binding Brr2p, allowing formation of a ternary complex. (F–H) Gel filtration analysis of Aar2p–Prp8p interactions. Details and labeling are as in Figure 1, C–I. (F) Aar2-His6p sequesters Prp8pJab1/MPN after formation of an Aar2-His6p–Prp8pRNaseH complex. Aar2pSub–Prp8pRNaseH (G) or Aar2ΔC-His6p–Prp8pRNaseH (H) complexes are not able to sequester the Prp8pJab1/MPN domain, showing that Prp8pRNaseH and the C-terminal tail of Aar2p are required to bind Prp8pJab1/MPN.
Despite the preferred open structure of isolated Prp8pCTF, which might permit concomitant binding of both Aar2p and Brr2p, we could not assemble a ternary Aar2-His6p–Prp8pCTF–Brr2p complex. Mixing the three proteins stoichiometrically produced primarily a binary Aar2-His6p–Prp8pCTF complex and free Brr2p (Fig. 4D). Increasing amounts of Brr2p led to formation of a mixture of binary Brr2p–Prp8pCTF and Aar2-His6p–Prp8pCTF complexes but were not able to displace Aar2-His6p quantitatively (Supplemental Fig. S4).
These findings could be explained if a preformed Aar2-His6p–Prp8pRNaseH complex sequestered the Jab1/MPN domain, making it unavailable for Brr2p. This idea is consistent with the observation that a transposon insertion in the C-terminal Jab1/MPN domain of Prp8p (at residue 2173) interfered with Aar2p binding to U5 snRNP (Boon et al. 2006), suggesting an involvement of the Jab1/MPN domain in Aar2p binding to Prp8p. Indeed, we observed that addition of Prp8pJab1/MPN to a preformed Aar2p–Prp8pRNaseH complex assembled a ternary complex, as revealed in gel filtration (Fig. 4F) and by GST pull-down assays (Fig. 5A, lanes 22,23). Unlike full-length Aar2p, a Aar2pSub–Prp8pRNaseH complex did not bind Prp8pJab1/MPN (Fig. 4, cf. F and G).
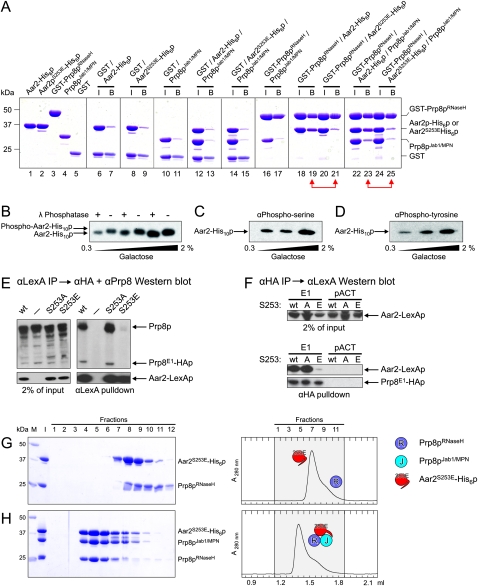
Figure 5.
Aar2p is phosphorylated. (A) SDS PAGE analysis of Aar2p–Prp8p interactions by GST pull-down. All experiments were analyzed on the same gel. Numbers on the left indicate the molecular mass of standard proteins (M) in kilodaltons. (I) Mixture added to the glutathione sepharose; (B) pulled-down (bead) fraction. Proteins or protein mixtures added to the beads are identified above the gel. Red double arrows connect pull-downs of wild-type Aar2-His6p compared with Aar2S253E-His6p. GST-Prp8pRNaseH brought down significantly reduced amounts of Aar2S253E-His6p (lane 21) compared with wild-type Aar2-His6p (lane 19). Similarly, significantly less Prp8pJab1/MPN was brought down by GST-Prp8pRNaseH in the presence of Aar2S253E-His6p (lane 25) compared with wild-type Aar2-His6p (lane 23). (B) Phosphatase treatment indicates that Aar2p is phosphorylated. AGY8 cells were grown in different concentrations of galactose to induce Aar2-His10p production to increasing levels. After incubation of extracts with Ni2+-NTA beads, the precipitated Aar2-His10p was treated with λ phosphatase (+) or not (−) as recommended by the manufacturer (New England Biolabs), fractionated by SDS PAGE, blotted, and probed with anti-Aar2 antibody (R5725). (C,D) Aar2p has phosphorylated serine(s) and tyrosine(s) in vivo. Aar2-His10p produced as in A was analyzed by SDS PAGE, blotted, and probed with anti-phospho-serine (mAb p5747, Sigma) (C) or anti-phospho-tyrosine (mAb 42H4; New England Biolabs) (D) antibodies. (E,F) The S253E variant of Aar2p does not interact with Prp8p. (E) Extracts from yeast cells producing Prp8E1-HAp and wild-type, S253A, or S253E variants of Aar2-LexA fusion protein or with only LexAp (−) were immunoprecipitated with anti-LexA antibodies. Western blot with anti-HA and anti-Prp8 (α8.6) (Boon et al. 2006) antibodies shows that Aar2pS253E pulls down neither full-length