Abstract
With regard to human induced pluripotent stem cells (hiPSCs), in which adult cells are reprogrammed into embryonic-like cells using defined factors, their functional and transcriptional expression pattern during endothelial differentiation has yet to be characterized. In this study, hiPSCs and human embryonic stem cells (hESCs) were differentiated using the embryoid body method, and CD31+ cells were sorted. Fluorescence activated cell sorting analysis of hiPSC-derived endothelial cells (hiPSC-ECs) and hESC-derived endothelial cells (hESC-ECs) demonstrated similar endothelial gene expression patterns. We showed functional vascular formation by hiPSC-ECs in a mouse Matrigel plug model. We compared the gene profiles of hiPSCs, hESCs, hiPSC-ECs, hESC-ECs, and human umbilical vein endothelial cells (HUVECs) using whole genome microarray. Our analysis demonstrates that gene expression variation of hiPSC-ECs and hESC-ECs contributes significantly to biological differences between hiPSC-ECs and hESC-ECs as well as to the “distances” among hiPSCs, hESCs, hiPSC-ECs, hESC-ECs, and HUVECs. We further conclude that hiPSCs can differentiate into functional endothelial cells, but with limited expansion potential compared with hESC-ECs; thus, extensive studies should be performed to explore the cause and extent of such differences before clinical application of hiPSC-ECs can begin.
Introduction
In recent years, human embryonic stem cells (hESCs) have gained popularity as a potentially ideal cell candidate for regenerative medicine. hESCs are derived from the inner cell mass of the human blastocyte and can be kept in an undifferentiated, self-renewing state indefinitely [1]. In contrast to adult stem cells, hESCs are pluripotent and can differentiate into virtually any cell type. However, the use of human embryos is controversial, and the problem of immune rejection after transplantation remains challenging. One way to circumvent these issues is to generate pluripotent cells directly from the patients' own cells. The introduction of defined transcription factors into mouse and human somatic cells has recently been shown to reprogram the developmental state of mature cells into that of pluripotent embryonic cells, generating so-called human “induced pluripotent stem cells” (hiPSCs). hiPSCs have been generated from multiple cell types by viral expression of Oct4 and Sox2, combined with either Klf4 and c-Myc or LIN28 and Nanog [2,3]. hiPSCs are believed to be molecularly and functionally similar to hESCs, which makes in vitro reprogramming an attractive approach to produce patient-specific stem cells for treating degenerative disease. Indeed, reprogrammed skin cells have recently been shown to alleviate the symptoms of Parkinson's disease and sickle cell anemia in mouse models [4,5], and hiPSCs have already been differentiated into various functional cell types, including endothelial cells and cardiomyocytes [6–8].
However, before clinical implementation of hiPSC-based therapy can safely commence, several issues should be addressed. Most of the hiPSCs made so for are based on lentivirus and retrovirus, which carries a potential risk of insertional mutagenesis. To realize the full therapeutic potential of hiPSC technology, it will be necessary to develop novel and improved quality assessments that can be readily used to determine the exact cellular state of reprogrammed cells. Further, major efforts are needed to generate all desired cell types. In addition, once such differentiation is possible, it remains to be determined whether the in vitro derived cell types are comparable to their in vivo counterparts and whether they can be isolated with sufficient purity. Finally, whether hiPSCs and hESCs are truly equivalent at the molecular and functional levels is another question that should be answered [9].
In this study, hiPSCs and hESCs were differentiated into endothelial cells in vitro by using the embryoid body (EB) method, and CD31+ cells were sorted. Further functional characterization of hiPSC-derived endothelial cells (hiPSC-ECs) were carried out by in vitro analysis and in vivo angiogenesis. We compared the gene profiles of hiPSCs, hESCs, hiPSC-ECs, hESC-derived endothelial cells (hESC-ECs), and human umbilical vein endothelial cells HUVECs using the whole genome microarray. Our analysis indicates that variation in gene expression of hiPSC-ECs and hESC-ECs contributes significantly to biological differences between hiPSC-ECs and hESC-ECs as well as to the “distances” among hiPSC-ECs, hESC-ECs, and HUVECs.
Methods
Maintenance of hiPSCs
We obtained hiPSCs from the James Thomson Lab (University of Wisconsin-Madison), which were originally derived from IMR90 fetal fibroblasts (ATCC) using the reprogramming factors OCT4, SOX2, NANOG, and LIN28 [2]. hESC (H9 from WiCell) and hiPSC were maintained on an inactivated mouse embryonic fibroblast feeder layer as previously described [10,11]. Before endothelial differentiation, hiPSCs were seeded onto Matrigel-coated plates in mTeSR1 medium (StemCell Technologies) as previously described [11]. hESCs (H9 line from WiCell, passages 35 to 45) were used as controls and cultured as described [10,12,13]. hESCs (control) and hiPSCs were stained for Oct-4 (Chemicon). The cell colonies were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min. Nonspecific binding was blocked with 4% normal goat serum for 30 min, after which the colonies were stained with antibodies to Oct-4, incubated with Alexa 594-conjugated rabbit anti-goat secondary antibodies (Invitrogen) for 30 min, and nuclear counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Images were obtained with a Zeiss Axiovert microscopy (Sutter Instrument).
In vitro differentiation of hiPSC-ECs and hESC-ECs
To induce hiPSC differentiation, undifferentiated hiPSCs were cultured in a differentiation medium containing Iscove's modified Dulbecco's medium and 15% defined fetal bovine serum (Hyclone), 0.1 mM nonessential amino acids, 2 mM L-glutamine, 450 μM monothioglycerol (Sigma), 50 U/mL penicillin, and 50 μg/mL streptomycin, in ultra-low attachment plates for the formation of suspended EBs as previously described [10,12]. Briefly, hiPSCs cultured on Matrigel coated plate with mTeSR™1 medium were treated by 2 mg/mL dispase (Invitrogen) for 15 min at 37°C to loosen the colonies. The colonies were then scraped off and transferred into ultra low-attachment plates (Corning Incorporated) for EB formation. hiPSC-EB sprouting differentiation in collagen type I was performed as described [11]. Briefly, 12 day-old EBs were harvested and then suspended into rat tail collagen type I (Becton Dickinson) at a final concentration of 1.5 mg/mL collagen. After thoroughly mixing hiPSC-EBs into collagen, 1.5 mL/well of the mixture was added into 6-well plates. The plates were incubated at 37°C for 30 min, allowing gel polymerization before the addition of medium. After gel formation, each dish was supplemented with EGM-2 medium (Lonza) and 5% Knockout™ SR with 50 ng/mL vascular endothelial growth factor (VEGF) and 20 ng/mL basic fibroblast growth factor. The cultures were then incubated for 3 days without media change.
