Abstract
Eukaryotic cell cycle transitions are driven by E3 ubiquitin ligases that catalyze the ubiquitylation and destruction of specific protein targets. For example, the anaphase-promoting complex/cyclosome (APC/C) promotes the exit from mitosis via destruction of securin and mitotic cyclins, whereas CRL1Skp2 allows entry into S phase by targeting the destruction of the cyclin-dependent kinase (CDK) inhibitor p27. Recently, an E3 ubiquitin ligase called CRL4Cdt2 has been characterized, which couples proteolysis to DNA synthesis via an unusual mechanism that involves display of substrate degrons on the DNA polymerase processivity factor PCNA. Through its destruction of Cdt1, p21, and Set8, CRL4Cdt2 has emerged as a master regulator that prevents rereplication in S phase. In addition, it also targets other factors such as E2F and DNA polymerase η. In this review, we discuss our current understanding of the molecular mechanism of substrate recognition by CRL4Cdt2 and how this E3 ligase helps to maintain genome integrity.
Keywords: Cul4, Ddb1, Cdt2, CRL4, CNA, PIP degron
The regulated destruction of proteins is integral to the physiology of all eukaryotic cells. Thus, cell cycle transitions, the maintenance of genome integrity, signaling, and many other cellular processes involve controlled proteolysis. Regulated proteolysis is carried out by the ubiquitin–proteasome system. Ubiquitin is attached to proteins destined for destruction via an isopeptide bond between the C-terminal glycine of ubiquitin and one or more lysines of the target. This ubiquitin is then modified by additional ubiquitins that are connected to a lysine on the foregoing ubiquitin, thereby forming “ubiquitin chains.” Most ubiquitin chains target the substrate for destruction by the 26S proteasome. However, some proteins are monoubiquitylated or diubiquitylated, while others are polyubiquitylated via Lys 63 chains that modulate protein function without causing destruction (Komander 2009; Ye and Rape 2009; Behrends and Harper 2011).
The attachment of ubiquitin to substrates is carried out by an enzymatic cascade. First, ubiquitin is attached via a high-energy thioester bond to an “E1” ubiquitin-activating enzyme in a reaction that consumes ATP. Next, the ubiquitin is transferred from E1 to the cysteine of an “E2” ubiquitin-conjugating enzyme. Finally, the E2 interacts with an “E3” ubiquitin ligase that also binds the substrate. The juxtaposition of the substrate and the charged E2 enzyme leads to ubiquitin transfer to the substrate. The specificity of ubiquitylation is encoded at the level of substrate recognition by the E3 enzymes (Ravid and Hochstrasser 2008); however, recently it has become clear that E2s can also contribute to processivity and specificity for ubiquitin chain nucleation and elongation (Jin et al. 2008; Ye and Rape 2009; Rodrigo-Brenni et al. 2010; Saha et al. 2011; Wickliffe et al. 2011). Generally, the E3–substrate interaction involves the binding of a “substrate receptor” subunit of the E3 to a short stretch of amino acids called a “degron” motif within the substrate. The best understood degrons are six to eight amino acids long, transferable, and necessary and sufficient for binding to the substrate.
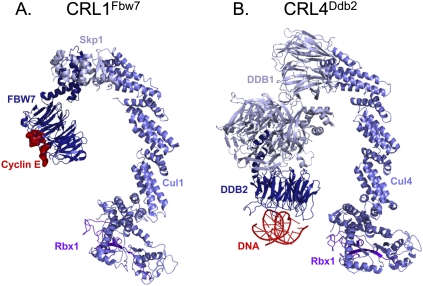
There are two major families of multiprotein E3 ubiquitin ligases in eukaryotes, which are characterized by the presence of RING or HECT domains (Ardley and Robinson 2005; Deshaies and Joazeiro 2009). The RING domain coordinates zinc ions in a protein fold that binds to an E2-conjugating enzyme. The cullin ring E3 ligases (CRLs) are the largest family of RING ubiquitin ligases. In CRLs, a cullin scaffold binds via its C terminus to the RING domain protein Rbx1, which recruits an E2 (Fig. 1A, E2 not shown). The N terminus of the cullin binds to an adaptor (e.g., Skp1), which in turn binds a substrate receptor that contacts the substrate, bringing it close to the E2 (Fig. 1A). In HECT domain ligases, the E2 enzyme transfers ubiquitin to a cysteine residue in the HECT domain, from which it is transferred to the substrate (data not shown).
Figure 1.
Models of CRL1Fbw7 and CRL4Ddb2 structures. (A) A ribbon diagram model depicting the CRL1Fbw7 ubiquitin ligase and its substrate, Cyclin E, was generated using PyMOL (http://www.pymol.org). Cul1 N terminus (slate), Cul1 C terminus (slate), and Rbx1 (purple) are from Zheng et al. (2002) (Protein Data Bank [PDB] accession no. 1LDK). Skp1 (light blue), Fbw7 (dark blue), and the substrate, Cyclin E C-terminal degron (red), are from Hao et al. (2007) (PDB 2OVQ). (B) A ribbon diagram model depicting CRL4Ddb2 bound to abasic DNA was generated using PyMOL. Cul4a (slate) and Rbx1 (purple) are from Angers et al. (2006) (PDB 2HYE) and aligned to the Cul1 N terminus (PDB 1LDK). Ddb1 (light blue), zebrafish Ddb2 (dark blue), and 16 base pairs of dsDNA containing an Abasic site (red) are from Scrima et al. (2008) (PDB 3EI2) and aligned to BPB (β propeller B) of Ddb1 (PDB 2HYE).
Regulation of substrate recognition by E3 ubiquitin ligases
Many substrates of the ubiquitin proteasome system are not destroyed constitutively, but rather become unstable only in response to an endogenous or exogenous signal. Often, signaling leads to post-translational modification of the degron, which triggers binding to the ligase (Ravid and Hochstrasser 2008). For example, the E3 ligase Cul1–Skp1–FBW7 (CRL1FBW7; also known as SCFFBW7) recognizes the Cyclin E degron by this mechanism. FBW7 contains eight WD40 repeats that fold into an eight-bladed β propeller. The bottom surface of the propeller connects to an F-box motif that interacts with Skp1. The top surface of the propeller forms a surface that interacts specifically with the doubly phosphorylated degron of Cyclin E. Interestingly, some substrates such as Cdc6 and Skp2 can be protected from destruction when their degrons are phosphorylated (Mailand and Diffley 2005; Gao et al. 2009). Phosphorylation is not the only post-translational modification that regulates degrons. For instance, the E3 ligase CRL2VHL targets HIF1 (hypoxia-inducible factor-1) only when its degron motif is hydroxylated, ensuring that HIF1 protein levels increase under hypoxic conditions (Ivan et al. 2001; Jaakkola et al. 2001; Hon et al. 2002; Min et al. 2002). In another example, CRL1Fbx2 recognizes glycosylated proteins that are retro-translocated from the ER into the cytosol and targets them for destruction (Yoshida et al. 2002; Mizushima et al. 2007). Finally, some ubiquitin ligases interact preferentially with substrates that are sumoylated (Perry et al. 2008). Thus, post-translational modification of substrates represents a common means to couple the activity of E3 ligases to signaling events that sense intra- or extracellular conditions.
Notably, E3–substrate interactions are not always dependent on post-translation modification of the substrate. The anaphase-promoting complex/cyclosome (APC/C), a RING E3 ubiquitin ligase that promotes the exit from mitosis by destroying mitotic cyclins and other proteins, is itself phosphorylated. Phosphorylation of APC/C core subunits is required to allow activation of the complex and the interaction between APC/C and Cdc20 (Kraft et al. 2003), the WD40 substrate receptor that initially recruits APC/C substrates during mitotic exit (Pfleger et al. 2001). Conversely, the other APC/C substrate receptor, Cdh1, is inactive when phosphorylated by CDKs, ensuring that it acts after APC/CCdc20 (Zachariae et al. 1998; Jaspersen et al. 1999; Kramer et al. 2000).
CRL1TIR1 (SCFTIR1) is another example of a ubiquitin ligase that is modified to regulate destruction of a substrate, but in this case, the modification is noncovalent. In the absence of the plant hormone auxin, repressors block transcription of auxin-responsive factors. However, in the presence of auxin, the repressors are targeted for ubiquitin-mediated degradation. Strikingly, auxin binds directly to a pocket in the substrate receptor F-box protein TIR1, which stabilizes the interaction between CRL1TIR1 and its transcription repressor substrates, allowing them to be ubiquitylated and destroyed (Mockaitis and Estelle 2008; Tan and Zheng 2009).
In summary, the interaction of E3 ubiquitin ligases with their substrates is regulated in a number of different ways, most of which involve post-translation modification of the substrate or ligase, or employment of small molecule cofactors. In this review, we discuss in detail the E3 ubiquitin ligase CRL4Cdt2, whose interaction with substrates depends on a novel strategy that involves display of a degron motif on a cell cycle-regulated protein scaffold, chromatin-bound PCNA.
The architecture of CRL4Cdt2
CRL4 ubiquitin ligases consist of a cullin scaffold (Cul4), an adaptor protein (Ddb1), and a substrate receptor (DCAF [Ddb1- and Cul4-associated factor]) that binds directly to Ddb1. At least 20 bona fide DCAFs likely exist in mammalian cells (Angers et al. 2006; He et al. 2006; Higa et al. 2006b; Jin et al. 2006; Higa and Zhang 2007; O'Connell and Harper 2007; Hu et al. 2008; Lee et al. 2008; McCall et al. 2008; Scrima et al. 2008; Choe et al. 2009; Jackson and Xiong 2009; Xu et al. 2010). CRL4Cdt2 contains the DCAF Cdt2 (Cdc10-dependent transcript 2), also known as DCAF2, DTL, L2DTL, or RAMP, which was first discovered in fission yeast (Hofmann and Beach 1994). The overall architecture of CRL4Cdt2 probably conforms to the basic modular structure established for cullin-based ligases (Fig. 1A; Zheng et al. 2002). Like Cul1, Cul4 is an elongated α-helical protein that interacts through its C terminus with the ring finger protein Rbx1. The N terminus of Cul4 binds to Ddb1, which contains three β propellers (BPA, BPB, and BPC) that each comprise seven WD40 repeats or “blades.” BPB binds Cul4, whereas BPA and BPC form a clam-shaped structure that points toward the C terminus of Cul4. Although the structure of Cdt2 has not been solved, its sequence suggests that between residues 40 and 397 it contains a seven-bladed β propeller, as seen for many other CRL substrate receptors (data not shown). The structure of CRL4Cdt2 is likely to be similar to that of CRL4Ddb2, which promotes ubiquitylation of the nucleotide excision repair factor XPC in the context of UV damage (Sugasawa et al. 2005) and whose architecture can be modeled from two previous crystal structures (Fig. 1B; Angers et al. 2006; Scrima et al. 2008). If Cdt2 is indeed structurally analogous to Ddb2 (Fig. 1B), an α helix near the “bottom” of its β propeller would bind the cleft formed by BPA and BPC, whereas the “top” would interact with substrates, positioning them for ubiquitin transfer from the E2 protein. The functions of the N-terminal and C-terminal domains of Cdt2 that reside outside the β propeller are unknown.
