Bcl11b is required for optimal FoxP3 expression and suppressor function by regulatory T cells and for the generation of inducible regulatory T cells.
Abstract
Dysregulated CD4+ T cell responses and alterations in T regulatory cells (Treg cells) play a critical role in autoimmune diseases, including inflammatory bowel disease (IBD). The current study demonstrates that removal of Bcl11b at the double-positive stage of T cell development or only in Treg cells causes IBD because of proinflammatory cytokine-producing CD4+ T cells infiltrating the colon. Provision of WT Treg cells prevented IBD, demonstrating that alterations in Treg cells are responsible for the disease. Furthermore, Bcl11b-deficient Treg cells had reduced suppressor activity with altered gene expression profiles, including reduced expression of the genes encoding Foxp3 and IL-10, and up-regulation of genes encoding proinflammatory cytokines. Additionally, the absence of Bcl11b altered the induction of Foxp3 expression and reduced the generation of induced Treg cells (iTreg cells) after Tgf-β treatment of conventional CD4+ T cells. Bcl11b bound to Foxp3 and IL-10 promoters, as well as to critical conserved noncoding sequences within the Foxp3 and IL-10 loci, and mutating the Bcl11b binding site in the Foxp3 promoter reduced expression of a luciferase reporter gene. These experiments demonstrate that Bcl11b is indispensable for Treg suppressor function and for maintenance of optimal Foxp3 and IL-10 gene expression, as well as for the induction of Foxp3 expression in conventional CD4+ T cells in response to Tgf-β and generation of iTreg cells.
Alterations in T regulatory cells (Treg cells) play a critical role in autoimmune disorders, including inflammatory bowel disease (IBD; Zenewicz et al., 2009; Wing and Sakaguchi, 2010). IBD is a chronic inflammatory disorder that includes Crohn’s disease and ulcerative colitis, which are both characterized by gut infiltration of highly reactive CD4+ T cells (Mottet et al., 2003; Izcue et al., 2006). Mutations in the Foxp3 gene, which encodes a critical transcription factor for Treg cell development and function, lead to lymphoproliferative disorders associated with fatal inflammation, both in humans and mice (Bennett et al., 2001; Brunkow et al., 2001; Fontenot et al., 2003, 2005; Wu et al., 2006). It has been proposed that other transcription factors act in concert with, downstream, or even upstream of Foxp3 in controlling Treg cell development and function (Wu et al., 2006; Chaudhry et al., 2009; Kitoh et al., 2009; Koch et al., 2009; Pan et al., 2009; Zheng et al., 2009). However, unlike deletion of Foxp3, lack of such transcription factors has not led to systemic lymphoproliferative diseases, but rather to more specific pathologies. For instance, removal of Stat3 from Treg cells resulted in the inability of these cells to suppress Th17 immune responses, which consequently caused intestinal inflammation with massive infiltration of CD4+ T cells (Chaudhry et al., 2009), whereas IRF4 deletion from Treg cells caused a Th2-predominant pathology. In addition, T-bet was found necessary for Treg cell function during Th1 response (Koch et al., 2009), whereas Eos was required for silencing of effector genes in Treg cells (Pan et al., 2009). Foxo1 and Foxo3 were also required for blocking the acquisition of effector T cell phenotype by Treg cells (Ouyang et al., 2010).
Bcl11b is a C2H2 zinc finger transcriptional regulator (Avram et al., 2000, 2002), previously demonstrated to act both as a transcriptional repressor and activator (Cismasiu et al., 2005, 2006, 2008, 2009). It is required for β and positive selection and survival of DN3 and double-positive (DP) thymocytes (Wakabayashi et al., 2003; Albu et al., 2007), in addition to controlling the commitment to T cell lineage of early thymic precursors (Ikawa et al., 2010; Li et al., 2010a,b) and antigen-specific clonal expansion and cytolytic function of CD8+ T lymphocytes (Zhang et al., 2010). Mice lacking Bcl11b starting with DP thymocytes displayed splenomegaly and enlarged mesenteric LNs (mLNs), and the CD4+ T lymphocytes presented an effector phenotype (Albu et al., 2007). In this study, we further show that these mice develop IBD with massive colon infiltrations of proinflammatory cytokine–producing CD4+ T cells. Additionally, this occurred also in mice solely lacking Bcl11b in Treg cells. The IBD was caused by alterations in Treg cells, as provision of WT Treg cells prevented the disease. Bcl11b-deficient Treg cells presented reduced suppressor function, with reduced levels of Foxp3 and IL-10, and increased CD4+ T cell proinflammatory cytokines Tnf, IFN-γ, and IL-17, as well as other alterations in Foxp3-dependent and -independent Treg cell genetic program. Mechanistic experiments demonstrate that Bcl11b bound to conserved noncoding regions in the Foxp3 and IL-10 genes, therefore participating in regulation of expression of these genes. Additionally, in the absence of Bcl11b, induction of Foxp3 expression in peripheral conventional CD4+ T cells, and consequently the generation of Treg cells, was affected. These results demonstrate critical roles of Bcl11b in Treg cells.
RESULTS
Bcl11bF/F/CD4-Cre mice develop wasting disease associated with IBD
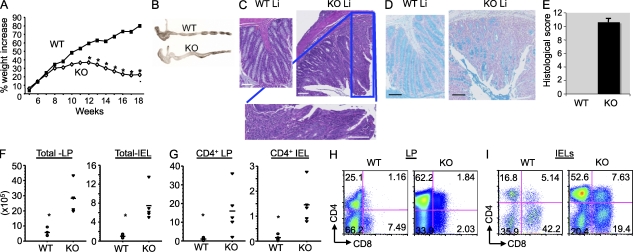
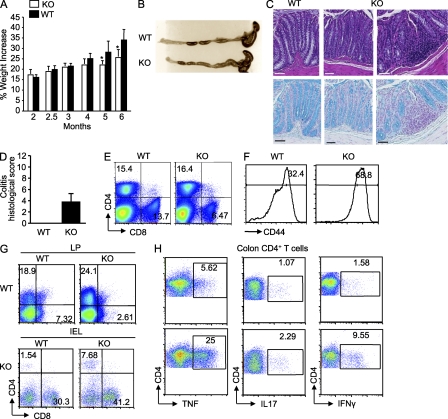
We previously observed that mice lacking Bcl11b starting with the DP thymocytes displayed splenomegaly and enlarged mLNs, starting at 5–7 wk of age. Additionally, the CD4+ T lymphocytes presented an effector phenotype (Albu et al., 2007). Further investigation of these mice demonstrated that they presented significantly lower weights starting with 10–12 wk of age (Fig. 1 A). Gross examination of the colons showed enlargement and thickening (Fig. 1 B), and hematoxylin and eosin (H&E) and Alcian blue staining indicated disruption of the crypt architecture, epithelial damage, massive infiltration of leukocytes, and loss of goblet cells (Fig. 1, C–E). Infiltrates in the liver were also observed (not depicted), which are all characteristics of IBD (Ito et al., 2006).
Figure 1.
Bcl11bF/F/CD4-Cre mice develop wasting disease, caused by IBD, associated with massive infiltrations of CD4+ T cells in the colon. (A) Weights of Bcl11bF/F/CD4-Cre and WT mice were recorded starting at 4 wk of age (n = 6 mice per group). Asterisks indicate statistical significance determined by unpaired two-tailed Student’s t test (*, P < 0.05). (B) Representative colon gross anatomy of Bcl11bF/F/CD4-Cre and WT mice 12–16 wk of age. (C) Representative H&E staining of colon sections of a similar area along the length of the colon from Bcl11bF/F/CD4-Cre and age- and sex-matched WT mice at 15–16 wk of age. The bottom panel shows a higher magnification image of the marked areas in the KO section. (D) Alcian blue counterstained with Nuclear Fast red of colon sections from Bcl11bF/F/CD4-Cre and WT mice, indicating in blue mucus-producing goblet cells. Bars, 100 µm. (B–D) Data are representative of at least 10 pairs of mice. (E) Histological scores of colitis at 15–16 wk of age, using the criteria described in Table S1. P (evaluated by unpaired two-tailed Student’s t test) = 4.52 × 10−8. The error bar indicates SD. (F) Absolute numbers of total cells in the colon LP and epithelium (IEL) of Bcl11bF/F/CD4-Cre versus WT mice. (G) Absolute numbers of CD4+ T cells present in the colon LP and epithelial compartment (IEL). (F and G) Asterisks indicate statistical significance as determined by unpaired two-tailed Student’s t test (*, P < 0.05). Horizontal bars indicate the mean. (H and I) Frequencies of CD4+ and CD8+ T cells among the colon LP lymphocytes (H) and IELs (I) evaluated by flow cytometry. (E–I) Data are derived from four to five mice per group.
