Abstract
Predation pressure represents a strong selective force that influences the development and evolution of living organisms. An increasing number of studies have shown that both environmental and social factors, including exposure to predators, substantially shape the structure and function of the brain. However, our knowledge about the molecular mechanisms underlying the response of the brain to environmental stimuli is limited. In this study, we used whole-genome comparative oligonucleotide microarrays to investigate the brain transcriptomic response to cues of a predator in the threespine stickleback, Gasterosteus aculeatus. We found that repeated exposure to olfactory, visual and tactile cues of a predator (rainbow trout, Oncorrhynchus mykiss) for 6 days resulted in subtle but significant transcriptomic changes in the brain of sticklebacks. Gene functional analysis and gene ontology enrichment revealed that the majority of the transcripts differentially expressed between the fish exposed to cues of a predator and the control group were related to antigen processing and presentation involving the major histocompatibility complex, transmission of synaptic signals, brain metabolic processes, gene regulation and visual perception. The top four identified pathways were synaptic long-term depression, RAN signaling, relaxin signaling and phototransduction. Our study demonstrates that exposure of sticklebacks to cues of a predator results in the activation of a wide range of biological and molecular processes and lays the foundation for future investigations on the molecular factors that modulate the function and evolution of the brain in response to stressors.
Key Words: Neurogenomics, Stress, Predation, Microarray, Gene expression, Gasterosteus aculeatus
Introduction
Physiological and behavioral reactions to predators are conserved across organisms, and predation pressure has played an important role in the evolution of brain structures that mediate responses to environmental cues [Cantor, 2009]. Thus, studying how the brain responds to the threat of predation could provide insights into the mechanisms underlying neural plasticity and how they have evolved. However, our understanding of the molecular and biological processes underlying the response to predators is in its infancy. Important recent studies have shown that exposure to a predator results in the activation of particular neural circuitry in the brain [e.g. Nanda et al., 2008] and that specific genes, including immediate early genes [e.g. Comoli et al., 2003; Blanchard et al., 2005; Motta et al., 2009], are upregulated in the brain in response to the threat of predation. For example, corticotropin-releasing factor genes were upregulated in the amygdala of rats 3 h after a 10-min exposure to a ferret predator [Roseboom et al., 2007].
We know less about changes in the brain that might occur in response to repeated exposure to predation risk over longer periods of time. Most studies have measured gene expression within minutes after a single exposure to predation risk [Wang et al., 2003; Nanda et al., 2008; Leder et al., 2009; Mori et al., 2009]. However, animals are often recurrently exposed to cues of predators, and other studies suggest that the response of the hypothalamic-pituitary-adrenal axis to acute versus chronic stress is different [McEwen, 2007]. Recent gene expression studies have supported this hypothesis by showing that an acute stressor activates different genes, and a different number of genes, compared to a chronic stressor. For example, different sets of genes are involved in the transcriptomic response to acute versus chronic stress in the brain [Krasnov et al., 2005; Cairns et al., 2008]. Other studies have shown that the number of differentially expressed genes in response to a stressor depends on the duration of the stressor. For example, there were more differentially expressed genes in proximal tubule cells after 1 day of exposure to cadmium compared to 13 days of exposure [Garrett et al., 2011]. On the other hand, another study found that there were more differentially expressed genes in the liver in response to confinement stress in sea bream that had been confined for 20 h compared to fish that had been confined for 120 h [Calduch-Giner et al., 2010]. We also know that exposure to predation risk can have long-term effects on antipredator behavior [Tulley and Huntingford, 1987; Magurran, 1990; Kelley and Magurran, 2003], which suggests that there might be changes in brain gene expression in response to predation risk that are relatively enduring.
The power of genomics has only recently been applied to understand the coordinated action of the entire transcriptome in response to predation [but see Wang et al., 2003; Roseboom et al., 2007]. Whole-genome surveys are an attractive strategy because it is likely that the response to threat of predation involves multiple interacting genes and pathways. In addition, transcriptomic profiling can reveal which genes display similar expression patterns and opens the possibility of more integrated analyses of whole-genome dynamics via techniques such as pathway, network and gene ontology (GO) analyses. Indeed, new insights have emerged from studies measuring the whole transcriptomic response to other stressors, e.g. pollution [Craig et al., 2009], disease [Ching et al., 2010] and confinement [Kennerly et al., 2008].
Threepined sticklebacks (Gasterosteus aculeatus) are small teleost fish whose evolution has been strongly influenced by predation pressure. Sticklebacks are subject to a wide range of predators including piscivorous fishes, birds, odonate larvae and snakes [Reimchen, 1994]. Much of the extensive phenotypic variation among freshwater stickleback populations, including morphology [Reimchen, 1980], behavior [Huntingford et al., 1994], physiology [Bell et al., 2010] and life history traits [Moodie, 1972; Baker et al., 1998] can be attributed to variation in predator pressure. For example, sticklebacks from populations that are subject to high levels of predation pressure have more exterior armor (lateral plates) [Reimchen, 1994], show stronger antipredator behavioral responses to the immediate risk of predation [Huntingford et al., 1994] and are more physiologically responsive to the threat of predation as assessed by respiration rate [Bell et al., 2010]. A recent study of the time course of the acute response to predation risk [Bell et al., 2007] showed that acute exposure to predation risk stimulated a glucocorticoid stress response within minutes, with levels of cortisol increasing up to 1 h after exposure to predation risk. While recent studies have successfully pinpointed the genes underlying the evolution of morphological defenses against predators in sticklebacks [Peichel et al., 2001; Cresko et al., 2004; Shapiro et al., 2004; Colosimo et al., 2005; Chan et al., 2010], we know less about the molecular basis of plastic behavioral and physiological reactions to predators within an individual's lifetime. Growing genomic resources for sticklebacks [Kingsley and Peichel, 2007], including a full genome sequence, make this organism a suitable model for investigating the molecular mechanisms that have evolved in response to predation pressure.
Therefore, we investigated the brain's response to cues of a predator using whole-genome microarrays in sticklebacks. We hypothesized that sticklebacks exposed to predation risk would exhibit differential transcriptomic responses compared to a control group.
Materials and Methods
Sticklebacks
We studied laboratory-reared F1 female sticklebacks derived from a wild population in Putah Creek, Calif., USA. We elected to study females in this experiment because males undergo dramatic changes in behavior over the reproductive period, some of which could influence an individual's response to predation risk [Wootton, 1976]. The fish were maintained in our laboratory in 104-liter tanks under 8:16-hour light/dark photoperiod. An average of 10% of the water in the tanks was exchanged each day. The adult fish were fed ad libitum with a mixture of bloodworms, brine shrimp and mysis shrimp.
Adult fish used in the study were sampled from a tank containing representatives of 33 different families. Fish were acclimated together in one tank for 2 weeks prior to the experiment. Fish were placed in six different 26-liter tanks with three fish per tank in a partially recirculating flow-through system. The water was filtered through particulate, UV, biological and charcoal filters, which remove olfactory cues. Half of the tanks were assigned to the control group and the other half to the experimental group. The position of the control and experimental tanks was randomized in the room and the tanks were isolated from external disturbances during the entire duration of the experiment by opaque covers on the back and front of the tanks. The fish were allowed to acclimate for 3 days during which the sides of the tanks were left open to allow the fish to visually interact with fish in neighboring tanks. Dividers were then inserted between tanks for the remainder of the experiment.
Applying Predator Cues
We designed a procedure to mimic the situation of sticklebacks that are repeatedly exposed to a high level of predation pressure in the field. Specifically, over 6 days, sticklebacks were exposed to olfactory, visual and tactile cues of rainbow trout, a natural predator, in order to simulate real predation risk. This procedure elicited enduring changes in stickleback growth and behavior [Dingemanse et al., 2009; Bell et al., 2011], and other studies using this population have shown that they exhibit strong antipredator responses to rainbow trout [Bell and Sih, 2007]. Nanda et al. [2008] used a similar procedure to measure the immediate brain transcriptomic response of rats to olfactory, visual and acoustic cues of predators. Sticklebacks were exposed to two types of olfactory cues. First, olfactory cues of rainbow trout were applied to simulate the presence of trout in the environment. Olfactory cues of trout were prepared by soaking 1 cm2 trout skin in 1 liter of distilled water for 2 h. Second, cues of dead stickleback were applied to simulate predation on nearby conspecifics. The dead-stickleback cues were prepared by soaking one sacrificed fish in 1 liter of distilled water for 2 h. Incisions were made in the skin on the sides of the fish with a clean razor to release cues. This procedure releases alarm pheromones in the skin of sticklebacks [Brown and Godin, 1997].
The infusions were frozen in trays and the ice cubes were thawed in a beaker for 1 h prior to use. Once a day for 6 days, at a random time of day, we turned the water off in the tank and added 50 ml of trout infusion and 50 ml of dead stickleback infusion. The olfactory cues were placed within 30 cm of the majority of sticklebacks in the tank. On days 2 and 5 of the treatment period, a trout model was added to the tank at the site of application of the olfactory cues, held still for 30 s, and then quickly moved toward the corner of the tank that contained the majority of the sticklebacks, then to the side and back again. We used a model of a rainbow trout constructed of clay and painted realistically (20.5 cm standard length). The model was thoroughly rinsed in distilled water before being reused. On day 6, the sticklebacks were chased by the model trout for an entire minute. Fish in the control tanks were treated exactly the same except that 100 ml clean water was added instead of olfactory cues, and they were not exposed to the model predator. To control for disruption caused by adding the predator, we splashed the surface of the water in the control tanks once within 30 cm of the majority of the sticklebacks on days 2 and 5 and on day 6, the water was splashed several times for 1 min. The water in the tank was turned on 3 h after the treatment was applied. On the 7th day, all the fish (n = 18) were quickly netted and sacrificed by decapitation using sharp scissors. All methods were approved by IACUC protocol (No. 06178) of the University of Illinois at Urbana-Champaign.