Cdt1 destruction by CRL4Cdt2 is coupled to DNA replication and repair via PCNA
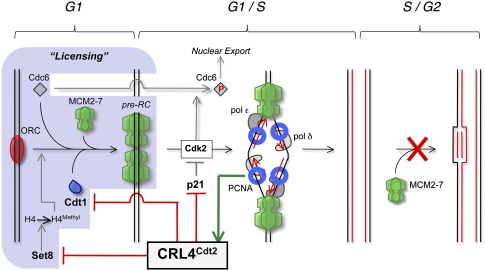
Among eukaryotes, there are six confirmed substrates of CRL4Cdt2 (see Fig. 2, bold names), but the mechanism of ubiquitylation has been most intensively studied for the replication licensing factor Cdt1 (Cdc10-dependent transcript 1, no structural relationship with Cdt2) (Hofmann and Beach 1994). Cdt1 is required for the recruitment of the MCM2–7 helicase to origins of DNA replication in G1 (the “licensing” reaction), but it is destroyed in S phase (Nishitani et al. 2004), which ensures that each origin of DNA replication undergoes only one initiation event per cell cycle (Fig. 3; for review, see Arias and Walter 2007). Early experiments in worms showed that the destruction of Cdt1 in S phase is dependent on Cul4 (Zhong et al. 2003). Soon thereafter, experiments in mammalian cells demonstrated that Cul4 and Ddb1 promote Cdt1 destruction after DNA damage (Higa et al. 2003; Hu et al. 2004). Subsequently, multiple groups showed that Cdt2 is also required for Cdt1 destruction (Higa et al. 2006a; Jin et al. 2006; Ralph et al. 2006; Sansam et al. 2006). It is now clear that CRL4Cdt2 ubiquitylates Cdt1 in S phase and after DNA damage in all metazoans and in fission yeast. Cdt2 likely functions as the substrate receptor for CRL4Cdt2 (Abbas et al. 2008), although this remains to be formally proven. While budding yeast contains Cul4 and Ddb1-like proteins (Zaidi et al. 2008), it lacks an identifiable Cdt2 ortholog, and Cdt1 is not unstable in S phase of these cells, suggesting that budding yeast lacks CRL4Cdt2.
Figure 2.
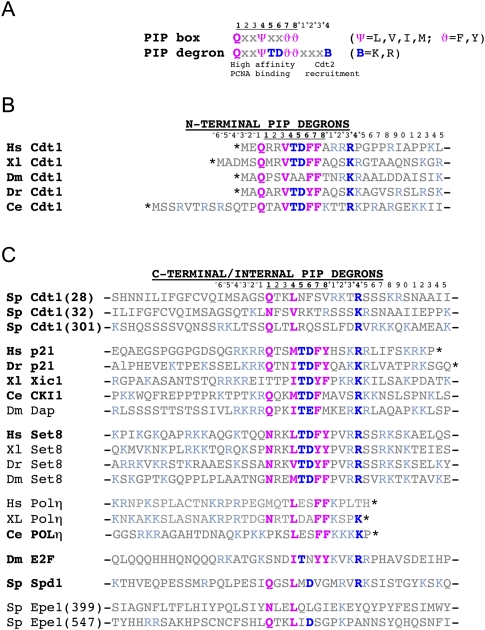
CRL4Cdt2 degrons. (A) PIP box and PIP degron consenuses. Canonical PIP box residues are shown in violet, and PIP degron-specific residues are shown in blue. The TD motif promotes high-affinity binding to PCNA, whereas the basic residue four amino acids downstream from the PIP box is required for docking of CRL4Cdt2 onto the PCNA–PIP degron complex. (B) The Cdt1 PIP degron sequence from various species. In all cases, the degron is at the extreme N terminus. Same color scheme as in A. (C) C-terminal and internal PIP degrons of various other substrates. Confirmed substrates are in bold. Asterisk denotes the C terminus.
Figure 3.
The role of CRL4Cdt2 in preventing rereplication. Origins of replication are primed for initiation in the G1 phase of the cell cycle via the ordered loading of ORC, Cdc6, Cdt1, and the MCM2–7 helicase (licensing), a process that requires histone H4 methylation on Lys 20 by Set8. In S phase, the MCM2–7 helicase is activated by Cdk2 and many additional factors, whereupon origins are unwound and two replisomes are assembled. Each replisome includes the MCM2–7 helicase, leading (pol ɛ) and lagging (pol δ) DNA polymerases, and the processivity factor PCNA. As replication proceeds, MCM2–7 travels away from the origin. Reinitiation is inhibited because once cells are in S phase, new MCM2–7 recruitment is not allowed, largely due to CRL4Cdt2, whose activity is coupled to chromatin-bound PCNA (green arrow). Thus, CRL4Cdt2 marks the licensing factors Cdt1 and Set8 for destruction (red arrows). In addition, it destabilizes p21, an inhibitor of Cdk2. Cdk2 activity in S phase promotes replication initiation but also phosphorylates Cdc6, leading to its export to the cytoplasm, where it is unavailable for licensing.
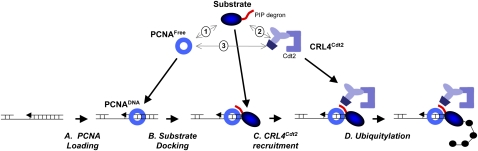
In a separate line of inquiry, experiments in Xenopus egg extracts showed that the ubiquitylation of Cdt1 during S phase occurs on chromatin and is strictly coupled to the process of DNA replication (Arias and Walter 2005). Analogously, after DNA damage, ubiquitylated Cdt1 also localizes to chromatin (Jin et al. 2006; Ishii et al. 2010; Roukos et al. 2011). Strikingly, all metazoan Cdt1 molecules contain a PCNA interaction protein motif (PIP box) at their extreme N termini (Fig. 2A,B), which binds to a hydrophobic pocket within PCNA that is composed of residues from the interdomain connector loop and the C terminus of PCNA (Fig. 4A). The PIP box is essential for Cdt1 destruction in S phase and after DNA damage (Arias and Walter 2006; Higa et al. 2006a; Hu and Xiong 2006; Nishitani et al. 2006; Senga et al. 2006). Importantly, CRL4Cdt2 itself also binds to chromatin during S phase and after DNA damage (Jin et al. 2006; Ishii et al. 2010; Roukos et al. 2011). This binding is dependent on prior interaction of a CRL4Cdt2 substrate with PCNA on chromatin (Jin et al. 2006; Ishii et al. 2010). If DNA replication or repair are inhibited upstream of PCNA loading, Cdt1 and CRL4Cdt2 do not bind to chromatin and Cdt1 is not destroyed (Arias and Walter 2005, 2006; Havens and Walter 2009; Guarino et al. 2011). Together, the data suggest a model wherein Cdt1 first uses its PIP box to dock onto PCNA that is engaged in replication or repair synthesis; subsequently, CRL4Cdt2 is recruited, whereupon Cdt1 is polyubiquitylated on chromatin and destroyed (Fig. 5).
Figure 4.
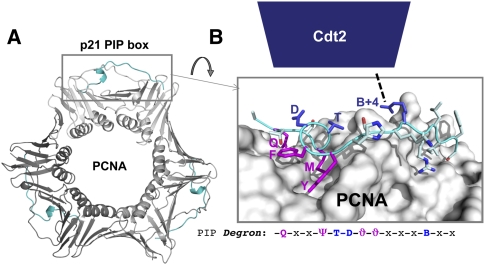
Model for docking of CRL4Cdt2 onto the PCNA–PIP degron complex. (A) Crystal structure of PCNA with the PIP degron of p21 (residues X to Y) (Gulbis et al. 1996). The p21 peptide is show in cyan. (B) Close-up of the PCNA–p21 complex, including the PIP degron sequence (same color coding as in Fig. 2A) and a putative contact between the B+4 residue and Cdt2 (dashed line).
Figure 5.
Model for assembly of the PCNA–PIP degron–CRL4Cdt2 complex on DNA. (A) RFC loads PCNA onto a primer–template junction during DNA replication or repair-associated gap filling. (B) A PIP degron-containing protein docks onto the PCNA–DNA complex. (C) CRL4Cdt2 recognizes the PIP–degron–PCNA complex. (D) CRL4Cdt2 promotes ubiquitin transfer to the substrate. The three double arrows depict possible interactions between PCNA, the PIP degron, and CRL4Cdt2 in the absence of DNA (see the text for details).
CRL4Cdt2 recognizes a ‘PIP degron'
The discovery that Cdt1 destruction is dependent on the binding of its PIP box to PCNA raised the question of why most PIP box-containing proteins, such as DNA polymerases and DNA ligases, are not targeted by CRL4Cdt2. The answer emerged from a comparison between known CRL4Cdt2 substrates and canonical PIP box-binding proteins, which showed that the former contain a specialized PIP box dubbed a “PIP degron” (Havens and Walter 2009; Michishita et al. 2011). The PIP degron contains at least two essential elements (Fig. 2A). As discussed above, the first is a PIP box, which is crucial for binding to chromatin-bound PCNA (PCNADNA). Interestingly, most CRL4Cdt2 substrates contain a TD motif at positions 5 and 6 of the PIP box (Fig. 2A), which forms a loop that protrudes off the surface of PCNA (Fig. 4A). The TD motif is not usually considered part of the PIP consensus (Moldovan et al. 2007). However, in combination with the classic PIP box residues (Fig. 2A, pink residues), the TD motif confers unusually high-affinity PCNA binding (Nakanishi et al. 1995; Warbrick et al. 1995; Chuang et al. 2005; Havens and Walter 2009; Michishita et al. 2011). Thus, effective CRL4Cdt2-dependent destruction of substrates requires that they bind with high affinity to PCNA, perhaps to ensure processive ubiquitylation. The high-affinity binding might also ensure that CRL4Cdt2 substrates are destroyed before other PCNA-dependent processes take place. The few CRL4Cdt2 substrates that lack the TD motif (e.g., Dm Cdt1, Sp Cdt1, Ce polH, Dm E2F) (Fig. 2C) likely compensate with other residues to achieve efficient PCNA binding. It is interesting to note that most PIP box proteins that are not CRL4Cdt2 substrates lack a TD motif and thus bind to PCNA with suboptimal affinity (data not shown). Indeed, addition of the TD motif to the PIP boxes of Fen1 or DNA ligase dramatically increases their affinities for PCNA (Havens and Walter 2009; Michishita et al. 2011). Suboptimal binding of canonical PIP box proteins to PCNA may be important to maintain appropriate exchange of PCNA binding proteins at the replication fork and during DNA repair (Fridman et al. 2010). In contrast, high-affinity binding of CRL4Cdt2 substrates to PCNA is clearly not deleterious, likely because they are destroyed.
The second essential element of the PIP degron, which is conserved in all known CRL4Cdt2 substrates, is a basic residue four amino acids downstream from the PIP box (Fig. 2A). This residue, called “B+4,” is surface-exposed in the p21–PCNA cocrystal structure (Fig. 4B; Gulbis et al. 1996). Importantly, mutation of B+4 abolishes docking of CRL4Cdt2 onto the PCNA–Cdt1 complex and inhibits Cdt1 destruction, but it has no effect on the binding of Cdt1 to PCNA (Havens and Walter 2009; Michishita et al. 2011). Thus, B+4 likely makes specific contact with acidic residues on the top face of Cdt2 (Fig. 4B, dashed line).