Bcl11bF/F/CD4-Cre mice present large numbers of CD4+ T cells infiltrating the colon lamina propria (LP) and epithelium
The percentage and absolute numbers of CD4+ and CD8+ T cells were reduced in the peripheral lymphoid organs of Bcl11bF/F/CD4-Cre mice, including mLNs and Peyer’s patches (Fig. S1 A; Albu et al., 2007), which is believed to be a consequence of the developmental block at the DP stage of T cell development (Albu et al., 2007). The CD4+ T cells in the peripheral lymphoid organs presented an effector/memory phenotype (Fig. S1 B; Albu et al., 2007). Conversely, the percentage and absolute numbers of CD4+ T cells were increased in the colon LP and intraepithelial lymphocyte (IEL) compartment (Fig. 1, G–I). Additionally, there was an increase in a CD4+CD8+ cell population in the IEL compartment (Fig. 1 I), which presented high levels of Tcr-β. This population presented a slightly higher frequency of CD8ab versus CD8aa, compared with WT (Fig. S2, A and B). Overall, the total number of lymphocytes was increased both in the colon LP and IEL compartment (Fig. 1 F), in agreement with the massive infiltrates observed by histological analysis. Of note, the percentages and absolute numbers of CD8+ T cells were variable (Fig. 1, H and I; and Fig. S2 C; see Fig. 5 F). Similarly to what was reported for immature Bcl11b-deficient T cells or Bcl11b-deficient CD8+ T cells differentiated in vitro with the ER-Cre system (Ikawa et al., 2010; Li et al., 2010a,b), a large population of Bcl11b-deficient CD8+ T cells of Bcl11bF/F/CD4-Cre mice up-regulated Nk1.1 (Fig. S2 D). To determine whether the CD8+ T cells contributed to IBD, we generated Bcl11bF/F/CD4-Cre/b2m−/− mice. b2m−/− mice are almost devoid of CD8+ T cells because of defective positive selection of these cells in the thymus (Koller et al., 1990). Our results show that even in the absence of CD8+ T cells, Bcl11bF/F/CD4-Cre mice continued to present wasting disease (Fig. S2 E), suggesting that it is unlikely that the CD8+ T cells, even though they up-regulated NK markers, have a critical contribution to the disease phenotype. In conclusion, these results collectively demonstrate that although the numbers of CD4+ T cells in the peripheral lymphoid organs of Bcl11bF/F/CD4-Cre mice were reduced, there was a profound increase of CD4+ T cells in the colon, likely to be associated with the IBD observed in these mice. In addition, our results suggest that the CD8+ T cells are unlikely to have a critical contribution to the disease phenotype.
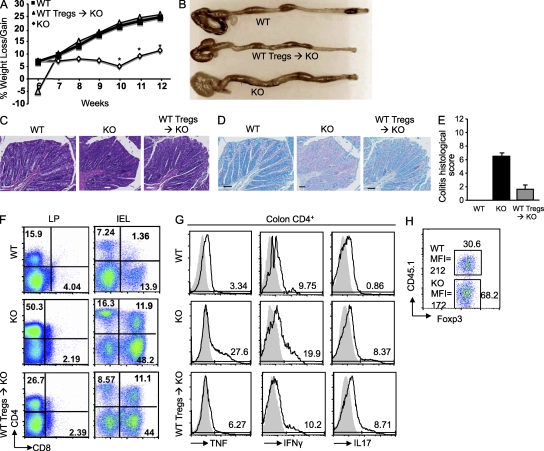
Figure 5.
Provision of WT Treg cells prevents IBD development in Bcl11bF/F/CD4-Cre mice. (A) Sorted WT Foxp3-GFP+ Treg cells were transferred into 4–5-wk-old Bcl11bF/F/CD4-Cre mice, as described in Materials and methods, and the weight of the mice was assessed weekly. Asterisks indicate statistical significance between the Treg cell transferred and untransferred groups (*, P < 0.05; determined by unpaired two-tailed Student’s t test). (B) Colon gross anatomy of Bcl11bF/F/CD4-Cre (KO), WT Treg cell–transferred Bcl11bF/F/CD4-Cre (WT Tregs → KO), and WT mice. (C) H&E staining of colon sections from the mice in B. (D) Alcian blue counterstained with Nuclear Fast red of colon sections from the mice in B. Goblet cells are blue. (C and D) Histological images are from similar areas along the length of the colon of the three groups of mice. Bars, 100 µm. (E) Histological scores of colitis, conducted as described in Table S1. (P < 0.05, determined by unpaired two-tailed Student’s t test.) Error bars indicate SD. (F) CD4+ and CD8+ T cell frequencies within the total lymphocyte population isolated from colon LP and epithelial compartment (IEL). (A–F) Data are representative of two independent experiments with three to five mice per group. (G) Intracellular cytokines in gated CD4+ T cells isolated from the colon of the indicated groups of mice. (H) Sorted WT CD45.1+/CD45.2+/Foxp3-GFP+ Treg cells were transferred into Bcl11bF/F/CD4-Cre/CD45.2+ mice, and WT (CD45.1+) and Bcl11b-deficient (CD45.1−) cells were evaluated 6–7 wk after the transfer. Splenocytes were stained for CD4, CD45.1, and Foxp3 and analyzed by flow cytometry. Numbers in boxes indicate the frequencies of the donor (CD45.1+) and recipient (CD45.1−) cells within the Foxp3+ population. Foxp3 mean fluorescence intensity (MFI) in Bcl11b-deficient and WT cells is indicated. (G and H) Data are representative of three independent experiments each with two to three mice per group.
Abnormal up-regulation of the gut-homing markers in Bcl11b-deficient T cells
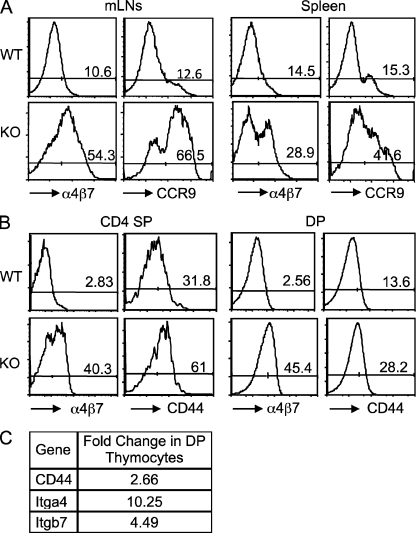
To further investigate why the CD4+ T cells of Bcl11bF/F/CD4-Cre mice accumulated in the colon, we evaluated several gut-homing markers and found that peripheral CD4+ T lymphocytes of Bcl11bF/F/CD4-Cre mice show considerable up-regulation of CCR9 and integrin α4β7 in the mLNs and spleen (Fig. 2 A), in addition to CD44 (Fig. S1 B). Up-regulation of α4β7 integrin and CD44 but not CCR9 was already observed in the DP and single-positive thymocytes, where the removal of Bcl11b was initiated (Fig. 2 B and not depicted). α4β7 integrins and CD44 messenger RNA (mRNA) levels were also up-regulated in DP thymocytes (Fig. 2 C). These results collectively demonstrate that Bcl11b-deficient CD4+ T cells show an abnormal and premature activation of gut-homing markers, which perhaps favors their accumulation in the colon.
Figure 2.
Bcl11bF/F/CD4-Cre mice show abnormal up-regulation of the gut-homing markers in T cells. (A) Frequencies of CD4+ T cells expressing the gut-homing markers α4β7 integrin and CCR9 in the mLNs and spleen of Bcl11bF/F/CD4-Cre and WT mice. (B) Frequencies of α4β7 integrin and CD44-expressing CD4 single-positive and DP thymocytes in Bcl11bF/F/CD4-Cre and WT mice. (A and B) Data are representative of at least three pairs of mice. (C) Fold change in mRNA levels for α4β7 integrins and CD44 in DP thymocytes from Bcl11bF/F/CD4-Cre mice. Data are representative of two independent microarrays.
Bcl11bF/F/CD4-Cre mice colons contain large numbers of infiltrating Th1 and Th17 cytokine–producing CD4+ T cells, neutrophils, dendritic cells, and macrophages
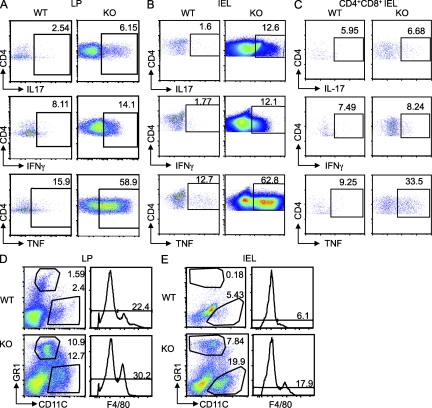
The proinflammatory Th1 and Th17 cytokines have a major impact on IBD development and progression (Zenewicz et al., 2009). We therefore investigated whether the accumulated CD4+ T cells in the colon produce proinflammatory cytokines. Our results demonstrated that there was a marked increase in the Th1 and Th17 cytokine–producing CD4+ T cells in the colon of diseased Bcl11bF/F/CD4-Cre mice (Fig. 3, A and B). Additionally, the increased IEL CD4+CD8+ cell population (Fig. 1 I) showed elevated levels of Tnf (Fig. 3 C). The H&E staining of the large intestine suggested that some of the infiltrating cells could be polymorphonuclear leukocytes. The flow cytometry data show that there was also a significant increase in the neutrophil, macrophage, and dendritic cell populations in the colon, both in the LP and IEL compartment (Fig. 3, D and E). The presence of neutrophils, macrophages, and dendritic cells in the IEL compartment is unusual even for diseased mice. We thus conducted immunohistochemistry to determine whether indeed such cells are present in the epithelium compartment of diseased mice or perhaps their presence in the IEL preparation was a consequence of contamination with cells from LP. Although it was clear that in the WT the presence of neutrophils, macrophages, and dendritic cells was confined to the LP, in the Bcl11bF/F/CD4-Cre mice, some of the cells seem to reach the epithelium layer area (Fig. S3). However, the colon structure in these mice became disorganized, therefore making it difficult to conclude on the precise location of these cells in the colon epithelium layer. These results clearly demonstrate that there was an accumulation of the CD4+ T cells producing proinflammatory cytokines in the colon, as well as of neutrophils, dendritic cells, and macrophages, which together with the Th1 and Th17 CD4+ T cells are likely to participate in the disease.
