A common Shp2 mutation leads to myeloproliferative disease and malignant acute leukemia in stem cells and committed progenitors, associated with Shp2 maintaining chromosomal stability
Abstract
Activating mutations in protein tyrosine phosphatase 11 (Ptpn11) have been identified in childhood acute leukemias, in addition to juvenile myelomonocytic leukemia (JMML), which is a myeloproliferative disorder (MPD). It is not clear whether activating mutations of this phosphatase play a causal role in the pathogenesis of acute leukemias. If so, the cell origin of leukemia-initiating stem cells (LSCs) remains to be determined. Ptpn11E76K mutation is the most common and most active Ptpn11 mutation found in JMML and acute leukemias. However, the pathogenic effects of this mutation have not been well characterized. We have created Ptpn11E76K conditional knock-in mice. Global Ptpn11E76K/+ mutation results in early embryonic lethality. Induced knock-in of this mutation in pan hematopoietic cells leads to MPD as a result of aberrant activation of hematopoietic stem cells (HSCs) and myeloid progenitors. These animals subsequently progress to acute leukemias. Intriguingly, in addition to acute myeloid leukemia (AML), T cell acute lymphoblastic leukemia/lymphoma (T-ALL) and B-ALL are evolved. Moreover, tissue-specific knock-in of Ptpn11E76K/+ mutation in lineage-committed myeloid, T lymphoid, and B lymphoid progenitors also results in AML, T-ALL, and B-ALL, respectively. Further analyses have revealed that Shp2 (encoded by Ptpn11) is distributed to centrosomes and that Ptpn11E76K/+ mutation promotes LSC development, partly by causing centrosome amplification and genomic instability. Thus, Ptpn11E76K mutation has non–lineage-specific effects on malignant transformation of hematopoietic cells and initiates acute leukemias at various stages of hematopoiesis.
Shp2, a ubiquitously expressed protein tyrosine phosphatase (PTP), is implicated in multiple cell signaling processes, such as the RAS–MAP kinase, JAK–STAT, PI3K–AKT, NF-κB, and NFAT pathways (Neel et al., 2003; Tonks, 2006; Xu and Qu, 2008). It contains two tandem SH2 domains and a PTP domain. The SH2 domains, in particular, the N-terminal SH2 (N-SH2) domain, mediate the binding of Shp2 to other signaling proteins via phosphorylated tyrosine (pY) residues in a sequence-specific fashion (Zhao et al., 2003; Pawson, 2004; Songyang and Cantley, 2004; Waksman and Kuriyan, 2004). This directs Shp2 to the appropriate subcellular location and helps determine the specificity of substrate–enzyme interactions. Shp2 is normally self-inhibited by hydrogen bonding of the backside of the N-SH2 domain loop to the deep pocket of the PTP domain (Eck et al., 1996; Hof et al., 1998). The self-inhibition leads to occlusion of the phosphatase catalytic site and a distortion of the pY-binding site of N-SH2. Ligands with pY residues activate Shp-2 by binding the SH2 domains (primarily the N-SH2 domain), thereby disrupting the interaction between N-SH2 and PTP domains and exposing the phosphatase catalytic site (Eck et al., 1996; Barford and Neel, 1998; Hof et al., 1998). Intriguingly, despite its direct function in protein dephosphorylation, Shp2 plays an overall positive role in transducing signals initiated from receptor and cytosolic kinases (Neel et al., 2003; Tonks, 2006; Xu and Qu, 2008). This is particularly the case for the RAS–ERK pathway. The underlying mechanism, however, remains elusive. Shp2 interacts with several cell signaling intermediates. Of these partners, some are the targets of Shp2 enzymatic activity. However, none of the putative substrates identified to date can fully account for the overall positive signaling effects of Shp2 on the many biological processes with which it has been implicated. It appears that Shp2 functions in growth factor and cytokine signaling in both catalytically dependent and independent manners (Bennett et al., 1994; Li et al., 1994; Yu et al., 2003).
Shp2 plays a vital role in embryogenesis and hematopoietic cell development. A null mutation of Ptpn11 resulted in periimplantation lethality in mice (Yang et al., 2006). Shp2-deficient blastocysts exhibited inner cell mass cell death and no trophoblast stem cells were developed in these embryos (Yang et al., 2006). Deletion of the N-SH2 domain generated a loss-of-function mutation in Shp2, which led to embryonic lethality at mid-gestation, with defects in mesodermal patterning (Saxton et al., 1997). Chimeric mouse analyses demonstrated that this loss-of-function mutation caused multiple developmental defects characterized by aberrant skeletal structures and pathological changes in the epithelial system, which were clearly associated with diminished growth factor signaling (Qu et al., 1998, 1999). Shp2 plays a positive role in hematopoietic cell development. In vitro erythroid lineage differentiation of embryonic stem (ES) cells with the N-SH2 deletion mutation of Shp2 was severely suppressed, and myeloid lineage differentiation was totally blocked (Qu et al., 1997). Moreover, the contribution from these mutant ES cells to erythroid, myeloid, or lymphoid cells in the chimeric mice was undetectable (Qu et al., 1998, 2001). Most recent studies (Chan et al., 2011; Zhu et al., 2011) have confirmed that Shp2 is critical for the survival and maintenance of hematopoietic stem cells (HSCs) and immature progenitors. Depletion of Shp2 from adult mice resulted in rapid loss of HSCs and immature progenitors of all hematopoietic lineages.
Notably, germline and somatic mutations (heterozygous) in Ptpn11 (encoding Shp2) have been identified in 50% of the children with the developmental disorder Noonan syndrome (Tartaglia et al., 2001) and in 35% of the patients with juvenile myelomonocytic leukemia (JMML; Tartaglia et al., 2003; Loh et al., 2004b), a childhood myeloproliferative disorder (MPD), both of which are associated with hyperactivation of the RAS–ERK pathway. Ptpn11 mutations found in Noonan syndrome are clustered in the PTP domain, whereas mutations seen in JMML are mainly localized in the N-SH2 domain. These mutations result in amino acid changes at the interphase formed between N-SH2 and PTP domains, disrupting the inhibitory intramolecular interaction, leading to hyperactivation of Shp2 catalytic activity (Tartaglia et al., 2003; Keilhack et al., 2005). In addition, Ptpn11 disease mutations, especially leukemia mutations, enhance the binding of mutant Shp2 to signaling partners (Araki et al., 2004; Fragale et al., 2004; Kontaridis et al., 2006; Yu et al., 2006). Nevertheless, as the biochemical basis for the positive role that Shp2 phosphatase plays in cell signaling and other cellular processes is unknown, the cellular and molecular mechanisms by which Ptpn11 gain-of-function (GOF) mutations induce JMML are not well understood. Furthermore, Ptpn11 GOF mutations are also found in myelodysplastic syndromes (also known as pre–acute myeloid leukemia (AML); 10%), B cell acute lymphoblastic leukemia/lymphoma (B-ALL; 7%), AML (4%; Loh et al., 2004a; Tartaglia et al., 2004), and sporadic solid tumors (Bentires-Alj et al., 2004). Although previous studies have shown that Ptpn11 GOF mutations induce cytokine hypersensitivity in myeloid progenitors (Chan et al., 2005; Schubbert et al., 2005; Yu et al., 2006) and MPD in mice (Araki et al., 2004; Mohi et al., 2005; Chan et al., 2009; Xu et al., 2010), it is unclear whether Ptpn11 GOF mutations also play a causal role in acute leukemias. If so, the underlying mechanisms remain to be determined.
To address these important questions and to further dissect the role of Shp2 in health and disease, we created a line of conditional knock-in mice with the most common and most active Ptpn11 mutation (E76K) found in JMML and acute leukemias (Tartaglia et al., 2003, 2006; Kratz et al., 2005; Aoki et al., 2008). Global Ptpn11E76K/+ mutation results in early embryonic lethality. Induced knock-in of this mutation in pan hematopoietic cells leads to MPD followed by malignant evolution into various acute leukemias. Furthermore, we have discovered that Ptpn11E76K/+ mutation induces leukemia-initiating stem cell (LSC) development not only in stem cells but also in lineage-committed progenitors. This non–lineage/stage-specific effect of Ptpn11E76K/+ mutation on hematopoietic cell transformation appears to be partially associated with the disturbance of a previously unrecognized function of Shp2 in centrosomes and maintenance of chromosomal stability.
RESULTS
Global Ptpn11E76K/+ mutation results in embryonic lethality
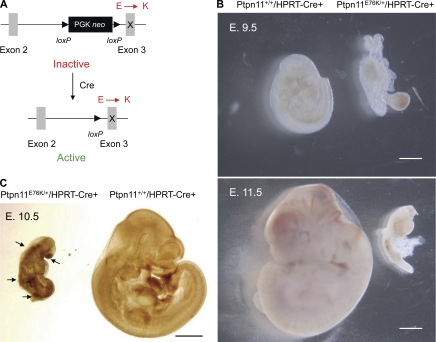
The pathogenic effects of the most common and most active Ptpn11 mutation (Ptpn11E76K/+) identified in JMML and acute leukemias (Tartaglia et al., 2003, 2006; Kratz et al., 2005; Aoki et al., 2008) have not been well characterized. To this end, we generated Ptpn11E76K knock-in mice (Fig. S1). In our gene-targeting strategy, a loxP-flanked neo cassette was inserted in intron 2 as the selective marker, which was followed by mutated exon 3 with E76K mutation. Intriguingly, we unexpectedly discovered that the inserted neo cassette with a stop codon prevented expression of the targeted allele (Ptpn11E76K neo; Supplemental text and Fig. S2, C and D) and Ptpn11E76K neo/+ mice appeared healthy without gross abnormalities (Fig. S2 B). Upon deletion of neo by Cre DNA recombinase, the mutant allele (Ptpn11E76K) was reactivated, producing Shp2 E76K at the physiological level (Fig. S2 D and Fig. 1 A). The mechanism for the inactivation of the targeted allele is unclear. Importantly, this unexpected conditional allele allows expression of mutant Shp2 under the endogenous Ptpn11 promoter in an inducible manner, preserving temporal and spatial transcriptional regulation. By crossing Ptpn11E76K neo/+ mice with global Cre transgenic (hypoxanthine-guanine phosphoribosyltransferase [HPRT]-Cre+) mice we found that induction of Shp2 E76K during early embryogenesis resulted in embryonic lethality at around embryonic day 11.5 (E. 11.5; Fig. 1 B, Supplemental text, and Fig. S2 A) that appeared to be associated with aberrantly enhanced ERK activation (Fig. 1 C and Supplemental text).