The PIP degron is portable, since canonical PIP box proteins such as Fen1 or DNA ligase can be converted into a CRL4Cdt2 substrate by introducing the TD and B+4 motifs into its PIP box (Havens and Walter 2009; Michishita et al. 2011). In addition, an ∼25-amino-acid peptide containing the PIP box of human Cdt1 is sufficient to confer CRL4Cdt2-mediated destruction on GST (Nishitani et al. 2006; Senga et al. 2006). Together, these results indicate that the PIP degron, which has the consensus Q-x-x-Ψ-T-D-ϑ-ϑ-x-x-x-B (Fig. 2A), is necessary and sufficient to interact with CRL4Cdt2 in the context of chromatin-bound PCNA and to promote protein destruction.
Other amino acids that are closely linked to the PIP box play roles in some CRL4Cdt2 targets. For example, several substrates including the CDK inhibitors (CKIs) contain basic residues at the +3 and +5 positions (Fig. 2C), which interact with acidic residues within PCNA in the p21–PCNA crystal structure (Gulbis et al. 1996). Indeed, mutation of B+5 in Xenopus Cdt1 and human p21 reduced destruction due to impaired PCNA binding (Nishitani et al. 2008; Havens and Walter 2009). However, in some CRL4Cdt2 targets, the absence of this residue can apparently be compensated for by other amino acids. Mutation of B+3 to alanine also inhibited HsCdt1 destruction (Michishita et al. 2011), and this is probably because only basic or polar amino acids appear to be tolerated at this position (Fig. 2). Finally, many substrates in which the PIP box is internal or C-terminal contain a cluster of positively charged amino acids upstream of the PIP box (Fig. 2C, light-blue residues). In the context of human p21 or Xenopus Xic1, it appears these residues can play a role in CRL4Cdt2-mediated destruction (Chuang and Yew 2005; Nishitani et al. 2008; Michishita et al. 2011). However, it is unclear whether this is due to impaired PCNA binding or defective CRL4Cdt2 recruitment.
Recently, the determinants that trigger SpCdt1 were investigated (Guarino et al. 2011). As seen for many other PCNA-binding proteins in yeast, it lacks aromatic residues (Fig. 2B). Interestingly, SpCdt1 contains three putative PIP boxes, which all contain a B+4 (Fig. 2C). Mutation of the B+4 in SpCdt1(28) alone does not appear to be required for destruction after induction of DNA damage; however, one of the other two PIP degrons may compensate. Indeed, when all three PIP degrons were inactivated at the same time, SpCdt1 was stabilized. Further work will be required to precisely delineate the PIP degrons in SpCdt1, as well as SpSpd1 and SpEpe1.
In summary, in higher eukaryotes, CRL4Cdt2 recognizes a PIP degron that must be displayed on chromatin-bound PCNA to be functional. Although most PIP degrons contain a canonical PIP box, a TD motif, and a B+4 residue, a small number of CRL4Cdt2 substrates lack the TD motif sometimes, the B+4 residue may not be essential (Abbas et al. 2010). This suggests that there may exist many substrates that are not identifiable by searching the protein database using the PIP degron consensus shown in Figure 2A.
Mechanisms that couple CRL4Cdt2 activity to S phase and DNA damage
CRL4Cdt2-mediated proteolysis occurs only in S phase and after DNA damage because this ubiquitin ligase promotes ubiquitylation of substrates only on the DNA-bound fraction of PCNA (You et al. 2002; Arias and Walter 2005, 2006; Chuang and Yew 2005; Havens and Walter 2009; Guarino et al. 2011; Jorgensen et al. 2011). What is the mechanistic basis for the coupling to PCNADNA, or, why does soluble PCNA not promote CRL4Cdt2 activity? In Xenopus egg extracts, endogenous Cdt1 does not interact with soluble PCNA (PCNAsoluble) (Fig. 5, double arrow #1) but only with PCNA that is bound to PCNADNA (Havens and Walter 2009). As a result, Cdt1 and CRL4Cdt2 are only able to interact on PCNADNA (Fig. 5). How the selective binding of Cdt1 to PCNADNA comes about is currently unclear. However, it does not require CDK, ATR, ATM, or DNA-PK, since none of these protein kinases are required for Cdt1 destruction (Higa et al. 2003; Arias and Walter 2006; Jin et al. 2006; Ralph et al. 2006). Interestingly, an artificial PIP degron (Fen1 protein mutated to contain the TD and B+4 motifs) was found to bind to PCNAsoluble. However, this interaction does not support CRL4Cdt2 recruitment, demonstrating that CRL4Cdt2 senses whether DNA is present in the PCNA–PIP degron complex (Havens and Walter 2009). The requirement for DNA is not understood, but it could reflect direct interactions of CRL4Cdt2 with DNA or the involvement of other DNA-bound proteins. In summary, CRL4Cdt2 activity is coupled to PCNADNA by at least two mechanisms. First, some substrates, like Cdt1, bind only to PCNADNA. Second, a substrate–PCNAsoluble complex does not serve as a docking site for CRL4Cdt2. In theory, steps even further downstream, such ubiquitin transfer to the substrate itself, might also be DNA-dependent, but this remains to be examined.
Mechanism of PCNA–PIP degron–CRL4Cdt2 complex assembly
The molecular architecture of the ternary PCNADNA–PIP degron–CRL4Cdt2 complex and the role of PCNA in promoting ubiquitin transfer are not well understood. As noted above, Xenopus Cdt1 does not interact with PCNAsoluble or CRL4Cdt2 in the absence of DNA (Fig. 5, gray double-headed arrows #1 and #2). Instead, Cdt1 docks onto PCNADNA, and its B+4 residue is then instrumental in recruiting CRL4Cdt2 (Fig. 5). Recently, we identified residues in the interdomain connector loop of PCNA that are essential for recruitment of CRL4Cdt2 to the PIP degron–PCNA complex but do not affect binding of the PIP degron to PCNA (CG Havens and JC Walter, unpubl.). This argues that CRL4Cdt2 contacts residues not only in the PIP degron but also in PCNA. As such, CRL4Cdt2 effectively recognizes a composite interface formed by two distinct polypeptides: PCNA and the PIP degron of the substrate. Since soluble Cdt1 and CRL4Cdt2 do not interact, PCNADNA functions as a matchmaker that brings these two components together.
Some studies have reported that certain substrates (like p21) interact with CRL4Cdt2 in the absence of PCNA (Fig. 5, gray double arrow #2; Hu et al. 2004; Liu et al. 2005; Higa et al. 2006b; Kim and Kipreos 2007; Abbas et al. 2008, 2010; Kim et al. 2008). Since destruction of all substrates requires PCNADNA, this result would imply that PCNADNA acts on the preformed substrate–ligase complex to facilitate ubiquitin transfer. For example, PCNADNA might induce an allosteric change in the substrate or CRL4Cdt2 that potentiates ubiquitin transfer. However, a caveat in virtually all experiments showing substrate–CRL4Cdt2 interaction in the absence of PCNA is that the binding is detected when one or both components are overexpressed. An alternative interpretation of the data is that the detected interactions contribute to the stability of the ternary complex, but only in the context of PCNADNA.
Recent biochemical experiments suggest that the C-terminal half of Xenopus Cdt2 interacts directly with PCNA, independently of substrate (Fig. 5, gray double arrow #3; Kim et al. 2010), and imaging of Cdt2 in mammalian cells is consistent with this idea (Roukos et al. 2011). Although deletion of the C-terminal 310 amino acids of Cdt2 disrupted the interaction with PCNA and inhibited CRL4Cdt2 function, this deletion likely caused large-scale disruption of the Cdt2 structure. In the future, targeted disruption of the CRL4Cdt2–PCNA interaction will be required to examine its relevance for CRL4Cdt2 function.
CRL4Cdt2 substrate ubiquitylation and beyond
The residues that are ubiquitylated by CRL4Cdt2 have been studied in three substrates: Cdt1, p21, and Xic1. A fragment of human Cdt1 containing amino acids 1–98 in which all the lysines were mutated to arginines was still destroyed, albeit slowly (Senga et al. 2006). These data suggest that in the absence of lysine acceptor sites for ubiquitin, the N-terminal methionine could be used. In contrast, when all of the lysines in p21 were mutated to arginines, p21 was stabilized (Bendjennat et al. 2003; Stuart and Wang 2009). In Xic1, mutation of five lysine residues just downstream from the PIP box reduced polyubiquitylation and destruction (Kim et al. 2010). Given the ubiquitylation data from Xic1 and the fact that short peptides of CRL4Cdt2 substrates containing the PIP boxes were destroyed (You et al. 2002; Chuang and Yew 2005; Nishitani et al. 2006; Senga et al. 2006), it is likely that multiple lysines upstream of or downstream from the PIP box can function as ubiquitin acceptors. Determining the preferred sites of CRL4Cdt2-dependent ubiquitylation requires more work.
Recently, it was discovered that CRL4Cdt2 uses two separate E2 ubiquitin-conjugating enzymes to target different substrates (Shibata et al. 2011). Thus, it cooperates with UBE2G to ubiquitylate Cdt1 and UBCH8 to ubiquitylate Set8 and p21. E2s can regulate the specificity for ubiquitin nucleation on the substrate and chain formation on the preceding ubiquitins by recognizing the surface of ubiquitin or the substrate near the lysine to be ubiquitylated (Jin et al. 2008; Ye and Rape 2009; Rodrigo-Brenni et al. 2010; Saha et al. 2011). We speculate that the location of the PIP box (N- vs. C-terminal) and possible contributions from surrounding residues in substrates might necessitate the use of two different E2 enzymes.
An important question is how ubiquitylated Cdt1 is removed from PCNADNA. A recent genome-wide siRNA screen identified the ATPase p97 (also known as VCP or Cdc48p) and its cofactor, UFD1, as being essential for Cdt1 destruction (Raman et al. 2011). Cells treated with siRNA against p97 or UFD1 accumulated polyubiquitylated Cdt1 and Set8 on chromatin. Therefore, p97-UFD1 likely functions downstream from CRL4Cdt2 ubiquitylation to extract substrates from the chromatin so they can be degraded by the proteasome.
The biology of CRL4Cdt2 targets
In addition to Cdt1, there are five confirmed substrates of CRL4Cdt2 (Fig. 2). Below, we discuss these substrates, focusing on the biological importance of their destruction in S phase and after DNA damage.
Spd1
The first confirmed substrate of CRL4Cdt2 was the Schizosaccharomyces pombe protein Spd1 (Liu et al. 2003, 2005; Holmberg et al. 2005), an inhibitor of the ribonucleotide reductase (RNR) enzyme that catalyzes the synthesis of dNTPs. Spd1 inhibits RNR through at least two mechanisms (Nestoras et al. 2010), and it is destroyed in S phase and after DNA damage (Liu et al. 2003). Importantly, mutations in CRL4Cdt2 induce mutagenesis, cause slow growth, and prevent double-strand break repair in S. pombe, and these defects are partially or completely alleviated if Spd1 is also deleted (Liu et al. 2005; Moss et al. 2010). Thus, a major function of CRL4Cdt2 in S. pombe is the elimination of Spd1 in S phase and after DNA damage so that dNTPs are available for replicative and repair DNA synthesis. Although Spd1 contains a sequence that conforms to the PIP degron consensus (Fig. 2C), its role in mediating the protein's destruction has not been reported. Interestingly, S-phase-dependent destruction of Spd1 does not require checkpoint signaling; however, damage-dependent Spd1 destruction does require the checkpoint (Liu et al. 2003). This is because transcriptional up-regulation of Cdt2 after DNA damage involves checkpoint kinases (Liu et al. 2005), but the underlying mechanism is unknown. It seems that this transcriptional regulation of Cdt2 function is unique to S. pombe.