Figure 3.
Colons of Bcl11bF/F/CD4-Cre mice accumulate proinflammatory cytokine–producing CD4+ T cells, neutrophils, dendritic cells, and macrophages. (A and B) Intracellular cytokine production by CD4+ T cells isolated from the colon LP (A) and epithelium (IEL; B) of Bcl11bF/F/CD4-Cre and WT mice. (C) Intracellular cytokine production by colon IEL CD4+ CD8+ T cells. (A–C) Cells were isolated and activated as described in Materials and methods, followed by staining for surface CD4 and CD8 and intracellularly for the indicated cytokines, and analyzed by flow cytometry. Frequencies of cytokine-producing cells are indicated within the gated CD4+ T or CD4+ CD8+ T cell population. (D and E) Frequencies of neutrophils, dendritic cells, and macrophages in the colon LP (D) and epithelium compartment (IEL; E) are shown. Cells were isolated as described in Materials and methods, stained for CD4, CD8, Gr1, CD11c, and F4/80, and analyzed by flow cytometry. The numbers in the dot plots and histograms indicate the percentage of Gr1hi, CD11c+, and F4/80+ populations, respectively, within the gated CD4−CD8− population. The data are representative of four to five pairs of mice.
Bcl11b-deficient Treg cells have reduced in vitro suppressor function
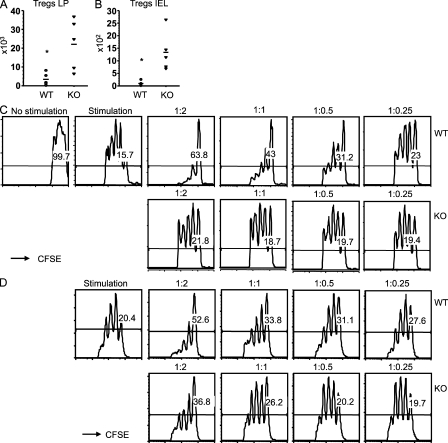
To further investigate why the cells in the colon produced uncontrolled cytokines, we evaluated the Treg cells of Bcl11bF/F/CD4-Cre mice. The percentages and absolute numbers of Treg cells in the peripheral lymphoid organs were reduced compared with WT (see Fig. 7 A and not depicted). However, their absolute numbers were significantly increased in the colon (Fig. 4, A and B), in agreement with the up-regulation of the integrin α4β7 and CCR9 in these cells as well (Fig. S4 A). Even though Treg cells accumulated in the colon, the conventional CD4+ T cells produced elevated levels of proinflammatory cytokines, raising the possibility that the Treg cells may not be able to suppress the conventional effector CD4+ T cells. To address this question, we conducted in vitro suppression assays, and the results demonstrated that Bcl11b-deficient Treg cells were unable to control the proliferation of effector CD4+ T cells with the same efficiency as WT Treg cells (Fig. 4 C). Furthermore, this was not caused by a significant defect in their maintenance in vitro (Fig. S4 B). Not only the Treg cells of Bcl11bF/F/CD4-Cre mice showed altered suppressor function, but Bcl11b-deficient Treg cells derived from mice in which Bcl11b was solely removed in Treg cells (Bcl11bF/F/Foxp3-Cre mice) also showed a similar phenotype (Fig. 4 D). These results collectively demonstrate that Bcl11b-deficient Treg cells are present in the colon; however, their suppressor function is altered.
Figure 7.
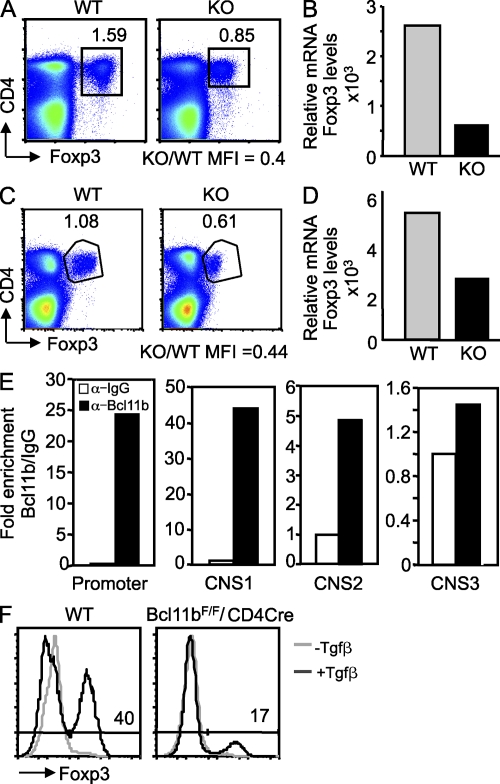
Altered expression of Foxp3 in Bcl11b-deficient Treg cells of Bcl11bF/F/CD4-Cre and Bcl11bF/F/Foxp3-Cre mice and altered induction of Foxp3 during generation of iTreg cells. (A and C) Evaluation of Foxp3 protein levels in Treg cells from Bcl11bF/F/CD4-Cre (A) and Bcl11bF/F/Foxp3-Cre (C) versus control mice. Splenocytes were surface stained for CD4, fixed and permeabilized, and further stained with anti-Foxp3 antibodies. Numbers indicate the percentages of CD4+Foxp3+ cells among all splenocytes, and the ratio between KO and WT Foxp3 mean fluorescence intensity (MFI) is indicated below. The results are representative for four pairs of mice. (B and D) Relative Foxp3 mRNA levels in sorted GFP+ Bcl11b-deficient Treg cells from Bcl11bF/F/CD4-Cre/Foxp3-GFP (B) and Bcl11bF/F/Foxp3-Cre (D) mice versus control levels evaluated by qRT-PCR. The data show one representative experiment of two with cells sorted from two to three mice, or multiple mice in the case of Bcl11bF/F/CD4-Cre/Foxp3-GFP mice. Bcl11bF/F/Foxp3-Cre mice and their controls were 5–6 mo of age. (E) ChIP assays with anti-Bcl11b or IgG antibodies. The immunoprecipitated DNA was amplified with primers corresponding to the indicated regulatory regions. Binding was normalized to the input, as described in Material and methods. Enrichment by Bcl11b is indicated as fold increase versus IgG. Data are derived from a representative experiment of two. (F) Foxp3-GFP−CD4+CD45rbhi cells from Bcl11bF/F/CD4-Cre/Foxp3-GFP+ and Foxp3-GFP+ WT mice were sorted and incubated in complete media in the presence of cross-linked anti-CD3e, 100 U IL-2, and 10 ng Tgf-β, for 3 d, and then stained for CD4 and Foxp3 and analyzed by FACS. Data are representative of three independent experiments.
Figure 4.
Treg cells of Bcl11bF/F/CD4-Cre and Bcl11bF/F/Foxp3-Cre mice show altered suppressor activity in vitro. (A and B) Evaluation of absolute numbers of Treg cells in the colon of Bcl11bF/F/CD4-Cre/Foxp3-GFP mice. Cells were isolated from the colon LP and epithelial compartment (IEL) stained for CD4 and analyzed by flow cytometry, and the percentage of Foxp3-GFP+ cells and their absolute numbers were evaluated. Four to five mice were used for each group. Asterisks indicate statistical significance determined by unpaired two-tailed Student’s t test (*, P = 0.04 in A; *, P = 0.02 in B). Horizontal bars indicate the mean. (C and D) Sorted CD4+CD45RbhiFoxp3-GFP− CFSE-labeled WT T cells were activated and co-cultured with sorted Foxp3-GFP+ Treg cells from Bcl11bF/F/CD4-Cre/Foxp3-GFP+ or Foxp3-GFP+ mice (C) or from Bcl11bF/F/Foxp3-Cre or Foxp3-Cre mice (D) at the indicated ratios of conventional T cells to WT or KO Treg cells (conventional/Treg cells). After 72 h, cells were stained with anti–CD4-APC-Cy7 and anti–Foxp3-APC antibodies and analyzed by flow cytometry. The number of cell divisions was evaluated in the CD4+/Foxp3− population based on gradual loss of CFSE fluorescence. The maintenance of Bcl11b-deficient and WT Treg cells during the suppression assay is shown in Fig. S4 B. The results are representative of three independent experiments, each with one pair of mice.
Provision of the WT Treg cells prevented the IBD in Bcl11bF/F/CD4-Cre mice
To further determine whether the IBD observed in Bcl11bF/F/CD4-Cre mice is indeed a consequence of altered Treg cell function, we transferred WT Treg cells in 4–5-wk-old Bcl11bF/F/CD4-Cre mice, before these mice showed any disease signs. Provision of WT Treg cells prevented the wasting disease in Bcl11bF/F/CD4-Cre mice (Fig. 5 A), and the colon did not present enlargement or thickening (Fig. 5 B). H&E and Alcian blue staining showed normal structure of the colon, with normal crypts containing abundant goblet cells, and only minor infiltrates (Fig. 5, C–E). Flow cytometry analysis demonstrated a marked reduction in the number of CD4+ T cells infiltrating the colon (Fig. 5 F), as well as in the CD4+ T cells producing IFN-γ and Tnf, but surprisingly not IL-17 (Fig. 5 G). Prevention of disease as a result of WT Treg cell provision was not caused by a defective maintenance of Bcl11b-deficient Treg cells in the presence of WT Treg cells (Fig. 5 H). These results demonstrate that provision of WT Treg cells prevented IBD, strongly suggesting that the alterations in Bcl11b-deficient Treg cells are very likely the cause of IBD.