Whole-mount immunostaining of hiPSC-EBs and hESC-EB sproutings
Whole-mount immunohistostaining of hiPSC-EBs and hESC-EB sproutings were performed as previously described [11,14] with minor modifications. The hiPSC-EB and hESC-EB sproutings were fixed in methanol and dimethyl sulfoxide (4:1) at 4°C overnight. For staining, the rehydrated hiPSC-EBs were first blocked by 2 incubations in PBSBT (2% bovine serum albumin and 0.2% Tween-20 in PBS), then with PBSBT containing mouse anti-human MoAb CD31 (Becton Dickinson) overnight at 4°C. The hiPSC-EBs were washed 5 times in PBSBT each for 1 h at 4°C for the initial 3 washes and at room temperature for the final 2 washes. The primary antibody was labeled by incubating the hiPSC-EBs with Alex 488/594-conjugated goat anti-mouse IgG (Invitrogen) in PBSBT overnight at 4°C and nuclear counterstained with DAPI.
Fluorescence activated cell sorting of hiPSC-ECs and hESC-ECs
Single cell suspensions from day 12 of differentiated hiPSC-EBs were obtained by treatment with 0.56 units/mL of Liberase Blendzyme IV (Roche) at 37°C for 20–30 min. Cells were passed through a 40-μm cell strainer (BD Falcon) [10,11]. They were incubated for 30 min at 4°C with mouse phycoerythrin conjugated anti-human CD31 (BD). The CD31+ cells were isolated using Vantoo (Becton Dickinson). To generate hiPSC-ECs, the isolated CD31+ cells from differentiated hiPSC-EBs were grown on 4 μg/cm2 human fibronectin (Calbiochem) coated plates in EGM-2 (Clonetics) with additional 5 ng/mL VEGF. The medium was changed every 2–3 days. A similar process was used for FACS of hESC-ECs.
Biological characteristics of hiPSC-ECs and hESC-ECs
Flow cytometry analysis, DiI-acetylated low-density lipoprotein (DiI-ac-LDL) uptake assay, immunostaining, and Matrigel assay were used to confirm endothelial cell phenotype within these CD31+ purified hiPSC-ECs. Antibodies used for flow cytometry analysis were phycoerythrin (conjugated anti-CD31 and anti-CD34 (BD Pharmingen, Mountain View, CA), Allophycocyanin conjugated anti-Flk-1 and anti-CD133 (R&D Systems), Allophycocyanin conjugated anti-mouse IgG2a, and rabbit anti-human CD144 (Abcam). The stained cells were analyzed using FACS Vantage (Becton-Dickinson). Dead cells stained by propidium-iodide (PI) were excluded from the analysis. Isotype-identical antibodies served as controls (BD Pharmingen). For immunostaining, the cells were fixed with 4% paraformaldehyde in PBS at room temperature for 15 min. The fixed cells were permeated by 1% Triton 100, incubated with 2% bovine serum albumin for 30 min to block nonspecific binding, and stained for 1 h with the primary antibodies CD31 (BD Pharmingen). The cells were then incubated for 30 min with Alexa 594-conjugated donkey anti-mouse secondary antibody and counter stained with DAPI. For DiI-ac-LDL uptake assay, hiPSC-ECs were incubated with 10 μg/mL of Dil-Ac-LDL (Molecular Probes) at 37°C for 6 h. After washing twice with PBS, DiI-ac-LDL uptake was detected with fluorescence microscopy as described [15]. The formation of endothelial tubes was assessed by seeding cells in 24-well plates coated with Matrigel (BD Pharmingen) and incubating them at 37°C for 12 h as described [16,17]. Cell cycle of CD31+ purified hESC-ECs and hiPSC-ECs was analyzed by PI Staining. Apoptosis analysis was performed by FITC Annexin V Apoptosis Detection Kit (BD Pharmingen). For detection of endothelial cell senescence, hESC-ECs and hiPSC-ECs were fixed between 14 and 16 days after initiation of endothelial differentiation and then stained for endogenous β-galactosidase using a Cell Senescence Kit (Cell Signaling Technology) according to the manufacturer's manual.
In vivo vasculogenic potential of hiPSC-ECs
To evaluate whether hiPSC-ECs are capable of forming functional blood vessels in vivo, we used Matrigel plug models in immunodeficient SCID mice that allow observation of blood vessels derived from implanted cells [18,19]. Briefly, 2 weeks after subcutaneous injection, Matrigel plugs were harvested, and histology was performed. Hematoxylin and eosin staining showed microvessels that contained murine blood cells in the vessel lumens. Harvested Matrigel plugs were also stained with anti-human CD31 and anti-human CD144 antibodies.
Microarray hybridization and data acquisition
Total RNA samples were isolated in Trizol (Invitrogen) followed by purification over a Qiagen RNeasy column (Qiagen) from undifferentiated hESCs, hiPSCs, hESC-ECs (after CD31 sort), hiPSC-ECs (after CD31 sort), and HUVECs. Four samples from each group (for a total of 20 unique samples) were harvested for RNA isolation. Using Agilent Low RNA Input Fluorescent Linear Amplification Kits, cDNA was reverse transcribed from each of the 16 RNA samples representing 4 biological quadruplicates as well as the pooled reference control, and cRNA was then transcribed and fluorescently labeled with Cy5/Cy3. cRNA was purified using an RNeasy kit (Qiagen). 825 ng of Cy3- and Cy5- labeled and amplified cRNA were hybridized to Agilent 4x44K whole human genome microarrays (G4112F) and processed according to the manufacturer's instructions. The array was scanned using Agilent G2505B DNA microarray scanner. The image files were extracted using Agilent Feature Extraction software version 9.5.1 applying LOWESS background subtraction and dye-normalization.