E2F
Most metazoan cells express two or more E2F family transcription factors. The “activator E2Fs” promote transcription of genes in G1 whose expression is essential to enter S phase. Activator E2F is negatively regulated in early G1 phase by the retinoblastoma gene product Rb. In addition, “repressor E2Fs” inhibit the activator E2F by competing for its binding partner, Dp, which is essential for E2F DNA binding. In Drosophila, the activator E2F, E2f1, is antagonized by Rb and the repressor E2F, E2f2, but neither inhibitory pathway is essential for development, suggesting the existence of additional mechanisms that restrain E2f1 activity. Indeed, it was noted that E2f1 is specifically destroyed in S phase (Asano et al. 1996). Duronio and colleagues (Shibutani et al. 2008) then discovered that this process requires a PIP degron in E2F1, as well as components of CRL4Cdt2. Interestingly, CRL4Cdt2-dependent E2f1 destruction also requires Dp, suggesting that assembly of the PCNADNA–E2f1–CRL4Cdt2 complex might require DNA binding by E2f1–Dp. This could help link destruction to the DNA-bound form of PCNA. Compared with E2f1, ectopic expression of E2f1ΔPIP caused increased expression of an E2f1 target gene, a reduced G1 population, developmental defects, and apoptosis, indicating that E2f1ΔPIP functions as a gain-of-function allele in vivo. Given that E2Fs in other species do not contain recognizable PIP degrons, it appears unlikely that CRL4Cdt2-dependent destruction of activator E2Fs is conserved in other species. Instead, mammalian cells appear to contain multiple pathways to restrain E2F activity once cells have entered S phase (Dimova and Dyson 2005). At present, it has not been examined whether Drosophila E2F is destroyed after DNA damage.
DNA polymerase (pol) η
Pol η is a translesion DNA polymerase that specializes in the bypass of UV-induced pyrimidine dimers. Interestingly, Caenorhabditis elegans pol η suppresses the DNA damage-induced cell cycle checkpoint during early embyrogenesis, presumably by promoting efficient bypass of lesions before they lead to ATR checkpoint activation, a function that is shared by a sumo E3 ligase called gei-17 (Kim and Michael 2008). The checkpoint is likely suppressed because embryogenesis is extremely sensitive to perturbations of cell cycle progression. Strikingly, in embryos lacking gei-17, DNA damage induces the efficient destruction of pol η by CRL4Cdt2 dependent on a PIP degron within pol η (Fig. 2). Gei-17 protects pol η from CRL4Cdt2 modification via direct, DNA damage-induced sumoylation. Importantly, when wild-type embryos are treated with high levels of DNA-damaging agents, CRL4Cdt2-dependent pol η destruction become detectable. Based on these results, it was proposed that in early C. elegans embryos, Gei-17 normally sumoylates pol η at sites of DNA damage and thereby protects it from CRL4Cdt2-mediated destruction. After lesion bypass, sumoylation is reversed, allowing targeting and subsequent eviction of the error-prone pol η by CRL4Cdt2. In the future, it will be important to test this interesting model explicitly.
Cdt1
As mentioned above, CRL4Cdt2 targets Cdt1 for destruction in S phase and after DNA damage. The function of the S-phase destruction is clearly to prevent rereplication. This is evident from the fact that overexpression of Cdt1 is sufficient to promote DNA rereplication in various systems (Vaziri et al. 2003; Arias and Walter 2005; Li and Blow 2005; Takeda et al. 2005; Yoshida et al. 2005; Kerns et al. 2007). Moreover, Cdt1 lacking a PIP box is more potent than wild-type Cdt1 in promoting rereplication (Arias and Walter 2006). Interestingly, expression of nondegradable Cdt1 in fly embryonic epidermal cells caused S-phase defects and failure of cells to enter mitosis, but in follicle cell endocycles, Geminin was able to restrain Cdt1 activity (Lee et al. 2010). This result suggests a differential requirement for CRL4Cdt2 in maintaining genome stability in different tissues.
When asynchronously growing cells are UV-irradiated, the G1-phase population of Cdt1 is destroyed within minutes via CRL4Cdt2 (Higa et al. 2003). It was proposed that this destruction represents a checkpoint to prevent S-phase entry when cells experience genotoxic stress. However, MCM2–7 complexes are already loaded onto DNA in telophase and Cdt1 is not required for initiation from loaded MCM2–7 complexes. Therefore, such a DNA damage-induced initiation checkpoint would only be effective if MCM complexes dissociate after Cdt1 destruction. Dissociation may not have to be complete to block S-phase entry given the proposal that cells contain a licensing checkpoint that blocks S phase when there are insufficient levels of chromatin-bound MCM2–7 complexes (Machida et al. 2005; Teer et al. 2006; Liu et al. 2009). An alternative role for DNA damage-induced Cdt1 destruction is in the prevention of rereplication in G2-phase cells (Arias and Walter 2007). Thus, when G2 cells experience replicative stress, CDKs are inhibited to block cell cycle progression. However, this likely removes an important barrier against rereplication. To compensate, cells might replace the CDK-dependent CRL1Skp2-mediated destruction of Cdt1 that occurs in G2 with CRL4Cdt2-mediated destruction, which is normally turned off at this time in the cell cycle (Nishitani et al. 2006). Unfortunately, there is no established way to separate the function of CRL4Cdt2-mediated destruction in S phase and after DNA damage, making it difficult to specifically address the function of UV-dependent Cdt1 destruction.
CKIs
Mammalian cells express three major CKIs, p21Cip1, p27Kip1, and p57Kip2, whose abundance during the cell cycle is carefully regulated. Thus, p21 is expressed in G1 and G2, but its level drops dramatically in S phase and after UV irradiation (Amador et al. 2007; Abbas et al. 2008; Kim et al. 2008). While several studies showed that p21 levels are negatively regulated by the APC/C in mitosis and by CRL1Skp2 (Yu et al. 1998; Bornstein et al. 2003; Wang et al. 2005; Amador et al. 2007), the disappearance of p21 in S phase remained unexplained. Moreover, conflicting theories accounted for the destruction of p21 after UV irradiation (for review, see Abbas et al. 2008). In 2008, four studies reported that human p21 is a CRL4Cdt2 target (Abbas et al. 2008; Kim et al. 2008; Nishitani et al. 2008; Stuart and Wang 2009). Thus, knockdown of Cul4, Ddb1, Cdt2, or PCNA inhibits p21 destruction in S phase and after damage, as does mutation of p21's PIP box, which conforms perfectly to the PIP degron consensus (Fig. 2C). Moreover, purified CRL4Cdt2 is able to ubiquitylate p21 in vitro and in vivo. The destruction of CKIs by CRL4Cdt2 is highly conserved, as this phenomenon is observed for Xic1 (a Xenopus CKI) in frogs (You et al. 2002; Chuang and Yew 2005; Chuang et al. 2005; Kim et al. 2010), and the CKI in worms (CKI-1) (Kim et al. 2007, 2008). Although it has not been confirmed, the Drosophila CKI Dacapo (Dap) is also a likely target, since it contains a nearly perfect PIP degron (Fig. 2C). While it appears that CRL4Cdt2 is solely responsible for the UV-induced destruction of p21, the relative roles of CRL4Cdt2 and CRL1Skp2 in the S-phase destruction of p21 remains to be fully resolved, although both ligases appear to make significant contributions (Abbas et al. 2008; Kim et al. 2008; Nishitani et al. 2008; Stuart and Wang 2009).
The first hints of why CKIs are targeted for destruction in S phase came from studies in worms. Kipreos and colleagues (Zhong et al. 2003; Kim and Kipreos 2007) showed that mutations in CRL4Cdt2 cause accumulation of Cdt1 and CKI-1, as well as massive rereplication. Importantly, this phenotype is largely reversed when CKI-1 or Cdt1 are cosilenced, suggesting that the destruction of either factor in S phase is sufficient to prevent rereplication. They then showed that CKI-1 exerts its effect through the licensing protein Cdc6. Thus, previous experiments had shown that Cdc6 is phosphorylated by Cdk2 in S phase, which leads to its nuclear export, but the relevance of this pathway in controlling replication had been unclear (for review, see Arias and Walter 2007). Kim et al. (2007) revealed that destruction of CKI-1 is required for the export of Cdc6, presumably because persistent CKI-1 inhibits Cdk2, blocking Cdc6 phosphorylation and export. Expressing nondegradable Cdt1 together with nonexportable Cdc6 was sufficient to cause rereplication (Kim et al. 2007). These experiments support the model that CRL4Cdt2 inhibits licensing by two largely redundant means: (1) destruction of the licensing factor Cdt1 and (2) destruction of CKI-1, which maintains CDK2 activity during S phase, thus promoting the nuclear export of Cdc6 (Fig. 3). The CRL4Cdt2-dependent destruction of CKIs in S phase has been confirmed in mammalian cells (Kim et al. 2008; Nishitani et al. 2008), where it also prevents rereplication by promoting the nuclear export of Cdc6 (Kim et al. 2008).
The reason for p21 destruction after UV damage is not fully understood. One possibility is that p21 must be destroyed to alleviate its inhibition of PCNA so that PCNA can participate in repair. It is clear in vitro that p21 can inhibit DNA replication and repair through direct binding to PCNA via the PIP box in p21 (Chen et al. 1995; Cooper et al. 1999; Bendjennat et al. 2003). However, what is unclear is whether this occurs in vivo. Some laboratories report inhibition of DNA repair synthesis or nucleotide excision repair in cultured cells upon overexpression of the p21 PIP box (Cayrol et al. 1998; Cooper et al. 1999; Bendjennat et al. 2003; Cazzalini et al. 2003), while others show no effect of p21 on nucleotide excision repair (Bates et al. 1998; Medema et al. 1998; Niculescu et al. 1998; Adimoolam et al. 2001; Soria et al. 2008). Other evidence indicates that the PIP box region of p21 can block pol η and PCNA foci from forming and therefore negatively regulates translesion DNA synthesis (Soria et al. 2008). Thus, although the CRL4Cdt2-dependent destruction of p21 after DNA damage is now firmly established, further work is needed to determine the precise role of this proteolysis event in cell cycle progression and DNA repair.
Interestingly, ATR is involved in DNA damage-induced p21 destruction through activation of GSKβ, which phosphorylates p21 on S114 (Bendjennat et al. 2003; Lee et al. 2007). While a phospho-mimetic substitution of S114 enhanced p21 ubiquitylation by CRL4Cdt2 in vitro (Abbas et al. 2008), this phosphorylation event has so far not been directly linked to CRL4Cdt2 function in vivo. If CRL4Cdt2-dependent p21 destruction requires substrate phosphorylation by ATR, it will represent an interesting exception, since Cdt1 and Set8 destruction are independent of checkpoint signaling (Higa et al. 2003; Arias and Walter 2006; Ralph et al. 2006; Centore et al. 2010).