Removal of Bcl11b solely in Treg cells causes IBD
We showed above that the absence of Bcl11b starting with DP thymocytes caused functional alterations in Treg cells, including reduced suppressor function (Fig. 4 C). Importantly, Bcl11b-deficient Treg cells from Bcl11bF/F/Foxp3-Cre mice also showed altered suppressor function (Fig. 4 D). We further evaluated Bcl11bF/F/Foxp3-Cre mice and found that they developed IBD; however, there was a delay in the onset, and the severity of the disease was reduced compared with the CD4-Cre model. Specifically, Bcl11bF/F/Foxp3-Cre mice failed to gain weight only at ∼20 wk of age (Fig. 6 A), whereas this occurred in Bcl11bF/F/CD4-Cre mice at ∼10–12 wk, and most of the Bcl11bF/F/CD4-Cre mice did not survive over 16–18 wk of age (Fig. 1 A and not depicted). In addition, 5-mo-old and older Bcl11bF/F/Foxp3-Cre mice started to have enlarged spleens and colon inflammation with cellular infiltrations and reduction in goblet cells (Fig. 6, B–D; and Fig. S5 A). We thus investigated the removal of Bcl11b in 5–6-wk-old Bcl11bF/F/Foxp3-Cre mice versus 20-wk-old mice and found that although removal of Bcl11b occurred in Treg cells from 20-wk-old mice, there was no removal of the gene in the Treg cells from young mice, whereas Bcl11b was removed in 5–6-wk-old CD4-Cre mice (Fig. S5 B). Of note, the levels of Bcl11b in WT and knockout cells should be evaluated only in relation to each other in each panel. We do not know at the moment whether there are variations in Bcl11b levels in WT cells related to age or whether these are experimental variations. Thus, these results suggest that removal of Bcl11b in the Bcl11bF/F/Foxp3-Cre mice is delayed compared with the CD4-Cre model, which is likely to be responsible for the late onset of disease. Indeed the Foxp3-Cre deleter used in this study is a low Cre-expressing line. Despite this fact, similarly to the 15-wk-old CD4-Cre mice, 5–6-mo-old Bcl11bF/F/Foxp3-Cre mice had CD4+ T cells infiltrating in larger numbers in the colon compared with age- and sex-matched controls (Fig. 6 G), although in the periphery their numbers were comparable with WT mice (Fig. 6 E). This occurred despite the fact that the conventional CD4+ T cells of Bcl11bF/F/Foxp3-Cre mice only minimally up-regulated CCR9 and α4β7 integrin (Fig. S5 C). The splenic conventional CD4+ T cells of Bcl11bF/F/Foxp3-Cre mice had a larger percentage of activated CD44hi population compared with WT (Fig. 6 F), similar to the Bcl11bF/F/CD4-Cre mice. Furthermore, the CD4+ T cells infiltrating the colons of 5–6-mo-old Bcl11bF/F/Foxp3-Cre mice produced higher levels of Th1 and Th17 cytokines compared with age-matched controls (Fig. 6 H) but less pronounced than in diseased Bcl11bF/F/CD4-Cre mice. These results demonstrate that conditional removal of Bcl11b solely in Treg cells causes IBD with infiltrations of Th1 and Th17 cytokine–producing CD4+ T cells in the colon, similar to Bcl11bF/F/CD4-Cre mice.
Figure 6.
Bcl11bF/F/Foxp3-Cre mice fail to gain weight only after 20 wk of age and show colon inflammation with infiltration of proinflammatory cytokine–producing CD4+ T cells. (A) Weight of Bcl11bF/F/Foxp3-Cre mice between 2 and 6 mo of age. 6–10 mice/group were evaluated. Asterisks indicate statistical significance determined by unpaired two-tailed Student’s t test (*, P < 0.05). (B) Colon gross anatomy representative for 5–6-mo-old Bcl11bF/F/Foxp3-Cre mice and age-matched controls. (C) H&E staining (top) and Alcian blue counterstained with Nuclear Fast red (bottom) of colon sections of 5–6-mo-old Bcl11bF/F/Foxp3-Cre and age-matched controls. Goblet cells are blue. Histological images are from similar areas along the length of the colon. Bars, 100 µm. (B and C) Data are representative of five pairs of mice. (D) Colitis histological scale of 5–6-mo-old Bcl11bF/F/Foxp3-Cre and age-matched controls, using the criteria in Table S1. (A and D) Error bars indicate SD. (E) Frequencies of CD4+ and CD8+ T cells in spleen of Bcl11bF/F/Foxp3-Cre and WT mice. (F) Frequencies of CD44+CD4+ T cells in spleen of Bcl11bF/F/Foxp3-Cre and WT mice. Data are representative of three pairs of mice. (G) Frequencies of CD4+ and CD8+ T cells in the colon LP and intraepithelial compartment (IEL) of Bcl11bF/F/Foxp3-Cre and WT mice. (H) Cytokine production by CD4+ T cells isolated from Bcl11bF/F/Foxp3-Cre and WT mice colons. Cells were isolated and activated as described in Materials and methods, followed by staining for surface CD4 and intracellularly for the indicated cytokines, and analyzed by flow cytometry. Frequencies of cytokine-producing CD4+ T cells are indicated. Data are representative of three pairs of mice.
Bcl11b is required for optimal expression of Foxp3 and binds to regulatory regions of the Foxp3 gene
We further investigated the mechanisms responsible for reduced suppressor function of Bcl11b-deficient Treg cells. The first observation was that the level of Foxp3 was reduced in Bcl11b-deficient Treg cells compared with controls both in the Bcl11bF/F/CD4-Cre mice and Bcl11bF/F/Foxp3-Cre mice (Fig. 7, A and C). The reduction was not only in the Foxp3 protein level, but also in the Foxp3 mRNA (Fig. 7, B and D; and Fig. S6, A and B). Bcl11b-deficient T cells have been previously shown to have altered TCR signaling and reduced expression of TCR complex components (Albu et al., 2007; Li et al., 2010a,b; Zhang et al., 2010). Therefore, Bcl11b might regulate Foxp3 expression as a consequence of altered TCR signaling in Bcl11b-deficient Treg cells. We first evaluated the CD3 levels and found a very modest reduction in mRNA levels (Fig. S6, A and B) and a minor reduction in the surface levels, and only in the CD4-Cre system (Fig. S7, A and C). We further evaluated the early activation marker CD69, which is normally up-regulated in response to TCR signaling, and the response of Bcl11b-deficient Treg cells was similar to WT, both for the CD4-Cre and Foxp3-Cre systems (Fig. S7, B and D). These results suggest that Bcl11b-deficient Treg cells respond to TCR stimulation similar to WT, and therefore it is unlikely that the reduction in the Foxp3 levels is caused by altered TCR signaling.
Considering that Bcl11b is a transcription factor, we next investigated whether Bcl11b binds to Foxp3 gene regulatory regions. The definition of these regions is given based on the information provided in Kim and Leonard (2007) and updated after Zheng et al. (2010). We specifically investigated the promoter and the conserved noncoding sequences CNS-1, CNS-2, and CNS-3, which are known to play a critical role in the control of this gene expression (Mantel et al., 2006; Kim and Leonard, 2007; Tone et al., 2008; Kitoh et al., 2009; Zheng et al., 2010). CNS-1 and CNS-2 are both localized in the intron after the untranslated exon 1 of Foxp3 gene (Zheng et al., 2010), whereas CNS-3 is localized in the intron after the first coding exon (Zheng et al., 2010). We conducted chromatin immunoprecipitations (ChIPs) followed by quantitative PCR (qPCR) and found that Bcl11b strongly associated with the promoter and CNS-1, less efficiently with the CNS-2, and did not associate with CNS-3 (Fig. 7 E). We then inspected the proximal region of the promoter, shown to be bound in the ChIP assays, and found a potential Bcl11b-binding site, located at position −89 from the transcription start site (TSS; Fig. S7 E). We mutated this site to AAA (Fig. S7 E), which is known not to be bound by Bcl11b (Avram et al., 2002; Cismasiu et al., 2006), in a construct that contains the Foxp3 promoter upstream of and the Foxp3 enhancer 1 (Smad enhancer) downstream of Firefly luciferase (Tone et al., 2008). The mutation significantly reduced the reporter activity in response to activation (Fig. S7 E), suggesting that this site is important for Foxp3 promoter activity. In conclusion, these results demonstrate that Bcl11b regulates expression of the Foxp3 gene and associates predominantly with the Foxp3 gene promoter and the CNS-1 and CNS-2 regulatory regions and that the potential Bcl11b-binding site in the Foxp3 promoter is important for its activation.
Bcl11b plays a critical role in the induction of Foxp3 gene during generation of induced Treg cells (iTreg cells) from conventional CD4+ T cells
It has been recently demonstrated that CNS-1 is indispensable for peripheral induction of the Foxp3 gene and generation of iTreg cells from conventional CD4+ T cells but not for thymic Treg cell differentiation (Zheng et al., 2010). Considering that Bcl11b associated with the CNS-1, we investigated whether the absence of Bcl11b causes alterations in the induction of Foxp3 gene expression in conventional CD4+ T cells in response to Tgf-β1 and iTreg cell generation. Our results demonstrate that Bcl11b is required for peripheral induction of Foxp3 in the conventional CD4+ T cells in response to Tgf-β1 (Fig. 7 F), which is in agreement with Bcl11b binding to the CNS-1 (Fig. 7 E). These results demonstrate that Bcl11b plays a critical role in Foxp3 induction in response to Tgf-β1, potentially through association with CNS-1.