Prp8p nor Prp8E1-HAp. The blot was reprobed with anti-LexA antibody, showing efficient pull-down of Aar2-LexAp. (F) Extracts from yeast cells producing Prp8E1-HAp or with Gal4AD-HAp (pACT) control as well as wild-type, S253A, or S253E variants of the Aar2-LexA fusion protein were immunoprecipitated with anti-HA antibodies, blotted, and probed with anti-LexA, indicating that Prp8E1-HAp interacts with Aar2-LexAp wild type and with S253A, but not with S253E. Reprobing with anti-HA antibody verified that Prp8E1-HAp was efficiently immunoprecipitated in the three extracts. (G,H) Gel filtration analysis of Aar2S253E-His6p–Prp8p interactions. Details and labeling are as in Figure 1, C–I. Aar2S253E-His6p does not form a stable complex with Prp8pRNaseH (G), but addition of Prp8pJab1/MPN rescues the ternary complex in gel filtration (H).
The very C terminus of Aar2p, not contained in our crystal structures, is well conserved among Aar2p orthologs (Supplemental Fig. S3). To test the importance of this C-terminal tail for binding of Prp8pJab1/MPN, we generated Aar2ΔC-His6p lacking only the last 23 amino acids, but, in contrast to Aar2pSub, retaining the flexible loop between NTD and CTD. While Aar2ΔC-His6p stably bound to Prp8pRNaseH, the preformed Aar2ΔC-His6p–Prp8pRNaseH complex failed to bind Prp8pJab1/MPN (Fig. 4H). Furthermore, unlike full-length Aar2-His6p, Aar2ΔC-His6p allowed formation of a ternary Aar2ΔC-His6p–Prp8pCTF–Brr2p complex, showing that in the Aar2ΔC-His6p–Prp8pCTF complex, the Jab1/MPN domain remains available for binding to Brr2p (Fig. 4E). These data support a model in which Aar2p bound to the RNaseH domain provides a binding platform for the Jab1/MPN domain. The C-terminal 23 amino acids of Aar2p are required for stable sequestration of Prp8pJab1/MPN and competition with Brr2p.
Prp8p residues involved in interface I with Aar2p (Fig. 3C, left) belong to a region (residues 2033–2067) in which amino acid substitutions led to enhanced interaction with Brr2p (van Nues and Beggs 2001). In light of the present Aar2pSub–Prp8pRNaseH structure, the direct competition between Aar2p and Brr2p for Prp8p suggests that this phenotype may be explained in part by reduced binding of Aar2p and thus easier access of Brr2p to Prp8p.
Aar2p is phosphorylated in vivo
In search of a mechanism that would allow replacement of Aar2p by Brr2p to form functional U5 snRNP, we speculated that post-translational modification of Aar2p might regulate the Aar2p–Prp8p interaction. To test whether Aar2p is phosphorylated in vivo, extracts from AGY8 yeast cells (PGAL1:AAR2-His10) were treated with λ phosphatase, which dephosphorylates phospho-serine (pSer), phospho-threonine (pThr), and phospho-tyrosine (pTyr) residues, the most frequently phosphorylated amino acids in eukaryotes (Zhuo et al. 1993). λ Phosphatase treatment caused Aar2p to migrate faster than nontreated Aar2p in an SDS denaturing gel (Fig. 5B), indicative of the removal of one or more phosphate groups. In addition, antibodies specific for pSer or pTyr detected Aar2p by Western blotting, with the signal increasing with the amount of Aar2p in the extracts (Fig. 5C,D). However, there was no signal with anti-pThr antibodies.
In order to identify the phosphorylated amino acids in Aar2-His10p, the protein was purified from AGY8 cell extracts, digested with proteases, and analyzed by mass spectrometry (MS). Five phosphorylated residues were found (S253, T274, Y328, S331, and T345) (Supplemental Fig. S3), confirming the immunodetection of pSer and pTyr residues and additionally detecting pThr residues, possibly due to superior sensitivity of the MS approach.