RNA Extraction and cDNA Synthesis
Whole fish brains were individually dissected and placed immediately on ice in 500 μl of Trizol®reagent (Invitrogen, Carlsbad, Calif., USA). Total RNA was isolated immediately from individual brains (n = 18) using Trizolreagent according to the manufacturer's recommendation and subsequently purified on columns with RNeasy kit (Qiagen GmbH, Hilden, Germany). RNA was eluted in a total volume of 60 μl in RNase-free water. In addition, to remove contaminating genomic DNA, all RNA samples were treated with DNase using the Turbo DNA free kit (Ambion, Austin, Tex., USA) following the manufacturer's instructions. The quantity of RNA was estimated using a NanoDrop 1000 (Thermo Fisher Scientific, Inc., Waltham, Mass., USA) and the quality was assessed using the Agilent Bioanalyzer 2100 (Agilent, Santa Clara, Calif., USA). RNA was immediately stored at –80°C for 1 week. To generate enough double-stranded cDNA for hybridization, we first amplified RNA (aRNA) from 1 μg of total RNA of 10 fish used in microarray using the MessageAmp II aRNA amplification kit (Applied Biosystems/Ambion, Austin, Tex., USA) following the manufacturer's instructions. Other studies have suggested that aRNA does not introduce significant bias in gene expression data [Feldman et al., 2002; Polacek et al., 2003]. Nonamplified RNA was preserved for microarray validation experiments. The aRNA was subsequently converted to double-stranded cDNA using the SuperScript® Double-Stranded cDNA Synthesis Kit (Invitrogen). cDNA was purified using the Qiaquick PCR Purification Kit (Qiagen). All samples for microarray were successfully amplified. The cDNA yields ranged from 3 to 6 μg. In accordance with NimbleGen (Madison, Wisc., USA) requirements, 2.5 μg of double-stranded cDNA were sent to the NimbleGen Microarray Service Department for hybridization.
Chip Design
The arrays were printed using the NimbleGen proprietary Maskless Array Synthesis technology on a 385 K high-density chip using both the 44,884 in silico Genscan predictions from the stickleback genome and the 27,633 gene transcripts obtained from ENSEMBL v.45.1c. Genscan predictions are based on transcriptional, translational and splicing signals as well as the length and compositional distributions of exons, introns and intergenic regions. They differ from the transcripts, which have been backed by some experimental evidence. 60-bp probes were designed from a total of 72,517 sequences. Of these genes, all except 12 sequences had 5 probes per gene. No probes were found for 44 sequences because all of them were shorter than the minimum 60-bp probe size. Probes were designed taking into account various criteria such as nonrepetitive elements, frequency, uniqueness and melting temperature according to the NimbleGen probe design procedure.
Microarray Design and Hybridization
Ten samples were used for microarray analysis. The samples comprised 5 biological replicates in the experimental group (fish exposed to cues of predator) and 5 biological replicates in the control group (fish not exposed to cues of predator). Samples were selected to ensure even distribution among the tanks and high RNA quality. Three tanks contributed 1 individual, 2 tanks contributed 2 individuals and 1 tank contributed 3 individuals to the experiment. The cDNA-labeling (single color), hybridization, washing and scanning steps were performed in the NimbleGen Microarray Gene Expression Service Department. In addition, NimbleGen also conducted the data feature extraction and generated the XYS file format that was used in data analysis. The microarray data have been deposited in NCBI's Gene Expression Omnibus (GEO, http://www.ncbi.nlm.gov/geo) under accession No. GSE21599, GPL10368.
Data Analysis
The microarray data were analyzed using various packages in R Bioconductor [R Development Core Team, 2009]. Although both the in silicoGenescan predictions and transcripts were included in the microarray chips, we focused the analysis only on the transcripts of confirmed function because some of the Genescan predictions do not have biological function. Genescan predictions were not further considered in the analysis. Differential expression of genes between the control group and the experimental group was assessed using the package Oligo in R Bioconductor [Carvalho et al., 2007]. As required for the package Oligo, we first built a package ‘pd.2007.07.05.g.aculeatus.expr’, which contains the annotation information of the arrays using pdInfoBuilder package [Carvalho et al., 2007]. This package was then installed in R and used for analysis. Background subtraction, quantile normalization and summarization were performed using the Robust Multichip Average Algorithm. This algorithm performs three steps in the following sequence: background correction, quantile normalization and log2-transformed perfect-match value summarization, a method that averages the probe's expression intensities for each transcript [Irizarry et al., 2003]. Before exploring the differences in gene expression between the two groups, we first applied a nonspecific filtering method to the data to remove the probe sets (transcripts) with no or very low expression. This approach is commonly used to improve the detection power in microarray data analysis, particularly when testing multiple hypotheses [Calza et al., 2007; Bourgon et al., 2010]. We applied the function rowSds in the package genefilter, which calculates the standard deviation of expression intensities for each row (probe set). We then estimated the ‘peak’ of the distribution using the function shorth (the shortest interval containing half of the data). Probes with standard deviation below the shorth value were filtered out of the data. The quality of the arrays was assessed by examining the boxplots and the density plots of the log2-transformed intensity values of the probes. Hierarchical clustering was done using the function hclust in the package stats in R Bioconductor by employing the complete linkage method as the agglomeration procedure.
We used principal components analysis as a dimension reduction method to visualize the data using the plotPCA function in R. The experimental treatment (predator-exposed fish and control group) were the variables and the gene expression measurements served as the observations.
The log2-fold changes (logFCs) and moderated t statistics were computed to infer the differential expression of genes between the control and experimental groups using the Bayesian moderated t test in the package LIMMA (LinearModels for Microarray Data) in R Bioconductor. We used a false discovery rate (FDR) to correct for multiple testing [Benjamini and Hochberg, 1995]. The function topTable was used to rank the p values.
Because some fish used in the experiment were from the same tank, we conducted a post-hoc analysis of the top differentially expressed genes to detect any possible tank effect; these results are shown graphically in online supplementary figure 1 (www.karger.com/doi/10.1159/000328221).
Gene Function, GO and Pathway Analysis
Gene functional studies and GO enrichment analysis were conducted in R Bioconductor using the ENSEMBL dataset Gasterosteus_aculeatus. BROADS1. 58. The annotation of the differentially expressed genes was determined using the package BiomaRt as implemented in R. Genes with no known function in the stickleback genome annotation were manually annotated using the BLASTX algorithm against the nonredundant protein database. The output of the file was parsed using a custom Perl script.
GO enrichment was carried out using the package GOstat. As a gene universe (background), we used the transcripts that were maintained after nonspecific filtering and we tested for overrepresentation of transcripts with raw p values ≤0.01 as determined by the Bayesian statistics. Both the gene universe and the target genes were first converted into ENTREZIDs using human homologues by the function getBML in BiomaRt because the stickleback genes have not been assigned ENTREZIDs, which are used in GOstat. The overrepresented biological processes (BPs), molecular functions (MFs) and cellular components (CCs) were determined by using the function hyperGtest with conditional testing, which removes the effect of child GO terms before testing parents. We also performed pathway and network analyses using the Ingenuity Pathways Analysis (IPA) software version 8.7 (Ingenuity Systems, Redwood City, Calif., USA). IPA is a proprietary application that uses data from its own repository of biological interactions and functional annotations from all species known as Ingenuity Knowledge Base, a manually curated database that is updated weekly and quarterly using annotations and findings from public and commercial databases and from information extracted from published biomedical literature. Third-party sources of database used by IPA include, for example, the NCBI Entrez Gene, Refseq and OMIM, the GWAS Database, GO annotation, Hazardous Substances Database, Human Metabolome Database, GNF Tissue Expression Body Atlas and KEGG metabolic pathway information. Since IPA does not accept stickleback transcript IDs, the differentially expressed genes were converted into ENTREZIDs using human homologues.
Microarray Validation
Microarray validation was conducted on samples used in microarray experiment and also on 4 samples that were part of the experiment but were not included in the microarray gene expression analysis. Total RNA from each sample was reverse transcribed using the SuperScript® III First-Strand Synthesis System (Invitrogen). Quantitative real-time PCR (qPCR) reactions were performed on the Applied Biosystems 7900HT (Applied Biosystems) using the FastStart Universal SYBR Green Master (Rox) (Roche Diagnostics GmbH, Mannheim, Germany) following the manufacturer's recommendations. The housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as internal reference because the average expression values of this gene did not vary significantly across samples in the microarray experiment. Genes with highest fold change and lowest p values, as determined in R Bioconductor, are more likely to be differentially expressed between conditions, and were therefore selected for validation. Primers for validation were designed from each gene transcript using the Primer Express® Software v1.0 (Applied Biosystems). Primers were redesigned each time amplification across samples was not consistent. This was the case for the rhodopsin (RHO) genes, which are represented by two differentially expressed splice variants (ENSGACT00000003757, ENSGACT00000003760). The genes selected for validation and their respective primers are listed in table 1. Each reaction was run in triplicate and a dissociation curve was used to assess a single amplicon. No-template controls were included in the reaction to ensure the specificity of amplification. Primers were redesigned whenever primer dimers were observed in the dissociation curve. A standard curve was computed using a 10× serial dilution of samples made by pooling equal quantities of all samples, including both the control and experimental samples. A standard curve was run for each gene separately. The qPCR data were analyzed using the Relative Expression Software tool (REST©) [Pfaffl et al., 2002]. To test for consistence between the samples used in microarray and the independent biological replicates, we compared the expression values of three most differentially expressed genes in these samples using a one-way ANOVA using StatPlusMac (AnalystSoft, Vancouver, B.C., Canada).
Table 1.