Figure 1.
Global Ptpn11E76K/+ mutation results in embryonic lethality. (A) Schematic diagram of the Ptpn11E76K neo conditional allele. The insertion of the neo cassette with a stop codon in intron 2 disrupts the targeted allele. This allele is reactivated and expresses Shp2 E76K upon deletion of the neo cassette by Cre DNA recombinase. (B) Representative E. 9.5 and E. 11.5 embryos of Ptpn11+/+/HPRT-Cre+ and Ptpn11E76K/+/HPRT-Cre+ genotypes produced from intercrosses between Ptpn11E76K neo/+ and HPRT-Cre+ mice. Bars, 1 mm. (C) Whole mounts of E. 10.5 embryos produced from intercrosses between Ptpn11E76K neo/+ and HPRT-Cre+ mice were immunostained with anti–phospho-ERK antibody following standard procedures. Front nasal processes, limb buds, and somites of Ptpn11E76K/+ embryos are indicated by arrows. Bar, 1 mm.
Induced Ptpn11E76K/+ mutation in hematopoietic cells quickly leads to MPD with full penetrance
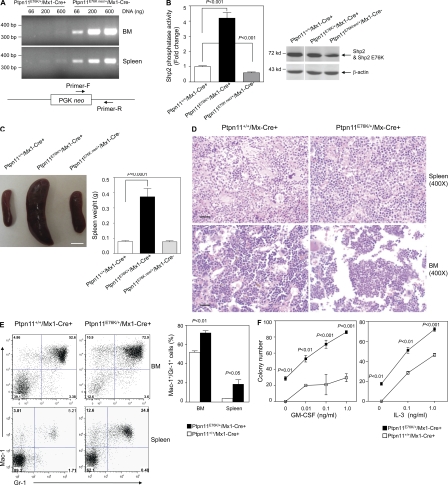
To determine the pathogenic effects of somatic Ptpn11E76K/+ mutation on hematopoietic cell development, we generated Ptpn11E76K neo/+/Mx1-Cre+ mice by crossing Ptpn11E76K neo/+ mice with Mx1-Cre transgenic mice that express Cre in pan-hematopoietic cells in response to polyinosinic-polycytidylic acid (pI-pC) treatment (Kühn et al., 1995). 4-wk-old Ptpn11E76K neo/+/Mx1-Cre+ mice were treated with pI-pC to induce Cre expression. Neo deletion efficiency in the BM cells and splenocytes of these mice was nearly complete (Fig. 2 A), and mutant Shp2 was expressed at a level comparable to that of WT Shp2 in Ptpn11+/+/Mx1-Cre+ mice (Fig. 2 B). Shp2 phosphatase activity in the spleen lysates of pI-pC-treated knock-in (referred as to Ptpn11E76K/+/Mx1-Cre+) mice was indeed substantially increased (Fig. 2 B), in agreement with previous observations that E76K mutation hyper-activates Shp2 (Tartaglia et al., 2003; Keilhack et al., 2005). Consequently, Ptpn11E76K/+/Mx1-Cre+ mice uniformly developed MPD. These mice showed high white blood cell (WBC; mainly neutrophils) counts (Table S2). Splenomegaly (Fig. 2 C) and hepatomegaly (unpublished data) were also prominent. Histopathological examination revealed hyperproliferation of myeloid cells in both the spleen and BM (Fig. 2 D). FACS analyses confirmed excess myeloid expansion in the BM and spleen. Mac-1+/Gr-1+ mature myeloid cells were markedly increased in Ptpn11E76K/+/Mx1-Cre+ mice (Fig. 2 E). Consistent with cell-autonomous signaling abnormalities in hematopoietic cells, myeloid progenitors from Ptpn11E76K/+/Mx1-Cre+ mice showed greatly increased responses to GM-CSF/IL-3 (Fig. 2 F). Furthermore, these mutant progenitors yielded growth factor–independent colony formation in cytokine-free medium (Fig. 2 F).
Figure 2.
Ptpn11E76K/+ mutation in hematopoietic cells initially induces MPD with full penetrance. (A) 4-wk-old Ptpn11E76K neo/+/Mx1-Cre+ and Ptpn11E76K neo/+/Mx1-Cre− mice were treated by i.p. injection of a total of 5 doses of pI-pC acid (350 µg/mouse) administered every other day over 10 d. 4 wk after pI-pC treatment, genomic DNA was extracted from the BM and spleen. PCR detection of the neo segment was performed using the primers shown in the diagram (bottom). (B) Spleen lysates prepared from Ptpn11+/+/Mx1-Cre+, Ptpn11E76K/+/Mx1-Cre+, and Ptpn11E76K neo/+/Mx1-Cre− mice 6 wk after pI-pC treatment were immunoprecipitated with anti-Shp2 antibody. Immune complexes were subjected to the phosphatase assay using 4-nitrophenyl phosphate disodium salt hexahydrate as the substrate following standard procedures. Shp2 levels in the tissue lysates were examined by anti-Shp2 immunoblotting. Blots were stripped and reprobed with β-actin antibody to check protein loading. (C) Mouse spleen weight was quantified 6–8 wk after pI-pC treatment (n = 8 per group). Bar, 5 mm. (D) Spleens and femurs isolated from Ptpn11E76K/+/Mx1-Cre+ and Ptpn11+/+/Mx1-Cre+ mice 8 wk after pI-pC injection were processed for histopathological examination (Hematoxylin and eosin staining). Bars, 50 µm. (E) BM cells and splenocytes isolated from Ptpn11E76K/+/Mx1-Cre+ and Ptpn11+/+/Mx1-Cre+ littermates (n = 5 per group) were assayed for Mac-1+/Gr-1+ cells by FACS analyses 8 wk after pI-pC injection. (F) BM cells (2 × 104/ml) harvested from Ptpn11E76K/+/Mx1-Cre+ and Ptpn11+/+/Mx1-Cre+ littermates (n = 3 per group) were assessed by myeloid colony assays 6 wk after pI-pC injection.
HSCs are hyperactivated by Ptpn11E76K/+ mutation
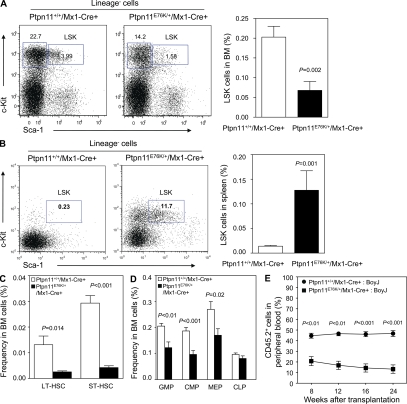
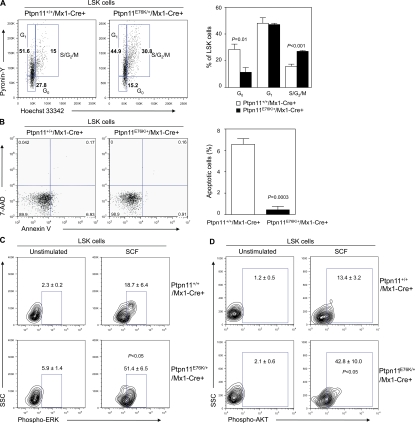
To further understand the mechanism by which Ptpn11E76K/+ mutation induces MPD, a clonal stem cell disorder, we assessed the effects of Ptpn11E76K/+ mutation on HSCs. We found that Lineage−Sca-1+c-Kit+ (LSK) cells that are enriched for HSCs were decreased by threefold in the BM of Ptpn11E76K/+/Mx1-Cre+ mice (Fig. 3 A and Table S3). LSK cells in the spleen, however, were greatly increased (Fig. 3 B), similar to those seen in PTEN-deficient mice (Yilmaz et al., 2006; Zhang et al., 2006), reflecting extramedullary hematopoiesis in the mutant mice and potential microenvironmental impact on mutant stem cell homeostasis. Additional multiparameter FACS analyses (Tothova et al., 2007; Fleming et al., 2008) showed that both long-term HSCs (LT-HSCs) and short-term HSCs (ST-HSCs; Fig. 3 C and Table S3) were significantly decreased in the BM of Ptpn11E76K/+ knock-in mice. Later stage progenitor populations, such as common myeloid progenitors (CMPs), granulocyte macrophage progenitors (GMPs), and megakaryocyte erythroid progenitors (MEPs), were also decreased in mutant mice (Fig. 3 D and Table S3). Common lymphoid progenitors (CLPs), however, did not show significant changes (Fig. 3 D and Table S3). The results of phenotypic HSC analyses were verified by the greatly decreased repopulating capabilities of mutant BM cells in competitive repopulation assays (Fig. 3 E). Because peripheral WBCs and BM Mac-1+/Gr-1+ mature myeloid cells were increased in mutant mice, this rightward shift in the BM hematopoietic compartment suggests an accelerated activation and differentiation of mutant stem/progenitor cells. In support of this notion, quiescent HSCs at the G0 phase in mutant mice were decreased by twofold, whereas HSCs at the S and G2/M phases were doubled (Fig. 4 A). The decrease in the stem cell pool in the BM of mutant mice is not caused by any defects in cell survival. In fact, programmed cell death in Ptpn11E76K/+ mutant LSK cells was decreased (Fig. 4 B). More likely, aberrant activation of mutant stem cells eventually led to the depletion of the stem cell population caused by exhaustion of the reserve capacity. HSC activities, such as cycling and survival, are tightly controlled by cytokines and growth factors. Assessment of cell signaling activities confirmed that in response to stem cell factor (SCF) stimulation, activation of ERK (Fig. 4 C) and AKT (Fig. 4 D) kinases in mutant LSK cells was greatly enhanced compared with that in control cells.