A multiple sequence alignment of Aar2p orthologs (Supplemental Fig. S3) showed the overall similarity in the sequences (23.0% identity and 38.3% similarity between the budding yeast and human proteins) and the locations of the five phosphorylated amino acids in the C-terminal third of Aar2p. Among those, only S253 is conserved in most organisms, including humans, but not in Arabidopsis thaliana, Caenorhabditis elegans, or Xenopus laevis. Thus, phosphorylation of S253 might be important for a widely conserved function of Aar2p.
The Aar2pS253E phospho-mimetic mutation inhibits binding of Aar2p to Prp8p
To test possible effects of Aar2p phosphorylation on the ability of Aar2p to bind Prp8p and compete with Brr2p, we mutated each of these phosphorylated residues to glutamate (mimicking phosphorylation) or to alanine (preventing phosphorylation) in the Aar2-LexA fusion protein, and tested the effect of these mutations in pull-down assays. Strikingly, the wild-type Aar2-LexA fusion protein and all of the mutant variants, except S253E, brought down endogenous Prp8p and the Prp8E1 fusion protein, whereas the S253E mutant protein reproducibly did not coprecipitate either protein (Figs. 5E; Supplemental Fig. S5A,B). Likewise, Prp8pE1 pulled down the S253A mutant and wild-type Aar2-LexA fusions but not the S253E variant (Fig. 5F). Furthermore, the smaller S253D phospho-mimetic substitution behaved like S253E (data not shown). These results show that the S253E or S253D phospho-mimetic substitutions inhibit interaction of Aar2p with Prp8p.
We also investigated the growth phenotypes of yeast strains overproducing the interacting proteins. Coproduction of the Aar2-LexA fusion protein and Prp8pE3 was slightly detrimental to growth at low temperature, and the coproduction of Aar2-LexAp and Prp8pE3H was even more so (Supplemental Fig. S5C). These results suggest that the C-terminal E3 and E3H fragments of Prp8p, containing the Jab1/MPN domain, might sequester Brr2p, preventing it from functioning normally. As the growth inhibition depended on the coproduction of the Aar2-LexA fusion (data not shown), the effect of sequestering Brr2p might be exacerbated by the excess Aar2p competing with endogenous Brr2p for binding to Prp8p. Mutating the S253 residue in the Aar2-LexA fusion protein to alanine or glutamate gave opposite effects, with S253A slightly exacerbating and S253E slightly suppressing the growth defect (Supplemental Fig. S5C). Thus, the inability of Aar2p with the S253E substitution to interact with Prp8p in vivo alleviates the growth inhibition caused by overproduction of the Prp8E3 or Prp8E3H proteins, supporting a model in which Aar2p competes with Brr2p for binding to Prp8p, unless residue S253 is phosphorylated.
We also tested whether the S253E mutation has a direct effect on binding of Aar2p to Prp8p in vitro. In GST pull-down assays, a strongly reduced amount of Aar2S253E-His6p was brought down by GST-Prp8pRNaseH compared with wild-type Aar2-His6p (Fig. 5A, lanes 18–21), and the fraction of Prp8pJab1/MPN pulled down with GST-Prp8pRNaseH in the presence of Aar2S253E-His6p was similarly reduced compared with wild-type Aar2-His6p (Fig. 5A, lanes 22–25). These data show that binding of Aar2S253E-His6p to Prp8p is strongly reduced compared with wild-type Aar2-His6p, and that this effect is exerted primarily through a reduced interaction with the Prp8p RNaseH domain. In analytical gel filtration, Aar2S253E-His6p separated from Prp8pRNaseH (Fig. 5G), but in the gentler conditions of this assay, addition of the Prp8pJab1/MPN fragment rescued the ternary complex (Fig. 5H). Similarly, in the reductionist in vitro system, Brr2p was not able to quantitatively titrate Prp8pCTF from Aar2S253E-His6p. However, reproducibly more Prp8pCTF was associated with Brr2p in the presence of Aar2S253E-His6p compared with wild-type Aar2-His6p (Supplemental Fig. S5D,E). These results qualitatively agree with the in vivo pull-down data but suggest that additional factors may be involved in regulating the Aar2p–Prp8p interaction in vivo.