List of primers used in the qPCR validation assays
| Gene | Forward sequence | Reverse sequence | Amplicon size bp |
|---|---|---|---|
| GAPDH | CAAACCGTTGGTGACAGTATTTG | GCACTGAGCATAAGGACACATCTAA | 69 |
| RHO_57 | ATCCCTTTCCTCTCAGGAAAAAA | TCAAGTCAGCCCACTACAGAGTTG | 95 |
| RHO 60 | CCCCGAGGGCATGCA | TGTAAGTGGCGAACATGTAGATGA | 95 |
| USP-like | TGACTCACAGCGCCACATG | TGAGAGAAGCCAAAAAAAGC | 71 |
| Tr_22897 | TGCGCTGGAACGAAGATG | GCCCTGAGGTCCGAAGCT | 60 |
| PLA2G12B | TCTCTCCTCAGCATGTGAAACG | AGGGTCTGCAGCCCAGAGT | 71 |
| GLUT1 | GTCATGTAACCCTGATGAAAAGGA | CGCGCCGCAGACTTTCT | 60 |
| C2 | AGGTGCTCACTGCGCTTACA | AAGTGCAACCGCTGTGCAT | 70 |
| CXCR1 | CCCTGAGCACTGCTTTGTTTT | GGTTGACGCAGCTGTGGAA | 60 |
| KCNK12 | CCACGGTCGCCCACAT | ACAATCTGCGGCCCTCAGTA | 67 |
| PCYT1B | CCTCCGTCCCTGAGCAGAA | TGGCGTAGGACAGCATTCG | 64 |
GADPH (glyceraldehyde 3-phosphate dehydrogenase): reference gene used for normalization of the qPCR results. Target genes were RHO_57 (rhodopsin transcript variant ENSGACT-00000003757), RHO_60 (rhodopsin transcript variant ENSGACT00000003760), USP-like (ubiquitin specific protease-like), Tr_22897 (transcript ‘ENSGACT00000022897’), PLA2G12B (phospholipase 2 group XIIB), GLUT1 (glucose transporter 1), C2 (complement precursor), CXCR1 (interleukin-8 receptor alpha), KCNK12 (potassium channel), and PCYT1B (phosphate cytidylyltransferase 1, choline, beta).
Results
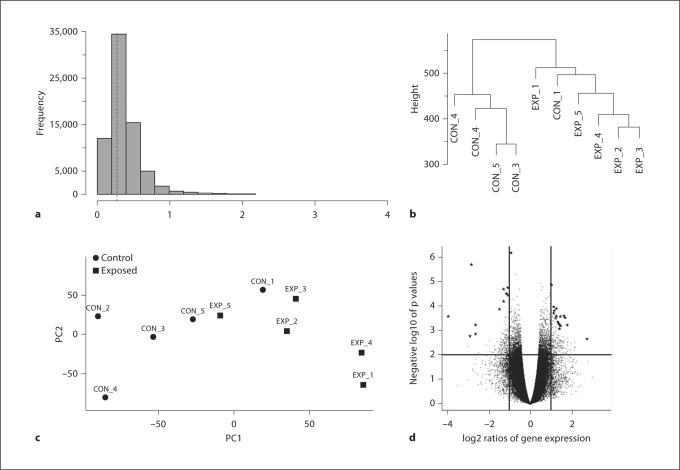
As a result of nonspecific filtering, 36% of probe sets (transcripts) with standard deviations below the shorth value (0.268) were removed because they had low expression levels (fig. 1a). The target genes of these probe sets were either not expressed in the samples or their expression was not significantly higher than above background level. In total, 17,681 transcripts were maintained for further analysis.
Fig. 1.
Methods used in preprocessing and quality control of the arrays. a Histogram of standard deviations. Genes with very low standard deviations are more likely not to be differentially expressed and were removed from the data. The dotted line represents the peak of the distribution (shorth value). b Hierarchical clustering of the samples (arrays). Control group: CON_1–5; experimental group: EXP_1–EXP_5. c Principal components plots used as dimension reduction of the samples (arrays). d Volcano plot highlighting the top 30 differentially expressed genes (★) between sticklebacks exposed to predator cues and the control group.
The hierarchical clustering based upon row expression values showed that all the samples clustered according to treatment, except 1 sample (CON_1), which grouped with the experimental samples (fig. 1b). However, this sample clustered with the control group based upon differential expression (fig. 2). The principal-component analysis shown in figure 1c revealed that 74% of the variation was explained by the first principal component (PC1), suggesting that the majority of observed changes in gene expression were caused by exposure to predator cues. PC2 and PC3 explained 8.6 and 6.0% of the variation, respectively.
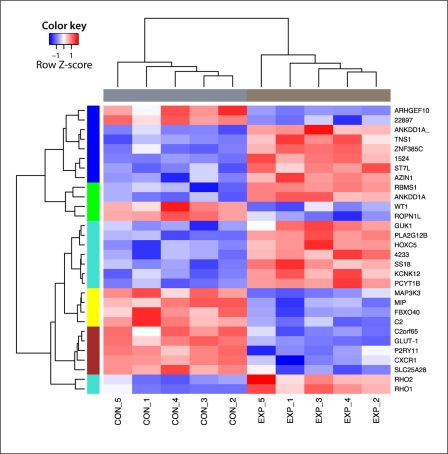
Fig. 2.
Heat map of top 30 differentially expressed genes between control (CON_1–5) and experimental (EXP_1–5) group with gene names. The Z-score represents the number of standard deviation units a specific gene expression value is from the mean (defined as 0). Red color in the heat map indicates upregulated genes, in blue are downregulated genes and in white are genes that do not change between conditions. Color-coded bars represent the clustering of samples per treatment (horizontal bar) and clusters of differentially expressed genes (vertical bar).
Differential Expression
There was no evidence that any of the treatment differences were driven by specific tanks within each treatment. Specifically, the random effect of tank on the expression of the top 15 genes was never statistically significant (p > 0.05); less than 0.04% of the variance could be attributed to tank effects (see online suppl. fig. 1).
A total of 735 transcripts had a logFC ≥2, of these, 30 had a logFC ≥4. Of the 17,681 transcripts, a total of 6,596 were upregulated and 11,085 were downregulated. Figure 1d shows a volcano plot highlighting the top 30 differentially expressed transcripts.
Based upon the empirical Bayesian t statistics, 909 transcripts were differentially expressed at p < 0.01. Multiple testing on transcripts using the method of Benjamini and Hochberg [1995] revealed that 30 transcripts were differentially expressed at FDR = 0.15 (table 2). While some differentially expressed genes had large fold changes (e.g. RHO, logFC = 3.96), others appeared to be expressed at lower levels (e.g. GLUT-1, logFC = –1.5). Figure 2 is a heatmap generated using the top 30 differentially expressed transcripts with lowest p values, and reveals a clear pattern that distinguishes between fish exposed to predator cues and the control group.
Table 2.
List and annotation of the top transcripts differentially expressed in the brain of sticklebacks exposed to predator cues using a p = 0.001 cutoff
| Transcript ID | logFC | Ave. Exprs | p value | Adj. P. Val | Gene name | Gene description |
|---|---|---|---|---|---|---|
| ENSGACT00000003757 | 3.96 | 6.81 | 2.00E-06 | 0.035 | RHO | rhodopsin |
| ENSGACT00000022897 | −1.71 | 8.93 | 1.99E-05 | 0.105 | NA | novel protein |
| ENSGACT00000024046 | 1.28 | 5.88 | 2.14E-05 | 0.105 | PLA2G12B | group XIIB secretory phospholipase A2-like protein |
| ENSGACT00000012440 | 1.47 | 5.65 | 2.38E-05 | 0.105 | HOXC5 | homeobox protein Hox-C5 |
| ENSGACT00000019166 | −1.82 | 7.59 | 3.21E-05 | 0.113 | CXCR1 | interleukin-8 receptor |
| ENSGACT00000003760 | 2.62 | 6.76 | 4.03E-05 | 0.115 | RHO | rhodopsin |
| ENSGACT00000025444 | 2.91 | 7.90 | 4.90E-05 | 0.115 | USP-like | ubiquitin-specific protease-like protein |
| ENSGACT00000003143 | −1.28 | 11.00 | 5.18E-05 | 0.115 | ROPN1L | ropoporin |
| ENSGACT00000019615 | −1.45 | 7.05 | 7.88E-05 | 0.132 | SLC25A28 | mitoferrin-2 (mitochondrial iron transporter 2) |
| ENSGACT00000003415 | 1.06 | 6.61 | 8.40E-05 | 0.132 | PCYT1B | phosphate cytidylyltransferase 1, choline, beta |
| ENSGACT00000016448 | −1.05 | 7.83 | 8.98E-05 | 0.132 | GLUT-1 | glucose transporter 1 |
| ENSGACT00000007884 | −1.16 | 5.22 | 9.01E-05 | 0.132 | C2 | complement C2 precursor (C2b fragment; C2a fragment) |
| ENSGACT00000026313 | −1.26 | 7.90 | 9.73E-05 | 0.132 | C2orf65 | uncharacterized protein C2orf65 |
| ENSGACT00000004329 | −1.44 | 10.22 | 0.00011 | 0.14 | WT1 | Wilm's tumor protein (WT33) |
| ENSGACT00000018879 | 1.17 | 9.61 | 0.00013 | 0.156 | TNS1 | tensin-1 |
| ENSGACT00000008104 | 1.06 | 8.14 | 0.00014 | 0.156 | ST7L | suppressor of tumorigenicity protein 7-like protein |
| ENSGACT00000020055 | 1.06 | 10.44 | 0.0002 | 0.157 | RBMS1 | RNA-binding motif, single-stranded-interacting protein 1 |
| ENSGACT00000018168 | −1.08 | 7.60 | 0.00022 | 0.157 | P2RY11 | P2Y purinoceptor 11 |
| ENSGACT00000019149 | 0.96 | 11.46 | 0.00025 | 0.157 | ANKDD1A | ankyrin repeat and death domain-containing protein 1A |
| ENSGACT00000012630 | −1.78 | 7.00 | 0.00025 | 0.157 | ALX1 | ALX homeobox protein 1 (cartilage homeoprotein 1) |
| ENSGACT00000009578 | −1.23 | 6.95 | 0.00026 | 0.157 | ZFP568 | zinc finger protein 568 |
| ENSGACT00000002994 | 1.18 | 9.16 | 0.00028 | 0.157 | NDRG4 | protein NDRG4 (brain development-related molecule) |
| ENSGACT00000012508 | −1.47 | 7.25 | 0.00035 | 0.157 | CLEC4M | C-type lectin domain family 4 member M |
| ENSGACT00000021759 | 1.39 | 9.09 | 0.00036 | 0.157 | CPLX4 | complexin-4 precursor |
| ENSGACT00000006879 | 0.97 | 8.57 | 0.00036 | 0.157 | AZIN1 | antizyme inhibitor 1 |
| ENSGACT00000002915 | −1.29 | 5.43 | 0.00037 | 0.157 | GAG | putative GAG protein |
| ENSGACT00000011207 | 1.22 | 7.23 | 0.00037 | 0.157 | TLCD2 | TLC domain-containing protein 2 |
| ENSGACT00000015180 | 1.72 | 5.55 | 0.00039 | 0.157 | MAPRE3 | microtubule-associated protein RP/EB family member 3 |
| ENSGACT00000027138 | −0.97 | 5.55 | 0.00039 | 0.157 | FBXO40 | F-box only protein 40 (muscle disease-related protein) |
Annotation of the transcripts based upon genome annotation in ensembl and BLASTX search. The moderated Bayesian statistics generated using the LIMMA package in R are included and the transcripts are ranked based upon their p values.
logFC = log2 of fold change; Ave. Exprs = average expression across all arrays; Adj. P. Val = p value adjusted; NA = information not available.