Figure 3.
Accelerated hematopoietic cell differentiation in Ptpn11E76K/+ knock-in mice during the MPD phase. BM cells freshly harvested from Ptpn11E76K/+/Mx1-Cre+ and Ptpn11+/+/Mx1-Cre+ littermates 8 wk after pI-pC treatment were assayed by multiparameter FACS analyses to determine frequencies of hematopoietic cell populations of various stages and lineages. (A) Frequency of HSC-enriched LSK cells in the BM (n = 8 per group). (B) Frequency of LSK cells in the spleen (n = 6 per group). (C) Frequencies of LT-HSCs and ST-HSCs in the BM (n = 4 per group). (D) Frequencies of CMP, GMP, and MEP populations in the BM (n = 7 per group). (E) BM cells (test cells) harvested from pI-pC–treated Ptpn11E76K/+/Mx1-Cre+ and Ptpn11+/+/Mx1-Cre+ littermates (CD45.2+) were transplanted with the same number of BoyJ (CD45.1+) BM cells (competitor cells) into lethally irradiated BoyJ (CD45.1+) recipients (n = 7 per group). Test cell reconstitution was determined 8, 12, 16, and 24 wk after transplantation by FACS analyses of peripheral blood leukocytes.
Figure 4.
HSCs are hyperactivated by Ptpn11E76K/+ mutation. (A) BM cells freshly harvested from Ptpn11E76K/+/Mx1-Cre+ and Ptpn11+/+/Mx1-Cre+ mice (n = 5 per group) were assayed by multiparameter FACS analyses to determine cell cycle status of HSC-enriched LSK cells. Percentages of LSK cells at G0, G1, and S/G2/M phases identified based on Pyronin Y and Hoechst staining profiles were quantified. (B) BM cells from Ptpn11E76K/+/Mx1-Cre+ and Ptpn11+/+/Mx1-Cre+ mice (n = 4 per group) were analyzed by multiparameter FACS analyses to determine apoptotic cells (Annexin V+ cells) in the LSK population. (C and D) Lineage− BM cells from Ptpn11E76K/+/Mx1-Cre+ and Ptpn11+/+/Mx1-Cre+ mice (n = 3 per group) were purified and starved for 1 h in IMDM medium. Cells were then stimulated with SCF (50 ng/ml) for 5 min, fixed, permeabilized, and stained with antibodies against Sca-1, c-Kit, and phospho-ERK or phospho-AKT. Percentages of the cells stained positive for phospho-ERK (C) or phospho-AKT (D) in the gated LSK population were measured by multiparameter FACS analyses.
Various types of leukemias are evolved in Ptpn11E76K/+ mice
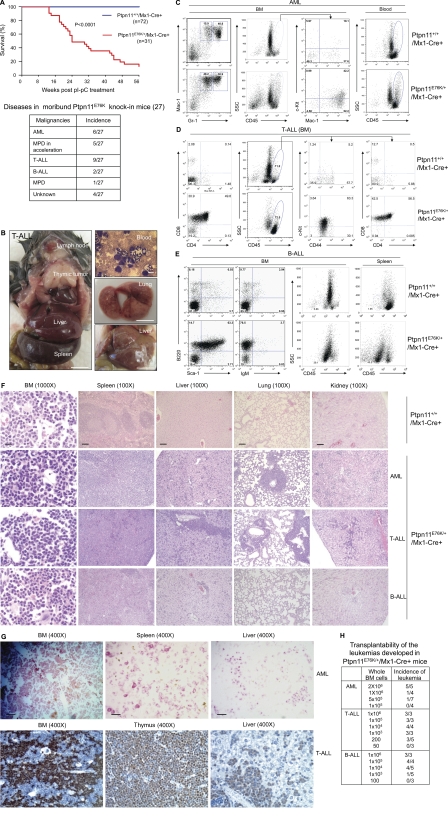
Strikingly, after 12–32 wk of chronic MPD where differentiation/maturation of myeloid cells was essentially normal, Ptpn11E76K/+ knock-in mice progressed to the accelerated phase, resembling acute leukemias where there was profound impairment of hematopoietic differentiation. These mice presented with extremely high WBC counts, progressive anemia, and leukemia infiltration in nonhematopoietic organs, and then became moribund (Fig. 5, A and B). Intriguingly, various types of leukemias were evolved, although the mutant mice initially manifested MPD (Fig. 5, A and B; Table S1 ; and Fig. S3 A). In the mice that progressed to AML, Mac-1+/Gr-1low, or Mac-1+/Gr-1−, poorly differentiated cells (Yilmaz et al., 2006; Guo et al., 2008) were substantially increased (Fig. 5 C). Mac-1+/c-Kit+ myeloblasts (Cozzio et al., 2003; So et al., 2003; Rosenbauer et al., 2004; Kirstetter et al., 2008) were >20% in the BM (Fig. S3 A). In the mice with T cell acute lymphoblastic leukemia/lymphoma (T-ALL) (Fig. 5 D), 50–70% of BM cells were CD4+/CD8+ and CD44+ cells, indicative of a T cell differentiation block near the pro–T stage. A substantial portion of these cells were also positive for c-Kit (Fig. 5 D), an early stage stem/progenitor cell marker. T lymphoma cells also expressed the myeloid marker Mac-1 (Fig. S3 B), which is consistent with lineage infidelity/promiscuity of leukemia cells (Smith et al., 1983). In addition, CD3+/c-Kit+ cells, defined as T-ALL LSCs in PTEN knockout mice (Yilmaz et al., 2006; Guo et al., 2008), were detected within the blast compartment (Fig. S3 A). In the mice that progressed to B-ALL, 60–70% of BM cells were B220+ (Fig. 5 E). Of these B220+ cells, 70–80% cells were also positive for an early B lineage marker CD43 and the stem cell marker Sca-1, but negative for the mature B cell marker IgM, indicating a B lineage maturation arrest at around the pro–B stage. Pathological examination revealed leukemic cell invasion into hematopoietic and nonhematopoietic organs (Fig. 5 F). This was further characterized by a large number of chloroacetate-esterase-positive myeloid leukemic cells in the BM and other organs in AML and large numbers of terminal deoxynucleotidyl transferase (TdT)-positive lymphoid blasts throughout the BM and other organs in T-ALL (Fig. 5 G). Transplantation of BM cells from terminally ill mice reproduced leukemias of the same type in recipient mice (Fig. 5 H). In addition, limiting dilution transplantation experiments revealed that the oncogenic Ptpn11-induced hematologic malignancies were prorogated by LSCs and that the frequency of LSCs in AML was low, whereas the frequencies of LSCs in T-ALL and B-ALL were very high (Fig. 5 H). This suggests that concentrations of LSCs vary depending on leukemia types, although they were initiated by the same oncogene Ptpn11E76K.
Figure 5.
Various leukemias with differentiation block are subsequently evolved in Ptpn11E76K/+ mice. (A) Kaplan–Meier survival curve in a cohort of Ptpn11E76K/+/Mx1-Cre+ mice (n = 31) and a cohort of Ptpn11+/+/Mx1-Cre+ (n = 72) mice after pI-pC treatment. Disease distribution in the 27 moribund/dead Ptpn11E76K/+/Mx1-Cre+ mice is shown on the bottom. (B) A representative Ptpn11E76K/+ knock-in mouse with thymic lymphoma is shown on the left. Peripheral blood smear, lung, and liver from another leukemic Ptpn11E76K/+ knock-in mouse are shown on the right. Bars on blood smear and organ pictures represent 10 µm and 10 mm, respectively. (C) BM and peripheral blood cells harvested from terminally ill Ptpn11E76K/+/Mx1-Cre+ mice and Ptpn11+/+/Mx1-Cre+ control mice were immunostained with antibodies against CD45, Mac-1, Gr-1, and c-Kit. The SSCmedCD45high population was subfractioned according to c-Kit and Mac-1 expressions to determine Mac-1+/c-Kit+ blasts. (D) BM cells collected from moribund Ptpn11E76K/+/Mx1-Cre+ mice and Ptpn11+/+/Mx1-Cre+ control mice were immunostained with antibodies against CD45, CD4, CD8, CD44, and c-Kit. The side scatter (SSC)lowCD45high/med blast population was subfractioned according to CD44 and c-Kit or CD4 and CD8 expressions to determine CD4+/CD8+ cells and CD44+/c-Kit+ blasts. (E) BM cells and splenocytes collected from moribund Ptpn11E76K/+/Mx1-Cre+ mice and Ptpn11+/+/Mx1-Cre+ control mice were immunostained with antibodies against CD45, B220, IgM, and Sca-1. B220+/Sca-1+ cells, B220+/IgM+ cells, and the blast (SSClowCD45low) population were shown. (F) Tissues harvested from terminally diseased Ptpn11E76K/+/Mx1-Cre+ mice and Ptpn11+/+/Mx1-Cre+ controls were processed for histopathological examination (hematoxylin and eosin staining). Bars on 1000× and 100× pictures represent 20 and 200 µm, respectively. (G) BM, spleen, and liver harvested from a Ptpn11E76K/+/Mx1-Cre+ mouse that had progressed to AML were processed for chloroacetate esterase staining. BM, thymus, and liver harvested from a Ptpn11E76K/+/Mx1-Cre+ mouse in which T-ALL was evolved were processed for terminal deoxynucleotidyl transferase (TdT) staining. Bars, 50 µm. (H) BM cells collected from Ptpn11E76K/+/Mx1-Cre+ mice with AML, T-ALL, or B-ALL were transplanted into sublethally irradiated BoyJ (600 rads) or NOD-SCID (200 rads) mice at the indicated cell doses. Recipient mice were monitored on a daily basis for ∼100 d. Moribund mice were sacrificed. Diagnosis for each mouse was made by multiparameter FACS analyses, blood smear, and tissue histopathological examinations as described.