The Aar2pS253E phospho-mimetic mutation allows binding of Brr2p to Prp8p and leads to conformational changes in Aar2p
In order to investigate the effect of the Aar2p mutants on the ability of Prp8p to bind to Brr2p in vivo, extracts from cells producing wild-type or mutant Aar2-LexA fusions were incubated with anti-Prp8 antibodies. Endogenous Prp8p interacted with wild-type Aar2p as well as with the S253A mutant, but not with the S253E mutant protein (Figs. 6A; Supplemental Fig. S5F). Concomitantly, the amount of Prp8p-associated Brr2p increased in extract containing the S253E variant of the Aar2-LexA protein (Fig. 6A,B; Supplemental Fig. S5F). These results are consistent with the idea that phosphorylation of Aar2p at S253 (mimicked by Aar2pS253E) reduces Aar2p affinity for Prp8p, allowing Brr2p to interact with Prp8p.
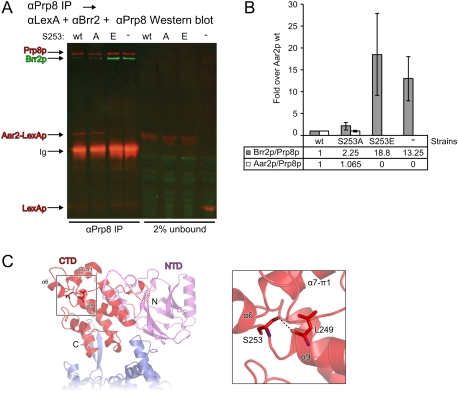
Figure 6.
Influence of Aar2p and variants on the Brr2p–Prp8p interaction. (A,B) Prp8p interacts with more Brr2p when bound by less Aar2p (S253E). (A) Full-length Prp8p was immunoprecipitated from yeast cell extracts containing the highly expressed wild-type, S253A, or S253E variants of Aar2-LexAp, or with LexAp as a control; blotted; and probed with rat anti-Brr2, rabbit anti-LexA, and rabbit anti-Prp8 primary antibodies, followed by goat anti-rat IRDye 680LT and donkey anti-rabbit IRDye 800CW secondary antibodies. The results were visualized and quantified using a LI-COR Odyssey imaging system. (B) Average ratios of precipitated Brr2p/Prp8p and Aar2p/Prp8p are shown for two experiments. Error bars indicate the maximum and minimum values obtained. A similar analysis was also performed in triplicate using detection by standard Western blot (see Supplemental Fig. S5F). (C) Positioning of S253 in the Aar2pSub–Prp8pRNaseH complex structure. (Left) Structural overview. The region surrounding Aar2p S253 is boxed. Protein termini, domains, and selected secondary structure elements are labeled. (Right) Closeup view of the boxed region. Selected residues are shown as sticks and colored by atom type as in Figure 3C. (Dashed line) Hydrogen bond.
S253 lies directly C-terminal of helix α9 in the Aar2p CTD (Fig. 6C). Its side chain hydroxyl engages in a direct hydrogen bond to the backbone carbonyl of L249. Clearly, conformational adjustments would be required to accommodate a phosphate group on S253 and similar conformational rearrangements are expected upon replacement of S253 with the larger, negatively charged glutamate or aspartate. In the present conformation, the larger side chains would lead to steric clashes and the negatively charged moieties would face a hydrophobic pocket formed by F183, L205, and L249. The structural changes are expected to affect the relative positioning of the first segment of the VHS-like domain, encompassing helices α6, α7, and π1, which lie on top of S253 (Fig. 6C). Such conformational changes may explain how S253 phosphorylation can affect binding of Prp8pRNaseH, which occurs remote from the S253 position (Fig. 6C).
Indeed, when subjected to comparative limited proteolysis as a measure of conformational differences, Aar2S253E-His6p yielded different digestion patterns compared with wild-type Aar2-His6p with a number of proteases tested. For example, upon treatment with trypsin, fragments of ∼24-kDa, 20-kDA, and 18-kDa as well as several smaller fragments were observed with both mutant and wild-type proteins, but these fragments appeared at lower trypsin concentrations with the mutant protein, which also gave rise to a novel band at ∼22 kDa (Supplemental Fig. S6A).
Circular dichroism (CD) spectra of Aar2-His6p and Aar2S253E-His6p reproducibly showed small differences (Supplemental Fig. S6B), indicating some minor changes in the secondary structure contents (calculated from CD: 35.3/33.2% helix, 18.3/17.8% β strand for wild-type/S253E Aar2-His6p; crystal structure of Aar2pSub relative to full-length: 33.5% helix, 15.2% β strand). CD melting profiles showed that both proteins exhibit cooperative unfolding with a similar melting temperature (46.1°C–46.5°C) for the main transition (Supplemental Fig. S6C). Reproducibly, wild-type Aar2-His6p showed a second minor transition above 60°C. Qualitatively, these results are in line with differences in the protease digestion pattern and suggest that upon phosphorylation at S253, Aar2p would undergo a limited conformational change, which most likely affects the VHS-like domain and possibly the C-terminal tail, and that this structural change leads to reduced affinity for Prp8p.