Microarray data analysis
The data were analyzed using GeneSpring GX 10.0. (Agilent Technologies). Summarization of gene expression data was performed by implementing the robust multichip averaging algorithm, with subsequent baseline normalization of the log-summarized values for each probe set to that of the median log summarized value for the same probe set in the control group. Expression data were then filtered to remove probe sets for which the signal intensities for all the treatment groups were in the lowest 20 percentile of all intensity values. The data were then subjected to analysis of variance, incorporating the Benjamini–Hochberg FDR multiple testing correction, with a significance level of P value<0.05 to get the differentially expressed genes between different groups. Probe sets were further filtered on the basis of a fold-change cut off of 2.0 unless specified. Hierarchical clustering was performed by average linkage and uncentered correlation using the open source clustering software Cluster 3.0; results were visualized using Java TreeView. In order to perform a further detailed functional annotation of the differentially expressed genes between hiPSC-EC and hESC-EC, we used Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems).
Real-time quantitative polymerase chain reaction analysis
Total RNA from each cell sample was isolated by RNeasy Mini Kit (QIAGEN), then it was reversed transcribed with High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative real-time polymerase chain reaction (PCR) was performed in triplicate on a StepOnePlus Real-time PCR system (Applied Biosystems) using Taqman primer probe sets (Applied Biosystems) for each gene of interest and glyceraldehyde 3-phosphate dehydrogenase control primer probe set for normalization.
Statistical analysis
Analysis of variance and 2-tailed Student's t-test were used. Differences were considered significant at P values of <0.05. Unless otherwise specified, data are expressed as mean±standard deviation.
Results
Endothelial differentiation of hiPSCs
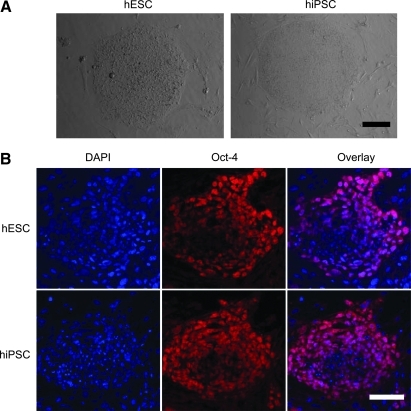
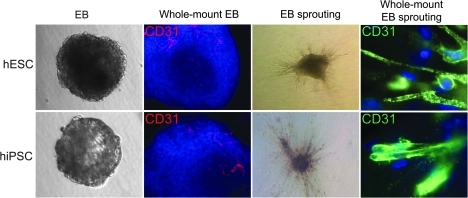
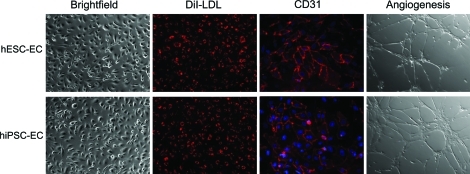
hiPSC colonies were found to be morphologically indistinguishable from hESCs (Fig. 1A). Both hESCs and hiPSCs showed similar expression patterns of stem cell markers Oct-4 on immunostaining (Fig. 1B). Using our previously optimized methods [10,11], we first carried out a series of experiments to determine whether the hiPSCs could generate endothelial cells. To isolate endothelial cells from hiPSCs, undifferentiated hiPSCs cultured on Matrigel-coated plated were placed into Petri dishes with differentiation medium for induction of EB formation. Previous studies have shown that differentiated hEBs from hESCs contain endothelial cells which can be isolated by CD31 [20] or CD34 [15] markers. Whole-mount immunostaining showed that within day-12 hiPSC-EBs, the CD31+ endothelial cells were organized in specific channel-like structures (Fig. 2). These data confirmed that hiPSCs cultivated as hiPSC-EBs spontaneously differentiated into endothelial cells and formed blood vessel-like structures. After embedding into collagen I and subculturing for an additional 3 days, hiPSC-EBs were observed to sprout channel-like outgrowths from these EBs in collagen I that stained positive for CD31 (Fig. 2). The population of CD31+ in hiPSC-EBs was further sorted, cultured, and propagated for further analysis to confirm endothelial cell traits. After sorting, CD31+ cells were further expanded as a nearly pure population (98%±1%), and these cells were used for subsequent experiments. To determine whether hiPSC-ECs resemble hESC-ECs in endothelial markers and angiogenesis potential, DiI-ac-LDL uptake assay, immunostaining, and Matrigel assay were used to confirm endothelial cell phenotype within these CD31+ purified cells. hiPSC-ECs morphologically resembled hESC-ECs, which were uniformly flat, adherent, and cobblestone-like in appearance. Both hiPSC-ECs and hESC-ECs expressed endothelial markers CD31 at the adherent junctions (Fig. 3). hiPSC-ECs also took up DiI-ac-LDL and rapidly formed vascular network-like structures when placed on Matrigel (Fig. 3). Taken together, these data confirm that these differentiated cells were endothelial lineage.
FIG. 1.
Morphology and pluripotent markers of hESC and hiPSCs. (A) hiPSCs exhibit colony formation similar to hESCs. (B) hESCs and hiPSCs showed similar expression pattern of Oct-4 under fluorescence microscopy. DAPI staining is used as a nuclear marker. Scale bar=50 μm. hESC, human embryonic stem cell; hiPSC, human induced pluripotent stem cells; DAPI, 4′,6-diamidino-2-phenylindole. Color images available online at www.liebertonline.com/scd
FIG. 2.