Non–lineage-/stage-specific effects of Ptpn11E76K/+ mutation on LSC development
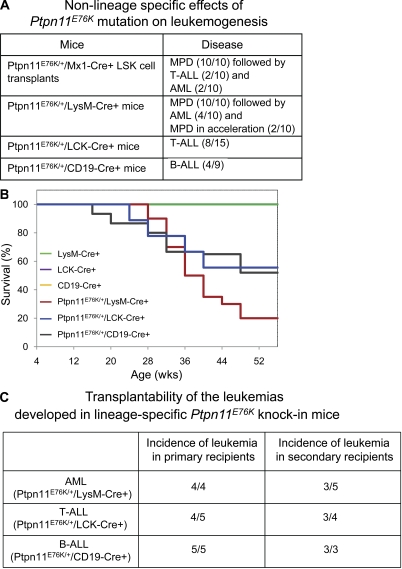
To determine at what stage in hematopoiesis initial LSCs were derived in Ptpn11E76K/+ knock-in mice during the progression from MPD to full blown leukemias, we purified HSC-enriched LSK cells from Ptpn11E76K/+/Mx1-Cre+ mice in the MPD stage (8–10 wk after pI-pC treatment), mixed these cells with 105 lineage+ cells (to radioprotect recipients), and transplanted them into BoyJ mice. As shown in Fig. 6 A, all of Ptpn11E76K/+ mutant LSK cell transplants developed MPD; 40% of them subsequently transformed into acute leukemias. These data suggest that the leukemogenic effects of Ptpn11E76K/+ mutation are cell intrinsic and that LSCs can be developed from early stage stem cells. To further test whether Ptpn11E76K/+ mutation can also cause leukemias in lineage-committed progenitors, we generated lineage-specific knock-in mice, i.e., Ptpn11E76K/+/LysM-Cre+ (in GMPs), Ptpn11E76K/+/LCK-Cre+ (in CD4−/CD8− stage T cells), and Ptpn11E76K/+/CD19-Cre+ (in pro–/pre–B stage B cells) mice using LysM-Cre, LCK-Cre, and CD19-Cre transgenic mice (Jude et al., 2007), respectively. Ptpn11E76K/+/LysM-Cre+ mice invariably manifested MPD as early as at the time when the mice were weaned because of apparently enhanced myeloid cell proliferation and differentiation (Fig. 6 A). In contrast, no T or B cell developmental abnormalities were detected in Ptpn11E76K/+/LCK-Cre+ and Ptpn11E76K/+/CD19-Cre+ mice during the first 4 mo of life (Fig. S4). Surprisingly, 40% of Ptpn11E76K/+/LysM-Cre+, 53% of Ptpn11E76K/+/LCK-Cre+, and 44% of Ptpn11E76K/+/CD19-Cre+ mice subsequently developed AML, T-ALL, and B-ALL, respectively (Fig. 6, A and B). Similar to the acute leukemias in Ptpn11E76K/+/Mx1-Cre+ mice, AML, T-ALL, and B-ALL developed in lineage-specific knock-in mice were also transplantable in primary and secondary recipient mice (Fig. 6 C). The fact that Ptpn11E76K/+ mutation causes leukemias in lineage progenitors strongly suggests that preexisting self-renewal program in target cells is not required for Ptpn11E76K/+ mutation to induce LSC development.
Figure 6.
Non–lineage/stage-specific effects of Ptpn11E76K/+ mutation on malignant transformation of hematopoietic cells. (A) LSK cells were purified from Ptpn11E76K/+/Mx1-Cre+ mice at the MPD stage (8 wk after pI-pC treatment). These cells (2.5 × 103/per mouse) were mixed with 105 lineage+ cells and then transplanted into lethally irradiated (1,100 rads) BoyJ mice. Recipient mice were monitored on a daily basis for 12 mo. Leukemias developed in Ptpn11E76K/+/LysM-Cre+, Ptpn11E76K/+/LCK-Cre+, and Ptpn11E76K/+/CD19-Cre+ mice were summarized. (B) Kaplan–Meier survival curves of lineage-specific Ptpn11E76K/+ knock-in mice. Ptpn11E76K neo/+ mice were used to cross LysM-Cre, LCK-Cre, or CD19-Cre transgenic mice of C57BL6 genetic background (The Jackson Laboratory) to generate Ptpn11E76K/+/LysM-Cre+ (n = 10), Ptpn11E76K/+/LCK-Cre+ (n = 15), and Ptpn11E76K/+/CD19-Cre+ (n = 9) mice, respectively. These mice and three control lines of mice (n = 15 each per group) were monitored for 12 mo. Moribund mice were euthanized and diagnosed as described. (C) BM cells collected from moribund Ptpn11E76K neo/+/LysM-Cre+, Ptpn11E76K/+/LCK-Cre+, and Ptpn11E76K/+/CD19-Cre+ mice with AML (2 × 106 cells), T-ALL (103 cells), or B-ALL (104 cells) were transplanted into sublethally irradiated BoyJ (600 rads) mice. BM cells harvested from moribund recipient mice with leukemias were transplanted into sublethally irradiated secondary recipients at the same doses as described.
Ptpn11E76K/+ mutation also induces centrosome amplification and chromosomal instability
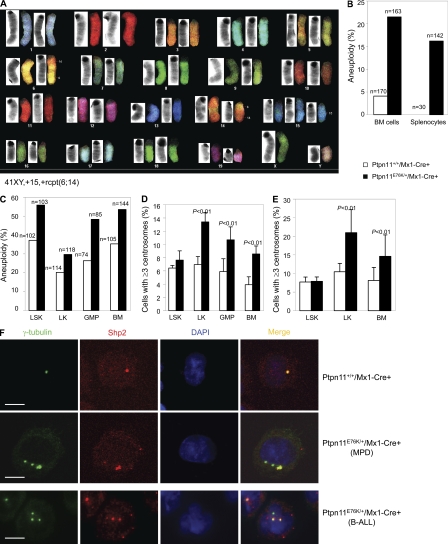
During the MPD-to-leukemia progression in Ptpn11E76K/+/Mx1-Cre+ mice, additional genetic alterations necessary for malignant transformation might have acquired. Indeed, spectral karyotype analyses for T-ALL cells showed gain or loss of whole chromosomes, or chromosomal structural (translocations and deletion) changes (Fig. 7 A). Common abnormalities across multiple karyotypes from each mouse additionally suggest that the malignancies that evolved were clonal or oligoclonal. Genomic abnormalities are thought to arise from enhanced cell proliferation and differentiation. However, T and B cell development was normal in Ptpn11E76K/+/LCK-Cre+ and Ptpn11E76K/+/CD19-Cre+ mice before the onset of T-ALL and B-ALL (Fig. S4). The non–lineage-specific effects of Ptpn11E76K/+ mutation on hematopoietic cell transformation prompted us to test for a potential general impact of this mutation on genomic stability independent of cell growth rate. We examined karyotypes of mutant hematopoietic cells before acute leukemia transformation. BM cells (nonmegakaryocytes) and splenocytes from Ptpn11E76K/+/Mx1-Cre+ mice during the MPD phase readily displayed aneuploidy, a state of having abnormal numbers of chromosomes (Fig. 7 B). Moreover, this genomic instability in purified mutant LSK, LK (Lineage−c-Kit+, early myeloid progenitors), GMP, and whole BM cells was accelerated by in vitro culture stress (Fig. 7 C). Genome integrity is maintained by various cellular surveillance mechanisms (Kastan and Bartek, 2004). Because Ptpn11E76K/+ mutant cells overall displayed increased aneuploidy that is known to be associated with defects in mitosis, we surveyed centrosomes in hematopoietic cells and found marked centrosome amplification in preleukemic Ptpn11E76K/+/Mx1-Cre+ mice. Percentages of the cells with ≥3 centrosomes in fresh (Fig. 7 D and Fig. S5 A) or cultured (Fig. 7 E) mutant LK, GMP, and unsorted BM cells (nonmegakaryocytes) were significantly increased, although centrosome amplification in mutant LSK cells was not significant. More interestingly, during the course of these experiments, we surprisingly found that Shp2 (encoded by Ptpn11) was distributed to centrosomes. Shp2 co-localized with γ-tubulin, a centrosome-specific structural protein (Fig. 7 F). Intriguingly, in Ptpn11E76K/+ hematopoietic cells (at both MPD and acute leukemia phases) with multiple centrosomes, Shp2 was only localized to part, but not all amplified centrosomes (Fig. 7 F and Fig. S5 B), indicating that centrosome amplification and genomic instability in the mutant cells are associated at least in part with Shp2 E76K mutation.