Gene Functional Analysis: Upregulated Genes
RHO was the most upregulated gene based on p values and fold changes in fish exposed to cues of a predator. RHO is a complex of vision protein opsin and the chromofore 11-cis-retinal that mediates vision at low illumination. Phospholipase A2, group XIIB (PLA2G12B) was also significantly upregulated. This gene is a secreted phospholipase (sPLA2) from a family of small secreted proteins with the ability to catalyze the hydrolysis of glycolipids and release free fatty acids and lysophospholipids. The upregulated gene homeobox C5 (HOXC5) is a member of the homeobox family of genes which encodes a highly conserved family of transcription factors that provide cells with specific positional identities on the anterior-posterior axis during morphogenesis in all multicellular organisms. The ubiquitin-specific protease-like protein (USP-like), a group of serine proteases that plays a pivotal role in protein homeostasis by tagging the ubiquitin complex for degradation was also upregulated. The upregulated gene phosphate cytidylyltransferase 1, choline, beta gene (PCYT1B) is an enzyme that controls the phosphatidylcholine synthesis and is a major component of cell membranes with an important role in membrane signaling and cell growth. Other upregulated genes included the suppressor of tumorigenicity protein 7-like protein (STL7), the RNA-inding motif (RBSMS1), the ankyrin repeat (ANKDD1A), tensin (TNS1) and the potassium channel subfamily K (KCNK12) gene (table 2).
Downregulated Genes
The most downregulated gene in fish exposed to predator cues was the transcript ENSGACT00000022897, a novel unnamed gene of undetermined function. Also downregulated was ropoporin (ROPN1), a gene most likely involved in regulating the growth, development and/or maintenance of motile cilia. The downregulation of mitoferrin-2 (MFRN2), a solute ion carrier essential for red-blood iron uptake, is also noteworthy. Of particular interest is the downregulation of the glucose transporter (GLUT1). GLUT1 is the first member of the solute carrier family (SLC2) that facilitates the transport of glucose from the bloodstream across the blood-brain barrier to the nervous system. Other downregulated genes included the complement precursor (C2), an immune gene which is part of the classical pathway of the complement system; interleukin-8 receptor alpha (CXCR1), a signal transduction protein that plays a role in inflammatory responses; the purinergic receptor P2Y G-protein coupled receptor and other genes, such as zinc finger proteins (ZFP568); the peroxisomal biogenesis factor 19 (Peroxin-19); and C-type lectin domain family 4 member M (CD209 antigen-like protein 1) (table 2).
GO and Pathway Enrichment Analysis
A hypergeometric test was used to determine the GO terms overrepresentated in the brains of fish exposed to predator cues. Our study showed that 41 BPs, 17 CCs and 20 MFs were significantly overrepresented (p < 0.05) in transcripts differentially expressed at the p ≤ 0.01 cutoff. The fraction of genes assigned to these GO terms among the differentially expressed transcripts is higher than expected by chance alone when they are compared to the fraction of genes that are in the universe or the background (all the transcripts after nonspecific filtration). As shown in tables 3 and 4, a wide range of biological GO terms was enriched in the brain. These terms can be grouped in several categories such as neuroplastic processes that include neurological system processes, phototransduction and detection of light stimulus involved in sensory perception. GO terms in immune response categories involved antigen processing and presentation, detection of external stimulus while biosynthetic processes overrepresented included neutral lipid biosynthetic process, glycerol ether biosynthetic process and negative regulation of biosynthetic process. Terms associated with gene regulation were negative regulation of tissue remodeling and positive regulation of protein kinase activity. GO terms associated with development were embryonic skeletal system morphogenesis, anterior/posterior pattern formation and dentate gyrus development. Other GO terms were, for example, signal transduction resulting in induction of apoptosis. MHC class I protein complex was the most overrepresented cellular GO term with 46% genes enriched for this term followed by the postsynaptic membrane mitochondrial respiratory chain complex IV, the synaptosome and the voltage-gated calcium channel complex.
Table 3.
List of GO BPs overrepresented in a set of genes differentially expressed in the brain of sticklebacks exposed to predator cues at p< 0.01
| GOBPID | p value | Odds ratio | Exp. count | Count | Size | Term |
|---|---|---|---|---|---|---|
| GO:0019882 | 0.0008 | 5.99 | 1.64 | 7 | 21 | antigen processing and presentation |
| GO:0050962 | 0.0009 | 7.13 | 1.26 | 6 | 16 | detection of light stimulus involved in sensory perception |
| GO:0007602 | 0.0014 | 8.48 | 0.94 | 5 | 12 | phototransduction |
| GO:0050877 | 0.0016 | 1.85 | 21.05 | 35 | 270 | neurological system process |
| GO:0048704 | 0.0019 | 5.94 | 1.42 | 6 | 18 | embryonic skeletal system morphogenesis |
| GO:0009584 | 0.0022 | 7.42 | 1.02 | 5 | 13 | detection of visible light |
| GO:0009952 | 0.0027 | 3.97 | 2.52 | 8 | 32 | anterior/posterior pattern formation |
| GO:0009416 | 0.0032 | 3.82 | 2.59 | 8 | 33 | response to light stimulus |
| GO:0009581 | 0.0044 | 4.75 | 1.65 | 6 | 21 | detection of external stimulus |
| GO:0009582 | 0.0044 | 4.75 | 1.65 | 6 | 21 | detection of abiotic stimulus |
| GO:0046460 | 0.0062 | Inf | 0.16 | 2 | 2 | neutral lipid biosynthetic process |
| GO:0046504 | 0.0062 | Inf | 0.16 | 2 | 2 | glycerol ether biosynthetic process |
| GO:0032868 | 0.0081 | 11.80 | 0.47 | 3 | 6 | response to insulin stimulus |
| GO:0009890 | 0.0108 | 1.75 | 15.50 | 25 | 197 | negative regulation of biosynthetic process |
| GO:0045666 | 0.0133 | 8.84 | 0.55 | 3 | 7 | positive regulation of neuron differentiation |
| GO:0006816 | 0.0149 | 2.80 | 3.30 | 8 | 42 | calcium ion transport |
| GO:0032940 | 0.0156 | 3.42 | 2.10 | 6 | 27 | secretion by cell |
| GO:0043405 | 0.0165 | 2.55 | 4.01 | 9 | 51 | regulation of MAP kinase activity |
| GO:0007168 | 0.0176 | 23.54 | 0.24 | 2 | 3 | receptor guanylyl cyclase signaling pathway |
| GO:0021542 | 0.0176 | 23.54 | 0.24 | 2 | 3 | dentate gyms development |
| GO:0050847 | 0.0176 | 23.54 | 0.24 | 2 | 3 | progesterone receptor signaling pathway |
| GO:0015980 | 0.0178 | 23.35 | 0.24 | 2 | 3 | energy derivation by oxidation of organic compounds |
| GO:0007268 | 0.0192 | 1.95 | 8.41 | 15 | 108 | synaptic transmission |
| GO:0045909 | 0.0201 | 7.07 | 0.63 | 3 | 8 | positive regulation of vasodilation |
| GO:0032774 | 0.0242 | 1.38 | 44.39 | 57 | 564 | RNA biosynthetic process |
| GO:0007623 | 0.0255 | 4.30 | 1.18 | 4 | 15 | circadian rhythm |
| GO:0006607 | 0.0284 | 5.89 | 0.71 | 3 | 9 | NLS-bearing substrate import into nucleus |
| GO:0034104 | 0.0284 | 5.89 | 0.71 | 3 | 9 | negative regulation of tissue remodeling |
| GO:0003001 | 0.0298 | 3.30 | 1.80 | 5 | 23 | generation of a signal involved in cell-cell signaling |
| GO:0006533 | 0.0333 | 11.77 | 0.31 | 2 | 4 | aspartate catabolic process |
| GO:0009405 | 0.0333 | 11.77 | 0.31 | 2 | 4 | pathogenesis |
| GO:0006334 | 0.0359 | 3.11 | 1.89 | 5 | 24 | nucleosome assembly |
| GO:0015674 | 0.0366 | 1.99 | 6.05 | 11 | 77 | di-, tri-valent inorganic cation transport |
| GO:0045860 | 0.0371 | 1.98 | 6.06 | 11 | 77 | positive regulation of protein kinase activity |
| GO:0051347 | 0.0371 | 1.98 | 6.06 | 11 | 77 | positive regulation of transferase activity |
| GO:0000122 | 0.0377 | 1.91 | 6.83 | 12 | 87 | negative regulation of transcription from RNA polymerase II promoter |
| GO:0008630 | 0.0382 | 5.05 | 0.79 | 3 | 10 | DNA damage response, signal transduction resulting in apoptosis |
| GO:0046530 | 0.0382 | 5.05 | 0.79 | 3 | 10 | photoreceptor cell differentiation |
| GO:0007601 | 0.0388 | 1.85 | 7.62 | 13 | 97 | visual perception |
| GO:0000187 | 0.0414 | 2.97 | 1.96 | 5 | 25 | activation of MAPK activity |
| GO:0002504 | 0.0496 | 4.42 | 0.87 | 3 | 11 | antigen processing and presentation via MHC class II |
GOBPID: GO ID of BP term; p value is the significance of enrichment; Odds ratio is the ratio of odds that a GO term is enriched in the selected category; Exp. count represents the number expected; Count represents the number of transcripts annotated for the related GO term; Size is the number of probe sets overrepresented in the universe (background).