Set8
Set8 (PR-Set7 or KMT5A) is the sole histone methyltransferase that monomethylates histone H4 on Lys 20. In the absence of Set8, cells arrest in G2 and fail to condense their chromosomes (Houston et al. 2008). Set8 expression is normally low in G1, ceases just before S-phase entry, remains absent in S phase, and then rises to high levels in the G2 phase and in mitosis (Rice et al. 2002; Yin et al. 2008; Oda et al. 2010). Consistent with this expression profile of Set8, H4K20me1 levels are also low in S phase and rise late in G2/M (Rice et al. 2002). The Set8 expression profile appears to result from the action of at least three distinct E3 ubiquitin ligases (for review, see Wu and Rice 2011). Thus, APC/CCdh1 targets Set8 during G1 (Wu et al. 2010), and CRL1Skp2 further reduces its levels in late G1 (Yin et al. 2008). The absence of Set8 in S phase and after DNA damage is due to the action of CRL4Cdt2 (Abbas et al. 2010; Centore et al. 2010; Oda et al. 2010; Tardat et al. 2010; Jorgensen et al. 2011), which recognizes a highly conserved PIP degron in Set8.
To determine why Set8 is eliminated in S phase by CRL4Cdt2, several groups overexpressed a PIP box mutant of Set8 (Set8ΔPIP) that cannot be destroyed in S phase. Set8ΔPIP expression caused elevated H4K20me1 in S phase, DNA damage, and checkpoint activation, as well as G2 arrest and loss of cell proliferation (Abbas et al. 2010; Centore et al. 2010; Tardat et al. 2010; Jorgensen et al. 2011). Various explanations for these phenotypes were put forward. Two reports observed that Set8ΔPIP caused premature chromatin compaction in S phase, which was attributed to abnormal H4 methylation, and this compacted chromatin structure was proposed to underlie the observed DNA damage and G2 arrest (Centore et al. 2010; Jorgensen et al. 2011). In contrast, another study found that Set8ΔPIP expression causes loss of histone gene expression and chromatin decondensation, and these defects were invoked to explain the effect of Set8 on cell cycle progression (Abbas et al. 2010). Interestingly, two groups also observed that Set8ΔPIP caused substantial rereplication, as seen by the accumulation of cells with >4N DNA content (Abbas et al. 2010; Tardat et al. 2010). Notably, Tardat et al. (2010) showed that Set8 binds to origins of replication in G1 and promotes local H4 monomethylation. Furthermore, silencing of Set8 inhibited MCM2–7 recruitment to origins. Thus, Set8 might be a bona fide licensing factor, which must be down-regulated in S phase to prevent rereplication. In this view, the primary defect of Set8ΔPIP expression is rereplication, which is known to cause DNA damage and G2 arrest (Hook et al. 2007). A potential complication with all of the above experiments that might explain some of the variability observed is that Set8ΔPIP was expressed ectopically, in many cases from very strong promoters. Ideally, Set8ΔPIP should be knocked into the endogenous Set8 locus and the resulting phenotype examined.
Like Spd1, E2F, Cdt1, and p21, Set8 is also targeted for destruction by CRL4Cdt2 after DNA damage (Abbas et al. 2010; Centore et al. 2010; Oda et al. 2010; Jorgensen et al. 2011). Interestingly, Set8 is proposed to play a positive role in the DNA damage response, as Set8 localizes to laser-induced DNA double-stranded breaks in a PCNA-dependent manner, and its catalytic activity is required for the recruitment of 53BP1 to the breaks (Oda et al. 2010). This raises the question of why Set8 might be destroyed after damage. Notably, Set8 destruction occurs well after Set8 and 53BP1 are recruited to damage foci (Oda et al. 2010). Thus, Set8 proteolysis might contribute to the completion of DNA repair and/or help turn off DNA damage signaling, but this model would have to be tested explicitly. It should be noted that for some CRL4Cdt2 substrates that are destroyed in S phase and after DNA damage, only the S phase or DNA damage pathway may have functional consequences.
The role of Cdt2 in cell physiology
The disruption of Cdt2 expression in various systems has shed light on the role of CRL4Cdt2 in cell physiology. Homozygous Cdt2−/− mouse embryos die at the two- to four-cell stage with an abnormal nuclear morphology whose cause is unknown (Liu et al. 2007). This observation shows that Cdt2 is essential for cell proliferation and development, possibly due to critical roles in chromosome duplication. In tissue culture cells, silencing of Cdt2 via siRNA leads to G2 arrest, rereplication, and centrosome amplification (Jin et al. 2006; Sansam et al. 2006; Kim et al. 2008). The G2 arrest and rereplication defect was also seen in fish embryos carrying a Cdt2 mutation (Sansam et al. 2006). Since G2 arrest and DNA damage are observed during rereplication (Hook et al. 2007), the primary cause of these defects may be rereplication. This interpretation is bolstered by the fact that all three vertebrate CRL4Cdt2 substrates identified (Cdt1, CKIs, and Set8) are implicated in preventing rereplication (see above). Importantly, cosilencing of p21 with Cdt2 completely suppresses rereplication (Kim et al. 2008), likely because p21 inhibition allows Cdk2-mediated export of the licensing factor Cdc6 (see above). Therefore, the destruction of p21 in S phase is sufficient to suppress measurable rereplication. Similarly, cosilencing of Cdt1 suppresses some of the phenotypes exhibited by Cdt2 mutant fish embryos (Sansam et al. 2006). It will be interesting to determine the effect of cosilencing Set8 with Cdt2, as this will test the idea that CRL4Cdt2-mediated destruction of p21, Cdt1, or Set8 in S phase is sufficient to block rereplication. In summary, by targeting these three distinct licensing activities, vertebrate CRL4Cdt2 functions as a master regulator of rereplication. Importantly, as illustrated by E2F and DNA pol η, CRL4Cdt2 likely also has important roles outside the suppression of rereplication.
Interestingly, aspects of CRL4Cdt2 inhibition are phenocopied by MLN4924, a new small molecule that inhibits the CRL NEDD8-activating enzyme (NAE) (Soucy et al. 2009). Conjugation of the ubiquitin-like protein NEDD8 is required to activate CRL ligases (Duda et al. 2008; Deshaies et al. 2010). Thus, inhibiting the NAE by MLN4924 prevents destruction of numerous CRL substrates involved in cell proliferation and cancer pathways, including Cdt1, p21, Cyclin D, and Cyclin E (Soucy et al. 2009; Lin et al. 2010; Milhollen et al. 2011). Strikingly, treatment of cancer cells with MLN4924 triggers rereplication, DNA damage, G2 arrest, and apoptosis. However, when the cells were also treated with siRNA against Cdt1, rereplication is prevented (Lin et al. 2010; Milhollen et al. 2011). MLN4924 is currently in clinical trials as an anti-cancer agent. In the future, it will be interesting to determine whether the stabilization of other CRL4Cdt2 targets, principally p21 and Set8, contributes to the rereplication and anti-proliferative effects of MLN4924.
New substrates of CRL4Cdt2?
To fully understand the biology and mechanism of CRL4Cdt2, it will be important to determine all the substrates that are regulated by this E3 ubiquitin ligase. In addition to the six proteins discussed in the previous section that are confirmed targets of CRL4Cdt2, there are a few other likely substrates.
S. pombe Epe1 is an anti-silencing factor that contains a catalytically inactive JmjC-type demethylase domain. Epe1 is concentrated at the boundaries between heterochromatin and euchromatin and prevents the ectopic spread of heterochromatin (Braun et al. 2011). Interestingly, CRL4Cdt2 is required to prevent the accumulation of Epe1 in the body of heterochromatin and to maintain gene silencing. In the absence of CRL4Cdt2, the turnover of Epe1 decreased and its steady-state levels increased, including during S phase. Epe1 ubiquitylation also depended on Ddb1 (Cdt2 was not tested), and a two-hybrid interaction was detected between Epe1 and Cdt2. While these observations provide strong evidence that Epe1 is a CRL4Cdt2 substrate, it will be important to identify the PIP degron in Epe1 and confirm that Epe1 proteolysis depends on PCNA. Interestingly, Epe1 contains two QxxL PIP boxes (Fig. 2C). While they lack a B+4 residue, they both contain lysines at the +2 position, which is also observed for one S. pombe Cdt1 PIP box and for a putative Spd1 PIP box (Fig. 2C). We speculate that in fission yeast, CRL4Cdt2 recognizes a B+2 and/or B+4 residue. If Epe1 is indeed targeted by the canonical PCNA and CRL4Cdt2 pathway, it will be interesting to determine how Epe1 destruction is confined to heterochromatic regions.
In zebrafish and humans, deletion of Cdt2 causes failure of the G2/M checkpoint after ionizing radiation. In fish, this defect is not rescued when Cdt1 is also silenced (Sansam et al. 2006). These data raise the interesting possibility that Cdt2 is directly involved in initiating G2/M arrest, or that a protein other than Cdt1 must be destroyed by CRL4Cdt2 to activate the checkpoint. Since it is not clear how destruction of any known CRL4Cdt2 substrates would accomplish this function, there might exist a new CRL4Cdt2 target that participates in regulation of the G2/M checkpoint.
Interestingly, when Cdt2 is silenced in mammalian tissue culture cells, the basal level of PCNA monoubiquitylation at K164 that is normally seen in the absence of DNA damage is diminished, and PCNA-dependent translesion DNA synthesis is reduced (Terai et al. 2010). One interpretation of these data is that PCNA is ubiquitylated by CRL4Cdt2 that has docked onto PCNA via a bona fide PIP degron substrate. However, based on biochemical experiments, CRL4Cdt2 might also recognize PCNA directly (Terai et al. 2010). If this is the case, it raises the intriguing possibility that there exists a class of CRL4Cdt2 substrates that lack PIP degrons.
One way to find new substrates of CRL4Cdt2 is to search the database for matches to the PIP degron. Indeed, two groups used this approach to identify Set8 (Abbas et al. 2010; Centore et al. 2010), which contains a PIP degron that conforms almost perfectly to the consensus in all metazoan organisms and was therefore readily identifiable. However, this approach has limitations, since some substrates exhibit substantial deviations from the ideal PIP degron. For example, fly Cdt1 and E2F and worm pol η lack the TD motif at positions 5 and 6 of the PIP box (Fig. 2). In addition, Set8 is destroyed, albeit slowly, when the B+4 residue is mutated (Abbas et al. 2010; CG Havens, RC Centore, L Zou, and JC Walter, unpubl.). It therefore seems likely that numerous proteins with nonideal PIP degrons are targeted for destruction by CRL4Cdt2. One potential approach to identify these is to isolate CRL4Cdt2-interacting proteins. However, this method suffers from the fact that key substrates like Cdt1 appear to bind CRL4Cdt2 only in the context of PCNADNA and are therefore unlikely to be isolated in this manner. Therefore, identification of new CRL4Cdt2 substrates will likely require more sensitive methods (Yen and Elledge 2008; Yen et al. 2008).