Figure 7.
Centrosome amplification and chromosomal instability in Ptpn11E76K/+ hematopoietic cells. (A) T-ALL tumor cells from pI-pC–treated Ptpn11E76K/+/Mx1-Cre+ mice were examined by spectral karyotype analyses. 12 metaphase spreads were examined for each sample. A representative result is shown. (B) BM cells and splenocytes freshly isolated from Ptpn11E76K/+/Mx1-Cre+ mice at the MPD stage and Ptpn11+/+/Mx1-Cre+ control mice (6–8 wk after pI-pC treatment) were assayed by karyotyping analyses. Cells (nonmegakaryocytes) with more or <40 chromosomes were counted as aneuploid cells. Results shown are a summary of the data obtained from five mice in each group. (C) LSK cells, LK cells, and GMPs were sorted from the BM of Ptpn11E76K/+/Mx1-Cre+ mice at the MPD stage and Ptpn11+/+/Mx1-Cre+ control mice. Purified cells, along with whole BM cells, were cultured in IMDM medium containing Tpo (20 ng/ml), FIt3 ligand (50 ng/ml), SCF (50 ng/ml), IL-3 (20 ng/ml), IL-6 (20 ng/ml), and 10% FBS (for LSK cells) or SCF (50 ng/ml), IL-3 (20 ng/ml), IL-6 (20 ng/ml), and 10% FBS (for other cell types) for 60 h (LSK and LK cells), 48 h (BM cells), and 16 h (GMPs) hours. Cells (nonmegakaryocytes) were then examined by karyotyping analyses, as described. (D) LSK cells, LK cells, and GMPs were sorted from the BM of Ptpn11E76K/+/Mx1-Cre+ mice at the MPD stage and Ptpn11+/+/Mx1-Cre+ control mice. Purified cells along with whole BM cells were immunostained with anti–γ-tubulin antibody. Centrosomes were then examined under a fluorescence microscope. For LSK cells, at least 100 cells were surveyed in each experiment. For the other cell types, at least 300 cells were examined in each experiment. Results shown are mean ± SEM of three independent experiments. (E) Purified LSK cells, LK cells, and unsorted BM cells were cultured in the medium (as described) for 48 h. Cells were then immunostained with anti–γ-tubulin antibody. Centrosomes were examined under a fluorescence microscope. Data shown are mean ± SEM of three independent experiments. (F) LK cells purified from Ptpn11E76K/+/Mx1-Cre+ mice at the MPD stage and Ptpn11+/+/Mx1-Cre+ control mice (top), and BM cells from Ptpn11E76K/+/Mx1-Cre+ mice with B-ALL (bottom) were immunostained with anti–γ-tubulin and Shp2 antibodies. Nuclei were counterstained with DAPI. The images were captured and analyzed using a laser-scanning confocal microscope. Bars, 5 µm.
DISCUSSION
Hematopoietic cell development and function are tightly controlled by environmental cues, such as cytokines and growth factors. Dysregulation of cytokine/growth factor signaling may result in hematologic disorders. Shp2 phosphatase plays a positive role in the signal transduction of multiple cytokines and growth factors, although the underlying mechanisms are still unclear (Neel et al., 2003; Tonks, 2006; Xu and Qu, 2008). Shp2 functions in cytokine (IL-3) signaling in both catalytically dependent and independent manners (Yu et al., 2003). The E76K mutation in the N-SH2 domain of Shp2 enhances IL-3 signaling through both elevated catalytic activity and increased binding to signaling partners (Yu et al., 2006). Myeloid progenitors in Ptpn11E76K/+ knock-in mice are hypersensitive to GM-CSF and IL-3 (Fig. 2 F), which is reminiscent of JMML (Birnbaum et al., 2000; Zhang et al., 1998). This is obviously caused by the enhancing effect of Shp2 E76K mutant on cytokine signaling. Notably, stem cells are also aberrantly activated in Ptpn11E76K/+ knock-in mice. The percentage of stem cells (LSK cells) in G1 or S/G2/M phases is increased in these animals, whereas quiescent stem cells at the G0 phase are decreased (Fig. 4 A), and growth factor (SCF)–induced signaling activities are much stronger in Ptpn11E76K/+ LSK cells (Fig. 4, C and D). Thus, the MPD phenotypes caused by Ptpn11E76K/+ mutation are largely attributed to the enhanced cytokine/growth factor signaling in stem cells and myeloid progenitors. Because Ptpn11E76K/+ LSK cells reproduce MPD in recipients (Fig. 6 A), whereas CMPs and CLPs purified from Ptpn11E76K/+ mice with MPD do not generate any diseases in recipients (unpublished data), it appears that MPD can only be initiated when Ptpn11E76K/+ mutation occurs in HSCs but not in lineage progenitors lacking self-renewal capabilities, although persistent induction of Ptpn11E76K/+ mutation in myeloid progenitors also causes overproduction of myeloid cells in Ptpn11E76K/+/LysM-Cre+ mice (Fig. 6 A).
It is interesting that Ptpn11E76K/+ mutation causes acute leukemias in both stem cells and lineage-committed progenitors. Ptpn11E76K/+ mutation in pan-hematopoietic cells serves as the first hit, resulting in MPD initially. During this chronic phase, apparently, additional genetic alterations are evoked, which then cooperatively transform hematopoietic cells, leading to the onset and full development of acute leukemias. Remarkably, Ptpn11E76K/+ mutation also causes acute leukemias in lineage-committed progenitors (Fig. 6). Progenitors do not possess self-renewal capabilities, which is a characteristic feature of LSCs, thus a preexisting self-renewal program does not seem to be required for Ptpn11E76K/+ mutation to transform the target cells to LSCs. Likely, it is the additional genetic alterations subsequently evoked that reprogram these cells, conferring self-renewal capabilities to sustain leukemia growth. Indeed, centrosome amplification and aneuploidy that are associated with genetic alterations are detected much earlier than the emergence of acute leukemias in Ptpn11E76K/+ knock-in mice (Fig. 7, B and D). The fact that Ptpn11E76K/+ mutation has non–lineage/stage-specific effects on the development of LSCs suggests that Ptpn11E76K mutation has a cell-transforming capability.
Genetic abnormalities are thought to arise from enhanced cell proliferation and differentiation. Uncontrolled activation of Ptpn11E76K/+ stem cells or myeloid progenitors may lead to acquisition of additional genetic alterations. Thus, it is not unexpected that MPD subsequently progress to AML in Ptpn11E76K/+/MX1-Cre+ mice. However, T-ALL and B-ALL are also evolved in these animals. Furthermore, persistent induction of Ptpn11E76K/+ mutation in T and B lymphoid progenitors in lineage-specific knock-in mice also results in T-ALL and B-ALL, respectively (Fig. 6). Because no T or B lymphoid developmental abnormalities are detected before the onset of T-ALL and B-ALL in these mice (Fig. S4), these observations argue that the acquisition of the secondary genetic lesions in T and B progenitors conferring self-renewal properties is not related to uncontrolled cell proliferation/differentiation. Rather, the secondary genetic abnormalities are evoked by Ptpn11E76K/+ mutation through other mechanisms.
Ptpn11E76K/+ mutation may induce genetic alterations by disturbing centrosome function among other mechanisms. In support of this hypothesis, Shp2 is found to be distributed to centrosomes that play a critical role in chromosome segregation during mitosis (Draviam et al., 2004; Ruchaud et al., 2007), and centrosomes are amplified in Ptpn11E76K/+ mutant hematopoietic cells (Fig. 7, D and F). As centrosome amplification is known to result in aberrant multipolar mitosis and missegregation of chromosomes, leading to aneuploidy and proneness to cancer (Draviam et al., 2004; Ruchaud et al., 2007), the overall correlation of centrosome amplification with aneuploidy in Ptpn11E76K/+ hematopoietic cells indicates that centrosome amplification is likely responsible for aneuploidy in Ptpn11E76K/+/Mx1-Cre+ mice (Fig. 7, B and D). Nevertheless, the detailed molecular mechanisms by which Shp2 E76K mutation causes centrosome amplification remain to be further determined. Also, it is important to verify whether aneuploidy is a characteristic of Ptpn11-mutation-associated leukemias in humans and whether aneuploidy in these leukemias correlates with centrosome amplification. Shp2 is only localized to some, but not all, amplified centrosomes in Ptpn11E76K/+ cells in both MPD and acute leukemia phases (Fig. 7 F and Fig. S5 B), raising the possibility that displacement of Shp2 from centrosomes may be associated with the centrosome amplification although it is uncertain how this displacement occurs and how it disturbs centrosome functions. Clearly, identification of Shp2 substrates or interacting proteins in centrosomes may shed the light on detailed mechanisms. As other Shp2 GOF mutations (D61G and D61Y) only induce MPD (Araki et al., 2004; Chan et al., 2009; Xu et al., 2010) and the E76K mutation is more potent than D61G/Y mutations in enhancing the catalytic activity of Shp2 (Tartaglia et al., 2003; Keilhack et al., 2005), the role of Shp2 GOF mutations in inducing additional genetic abnormalities driving LSC development seems to be attributable to the enhanced catalytic activity of mutant Shp2. Finally, our finding that Ptpn11E76K/+ mutation is sufficient to induce acute leukemias implicates that Ptpn11E76K/+ mutation might be the initiating genetic lesion in pediatric B-ALL and AML that have Ptpn11E76K/+ mutation, highlighting a broader causative role for Ptpn11 GOF mutations in the pathogenesis of childhood leukemias. Accordingly, Shp2 may be a useful therapeutic target for the treatment of these acute leukemias and for the prevention of malignant transformation in JMML.