Table 4.
List of GO terms overrepresented in a set of genes differentially expressed in the brain of sticklebacks exposed to predator cues at p < 0.01
| GOCCID | p value | Odds ratio | Exp. count | Count | Size | Term |
|---|---|---|---|---|---|---|
| Cellular component | ||||||
| GO:0042612 | 0.0009 | 9.93 | 0.86 | 5 | 11 | MHC class I protein complex |
| GO:0045211 | 0.0045 | 2.67 | 5.17 | 12 | 66 | postsynaptic membrane |
| GO:0005751 | 0.0061 | Infinity | 0.16 | 2 | 2 | mitochondrial respiratory chain complex IV |
| GO:0042622 | 0.0061 | Infinity | 0.16 | 2 | 2 | photoreceptor outer segment membrane |
| GO:0005625 | 0.0077 | 2.14 | 8.30 | 16 | 106 | soluble fraction |
| GO:0019717 | 0.0086 | 3.48 | 2.43 | 7 | 31 | synaptosome |
| GO:0001917 | 0.0174 | 23.67 | 0.23 | 2 | 3 | photoreceptor inner segment |
| GO:0000159 | 0.0198 | 7.11 | 0.63 | 3 | 8 | protein phosphatase type 2 A complex |
| GO:0042613 | 0.0198 | 7.11 | 0.63 | 3 | 8 | MHC class II protein complex |
| GO:0005834 | 0.0252 | 4.32 | 1.17 | 4 | 15 | heterotrimeric G-protein complex |
| GO:0005640 | 0.0280 | 5.93 | 0.70 | 3 | 9 | nuclear outer membrane |
| GO:0005891 | 0.0316 | 3.96 | 1.25 | 4 | 16 | voltage-gated calcium channel complex |
| GO:0042825 | 0.0330 | 11.83 | 0.31 | 2 | 4 | TAP complex |
| GO:0045202 | 0.0351 | 1.72 | 10.64 | 17 | 136 | synapse |
| GO:0019898 | 0.0377 | 2.45 | 3.21 | 7 | 41 | extrinsic to membrane |
| GO:0034707 | 0.0480 | 2.83 | 2.04 | 5 | 26 | chloride channel complex |
| Molecular functions | ||||||
| GO:0004918 | 0.0063 | Infinity | 0.16 | 2 | 2 | interleukin-8 receptor activity |
| GO:0005546 | 0.0063 | Infinity | 0.16 | 2 | 2 | phosphatidylinositol-4,5-bisphosphate binding |
| GO:0048038 | 0.0063 | Infinity | 0.16 | 2 | 2 | quinone binding |
| GO:0004691 | 0.0082 | 11.72 | 0.48 | 3 | 6 | cAMP-dependent protein kinase activity |
| GO:0004871 | 0.0139 | 1.94 | 9.60 | 17 | 122 | signal transducer activity |
| GO:0015226 | 0.0178 | 23.38 | 0.24 | 2 | 3 | carnitine transporter activity |
| GO:0005388 | 0.0204 | 7.03 | 0.63 | 3 | 8 | calcium-transporting ATPase activity |
| GO:0008601 | 0.0204 | 7.03 | 0.63 | 3 | 8 | protein phosphatase type 2 A regulator activity |
| GO:0008565 | 0.0271 | 2.66 | 3.00 | 7 | 38 | protein transporter activity |
| GO:0022890 | 0.0306 | 3.27 | 1.82 | 5 | 23 | inorganic cation transmembrane transporter activity |
| GO:0003713 | 0.0327 | 1.90 | 7.44 | 13 | 94 | transcription coactivator activity |
| GO:0019905 | 0.0335 | 11.72 | 0.32 | 2 | 4 | syntaxin binding |
| GO:0003711 | 0.0336 | 11.71 | 0.32 | 2 | 4 | transcription elongation regulator activity |
| GO:0004571 | 0.0337 | 11.69 | 0.32 | 2 | 4 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity |
| GO:0008131 | 0.0337 | 11.69 | 0.32 | 2 | 4 | amine oxidase activity |
| GO:0004674 | 0.0372 | 1.61 | 13.97 | 21 | 177 | protein serine/threonine kinase activity |
| GO:0005245 | 0.0387 | 5.02 | 0.79 | 3 | 10 | voltage-gated calcium channel activity |
| GO:0004112 | 0.0403 | 3.61 | 1.35 | 4 | 17 | cyclic-nucleotide phosphodiesterase activity |
| GO:0005216 | 0.0485 | 2.51 | 2.70 | 6 | 34 | ion channel activity |
GOCCID: GO ID of the CC term, GOMFID: GO ID of MF term; p value is the significance of enrichment; Odds ratio is the ratio of odds that a GO term is enriched in the selected category; Exp. count represents the number expected; Count represents the number of transcripts annotated for the related GO term; GO term size is the number of probe sets overrepresented in the universe (background).
Network analysis showed that out of the 5,016 network eligible genes (genes mapped to a pathway by IPA), 348 transcripts were mapped to a network using the IPA system (p = 0.01). For example, 35 genes in a network involving cell signaling, nucleic acid metabolism, cellular assembly and organization were differentially expressed in our experiment (IPA score = 35). Similarly, 33 genes in a network involving neurological disease, developmental disorder and psychological disorders were also differentially expressed (IPA score = 30). Pathway analysis revealed that the synaptic long-term depression pathways was the most overrepresented [–log(p) = 2.84E-04], followed by the RAN signaling pathway [–log(p) = 2.62 E-04], the relaxin-signaling pathway [–log(p) = 1.98E-04] and the phototransduction pathways [-log(p) = 1.98E-04].
Microarray Validation
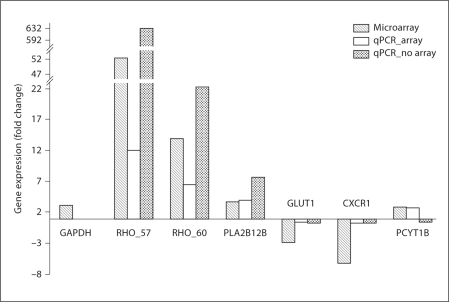
All the 10 genes selected for validation of the microarray results, in addition to the reference gene GAPDH, were successfully amplified by qPCR. As a first round of amplification of RHO was not consistent across samples, new primers were designed based upon the sequences of the transcript variants. For the samples that had been included on the microarray, the 10 genes were regulated in the same direction as in the microarray (table 5). Of the 10 genes selected for validation, 7 genes were significantly differentially expressed. Both RHO variants (RHO_57 and RHO_60), USP-like and PCYB12 were upregulated in qPCR as in the microarray, while GLUT1, C2, and CXCR1 were significantly downregulated at 95% confidence interval. PLA2G12B, Tr_22897 and KCNK12, which were significantly upregulated in the microarray, were also upregulated in qPCR; however, the difference between treatments was not statistically significant. Other studies have also failed to validate all genes by qPCR, particularly when the genes have low fold changes [Mukai et al., 2009]. There was no detectable difference between the samples included in the microarray experiment and in the independent biological replicates that were not included in the microarray analysis. For example, the expression values of the top differentially expressed genes did not differ significantly between these two groups (d.f. = 1, F = 0.04, p = 0.8). We also compared the expression of 6 genes in samples that had not been included on the microarray and found that the microarray and independent biological replicates showed similar patterns: 5 of the 6 genes showed the same direction of expression change (up- or downregulated) (fig. 3).
Table 5.
qPCR results of microarray data validation
| Gene | Type | Fold change | STD error | P value | Regulation |
|---|---|---|---|---|---|
| GAPDH | reference | 1.0 | |||
| RHO_57 | target | 12.1 | 0.573 | 0.038 | up |
| RHO 60 | target | 6.4 | 0.463 | 0.006 | up |
| USP-like | target | 5.9 | 0.443 | 0.002 | up |
| Tr_22897 | target | 1.5 | 1.231 | 0.25 | NS |
| PLA2G12B | target | 4.0 | 0.784 | 0.207 | NS |
| GLUT1 | target | 0.5 | 0.354 | 0.006 | down |
| C2 | target | 0.6 | 0.452 | 0.03 | down |
| CXCR1 | target | 0.3 | 0.139 | 0.002 | down |
| KCNK12 | target | 1.2 | 0.784 | 0.312 | NS |
| PCYB12 | target | 2.7 | 1.289 | 0.046 | up |
GADPH (glyceraldehyde 3-phosphate dehydrogenase): reference gene used for normalization of the expression values of the target genes. Target genes were RHO_57 (rhodopsin transcript variant ENSGACT00000003757), RHO_60 (rhodopsin transcript variant ENSGACT00000003760), USP (ubiquitin-spe-cific protease), Tr_22897 (transcript ‘ENSGACT00000022897’), PLA2G12B (phospholipase 2 group XIIB), GLUTl (glucose transporter 1), C2 (complement precursor), CXCR1 (interleukin-8 receptor alpha), KCNK12 (potassium channel), PCYT1B (phosphate cytidylyltransferase 1, choline, beta). Fold changes of the target genes are expression changes relative to the expression of the reference gene, which is by default 1. The direction of the changes in gene expression is indicated in the ‘Regulation’ column, up are upregulated genes, down are downregulated genes and NS represents genes whose expression is not statistically different from the reference gene.
Fig. 3.
qPCR validation of microarray gene expression in the brains of sticklebacks exposed to predator cues. The microarray results are plotted next to the qPCR results for the samples used in the microarray (n = 7 experimental and n = 7 control), and the qPCR results for the independent biological replicates not on the array (n = 2 experimental and n = 2 control).
