Abstract
The U5 snRNP (small ribonucleoprotein) contains several functionally crucial splicing factors that form an extensive interaction network both in the snRNP and within the spliceosome. In this issue of Genes & Development, Weber and colleagues (pp. 1601–1612) shed light on the dynamic assembly of this critical spliceosomal component and elucidate the molecular interactions underlying the ordered addition of Brr2, a pivotal spliceosomal helicase, to the U5 snRNP.
Keywords: pre-mRNA splicing, protein interaction, protein phosphorylation, protein structure, spliceosome, yeast
A fascinating feature of eukaryotic gene expression is the presence of intervening sequences or introns in the majority of primary genetic transcripts that need to be accurately removed or “spliced” to generate a functional transcript. Commensurate with its critical importance, splicing is performed through a highly elaborate, stepwise process involving the assembly of a large number of molecular complexes and myriad conformational rearrangements that serve as checkpoints to ensure accuracy (Wahl et al. 2009; Valadkhan and Jaladat 2010; Will and Lührmann 2011). Once splicing is performed, the assembled splicing machinery, called the spliceosome, disassembles, and its components are prepared for the next round of assembly and splicing. While three decades of intense research have elucidated the broad outline of the spliceosomal assembly and disassembly steps, our understanding of the molecular events involved in the orchestration of the spliceosomal conformational changes has remained very limited (Nilsen 1998; Will and Luhrmann 2006). Smith et al. 2008; Newman and Nagai 2010).
A large fraction of spliceosomal components are preassembled into snRNPs (small ribonucleoproteins), each of which contains one of the five spliceosomal small nuclear RNAs (U1, U2, U4, U5, or U6 snRNAs) and several associated proteins. Of the five spliceosomal snRNPs, U5 snRNP is the largest and, in addition to U5 snRNA, carries a number of highly critical spliceosomal proteins to the assembling spliceosomes (Wahl et al. 2009; Valadkhan and Jaladat 2010; Will and Lührmann 2011). The U5-associated proteins include Prp8, a large and highly conserved protein that is thought to have a pivotal role in coordination of the spliceosomal cycle (Grainger and Beggs 2005). The mode of function of Prp8 is poorly understood, and it lacks well-defined, conserved functional motifs. However, Prp8 contains a number of degenerate nucleic acid-binding domains, including reverse transcriptase, RNase H, RNA-binding motif (RRM), and likely a bromodomain (Grainger and Beggs 2005; Dlakić and Mushegian 2011). In addition, close to its C terminus it contains a degenerate MPN/Jab1 domain that is found in deubiquitinating enzymes (Grainger and Beggs 2005; Bellare et al. 2006; Pena et al. 2007; Zhang et al. 2007). In these degenerate domains, most of the functionally critical residues are missing, and it is likely that they are remnants of an ancient mobile element from which Prp8 has evolved (Dlakić and Mushegian 2011). Instead of performing their original functions, these degenerate domains seem to be used as protein–nucleic acid and protein–protein interaction domains in Prp8.
Another functionally essential U5-associated protein is Brr2, a DExD/H-box helicase that is involved in remodeling the RNA–RNA interactions during both the spliceosomal activation and the disassembly steps (Fig. 1; Wahl et al. 2009; Hahn and Beggs 2010; Valadkhan and Jaladat 2010). An important step in the final stages of spliceosomal activation involves the formation of a catalytically required base-pairing interaction between U2 and U6 snRNAs (Valadkhan 2010). For this to happen, an existing base-pairing interaction between U6 and U4 snRNAs has to be disrupted, an important conformational change known to be mediated by Brr2 (Kim and Rossi 1999; Hahn and Beggs 2010). Furthermore, after completion of the splicing reaction, Brr2 disrupts the base-pairing interaction of U2 and U6 snRNAs as one of the first steps in the disassembly of the spliceosome and the recycling of its components for a new round of splicing (Hahn and Beggs 2010; Valadkhan and Jaladat 2010; Will and Lührmann 2011). These important functions of Brr2 are regulated by the C-terminal domain (CTD) of Prp8, which has been shown to stimulate the unwinding activity of Brr2 (Maeder et al. 2009), and by Snu114, another U5-associated protein that acts as a classic regulatory G protein and regulates the unwinding function of Brr2 in a GTP-dependent manner (Small et al. 2006; Valadkhan and Jaladat 2010; Will and Lührmann 2011). Furthermore, there is extensive in vitro and in vivo evidence for direct interaction between Snu114 and the CTD of Prp8 with Brr2 (Bottner et al. 2005; Grainger and Beggs 2005; Boon et al. 2006; Liu et al. 2006; Will and Lührmann 2011).