MATERIALS AND METHODS
Generation of Ptpn11E76K knock-in mice.
Ptpn11 allele was targeted by homologous recombination. The targeting vector was constructed using the recombineering technique, as previously described (Liu et al., 2003). In brief, a mini–targeting vector with a loxP-flanked neo cassette and the mutation GAA (E) → AAA (K) at the amino acid 76 encoding position in exon 3 of Ptpn11 was first generated. This mini vector was then used to construct the targeting vector. The targeting vector was linearized and electroporated into D1 mouse ES cells derived from F1 hybrid blastocysts of 129S6 x C57BL/6J. G418-resistant ES cell clones were isolated and screened for homologous recombination by nested PCR using primers outside the construct paired with primers inside the neo cassette (Fig. S1 A). Positive clones were further confirmed by PCR genotyping (Fig. S1 B) and sequencing for the mutation (GAA → AAA) in genomic DNA (Fig. S1 C). Two individual ES cell clones, containing a correctly targeted Ptpn11 allele, were used to generate chimeric mice. Germline transmitted chimeric mice were obtained and used to cross C57BL6/J mice to generate heterozygous mice with the neo cassette (Ptpn11E76K neo/+). E76K mutation in F1 Ptpn11E76K neo/+ mice was verified by sequencing the targeted site of genomic DNA. These mice were backcrossed with C57BL6/J mice for 3–6 generations for experiments. Mice used for transplantation analyses were 8th to 10th generation backcross to C57BL6/J background. No differences in the two lines of mutant mice derived from the original two ES cell clones were observed. All mice were kept under specific pathogen–free conditions in the Animal Resources Center at Case Western Reserve University. All animal procedures complied with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee.
Flow cytometric analysis and cell sorting.
Multiparameter FACS analysis was performed to determine populations of HSC-enriched Lineage−Sca-1+c-Kit+ (LSK) cells, LT-HSCs (Lineage−Sca-1+c-Kit+Flk2−CD34−), ST-HSCs (Lineage−Sca-1+c-Kit+Flk2−CD34+), and lineage progenitors, such as CMPs (Lineage−c-Kit+Sca-1−CD16/32medCD34+), CLPs (Lineage−c-KitlowSca-1lowCD127+), GMPs (Lineage−c-Kit+Sca-1−CD16/32highCD34+), and MEPs (Lineage−c-Kit+Sca-1−CD16/32med/lowCD34−). BM cells freshly harvested from femurs and tibias were first stained with anti–Flk2-biotin and subsequently stained with antibodies labeled with various fluorochromes: streptavidin-APC-Cy7, c-Kit-APC, Sca-1-PE, CD34-Pacific blue, CD16/32-PE-Cy7, and CD127 (IL-7Rα)-PE-Cy5 (eBioscience); and FITC-labeled antibodies for lineage markers Mac-1, Gr-1, Ter119, CD4, CD8a, CD3e, and B220 (BD). Specific cell populations were gated based on immunophenotypes for quantification or cell sorting, as previously reported (Kiel et al., 2005; Tothova et al., 2007; Fleming et al., 2008; Schindler et al., 2009; Xu et al., 2010). Fluorescence minus one (FMO) was used for setting the gating on control samples. For intracellular signaling analysis, Lineage− cells were purified from BM and starved for 1 h in IMDM medium. Cells were then stimulated with SCF (50 ng/ml) for 5 min, fixed, permeabilized, and stained with antibodies against Sca-1, c-Kit, and phospho-ERK or phospho-AKT (Cell Signaling Technology) as previously reported (Kalaitzidis and Neel, 2008; Chan et al., 2009). Percentages of the cells stained positive for phospho-ERK or phospho-AKT in the gated LSK population were quantified by multiparameter FACS analyses.
Apoptosis and cell cycle analysis.
Fresh BM cells were stained with biotin-labeled antibodies against lineage markers (Gr-1, Mac-1, B220, Ter119, CD4, CD8, and CD3e), followed by staining with streptavidin-conjugated APC-Cy7, anti–c-Kit-APC, and anti–Sca-1-FITC. The cells were then stained with anti–Annexin V-PE and 7-amino-actinomycin D using Annexin V-PE apoptosis Detection kit I (BD). Apoptotic (Annexin V+) cells in the gated LSK cell population were quantified by FACS. For LSK cell cycle analysis, fresh BM cells were stained with Pyronin Y (1 µg/ml) and Hoechst 33342 (10 µg/ml). The cells were washed and then stained with the previously described antibodies. Subsequent LSK population gating and quantification of the cells at G0, G1, and S/G2/M phases by FACS were performed as previously reported (Cheng et al., 2000; Xu et al., 2010).
Competitive repopulation assay.
In brief, 106 BM cells (test cells) freshly harvested from Ptpn11E76K/+/Mx1-Cre+ and Ptpn11+/+/Mx1-Cre+ littermates (CD45.2+) 8–10 wk after pI-pC treatment were transplanted with the same number of BoyJ (CD45.1+) BM cells (competitor cells) into lethally irradiated (11.0 Gy) BoyJ (CD45.1+) recipients. Test cell reconstitution was determined at 8, 12, 16, and 24 wk after transplantation by FACS analyses of peripheral blood, as we previously described (Xu et al., 2010).
CFU assay.
For the myeloid progenitor assay, freshly harvested BM cells (2 × 104 cells/ml) were assayed for CFUs in 0.9% methylcellulose IMDM medium containing 30% FBS, glutamine (10−4 M), β-mercaptoethanol (3.3 × 10−5 M), and IL-3 or GM-CSF at the indicated concentrations. After 7 d of culture at 37°C in a humidified 5% CO2 incubator, myeloid colonies (CFU-GM and CFU-M) were counted under an inverted microscope.
Karyotype and spectral karyotype analyses.
To prepare metaphase spreads, fresh or cultured LSK cells or purified progenitors were treated with colcemid (0.05 µg/ml) at 37°C for 2 h. Cells were harvested, suspended in prewarmed 75 mM KCl hypotonic solution, and incubated at 37°C for 10 min. The cells were then fixed in Carnoy’s solution (75% methanol and 25% acetic acid) at room temperature for 15 min, washed twice with fixative, and dropped onto prechilled microscope slides. The slides were dried and stained in 1 µg/ml DAPI for 10 min. Chromosomes in each metaphase cell (nonmegakaryocyte) were enumerated under a fluorescence microscope using an 100× oil objective. For spectral karyotype analysis, metaphase spreads were incubated with a mouse SKY Paint kit probe (Applied Spectral Imaging), followed by counterstaining with DAPI. Chromosomes were identified using Spectral Karyotyping (SKY) SD300VDS workstation equipped with SKY View software (Applied Spectral Imaging).
Immunostaining and confocal microscopy.
Freshly isolated or cultured hematopoietic cells were spun onto microscope slides by cytospin, fixed in 100% methanol at −20°C for 10 min, permeabilized with 0.1% Triton X-100 in PBS, and blocked with 2% BSA in PBS at room temperature for 1 h. Cells were incubated with primary antibodies at room temperature for 1 h or 4°C overnight. The cells were then washed three times with PBS and incubated with Alexa Fluor 488– or Alexa Fluor 568–conjugated secondary antibodies for 1 h. Nuclei were counterstained with DAPI. After washing, slides were mounted by coverslips. All confocal images were acquired using an LSM 510 inverted laser-scanning confocal microscope (Carl Zeiss, Inc.). Images were analyzed with MetaMorph software. Whole-mount phospho-Erk immunostaining was performed as previously described (Corson et al., 2003).
Statistical analysis.
All studies were repeated at least twice with consistent results and with a minimum of three mice per group, although typically more (as indicated in figure legends). Data are presented as mean ± SEM. Statistical significance was determined using unpaired two-tailed Student’s t test. P-values <0.05 were considered to be significant.
Online supplemental material.
Fig. S1 shows the gene targeting strategy, verification of the targeted Ptpn11 allele, and heterozygous E76K mutation in chimeric mice. Fig. S2 shows that global Ptpn11E76K/+ mutation results in embryonic lethality and that insertion of neo in intron 2 of the targeted Ptpn11 allele prevents expression of Shp2 E76K. Fig. S3 shows disease progression in Ptpn11E76K/+ knock-in mice. Fig. S4 shows that Ptpn11E76K/+ mutation in T or B lymphoid progenitors does not significantly disturb T or B cell development. Fig. S5 shows centrosome amplification in Ptpn11E76K/+ BM cells and that Shp2 is localized to part, but not all, of amplified centrosomes in these cells. The Supplemental text describes the pathogenic effects of Ptpn11E76K/+ mutation on embryonic development and the inducible nature of the Ptpn11E76K allele created. Table S1 summarizes the features of T-ALL, B-ALL, AML, MPD in acceleration, and MPD developed in Ptpn11E76K/+ knock-in mice. Table S2 shows peripheral blood cell counts of Ptpn11E76K knock-in and control mice. Table S3 shows the absolute numbers of HSCs and progenitors of various stages in the BM from Ptpn11E76K knock-in and control mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20110450/DC1.