Discussion
Assembly of snRNPs depends on chaperones and recycling factors
Assembly of functional macromolecular complexes is a challenging task in the crowded cellular environment and is frequently assisted by trans-acting factors (for review, see Chari and Fischer 2010). Among such factors, assembly chaperones associate with subunits or partial complexes to promote the formation of native-like subassemblies, prevent the formation of unproductive interactions, and/or avoid premature contacts. Frequently, assembly chaperones impose kinetic or thermodynamic traps that have to be resolved by the action of additional assembly factors. Furthermore, different assembly steps often take place in different cellular or subcellular compartments. These principles are nicely illustrated in the biogenesis of Sm core RNPs in metazoans (Chari and Fischer 2010). However, it is presently unknown to what extent and how the de novo assembly of snRNP-specific proteins is also regulated.
Continued pre-mRNA splicing requires snRNP recycling and reassembly, since several snRNPs are compositionally and structurally remodeled during the splicing process (Wahl et al. 2009). As the Sm core RNPs apparently remain intact during the splicing cycle, the main task in recycling of snRNPs may be the reorganization of snRNAs and reassociation of snRNP-specific proteins. While Prp24p and the U6-associated Lsm2–8 protein complex indeed assist reassembly of U4 and U6 snRNAs after each round of splicing (Raghunathan and Guthrie 1998; Verdone et al. 2004), no recycling factors for snRNP-specific proteins have so far been characterized.
Aar2p—an assembly factor for a U5 snRNP-specific protein
Like the U4/U6 di-snRNP, U5 snRNP is also profoundly remodeled during splicing (Makarov et al. 2002). Based on the documented precursor–product relationship between Aar2p-U5 snRNP and mature U5 snRNP in yeast (Boon et al. 2007), and since Aar2p depletion interferes with repeated rounds of splicing in vitro (Gottschalk et al. 2001), we investigated Aar2p as a candidate assembly and recycling factor for U5 snRNP-specific proteins.
We show that Aar2p forms a stable complex with the C-terminal region of the U5-specific Prp8p, binding its RNaseH domain and sequestering its Jab1/MPN domain via Aar2p's C terminus. It thereby hinders Brr2p from interacting with the Jab1/MPN domain. Our in vitro binding analyses demonstrate that Brr2p alone is inefficient in displacing Aar2p from Prp8p (Supplemental Fig. S4), suggesting that Aar2p imposes a kinetic or thermodynamic trap on U5 snRNP assembly, possibly functioning as an assembly chaperone. Phosphorylation mimicry at residue S253 of Aar2p reduces the affinity of the protein for Prp8p, and our data indicate structural changes in Aar2pS253E compared with wild-type Aar2p. Similar structural changes can be expected upon phosphorylation at position 253. Therefore, a presently unidentified Ser/Thr protein kinase may act as another U5 snRNP assembly factor that overcomes the block imposed by Aar2p. Taken together, our results demonstrate that Aar2p is a veritable U5 snRNP assembly factor, which, unlike previously characterized snRNP assembly factors, is required for the ordered binding of a U5 snRNP-specific protein, Brr2p (Fig. 7). Further experiments will be required to determine whether Aar2p actually functions as a molecular chaperone.
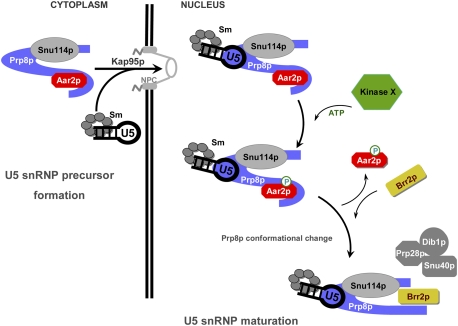
Figure 7.
Model for the U5 snRNP maturation. New elements added to the U5 snRNP maturation model proposed by Boon et al. (2007). Aar2p-U5 snRNP is assembled in the cytoplasm and transported to the nucleus. In the nucleus, Aar2p is phosphorylated by an unidentified kinase (Kinase X). Phosphorylated Aar2p exhibits reduced affinity for Prp8p, leaves the pre-U5 snRNP, and allows Brr2p entry. In view of our finding that the RNaseH and Jab1/MPN domains, which are joined by a flexible linker region, do not interact, we propose that the C terminus of Prp8p takes on a different conformation when not bound by Aar2p. Additional U5 snRNP proteins lacking from Aar2p-U5 snRNP are presumably assembled concomitantly with Brr2p (Boon et al. 2007). Phosphorylated Aar2p has a possible active role in recruiting Brr2p (not shown).