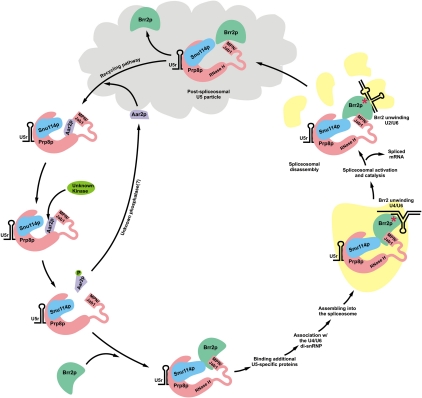
Figure 1.
The cycle of U5 snRNP maturation and function. The interactions of the main players in the maturation and function of U5 have been shown. (U5r) U5 snRNA. The snRNAs are shown as thick black lines. The steps shown in the gray cloud are hypothetical and are not based on experimental evidence. Due to space constraints, only the steps relevant to this work have been shown.
Thus, in the light of the critical function of Brr2, it was a surprise when it was discovered that a fraction of U5 complexes lack Brr2 (Gottschalk et al. 2001). Further work indicated that these Brr2-less U5 snRNPs corresponded to immature snRNPs. Instead of Brr2, these immature U5 snRNPs contained Aar2, a spliceosomal protein that was shown to be dispensable for a single round of in vitro splicing but required for multiple cycles of splicing, which suggested a role in spliceosomal recycling (Gottschalk et al. 2001; Stevens et al. 2001). Since analysis of U5 snRNPs at various stages of maturity suggested that the presence of Brr2 and Aar2 was mutually exclusive, it was hypothesized that they may be competing for interaction with the same binding motif as a potential regulatory step during the maturation of the U5 snRNPs (Boon et al. 2007).
Through an elegant series of biochemical and structural biology studies, Weber et al. (2011) addressed this possibility by unraveling the network of interactions between Aar2 and Prp8 and its subsequent replacement by Brr2 during the maturation of the U5 snRNP particle. By elucidating the molecular details of the interaction between Aar2 and Prp8, Weber et al. (2011) provide evidence for a previously unknown regulated step in the maturation of U5 snRNPs, which involves phosphorylation of Aar2 that leads to its release from Prp8 and replacement by Brr2. Furthermore, this work reveals the presence of yet another mechanism for ensuring the ordered, stepwise assembly of the spliceosomal components.
Sequestration of the Brr2-binding site on Prp8 by Aar2
To gain insight into the interaction of Aar2 and Prp8, Weber et al. (2011) first used pull-down assays to show that Aar2 can bind a peptide containing the RNase H and MPN/Jab1 domains of Prp8. While the RNase H domain in isolation was able to bind Aar2, the MPN/Jab1 domain by itself could not bind Aar2. Crystal structure of a polypeptide containing the RNase H and MPN/Jab1 domains of Prp8 (but lacking 16 amino acids from its very C terminus) indicated that the two domains folded independently with a flexible linker joining them, and in vitro analyses confirmed that the two domains did not interact with each other.
Next, by subjecting Aar2 to limited proteolysis, Weber et al. (2011) succeeded in obtaining crystal structures for both the isolated Aar2 and the complex of Aar2 and the RNase H domain of Prp8. While the N-terminal domain of Aar2 contained a novel fold, the CTD showed similarities to structural motifs found in Golgi–ER transport adapters and endocytic adapter proteins. Interestingly, compared with the isolated Aar2 and Prp8 RNase H domain structures (Pena et al. 2008; Ritchie et al. 2008; Yang et al. 2008), the binary complex showed little conformational change in either Aar2 or the RNase H domain, indicating that the impact of binding of Aar2 was limited to physically blocking the binding interface on the RNase H domain.