Regulation of CRL4Cdt2 function by post-translational modifications
An important question is whether any post-translational modifications are required for CRL4Cdt2 function. While the destruction of Cdt1 and Set8 does not depend on the checkpoint protein kinases ATR and ATM (Higa et al. 2003; Arias and Walter 2006; Centore et al. 2010), p21 ubiquitylation and destruction after UV irradiation requires ATR-dependent phosphorylation on Ser 114 (Bendjennat et al. 2003; Lee et al. 2007; Abbas et al. 2008) for reasons that are currently unknown. Cdt2 is another potential target of modification, as Cdt2 is phosphorylated in S phase, and a hyperphosphorylated form of Cdt2 is enriched on chromatin after UV irradiation (Ishii et al. 2010). However, the functional significance of this phosphorylation, as well as the relevant protein kinase(s), are unknown. Finally, chromatin-bound PCNA is phosphorylated on Tyr 211 (Wang et al. 2006) and ubiquitylated/sumoylated on K164 (Hoege et al. 2002). At present, there is no evidence that these modifications are required for CRL4Cdt2 function. In summary, with the exception of p21 modification, there is no compelling evidence so far that other substrates, CRL4Cdt2, or PCNA must be modified to support this proteolysis pathway, but this picture could change upon further investigation.
There are a few instances in which post-translational modifications function to inhibit the CRL4Cdt2 pathway. Thus, p21 phosphorylation on Thr 145 or Ser 146 within the PIP box (positions 2 and 3) stabilizes the protein after UV irradiation, and this effect is explained by the fact that this modification disrupts the interaction with PCNA (Scott et al. 2000; Li et al. 2002).
In worms, pol η is SUMOylated by GEI-17 after DNA damage (Kim and Michael 2008). This modification stabilizes pol η and protects it from CRL4Cdt2-mediated destruction so the translesion polymerase can repair damaged DNA. A recent study in S. pombe suggests that the presence of ubiquitylated PCNA may interfere with Cdt1 destruction (Guarino et al. 2011).
Temporal regulation of CRL4Cdt2-dependent ubiquitylation
An intrinsic feature of CRL4Cdt2-mediated proteolysis is that the process is only initiated after DNA replication has begun, since PCNA loading requires new DNA synthesis. This temporal delay in the destruction of CRL4Cdt2 substrates upon S-phase entry has interesting implications for the destruction of different substrates. For example, immediate destruction of Spd1 upon S-phase entry may not be essential. Thus, S. pombe cells likely use the existing pool of dNTPs to begin DNA synthesis, whereupon CRL4Cdt2-dependent destruction of Spd1 stimulates the production of additional dNTPs to enable progression through S phase. Similarly, the precise timing of E2F destruction may not be crucial, as long as E2F activity is attenuated in S phase. In contrast, a delay in Cdt1, Set8, or p21 destruction in S phase would not be ideally suited to achieve an absolute injunction against rereplication. Thus, there might exist a temporal window at the start of S phase when DNA replication has begun but before Cdt1, Set8, and p21 levels have dropped. During this window, licensing on the nascent DNA should be possible. There are several potential solutions to this dilemma. First, in humans, CRL1Skp2 might help destroy Cdt1, Set8, and/or p21 in late G1 or S early phase (Li et al. 2003; Kondo et al. 2004; Takeda et al. 2005; Nishitani et al. 2006). Second, other inhibitors of rereplication, such as Geminin, might prevent licensing in early S phase when Cdt1 is still present (Arias and Walter 2007). Third, we speculate that CRL4Cdt2 might protect replicated DNA from reinitiation locally. This idea is based on the fact that a new PCNA molecule is loaded every 150 nucleotides during Okazaki fragment synthesis. If PCNA unloading is slow after Okazaki fragment maturation, a high concentration of PCNA molecules would accumulate on the lagging strand (Arias and Walter 2006), protecting it from relicensing. The high density of PCNA would promote destruction of Cdt1 near replicated DNA even before the total pool of Cdt1 has disappeared from the nucleus. This mechanism is plausible because in metazoans, there is a lag phase between Cdt1 binding to ORC and recruitment of MCM2–7 (Waga and Zembutsu 2006), allowing time for CRL4Cdt2-dependent destruction of any Cdt1 bound to replicated DNA. This mechanism might also protect the leading strand, which is held in close association with the lagging strand by cohesion. As such, PCNA might function as a “body armor” for replicated DNA. For individual substrates, it is interesting to consider how well the timing and function of CRL4Cdt2-dependent destruction are aligned, and in cases of misalignment, whether compensatory mechanisms might exist.
Conclusion and future perspectives
As discussed in this review, the study of CRL4Cdt2 is establishing new concepts in the field of proteolysis. CRL4Cdt2 appears to be unique among E3 ubiquitin ligases because there is little evidence that its interaction with substrates depends on post-translational modifications. Rather, binding to substrates depends on their prior interaction, through a PIP degron, with a cell cycle-regulated structure, chromatin-bound PCNA. The coupling of ubiquitin ligase function to a transient interaction between the degron and another protein is a novel concept in ubiquitin-mediated proteolysis. This strategy represents an elegant means to couple proteolysis to other metabolic events, and it will be surprising if this mechanism is not used by other E3 enzymes.
The analysis of CRL4Cdt2 substrates has also revealed interesting new concepts in DNA replication and cell cycle regulation. Thus, the discovery of p21 as a CRL4Cdt2 target has provided the most compelling evidence to date that Cdc6 export is important to prevent relicensing in S phase. Similarly, the targeting of Set8 by CRL4Cdt2 led to the insight that Set8 participates in licensing and that certain forms of chromatin are incompatible with the G1- and S-phase programs. The identification of Epe1 as a likely CRL4Cdt2 target represents a fascinating example in which proteolysis sculpts chromatin structure. Given the crucial role of CRL4Cdt2 in chromosome maintenance, particularly the control of licensing, it is possible that this E3 ubiquitin ligase has tumor suppressor activity (Abbas and Dutta 2011).
Several important questions remain to be answered. First, how many CRL4Cdt2 substrates exist? Identification of new targets is of prime importance, since it will help refine our understanding of the PIP degron and teach us further lessons about the regulation of S phase, DNA repair, and other chromatin-related processes. Second, does PCNA function solely as a platform to bring together the substrate and ligase, or does it also modulate the activity of the ligase? Third, why is CRL4Cdt2 activity strictly dependent on chromatin-bound PCNA? This dependency is the crux of CRL4Cdt2's temporal regulation during the cell cycle. Finally, are DNA-bound PCNA and a PIP degron sufficient to harness the activity of CRL4Cdt2 or are additional proteins and/or post-translational modifications required? Given the intense interest that has focused on CRL4Cdt2 in recent years, we hope the answers to these questions will soon emerge.
Acknowledgments
We thank Benjamin Morris for preparing Figure 1. We also thank Tammy Slenn and Benjamin Morris for critical reading of the manuscript. Our work on CRL4Cdt2 is funded by NIH grant GM80676.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2068611.
References
- Abbas T, Dutta A 2011. CRL4Cdt2: master coordinator of cell cycle progression and genome stability. Cell Cycle 10: 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A 2008. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev 22: 2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A 2010. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell 40: 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adimoolam S, Lin CX, Ford JM 2001. The p53-regulated cyclin-dependent kinase inhibitor, p21 (cip1, waf1, sdi1), is not required for global genomic and transcription-coupled nucleotide excision repair of UV-induced DNA photoproducts. J Biol Chem 276: 25813–25822 [DOI] [PubMed] [Google Scholar]
- Amador V, Ge S, Santamaria PG, Guardavaccaro D, Pagano M 2007. APC/C(Cdc20) controls the ubiquitin-mediated degradation of p21 in prometaphase. Mol Cell 27: 462–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers S, Li T, Yi X, MacGoss MJ, Moon RT, Zheng N 2006. Molecular architecture and assembly of the Ddb1–Cul4A ubiquitin ligase machinery. Nature 443: 590–593 [DOI] [PubMed] [Google Scholar]
- Ardley HC, Robinson PA 2005. E3 ubiquitin ligases. Essays Biochem 41: 15–30 [DOI] [PubMed] [Google Scholar]
- Arias EE, Walter JC 2005. Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev 19: 114–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias EE, Walter JC 2006. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol 8: 84–90 [DOI] [PubMed] [Google Scholar]
- Arias EE, Walter JC 2007. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev 21: 427–518 [DOI] [PubMed] [Google Scholar]
- Asano M, Nevins JR, Wharton RP 1996. Ectopic E2F expression induces S phase and apoptosis in Drosophila imaginal discs. Genes Dev 10: 1422–1432 [DOI] [PubMed] [Google Scholar]
- Bates S, Ryan KM, Phillips AC, Vousden KH 1998. Cell cycle arrest and DNA endoreduplication following p21Waf1/Cip1 expression. Oncogene 17: 1691–1703 [DOI] [PubMed] [Google Scholar]
- Behrends C, Harper JW 2011. Constructing and decoding unconventional ubiquitin chains. Nat Struct Mol Biol 18: 520–528 [DOI] [PubMed] [Google Scholar]
- Bendjennat M, Boulaire J, Jascur T, Brickner H, Barbier V, Sarasin A, Fotedar A, Fotedar R 2003. UV irradiation triggers ubiquitin-dependent degradation of p21(WAF1) to promote DNA repair. Cell 114: 599–610 [DOI] [PubMed] [Google Scholar]
- Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A 2003. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J Biol Chem 278: 25752–25757 [DOI] [PubMed] [Google Scholar]
- Braun S, Garcia JF, Rowley M, Rougemaille M, Shankar S, Madhani HD 2011. The Cul4-Ddb1(Cdt)(2) ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell 144: 41–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrol C, Knibiehler M, Ducommun B 1998. p21 binding to PCNA causes G1 and G2 cell cycle arrest in p53-deficient cells. Oncogene 16: 311–320 [DOI] [PubMed] [Google Scholar]
- Cazzalini O, Perucca P, Riva F, Stivala LA, Bianchi L, Vannini V, Ducommun B, Prosperi E 2003. p21CDKN1A does not interfere with loading of PCNA at DNA replication sites, but inhibits subsequent binding of DNA polymerase δ at the G1/S phase transition. Cell Cycle 2: 596–603 [PubMed] [Google Scholar]
- Centore RC, Havens CG, Manning AL, Li JM, Flynn RL, Tse A, Jin J, Dyson NJ, Walter JC, Zou L 2010. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol Cell 40: 22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Jackson PK, Kirschner MW, Dutta A 1995. Separate domains of p21 involved in the inhibition of Cdk kinase and PCNA. Nature 374: 386–388 [DOI] [PubMed] [Google Scholar]
- Choe KP, Przybysz AJ, Strange K 2009. The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol Cell Biol 29: 2704–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang LC, Yew PR 2005. PCNA recruits CDK inhibitor Xic1 to DNA and couples its proteolysis to DNA polymerase switching. J Biol Chem 280: 35299–35309 [DOI] [PubMed] [Google Scholar]
- Chuang LC, Zhu XN, Herrera CR, Tseng HM, Pfleger CM, Block K, Yew PR 2005. The C-terminal domain of the Xenopus cyclin-dependent kinase inhibitor, p27Xic1, is both necessary and sufficient for phosphorylation-independent proteolysis. J Biol Chem 280: 34290–35285 [DOI] [PubMed] [Google Scholar]