Discussion
Exposure to olfactory, visual and tactile cues of a predator for 6 days resulted in significant transcriptomic changes in the brain of sticklebacks. We found that 10% of the transcripts were differentially expressed (p ≤ 0.01) between fish exposed to cues of a predator and a control group. These patterns are consistent with other studies involving predator cues [Nanda et al., 2008; Leder et al., 2009]. For example, a 15-min exposure to ferret olfactory, visual and acoustic cues increased stress-like behavior in rats and resulted in the differential expression of 7% of genes in the amygdala 3 h after exposure to a predator [Nanda et al., 2008]. Another study found that of the 5,931 cDNA clones used in cDNA microarray, a total of 52 genes (8%) were differentially expressed in the brain of rats exposed to cats when gene expression was assessed 20 min after exposure to a predator [Wang et al., 2003].
Some of the genes and pathways that were identified in this study have also been implicated in related studies. For example, 17 out of 909 differentially expressed genes (at p < 0.01 cutoff) have been identified in previous studies. These include, for example, SAMD4A, FGFR10P2, and CD99LD, which were identified during a selection response for contextual fear conditioning in rats using quantitative trait locus (QTL) and gene expression studies [Ponder et al., 2008]; KEK4, MUB and HSPB1 were identified in fruit flies selected for aggressive behavior [Dierick and Greenspan, 2006] and PEX19 and DEDD were identified proximal to a QTL hotspot on mouse chromosome 1 that modulates behavior [Mozhui et al., 2008]. Similarly, the long-term synaptic depression (LTD) pathway, one of the most overrepresented pathways revealed in our study, has been associated with predator threat. LTD is an electrical correlate of the weakening of the connection between two neurons, thus leading to long-term reduction in the efficacy of neuronal synapsis as a consequence of a decrease in the density of postsynaptic receptors. Studies have shown that both acute and chronic stress [Yang et al., 2004; Holderbach et al., 2007], including exposure to predators [Kuzmiski et al., 2010; Kim and Diamond, 2002], can lead to LTD. LTD has also been associated with synaptic plasticity and memory formation [Bear, 1999; Malleret et al., 2010; Kim and Diamond, 2002].
Importantly, our study also identified many genes and biological pathways that have not been previously reported. For example, to the best of our knowledge PLA2G12B, RHO and GLUT1 have not yet been associated with exposure to predator stress. Similarly, our study is one of the first to report the activation of the RAN and the relaxin pathways in response to long-term exposure to predator cues. Different sets of genes in the brain respond to acute versus chronic stress [Krasnov et al., 2005], which might explain why there was some, but not complete, overlap between the differentially expressed genes found in this experiment compared to other experiments that have examined the transcriptomic response to a single, brief exposure to predation risk [e.g. Wang et al., 2003].
Although sticklebacks mounted a transcriptomic response to repeated exposure to a predator, the overall magnitude of the response, as assessed by fold change, was relatively low. While some genes, such as RHO, had a high fold change, most genes were weakly differentially expressed. Other studies have also detected subtle transcriptomic responses to behavioral challenges such as predator threat [Wang et al., 2003] or aggression [Mukai et al., 2009]. One possible explanation for the relatively small changes in expression is that transcription profiling was conducted 24 h after repeated exposure to predation risk. It is possible that the patterns we observed reflect relatively long-term, enduring changes in the brain rather than transitory responses. Alternatively, or in addition, it is possible that we measured the downstream consequences of exposure to predation risk 24 h earlier. Detailed time-course experiments are needed to distinguish the response to single (acute) versus repeated (chronic) exposures to predation risk across short (over the course of minutes) versus long (over the course of days to months) timescales. The stickleback system offers the opportunity to frame this question from an evolutionary perspective by comparing the genes that are involved in the plastic response to predation risk within an individual's lifetime with the genes that are involved in evolutionary divergence between different stickleback populations in response to variation in predation pressure [Giles and Huntingford, 1984; Bell, 2005].
The wide range in GO terms that were overrepresented in the stickleback brain suggests that exposure to cues of a predator triggers several biological processes in the prey. Many genes related to neurological system process (n = 270), negative regulation of neurological processes (n = 197 genes), synaptic transmission (n = 108) and visual perception (n = 97) were represented in the list of differentially expressed genes; however, these genes were not necessarily the most overrepresented based upon p values of the GO terms. Intriguingly, the most overrepresented biological GO terms in the brain of sticklebacks following exposure to predator cues were related to antigen processing and presentation (p < 0.0008), which also corresponded to the top cellular component GO term: MHC class I protein complex (p < 0.0009). MHC is a cluster of genes that encode a complex of transmembrane glucoproteins expressed on the surface of all somatic cells, including neurons [Corriveau et al., 1998]. They are involved in immune response and are critical in the olfactory-based discrimination of self/nonself in many vertebrate species, including humans [Wedekind et al., 1995], mice [Egid and Brown, 1989] and fish [Milinski et al., 2005]. In sticklebacks, while MHC class II gene has been shown to play a role in mate choice [Milinski et al., 2005] and kin recognition [Olsen et al., 1998], the role of MHC class I in sticklebacks is less clear. Intriguingly, studies have shown that MHCI plays a key role in synaptic plasticity and neuron regeneration [Oliveira et al., 2004]. Other studies have demonstrated that MHCI is critical in neuronal plasticity [Goddard et al., 2007]. In addition, recent studies have implicated MHCI in memory formation and behavior [Shatz, 2009]. Some studies have shown that acute predator stress can lead to memory impairment [Diamond and Park, 2000]. The involvement of MHCI in many biological functions is correlated with its high diversity observed both intra- and interspecifically [Shiina et al., 2010]. It has been postulated that this diversity is driven by selection pressure that acts on MHC genes under various environmental pressures such as microbial threats or environmental sensory cues [Apanius et al., 1997; Bernatchez and Landry, 2003; Spurgin and Richardson, 2010]. Understanding the interactions between MHC genes, olfaction, memory and predator stress is a promising area for future research.
Our data suggest that sticklebacks strongly responded to visual cues of a predator. Both RHO splice variants were successfully validated by qPCR. This gene is expressed not only in the retina but also in the deep brain, including the pineal gland where it plays an important role in photoreception and photoperiodism [Masuda et al., 2003]. Since the eyes were separated from the brain during sample preparation, the high expression of this gene suggests its involvement in brain function. This is further supported by the overrepresentation of the GO terms related to detection of light stimulus involved in sensory perception and phototransduction and supports the hypothesis that processes associated with visual perception were important in the response to predation risk. This is consistent with previous observations that vision is an important modality for communication in sticklebacks [Huntingford and Ruiz-Gomez, 2009].
Sticklebacks in this experiment were exposed to olfactory cues of trout and dead conspecifics that contained alarm pheromones. Alarm pheromones are released from club cells when the skin is punctured and cause sticklebacks to exhibit increased antipredator behaviors [Brown and Godin, 1997] and may have originally evolved as an immune response [Chivers et al., 2007]. Interestingly, PLA2G12B, a gene that has been shown to play a regulatory role in the induction of olfactory structures in frogs [Munoz-Sanjuan and Brivanlou, 2005], was upregulated in response to our treatment, highlighting the importance of olfaction in sticklebacks.
The animals in this experiment were socially housed. Therefore, it is possible that some of the response of the sticklebacks in the experimental group was a reaction to stressed or frightened conspecifics. Observations in mammals, including humans, have shown that exposure to conspecific emotional stress elicits fear response in the amygdala [Mujica-Parodi et al., 2009]. Studies have also demonstrated that exposure to conspecific alarm hormones impedes the humoral and cellular immune response in male BALB/cJ mice [Cocke et al., 1993].
Further investigations of the gene modules and pathways herein identified might provide insight into the molecular mechanisms underlying the response of sticklebacks to predator-induced stress. Network and pathway analyses revealed that genes differentially expressed as a result of exposure of sticklebacks to predator cues are clustered in gene modules. This was also supported by the clusters observed in the hierarchical clustering in the heat map (fig. 2). For example, gene regulatory networks that involve neurological diseases, developmental disorders and psychological disorders as well as cell signaling, nucleic acid metabolism, cellular assembly and organization were revealed using network analysis. Future studies will compare the expression of these genes across different populations of sticklebacks that vary in predation pressure, and will pharmacologically manipulate the pathways in order to infer a causal relationship between exposure to predators and behavior.
In conclusion, this study examined the gene expression pattern in the brain of sticklebacks under conditions that mimic naturally high predation pressure in the wild. We showed that repeated exposure to cues of a predator results in significant differential expression of genes involved in immune response, synaptic processes, brain metabolic processes and visual perception. Gene functional annotation and gene ontology and pathway analyses identified several biological and molecular processes that might be activated as a response to predation risk. The radiation of sticklebacks along with growing genomic resources [Kingsley and Peichel, 2007; Brown et al., 2008; Geoghegan et al., 2008; Williams et al., 2008; Leder et al., 2009] and its well-characterized biology, make this organism a suitable model system for investigating the molecular and physiological responses of the brain to ecologically relevant selection pressures that have influenced their evolution.
Supplementary Material
supplementary figure
Acknowledgements
We would like to thank Jenny Drnevich for help with statistical analysis in R Bioconductor, and Tom Newman and Gene Robinson for access to their qPCR machine. This study was supported by a grant from the National Institutes of Health, R01 GM082937, to A.M.B. and M.B.