In vitro endothelial differentiation of hESCs and hiPSCs. Undifferentiated hESCs and hiPSCs subcultured in a low-attachment dish with differentiation medium supplement vascular endothelial growth factor and basic fibroblast growth factor can form EBs. Whole-mount immunochemistry of day-12 EBs show areas of CD31 (red) expression within EBs are organized in elongated clusters and channels in both groups. After 3 days of culturing in collagen matrix, hiPSC-EB and hESC-EB showed sprouting angiogenesis. Whole-mount immunostaining showed CD31 sprouting with channel-like vessel structures (green). Cell nuclei stained with DAPI (blue). EB, embryoid body. Color images available online at www.liebertonline.com/scd
FIG. 3.
In vitro characterization of hiPSC-ECs. Morphogenesis shows that the hiPSC-ECs are cobblestone-like. Similar to hESC-ECs, hiPSC-ECs can uptake DiI-ac-LDL (red). Histology shows that CD31 are expressed in cell membranes (red). Cell nuclei were stained with DAPI (blue). Angiogenesis assay reveals that hiPSC-ECs can form tube-like structure on Matrigel. DiI-ac-LDL, DiI-acetylated low-density lipoprotein; hiPSC-ECs, hiPSC-derived endothelial cells; hESC-ECs, hESC-derived endothelial cells. Color images available online at www.liebertonline.com/scd
Characterization of hiPSC-ECs
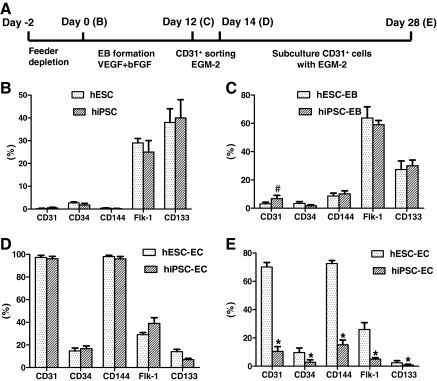
To further determine whether hiPSC-ECs resemble hESC-EC in endothelial markers and angiogenesis potential, we expanded FACS sorted CD31+ cells on fibronectin-coated plates supplemented with endothelial cell medium EGM-2 (Fig. 4A). For FACS analysis, we examined CD31, CD34, CD144, Flk-1, and CD133, which are known markers for endothelial differentiation of hESCs [10,20]. There are striking similarities between these 2 types of undifferentiated pluripotent cells (Fig. 4B). Initial FACS analysis showed that 6.8%±2.3% cells expressed CD31 marker in hiPSC-EBs, but 3.1%±1.2% did so in hESC-EBs (P<0.05) (Fig. 4C). This population of CD31+ was sorted, cultured, and propagated for further analysis to confirm endothelial cell traits. The CD31+ hiPSC-ECs initially expressed similar levels of CD34, CD144, Flk-1, and CD133 (Fig. 4D), but gradually lost expression of these surface markers after 2 weeks of subculturing (Fig. 4E). Interestingly, Flk-1 and CD133 were continuously expressed in both undifferentiated and differentiated hESCs as well as hiPSCs as shown by FACS. This pattern of expression is unlike mouse ESCs or umbilical cord blood in which Flk-1 and CD133 are specifically expressed on hemagioblasts [14,21].
FIG. 4.
Kinetic expression of endothelial markers during hESC and hiPSC differentiation and maintenance procedures. (A) An outline of the protocol used for differentiation of hESCs and hiPSCs into endothelial cell lineage. (B–E) indicate the time points used for flow cytometry analysis. (B) Flow cytometric analysis of endothelial markers for hiPSCs and hESCs. The expression pattern of hiPSCs was similar to hESCs. (C) CD31/CD144 expression was observed after 12 days subculturing by EB formation in both hiPSCs and hESCs. (D) hiPSC-ECs and hESC-ECs expressed robust endothelial markers CD31 and CD144 immediately after sorting. (E) After 2 weeks of subculture, endothelial marker expression downregulated significantly in hiPSC-ECs. Isotype-matched antibodies were used in flow cytometry for background fluorescence. Experiments were performed in triplicates. #P<0.05, *P<0.01 compared with hESC-EB or hESC-ECs. VEGF, vascular endothelial growth factor; bFGF, basic fibroblast growth factor.
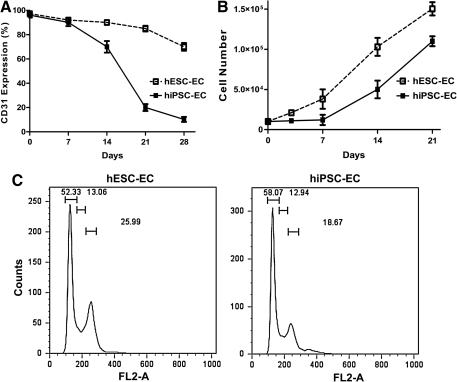
To further explore kinetic expression of CD31 on hiPSC-ECs during prolonged in vitro subculturing, repeated FACS analysis was carried out, and the results revealed that CD31 expression was downregulated significantly from week 1 to week 4 (Fig. 5A). Further, cell proliferation analysis demonstrated that hiPSC-ECs proliferated at a lower rate compared with hESC-ECs, especially during first few days (P<0.05 at day 3 and day 7) (Fig. 5B), which is consistent with a previous report [7]. By contrast, CD31 expression in hiPSC-ECs was downregulated swiftly after 10 days culture, which indicates that CD31+ endothelial cells from hiPSCs cannot be readily expanded in vitro. To further investigate the nature and extent of these biological differences, we carried out cell senescence and cell cycle analysis. Interestingly, neither hESC-ECs nor hiPSC-ECs expressed significant levels of β-galactosidase (Supplementary Fig. S1; Supplementary Data are available online at www.liebertonline.com/scd), which has been shown to be a reliable marker for cellular senescence [7]. Consistent with these results, apoptosis analysis revealed no obvious cell death in hiPSC-ECs and hESC-ECs (Supplementary Fig. S2). However, results obtained by PI stained cell cycle analysis with FACS for cell proliferation revealed 26.7%±2.1% and 17.6%±3.2% at the G2/M phase of hiPSC-EC and hESC-EC, respectively (P<0.05) (Fig. 5C), which is consistent with cell proliferation results (Fig. 5B). Finally, hiPSC-ECs displayed a large, fibroblast-like phenotype after subculture. In contrast, hESC-EC exhibited normal endothelial cell morphology (Supplementary Fig. S1).