Bcl11b-deficient Treg cells show reduced expression of IL-10 gene and elevated levels of proinflammatory cytokine genes
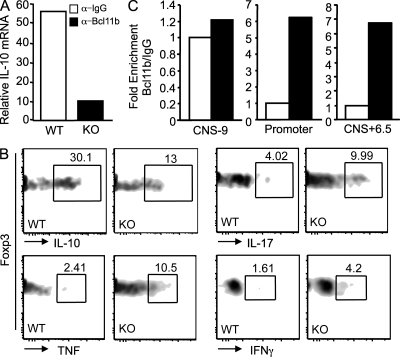
To further determine whether other alterations in the Bcl11b-deficient Treg cells contribute to their affected function, we profiled expression of genes in Bcl11b-deficient Treg cells versus WT, both in the CD4-Cre and Foxp3-Cre systems (Fig. S6, A and B). Importantly, Foxp3 itself was found down-regulated, and together with it, several Treg signature genes, some dependent and some independent of Foxp3 (Fig. S6, A and B). However, CD25, Gitr, CTLA4, Icos, and CD103 were not found altered at the expression level (not depicted), and their surface levels were either equal or elevated in Treg cells from Bcl11bF/F/CD4-Cre mice and Foxp3-Cre mice (Fig. S8, A and B), suggesting that it is unlikely that Bcl11b-deficient Treg cells have poor suppressor activity related to these markers. Interestingly, there was a major reduction in the IL-10 mRNA levels (Fig. 8 A and Fig. S6). IL-10 is known to play an important role in Treg suppressive function, particularly in colitis (Miyara and Sakaguchi, 2007; Vignali et al., 2008; Wing and Sakaguchi, 2010). Mice deficient in IL-10 develop spontaneous inflammation of the large intestine (Kühn et al., 1993). Removal of the IL-10 gene only in CD4+ T cells also results in inflammation of the colon (Roers et al., 2004), whereas conditional removal only in Treg cells causes IBD, though less severe (Rubtsov et al., 2008). Not only the mRNA levels of IL-10 were down-regulated in Bcl11b-deficient Treg cells, but importantly, the protein levels were also reduced (Fig. 8 B and Fig. S6 C). In addition, several effector CD4+ T cell genes, including IL-17 and IFN-γ, as well as IL-22 were found up-regulated (Fig. S6). Additionally the intracellular levels of the proinflammatory cytokines IL-17 and IFN-γ and TNF were found elevated as well (Fig. 8 B). Interestingly, Bcl11b-deficient Treg cells showed a modest up-regulation of several NK marker mRNAs (Fig. S6), including NK1.1; however, this was not reflected in an increased Nk1.1+ population in Treg cells from Bcl11bF/F/CD4-Cre mice or Bcl11bF/F/Foxp3-Cre mice (Fig. S8, C and D). These results demonstrate that Bcl11b-deficient Treg cells showed reduced levels of IL-10, which is important in suppressor function, and also elevated levels of several effector cytokines, suggesting that Bcl11b contributes to the repression of proinflammatory cytokine gene expression in Treg cells, either directly or indirectly, and overall demonstrate that Bcl11b is a critical transcriptional regulator of Treg cell genetic program.
Figure 8.
Treg cells from Bcl11bF/F/CD4-Cre mice show reduced IL-10 expression and up-regulation of proinflammatory cytokines, and Bcl11b binds to the IL-10 promoter and noncoding sequence CNS+6.5. (A) IL-10 mRNA levels in Treg cells from Bcl11bF/F/CD4-Cre and WT mice evaluated by qRT-PCR analysis. (B) Flow cytometry analysis of IL-10, IL-17, Tnf, and Ifn-γ in Treg cells from Bcl11bF/F/CD4-Cre/Foxp3-GFP and Foxp3-GFP colon LP. Data are representative of four pairs of mice. (C) ChIP assays with anti-Bcl11b or IgG antibodies. The immunoprecipitated DNA was amplified with primers corresponding to the indicated regulatory regions. Binding was normalized to the input, as described in Materials and methods. Enrichment by Bcl11b is indicated as fold increase versus IgG. This is one representative experiment out of two.
Bcl11b associates with critical control regions of the IL-10 gene
Considering the finding that IL-10 mRNA levels were reduced in Bcl11b-deficient Treg cells, we further investigated whether Bcl11b associated with the IL-10 gene promoter and/or other previously characterized regulatory regions of these genes, such as CNS+6.5 and CNS-9 (Jones and Flavell, 2005). ChIP assays demonstrated that Bcl11b associated with the IL-10 promoter and with CNS+6.5, a conserved noncoding potent regulatory sequence localized 6.5 kb downstream of IL-10 TSS (Jones and Flavell, 2005), but not with CNS-9, positioned 9 kb upstream of the TSS (Fig. 8 C; Jones and Flavell, 2005). These results demonstrate that Bcl11b associates with regulatory regions in the IL-10 gene localized in the promoter, as well as at the 3′ end of the gene, likely participating in the regulation of this gene expression.
DISCUSSION
Our results demonstrate that the absence of Bcl11b, either starting with DP thymocytes or only in Treg cells, results in IBD, associated with marked infiltration of proinflammatory cytokine–producing CD4+ T cells in the colon. In addition to the proinflammatory cytokine–producing CD4+ T cells, Bcl11bF/F/CD4-Cre mice showed a noticeable increase of a CD4+CD8+ T cell population in the IEL compartment, which produced elevated levels of Tnf. Based on the expression of Tcr-β, these cells seem to be mature and presented a slight increase in the frequencies of CD8ab versus CD8aa, compared with WT. This population is unlikely to be related to the CD4+CD8aa suppressive population, previously identified in the small intestine IEL compartment (Das et al., 2003), and may rather participate in the IBD pathology through the production of Tnf. Even though effector CD4+ T cells are responsible for the production of proinflammatory cytokines that cause tissue damage, Treg cells are responsible for maintaining peripheral tolerance and keeping these cells in check (Vignali et al., 2008). Removal of Bcl11b at the DP stage of T cell development caused premature up-regulation of the gut-homing markers integrin α4β7 and CCR9 in both effector CD4+ T cells and Treg cells, conferring ability to both types of cells to accumulate into the gut. Nevertheless, our results demonstrate that Bcl11b-deficient Treg cells have reduced suppressor activity, both when the gene is removed starting with DP thymocytes or solely in Treg cells. These results suggest that, although present in the colon, Bcl11b-deficient Treg cells were unable to suppress the reactive CD4+ T cells. Provision of WT Treg cells prevented the disease, demonstrating that it is mainly the defect in the Treg cells that is responsible for the disease in Bcl11bF/F/CD4-Cre mice. Moreover, supporting our findings, conditional removal of Bcl11b only in Treg cells with the use of the Foxp3-Cre strain also resulted in inflamed intestine and failure to gain weight, albeit much later in life. The late onset of the disease in Bcl11bF/F/Foxp3-Cre mice is caused by the delayed removal of Bcl11b in this model because the Foxp3-Cre deleter used in this study is a low Cre-expressing line. Even if later and milder, the disease of Foxp3-Cre mice mirrored that of the Bcl11bF/F/CD4-Cre mice. In support of the common defective function of Bcl11b-deficient Treg cells of Bcl11bF/F/CD4-Cre and Bcl11bF/F/Foxp3-Cre mice comes also the fact that their genetic programs were similarly altered compared with WT, and the Treg cells of both models had affected suppressive function. Specifically, expression profiling of Bcl11b-deficient Treg cells from Bcl11bF/F/CD4-Cre and Bcl11bF/F/Foxp3-Cre mice showed similar deregulated genes, which provided valuable insights on defects in their function that lead to the pathology observed in these mice. In both models, Foxp3 protein and mRNA levels were reduced, and we found that Bcl11b associated with the Foxp3 promoter, as well as with CNS-1 and CNS-2, but not with CNS-3 (Kim and Leonard, 2007; Zheng et al., 2010). Foxp3 promoter was shown to contain NFAT and AP1 motifs, in addition a GC box (Kim and Leonard, 2007). We found that mutation of a potential Bcl11b-binding site (Avram et al., 2002; Cismasiu et al., 2006) located at −89 from the TSS in the Foxp3 promoter attenuated the activity of the promoter, suggesting that this site is important. In addition, we found that in the absence of Bcl11b, there was a significant reduction in the generation of iTreg cells from conventional CD4+ T cells in response to Tgf-β1. CNS-1 plays an important role in the induction of the Foxp3 gene expression and the generation of iTreg cells (Zheng et al., 2010), and our data show that Bcl11b bound to CNS-1. Thus, Bcl11b is likely to also play an important contribution in the induction of Foxp3 expression in peripheral conventional CD4+ T cells and generation of iTreg cells in response to Tgf-β through CNS-1–mediated control. We also found that Bcl11b bound to CNS-2 but not to CNS-3. CNS-2 was demonstrated to play a critical role in expression of Foxp3, being induced by IL-2 through Stat5 (Zorn et al., 2006), and additionally by TCR signaling through CREB (Kim and Leonard, 2007). We thus investigated whether reduced expression of Foxp3 in the absence of Bcl11b is a consequence of altered TCR signaling, considering that Bcl11b-deficient T cells have been shown to have altered TCR signaling (Albu et al., 2007; Li et al., 2010a,b; Zhang et al., 2010). However, our results indicated that the up-regulation of CD69 after TCR activation was similar to WT, thus suggesting that altered TCR signaling is not the cause for reduced Foxp3 expression. The Runx1–Cbfb complex has been recently demonstrated to be required for optimal expression of Foxp3 by association with both Foxp3 promoter and with CNS-2 (Kitoh et al., 2009; Rudra et al., 2009). One of the two Runx1-binding sites in the CNS-2 are in the vicinity of a GC-rich sequence, raising the possibility that Bcl11b might associate with the Runx1–Cbfb complex and cooperate in Foxp3 expression maintenance. Foxp3 itself was recently found to be in a large complex with Runx–Cbfb and to bind in a Runx–Cbfb-dependent manner to CNS-2 (Kitoh et al., 2009; Zheng et al., 2010). It is therefore possible that Runx–Cbfb and Foxp3 associate in a common complex with Bcl11b to regulate Foxp3 expression and Treg cell lineage stability.