- Cooper MP, Balajee AS, Bohr VA 1999. The C-terminal domain of p21 inhibits nucleotide excision repair in vitro and in vivo. Mol Biol Cell 10: 2119–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA 2009. RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Emberley ED, Saha A 2010. Control of cullin-ring ubiquitin ligase activity by nedd8. Subcell Biochem 54: 41–56 [DOI] [PubMed] [Google Scholar]
- Dimova DK, Dyson NJ 2005. The E2F transcriptional network: old acquaintances with new faces. Oncogene 24: 2810–2826 [DOI] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA 2008. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134: 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman Y, Palgi N, Dovrat D, Ben-Aroya S, Hieter P, Aharoni A 2010. Subtle alterations in PCNA–partner interactions severely impair DNA replication and repair. PLoS Biol 8: e1000507 doi: 10.1371/journal.pbio.1000507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Inuzuka H, Tseng A, Chin RY, Toker A, Wei W 2009. Phosphorylation by Akt1 promotes cytoplasmic localization of Skp2 and impairs APCCdh1-mediated Skp2 destruction. Nat Cell Biol 11: 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino E, Shepherd ME, Salguero I, Hua H, Deegan RS, Kearsey SE 2011. Cdt1 proteolysis is promoted by dual PIP degrons and is modulated by PCNA ubiquitylation. Nucleic Acids Res doi: 10.1093/nar/gkr222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J 1996. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 87: 297–306 [DOI] [PubMed] [Google Scholar]
- Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP 2007. Structure of a Fbw7–Skp1–cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell 26: 131–143 [DOI] [PubMed] [Google Scholar]
- Havens CG, Walter JC 2009. Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol Cell 35: 93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YJ, McCall CM, Hu J, Zeng Y, Xiong Y 2006. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4–ROC1 ubiquitin ligases. Genes Dev 20: 2949–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa LA, Zhang H 2007. Stealing the spotlight: CUL4–DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell Div 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa LA, Mihaylov IS, Banks DP, Zheng J, Zhang H 2003. Radiation-mediated proteolysis of CDT1 by CUL4–ROC1 and CSN complexes constitutes a new checkpoint. Nat Cell Biol 5: 1008–1015 [DOI] [PubMed] [Google Scholar]
- Higa LA, Banks D, Wu M, Kobayashi R, Sun H, Zhang H 2006a. L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle 5: 1675–1680 [DOI] [PubMed] [Google Scholar]
- Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H 2006b. CUL4–DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol 8: 1277–1283 [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Hofmann JF, Beach D 1994. cdt1 is an essential target of the Cdc10/Sct1 transcription factor: requirement for DNA replication and inhibition of mitosis. EMBO J 13: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg C, Fleck O, Hansen HA, Liu C, Slaaby R, Carr AM, Nielsen O 2005. Ddb1 controls genome stability and meiosis in fission yeast. Genes Dev 19: 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon WC, Wilson MI, Harlos K, Claridge TD, Schofield CJ, Pugh CW, Maxwell PH, Ratcliffe PJ, Stuart DI, Jones EY 2002. Structural basis for the recognition of hydroxyproline in HIF-1α by pVHL. Nature 417: 975–978 [DOI] [PubMed] [Google Scholar]
- Hook SS, Lin JJ, Dutta A 2007. Mechanisms to control rereplication and implications for cancer. Curr Opin Cell Biol 19: 663–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston SI, McManus KJ, Adams MM, Sims JK, Carpenter PB, Hendzel MJ, Rice JC 2008. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem 283: 19478–19488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Xiong Y 2006. An evolutionarily conserved function of proliferating cell nuclear antigen for cdt1 degradation by the cul4–ddb1 ubiquitin ligase in response to DNA damage. J Biol Chem 281: 3753–3756 [DOI] [PubMed] [Google Scholar]
- Hu J, McCall CM, Ohta T, Xiong Y 2004. Targeted ubiquitination of CDT1 by the DDB1–CUL4A–ROC1 ligase in response to DNA damage. Nat Cell Biol 6: 1003–1009 [DOI] [PubMed] [Google Scholar]
- Hu J, Zacharek S, He YJ, Lee H, Shumway S, Duronio RJ, Xiong Y 2008. WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1–CUL4–ROC1 ligase. Genes Dev 22: 866–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Shiomi Y, Takami T, Murakami Y, Ohnishi N, Nishitani H 2010. Proliferating cell nuclear antigen-dependent rapid recruitment of Cdt1 and CRL4Cdt2 at DNA-damaged sites after UV irradiation in HeLa cells. J Biol Chem 285: 41993–42000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG Jr 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292: 464–468 [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. 2001. Targeting of HIF-α to the von Hippel–Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292: 468–472 [DOI] [PubMed] [Google Scholar]
- Jackson S, Xiong Y 2009. CRL4s: the CUL4–RING E3 ubiquitin ligases. Trends Biochem Sci 34: 562–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Morgan DO 1999. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol 9: 227–236 [DOI] [PubMed] [Google Scholar]
- Jin J, Arias EE, Chen J, Harper JW, Walter JC 2006. A family of diverse Cul4–Ddb1–interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell 23: 709–721 [DOI] [PubMed] [Google Scholar]
- Jin L, Williamson A, Banerjee S, Philipp I, Rape M 2008. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 133: 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen S, Eskildsen M, Fugger K, Hansen L, Larsen MS, Kousholt AN, Syljuasen RG, Trelle MB, Jensen ON, Helin K, et al. 2011. SET8 is degraded via PCNA-coupled CRL4(CDT2) ubiquitylation in S phase and after UV irradiation. J Cell Biol 192: 43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns SL, Torke SJ, Benjamin JM, McGarry TJ 2007. Geminin prevents rereplication during Xenopus development. J Biol Chem 282: 5514–5521 [DOI] [PubMed] [Google Scholar]
- Kim Y, Kipreos ET 2007. The Caenorhabditis elegans replication licensing factor CDT-1 is targeted for degradation by the CUL-4/DDB-1 complex. Mol Cell Biol 27: 1394–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Michael WM 2008. Regulated proteolysis of DNA polymerase η during the DNA-damage response in C. elegans. Mol Cell 32: 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Feng H, Kipreos ET 2007. C. elegans CUL-4 prevents rereplication by promoting the nuclear export of CDC-6 via a CKI-1-dependent pathway. Curr Biol 17: 966–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Starostina NG, Kipreos ET 2008. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev 22: 2507–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Budhavarapu VN, Herrera CR, Nam HW, Kim YS, Yew PR 2010. The CRL4Cdt2 ubiquitin ligase mediates the proteolysis of cyclin-dependent kinase inhibitor Xic1 through a direct association with PCNA. Mol Cell Biol 30: 4120–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D 2009. The emerging complexity of protein ubiquitination. Biochem Soc Trans 37: 937–953 [DOI] [PubMed] [Google Scholar]
- Kondo T, Kobayashi M, Tanaka J, Yokoyama A, Suzuki S, Kato N, Onozawa M, Chiba K, Hashino S, Imamura M, et al. 2004. Rapid degradation of Cdt1 upon UV-induced DNA damage is mediated by SCFSkp2 complex. J Biol Chem 279: 27315–27319 [DOI] [PubMed] [Google Scholar]
- Kraft C, Herzog F, Gieffers C, Mechtler K, Hagting A, Pines J, Peters JM 2003. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J 22: 6598–6609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters JM 2000. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell 11: 1555–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Yu SJ, Park YG, Kim J, Sohn J 2007. Glycogen synthase kinase 3β phosphorylates p21WAF1/CIP1 for proteasomal degradation after UV irradiation. Mol Cell Biol 27: 3187–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Terzaghi W, Gusmaroli G, Charron JB, Yoon HJ, Chen H, He YJ, Xiong Y, Deng XW 2008. Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4–RING E3 ubiquitin ligases. Plant Cell 20: 152–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HO, Zacharek SJ, Xiong Y, Duronio RJ 2010. Cell type-dependent requirement for PIP box-regulated Cdt1 destruction during S phase. Mol Biol Cell 21: 3639–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Blow JJ 2005. Cdt1 downregulation by proteolysis and geminin inhibition prevents DNA re-replication in Xenopus. EMBO J 24: 395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Dowbenko D, Lasky LA 2002. AKT/PKB phosphorylation of p21Cip/WAF1 enhances protein stability of p21Cip/WAF1 and promotes cell survival. J Biol Chem 277: 11352–11361 [DOI] [PubMed] [Google Scholar]
- Li X, Zhao Q, Liao R, Sun P, Wu X 2003. The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J Biol Chem 278: 30854–30858 [DOI] [PubMed] [Google Scholar]
- Lin JJ, Milhollen MA, Smith PG, Narayanan U, Dutta A 2010. NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer Res 70: 10310–10320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Powell KA, Mundt K, Wu L, Carr AM, Caspari T 2003. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev 17: 1130–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Poitelea M, Watson A, Yoshida SH, Shimoda C, Holmberg C, Nielsen O, Carr AM 2005. Transactivation of Schizosaccharomyces pombe cdt2+ stimulates a Pcu4–Ddb1–CSN ubiquitin ligase. EMBO J 24: 3940–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CL, Yu IS, Pan HW, Lin SW, Hsu HC 2007. L2dtl is essential for cell survival and nuclear division in early mouse embryonic development. J Biol Chem 282: 1109–1118 [DOI] [PubMed] [Google Scholar]
- Liu P, Slater DM, Lenburg M, Nevis K, Cook JG, Vaziri C 2009. Replication licensing promotes cyclin D1 expression and G1 progression in untransformed human cells. Cell Cycle 8: 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida YJ, Teer JK, Dutta A 2005. Acute reduction of an origin recognition complex (ORC) subunit in human cells reveals a requirement of ORC for Cdk2 activation. J Biol Chem 280: 27624–27630 [DOI] [PubMed] [Google Scholar]
- Mailand N, Diffley JF 2005. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell 122: 915–926 [DOI] [PubMed] [Google Scholar]
- McCall CM, Miliani de Marval PL, Chastain PD 2nd, Jackson SC, He YJ, Kotake Y, Cook JG, Xiong Y 2008. Human immunodeficiency virus type 1 Vpr-binding protein VprBP, a WD40 protein associated with the DDB1–CUL4 E3 ubiquitin ligase, is essential for DNA replication and embryonic development. Mol Cell Biol 28: 5621–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema RH, Klompmaker R, Smits VA, Rijksen G 1998. p21waf1 can block cells at two points in the cell cycle, but does not interfere with processive DNA-replication or stress-activated kinases. Oncogene 16: 431–441 [DOI] [PubMed] [Google Scholar]
- Michishita M, Morimoto A, Ishii T, Komori H, Shiomi Y, Higuchi Y, Nishitani H 2011. Positively charged residues located downstream of PIP box, together with TD amino acids within PIP box, are important for CRL4(Cdt2)-mediated proteolysis. Genes Cells 16: 12–22 [DOI] [PubMed] [Google Scholar]
- Milhollen MA, Narayanan U, Soucy TA, Veiby PO, Smith PG, Amidon B 2011. Inhibition of NEDD8-activating enzyme induces rereplication and apoptosis in human tumor cells consistent with deregulating CDT1 turnover. Cancer Res 71: 3042–3051 [DOI] [PubMed] [Google Scholar]
- Min JH, Yang H, Ivan M, Gertler F, Kaelin WG Jr, Pavletich NP 2002. Structure of an HIF-1α -pVHL complex: hydroxyproline recognition in signaling. Science 296: 1886–1889 [DOI] [PubMed] [Google Scholar]