References
- Apanius V, Penn D, Slev PR, Ruff LR, Potts WK. The nature of selection on the major histocompatibility complex. Crit Rev Immunol. 1997;17:179–224. doi: 10.1615/critrevimmunol.v17.i2.40. [DOI] [PubMed] [Google Scholar]
- Baker JA, Foster SA, Heins DC, Bell MA, King RW. Variation in female life-history traits among Alaskan populations of the threespine stickleback, Gasterosteus aculeatus L. (Pisces: Gasterosteidae) Biol J Linn Soc Lond. 1998;63:141–159. [PubMed] [Google Scholar]
- Bear MF. Homosynaptic long-term depression: a mechanism for memory? Proc Natl Acad Sci USA. 1999;96:9457–9458. doi: 10.1073/pnas.96.17.9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AM. Behavioural differences between individuals and two populations of stickleback (Gasterosteus aculeatus) J Evol Biol. 2005;18:464–473. doi: 10.1111/j.1420-9101.2004.00817.x. [DOI] [PubMed] [Google Scholar]
- Bell AM, Backstrom T, Huntingford FA, Pottinger TG, Winberg S. Variable neuroendocrine responses to ecologically-relevant challenges in sticklebacks. Physiol Behav. 2007;91:15–25. doi: 10.1016/j.physbeh.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Bell AM, Dingemanse NJ, Hankison S, Langenhof M, Rollins K. Early exposure to nonlethal predation risk by size-selective predators increases somatic growth and decreases size at adulthood in threespined sticklebacks. J Evol Biol. 2011;24:943–945. doi: 10.1111/j.1420-9101.2011.02247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AM, Henderson L, Huntingford FA. Behavioral and respiratory responses to stressors in multiple populations of three-spined sticklebacks that differ in predation pressure. J Comp Physiol B. 2010;180:211–220. doi: 10.1007/s00360-009-0395-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AM, Sih A. Exposure to predation generates personality in threespined sticklebacks. Ecol Lett. 2007;10:828–834. doi: 10.1111/j.1461-0248.2007.01081.x. [DOI] [PubMed] [Google Scholar]
- Bell MA, Foster SA. Introduction to the evolutionary biology of the threespine stickleback. In: Bell MA, Foster SA, editors. The Evolutionary Biology of the Threespine Stickleback. Oxford: Oxford University Press; 1994. pp. 1–27. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodological) 1995;57:289–300. [Google Scholar]
- Bernatchez L, Landry C. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J Evol Biol. 2003;16:363–377. doi: 10.1046/j.1420-9101.2003.00531.x. [DOI] [PubMed] [Google Scholar]
- Blanchard DC, Canteras NS, Markham CM, Pentkowski NS, Blanchard RJ. Lesions of structures showing FOS expression to cat presentation: effects on responsivity to a cat, cat odor, and nonpredator threat. Neurosci Biobehav Rev. 2005;29:1243–1253. doi: 10.1016/j.neubiorev.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Bourgon R, Gentleman R, Huber W. Independent filtering increases detection power for high-throughput experiments. Proc Natl Acad Sci USA. 2010;107:9546–9551. doi: 10.1073/pnas.0914005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GE, Godin JJ. Anti-predator responses to conspecific and heterospecific skin extracts by threespine sticklebacks: alarm pheromones revisited. Behaviour. 1997;134:1123–1134. [Google Scholar]
- Brown MM, Williams TD, Chipman JK, Katsiadaki I, Sanders M, Craft JA. Construction of subtracted EST and normalised cDNA libraries from liver of chemical-exposed three-spined stickleback (Gasterosteus aculeatus) containing pollutant-responsive genes as a resource for transcriptome analysis. Mar Env Res. 2008;66:127–130. doi: 10.1016/j.marenvres.2008.02.043. [DOI] [PubMed] [Google Scholar]
- Cairns MT, Johnson MC, Talbot AT, Pernmasani JK, McNeill RE, Houeix B, Sangrador-Vegas A, Pottinger TG. A cDNA microarray assessment of gene expression in the liver of rainbow trout (Oncorhynchus mykiss) in response to a handling and confinement stressor. Comp Biochem Physiol D Genomics Proteomics. 2008;3:51–66. doi: 10.1016/j.cbd.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Calduch-Giner JA, Davey G, Saera-Vila A, Houeix B, Talbot A, Prunet P, Cairns MT, Perez-Sanchez J. Use of microarray technology to assess the time course of liver stress response after confinement exposure in gilthead sea bream (Sparus aurata L.) BMC Genomics. 2010;11 doi: 10.1186/1471-2164-11-193. No. 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calza S, Raffelsberger W, Ploner A, Sahel J, Leveillard T, Pawitan J. Filtering genes to improve sensitivity in oligonucleotide microarray data analysis. Nucleic Acids Res. 2007;35:e102. doi: 10.1093/nar/gkm537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor C. Post-traumatic stress disorder: evolutionary perspectives. Aust NZJ Psychiatry. 2009;43:1038–1048. doi: 10.3109/00048670903270407. [DOI] [PubMed] [Google Scholar]
- Carvalho B, Bengtsson H, Speed TP, Irizarry RA. Exploration, normalization, and genotype calls of high-density oligonucleotide SNP array data. Biostatistics. 2007;8:485–499. doi: 10.1093/biostatistics/kxl042. [DOI] [PubMed] [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G, Jr, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, Myers RM, Petrov D, Jonsson B, Schluter D, Bell A, Kingsley DM. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching B, Jamieson S, Heath JW, Heath DD, Hubberstey A. Transcriptional differences between triploid and diploid Chinook salmon (Oncorhynchus tshawytscha) during live Vibrio anguillarum challenge. Heredity. 2010;104:224–234. doi: 10.1038/hdy.2009.108. [DOI] [PubMed] [Google Scholar]
- Chivers DP, Wisenden BD, Hindman CJ, Michalak TA, Kusch RC, Kaminskyj SG, Jack KL, Ferrari MC, Pollock RJ, Halbgewachs CF, Pollock MS, Alemadi S, James CT, Savaloja RK, Goater CP, Corwin A, Mirza RS, Kiesecker JM, Brown GE, Adrian JC, Jr, Krone PH, Blaustein AR, Mathis A. Epidermal ‘alarm substance’ cells of fishes maintained by non-alarm functions: possible defence against pathogens, parasites and UVB radiation. Proc Biol Sci. 2007;274:2611–2619. doi: 10.1098/rspb.2007.0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocke R, Moynihan JA, Cohen N, Grota LJ, Ader R. Exposure to conspecific alarm chemosignals alters immune responses in BALB/c mice. Brain Behav Immun. 1993;7:36–46. doi: 10.1006/brbi.1993.1004. [DOI] [PubMed] [Google Scholar]
- Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G, Jr, Dickson M, Grimwood J, Schmutz J, Myers RM, Schluter D, Kingsley DM. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- Comoli E, Ribeiro-Barbosa ER, Canteras NS. Predatory hunting and exposure to a live predator induce opposite patterns of Fos immunoreactivity in the PAG. Behav Brain Res. 2003;138:17–28. doi: 10.1016/s0166-4328(02)00197-3. [DOI] [PubMed] [Google Scholar]
- Corriveau RA, Huh GS, Shatz CJ. Regulation of class I MHC gene expression in the developing and mature CNS by neural activity. Neuron. 1998;21:505–520. doi: 10.1016/s0896-6273(00)80562-0. [DOI] [PubMed] [Google Scholar]
- Craig PM, Hogstrand C, Wood CM, McClelland GB. Gene expression endpoints following chronic waterborne copper exposure in a genomic model organism, the zebrafish, Danio rerio. Physiol Genomics. 2009;40:23–33. doi: 10.1152/physiolgenomics.00089.2009. [DOI] [PubMed] [Google Scholar]
- Cresko WA, Amores A, Wilson C, Murphy J, Currey M, Phillips P, Bell MA, Kimmel CB, Postlethwait JH. Parallel genetic basis for repeated evolution of armor loss in Alaskan threespine stickleback population. Proc Nat Acad Sci. 2004;101:6050–6055. doi: 10.1073/pnas.0308479101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond DM, Park CR. Predator exposure produces retrograde amnesia and blocks synaptic plasticity. Progress toward understanding how the hippocampus is affected by stress. Ann NY Acad Sci. 2000;911:453–455. doi: 10.1111/j.1749-6632.2000.tb06743.x. [DOI] [PubMed] [Google Scholar]
- Dierick HA, Greenspan J. Molecular analysis of flies selected for aggressive behavior. Nat Genet. 2006;38:1023–1031. doi: 10.1038/ng1864. [DOI] [PubMed] [Google Scholar]
- Dingemanse NJ, Van der Plas F, Wright J, Reale D, Schrama M, Roff DA, Van der Zee E, Barber I. Individual experience and evolutionary history of predation affect expression of heritable variation in fish personality and morphology. Proc Biol Sci. 2009;276:1285–1293. doi: 10.1098/rspb.2008.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egid K, Brown JL. The major histocompatibility complex and female mating preference in mice. Anim Behav. 1989;38:548–549. [Google Scholar]
- Feldman AJ, Costouros NG, Wang E, Qian M, Marincola FM, Alexander HR, Libutti SK. Advantages of mRNA amplification for microarray analysis. Biotechniques. 2002;33:906–914. doi: 10.2144/02334mt04. [DOI] [PubMed] [Google Scholar]
- Garrett SH, Somji S, Sens MA, Zhang K, Sens DA. Microarray analysis of gene expression patterns in human proximal tubule cells over a short and long time course of cadmium exposure. J Toxicol Environ Health. 2011;74:24–42. doi: 10.1080/15287394.2010.514230. [DOI] [PubMed] [Google Scholar]
- Geoghegan F, Katsiadaki I, Williams, TD, Chipman JK. A cDNA microarray for the three-spined stickleback, Gasterosteus aculeatus L., and analysis of the interactive effects of oestradiol and dibenzanthracene exposures. J Fish Biol. 2008;72:2133–2153. [Google Scholar]
- Giles N, Huntingford FA. Predation risk and inter-population variation in antipredator behaviour in the threespined stickleback. Anim Behav. 1984;32:264–275. [Google Scholar]
- Goddard CA, Butts DA, Shatz CJ. Regulation of CNS synapses by neuronal MHC class I. Proc Natl Acad Sci. 2007;104:6828–6833. doi: 10.1073/pnas.0702023104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holderbach R, Clark K, Moreau JL, Bischofberger J, Normann C. Enhanced long-term synaptic depression in an animal model of depression. Biol Psychiatry. 2007;62:92–100. doi: 10.1016/j.biopsych.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Huntingford FA, Ruiz-Gomez ML. Three-spined sticklebacks Gasterosteus aculeatus as a model for exploring behavioural biology. J Fish Biol. 2009;75:1943–1976. doi: 10.1111/j.1095-8649.2009.02420.x. [DOI] [PubMed] [Google Scholar]