Overexpressed Aar2p or Aar2pS253A fusion protein greatly reduced the amount of Brr2p bound to Prp8p compared with the control, in which Aar2p is present only at the endogenous level (minus sign [−] in Fig. 6A,B). Thus, the excess Aar2 fusion protein inhibits Brr2p binding to full-length Prp8p. In contrast, overexpressed Aar2pS253E does not inhibit Brr2p binding to Prp8p and may even facilitate this interaction, as the amount of Prp8p-associated Brr2p was reproducibly greater in the presence of Aar2pS253E than in control extract with no excess Aar2p (Figs. 6B; Supplemental Fig. S5F). By inference, the S253-phosphorylated variant, may have the ability to make Brr2p more accessible to Prp8p, possibly by sequestering an unknown factor that inhibits interaction of Brr2p with Prp8p. For a better understanding of the protein interactions, this should be further studied, with the mutant protein expressed at the endogenous level. Taken together, while our data do not provide ultimate proof of the mechanism of Aar2p/Brr2p exchange, they provide strong evidence for the involvement of an Aar2p kinase.
Although the RNaseH and Jab1/MPN domains of Prp8p do not stably interact in isolation (Fig. 2B), the close configuration of the Prp8pCTF* fragment (Fig. 2A, left) may still represent a biologically relevant interaction between these domains, which is too weak to survive gel filtration or pull-down assays with the separated domains. The surface of the RNaseH domain that interacts with Aar2p is still accessible in the close conformation of Prp8pCTF*. Furthermore, the C-terminal ∼ 35 residues of the Prp8p Jab1/MPN domain, which comprise a putative Brr2-binding region (Pena et al. 2007), are also surface-exposed in the close conformation of Prp8pCTF*. However, since in a combined model the Jab1/MPN C terminus is remote from the Aar2p binding site on the RNaseH domain (data not shown), additional structural and biochemical studies are required to clarify whether the close conformation of Prp8pCTF* is relevant for the sequestering of the Jab1/MPN domain by Aar2p.
Functional significance of the Aar2p-assisted U5 snRNP assembly
What could be the benefit of the complex Aar2p-mediated U5 snRNP assembly mechanism? Prp8p is an important scaffolding protein at the heart of the spliceosome and interacts with all functional elements of the pre-mRNA, several snRNAs, and other key protein splicing factors (for review, see Grainger and Beggs 2005). Structural and functional analyses of the C-terminal region of Prp8p have suggested that its RNaseH-like domain provides a platform for the handover of the pre-mRNA 5′-splice site from U1 snRNA to U6 snRNA and may be involved in stabilizing the catalytic RNA network (Pena et al. 2008; Ritchie et al. 2008; Yang et al. 2008). Consistent with a role during spliceosome catalytic activation, the C-terminal region of Prp8p encompassing its RNaseH and Jab1/MPN domains directly modulates the activity of Brr2p (Maeder et al. 2009; Pena et al. 2009; Zhang et al. 2009).
The C-terminal region of Prp8p directly binds the U4/U6 snRNA duplex (Zhang et al. 2009), and the Prp8p RNaseH-like domain also interacts with other RNAs, including a putative mimic of the catalytic RNA core of the spliceosome (Ritchie et al. 2008). Modeling studies have suggested that the groove between the thumb and fingers of the RNaseH-like domain serves as an RNA-binding site (Pena et al. 2008). Thus, Aar2p binding at the same site, as shown in the present Aar2pSub–Prp8pRNaseH cocrystal structure, may prevent premature binding of the U4/U6 duplex or binding of nonspecific RNAs at this domain during U5 snRNP assembly.
Upon U4/U6–U5 tri-snRNP formation, Brr2p must be regulated to avoid unwinding U4/U6 di-snRNA prematurely. We suggest that Aar2p regulates the incorporation of Brr2p into U5 snRNP in a manner that silences the Brr2p enzymatic activity during U4/U6–U5 tri-snRNP formation. This suggested role of Aar2p is supported by the observation that the D281N mutant of yeast AAR2 acts as a suppressor of the temperature-sensitive prp38-1 allele (Pandit et al. 2006) that causes slow release of U1 and U4 snRNAs during catalytic activation (Xie et al. 1998). D281 of Aar2p forms a salt bridge with K2066 of Prp8p in interface I (Fig. 3C, left), an interaction that is expected to be weakened upon mutation of D281 to an asparagine.