Acknowledgments
This work was supported by the National Institutes of Health grants HL068212 and HL095657 (to C.K. Qu) and Case Comprehensive Cancer Center Cancer Stem Cell Pilot Grant (to C.K. Qu).
The authors have no conflicting or competing financial interests.
Footnotes
Abbreviations used:
- AML
- acute myeloid leukemia
- B-ALL
- B cell acute lymphoblastic leukemia/lymphoma
- CLP
- common lymphoid progenitor
- CMP
- common myeloid progenitor
- ES
- embryonic stem
- GMP
- granulocyte macrophage progenitor
- GOF
- gain-of-function
- HSC
- hematopoietic stem cell
- JMML
- juvenile myelomonocytic leukemia
- LSC
- leukemia-initiating stem cell
- LSK
- Lineage−Sca-1+c-Kit+
- MEP
- megakaryocyte erythroid progenitor
- MPD
- myeloproliferative disorder
- N-SH2
- N-terminal SH2
- pI-pC
- polyinosine-polycyticyclic
- PTP
- protein tyrosine phosphatase
- pY
- phosphorylated tyrosine
- T-ALL
- T cell acute lymphoblastic leukemia/lymphoma
References
- Aoki Y., Niihori T., Narumi Y., Kure S., Matsubara Y. 2008. The RAS/MAPK syndromes: novel roles of the RAS pathway in human genetic disorders. Hum. Mutat. 29:992–1006 10.1002/humu.20748 [DOI] [PubMed] [Google Scholar]
- Araki T., Mohi M.G., Ismat F.A., Bronson R.T., Williams I.R., Kutok J.L., Yang W., Pao L.I., Gilliland D.G., Epstein J.A., Neel B.G. 2004. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat. Med. 10:849–857 10.1038/nm1084 [DOI] [PubMed] [Google Scholar]
- Barford D., Neel B.G. 1998. Revealing mechanisms for SH2 domain mediated regulation of the protein tyrosine phosphatase SHP-2. Structure. 6:249–254 10.1016/S0969-2126(98)00027-6 [DOI] [PubMed] [Google Scholar]
- Bennett A.M., Tang T.L., Sugimoto S., Walsh C.T., Neel B.G. 1994. Protein-tyrosine-phosphatase SHPTP2 couples platelet-derived growth factor receptor beta to Ras. Proc. Natl. Acad. Sci. USA. 91:7335–7339 10.1073/pnas.91.15.7335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentires-Alj M., Paez J.G., David F.S., Keilhack H., Halmos B., Naoki K., Maris J.M., Richardson A., Bardelli A., Sugarbaker D.J., et al. 2004. Activating mutations of the noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 64:8816–8820 10.1158/0008-5472.CAN-04-1923 [DOI] [PubMed] [Google Scholar]
- Birnbaum R.A., O’Marcaigh A., Wardak Z., Zhang Y.Y., Dranoff G., Jacks T., Clapp D.W., Shannon K.M. 2000. Nf1 and Gmcsf interact in myeloid leukemogenesis. Mol. Cell. 5:189–195 10.1016/S1097-2765(00)80415-3 [DOI] [PubMed] [Google Scholar]
- Chan R.J., Leedy M.B., Munugalavadla V., Voorhorst C.S., Li Y., Yu M., Kapur R. 2005. Human somatic PTPN11 mutations induce hematopoietic-cell hypersensitivity to granulocyte-macrophage colony-stimulating factor. Blood. 105:3737–3742 10.1182/blood-2004-10-4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan G., Kalaitzidis D., Usenko T., Kutok J.L., Yang W., Mohi M.G., Neel B.G. 2009. Leukemogenic Ptpn11 causes fatal myeloproliferative disorder via cell-autonomous effects on multiple stages of hematopoiesis. Blood. 113:4414–4424 10.1182/blood-2008-10-182626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan G., Cheung L.S., Yang W., Milyavsky M., Sanders A.D., Gu S., Hong W.X., Liu A.X., Wang X., Barbara M., et al. 2011. Essential role for Ptpn11 in survival of hematopoietic stem and progenitor cells. Blood. 117:4253–4261 10.1182/blood-2010-11-319517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T., Rodrigues N., Shen H., Yang Y., Dombkowski D., Sykes M., Scadden D.T. 2000. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 287:1804–1808 10.1126/science.287.5459.1804 [DOI] [PubMed] [Google Scholar]
- Corson L.B., Yamanaka Y., Lai K.M., Rossant J. 2003. Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development. 130:4527–4537 10.1242/dev.00669 [DOI] [PubMed] [Google Scholar]
- Cozzio A., Passegué E., Ayton P.M., Karsunky H., Cleary M.L., Weissman I.L. 2003. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 17:3029–3035 10.1101/gad.1143403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draviam V.M., Xie S., Sorger P.K. 2004. Chromosome segregation and genomic stability. Curr. Opin. Genet. Dev. 14:120–125 10.1016/j.gde.2004.02.007 [DOI] [PubMed] [Google Scholar]
- Eck M.J., Pluskey S., Trüb T., Harrison S.C., Shoelson S.E. 1996. Spatial constraints on the recognition of phosphoproteins by the tandem SH2 domains of the phosphatase SH-PTP2. Nature. 379:277–280 10.1038/379277a0 [DOI] [PubMed] [Google Scholar]
- Fleming H.E., Janzen V., Lo Celso C., Guo J., Leahy K.M., Kronenberg H.M., Scadden D.T. 2008. Wnt signaling in the niche enforces hematopoietic stem cell quiescence and is necessary to preserve self-renewal in vivo. Cell Stem Cell. 2:274–283 10.1016/j.stem.2008.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragale A., Tartaglia M., Wu J., Gelb B.D. 2004. Noonan syndrome-associated SHP2/PTPN11 mutants cause EGF-dependent prolonged GAB1 binding and sustained ERK2/MAPK1 activation. Hum. Mutat. 23:267–277 10.1002/humu.20005 [DOI] [PubMed] [Google Scholar]
- Guo W., Lasky J.L., Chang C.J., Mosessian S., Lewis X., Xiao Y., Yeh J.E., Chen J.Y., Iruela-Arispe M.L., Varella-Garcia M., Wu H. 2008. Multi-genetic events collaboratively contribute to Pten-null leukaemia stem-cell formation. Nature. 453:529–533 10.1038/nature06933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof P., Pluskey S., Dhe-Paganon S., Eck M.J., Shoelson S.E. 1998. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 92:441–450 10.1016/S0092-8674(00)80938-1 [DOI] [PubMed] [Google Scholar]
- Jude C.D., Climer L., Xu D., Artinger E., Fisher J.K., Ernst P. 2007. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 1:324–337 10.1016/j.stem.2007.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaitzidis D., Neel B.G. 2008. Flow-cytometric phosphoprotein analysis reveals agonist and temporal differences in responses of murine hematopoietic stem/progenitor cells. PLoS ONE. 3:e3776 10.1371/journal.pone.0003776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan M.B., Bartek J. 2004. Cell-cycle checkpoints and cancer. Nature. 432:316–323 10.1038/nature03097 [DOI] [PubMed] [Google Scholar]
- Keilhack H., David F.S., McGregor M., Cantley L.C., Neel B.G. 2005. Diverse biochemical properties of Shp2 mutants. Implications for disease phenotypes. J. Biol. Chem. 280:30984–30993 10.1074/jbc.M504699200 [DOI] [PubMed] [Google Scholar]
- Kiel M.J., Yilmaz O.H., Iwashita T., Yilmaz O.H., Terhorst C., Morrison S.J. 2005. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 121:1109–1121 10.1016/j.cell.2005.05.026 [DOI] [PubMed] [Google Scholar]
- Kirstetter P., Schuster M.B., Bereshchenko O., Moore S., Dvinge H., Kurz E., Theilgaard-Mönch K., Månsson R., Pedersen T.A., Pabst T., et al. 2008. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 13:299–310 10.1016/j.ccr.2008.02.008 [DOI] [PubMed] [Google Scholar]
- Kontaridis M.I., Swanson K.D., David F.S., Barford D., Neel B.G. 2006. PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J. Biol. Chem. 281:6785–6792 10.1074/jbc.M513068200 [DOI] [PubMed] [Google Scholar]
- Kratz C.P., Niemeyer C.M., Castleberry R.P., Cetin M., Bergsträsser E., Emanuel P.D., Hasle H., Kardos G., Klein C., Kojima S., et al. 2005. The mutational spectrum of PTPN11 in juvenile myelomonocytic leukemia and Noonan syndrome/myeloproliferative disease. Blood. 106:2183–2185 10.1182/blood-2005-02-0531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn R., Schwenk F., Aguet M., Rajewsky K. 1995. Inducible gene targeting in mice. Science. 269:1427–1429 10.1126/science.7660125 [DOI] [PubMed] [Google Scholar]
- Li W., Nishimura R., Kashishian A., Batzer A.G., Kim W.J., Cooper J.A., Schlessinger J. 1994. A new function for a phosphotyrosine phosphatase: linking GRB2-Sos to a receptor tyrosine kinase. Mol. Cell. Biol. 14:509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Jenkins N.A., Copeland N.G. 2003. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13:476–484 10.1101/gr.749203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh M.L., Reynolds M.G., Vattikuti S., Gerbing R.B., Alonzo T.A., Carlson E., Cheng J.W., Lee C.M., Lange B.J., Meshinchi S.; Children’s Cancer Group 2004a. PTPN11 mutations in pediatric patients with acute myeloid leukemia: results from the Children’s Cancer Group. Leukemia. 18:1831–1834 10.1038/sj.leu.2403492 [DOI] [PubMed] [Google Scholar]