- Huntingford FA, Wright PJ, Tierney JF. Adaptive variation and antipredator behaviour in threespine stickleback. In: Bell MA, Foster SA, editors. The Evolutionary Biology of the Threespine Stickleback. Oxford: Oxford University Press; 1994. pp. 277–295. [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JL, Magurran AE. Learned predator recognition and antipredator responses in fishes. Fish Fish. 2003;4:216–226. [Google Scholar]
- Kennerly E, Ballmann A, Martin S, Wolfinger R, Gregory S, Stoskopf M, Gibson G. A gene expression signature of confinement in peripheral blood of red wolves (Canis rufus) Mol Ecol. 2008;17:2782–2791. doi: 10.1111/j.1365-294X.2008.03775.x. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Diamond D. The stressed hippocampus, synaptic plasticity and lost memories. Nat Rev Neurosci. 2002;3:453–462. doi: 10.1038/nrn849. [DOI] [PubMed] [Google Scholar]
- Kingsley DM, Peichel CL. The molecular genetics of evolutionary change in sticklebacks. In: Östlund-Nilsson O, Mayer I, Huntingford FA, editors. Biology of the Three-Spined Stickleback. Boca Raton: CRC Press; 2007. [Google Scholar]
- Krasnov A, Koskinen H, Pehkonen P, Rexroad CE, Afanasyev S, Molsa H. Gene expression in the brain and kidney of rainbow trout in response to handling stress. BMC Genomics. 2005;6:3. doi: 10.1186/1471-2164-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmiski JB, Marty V, Baimoukhametova DV, Bains JS. Stress-induced priming of glutamate synapses unmasks associative short-term plasticity. Nat Neurosci. 2010;13:1257–1264. doi: 10.1038/nn.2629. [DOI] [PubMed] [Google Scholar]
- Leder EH, Merila J, Primmer CR. A flexible whole-genome microarray for transcriptomics in three-spine stickleback (Gasterosteus aculeatus) BMC Genomics. 2009;10:426. doi: 10.1186/1471-2164-10-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magurran AE. The inheritance and development of minnow antipredator behavior. Anim Behav. 1990;39:834–842. [Google Scholar]
- Malleret G, Alarcon JM, Martel G, Takizawa, S, Vronskaya S, Yin D, Chen IZ, Kandel ER, Shumyatsky GP. Bidirectional regulation of hippocampal long-term synaptic plasticity and its influence on opposing forms of memory. J Neurosci. 2010;30:3813–3825. doi: 10.1523/JNEUROSCI.1330-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchinko KB. Predation's role in repeated phenotypic and genetic divergence of armor in threespine stickleback. Evolution. 2009;63:127–138. doi: 10.1111/j.1558-5646.2008.00529.x. [DOI] [PubMed] [Google Scholar]
- Masuda T, Iigo M, Mizusawa K, Aida K. Retina-type rhodopsin gene expressed in the brain of a teleost, ayu (Plecoglossus altivelis) Zoolog Sci. 2003;20:989–997. doi: 10.2108/zsj.20.989. [DOI] [PubMed] [Google Scholar]
- McEwen BS. Physiology and neurobiology of stress and adaptation: central role of the brain. Physiol Rev. 2007;87:873–904. doi: 10.1152/physrev.00041.2006. [DOI] [PubMed] [Google Scholar]
- Milinski M, Griffiths S, Wegner KM, Reusch TB, Haas-Assenbaum A, Boehm T. Mate choice decisions of stickleback females predictably modified by MHC peptide ligands. Proc Natl Acad Sci. 2005;102:4414–4418. doi: 10.1073/pnas.0408264102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodie GEE. Predation, natural selection and adaptation in an unusual threespine stickleback. Evolution. 1972;28:167. [Google Scholar]
- Mori T, Kawachi H, Imai C, Sugiyama M, Kurata Y, Kishida O, Nishimura K. Identification of a novel uromodulin-like gene related to predator-induced bulgy morph in anuran tadpoles by functional microarray analysis. PLoS One. 2009;4:e5936. doi: 10.1371/journal.pone.0005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta SC, Goto M, Gouveia FV, Baldo MV, Canteras NS, Swanson LW. Dissecting the brain's fear system reveals the hypothalamus is critical for responding in subordinate conspecific intruders. Proc Natl Acad Sci. 2009;106:4870–4875. doi: 10.1073/pnas.0900939106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozhui K, Ciobanu DC, Schikorski T, Wang X, Lu L, Williams RW. Dissection of a QTL hotspot on mouse distal chromosome 1 that modulates neurobehavioral phenotypes and gene expression. PLoS Genet. 2008;4:e1000260. doi: 10.1371/journal.pgen.1000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujica-Parodi LR, Strey HH, Frederick B, Savoy R, Cox D, Botanov Y, Tolkunov D, Rubin D, Weber J. Chemosensory cues to conspecific emotional stress activate amygdala in humans. PLoS One. 2009;4:e6415. doi: 10.1371/journal.pone.0006415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai M, Replogle K, Drnevich J, Wang G, Wacker D, Band M, Clayton DF, Wingfield JC. Seasonal differences of gene expression profiles in song sparrow (Melospiza melodia) hypothalamus in relation to territorial aggression. PLoS One. 2009;4:e8182. doi: 10.1371/journal.pone.0008182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Sanjuan I, Brivanlou AH. Induction of ectopic olfactory structures and bone morphogenetic protein inhibition by Rossy, a group XII secreted phospholipase A2. Mol Cell Biol. 2005;25:3608–3619. doi: 10.1128/MCB.25.9.3608-3619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda SA, Qi C, Roseboom PH, Kalin NH. Predator stress induces behavioral inhibition and amygdala somatostatin receptor 2 gene expression. Genes Brain Behav. 2008;7:639–648. doi: 10.1111/j.1601-183X.2008.00401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira AL, Thams S, Lidman O, Piehl F, Hokfelt T, Karre K, Linda H, Cullheim S. A role for MHC class I molecules in synaptic plasticity and regeneration of neurons after axotomy. Proc Natl Acad Sci. 2004;101:17843–17848. doi: 10.1073/pnas.0408154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen KH, Grahn M, Lohm J, Langefors A. MHC and kin discrimination in juvenile arctic charr, Salvelinus alpinus (L.) Anim Behav. 1998;56:319–327. doi: 10.1006/anbe.1998.0837. [DOI] [PubMed] [Google Scholar]
- Peichel CL, Nereng KS, Ohgi KA, Cole BL, Colosimo PF, Buerkle CA, Schluter D, Kingsley DM. The genetic architecture of divergence between threespine stickleback species. Nature. 2001;414:901–905. doi: 10.1038/414901a. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacek DC, Passerini AG, Shi C, Francesco NM, Manduchi E, Grant GR, Powell S, Bischof H, Winkler H, Stoeckert CJ, Davies PF. Fidelity of enhanced sensitivity of differential transcription profiles following linear amplification of nanogram amounts of endothelial mRNA. Physiol Genomics. 2003;13:147–156. doi: 10.1152/physiolgenomics.00173.2002. [DOI] [PubMed] [Google Scholar]
- Ponder CA, Huded CP, Munoz MB, Gulden FO, Gilliam TC, Palmer AA. Rapid selection response for contextual fear conditioning in a cross between C57BL/6J and A/J: behavioral, QTL and gene expression analysis. Behav Genet. 2008;38:277–291. doi: 10.1007/s10519-008-9203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, (2009) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. ISBN 3-900051-07-0, URL http://www.R-Project.Org
- Reimchen TE. Spine deficiency and polymorphism in a population of Gasterosteus aculeatus – an adaptation to predators. Can J Zool. 1980;58:1232–124. [Google Scholar]
- Reimchen TE. Predators and morphological evolution in threespine stickleback. In: Bell MA, Foster SA, editors. The Evolutionary Biology of the Threespine Stickleback. Oxford: Oxford University Press; 1994. pp. 240–273. [Google Scholar]
- Roseboom PH, Nanda, SA, Bakshi VP, Trentani A, Newman SM, Kalin NH. Predator threat induces behavioral inhibition, pituitary-adrenal activation and changes in amygdala CRF-binding protein gene expression. Psychoneuroendocrinology. 2007;32:44–55. doi: 10.1016/j.psyneuen.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, Jonsson B, Schluter D, Kingsley DM. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- Shatz CJ. MHC class I: an unexpected role in neuronal plasticity. Neuron. 2009;64:40–45. doi: 10.1016/j.neuron.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina T, Ota M, Shimizu S, Katsuyama Y, et al. Rapid evolution of major histocompatibility complex class I genes in primates generates new disease alleles in humans via hitchhiking diversity. Genetics. 2006;173:1555–1570. doi: 10.1534/genetics.106.057034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurgin LG, Richardson DS. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc Biol Sci. 2010;277:979–988. doi: 10.1098/rspb.2009.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulley TJ, Huntingford FA. Paternal care and the development of adaptive variation in anti-predator responses in sticklebacks. Anim Behav. 1987;35:1570–1572. [Google Scholar]
- Wang H, Zhu YZ, Wong PT, Farook JM, Teo AL, Lee LK, Moochhala S. cDNA microarray analysis of gene expression in anxious PVG and SD rats after cat-freezing test. Exp Brain Res. 2003;149:413–421. doi: 10.1007/s00221-002-1369-1. [DOI] [PubMed] [Google Scholar]
- Wedekind C, Seebeck T, Bettens F, Paepke AJ. MHC-dependent mate preferences in humans. Proc Biol Sci. 1995;260:245–249. doi: 10.1098/rspb.1995.0087. [DOI] [PubMed] [Google Scholar]
- Williams TD, Brown M, Chipman JK, et al. Development of a stickleback (Gasterosteus aculeatus) cDNA microarray and gene expression responses to dibenzanthracene, ethinyl-estradiol and copper. Marine Environ Res. 2008;66:140–140. [Google Scholar]
- Wootton RJ. The Biology of the Stickleback. London: Academic Press; 1976. [Google Scholar]
- Yang Y, Zheng X, Wang Y, Cao J, Dong Z, Cai J, Sui N, Xu L. Stress enables synaptic depression in CA1 synapses by acoustic and chronic morphine: Possible mechanisms for corticosterone on opiate addiction. J Neurosci. 2004;24:2412–2420. doi: 10.1523/JNEUROSCI.5544-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary figure