As in vitro experiments have indicated a role for Aar2p in recycling snRNPs during extended splicing reactions (Gottschalk et al. 2001), Aar2p may have a reciprocal function, displacing Brr2p from Prp8p in a post-splicing complex, to facilitate the regeneration of functional U5 snRNPs. Thus, cycles of phosphorylation and dephosphorylation of Aar2p may regulate U5 snRNP assembly and recycling, and identifying the relevant kinase and phosphatase is the next important step in further elucidating the U5 snRNP (re)assembly mechanism.
Materials and methods
Yeast work
Details of yeast strains and plasmids are provided in Supplemental Tables S2 and S3. For MS analysis, yeast extract was prepared from AGY8 (PGAL1:AAR2-His10) cells grown in 2% (w/v) galactose, and Aar2-His10p was affinity-purified using Ni2+-NTA beads followed by SDS-PAGE and analyzed as described in the Supplemental Material.
Analysis of recombinant proteins
All proteins for biochemical, biophysical, and structural studies were from yeast and, except Brr2p, were produced in E. coli and purified to near homogeneity by chromatographic techniques. Brr2p was produced in insect cell culture. Structural integrity was checked by CD spectroscopy and CD melting analyses. Targets were crystallized by the sitting drop vapor diffusion method, and diffraction data were collected at beamline 14.2 of the BESSY storage ring (Berlin, Germany). The structures were solved by molecular replacement and refined by standard strategies. Details are given in the Supplemental Material.
Database deposition
Structure coordinates and diffraction data were deposited with the Protein Data Bank (http://www.pdb.org) under accession codes 3SBG (Prp8CTF*), 3SBS (Aar2pSub), and 3SBT (Aar2pSub–Prp8RNaseH) and will be released upon publication.
Acknowledgments
We thank Claudia Kipar, Claudia Alings, and Traudy Wandersleben (Freie Universität Berlin) for help with cloning, expression, protein preparation, in vitro interaction analyses, and crystallization. We acknowledge access to beamline BL14.2 of the BESSY II storage ring (Berlin, Germany) via the Joint Berlin MX-Laboratory, sponsored by the Helmholtz Zentrum Berlin für Materialien und Energie, the Freie Universität Berlin, the Humboldt-Universität zu Berlin, the Max-Delbrück Centrum, and the Leibniz-Institut für Molekulare Pharmakologie. This work was supported by the Deutsche Forschungsgemeinschaft (grants SFB 740/2 and WA 1126/4-1 to M.C.W.) and the Wellcome Trust (grant 087551 to J.D.B.), and benefited from European Commission funding LSGH-CT-2005-518238 for the EURASNET Network of Excellence (to J.D.B.). V.F.C. is a predoctoral student of the Gulbenkian PhD Programme in Biomedicine funded by Scholarship SFRH/BD/15854/2005 from the FCT of Portugal. J.D.B. holds the Royal Society Darwin Trust Research Professorship. J.R. is a Senior Research Fellow of the Wellcome Trust. G.W., K.F.S., and N.H. prepared and characterized recombinant proteins and conducted interaction studies in vitro. G.W. and M.C.W. carried out the crystallographic analyses and supervised the work performed by K.F.S. and N.H. V.F.C. performed the experiments to identify phosphorylated residues in Aar2p and to characterize the mutant variants of Aar2p in vivo and in yeast extracts. F.A. and J.R. performed the MS analyses. J.D.B. supervised the work performed by V.F.C. J.D.B. and M.C.W. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.635911.