Other transcription factors recently shown to regulate Foxp3 expression include E2a, found to bind to the Foxp3 promoter and induce its expression in response to Tgf-β. This required Id3 for the inhibition of the Gata3-mediated repressive activity on Foxp3 expression (Maruyama et al., 2011).
Our results show that CD25, Gitr, CTLA4, Icos, and CD103 surface levels were either equal or elevated in Bcl11b-deficient Treg cells, suggesting that it is unlikely that these cells have poor suppressive activity related to these markers. However, Bcl11b-deficient Treg cells showed reduced levels of IL-10, which may also contribute to the reduced suppressive function of Bcl11b-deficient Treg cells. Studies have shown that IL-10–deficient mice lack Treg cells that are capable to control inflammatory responses in the intestine (Kühn et al., 1993; Roers et al., 2004; Rubtsov et al., 2008). We found that Bcl11b binds to critical conserved noncoding regulatory sequences in the IL-10 locus, including the promoter, CNS+6.5, but not CNS-9. Interestingly, CNS+6.5 was found to contain c-Jun and JunB and regulate IL-10 expression in Th2 cells (Wang et al., 2005). It remains a question whether Jun proteins might be implicated in the regulation of IL-10 expression in Treg cells and eventually compete or cooperate with Bcl11b in this process.
Furthermore, Bcl11b-deficient Treg cells showed up-regulation of proinflammatory gene expression, including Tnf, Ifn-γ, and IL-17. Decreased Foxp3 levels were previously shown to cause immune disease by subverting the suppressor function of Treg cells and conferring them characteristics of effector cells (Wan and Flavell, 2007). At the moment, we do not know whether up-regulation of the proinflammatory cytokine genes is caused by reduced Foxp3 expression or Bcl11b is directly implicated in the repression of these genes’ expression in Treg cells. Other transcription factors such as Eos were recently shown to repress effector gene expression in Treg cells (Pan et al., 2009). In this regard, the idea that effector CD4+ T cells, as well as Treg cells, have the capacity to redirect their functional programs because their genomic plasticity has recently emerged (Xu et al., 2007; Zhou et al., 2009).
Another potential mechanism implicated in the reduced suppression of Bcl11b-deficient Treg cells may come from the fact that Bcl11b-deficient Treg cells had decreased Stat3 mRNA levels, which may contribute to their reduced ability to suppress Th17 immune responses, as has been recently shown for Stat3-deficient Treg cells (Chaudhry et al., 2009).
In relation to the reduced levels of Foxp3, Bcl11b-deficient Treg cells also had deregulated expression of multiple genes known to be Foxp3 dependent (Hill et al., 2007). Though the deregulated expression of these genes may be a consequence of lower Foxp3 levels, it is also possible that Bcl11b may associate with Foxp3 complexes and participate in regulation of Foxp3-dependent genes, being one of the transcription factors that provides help to Foxp3 to maintain the genetic program and keeps the identity of Treg cells. Bcl11b-deficient Treg cells also had deregulated expression of Treg cell signature genes that are not dependent of Foxp3 (Hill et al., 2007), raising the possibility that Bcl11b may participate in regulation of expression of Foxp3-independent genes. In addition to deregulation in the Treg cell–specific program, Bcl11b-deficient Treg cells showed up-regulation of several NK and Fc receptor mRNAs; however, NK1.1 was not up-regulated on the surface of Treg cell and only on the surface of Bcl11b-deficient CD8+ T cells of Bcl11bF/F/CD4-Cre mice. Our results demonstrate that the role of Bcl11b is not only to keep the identity of T cells, and not allow expression of genes specific to other lineages, but also to keep the identity of specific subtypes of T cells such as Treg cells, perhaps by working in association with T cell subset–specific transcription factors.
MATERIALS AND METHODS
Mice.
Bcl11bF/F/CD4-Cre mice have been previously described (Albu et al., 2007). Foxp3-Cre mice were purchased from The Jackson Laboratory and backcrossed on a C57BL/6 background for five generations using the Speed Congenics Technology provided by The Jackson Laboratory. Foxp3-GFP mice were a gift from A. Rudensky (Memorial Sloan-Kettering Cancer Center, New York, NY) and were crossed with Bcl11bF/F/CD4-Cre mice to generate Bcl11bF/F/CD4-Cre/Foxp3-GFP mice, and Foxp3-Cre–GFP mice were crossed with Bcl11bF/F mice to obtain Bcl11bF/F/Foxp3-Cre mice. Mice were maintained under specific pathogen-free conditions in the Animal Resource Facility of Albany Medical College. Mice were sacrificed between 6–14 wk of age, except for the Foxp3-Cre mice, which were sacrificed at 5–6 mo of age, unless otherwise specified. The experiments were conducted according to animal protocols approved by the Albany Medical Center Institutional Animal Care and Use Committee.
Cell isolation.
Lymphocytes isolated from mLNs, spleens, Peyer’s patches, or colons were manipulated in 10% FBS RPMI 1640 medium (complete medium [CM]). CD4+ T cells were purified using anti-CD4 magnetic beads (Miltenyi Biotec) to a purity of at least 95%. For in vitro suppression assays, conventional CD4+ T cells were isolated from WT, Bcl11bF/F/CD4-Cre/Foxp3-GFP, or Bcl11bF/F/Foxp3-Cre-GFP mice and sorted as CD4+GFP−CD45Rbhi. CD4+ Treg cells were sorted based on their GFP. Colon IELs were obtained by incubating cleaned and cut tissue pieces in a solution containing 1 mM DTT in PBS for 10 min, followed by two 10-min incubations in 10 ml of buffer containing 30 mM EDTA, pH 8.0, and 10 mM Hepes in 1× HBSS. Remaining tissue, containing LP lymphocytes, was digested for 90 min in CM containing 100 mg/ml collagenase D (Roche), 1 mg/ml Dispase II (Invitrogen), and 0.2–1 mg/ml DNase (Roche). This was followed by a gradient purification in 33% Percoll (Sigma-Aldrich).
Flow cytometry.
Surface and intracellular staining was performed on FcR-blocked lymphocytes using the antibodies listed in Supplemental materials and methods. For intracellular cytokine determination, up to 2 × 106 lymphocytes/well were stimulated with 50 ng/ml PMA, 500 ng/ml ionomycin, 10 µg/ml Brefeldin A, and 2 µM monensin in CM for 5 h at 37°C. Cells were first stained for surface markers, followed by fixation in 4% paraformaldehyde and permeabilization with 0.05% saposin, followed by intracellular staining for cytokines with the fluorescence-labeled antibodies listed in Supplemental materials and methods. Flow cytometry analysis was conducted on an upgraded (Cytek) five-color FACSCalibur, FACSCanto, or LSR II flow cytometer (BD), and data were analyzed using FlowJo software (Tree Star). Sorting of CD4+/Foxp3-GFP+ and CD4+GFP−CD45Rbhi cells was performed using a FACSAria instrument (BD).
In vitro suppression assay was performed by co-culture of 105 CFSE-labeled sorted CD4+CD45Rbhi T cells with sorted CD4+Foxp3-GFP+ Treg cells at various ratios. Cells were co-cultured for 72 h at 37°C in round-bottomed plates in the presence of irradiated TCRa−/− splenocytes (2 × 105 cells/well), 3 µg/ml anti-CD3, and 1.5 µg/ml anti-CD28, and the number of cell divisions based on gradual loss of CFSE fluorescence was assessed by flow cytometry. Foxp3-GFP+ cells were gated out based on staining anti–Foxp3-APC antibodies.
iTreg cell induction.
CD4+/CD45Rbhi/Foxp3-GFP− T cells were sorted and incubated at a density of 0.25 × 106/well in 96-well flat-bottom plates for 3 d in complete media containing cross-linked anti-CD3e, 100 U IL-2, and 10 ng Tgf-β. Cells were then stained for CD4 and Foxp3 and assessed by flow cytometry.
Adoptive transfer of cells.
0.3 × 106 sorted WT CD45.1/CD45.2/CD4+/Foxp3-GFP+ T cells were i.v. injected into 4–5-wk-old Bcl11bF/F/CD4-Cre mice. The weight of recipient mice was recorded weekly, and mice were sacrificed for analysis at 12 wk.
Histology.
Colons were removed from mice, washed with PBS, fixed overnight in 10% buffered formalin, placed in 70% ethanol, and embedded in paraffin. 5-mm sections were stained with H&E or Alcian blue. Microscopic examination of the sections was performed using a BX51 instrument (Olympus). A colitis histological score was established based on the criteria shown in Table S1.