- Loh M.L., Vattikuti S., Schubbert S., Reynolds M.G., Carlson E., Lieuw K.H., Cheng J.W., Lee C.M., Stokoe D., Bonifas J.M., et al. 2004b. Mutations in PTPN11 implicate the SHP-2 phosphatase in leukemogenesis. Blood. 103:2325–2331 10.1182/blood-2003-09-3287 [DOI] [PubMed] [Google Scholar]
- Mohi M.G., Williams I.R., Dearolf C.R., Chan G., Kutok J.L., Cohen S., Morgan K., Boulton C., Shigematsu H., Keilhack H., et al. 2005. Prognostic, therapeutic, and mechanistic implications of a mouse model of leukemia evoked by Shp2 (PTPN11) mutations. Cancer Cell. 7:179–191 10.1016/j.ccr.2005.01.010 [DOI] [PubMed] [Google Scholar]
- Neel B.G., Gu H., Pao L. 2003. The ‘Shp’ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem. Sci. 28:284–293 10.1016/S0968-0004(03)00091-4 [DOI] [PubMed] [Google Scholar]
- Pawson T. 2004. Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 116:191–203 10.1016/S0092-8674(03)01077-8 [DOI] [PubMed] [Google Scholar]
- Qu C.K., Shi Z.Q., Shen R., Tsai F.Y., Orkin S.H., Feng G.S. 1997. A deletion mutation in the SH2-N domain of Shp-2 severely suppresses hematopoietic cell development. Mol. Cell. Biol. 17:5499–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu C.K., Yu W.M., Azzarelli B., Cooper S., Broxmeyer H.E., Feng G.S. 1998. Biased suppression of hematopoiesis and multiple developmental defects in chimeric mice containing Shp-2 mutant cells. Mol. Cell. Biol. 18:6075–6082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu C.K., Yu W.M., Azzarelli B., Feng G.S. 1999. Genetic evidence that Shp-2 tyrosine phosphatase is a signal enhancer of the epidermal growth factor receptor in mammals. Proc. Natl. Acad. Sci. USA. 96:8528–8533 10.1073/pnas.96.15.8528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu C.K., Nguyen S., Chen J., Feng G.S. 2001. Requirement of Shp-2 tyrosine phosphatase in lymphoid and hematopoietic cell development. Blood. 97:911–914 10.1182/blood.V97.4.911 [DOI] [PubMed] [Google Scholar]
- Rosenbauer F., Wagner K., Kutok J.L., Iwasaki H., Le Beau M.M., Okuno Y., Akashi K., Fiering S., Tenen D.G. 2004. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat. Genet. 36:624–630 10.1038/ng1361 [DOI] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W.C. 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8:798–812 10.1038/nrm2257 [DOI] [PubMed] [Google Scholar]
- Saxton T.M., Henkemeyer M., Gasca S., Shen R., Rossi D.J., Shalaby F., Feng G.S., Pawson T. 1997. Abnormal mesoderm patterning in mouse embryos mutant for the SH2 tyrosine phosphatase Shp-2. EMBO J. 16:2352–2364 10.1093/emboj/16.9.2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler J.W., Van Buren D., Foudi A., Krejci O., Qin J., Orkin S.H., Hock H. 2009. TEL-AML1 corrupts hematopoietic stem cells to persist in the bone marrow and initiate leukemia. Cell Stem Cell. 5:43–53 10.1016/j.stem.2009.04.019 [DOI] [PubMed] [Google Scholar]
- Schubbert S., Lieuw K., Rowe S.L., Lee C.M., Li X., Loh M.L., Clapp D.W., Shannon K.M. 2005. Functional analysis of leukemia-associated PTPN11 mutations in primary hematopoietic cells. Blood. 106:311–317 10.1182/blood-2004-11-4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L.J., Curtis J.E., Messner H.A., Senn J.S., Furthmayr H., McCulloch E.A. 1983. Lineage infidelity in acute leukemia. Blood. 61:1138–1145 [PubMed] [Google Scholar]
- So C.W., Karsunky H., Passegué E., Cozzio A., Weissman I.L., Cleary M.L. 2003. MLL-GAS7 transforms multipotent hematopoietic progenitors and induces mixed lineage leukemias in mice. Cancer Cell. 3:161–171 10.1016/S1535-6108(03)00019-9 [DOI] [PubMed] [Google Scholar]
- Songyang Z., Cantley L.C. 2004. ZIP codes for delivering SH2 domains. Cell. 116(2, Suppl):S41–S43: 2: S48 10.1016/S0092-8674(04)00041-8 [DOI] [PubMed] [Google Scholar]
- Tartaglia M., Mehler E.L., Goldberg R., Zampino G., Brunner H.G., Kremer H., van der Burgt I., Crosby A.H., Ion A., Jeffery S., et al. 2001. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet. 29:465–468 10.1038/ng772 [DOI] [PubMed] [Google Scholar]
- Tartaglia M., Niemeyer C.M., Fragale A., Song X., Buechner J., Jung A., Hählen K., Hasle H., Licht J.D., Gelb B.D. 2003. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat. Genet. 34:148–150 10.1038/ng1156 [DOI] [PubMed] [Google Scholar]
- Tartaglia M., Martinelli S., Cazzaniga G., Cordeddu V., Iavarone I., Spinelli M., Palmi C., Carta C., Pession A., Aricò M., et al. 2004. Genetic evidence for lineage-related and differentiation stage-related contribution of somatic PTPN11 mutations to leukemogenesis in childhood acute leukemia. Blood. 104:307–313 10.1182/blood-2003-11-3876 [DOI] [PubMed] [Google Scholar]
- Tartaglia M., Martinelli S., Stella L., Bocchinfuso G., Flex E., Cordeddu V., Zampino G., Burgt I., Palleschi A., Petrucci T.C., et al. 2006. Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. Am. J. Hum. Genet. 78:279–290 10.1086/499925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonks N.K. 2006. Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 7:833–846 10.1038/nrm2039 [DOI] [PubMed] [Google Scholar]
- Tothova Z., Kollipara R., Huntly B.J., Lee B.H., Castrillon D.H., Cullen D.E., McDowell E.P., Lazo-Kallanian S., Williams I.R., Sears C., et al. 2007. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 128:325–339 10.1016/j.cell.2007.01.003 [DOI] [PubMed] [Google Scholar]
- Waksman G., Kuriyan J. 2004. Structure and specificity of the SH2 domain. Cell. 116(2, Suppl):S45–S48: 3: S48 10.1016/S0092-8674(04)00043-1 [DOI] [PubMed] [Google Scholar]
- Xu D., Qu C.K. 2008. Protein tyrosine phosphatases in the JAK/STAT pathway. Front. Biosci. 13:4925–4932 10.2741/3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Wang S., Yu W.M., Chan G., Araki T., Bunting K.D., Neel B.G., Qu C.K. 2010. A germline gain-of-function mutation in Ptpn11 (Shp-2) phosphatase induces myeloproliferative disease by aberrant activation of hematopoietic stem cells. Blood. 116:3611–3621 10.1182/blood-2010-01-265652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Klaman L.D., Chen B., Araki T., Harada H., Thomas S.M., George E.L., Neel B.G. 2006. An Shp2/SFK/Ras/Erk signaling pathway controls trophoblast stem cell survival. Dev. Cell. 10:317–327 10.1016/j.devcel.2006.01.002 [DOI] [PubMed] [Google Scholar]
- Yilmaz O.H., Valdez R., Theisen B.K., Guo W., Ferguson D.O., Wu H., Morrison S.J. 2006. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 441:475–482 10.1038/nature04703 [DOI] [PubMed] [Google Scholar]
- Yu W.M., Hawley T.S., Hawley R.G., Qu C.K. 2003. Catalytic-dependent and -independent roles of SHP-2 tyrosine phosphatase in interleukin-3 signaling. Oncogene. 22:5995–6004 10.1038/sj.onc.1206846 [DOI] [PubMed] [Google Scholar]
- Yu W.M., Daino H., Chen J., Bunting K.D., Qu C.K. 2006. Effects of a leukemia-associated gain-of-function mutation of SHP-2 phosphatase on interleukin-3 signaling. J. Biol. Chem. 281:5426–5434 10.1074/jbc.M507622200 [DOI] [PubMed] [Google Scholar]
- Zhang Y.Y., Vik T.A., Ryder J.W., Srour E.F., Jacks T., Shannon K., Clapp D.W. 1998. Nf1 regulates hematopoietic progenitor cell growth and ras signaling in response to multiple cytokines. J. Exp. Med. 187:1893–1902 10.1084/jem.187.11.1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Grindley J.C., Yin T., Jayasinghe S., He X.C., Ross J.T., Haug J.S., Rupp D., Porter-Westpfahl K.S., Wiedemann L.M., et al. 2006. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 441:518–522 10.1038/nature04747 [DOI] [PubMed] [Google Scholar]
- Zhao R., Fu X., Teng L., Li Q., Zhao Z.J. 2003. Blocking the function of tyrosine phosphatase SHP-2 by targeting its Src homology 2 domains. J. Biol. Chem. 278:42893–42898 10.1074/jbc.M306136200 [DOI] [PubMed] [Google Scholar]
- Zhu H.H., Ji K., Alderson N., He Z., Li S., Liu W., Zhang D.E., Li L., Feng G.S. 2011. Kit-Shp2-Kit signaling acts to maintain a functional hematopoietic stem and progenitor cell pool. Blood. 117:5350–5361 10.1182/blood-2011-01-333476 [DOI] [PMC free article] [PubMed] [Google Scholar]