References
- Boon KL, Norman CM, Grainger RJ, Newman AJ, Beggs JD 2006. Prp8p dissection reveals domain structure and protein interaction sites. RNA 12: 198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon KL, Grainger RJ, Ehsani P, Barrass JD, Auchynnikava T, Inglehearn CF, Beggs JD 2007. prp8 mutations that cause human retinitis pigmentosa lead to a U5 snRNP maturation defect in yeast. Nat Struct Mol Biol 14: 1077–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari A, Fischer U 2010. Cellular strategies for the assembly of molecular machines. Trends Biochem Sci 35: 676–683 [DOI] [PubMed] [Google Scholar]
- Chari A, Paknia E, Fischer U 2009. The role of RNP biogenesis in spinal muscular atrophy. Curr Opin Cell Biol 21: 387–393 [DOI] [PubMed] [Google Scholar]
- Gottschalk A, Kastner B, Lührmann R, Fabrizio P 2001. The yeast U5 snRNP coisolated with the U1 snRNP has an unexpected protein composition and includes the splicing factor Aar2p. RNA 7: 1554–1565 [PMC free article] [PubMed] [Google Scholar]
- Grainger RJ, Beggs JD 2005. Prp8 protein: at the heart of the spliceosome. RNA 11: 533–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb SJ, Battle DJ, Dreyfuss G 2007. Molecular functions of the SMN complex. J Child Neurol 22: 990–994 [DOI] [PubMed] [Google Scholar]
- Liu S, Rauhut R, Vornlocher HP, Lührmann R 2006. The network of protein–protein interactions within the human U4/U6. U5 tri-snRNP. RNA 12: 1418–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder C, Kutach AK, Guthrie C 2009. ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase requires the C terminus of Prp8. Nat Struct Mol Biol 16: 42–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Lührmann R 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298: 2205–2208 [DOI] [PubMed] [Google Scholar]
- Meinhart A, Cramer P 2004. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature 430: 223–226 [DOI] [PubMed] [Google Scholar]
- Misra S, Puertollano R, Kato Y, Bonifacino JS, Hurley JH 2002. Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains. Nature 415: 933–937 [DOI] [PubMed] [Google Scholar]
- Nakazawa N, Harashima S, Oshima Y 1991. AAR2, a gene for splicing pre-mRNA of the MATa1 cistron in cell type control of Saccharomyces cerevisiae. Mol Cell Biol 11: 5693–5700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesic D, Tanackovic G, Krämer A 2004. A role for Cajal bodies in the final steps of U2 snRNP biogenesis. J Cell Sci 117: 4423–4433 [DOI] [PubMed] [Google Scholar]
- Pandit S, Lynn B, Rymond BC 2006. Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc Natl Acad Sci 103: 13700–13705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena V, Liu S, Bujnicki JM, Lührmann R, Wahl MC 2007. Structure of a multipartite protein–protein interaction domain in splicing factor prp8 and its link to retinitis pigmentosa. Mol Cell 25: 615–624 [DOI] [PubMed] [Google Scholar]
- Pena V, Rozov A, Fabrizio P, Lührmann R, Wahl MC 2008. Structure and function of an RNase H domain at the heart of the spliceosome. EMBO J 27: 2929–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena V, Jovin SM, Fabrizio P, Orlowski J, Bujnicki JM, Lührmann R, Wahl MC 2009. Common design principles in the spliceosomal RNA helicase Brr2 and in the Hel308 DNA helicase. Mol Cell 35: 454–466 [DOI] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K 2009. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 458: 475–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan PL, Guthrie C 1998. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science 279: 857–860 [DOI] [PubMed] [Google Scholar]
- Ritchie DB, Schellenberg MJ, Gesner EM, Raithatha SA, Stuart DT, Macmillan AM 2008. Structural elucidation of a PRP8 core domain from the heart of the spliceosome. Nat Struct Mol Biol 15: 1199–1205 [DOI] [PubMed] [Google Scholar]
- Shiba T, Takatsu H, Nogi T, Matsugaki N, Kawasaki M, Igarashi N, Suzuki M, Kato R, Earnest T, Nakayama K, et al. 2002. Structural basis for recognition of acidic-cluster dileucine sequence by GGA1. Nature 415: 937–941 [DOI] [PubMed] [Google Scholar]
- Small EC, Leggett SR, Winans AA, Staley JP 2006. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol Cell 23: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- van Nues RW, Beggs JD 2001. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics 157: 1451–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdone L, Galardi S, Page D, Beggs JD 2004. Lsm proteins promote regeneration of pre-mRNA splicing activity. Curr Biol 14: 1487–1491 [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Weber G, Trowitzsch S, Kastner B, Lührmann R, Wahl MC 2010. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J 29: 4172–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Beickman K, Otte E, Rymond BC 1998. Progression through the spliceosome cycle requires Prp38p function for U4/U6 snRNA dissociation. EMBO J 17: 2938–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Zhang L, Xu T, Heroux A, Zhao R 2008. Crystal structure of the β-finger domain of Prp8 reveals analogy to ribosomal proteins. Proc Natl Acad Sci 105: 13817–13822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Xu T, Maeder C, Bud LO, Shanks J, Nix J, Guthries C, Pleiss JA, Zhao R 2009. Strutural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat Struct Mol Biol 16: 731–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo S, Clemens JC, Hakes DJ, Barford D, Dixon JE 1993. Expression, purification, crystallization, and biochemical characterization of a recombinant protein phosphatase. J Biol Chem 268: 17754–17761 [PubMed] [Google Scholar]