RNA extraction and microarray analysis.
Sorted Treg cells from Bcl11bF/F/CD4-Cre or Bcl11bF/F/Foxp3-Cre mice were used for RNA extraction performed as previously described (Cismasiu et al., 2006). Two independent sets of Treg cell mRNAs were analyzed for gene expression profiling using the Agilent platform (MOgene), available under GEO DataSets accession no. GSE30523. For the Bcl11bF/F/CD4-Cre mice, sorted Treg cells were pooled from 6–8 mice, whereas WT Treg cells were pooled from two to three mice. For Bcl11bF/F/Foxp3-Cre mice and their age-matched WT controls, sorted Treg cells were pooled from two to three mice. The functional annotation and classification were conducted using DAVID (Database for Annotation, Visualization, and Integrated Discovery) version 6.7 (Huang et al., 2009).
qRT-PCR.
RNA and qRT-PCR was conducted as previously described (Cismasiu et al., 2006). The primers for qPCR were chosen such as to extend products under 200 bp, with no formation of primer dimmers and cross introns. The relative abundance of each message was normalized to actin and calculated as 2−(Ct gene − Ct actin), where Ct represents the threshold cycle for each transcript.
ChIP.
ChIP assays were conducted using sorted Foxp3-GFP Treg cells or Dynabead-enriched CD4+CD25+ Treg cells, according to our previously published protocol (Cismasiu et al., 2006), with the difference that anti-Bcl11b antibodies were preloaded on anti–rabbit IgG-Dynabeads, and the precipitated DNA was analyzed by qPCR. The primers for Foxp3 promoter and CNS-1, CNS-2, and CNS-3 regions were previously described (Kim and Leonard, 2007; Kitoh et al., 2009; Zheng et al., 2010). The primers for IL-10 promoter and IL-10 gene regulatory regions CNS-9 and CNS+6.45 were also previously described (Ahyi et al., 2009; Lee et al., 2009). The transcription factor– or IgG-bound DNA was evaluated based on the following formula: 2−ct of the immunoprecipitated sample divided by 2−ct of the input, where ct is the threshold cycle, followed by corrections for the input dilution. The IgG values were represented as 1, and the transcription factor–bound DNA was represented as fold enrichment.
Statistical analysis.
Differences between Bcl11bF/F/CD4-Cre, Bcl11bF/F/Foxp3-Cre, and control mice under all experimental conditions were analyzed by the two-tailed Student’s t test and expressed as the mean ± SD. P ≤ 0.05 was considered significant.
Online supplemental material.
Fig. S1 shows FACS analysis of mLNs and Peyer’s patch lymphocytes, including CD62L and CD44 in Bcl11bF/F/CD4-Cre mice. Fig. S2 shows FACS analysis of the CD4+CD8+ T cell populations within the colon epithelial compartment, absolute numbers of CD8+ T cell populations within the colon, and FACS analysis of NK1.1 markers within the spleens of Bcl11bF/F/CD4-Cre mice, as well as weight experiments of Bcl11bF/F/CD4-Cre/b2−/− and b2−/− mice. Fig. S3 shows immunofluorescence microscopy of the large intestine in Bcl11bF/F/CD4-Cre mice. Fig. S4 shows FACS analysis of gut-homing markers in splenocytes and in vitro maintenance of WT and Bcl11bF/F/CD4-Cre Treg cells during the in vitro suppression assay showed in Fig. 4. Fig. S5 shows spleen gross anatomy and gut-homing markers of splenocytes in Bcl11bF/F/Foxp3-Cre mice, as well as removal of Bcl11b in splenocytes of Bcl11bF/F/Foxp3-Cre mice at 5 and 20 wk of age compared with Bcl11bF/F/CD4-Cre mice. Fig. S6 shows gene expression profiles of Treg cells for Bcl11bF/F/CD4-Cre and Bcl11bF/F/Foxp3-Cre mice and FACS analysis of IL-10 in Treg cells from Bcl11bF/F/Foxp3-Cre versus WT mice. Fig. S7 shows FACS analysis of CD3 within Treg cells from splenocytes in WT, Bcl11bF/F/CD4-Cre, and Bcl11bF/F/Foxp3-Cre mice, as well as the response of Bcl11b-deficient Treg cells from WT, Bcl11bF/F/CD4-Cre, and Bcl11bF/F/Foxp3-Cre mice to TCR stimulation. Fig. S8 shows FACS analysis of maturation and activation Treg cell markers and NK1.1 levels in splenocyte Treg cells of WT, Bcl11bF/F/CD4-Cre, and Bcl11bF/F/Foxp3-Cre mice. Table S1 shows colitis histological scores. The Supplemental materials and methods provide information in regard to antibodies used in the study, immunohistochemistry, in vitro stimulation of Treg cells, and luciferase reporter assays. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20102683/DC1.
Acknowledgments
We greatly acknowledge Dr. Paul Feustel (Albany Medical Center, Albany, NY) for assistance with the microarray analysis, Drs. Raj Verma and Amudhan Maniar (Albany Medical Center) for assistance with gut tissue isolation, and Dr. Hung Le for help with site-directed mutagenesis. We thank Dr. Alexander Rudensky for Foxp3-GFP mice, Dr. Ye Zheng and Dr. Masahide Tone for Foxp3 promoter constructs and for EL4 LAF cells, and Dr. Axel Kallies (Walter and Eliza Hall Institute for Medical Research, Parkville, Victoria, Australia) for information about IL-10 ChIP primers. We greatly acknowledge Ms. Debbie Moran for secretarial assistance and help with the figures, Adrian Avram for graphical presentation, Ms. Yili Lin and Ms. Bibiana Iglesias, and the Flow Cytometry Core for the help with sorting.
This work was supported by National Institutes of Health grant R01AI078273 to Dorina Avram.
The authors have no financial conflict of interests.
Footnotes
Abbreviations used:
- ChIP
- chromatin immunoprecipitation
- CM
- complete medium
- DP
- double positive
- IBD
- inflammatory bowel disease
- IEL
- intraepithelial lymphocyte
- iTreg cell
- induced Treg cell
- LP
- lamina propria
- mLN
- mesenteric LN
- mRNA
- messenger RNA
- TSS
- transcription start site
References
- Ahyi A.N., Chang H.C., Dent A.L., Nutt S.L., Kaplan M.H. 2009. IFN regulatory factor 4 regulates the expression of a subset of Th2 cytokines. J. Immunol. 183:1598–1606 10.4049/jimmunol.0803302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albu D.I., Feng D., Bhattacharya D., Jenkins N.A., Copeland N.G., Liu P., Avram D. 2007. BCL11B is required for positive selection and survival of double-positive thymocytes. J. Exp. Med. 204:3003–3015 10.1084/jem.20070863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram D., Fields A., Pretty On Top K., Nevrivy D.J., Ishmael J.E., Leid M. 2000. Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors. J. Biol. Chem. 275:10315–10322 10.1074/jbc.275.14.10315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram D., Fields A., Senawong T., Topark-Ngarm A., Leid M. 2002. COUP-TF (chicken ovalbumin upstream promoter transcription factor)-interacting protein 1 (CTIP1) is a sequence-specific DNA binding protein. Biochem. J. 368:555–563 10.1042/BJ20020496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C.L., Christie J., Ramsdell F., Brunkow M.E., Ferguson P.J., Whitesell L., Kelly T.E., Saulsbury F.T., Chance P.F., Ochs H.D. 2001. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 27:20–21 10.1038/83713 [DOI] [PubMed] [Google Scholar]
- Brunkow M.E., Jeffery E.W., Hjerrild K.A., Paeper B., Clark L.B., Yasayko S.A., Wilkinson J.E., Galas D., Ziegler S.F., Ramsdell F. 2001. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 27:68–73 10.1038/83784 [DOI] [PubMed] [Google Scholar]
- Chaudhry A., Rudra D., Treuting P., Samstein R.M., Liang Y., Kas A., Rudensky A.Y. 2009. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 326:986–991 10.1126/science.1172702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cismasiu V.B., Adamo K., Gecewicz J., Duque J., Lin Q., Avram D. 2005. BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene. 24:6753–6764 10.1038/sj.onc.1208904 [DOI] [PubMed] [Google Scholar]
- Cismasiu V.B., Ghanta S., Duque J., Albu D.I., Chen H.M., Kasturi R., Avram D. 2006. BCL11B participates in the activation of IL2 gene expression in CD4+ T lymphocytes. Blood. 108:2695–2702 10.1182/blood-2006-05-021790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cismasiu V.B., Paskaleva E., Suman Daya S., Canki M., Duus K., Avram D. 2008. BCL11B is a general transcriptional repressor of the HIV-1 long terminal repeat in T lymphocytes through recruitment of the NuRD complex. Virology. 380:173–181 10.1016/j.virol.2008.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cismasiu V.B., Duque J., Paskaleva E., Califano D., Ghanta S., Young H.A., Avram D. 2009. BCL11B enhances TCR/CD28-triggered NF-kappaB activation through up-regulation of Cot kinase gene expression in T-lymphocytes. Biochem. J. 417:457–466 10.1042/BJ20080925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das G., Augustine M.M., Das J., Bottomly K., Ray P., Ray A. 2003. An important regulatory role for CD4+CD8 alpha alpha T cells in the intestinal epithelial layer in the prevention of inflammatory bowel disease. Proc. Natl. Acad. Sci. USA. 100:5324–5329 10.1073/pnas.0831037100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot J.D., Gavin M.A., Rudensky A.Y. 2003. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4:330–336 10.1038/ni904 [DOI] [PubMed] [Google Scholar]