Similar binding studies on Brr2 showed that, as suggested by previous studies (Grainger and Beggs 2005; Boon et al. 2007; Hahn and Beggs 2010), Brr2 could bind the MPN/Jab1 domain of Prp8 in isolation and in the context of a larger fragment of Prp8 that contained both the RNase H and the MPN/Jab1 domains, but was unable to interact with the isolated Prp8 RNase H domain. While, based on these results, the formation of a ternary complex between Prp8, Brr2, and Aar2 seemed feasible, when stoichiometric amounts of Aar2, Brr2, and a peptide carrying the RNase H–MPN/Jab1 domains of Prp8 were mixed, no ternary complexes were observed. Instead, Aar2–Prp8 complexes and free Brr2 were the dominant species in the mixture. Even at very high concentrations, Brr2 was not able to efficiently displace Aar2 from its complex with Prp8, and no ternary complexes were observed.
Why was Brr2 unable to interact with the MPN/Jab1 domain in the presence of Aar2? Although neither the RNase H domain nor Aar2 showed significant binding to the MPN/Jab1 domain, it was possible that the complex of Aar2 and Prp8 RNase H domain could bind the MPN/Jab1 domain, thus blocking the binding of Brr2 to this region of the molecule, which indeed proved to be the case. Interestingly, while the complex of the RNase H domain and full-length Aar2 could bind the MPN/Jab1 domain, limited proteolytic digestion of Aar2 that had been used in the crystallographic studies resulted in loss of MPN/Jab1 domain binding, although the interaction between the partially digested Aar2 and the RNase H domain was intact. Weber et al. (2011) could show that the loss of binding to the MPN/Jab1 domain resulted from the digestion of a conserved, 31-amino-acid-long region at the C terminus of Aar2 during the limited proteolytic treatment, which likely indicated the inherent flexibility of this part of the molecule. Thus, although the crystallographic studies did not show a significant conformational change in Aar2 upon complex formation with the RNase H domain, this C-terminal region that was not present in the crystallized construct was likely to be organized upon binding to the RNase H domain. Alone or together with part of the RNase H domain, it could form a binding platform for the MPN/Jab1 domain, which would then effectively compete with Brr2 for binding to this region. Consistent with this possibility, addition of Brr2 to the complex of RNase H–MPN/Jab1 peptide and partially digested Aar2 resulted in the formation of a ternary complex. Thus, although Aar2 and Brr2 primarily bind to different regions on Prp8, binding of Aar2 results in sequestration of the binding site of Brr2 that effectively prohibits the simultaneous binding of the two proteins to Prp8. In addition to providing a satisfactory explanation for the mutually exclusive presence of Aar2 and Brr2 in maturing U5 particles, these data also explain previously observed improvements in Brr2 binding to Prp8 that resulted from mutations within the RNase H domain of this protein (van Nues and Beggs 2001; Kuhn et al. 2002).
Regulation of U5 snRNP assembly by phosphorylation
An important question that arises from the above observations is how Brr2 ultimately replaces Aar2 in the maturing U5 snRNPs. To answer this question, Weber et al. (2011) looked for any post-translational modifications that may serve to weaken the affinity of Aar2 for binding to Prp8. Indeed, they found that Aar2 contained several phosphorylated residues, one of which (S253 in yeast Aar2) was conserved in most organisms. To test the functional impact of phosphorylation at this position, Weber et al. (2011) mutated this residue to either glutamate or aspartate, which mimic a phosphorylated residue. The mutants not only showed a markedly reduced binding to the RNase H domain of Prp8, they also were less efficient in sequestering the MPN/Jab1 domain, as shown by in vitro pull-down assays. To analyze the impact of phosphorylation in vivo, wild-type or mutant Aar2s containing a glutamate in place of S253 were overexpressed along with a peptide corresponding to the Prp8 MPN/Jab1 domain. While overexpression of either wild-type Aar2 or the Prp8 fragment did not have a significant effect, their co-overexpression led to a cold-sensitive growth phenotype in yeast. The most plausible scenario leading to such a phenotype would be the titration of the free Brr2 by the overexpressed MPN/Jab1 domain along with increased competition for binding to the assembling U5 snRNPs due to Aar2 overexpression, which likely prevented Brr2 from efficiently integrating into U5 snRNPs. Interestingly, replacing the wild-type Aar2 with the glutamate-substituted mutant in the co-overexpression experiments led to a partial suppression of the growth defect. Furthermore, immunoprecipitation assays indicated that more Brr2 was associated with Prp8 when the glutamate-substituted Aar2 mutant was overexpressed compared with wild-type Aar2-overexpressing cells. Together with the results obtained from in vitro experiments, these data suggest that the glutamate substitution weakens the ability of Aar2 to compete with Brr2 for binding to Prp8 in vivo.