FIG. 5.
Kinetic expression of CD31 and cell proliferation during iPSC-EC maintenance. (A) Kinetic expression of CD31 during hiPSC-EC in vitro subculturing. CD31 expression on hiPSC-EC downregulated significantly after 4 weeks of culturing when compared with hESC-EC. (B) Cell proliferation assay shows that hiPSC-ECs proliferated slowly, especially at first few days. Experiments were performed in triplicates. (C) Cell cycle analysis of hiPSC-ECs and hESC-ECs. FACS analysis of propidium iodide staining for cell proliferation revealed significant difference between hiPSC-ECs and hESC-ECs (P<0.05). The 3 gates from left to right represent G1, S, and G2/M cell cycle phases, respectively. Data are representative of 3 independent experiments. FACS, fluorescence activated cell sorting.
In vivo vasculogenic potential of hiPSC-ECs
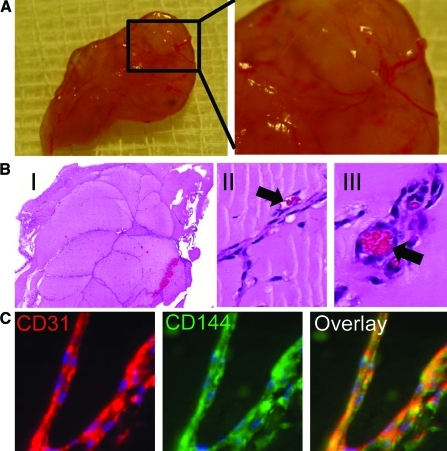
To evaluate whether hiPSC-ECs are capable of forming functional blood vessels in vivo, we used Matrigel plug and tissue-engineered vessel models in immunodeficient SCID mice that allow dynamic and long-term observation of blood vessels derived from implanted cells [18,19]. For Matrigel plug assay, 2 weeks after subcutaneous injection, Matrigel plugs were harvested, and histology was performed. Hematoxylin and eosin staining showed microvessels that contained murine blood cells in the vessel lumens. Harvested Matrigel plugs were also stained with anti-human CD31 and CD144 antibodies (Fig. 6).
FIG. 6.
Demonstration of functional vessels in vivo using Matrigel plug. (A) Matrigel plug with hiPSC-ECs were implanted via subcutaneous injection in the dorsal region of 8-week-old SCID mice. The plugs were harvested 2 weeks later. (B) (I) HE stain of Matrigel plug. (II, III) Some of these microvessels have mouse blood cells in their lumen (arrow). (C) 14-day Matrigel plugs were stained with anti-human CD31 (red) and anti-CD144 (green) antibodies, showing microvessels that are immunoreactive with these human-specific antibodies. Color images available online at www.liebertonline.com/scd
Gene profiles of hiPSC-EC differentiation
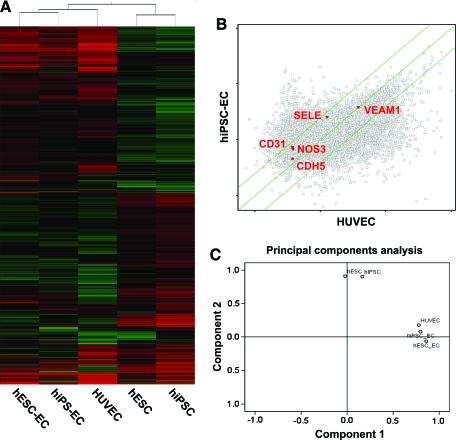
In order to define at a molecular level the changes occurring at each stage of hiPSC differentiation into endothelial cells, we next performed transcriptional profiling using whole human genome microarrays on (i) undifferentiated hiPSCs, (ii) undifferentiated hESCs, (iii) hiPSC-ECs after CD31 sorting, (iv) hESC-ECs after CD31 sorting, and (v) HUVECs as positive controls (n=4/group). The resulting data were analyzed using GeneSpring GX 10.0 to identify genes that had changed expression significantly between stages. A hierarchical clustering overview of the microarray experiments as a whole (Fig. 7A) likewise shows that the overall gene expressions among replicates of each stage are very similar, with progressive differences between more distantly separated stages. Of particular interest is the close clustering of hESC-ECs and hiPSC-ECs, indicating a high degree of similarity between their respective expression profiles immediately after CD31 sorting. Scatter plots showed a similar endothelial gene expression pattern between hESC-ECs and hiPSC-ECs (Fig. 7B). To obtain an overview of the transcriptional landscape, we looked at the data using principal components analysis, a dimensional reduction technique that identifies “principal components” or major trends in gene expression in the overall data (Fig. 7C). Principal components analysis demonstrates that differentiated cells and undifferentiated cells were significantly different between stages, as expected. Although hiPSC-ECs and hESC-ECs express endothelial cell genes similar to HUVECs, further investigation will be necessary to determine the nature and extent of the biological variances among these cell types, especially with regard to loss of endothelial phenotype after further subculturing of hiPSC-ECs as shown in Figs. 4 and 5.
FIG. 7.