- Mizushima T, Yoshida Y, Kumanomidou T, Hasegawa Y, Suzuki A, Yamane T, Tanaka K 2007. Structural basis for the selection of glycosylated substrates by SCF(Fbs1) ubiquitin ligase. Proc Natl Acad Sci 104: 5777–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis K, Estelle M 2008. Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol 24: 55–80 [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S 2007. PCNA, the maestro of the replication fork. Cell 129: 665–679 [DOI] [PubMed] [Google Scholar]
- Moss J, Tinline-Purvis H, Walker CA, Folkes LK, Stratford MR, Hayles J, Hoe KL, Kim DU, Park HO, Kearsey SE, et al. 2010. Break-induced ATR and Ddb1–Cul4(Cdt)(2) ubiquitin ligase-dependent nucleotide synthesis promotes homologous recombination repair in fission yeast. Genes Dev 24: 2705–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi M, Robetorye RS, Pereira-Smith OM, Smith JR 1995. The C-terminal region of p21SDI1/WAF1/CIP1 is involved in proliferating cell nuclear antigen binding but does not appear to be required for growth inhibition. J Biol Chem 270: 17060–17063 [DOI] [PubMed] [Google Scholar]
- Nestoras K, Mohammed AH, Schreurs AS, Fleck O, Watson AT, Poitelea M, O'Shea C, Chahwan C, Holmberg C, Kragelund BB, et al. 2010. Regulation of ribonucleotide reductase by Spd1 involves multiple mechanisms. Genes Dev 24: 1145–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu AB III, Chen X, Smeets M, Hengst L, Prives C, Reed SI 1998. Effects of p21(Cip1/Waf1) at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication. Mol Cell Biol 18: 629–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H, Lygerou Z, Nishimoto T 2004. Proteolysis of DNA replication licensing factor Cdt1 in S-phase is performed independently of Geminin through its N-terminal region. J Biol Chem 279: 30807–30816 [DOI] [PubMed] [Google Scholar]
- Nishitani H, Sugimoto N, Roukos V, Nakanishi Y, Saijo M, Obuse C, Tsurimoto T, Nakayama KI, Nakayama K, Fujita M, et al. 2006. Two E3 ubiquitin ligases, SCF–Skp2 and DDB1–Cul4, target human Cdt1 for proteolysis. EMBO J 25: 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H, Shiomi Y, Iida H, Michishita M, Takami T, Tsurimoto T 2008. CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4–DDB1Cdt2 pathway during S phase and after UV irradiation. J Biol Chem 283: 29045–29052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell BC, Harper JW 2007. Ubiquitin proteasome system (UPS): what can chromatin do for you? Curr Opin Cell Biol 19: 206–214 [DOI] [PubMed] [Google Scholar]
- Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D 2010. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell 40: 364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JJ, Tainer JA, Boddy MN 2008. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci 33: 201–208 [DOI] [PubMed] [Google Scholar]
- Pfleger CM, Lee E, Kirschner MW 2001. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev 15: 2396–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph E, Boye E, Kearsey SE 2006. DNA damage induces Cdt1 proteolysis in fission yeast via a pathway dependent on Cdt2 and Ddb1. EMBO Rep 7: 1134–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman M, Havens CG, Walter JC, Harper JW 2011. A genome wide screen identifies p97 as an essential regulator of DNA damage-dependent CDT1 destruction. Mol Cell (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T, Hochstrasser M 2008. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol 9: 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JC, Nishioka K, Sarma K, Steward R, Reinberg D, Allis CD 2002. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev 16: 2225–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Foster SA, Morgan DO 2010. Catalysis of lysine 48-specific ubiquitin chain assembly by residues in E2 and ubiquitin. Mol Cell 39: 548–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roukos V, Kinkhabwala A, Colombelli J, Kotsantis P, Taraviras S, Nishitani H, Stelzer E, Bastiaens P, Lygerou Z 2011. Dynamic recruitment of licensing factor Cdt1 to sites of DNA damage. J Cell Sci 124: 422–434 [DOI] [PubMed] [Google Scholar]
- Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ 2011. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol Cell 42: 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansam CL, Shepard JL, Lai K, Ianari A, Danielian PS, Amsterdam A, Hopkins N, Lees JA 2006. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes & Dev 20: 3117–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MT, Morrice N, Ball KL 2000. Reversible phosphorylation at the C-terminal regulatory domain of p21(Waf1/Cip1) modulates proliferating cell nuclear antigen binding. J Biol Chem 275: 11529–11537 [DOI] [PubMed] [Google Scholar]
- Scrima A, Konickova R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, Nakatani Y, Iwai S, Pavletich NP, Thoma NH 2008. Structural basis of UV DNA-damage recognition by the DDB1–DDB2 complex. Cell 135: 1213–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senga T, Sivaprasad U, Zhu W, Park JH, Arias EE, Walter JC, Dutta A 2006. PCNA is a co-factor for Cdt1 degradation by CUL4/DDB1 mediated N-terminal ubiquitination. J Biol Chem 281: 6246–6252 [DOI] [PubMed] [Google Scholar]
- Shibata E, Abbas T, Huang X, Wohlschlegel JA, Dutta A 2011. Selective ubiquitylation of p21 and Cdt1 by UBCH8 and UBE2G ubiquitin conjugating enzymes via the CRL4Cdt2 ubiquitin ligase complex. Mol Cell Biol doi: 10.1128/MCB.05496-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani ST, de la Cruz AF, Tran V, Turbyfill WJ III, Reis T, Edgar BA, Duronio RJ 2008. Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev Cell 15: 890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G, Speroni J, Podhajcer OL, Prives C, Gottifredi V 2008. p21 differentially regulates DNA replication and DNA-repair-associated processes after UV irradiation. J Cell Sci 121: 3271–3282 [DOI] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. 2009. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458: 732–736 [DOI] [PubMed] [Google Scholar]
- Stuart SA, Wang JY 2009. Ionizing radiation induces ATM-independent degradation of p21Cip1 in transformed cells. J Biol Chem 284: 15061–15070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Tanaka K, et al. 2005. UV-induced ubiquitylation of XPC protein mediated by UV–DDB–ubiquitin ligase complex. Cell 121: 387–400 [DOI] [PubMed] [Google Scholar]
- Takeda DY, Parvin JD, Dutta A 2005. Degradation of Cdt1 during S phase is Skp2-independent and is required for efficient progression of mammalian cells through S phase. J Biol Chem 280: 23416–23423 [DOI] [PubMed] [Google Scholar]
- Tan X, Zheng N 2009. Hormone signaling through protein destruction: a lesson from plants. Am J Physiol Endocrinol Metab 296: E223–E227 doi: 10.1152/ajpendo.90807.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, Julien E 2010. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol 12: 1086–1093 [DOI] [PubMed] [Google Scholar]
- Teer JK, Machida YJ, Labit H, Novac O, Hyrien O, Marheineke K, Zannis-Hadjopoulos M, Dutta A 2006. Proliferating human cells hypomorphic for origin recognition complex 2 and pre-replicative complex formation have a defect in p53 activation and Cdk2 kinase activation. J Biol Chem 281: 6253–6260 [DOI] [PubMed] [Google Scholar]
- Terai K, Abbas T, Jazaeri AA, Dutta A 2010. CRL4(Cdt2) E3 ubiquitin ligase monoubiquitinates PCNA to promote translesion DNA synthesis. Mol Cell 37: 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri C, Saxena S, Jeon Y, Lee C, Murata K, Machida Y, Wagle N, Hwang DS, Dutta A 2003. A p53-dependent checkpoint pathway prevents rereplication. Mol Cell 11: 997–1008 [DOI] [PubMed] [Google Scholar]
- Waga S, Zembutsu A 2006. Dynamics of DNA binding of replication initiation proteins during de novo formation of pre-replicative complexes in Xenopus egg extracts. J Biol Chem 281: 10926–10934 [DOI] [PubMed] [Google Scholar]
- Wang W, Nacusi L, Sheaff RJ, Liu X 2005. Ubiquitination of p21Cip1/WAF1 by SCFSkp2: substrate requirement and ubiquitination site selection. Biochemistry 44: 14553–14564 [DOI] [PubMed] [Google Scholar]
- Wang SC, Nakajima Y, Yu YL, Xia W, Chen CT, Yang CC, McIntush EW, Li LY, Hawke DH, Kobayashi R, et al. 2006. Tyrosine phosphorylation controls PCNA function through protein stability. Nat Cell Biol 8: 1359–1368 [DOI] [PubMed] [Google Scholar]
- Warbrick E, Lane DP, Glover DM, Cox LS 1995. A small peptide inhibitor of DNA replication defines the site of interaction between the cyclin-dependent kinase inhibitor p21WAF1 and proliferating cell nuclear antigen. Curr Biol 5: 275–282 [DOI] [PubMed] [Google Scholar]
- Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M 2011. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 144: 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Rice JC 2011. A new regulator of the cell cycle: the PR-Set7 histone methyltransferase. Cell Cycle 10: 68–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Wang W, Kong X, Congdon LM, Yokomori K, Kirschner MW, Rice JC 2010. Dynamic regulation of the PR-Set7 histone methyltransferase is required for normal cell cycle progression. Genes Dev 24: 2531–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Wang J, Hu Q, Quan Y, Chen H, Cao Y, Li C, Wang Y, He Q 2010. DCAF26, an adaptor protein of Cul4-based E3, is essential for DNA methylation in Neurospora crassa. PLoS Genet 6: e1001132 doi: 10.1371/journal.pgen.1001132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Rape M 2009. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 10: 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HC, Elledge SJ 2008. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science 322: 923–929 [DOI] [PubMed] [Google Scholar]
- Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ 2008. Global protein stability profiling in mammalian cells. Science 322: 918–923 [DOI] [PubMed] [Google Scholar]
- Yin Y, Yu VC, Zhu G, Chang DC 2008. SET8 plays a role in controlling G1/S transition by blocking lysine acetylation in histone through binding to H4 N-terminal tail. Cell Cycle 7: 1423–1432 [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Chiba T, Tokunaga F, Kawasaki H, Iwai K, Suzuki T, Ito Y, Matsuoka K, Yoshida M, Tanaka K, et al. 2002. E3 ubiquitin ligase that recognizes sugar chains. Nature 418: 438–442 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Takisawa H, Kubota Y 2005. Intrinsic nuclear import activity of geminin is essential to prevent re-initiation of DNA replication in Xenopus eggs. Genes Cells 10: 63–73 [DOI] [PubMed] [Google Scholar]
- You Z, Harvey K, Kong L, Newport J 2002. Xic1 degradation in Xenopus egg extracts is coupled to initiation of DNA replication. Genes Dev 16: 1182–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ZK, Gervais JL, Zhang H 1998. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21(CIP1/WAF1) and cyclin D proteins. Proc Natl Acad Sci 95: 11324–11329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W, Schwab M, Nasmyth K, Seufert W 1998. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science 282: 1721–1724 [DOI] [PubMed] [Google Scholar]
- Zaidi IW, Rabut G, Poveda A, Scheel H, Malmstrom J, Ulrich H, Hofmann K, Pasero P, Peter M, Luke B 2008. Rtt101 and Mms1 in budding yeast form a CUL4(DDB1)-like ubiquitin ligase that promotes replication through damaged DNA. EMBO Rep 9: 1034–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. 2002. Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF ubiquitin ligase complex. Nature 416: 703–709 [DOI] [PubMed] [Google Scholar]
- Zhong W, Feng H, Santiago FE, Kipreos ET 2003. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 423: 885–889 [DOI] [PubMed] [Google Scholar]