A remaining question was why phosphorylation at S253, which is not located in the binding interface of Aar2 and the Prp8 RNase H domain, weakens this interaction. One possibility was that S253 phosphorylation would result in a conformational change in Aar2 that would affect its Prp8-binding interface. Analysis of the high-resolution structure of Aar2 suggested that this might indeed be the case, since addition of a phosphate group at residue S253 would likely lead to steric clashes and require conformational changes to accommodate this negatively charged group, which would be also facing a hydrophobic pocket. To obtain experimental evidence for such a conformational change, Weber et al. (2011) analyzed the pattern of digestion of wild-type and glutamate-substituted Aar2 after subjecting them to limited proteolysis. Interestingly, the mutant Aar2 yielded a different digestion pattern, consistent with a conformational change resulting from the glutamate substitution at S253. Furthermore, circular dichroism (CD) spectra of the wild-type and mutant Aar2 species showed differences consistent with changes in the secondary structure content, further suggesting that phosphorylation at S253 led to a conformational change in Aar2, which in turn changed the affinity of this protein for binding to Prp8.
An ordered assembly pathway for U5 snRNP: a role in retinitis pigmentosa?
The work presented by Weber et al. (2011) provides detailed insight into the assembly of the U5 snRNP particle and suggests a model in which the initial interaction of Aar2 with Prp8 blocks the integration of Brr2 into the assembling snRNP until, through the action of an unknown kinase, Aar2 is phosphorylated (Fig. 1). This phosphorylation event results in a conformational change that weakens the binding of Aar2 to Prp8 in the immature U5 snRNP particles, leading to the release of Aar2 from the U5 snRNP and recruitment of Brr2 to the maturing snRNP. Once U5 has joined the U4/U6 snRNP and has entered the assembling spliceosome, Brr2 forms extensive interactions with the MPN/Jab1 domain of Prp8 and Snu114. Under the regulatory control of Prp8 and Snu114, Brr2 performs its critical function by mediating the release of the base-pairing partners of U6 snRNA both during spliceosomal assembly and its disassembly (Fig. 1).
According to this model, the timing of the recruitment of Brr2 to the U5 snRNP is regulated by a kinase that mediates the release of Aar2 from U5 during the last stages of the assembly of this snRNP. The functional significance of this highly ordered assembly pathway remains to be discovered. However, it is plausible that in the light of the critical role of Brr2 and to ensure its optimal regulation by Prp8 and Snu114, the recruitment of Brr2 to U5 snRNP is delayed until a stage where the rest of the U5 components are ideally positioned for making the appropriate contacts with Brr2 upon its joining the snRNP. Prp8, Snu114, and Brr2 extensively interact with each other and the rest of the spliceosomal components, and it is possible that during the spliceosomal cycle, their interaction network is remodeled according to their required function at each stage of the splicing cycle (Liu et al. 2006; Valadkhan 2007; Hahn and Beggs 2010). Such a scenario may also explain why Aar2 was originally described as a factor required for performing multiple rounds of splicing (Gottschalk et al. 2001). Through the action of a phosphatase, Aar2 can be dephosphorylated, and thus can bind to the post-splicing U5 snRNPs and promote the release of Brr2 to allow the resetting of the U5 snRNP conformation to the one needed for function during the spliceosomal assembly process (Fig. 1).