Major themes in gene expression profiles at each stage of differentiation. (A) Hierarchical Clustering Analysis shows cells from each developmental stage cluster relatively close to each other, with the most distance between hESCs and HUVECs. (B) Scatter plots depicted gene expression fold changes between hiPSC-EC and HUVEC. Highlighted are endothelial genes PECAM-1, NOS3, VACAM1, SELE, and CDH5 expression (red). Green lines indicate fivefold changes in expression between samples. (C) Principal Component Analysis shows that hiPSC-EC, hESC-EC, and HUVEC are very similar, whereas undifferentiated groups (hESC and hiPSC) and differentiated groups (hiPSC-EC, hESC-EC, and HUVEC) separate significantly along components 1 and 2. Color images available online at www.liebertonline.com/scd
Essential differences between hESC-ECs and hiPSC-ECs
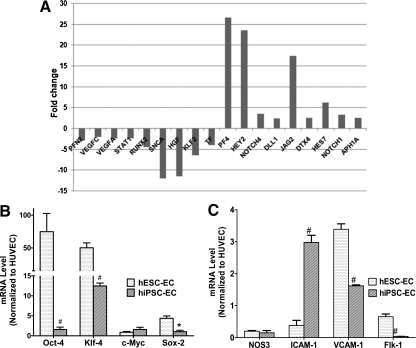
Although similarities exist between hiPSC-ECs and hESC-ECs, there are also considerable differences between them, especially with regard to their capability of cellular proliferation. To further investigate this difference, we performed a comparative analysis of the gene expression data of hiPSC-EC versus hESC-EC and performed a functional annotation of the differentially expressed genes between them (Supplementary Tables S1–S4). Interestingly, we found that a set of cellular proliferation-specific genes have significant difference in expression between the 2 groups. Figure 8 shows the expression fold-change of those genes. PF4 is a potent inhibiter of endothelial cell proliferation that is 26.5-fold upregulated in hiPSC-ECs compared with hESC-ECs [22]. Further, there is a significant upregulation of the genes related to Notch signaling pathway such as NOTCH4, DLL1, JAG2, DTX4, HEY2, HES7, NOTCH1, and APH1A. It has been previously reported that stimulation of Notch pathway suppresses endothelial cell proliferation [23,24] by limiting the number of endothelial progenitors within the mesoderm. Among the highly downregulated genes in hiPSC-ECs, SNCA and KLF2 play a positive modulatory role in endothelial cell proliferation [25,26]. Hence, insufficient induction of these genes in hiPSC-ECs might be a cause of their lower rate of proliferation. HGF is another highly downregulated gene that has been reported to play a role toward enhancing the proliferative, migratory, and angiogenic capabilities of endothelial progenitor cells [27]. PFN2 is one of the profilin group of proteins, the deletion of which causes defects in cellular migration and proliferation [28]. It has been reported that STATs contribute towards cellular proliferation. The signaling pathway of type I interferons activates STAT1 and STAT2, which promote endothelial cell proliferation [29]. Here we find that STAT1 is significantly downregulated in hiPSC-ECs, which might contribute to lowering the proliferative potential of these cells. RUNX2, a member of the RUNX family of transcription factors, contains a highly conserved DNA-binding runt domain and serves as a developmental regulator. Qiao et al. have shown the potential of RUNX2 in endothelial cell proliferation and cell cycle progression [30], which are downregulated in these cells. Further, we observed that both VEGFs, VEGFC and VEGFA, which induce endothelial cell proliferation and migration [31], are downregulated in hiPSC-ECs. Taken together, these data indicate an incomplete induction of cellular proliferation genes and higher expression of the genes inhibiting proliferation in hiPSC-ECs, which might explain the biological differences between the 2 cell populations. In order to further confirm microarray results and to measure viral transgenes together with a variety of endothelial markers, we performed real-time PCR analysis. We found that hiPSC-ECs expressed lower-level viral transgenes Oct-4 (P<0.01), Sox-2 (P<0.05), and Klf4 (P<0.01) compared with hESC-ECs (Fig. 8B). This indicates that the lentiviral transduction of reprogramming genes was silenced during the differentiation process and hence excludes reactivation of the viral transgenes on hiPSC-ECs as a potential cause of their biological differences compared with hESC-ECs. Interestingly, embryonic genes such as Oct-4 and Klf4 can still be detected in these differentiated cells, which is consistent with a previous report [32], indicating that hiPSC-ECs and hESC-ECs are not equivalent to HUVEC. Further, both hESC-ECs and hiPSC-ECs expressed endothelial constructive marker eNOS at a lower level compared with HUVEC, indicating that the 2 types of cells were still premature and probably did not fully represent somatic endothelial cells (Fig. 8C). Finally, hiPSC-ECs expressed a higher level of ICAM-1 compared with hESC-ECs (P<0.01), but a lower level of VCAM-1 and Flk-1 (P<0.01) (Fig. 8C).
FIG. 8.
Expression fold change of the genes that are differentially expressed in hiPSC-ECs compared with hESC-ECs and real-time polymerase chain reaction analysis. (A) Selected genes that play important modulatory roles in hiPSC-EC proliferation. (B) Real-time PCR of 4 viral transgenes normalized to glyceraldehyde 3-phosphate dehydrogenase shows significant upregulation of Oct-4, Klf-4, and Sox-2 in hESC-ECs compared with hiPSC-ECs. (C) Both hiPSC-ECs and hESC-ECs expressed similar levels of NOS3 but different levels of endothelial markers such as ICAM-1, VCAM-1, and Flk-1. #P<0.01, *P<0.05 compared with hESC-ECs.
Discussion
At present, hESCs are approved by the Food and Drug Administration for use in the treatment of acute spinal cord injury (www.geron.com) and Stargardt's macular dystrophy (www.advancedcell.com). The potential of hiPSC-based regenerative medicine is considered to be similar to that of hESCs, but largely free from the problems of ethical dilemmas and potential immune response [33,34]. For hiPSCs to be clinically useful, however, they should be free from genetic aberrations and capable of differentiating into fully committed cells. A better understanding of the gene profiling that guides their development and differentiation would allow the development of new techniques for hiPSC derivation. Moreover, researchers need to understand how best to evaluate the properties of hiPSC-derived differentiated cells and compare them with their natural counterparts in vivo. In this study, we have demonstrated that endothelial cells could be directionally induced from hiPSCs, recapitulating vasculogenesis in vitro and in vivo. However, our in vitro characterization and transcriptional analysis revealed broad similarities as well as significant differences between hESC-ECs and hiPSC-ECs.