- Fontenot J.D., Rasmussen J.P., Williams L.M., Dooley J.L., Farr A.G., Rudensky A.Y. 2005. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 22:329–341 10.1016/j.immuni.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Hill J.A., Feuerer M., Tash K., Haxhinasto S., Perez J., Melamed R., Mathis D., Benoist C. 2007. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 27:786–800 10.1016/j.immuni.2007.09.010 [DOI] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Ikawa T., Hirose S., Masuda K., Kakugawa K., Satoh R., Shibano-Satoh A., Kominami R., Katsura Y., Kawamoto H. 2010. An essential developmental checkpoint for production of the T cell lineage. Science. 329:93–96 10.1126/science.1188995 [DOI] [PubMed] [Google Scholar]
- Ito R., Shin-Ya M., Kishida T., Urano A., Takada R., Sakagami J., Imanishi J., Kita M., Ueda Y., Iwakura Y., et al. 2006. Interferon-gamma is causatively involved in experimental inflammatory bowel disease in mice. Clin. Exp. Immunol. 146:330–338 10.1111/j.1365-2249.2006.03214.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izcue A., Coombes J.L., Powrie F. 2006. Regulatory T cells suppress systemic and mucosal immune activation to control intestinal inflammation. Immunol. Rev. 212:256–271 10.1111/j.0105-2896.2006.00423.x [DOI] [PubMed] [Google Scholar]
- Jones E.A., Flavell R.A. 2005. Distal enhancer elements transcribe intergenic RNA in the IL-10 family gene cluster. J. Immunol. 175:7437–7446 [DOI] [PubMed] [Google Scholar]
- Kim H.P., Leonard W.J. 2007. CREB/ATF-dependent T cell receptor-in 1543–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitoh A., Ono M., Naoe Y., Ohkura N., Yamaguchi T., Yaguchi H., Kitabayashi I., Tsukada T., Nomura T., Miyachi Y., et al. 2009. Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity. 31:609–620 10.1016/j.immuni.2009.09.003 [DOI] [PubMed] [Google Scholar]
- Koch M.A., Tucker-Heard G., Perdue N.R., Killebrew J.R., Urdahl K.B., Campbell D.J. 2009. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat. Immunol. 10:595–602 10.1038/ni.1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller B.H., Marrack P., Kappler J.W., Smithies O. 1990. Normal development of mice deficient in beta 2M, MHC class I proteins, and CD8+ T cells. Science. 248:1227–1230 10.1126/science.2112266 [DOI] [PubMed] [Google Scholar]
- Kühn R., Löhler J., Rennick D., Rajewsky K., Müller W. 1993. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 75:263–274 10.1016/0092-8674(93)80068-P [DOI] [PubMed] [Google Scholar]
- Lee C.G., Kang K.H., So J.S., Kwon H.K., Son J.S., Song M.K., Sahoo A., Yi H.J., Hwang K.C., Matsuyama T., et al. 2009. A distal cis-regulatory element, CNS-9, controls NFAT1 and IRF4-mediated IL-10 gene activation in T helper cells. Mol. Immunol. 46:613–621 10.1016/j.molimm.2008.07.037 [DOI] [PubMed] [Google Scholar]
- Li L., Leid M., Rothenberg E.V. 2010a. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 329:89–93 10.1126/science.1188989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Burke S., Wang J., Chen X., Ortiz M., Lee S.C., Lu D., Campos L., Goulding D., Ng B.L., et al. 2010b. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 329:85–89 10.1126/science.1188063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel P.Y., Ouaked N., Rückert B., Karagiannidis C., Welz R., Blaser K., Schmidt-Weber C.B. 2006. Molecular mechanisms underlying FOXP3 induction in human T cells. J. Immunol. 176:3593–3602 [DOI] [PubMed] [Google Scholar]
- Maruyama T., Li J., Vaque J.P., Konkel J.E., Wang W., Zhang B., Zhang P., Zamarron B.F., Yu D., Wu Y., et al. 2011. Control of the differentiation of regulatory T cells and T(H)17 cells by the DNA-binding inhibitor Id3. Nat. Immunol. 12:86–95 10.1038/ni.1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyara M., Sakaguchi S. 2007. Natural regulatory T cells: mechanisms of suppression. Trends Mol. Med. 13:108–116 10.1016/j.molmed.2007.01.003 [DOI] [PubMed] [Google Scholar]
- Mottet C., Uhlig H.H., Powrie F. 2003. Cutting edge: cure of colitis by CD4+CD25+ regulatory T cells. J. Immunol. 170:3939–3943 [DOI] [PubMed] [Google Scholar]
- Ouyang W., Beckett O., Ma Q., Paik J.H., DePinho R.A., Li M.O. 2010. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat. Immunol. 11:618–627 10.1038/ni.1884 [DOI] [PubMed] [Google Scholar]
- Pan F., Yu H., Dang E.V., Barbi J., Pan X., Grosso J.F., Jinasena D., Sharma S.M., McCadden E.M., Getnet D., et al. 2009. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. 325:1142–1146 10.1126/science.1176077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roers A., Siewe L., Strittmatter E., Deckert M., Schlüter D., Stenzel W., Gruber A.D., Krieg T., Rajewsky K., Müller W. 2004. T cell–specific inactivation of the interleukin 10 gene in mice results in enhanced T cell responses but normal innate responses to lipopolysaccharide or skin irritation. J. Exp. Med. 200:1289–1297 10.1084/jem.20041789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov Y.P., Rasmussen J.P., Chi E.Y., Fontenot J., Castelli L., Ye X., Treuting P., Siewe L., Roers A., Henderson W.R., Jr, et al. 2008. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 28:546–558 10.1016/j.immuni.2008.02.017 [DOI] [PubMed] [Google Scholar]
- Rudra D., Egawa T., Chong M.M., Treuting P., Littman D.R., Rudensky A.Y. 2009. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 10:1170–1177 10.1038/ni.1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tone Y., Furuuchi K., Kojima Y., Tykocinski M.L., Greene M.I., Tone M. 2008. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 9:194–202 10.1038/ni1549 [DOI] [PubMed] [Google Scholar]
- Vignali D.A., Collison L.W., Workman C.J. 2008. How regulatory T cells work. Nat. Rev. Immunol. 8:523–532 10.1038/nri2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi Y., Watanabe H., Inoue J., Takeda N., Sakata J., Mishima Y., Hitomi J., Yamamoto T., Utsuyama M., Niwa O., et al. 2003. Bcl11b is required for differentiation and survival of alphabeta T lymphocytes. Nat. Immunol. 4:533–539 10.1038/ni927 [DOI] [PubMed] [Google Scholar]
- Wan Y.Y., Flavell R.A. 2007. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 445:766–770 10.1038/nature05479 [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Sato H., Kusam S., Sehra S., Toney L.M., Dent A.L. 2005. Regulation of IL-10 gene expression in Th2 cells by Jun proteins. J. Immunol. 174:2098–2105 [DOI] [PubMed] [Google Scholar]
- Wing K., Sakaguchi S. 2010. Regulatory T cells exert checks and balances on self tolerance and autoimmunity. Nat. Immunol. 11:7–13 10.1038/ni.1818 [DOI] [PubMed] [Google Scholar]
- Wu Y., Borde M., Heissmeyer V., Feuerer M., Lapan A.D., Stroud J.C., Bates D.L., Guo L., Han A., Ziegler S.F., et al. 2006. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 126:375–387 10.1016/j.cell.2006.05.042 [DOI] [PubMed] [Google Scholar]
- Xu L., Kitani A., Fuss I., Strober W. 2007. Cutting edge: regulatory T cells induce CD4+CD25−Foxp3− T cells or are self-induced to become Th17 cells in the absence of exogenous TGF-beta. J. Immunol. 178:6725–6729 [DOI] [PubMed] [Google Scholar]
- Zenewicz L.A., Antov A., Flavell R.A. 2009. CD4 T-cell differentiation and inflammatory bowel disease. Trends Mol. Med. 15:199–207 10.1016/j.molmed.2009.03.002 [DOI] [PubMed] [Google Scholar]
- Zhang S., Rozell M., Verma R.K., Albu D.I., Califano D., VanValkenburgh J., Merchant A., Rangel-Moreno J., Randall T.D., Jenkins N.A., et al. 2010. Antigen-specific clonal expansion and cytolytic effector function of CD8+ T lymphocytes depend on the transcription factor Bcl11b. J. Exp. Med. 207:1687–1699 10.1084/jem.20092136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Chaudhry A., Kas A., deRoos P., Kim J.M., Chu T.T., Corcoran L., Treuting P., Klein U., Rudensky A.Y. 2009. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 458:351–356 10.1038/nature07674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Josefowicz S., Chaudhry A., Peng X.P., Forbush K., Rudensky A.Y. 2010. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 463:808–812 10.1038/nature08750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Chong M.M., Littman D.R. 2009. Plasticity of CD4+ T cell lineage differentiation. Immunity. 30:646–655 10.1016/j.immuni.2009.05.001 [DOI] [PubMed] [Google Scholar]
- Zorn E., Nelson E.A., Mohseni M., Porcheray F., Kim H., Litsa D., Bellucci R., Raderschall E., Canning C., Soiffer R.J., et al. 2006. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood. 108:1571–1579 10.1182/blood-2006-02-004747 [DOI] [PMC free article] [PubMed] [Google Scholar]