In addition to elucidating the dynamic interactions involved in the maturation of the U5 snRNP, the data presented by Weber et al. (2011) offer insights into yet another aspect of function of Prp8. Widely thought to be the master regulator of the spliceosomal cycle, so far only bits and pieces of the way this crucial and mysterious spliceosomal protein performs its function have been uncovered. Interestingly, several point mutations in the very C-terminal amino acids of Prp8 have been associated with the hereditary blindness retinitis pigmentosa (McKie et al. 2001; Grainger and Beggs 2005). While the exact mechanism by which point mutations in Prp8 lead to this disease remains to be discovered, it has been shown that deletion of these residues or the point mutations that cause the disease result in loss or weakening of functionally critical interactions between Prp8, Snu114, and Brr2 (Boon et al. 2007; Pena et al. 2007; Maeder et al. 2009). Furthermore, it has been shown that these mutations result in accumulation of a U5 snRNP species that lacks Brr2 and instead contains Aar2 (Boon et al. 2007). Together, the existing data suggest that by weakening the interaction of Brr2 with the U5 snRNP, the retinitis pigmentosa-causing mutations may interfere with the efficient maturation and recycling of this snRNP. This, in turn, may reduce the efficiency of splicing, leading to splicing defects and disruption of function in tissues that have a very high level of transcriptional activity, such as the retina (Mordes et al. 2006). While defining the details of the molecular basis of this disease awaits future studies, the existing data point to the physiological importance of the dynamic events involved in the assembly of the spliceosomal components. Further analysis of the highly complex spliceosomal dynamics promises to provide novel insights into the mechanisms used for dynamic control of the cellular processes for the years to come.
Acknowledgments
I thank Tim Nilsen for critical reading of the perspective, and the NIH (grant no. GM078572) for funding.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.17311211.
References
- Bellare P, Kutach AK, Rines AK, Guthrie C, Sontheimer EJ 2006. Ubiquitin binding by a variant Jab1/MPN domain in the essential pre-mRNA splicing factor Prp8p. RNA 12: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon K-L, Norman CM, Grainger RJ, Newman AJ, Beggs JD 2006. Prp8p dissection reveals domain structure and protein interaction sites. RNA 12: 198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon K-L, Grainger RJ, Ehsani P, Barrass JD, Auchynnikava T, Inglehearn CF, Beggs JD 2007. prp8 mutations that cause human retinitis pigmentosa lead to a U5 snRNP maturation defect in yeast. Nat Struct Mol Biol 14: 1077–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottner CA, Schmidt H, Vogel S, Michele M, Käufer NF 2005. Multiple genetic and biochemical interactions of Brr2, Prp8, Prp31, Prp1 and Prp4 kinase suggest a function in the control of the activation of spliceosomes in Schizosaccharomyces pombe. Curr Genet 48: 151–161 [DOI] [PubMed] [Google Scholar]
- Dlakić M, Mushegian A 2011. Prp8, the pivotal protein of the spliceosomal catalytic center, evolved from a retroelement-encoded reverse transcriptase. RNA 17: 799–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A, Kastner B, Lührmann R, Fabrizio P 2001. The yeast U5 snRNP coisolated with the U1 snRNP has an unexpected protein composition and includes the splicing factor Aar2p. RNA 7: 1554–1565 [PMC free article] [PubMed] [Google Scholar]
- Grainger RJ, Beggs JD 2005. Prp8 protein: at the heart of the spliceosome. RNA 11: 533–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn D, Beggs JD 2010. Brr2p RNA helicase with a split personality: insights into structure and function. Biochem Soc Trans 38: 1105–1109 [DOI] [PubMed] [Google Scholar]