Since the first description of hiPSCs [2,3], multiple laboratories have collectively made tremendous strides in developing more clinically acceptable hiPSCs and inducing in vitro differentiation of hiPSCs into a variety of cell types to suit human disease models. hiPSCs are molecularly and functionally similar to hESCs, which makes in vitro reprogramming an attractive approach to produce patient-specific stem cells for studying and potentially treating degenerative disease. In this study, we demonstrated that hiPSCs can differentiate into endothelial lineage after CD31+ sorting. Although hiPSC-ECs were able to recapitulate vasculogenesis, the in vitro subculture revealed limited growth rate compared with hESC-ECs as indicated by a lower G2/M phase population. We did not find early senescence or apoptosis in hiPSC-ECs as reported before [7], which might be explained by the different differentiation methods that were used. To investigate the mechanism of the slower growth and phenotype shift of hiPSC-ECs, we performed real-time PCR analysis, which excluded reactivation of the viral transgene as a potential cause. Interestingly, recent studies have revealed that somatic memory does exist [35–37]. This may explain the slower growth and fast loss of endothelial phenotype, as the donor cells for our hiPSC-ECs were derived from fetal fibroblasts. Further improvements in the derivation and differentiation processes are needed to facilitate clinical application of hiPSC-ECs in the future.
To help resolve these problems, our microarray analysis is designed to provide a greater understanding of the patterns of activation for specific genes during endothelial differentiation, by identifying novel gene targets that may be used to enhance endothelial differentiation, and developing more efficient induction strategies for hiPSCs derivation in the future. Our microarray data not only demonstrated that hESCs and hiPSCs are identical but also revealed substantial differences between hiPSC-ECs and hESC-ECs. These differences are explained by the higher expression of the genes inhibiting proliferation in hiPSC-ECs as compared with hESC-ECs. Comparative functional studies of the 2 cell types in the future will provide an opportunity to fine-tune our understanding of the molecular mechanisms underlying endothelial differentiation and somatic cell reprogramming.
In summary, we have shown that hiPSC-ECs are similar to hESC-ECs, but the 2 cell types also possess important biological differences with potential clinical significance. This work represents a comprehensive analysis of available microarray data on hiPSCs and hESCs and has produced interesting observations that should help guide future clinical applications of hiPSC-ECs.
Supplementary Material
Acknowledgments
This work was funded by NIH HL091453, HL089027, DP2OD04437, AG036142, Mallinckrodt Foundation, Burroughs Wellcome Foundation (J.C.W.), National Key Scientific Program of China (2011CB964903), and National Natural Science Foundation of China (31071308) (Z.L.).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Thomson JA. Itskovitz-Eldor J. Shapiro SS. Waknitz MA. Swiergiel JJ. Marshall VS. Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Yu J. Vodyanik MA. Smuga-Otto K. Antosiewicz-Bourget J. Frane JL. Tian S. Nie J. Jonsdottir GA. Ruotti V. Stewart R. Slukvin II. Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K. Tanabe K. Ohnuki M. Narita M. Ichisaka T. Tomoda K. Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Soldner F. Hockemeyer D. Beard C. Gao Q. Bell GW. Cook EG. Hargus G. Blak A. Cooper O. Mitalipova M. Isacson O. Jaenisch R. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye L. Chang JC. Lin C. Sun X. Yu J. Kan YW. Induced pluripotent stem cells offer new approach to therapy in thalassemia and sickle cell anemia and option in prenatal diagnosis in genetic diseases. Proc Natl Acad Sci U S A. 2009;106:9826–9830. doi: 10.1073/pnas.0904689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi KD. Yu J. Smuga-Otto K. Salvagiotto G. Rehrauer W. Vodyanik M. Thomson J. Slukvin I. Hematopoietic and endothelial differentiation of human induced pluripotent stem cells. Stem Cells. 2009;27:559–567. doi: 10.1634/stemcells.2008-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng Q. Lu SJ. Klimanskaya I. Gomes I. Kim D. Chung Y. Honig GR. Kim KS. Lanza R. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells. 2010;28:704–712. doi: 10.1002/stem.321. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J. Wilson GF. Soerens AG. Koonce CH. Yu J. Palecek SP. Thomson JA. Kamp TJ. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. 2009;104:e30–e41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stadtfeld M. Hochedlinger K. Induced pluripotentcy: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z. Suzuki Y. Huang M. Cao F. Xie X. Connolly AJ. Yang PC. Wu JC. Comparison of reporter gene and iron particle labeling for tracking fate of human embryonic stem cells and differentiated endothelial cells in living subjects. Stem Cells. 2008;26:864–873. doi: 10.1634/stemcells.2007-0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z. Wilson KD. Smith B. Kraft DL. Jia F. Huang M. Xie X. Robbins RC. Gambhir SS. Weissman IL. Wu JC. Functional and transcriptional characterization of human embryonic stem cell-derived endothelial cells for treatment of myocardial infarction. PLoS One. 2009;4:e8443. doi: 10.1371/journal.pone.0008443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ueland J. Yuan A. Marlier A. Gallagher AR. Karihaloo A. A novel role for the chemokine receptor Cxcr4 in kidney morphogenesis: an in vitro study. Dev Dyn. 2009;238:1083–1091. doi: 10.1002/dvdy.21943. [DOI] [PubMed] [Google Scholar]
- 13.Su W. Zhou M. Zheng Y. Fan Y. Wang L. Han Z. Kong D. Zhao RC. Wu JC. Xiang R. Li Z. Bioluminescence reporter gene imaging characterize human embryonic stem cell-derived teratoma formation. J Cell Biochem. 2010 doi: 10.1002/jcb.22982. [Epub ahead of print]; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li ZJ. Wang ZZ. Zheng YZ. Xu B. Yang RC. Scadden DT. Han ZC. Kinetic expression of platelet endothelial cell adhesion molecule-1 (PECAM-1/CD31) during embryonic stem cell differentiation. J Cell Biochem. 2005;95:559–570. doi: 10.1002/jcb.20436. [DOI] [PubMed] [Google Scholar]
- 15.Chen T. Bai H. Shao Y. Arzigian M. Janzen V. Attar E. Xie Y. Scadden DT. Wang ZZ. Stromal cell-derived factor-1/CXCR4 signaling modifies the capillary-like organization of human embryonic stem cell-derived endothelium in vitro. Stem Cells. 2007;25:392–401. doi: 10.1634/stemcells.2006-0145. [DOI] [PubMed] [Google Scholar]
- 16.Lin HT. Kao CL. Lee KH. Chang YL. Chiou SH. Tsai FT. Tsai TH. Sheu DC. Ho LL. Ku HH. Enhancement of insulin-producing cell differentiation from embryonic stem cells using pax4-nucleofection method. World J Gastroenterol. 2007;13:1672–1679. doi: 10.3748/wjg.v13.i11.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamashita J. Itoh H. Hirashima M. Ogawa M. Nishikawa S. Yurugi T. Naito M. Nakao K. Nishikawa S. Flk1-positive cells derived from embryonic stem cells serve as vascular progenitors. Nature. 2000;408:92–96. doi: 10.1038/35040568. [DOI] [PubMed] [Google Scholar]
- 18.Malinda KM. In vivo matrigel migration and angiogenesis assays. Methods Mol Med. 2003;78:329–335. doi: 10.1385/1-59259-332-1:329. [DOI] [PubMed] [Google Scholar]
- 19.Wang ZZ. Au P. Chen T. Shao Y. Daheron LM. Bai H. Arzigian M. Fukumura D. Jain RK. Scadden DT. Endothelial cells derived from human embryonic stem cells form durable blood vessels in vivo. Nat Biotechnol. 2007;25:317–318. doi: 10.1038/nbt1287. [DOI] [PubMed] [Google Scholar]
- 20.Levenberg S. Golub JS. Amit M. Itskovitz-Eldor J. Langer R. Endothelial cells derived from human embryonic stem cells. Proc Natl Acad Sci U S A. 2002;99:4391–4396. doi: 10.1073/pnas.032074999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang C. Zhang ZH. Li ZJ. Yang RC. Qian GQ. Han ZC. Enhancement of neovascularization with cord blood CD133+ cell-derived endothelial progenitor cell transplantation. Thromb Haemost. 2004;91:1202–1212. doi: 10.1160/TH03-06-0378. [DOI] [PubMed] [Google Scholar]
- 22.Gupta SK. Hassel T. Singh JP. A potent inhibitor of endothelial cell proliferation is generated by proteolytic cleavage of the chemokine platelet factor 4. Proc Natl Acad Sci U S A. 1995;92:7799–7803. doi: 10.1073/pnas.92.17.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer A. Schumacher N. Maier M. Sendtner M. Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 2004;18:901–911. doi: 10.1101/gad.291004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee CY. Vogeli KM. Kim SH. Chong SW. Jiang YJ. Stainier DY. Jin SW. Notch signaling functions as a cell-fate switch between the endothelial and hematopoietic lineages. Curr Biol. 2009;19:1616–1622. doi: 10.1016/j.cub.2009.07.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee SS. Kim YM. Junn E. Lee G. Park KH. Tanaka M. Ronchetti RD. Quezado MM. Mouradian MM. Cell cycle aberrations by alpha-synuclein over-expression and cyclin B immunoreactivity in Lewy bodies. Neurobiol Aging. 2003;24:687–696. doi: 10.1016/s0197-4580(02)00196-3. [DOI] [PubMed] [Google Scholar]
- 26.Huang P. HDAC5: going with the flow. Blood. 2010;115:2728–2729. doi: 10.1182/blood-2010-01-263681. [DOI] [PubMed] [Google Scholar]
- 27.Song MB. Yu XJ. Zhu GX. Chen JF. Zhao G. Huang L. Transfection of HGF gene enhances endothelial progenitor cell (EPC) function and improves EPC transplant efficiency for balloon-induced arterial injury in hypercholesterolemic rats. Vascul Pharmacol. 2009;51:205–213. doi: 10.1016/j.vph.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Ding Z. Lambrechts A. Parepally M. Roy P. Silencing profilin-1 inhibits endothelial cell proliferation, migration and cord morphogenesis. J Cell Sci. 2006;119:4127–4137. doi: 10.1242/jcs.03178. [DOI] [PubMed] [Google Scholar]
- 29.Gomez D. Reich NC. Stimulation of primary human endothelial cell proliferation by IFN. J Immunol. 2003;170:5373–5381. doi: 10.4049/jimmunol.170.11.5373. [DOI] [PubMed] [Google Scholar]
- 30.Qiao M. Shapiro P. Fosbrink M. Rus H. Kumar R. Passaniti A. Cell cycle-dependent phosphorylation of the RUNX2 transcription factor by cdc2 regulates endothelial cell proliferation. J Biol Chem. 2006;281:7118–7128. doi: 10.1074/jbc.M508162200. [DOI] [PubMed] [Google Scholar]
- 31.Bernatchez PN. Rollin S. Soker S. Sirois MG. Relative effects of VEGF-A and VEGF-C on endothelial cell proliferation, migration and PAF synthesis: role of neuropilin-1. J Cell Biochem. 2002;85:629–639. doi: 10.1002/jcb.10155. [DOI] [PubMed] [Google Scholar]
- 32.Jozefczuk J. Prigione A. Chavez L. Adjaye J. Comparative analysis of human embryonic stem cell and induced pluripotent stem cell-derived hepatocyte-like cells reveals current drawbacks and possible strategies for improved differentiation. Stem Cells Dev. 2011 doi: 10.1089/scd.2010.0361. [Epub ahead of print]; [DOI] [PubMed] [Google Scholar]
- 33.Takahashi K. Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 34.Sun N. Longaker MT. Wu JC. Human iPS cell-based therapy: considerations before clinical applications. Cell Cycle. 2010;9:880–885. doi: 10.4161/cc.9.5.10827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh Z. Wilson KD. Wu Y. Hu S. Quertermous T. Wu JC. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS One. 2010;5:e8975. doi: 10.1371/journal.pone.0008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stadtfeld M. Apostolou E. Akutsu H. Fukuda A. Follett P. Natesan S. Kono T. Shioda T. Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urbach A. Bar-Nur O. Daley GQ. Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6:407–411. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.