- Kim DH, Rossi JJ 1999. The first ATPase domain of the yeast 246-kDa protein is required for in vivo unwinding of the U4/U6 duplex. RNA 5: 959–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn AN, Reichl EM, Brow DA 2002. Distinct domains of splicing factor Prp8 mediate different aspects of spliceosome activation. Proc Natl Acad Sci 99: 9145–9149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Rauhut R, Vornlocher H-P, Lührmann R 2006. The network of protein–protein interactions within the human U4/U6·U5 tri-snRNP. RNA 12: 1418–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder C, Kutach AK, Guthrie C 2009. ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase requires the C terminus of Prp8. Nat Struct Mol Biol 16: 42–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKie AB, McHale JC, Keen TJ, Tarttelin EE, Goliath R, van Lith-Verhoeven JJ, Greenberg J, Ramesar RS, Hoyng CB, Cremers FP, et al. 2001. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13). Hum Mol Genet 10: 1555–1562 [DOI] [PubMed] [Google Scholar]
- Mordes D, Luo X, Kar A, Kuo D, Xu L, Fushimi K, Yu G, Sternberg P Jr, Wu JY 2006. Pre-mRNA splicing and retinitis pigmentosa. Mol Vis 12: 1259–1271 [PMC free article] [PubMed] [Google Scholar]
- Newman AJ, Nagai K 2010. Structural studies of the spliceosome: blind men and an elephant. Curr Opin Struct Biol 20: 82–89 [DOI] [PubMed] [Google Scholar]
- Nilsen TW 1998. RNA–RNA interactions in nuclear pre-mRNA splicing. In RNA structure and function (ed. Simons RW, Grunberg-Manago M), pp. 279–307 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- Pena V, Liu S, Bujnicki JM, Lührmann R, Wahl MC 2007. Structure of a multipartite protein–protein interaction domain in splicing factor prp8 and its link to retinitis pigmentosa. Mol Cell 25: 615–624 [DOI] [PubMed] [Google Scholar]
- Pena V, Rozov A, Fabrizio P, Lührmann R, Wahl MC 2008. Structure and function of an RNase H domain at the heart of the spliceosome. EMBO J 27: 2929–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie DB, Schellenberg MJ, Gesner EM, Raithatha SA, Stuart DT, Macmillan AM 2008. Structural elucidation of a PRP8 core domain from the heart of the spliceosome. Nat Struct Mol Biol 15: 1199–1205 [DOI] [PubMed] [Google Scholar]
- Small EC, Leggett SR, Winans AA, Staley JP 2006. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol Cell 23: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Query CC, Konarska MM 2008. ‘Nought may endure but mutability': spliceosome dynamics and the regulation of splicing. Mol Cell 30: 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens SW, Barta I, Ge HY, Moore RE, Young MK, Lee TD, Abelson J 2001. Biochemical and genetic analyses of the U5, U6, and U4/U6·U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae. RNA 7: 1543–1553 [PMC free article] [PubMed] [Google Scholar]
- Valadkhan S 2007. The spliceosome: caught in a web of shifting interactions. Curr Opin Struct Biol 17: 310–315 [DOI] [PubMed] [Google Scholar]
- Valadkhan S 2010. Role of the snRNAs in spliceosomal active site. RNA Biol 7: 345–353 [DOI] [PubMed] [Google Scholar]
- Valadkhan S, Jaladat Y 2010. The spliceosomal proteome: at the heart of the largest cellular ribonucleoprotein machine. Proteomics 10: 4128–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nues RW, Beggs JD 2001. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics 157: 1451–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Weber G, Cristão VF, de L. Alves F, Santos KF, Holton N, Rappsilber J, Beggs JD, Wahl MC 2011. Mechanism for Aar2p function as a U5 snRNP assembly factor. Genes Dev (this issue). doi: 10.1101/gad.635911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R 2006. Spliceosome structure and function. In The RNA world (ed. Gesteland RF et al. ), pp. 369–400 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Will CL, Lührmann R 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707. doi: 10.1101/cshperspect.a003707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Zhang L, Xu T, Heroux A, Zhao R 2008. Crystal structure of the β-finger domain of Prp8 reveals analogy to ribosomal proteins. Proc Natl Acad Sci 105: 13817–13822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Shen J, Guarnieri MT, Heroux A, Yang K, Zhao R 2007. Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants. Protein Sci 16: 1024–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]